Abstract
More than 3000 clinical trials related to COVID-19 have been registered through clinicaltrials.gov. With so many trials, there is a risk that many will be inconclusive due to being underpowered or due to an inability to recruit patients. At academic medical centers, multiple trials are competing for the same resources; the success of one may come at the expense of another.
The COVID-19 Outpatient Pragmatic Protocol Study (COPPS) is a flexible phase 2, multi-site, randomized, blinded trial based at Stanford University designed to overcome these issues by simultaneously evaluating multiple COVID-19 treatments in the outpatient setting in one common platform with shared controls. This approach reduces the overall number of patients required for statistical power, while improving the likelihood that any enrolled patient receives active treatment.
The platform study has two main domains designed to evaluate COVID-19 treatments by assessing their ability to reduce viral shedding (Viral Domain), measured with self-collected nasal swabs, or improve clinical outcomes (Clinical Domain), measured through self-reported symptomology data. Data are collected on both domains for all participants enrolled. Participants are followed over a 28-day period.
COPPS has the advantage of pragmatism created around its workflow that is also appealing to potential participants because of a lower probability of inactive treatment. At the conclusion of this clinical trial we expect to have identified potentially effective therapeutic strategy/ies for treating COVID-19 in the outpatient setting, which will have a transformative impact on medicine and public health.
Keywords: COVID-19, SARS-CoV-2, Platform, Master, Adaptive, Phase 2
1. Introduction
The response to solving the COVID-19 crisis has been unprecedented [1]. 2020 included over 3000 registered COVID-19 trials on clinicaltrials.gov [2]. However, we currently lack a robust standard of care therapeutic agent for active COVID-19 in the outpatient setting to improve outcomes. This massive response from researchers worldwide has pushed the existing clinical trial infrastructure to its limits and requires changes to the existing clinical trial system [3]. At Stanford University alone, 40 trials – including 10 for the outpatient population – have begun or are pending launch.
With so many trials being launched, however, there is much at stake. At academic medical centers, multiple trials compete against one another for the same resources, where the success of one may come at the cost of the other. The thin spread of resources over multiple trials increases the risk launched trials will not be successful in addressing their objectives. Factors leading to possible failure include reduced enrollment, missing data (e.g., through attrition of participants), data integrity issues, or poor study design. We addressed several of these issues by creating a streamlined infrastructure within our Data Coordinating Center [4]; however, more is needed.
The need to consolidate effort has been recognized by several national entities, resulting in the ACTIV and ACTT trials in the US, and RECOVERY in the UK [5]. These trials are examples of a type of efficient study design known as a master, platform, or umbrella design, common in the oncology field, to which the Food and Drug Administration (FDA) is receptive and for which they have developed guidelines [6,7]. We also identified a platform protocol as a key element of our shared infrastructure that we established to streamline processes [4]. Specifically, the design allows for the simultaneous study of multiple agents and often includes flexible features to adapt given rapidly changing internal or external evidence [8]. Early in the pandemic (April 2020), Dean and authors – members of the WHO-sponsored R&D Blueprint or plan to design trials during public health emergencies [9] – raised the concept of such a protocol to address the virus [10].
The objective of this paper is to describe a platform protocol that provides a framework around multiple single trials to gain efficiency and increase flexibility thus mitigating some of the problems raised by the statistical and medical community for the proper conduct of trials during a pandemic. The COVID-19 Outpatient Pragmatic Protocol Study (COPPS) is a flexible, multi-site, randomized, blinded study based at Stanford University designed to evaluate COVID-19 treatments by assessing their ability to either reduce viral load or improve clinical outcomes. There are numerous strengths to the study that we describe here, including a shared control group that increases efficiency, a shared database, and pragmatic engagement with other sites. At the conclusion of this clinical trial we expect to have identified potentially effective therapeutic strategy/ies for treating COVID-19 that will have a transformative impact on precision medicine and public health.
2. Materials and methods
2.1. Trial design
COPPS is a multi-site adaptive platform protocol designed to evaluate COVID-19 treatments by assessing their ability to reduce viral shedding (Viral Domain) or improve clinical outcomes (Clinical Domain) across multiple sites. Treatment arms may be dropped or added during the trial period. For an investigational product to be included in the platform, data are collected on both Domains, with granular details on the evaluation of each treatment arm provided in a separate sub-protocol.
Overall, the study is designed to primarily evaluate efficacy. The platform trial allows one of two primary objectives when evaluating a treatment arm: one evaluating viral shedding (A. Viral Domain); and another evaluating clinical outcomes (B. Clinical Domain). The investigational product of interest, likely anti-viral or anti-inflammatory in nature, and its mechanism of action and scientific rationale will dictate which objective is primary under its sub-protocol (i.e., either A. Viral Domain or B. Clinical Domain) with the non-assigned Domain serving as a secondary objective. The platform trial will include a common control arm that will be used as a comparator for all arms in both Domains thus reducing the proportion of subjects not on an active agent and expediting recruitment for simultaneously run trials. Investigational products will not be compared to each other.
2.2. Primary, secondary, and exploratory objectives
The platform study allows objectives either evaluating viral shedding (Virology Domain) or COVID-19 related Clinical Outcomes (Clinical Domain).
The primary objective for investigational products within the Viral Domain is:
-
A.
To evaluate the efficacy of each therapeutic intervention in addition to standard supportive care compared with standard supportive care in reducing viral shedding of SARS-CoV-2 virus in outpatients with COVID-19 disease.
The primary objective for investigational products within for the Clinical Domain is:
-
B.
To evaluate the efficacy of each therapeutic intervention in addition to standard supportive care as compared to standard supportive care in improving clinical outcomes in outpatients with COVID-19 disease.
Secondary objectives are:
-
1)
The primary objective of the non-assigned domain
-
2)
To evaluate the efficacy of each therapeutic intervention in reducing SARS-CoV-2 related hospitalizations, Emergency Department (ED) visits, or death in outpatients with COVID-19 disease
-
3)
To assess the development of antibodies against SARS-CoV-2
-
4)
To evaluate the safety and tolerability of each therapeutic intervention compared with placebo plus supportive care.
Exploratory objectives are specific to each sub-protocol and are specified in their respective statistical analysis plans.
2.3. Eligibility criteria and exclusions
COPPS is designed to study adults with COVID-19 diagnosed as outpatients. Specifically, adults 18 through 80 years of age, with an initial diagnosis of COVID-19 as defined by an FDA-cleared molecular diagnostic assay based on reverse transcription polymerase chain reaction that provides evidence the patient is positive for SARS-CoV-2 within 72 h from initial swab from the time of informed consent, with onset of symptoms no more than 7 days at time of enrollment are potentially eligible for participation. Patients are not considered eligible if they need to be admitted to the hospital or are being evaluated for potential admission at enrollment. Exclusion criteria are tailored to each investigational drug. However, some exclusion criteria are shared among all arms in the platform in both domains. The general exclusion criteria for the platform protocol is as follows:
-
1.
At screening, the subject needs to be admitted to the hospital or is being evaluated for potential admission.
-
2.
Previous use of drugs that may be active against COVID-19 in the eyes of the investigators. Subject is using adrenocorticosteroids (except topical or inhaled preparations or oral preparations equivalent to or less than 10 mg of oral prednisone) or immunosuppressive or immunomodulatory drugs (e.g., immunosuppressants, anticancer drugs, interleukins, interleukin antagonists or interleukin receptor blockers).
-
3.
At screening, subject tests positive by a urine pregnancy test.
-
4.
Subject has a serious chronic disease (e.g., uncontrolled human immunodeficiency virus [HIV], cancer requiring chemotherapy within the preceding 6 months, and/or moderate or severe hepatic insufficiency).
-
5.
Has renal insufficiency requiring hemodialysis or continuous ambulatory peritoneal dialysis (CAPD).
-
6.
Has liver impairment greater than Child Pugh A.
-
7.
Has a history of alcohol or drug abuse in the previous 6 months.
-
8.
Has a psychiatric disease that is not well controlled where controlled is defined as: stable on a regimen for more than one year.
-
9.
Has taken another investigational drug within the past 30 days.
-
10.
Is deemed by the Investigator to be ineligible for any reason.
2.4. Randomization and blinding
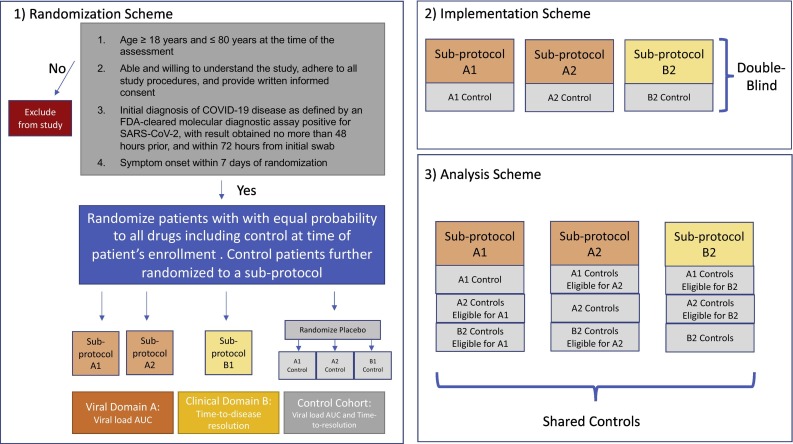
Patients will be randomized to one of the investigational therapies that are available at the time of the enrollment and for which the patients are eligible or to the control arm (See Fig. 1 ). A biased coin randomization using age and sex as strata will be used to balance the mix of older/younger (stratified by 50 years of age) and female/male randomized to each arm. Patients randomized to the control arm will be further randomized to a sub-protocol (among the set of sub-protocols currently available and for which that patient is eligible) that will determine the type of placebo treatment the patient receives and study procedures for any minor processes that may differ across sub-protocols to maintain blinding. Specifically, each investigational therapy will have a corresponding matching placebo that will mimic the active therapy. For example, if the drug is to be taken orally, the tablets will look similar to the investigational therapy for the assigned sub-protocol and intended to be taken on the same schedule as the active therapy. If delivered subcutaneously, the placebo will be a saline injection. Thus, as study arms are added, placebos that look like that study arm will also be added. Patients and investigators will be blinded to whether a patient receives an active or placebo version of the treatment, although study investigators will know which sub-protocol a patient is randomized to. In this manner, all sub-protocols will be double-blinded.
Fig. 1.
Schema for study inclusion.
2.5. Initial drugs of interest and common control arm
COPPS will be initiated with two candidate therapies: one under each Domain. Camostat/foipan will be studied under the Viral Domain, and acebilustat will be studied under the Clinical Domain.
Camostat/foipan has been extensively used to treat pancreatitis patients for more than 20 years in Japan and was based on evidence from multiple clinical trials. In brief, the hypothesis is that the transmembrane serine protease, TMPRSS2, plays a critical role in the life cycle of SARS-CoV-2. Using a panel of established human cell lines, Matsuyama et al. (2020) showed that SARS-CoV-2 infection is enhanced by TMPRSS2 expression in a host cell[15]. Independently, Hoffmann et al. (2021) demonstrated that SARS-CoV-2 cell entry depends on the angiotensin converting enzyme II receptor and TMPRSS2 activity, the latter resulting from the ability of the protease to prime the viral spike protein[16]. The authors further observed that camostat could block viral entry in vitro.
Acebilustat (CTX-4430) is a novel, synthetic, small molecule developed for the treatment of inflammatory conditions with potential to reduce the morbidity and mortality associated with COVID-19. It is a potent inhibitor of leukotriene A4 hydrolase (LTA4H), the rate-limiting enzyme in the production of LTB4. In cystic fibrosis patients, a 100 mg once-daily oral dose of acebilustat reduced white blood cell counts in the lungs by up to 65% and reduced the occurrence of pulmonary exacerbations that frequently lead to hospitalization by up to 48% in patients with early stage lung disease. The safety of 100 mg once daily oral acebilustat has been profiled in over 321 human adults (3 Phase 1 studies and 2 Phase 2 studies), including over 100 cystic fibrosis patients receiving 50 mg or 100 mg once daily oral acebilustat for 48 weeks. No suspected unexpected serious adverse reactions (SUSARs) have been reported in healthy volunteer studies to date.
To address primary objectives, a control arm will be included. The control arm is referred to as a common control arm because the participants in this arm will be shared to address objectives across multiple sub-protocols. More specifically, a subset of the common control arm will be used as the comparator for each individual treatment, where the subset of participants is defined by those participants in a control arm who could have been randomized to the active treatment at the time of randomization (i.e., the patient was eligible for the active treatment and the drug was being studied at the time of randomization). Similar to other platform trials including the United Kingdom-based RECOVERY trial, patients in the common control arm will be included in the comparison group of any treatment that the patient could have been randomized to at the time of randomization, regardless of the patient's assigned sub-protocol [11]. This maintains comparability between the study arm of interest and the controls used as comparators with respect to secular time and eligibility criteria.
2.6. Participant timelines, assessments, and measures
Under each Domain, a set of assessments will be performed (Table 1 ). Assessments can be specific to a sub-protocol, with the exception of the assessments required to evaluate the Viral and Clinical Domain primary endpoints. Both primary assessments are required to be collected for each sub-protocol in the platform in order to share controls across the sub-protocols.
Table 1.
Domain assessments (X is all domains, O is viral domain only, & is clinical domain only).
| Day of study | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 14 | 21 | 28 | 35 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Assessments in the clinic | X | X | O | X | ||||||||||
| Physical exam | X | X | O | X | ||||||||||
| Vitals | X | X | O | X | ||||||||||
| Clinical status | X | X | O | X | ||||||||||
| SpO2 | X | X | O | X | ||||||||||
| Self-collected nasal swab and self-assessmenta | X | X | X | X | X | X | X | X | X | O | X | X | X | |
| Oropharyngeal swab | X | X | O | X | ||||||||||
| Blood collected by phlebotomy for clinical labs (CBC, Chem 7, LFTs) | X | X | O | X | ||||||||||
| Blood collected by phlebotomy for biobanking/immunology studies | X | X | ||||||||||||
| Blood collected by phlebotomy for PK (if applicable) | X | X | O | |||||||||||
| Telehealth visit | & | X | X | X |
Note: Symptom self-assessment through COSS occurs every day from day 1 through 28.
The primary outcome for the Viral Domain is the AUC of viral load over a 10-day period. AUC of shedding of SARS-CoV-2 virus will be defined using the linear trapezoidal method through day 10 from self-collected nasal swab RT-PCR data. Mean AUC levels will be compared between groups using a linear regression model regressing log(AUC) on treatment arm, age, and sex.
The primary outcome measure for the Clinical Domain is time from randomization to sustained resolution of symptoms resolution, defined as absence of self-reported symptoms for at least 48 h not including sense of taste or smell, and defining recovery for fatigue and cough as mild or none [12]. Data are based on the COVID-19 Outpatient Symptom Survey (COSS) (See Appendix) and are considered right-censored. Specifically, if data on symptoms are only partially completed, resolution is undetermined and considered censored. For situations where the symptom survey indicates resolution, but the participant skips one or more surveys prior to a survey that indicates sustained resolution, we will employ multiple imputation techniques that assume a missing at random (MAR) mechanism to fill in the data. Any participant who drops out prior to having one survey indicating mild to no symptoms will be censored at Day 28. Death prior to providing evidence of resolution will be considered a competing risk.
Secondary outcomes include the primary outcome measure for the non-assigned Domain, as well as:
-
1)
An indicator for whether the patient was evaluated in either the Urgent Care setting or the ED for a SARS-CoV-2-related complication
-
2)
An indicator for whether the patient was admitted for a SARS-CoV-2-related complication
-
3)
An indicator for whether the patient was admitted to the ICU
-
4)
An indicator for whether the patient died due to a SARS-CoV-2-related complication
-
5)
Antibody titers
-
6)
The occurrence of adverse events during the study period
3. Statistical considerations
The analytic approach described below describes how we will address the primary and secondary objectives of each specific sub-protocol and will be performed at pre-specified timepoints listed in the Statistical Analysis Plan (e.g., at interim and final analyses).
3.1. Descriptive analyses
Descriptive statistics (proportions for categorical variables, means, medians, standard deviations and interquartile ranges for continuous variables) will be reported for all key patient characteristics, including baseline and demographic characteristics, use of medications, compliance, and study completion status. Data that are missing on key patient characteristics and the outcome will be fully described, including any patterns of missingness (i.e., any relationships between missingness of a variable and patient characteristics). A CONSORT diagram displaying the number of patients screened, eligible, and consented along with reasons for ineligibility will be provided for the overall trial as well as for the specific sub-protocol of interest. Graphical tools such as histograms, boxplots, and scatterplots will be created to assess quality of data and to display patterns over time.
3.2. Primary analyses
We rely on intention-to-treat (ITT) principles when addressing the primary objective. Specifically, we will address the primary objective of a given sub-protocol using the ITT population. The ITT population includes all patients randomized to that treatment and all control patients that could have been randomized to the treatment and that were eligible for the active treatment at the time of the patients' randomization. Patients will be analyzed according to their assigned treatment arm. All efficacy analyses will be completed in the ITT population. The per-protocol (PP) population will include all randomized patients who completed follow-up and adhered to study procedures. All efficacy analyses will be also be completed in the PP population as supportive evidence for the primary efficacy analysis.
For sub-protocols in the Viral Domain, the AUC SARS-CoV2 viral RNA levels during follow-up will be estimated using the linear trapezoidal method, and mean AUC levels compared between groups using a linear regression model regressing log(AUC) on treatment arm, age, and sex. We assess significance of differences using a two-sided Wald test.
For sub-protocols in the Clinical Domain, if no deaths are observed as anticipated, time to sustained symptom resolution will be compared between the treatment and control arms using a two-sided Wald test derived from a Cox proportional hazards model adjusted for age and sex. The hazard ratio for symptom resolution will be estimated, along with its 95% confidence interval, from a Cox proportional hazards model. If the proportional hazards assumption is not met, we will consider an extended Cox model that relaxes the proportional hazards assumption. The distribution of symptom resolution will be estimated using the Kaplan-Meier method, and Kaplan-Meier curves will be presented for each treatment arm. Median time to symptom resolution at the end of the study period along with 95% confidence intervals will be presented for each treatment arm. In the presence of any deaths, a Fine-Gray model will be used to compare symptom resolution between arms with death considered a competing risk using a subdistribution hazard ratio. [13]
3.3. Interim efficacy and futility analyses
There will be interim analyses for efficacy and futility based on the primary outcome measure in each experimental arm after 50% of the patients have initiated treated. An arm will stop early for superior efficacy if the two-sided Wald test using the investigational therapy and control arms is less than alpha = 0.00001, where a low level of significance was intentionally chosen to set the bar high for stopping. This decision was motivated by the low probability of mortality in this outpatient population and the critical need to learn about the full trajectory of virology and symptomatology in this low-risk population. The test will be performed using the same methods as described above for the primary endpoint at the final analysis (performed at alpha = 0.04999). A futility analysis that evaluates conditional power will indicate whether the study should be stopped early for futility, where a sub-protocol will be terminated if the conditional power is less than 0.25.
3.4. The handling of missing data
Our analyses will assume data are missing at random. Area under the curve (AUC) will be defined by a single participant's logarithmic base 10 viral load from self-collected nasal swab tests collected on days 1 through 10. AUC will be calculated by the trapezoidal rule, using exact times of collection of each nasal swab. Multiple imputation using chained equations will be used to impute missing viral load data prior to AUC calculation. Five data sets will be imputed, and imputed values will be calculated using non-missing viral load on each of the 13 sample collection days (days 1:10,14,21,28), treatment arm, age, sex, vaccination status, and whether or not a participant was hospitalized. A pooled linear regression model will be fit to log-transformed AUC as a function of treatment, age, and sex using the 5 imputed data sets. Difference in AUC by treatment arm and corresponding 95% confidence intervals will be reported using Rubin's Rules. The difference in AUC by treatment arm will also be done after refitting the model without participants who were hospitalized to assess sensitivity of results.
For the primary endpoint of time to sustained symptom resolution (Clinical Domain), data are considered right-censored. Specifically, if data on symptoms are only partially completed, resolution is undetermined and considered censored. For situations where the symptom survey indicates resolution, but the participant skips one or more surveys prior to a survey that indicates sustained resolution, we will employ multiple imputation techniques that assume a missing at random (MAR) mechanism to fill in the data. Any participant who drops out prior to having one survey indicating mild to no symptoms will be censored at Day 28. Sensitivity to assumptions regarding missingness will be addressed through sensitivity analyses. Deaths that occur prior to evidence of symptom resolution will not be censored and instead be considered a competing risk.
If more than 5% of participants are missing data on 20% of the measures required to derive their primary endpoint, we will perform a sensitivity analysis to evaluate the robustness of our findings to missing data assumptions. In the sensitivity analysis, among those missing timing of resolution, we will assume varying proportions (e.g. 25%, 50%) of participants in each arm who would have resolved symptoms by Day 28. The proportions will differ by arm so we can evaluate whether our findings stand if it were true that people who dropped out had symptom resolution earlier in the placebo arm vs the active arm. Additionally we will employ sensitivity analyses for the worst case scenario (not MAR or NMAR mechanism) where those under treatment are assumed to not have resolved symptoms, while those under the control arm follow a MAR assumption.
3.5. Operating characteristics
For a given sub-protocol under the Viral Domain, a two-sided Wald test at the alpha = 0.04999 level of significance will be utilized for the final analysis. 120 patients (60 per arm) will provide 80% power to detect an effect size of 0.5 (Cohen's D). This leaves alpha = 0.00001 to check for overwhelming efficacy after 50% of participants have completed 24 h of follow-up.
For a given sub-protocol under the Clinical Domain, A two-sided Wald test of the hazard ratio from a Cox proportional hazards model with 60 in the control group and 60 in the treatment group, achieves 80% power at a 5% significance level, to detect a hazard ratio of 1.9 when the control group median time-to-resolution is 10 days. This assumes the study lasts for 118 days for which subject accrual occurs in the first 90 days. For calculations, the accrual pattern across time periods is assumed to be uniform (all periods equal) and the proportion dropping out for each the control and treatment groups is assumed to be 5%.
Operating characteristics were further explored through simulation to provide insight into the impact of sharing a control group across sub-protocols on inferential errors (Table 2, Table 3 ). For simplicity we focused on only one outcome type – time to symptom resolution – where four sub-protocols or treatments were considered. Time-to-resolution was generated from an exponential distribution assuming an underlying hazard rate of 0.05. The simulation study further assumed one interim and one final analysis, a 10% censoring rate, and 5000 iterations per scenario. Two scenarios were considered: none of the four treatments were effective in Scenario A (HR = 1 for all drugs) (Table 2), and 2 of the 4 treatments were effective in Scenario B (HR = 2 for 2 drugs and HR = 1 for 2 drugs) (Table 3). Type I error and power were evaluated using Scenarios A and B, respectively, when a low (3.3%), medium (50%), and high (96.6%) proportion of the controls were shared. The conditions where the proportion of shared controls were low should closely mimic conditions of 4 independently run trials where only the joint infrastructure (e.g., database, electronic case report forms, type and timing of measurements, standard operating procedures) – and not controls – were shared. In this situation, our simulations showed that the type I error rate or probability that at least 1 drug was identified as interesting when none of the drugs were effective was 0.184 (close to the theoretical value of 0.187). The type I error rate decreased as the proportion of shared controls increased (from 0.184 when 3.3% were shared to 0.159 when 96.6% of the controls were shared). Not surprisingly, however, the probability that at least 2 drugs were incorrectly identified as effective increased as the proportion of shared controls increased. This phenomena has been described previously in the literature [14] and makes intuitive sense as the correlation increases with the proportion of shared controls, which would imply that errors of this type would also increase. While sharing the controls at a level of 96.6% led to a 3-fold increase in probability that at least 2 drugs were incorrectly identified as effective, the absolute probability of such an error remained low (0.033). Power (Table 3), i.e. the probability of correctly identifying the two effective drugs, is as expected (0.839 in simulations when sharing a low proportion of controls and 0.836 when conducting 4 independent trials with no shared controls). The power of correctly identifying the two effective drugs increased as the proportion of shared controls increased (from 0.836 to 0.858).
Table 2.
Operating characteristics for Scenario A when 4 drugs are assessed and none are effective: Prob 1 (probability that at least 1 drug incorrectly identified as effective) and Prob 2 (probability that at least 2 drugs incorrectly identified as effective) by proportion of shared controls under COPPS (estimated from simulation) relative to 4 independently run trials with no shared controls (theoretically derived probabilities).
| COPPS |
Four independent trials No shared controls |
|||
|---|---|---|---|---|
| Proportion of shared controls | Prob 1 | Prob 2 | Prob 1 | Prob 2 |
| 3.3% (2/60) | 0.184 | 0.013 | 0.188 | 0.013 |
| 50% (30/60) | 0.178 | 0.017 | ||
| 96.6% (58/60) | 0.159 | 0.033 | ||
Table 3.
Operating characteristics for Scenario B when 4 drugs are assessed and two are effective: Power 1 (probability that at least 1 of the 2 drugs correctly identified as effective) and Power 2 (probability that both drugs correctly identified as effective) by proportion of shared controls under COPPS (estimated from simulation) relative to 4 independently run trials with no shared controls (theoretically derived probabilities).
| COPPS |
Four independent trials No Shared Controls |
|||
|---|---|---|---|---|
| Proportion of shared controls | Power 1 | Power 2 | Power 1 | Power 2 |
| 3% (2/60) | 0.993 | 0.839 | 0.993 | 0.836 |
| 50% (30/60) | 0.986 | 0.849 | ||
| 96.6% (58/60) | 0.974 | 0.858 | ||
3.6. Data management and monitoring
Any paper data (e.g. informed consent documents, screening, and contact information) will be maintained in a research chart. The remaining clinical data– daily questionnaires, case record forms (CRFs), and laboratory results–will be entered and maintained in Stanford's REDCap database (https://www.project-redcap.org/) using the minimum number of personal identifiers (date of birth, dates of visit). Physical copies of laboratory data from point of care devices will be stored in a file. Specimen containers and blood tubes will be labeled by the clinical research coordinator only with study ID and date. Virologic measurements will be entered into a database stored on secured servers and provided from the clinical laboratory to the study team. Biobanked specimens (e.g. PaxGene, plasma, peripheral blood mononuclear cells (PBMC)) will be entered into the Stanford Biobank database stored on secure servers.
All subjects will be given a study ID. Symptom questionnaires and case report forms used at clinic visits will be completed by subjects and or study personnel using only study ID. Specimen containers will be labeled by the Clinical Research Coordinator (CRC) only with study ID and date, time of collection. Virologic measurements and other biomarkers will be provided back to the study team electronically once available, and entered into the database. In order to ensure data quality, the study CRC will perform a regular data quality audit. For this audit all study forms entered into the data management system will be assessed for accuracy with source documents. In addition, the study Data Manager will perform weekly reviews of the checks described in the data management plan to identify potential data quality issues. The data will be owned by Stanford University.
Electronic data including all study databases and supporting electronic documentation will be archived to cloud-based servers on a daily basis. All data will be kept in secured REDCap and Box servers. Only the research team will have access to the data.
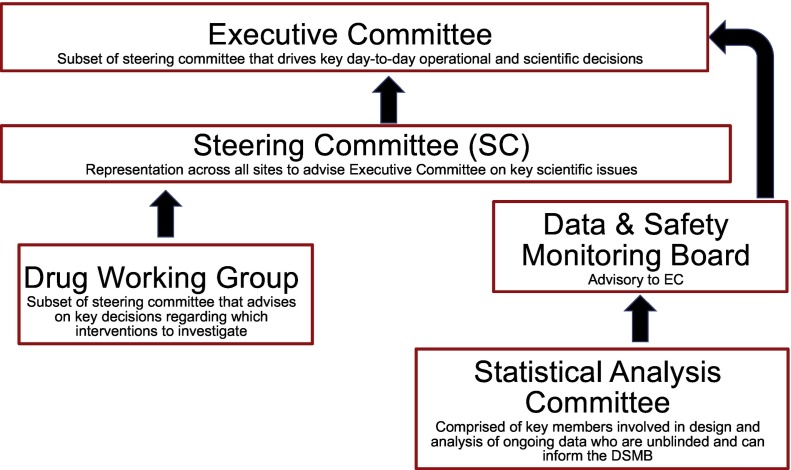
4. Governance
The trial will be governed by an Executive Committee (EC) that will drive operational and scientific decisions and a larger Steering Committee (SC) – comprised of the EC and other members including the sub-protocol principal investigators – who will meet on a quarterly basis and advise on key scientific decisions (Fig. 2 ). A subset of the SC will form the Drug Working Group that will advise the SC on drugs for consideration. An independent data and safety monitoring board (DSMB) will serve as advisory to the EC on issues regarding safety, ethical issues, and prompt dissemination of information important to the public. A Statistical Analysis Committee (SAC) of key unblinded members from the study team will support and inform the DSMB.
Fig. 2.
Governance structure for COPPS.
The EC will be the primary decision-making body for day-to-day operations of COPPS. The EC will meet weekly in order to review and have final sign-off on the trial protocol, manual of operations, risk-based monitoring plan, data management plan, and statistical analysis plan. The SC will advise on key scientific decisions and meet quarterly to address high-level trial policy, protocol and operational issues, and dissemination of findings. In particular, the SC will be responsible for introducing additional treatment arms for inclusion in the protocol and successful study completion.
5. Discussion
Coordinated effort is needed to address the COVID-19 pandemic. Initiatives by the NIH and the WHO to do so will ensure that findings from resulting efforts will be complementary and conclusive. We recognize the importance of such principles and our own institution's participation in these studies demonstrates its enthusiasm for contributing to such solutions. Even at the institutional level, however, consolidated effort is needed.
The COVID-19 Outpatient Pragmatic Platform Study (COPPS) is a flexible platform trial aligned with such principles and includes important goals to provide a shared infrastructure for leveraging resources across individual trials initiated at Stanford. Indeed, the motivation behind COPPS was the independent launch of numerous trials targeting similar populations. COPPS can therefore serve as a blueprint for other academic medical centers facing similar demands.
There are numerous strengths to the platform protocol that we describe here, including efficiency of shared controls, a shared database, and engagement with other sites that can benefit the objectives of multiple sub-protocols within the platform, to name a few. Having a common control group is more efficient and allows each primary objective of a sub-protocol to be addressed faster and with fewer participants. The benefits of the common control group go beyond efficiency. The sharing of the controls makes participation more appealing to a potential participant because assignment to a placebo arm has a probability less than 0.50 (typical of an individual trial), thus increasing the probability of participation in the trial. This is particularly relevant in an outpatient trial where patients may be less motivated to join a trial than in the hospitalized setting where the situation is known to be more dire. Finally, the platform provides a mechanism for introducing arms that is far more efficient than starting an entirely new trial.
The shared infrastructure of COPPS eliminates competition of individual trials launched at the same institution that require access to the same population. For this purpose, most academic medical centers have established committees to prioritize which trials can proceed given the demand to launch trials in the same space and their promise. A study like COPPS helps to solve this issue by randomizing across all potential therapies being evaluated on the same patient population. Indeed, with a unified study team, we can ensure that the quality of data to study each agent of interest is consistently high. In addition, long-term benefits that include one large cohort of outpatients with COVID-19 with common measures can be used to address many secondary analyses that can give further insight into the disease beyond the primary and secondary objectives listed here.
COPPS was designed to accommodate a busy workflow amidst launch of numerous trials in the face of a pandemic where answers were needed quickly, and therefore much flexibility has been allowed. For example, sub-protocols can derive a primary outcome under one of two Domains: Viral or Clinical using the required data. An important restriction is that the virology and symptomology data be collected for every sub-protocol in the platform. This allows investigators freedom to tailor the outcome to their specific question but also allows for secondary analyses that potentially pool across all sub-protocols made possible by the common measures. It is conceivable that we may add additional domains in the future; the only requirement is that the primary endpoint for the domain's objectives be consistent with the study's assessment schedule to make the sharing of controls possible.
There are some weaknesses to the COPPS design, primarily stemming from the pragmatic nature of its design. For example, the operating characteristics are only modestly improved with the sharing of controls relative to conducting multiple independent trials. Further, there is an increase in probability of falsely identifying multiple treatments as effective due to the correlation induced by the shared controls. We note, however, that the probability of such an error still remains small. The benefit of the shared controls in increasing operational efficiency and in obtaining an answer faster and with fewer resources outweighed this negative characteristic. In addition, the assessment schedules differ between the domains because the platform trial is designed to study a broad range of interventions. The flexibility of the design, however, allows tailored primary endpoints that may differ across sub-protocols. While this flexibility enhances inclusion of multiple types of interventions, it may compromise comparability across published study findings. However, the possibility of pooling across sub-protocols is enabled through the common collection of variables. Because COPPS was designed to incorporate multiple individual trials, a more comprehensive strategy as to which drugs and therapies to include along with a Bayesian approach to leverage information across trials was not taken. The established governance structure we established, however, will consider individual outpatient trials for eligibility based on the complementary nature of the agent being considered. However, we anticipate that some monotherapies that are studied within COPPS may present opportunities to investigate their activity in a combination therapy within COPPS. Blinding with respect to sub-protocol was not possible in COPPS overall, although double-blinding is present within each sub-protocol. Because each placebo generated by the company was designed to look like the drug of interest, we decided to randomly select a placebo type for those assigned to the placebo arm. Thus, the sub-protocol to which a participant was assigned (although not active or placebo) will be known. Each therapeutic agent is assigned to only one domain even though data are collected for both domains, as only one primary endpoint is assumed for each sub-protocol in this phase 2 study. However, it is possible for a sub-protocol to opt for multiple primary endpoints. Such a decision would require adjustments to the analysis plan that appropriately controls for the type I error. While the common control arm increases efficiency, it may bias our estimates of treatment toward the null. For example, if A1 is an oral drug and A2 is a drug administered through injection, it is likely that A2 will have higher placebo effect than A1. Including A2 controls eligible for A1 may bias our estimates of the treatment effect toward the null. However, we assume in general that the placebo effects are homogenous across treatment strategies and therefore the impact of this bias is likely negligible.
There were considerable challenges in establishing a platform protocol during a pandemic. One issue is the fast-changing landscape of therapeutics. For example, ongoing trials external to ours may provide sufficient evidence to warrant changes to standard of care for the outpatient. We have observed this with the recent Emergency Use Authorization for monoclonal antibodies for a subset of outpatients deemed at higher risk of mortality. For drugs where it makes sense to combine the experimental therapy with a monoclonal antibody treatment, participants may still be eligible for study under that drug's sub-protocol, but for others, such high-risk patients may be excluded. In cases, where such participants are included, they can serve as shared controls only among those sub-protocols for which they could be eligible. This exemplifies the additional rationale behind sharing controls across sub-protocols enrolled during contemporaneous time periods and only for those drugs for which they could have been eligible at the time of study entry. In addition, it is logistically easier to plan the study of one experimental therapy with one study team than to join multiple study teams already in motion and get consensus on primary and secondary outcomes, sample types, timing of measurements, follow-up period, in-clinic visits, biomarkers, and monitoring procedures. In addition to gaining consensus among study investigators, regulatory approval must be obtained from the FDA and IRB, here with the added complexity of multiple drugs that may deviate from shared inclusion/exclusion criteria or that may come with a unique set of contraindications that need to be considered. However, once established, the platform protocol can greatly facilitate entry and regulatory clearance of additional sub-protocols. Finally, the pharmaceutical company sponsoring the study of the drug must embrace participation in the platform. COPPS is designed such that there are no head-to-head comparisons between drugs and such that each pharmaceutical company can have access to the data needed to address their set of objectives only and not the data to study the other agents. We believe from this standpoint there is no downside from the company's standpoint to joining COPPS.
As Dean and others advocated [10], such a platform can be extended to accommodate the study of COVID-19 in the future as well as other types of disease outbreaks. We therefore see this platform as a longer-term investment of critical infrastructure that extends beyond COVID-19.
Funding
This work was partially supported by the National Institutes of Health Grant UL1-TR003142, Grant P30-CA124435, and Grant P30-DK116074.
Declaration of Competing Interest
Eric Springman: Dr. Springman is a paid consultant for Celltaxis LLC.
Yvonne Maldonado: Stanford University School of Medicine has received funds from an anonymous donor for Dr. Maldonado to conduct a Phase 2 favipiravir study which should be completed by spring 2021.
No disclosure from other authors.
Acknowledgments
Acknowledgement
The Stanford REDCap platform (http://redcap.stanford.edu) is developed and operated by Stanford Medicine Research IT team. The REDCap platform services at Stanford are subsidized by a) Stanford School of Medicine Research Office, and b) the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant UL1TR001085.
Appendix A. COVID Outpatient Symptom Score (COSS)
| Please take your oral temperature and assess your oxygen saturation with your pulse oximeter. | Oral temperature. | ________ F |
| Time temperature was obtained. | ________ AM/PM | |
| Blood oxygen saturation. | ________ % | |
| Did you take a nasal swab today (study day 1–10,14,21,28)? | Yes / No | |
| Have you taken your study drug as prescribed in the last 24 h? (study day 1–10) | Yes / No | |
| If it is written on your schedule (clinic visit day 3, 14, 28): did you collect your stool sample for your next visit? | Yes / No |
COVID-19 Outcome Symptom Scale (COSS) Daily Questionnaire
The following questions are about how you feel and how things have been during the past 24 h compared to your typical health. Give the one answer that comes closest to the way you have been feeling. Select “None” if you have not had this symptom.
| How do you feel compared to yesterday? |
|
||||
| How do you feel compared to last week? |
|
||||
| Cough? | |||||
|
|
|
|
|
|
| Shortness of breath (difficulty breathing)? | |||||
|
|
|
|
|
|
| Fatigue (low energy)? | |||||
|
|
|
|
|
|
| Headache? | |||||
|
|
|
|
|
|
| Body ache? | |||||
|
|
|
|
|
|
| Joint pain? | |||||
|
|
|
|
|
|
| Chest pressure? | |||||
|
|
|
|
|
|
| Abdominal pain? | |||||
|
|
|
|
|
|
| Sore throat? | |||||
|
|
|
|
|
|
| Nasal congestion? | |||||
|
|
|
|
|
|
| Chills? | |||||
|
|
|
|
|
|
| Feeling hot or feverish? | |||||
|
|
|
|
|
|
| Runny nose? | |||||
|
|
|
|
|
|
| Taste? | |||||
|
|
|
|
|
|
| Smell? | |||||
|
|
|
|
|
|
| Diarrhea? (loose or watery stools in 24 h) | |||||
|
|
|
|
|
|
| Nausea (feeling like you want to throw up)? | |||||
|
|
|
|
|
|
| Vomiting? | |||||
|
|
|
|
|
|
| How many times have you vomited today? | ______ | ||||
| Do you have a rash? | Yes / No | ||||
| The rash is (check all that apply): |
|
||||
| In the past 24 h, have you returned to your usual health (before your COVID-19 illness)? | Yes / No | ||||
References
- 1.Herper Matthew, Riglin Erin. Data shows panic and disorganization dominate the study of Covid-19 drugs. STAT. July 6, 2020 https://www.statnews.com/2020/07/06/data-show-panic-and-disorganization-dominate-the-study-of-covid-19-drugs/ (accessed on September 20, 2020) [Google Scholar]
- 2.Thorlund K., Dron L., Park J., Hsu G., Forrest J.I., Mills E.J. A real-time dashboard of clinical trials for COVID-19. Lancet Digit. Health. 2020 Jun 1;2(6):e286–e287. doi: 10.1016/S2589-7500(20)30086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn A. Business Insider; 2020 Apr 24. There are Already 72 Drugs in Human Trials for Coronavirus in the US. With Hundreds More on the Way, A Top Drug Regulator Warns We Could Run Out of Researchers to Test Them All.https://www.businessinsider.com/fda-woodcock-overwhelming-amount-of-coronavirus-drugs-in-the-works-2020-4 (accessed on July 20, 2020) [Google Scholar]
- 4.Hedlin H., Garcia A., Weng Y., et al. Clinical trials in a COVID-19 pandemic: shared infrastructure for continuous learning in a rapidly changing landscape. Clin. Trials. 2021;18(3):324–334. doi: 10.1177/1740774520988298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herper Matthew. NIH to start “flurry” of large studies of potential Covid-19 treatment. STAT. July 23 2020 https://www.statnews.com/2020/07/23/nih-to-start-flurry-of-large-studies-of-potential-covid-19-treatments/ (accessed on September 20, 2020) [Google Scholar]
- 6.Woodcock J., LaVange L.M. Master protocols to study multiple therapies, multiple diseases, or both. N. Engl. J. Med. 2017;377:62–70. doi: 10.1056/NEJMra1510062. [DOI] [PubMed] [Google Scholar]
- 7.Food and Drug Administration . U.S. Food & Drug Administration; October 2018. Master Protocols: Efficient Clinical Trial Design Strategies to Expedite Development of Oncology Drugs And Biologics. Draft Guidance for Industry.https://www.fda.gov/regulatory-information/search-fda-guidance-documents/master-protocols-efficient-clinical-trial-design-strategies-expedite-development-oncology-drugs-and (accessed on September 20, 2020) [Google Scholar]
- 8.The Adaptive Platform Trials Coalition, Angus D.C., Alexander B.M., et al. Adaptive platform trials: definition, design, conduct and reporting considerations. Nat. Rev. Drug Discov. 2019;18:797–807. doi: 10.1038/s41573-019-0034-3. [DOI] [PubMed] [Google Scholar]
- 9.Kieny M.P., Salama P. WHO R&D blueprint: a global coordination mechanism for R&D preparedness. Lancet. 2017;389:2469–2470. doi: 10.1016/S0140-6736(17)31635-5. [DOI] [PubMed] [Google Scholar]
- 10.Dean N.E., Gsell P.S., Brookmeyer R., et al. Creating a framework for conducting randomized clinical trials during disease outbreaks. N. Engl. J. Med. 2020;382:1366–1369. doi: 10.1056/NEJMsb1905390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Recovery Randomised Evaluation of COVID-19 Therapy. 2020. https://www.recoverytrial.net (accessed on September 20, 2020)
- 12.Food and Drug Administration . U.S. Food & Drug Administration; September 2020. Assessing COVID-19-Related Symptoms in Outpatient Adult and Adolescent Subjects in Clinical Trials of Drugs and Biological Products for COVID-19 Prevention or Treatment. Guidance for Industry.https://www.fda.gov/regulatory-information/search-fda-guidance-documents/assessing-covid-19-related-symptoms-outpatient-adult-and-adolescent-subjects-clinical-trials-drugs (accessed on September 20, 2020) [Google Scholar]
- 13.Fine J.P., Gray R.J. A proportional hazards model for the subdistribution of a competing risk. J. Am. Stat. Assoc. 1999;94:496–509. [Google Scholar]
- 14.Freidlin B., Korn E.L., Gray R., Martin A. Multi-arm clinical trials of new agents: some design considerations. Clin. Cancer Res. 2008 Jul 15;14(14):4368–4371. doi: 10.1158/1078-0432.CCR-08-0325. [DOI] [PubMed] [Google Scholar]
- 15.Matsuyama S., Nao N., Shirato K., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U.S.A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., Hofmann-Winkler H., Smith J.C., et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65:103255. doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]