Abstract
Polymerase chain reaction (PCR) testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is necessary for confirming a diagnosis of Coronavirus disease 2019 (COVID-19). Here we present a COVID-19 case of an elderly woman whose SARS-CoV-2 PCR tests showed false negative repeatedly by evaluating with different sampling sites and procedures. Nasopharyngeal swabs, suctioned sputum, and tongue swabs were collected for SARS-CoV-2-PCR. As for tongue swabs, we compared between two different sample conditions; one obtained with dry condition and the other obtained with moistened condition inside the oral cavity. SARS-CoV-2-PCR showed positive for an extended period with suctioned sputum samples compared with nasopharyngeal swabs and tongue swabs. No SARS-CoV-2 from a nasopharyngeal swab sample obtained on day 46 after symptoms onset was isolated despite high viral load (183740.5 copies/5μL). An adequate production of neutralizing antibody in a serum sample on day 46 was also confirmed. The number of RNA copies of the tongue swab samples was higher with moistened condition than with dry condition.
The present case suggests that the difference of sampling site or sample condition can affect PCR results. High loads viral RNA detection does not always correlate with infectivity.
Keywords: SARS-CoV-2, COVID-19, PCR, False-negative, Infectivity
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The disease has spread worldwide, and the number of deaths due to this infection continues to increase. Due to its novelty, there are still several unknown aspects regarding the pathology, diagnosis, and treatment. The gold standard for COVID-19 diagnosis is viral RNA detection through quantitative reverse-transcription-polymerase chain reaction (RT-PCR). The sensitivity of nucleic acid amplification was observed to be 89% in a meta-analysis with COVID-19 confirmed patients [1], and was as high as 86% for nasopharyngeal specimens and 92% for saliva samples in asymptomatic persons including close contacts [2]. Additionally, our study also showed higher sensitivity with saliva samples than with nasopharyngeal samples in asymptomatic or mild patients in the late phase of infection around 4 weeks after diagnosis as 38.2% and 19.5%, respectively [3]. Therefore, although infrequent, false-negative results can lead to misdiagnosis of COVID-19. Inadequate sampling is a well-known reason for false-negative PCR results [4,5]. The duration of SARS-CoV-2 infectivity remains unclear, and it is crucial for understanding transmission dynamics and implementing effective infection control mechanisms. However, positive PCR tests in the late phase of SARS-CoV-2 infection are not always considered infectious [6].
2. Case report
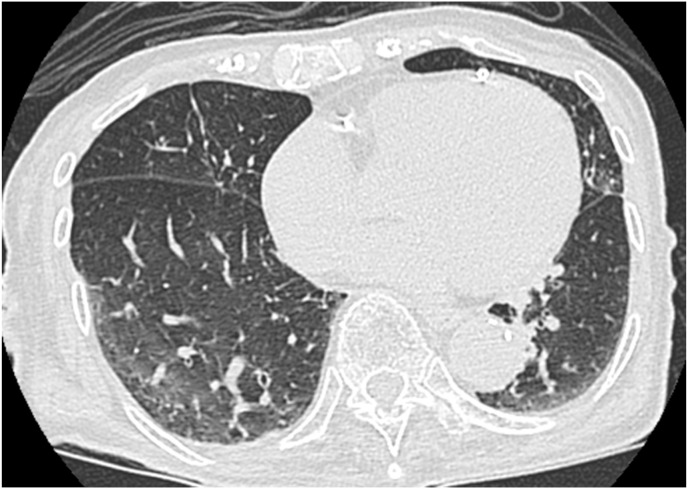
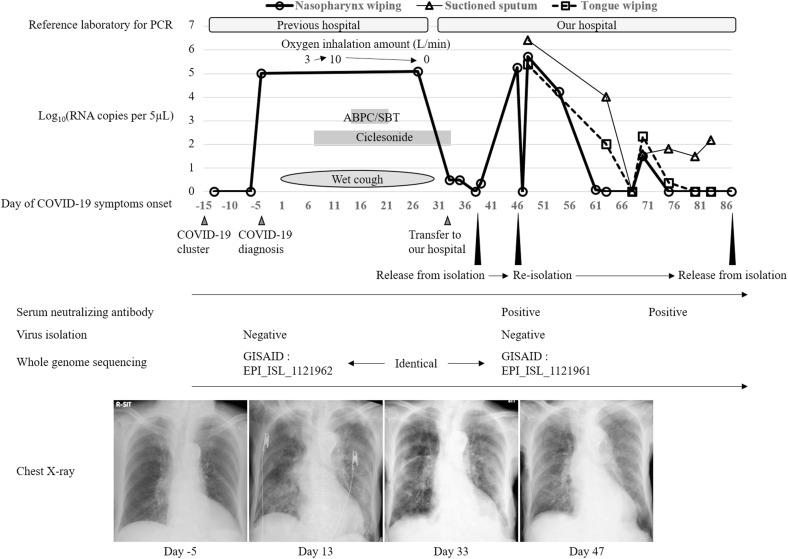
A 97-year-old woman presented to a nearby hospital with exacerbation of heart failure from a nursing home for the elderly. She had no history of traceable contact with COVID-19 patients. Diuretic treatment was administered, and symptoms were improved. A cluster of COVID-19 cases was identified in the hospital on the fifth day of admission. Eleven days after the recognition of the cluster, PCR test for SARS-CoV-2 with nasopharyngeal swab sample from the patient was positive, leading to the diagnosis of COVID-19 while asymptomatic. Computed tomography showed slight ground-glass opacity in the right lower lung lobe (Fig. 1 ). A wet cough developed at 4 days after the diagnosis of COVID-19 which was compatible with its symptom, and the patient required oxygen administration and ciclesonide inhalation, six days after COVID-19 symptoms onset. She also developed aspiration pneumonia and her respiratory state deteriorated. Her overall condition improved with ampicillin-sulbactam administration. However, a SARS-CoV-2-PCR on a nasopharyngeal swab sample on day 27 after symptoms onset was positive. The patient was transferred to our hospital on day 33 for subsequent medical care. We repeated the PCR test using a nasopharyngeal swab after admission to our hospital and found that the viral RNA copy numbers were low. We performed SARS-CoV-2 PCR examination at each laboratory unit inside the hospital where the samples were obtained. All the PCR examination of nasopharyngeal swab samples was performed in the same procedure using flocked swabs without any transport media promptly after sampling. We preserved the samples at 2–8 °C if it takes more than a few hours. Since the patient was asymptomatic, she was transferred from the negative-pressure isolation ward to the general ward and was released from contact and droplet precaution on day 39 after COVID-19 symptoms onset. However, 46 days after symptoms onset, SARS-CoV-2 PCR result with a nasopharyngeal sample displayed a high number of viral RNA copies with 183740.5 copies/5μL. The patient was transferred back to the COVID-19 isolation ward and contact and droplet precaution were applied on that day. We performed a SARS-CoV-2-PCR tests on a nasopharyngeal swab, suctioned sputum, and tongue swab samples from the patient promptly after sampling. As for tongue swab samples, we examined the number of viral RNA copies in two different sampling conditions which were both obtained with flocked swabs without any transport media on day 48. One sample was obtained following the usual procedure (dry), where the conditions inside the oral cavity are unchanged, and the other was obtained immediately after using saline solution to moisten the patient's tongue (wet) (Fig. 2 ), which resulted 13361 copies/5μL, and 245830 copies/5μL, respectively. Although the three different types of sample of the patient obtained on day 68 after symptoms onset were negative, they were positive again on day 70. The suctioned sputum sample remained PCR-positive for an extended period until day 83. However, we confirmed adequate neutralizing antibody titer of 1:640 with a serum sample obtained on day 46 when a high number of viral RNA copies of PCR was detected, with the method described previously [7]. Whole-genome sequencing of the nasopharyngeal swab sample obtained on day 46 displayed identical with the sample obtained 49 days before (GISAID accession ID: EPI_ISL_1121961 and ID: EPI_ISL_1121962 respectively). The virus could not be isolated with samples obtained on days −4 and 46 despite high viral loads. Possible reasons for this with sample obtained on day −4 is that nearly 50 days had passed when we started an analysis of virus isolation, and that the quality of sample might have been deteriorated during storage and transportation from the previous hospital. We analyzed neutralizing antibody, whole-genome sequencing, and virus isolation at Institute of Tropical Medicine, Nagasaki University. The patient's general condition improved, a nasopharyngeal sample yielded a negative SARS-CoV-2-PCR result (Fig. 3 ), and she was discharged back to the nursing home on day 108 after symptoms onset.
Fig. 1.
Chest computed tomography scan performed on the day of COVID-19 diagnosis. Slight ground-glass opacity in the right lower lung lobe.
Fig. 2.
Tongue wiping for PCR examination with dry condition (A), and wet condition after moistened with saline (B).
Fig. 3.
Clinical course of the present case. PCR, polymerase chain reaction; RNA, ribonucleic acid; ABPC/SBT, ampicillin-sulbactam.
3. Discussion
We present a COVID-19 case of an elderly woman whose SARS-CoV-2 PCR tests showed false negative repeatedly. This COVID-19 case highlights several important clinical issues. First, the results of SARS-CoV-2 PCR testing can be greatly affected by its sampling condition and infection cannot be completely ruled out by a single negative PCR result from a nasopharyngeal swab. Second, high viral RNA concentrations detected by PCR do not necessarily imply SARS-CoV-2 infectivity.
We concluded that the negative PCR results were false-negative despite the prolonged SARS-CoV-2 RNA presence in this case. False-negative [5,8,9] or test-retest positive [[9], [10], [11], [12]] results for SARS-CoV-2 PCRs have been previously reported. We ruled out the possibility of COVID-19 recurrence in this case because the patient was asymptomatic during her stay in our hospital. Additionally, we found an adequate production of neutralizing antibody in a serum sample. Re-infection was also considered unlikely because the patient had no traceable contact with COVID-19 confirmed or suspected cases during hospitalization. And more, the results of whole-genome sequencing were identical between the nasopharyngeal swab samples obtained at the previous hospital and our hospital. The well-known reasons for false-negative PCR results are inadequate sampling timing, sampling techniques, storage, transportation, processing of specimens, and technical problems related to the PCR [4,5]. We confirmed that the results of PCR tests varied depending on the type of samples among nasopharynx wiping, suctioned sputum, and tongue wiping samples even obtained on the same day. We collected two different tongue-wiping samples to clarify the cause of the poor reproducibility of the PCR test results and confirmed that the number of RNA copies of the sample obtained under moistened oral condition was approximately 20 times higher than in the sample obtained under dry conditions. This finding suggests that the sample quality can be affected by interventions, such as increasing humidity, and alter the results of the PCR test. We speculated that the false-negative PCR results in the present case were mostly due to inconsistent sampling conditions. We should be aware that SARS-CoV-2-PCR sensitivity varies [[1], [2], [3]] and decreases with time after symptoms onset [12,13].
Viable viral particles were successfully isolated on days seven [14], nine [15,16], 18 [17], or 20 [18] after symptoms onset, while no isolate was confirmed after a certain period since onset. The present case suggests that positive PCR testing in the later phase of SARS-CoV-2 infection does not necessarily imply infectivity.
The correlation between SARS-CoV-2 RNA detection and COVID-19 infectivity was not fully recognized as it is today when we were treating this patient. We continued isolation and precautions even after recognizing the results of adequate production of serum neutralizing antibody and negative virus isolation, and examined the PCR testing repeatedly because confirmation of negative result was requested from the nursing home.
However, the timing of discontinuation of precaution for COVID-19 patients is now proposed to decide based on the duration after onset but not with a test-based strategy. PCR testing for SARS-CoV-2 RNA cannot distinguish infectious from non-infectious virus. Therefore, PCR testing is not recommended for deciding discontinuation of infection precaution, and it should be mainly performed for the COVID-19 diagnosis purpose.
In conclusion, the present case showed false negative PCR testing for SARS-CoV-2 RNA repeatedly. Our work highlights that, although PCR is the gold standard for COVID-19 diagnosis, we should pay attention to avoid sampling error which can lead to false-negative results. This report also shows that SARS-CoV-2 RNA detection does not always imply infectivity or the dissemination of viable viral particles.
Authorship statement
Contributors SI, NA, TT, and TS managed the case as the physicians. PM, TN, MMNT, and KO were in charge of tests related to COVID-19 in this report. All authors contributed to the writing of the final manuscript. All authors meet the ICMJE authorship criteria.
Funding
None.
Consent for publication
Informed consent was obtained from the patient's family because the patient was incapable of giving consent.
Declaration of competing interest
The authors declare no conflicts of interest associated with this manuscript.
Acknowledgments
None.
References
- 1.Kim H., Hong H., Yoon S.H. Diagnostic performance of CT and reverse transcriptase polymerase chain reaction for coronavirus Disease 2019: a meta-analysis. Radiology. 2020;296(3):E145–E155. doi: 10.1148/radiol.2020201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokota I., Shane P.Y., Okada K., Unoki Y., Yang Y., Inao T., et al. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ota K, Yanagihara K, Sasaki D, Kaku N, Uno N, Sakamoto K, et al. Detection of SARS-CoV-2 using qRT-PCR in saliva obtained from asymptomatic or mild COVID-19 patients, comparative analysis with matched nasopharyngeal samples. PloS One. 2021;16(6) doi: 10.1371/journal.pone.0252964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abud B.T., Hajnas N.M., Redleaf M., Kerolus J.L., Lee V. Assessing the impact of a training initiative for nasopharyngeal and oropharyngeal swabbing for COVID-19 testing. OTO Open. 2020;4(3) doi: 10.1177/2473974X20953094. 2473974X20953094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piras A., Rizzo D., Uzzau S., De Riu G., Rubino S., Bussu F. Inappropriate nasopharyngeal sampling for SARS-CoV-2 detection is a relevant cause of false-negative reports. Otolaryngol Head Neck Surg. 2020;163(3):459–461. doi: 10.1177/0194599820931793. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention . 2020. Science Brief: options to reduce quarantine for contacts of persons with SARS-CoV-2 infection using symptom monitoring and diagnostic testing.https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-options-to-reduce-quarantine.html accessed 7 June 2021. [PubMed] [Google Scholar]
- 7.Ngwe Tun M.M., Muta Y., Inoue S., Morita K. Persistence of neutralizing antibody against dengue virus 2 after 70 years from infection in Nagasaki. Biores Open Access. 2016;5(1):188–191. doi: 10.1089/biores.2016.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D., Wang D., Dong J., Wang N., Huang H., Xu H., et al. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean J Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao A.T., Tong Y.X., Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020;92(10):1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang H., Wang Y., Tong Z., Liu X. Retest positive for SARS-CoV-2 RNA of “recovered” patients with COVID-19: persistence, sampling issues, or re-infection? J Med Virol. 2020;92(11):2263–2265. doi: 10.1002/jmv.26114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao X.M., Xu X.F., Zi H., Liu G.X., Li B.H., Du X., et al. Re-positive cases of nucleic acid tests in discharged patients with COVID-19: a follow-up study. Front Med. 2020;7:349. doi: 10.3389/fmed.2020.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widders A., Broom A., Broom J. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health. 2020;25(3):210–215. doi: 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 16.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W.D., Chang S.Y., Wang J.T., Tsai M.J., Hung C.C., Hsu C.L., et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee C., Kanjilal S., Baker M., Klompas M. Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation? Clin Infect Dis. 2020:ciaa1249. doi: 10.1093/cid/ciaa1249. [DOI] [PMC free article] [PubMed] [Google Scholar]