Abstract
Background
With virtually dried out new antibiotic discovery pipeline, emergence and spread of antimicrobial resistance is a cause for global concern. Colistin, a cyclic polypeptide antibiotic, often regarded as last resort for multi drug resistance gram-negative bacteria, is also rendered ineffective by horizontal transfer of resistance genes. Surveillance of colistin resistance in GNB is essential to ascertain molecular epidemiology.
Methods
Whole genome sequencing (WGS) of an unusual colistin resistant urinary isolate of Escherichia coli was performed using Illumina MiSeq platform using 2x250bp V2 chemistry by following the manufactures protocol (Illumina Inc. USA). Multiple web-based bio-informatic tools were utilized to ascertain antibiotic resistant genes.
Results
An approximate 5.4 Mb of genome of the urinary isolate AFMC_UC19 was sequenced successfully. Mobile colistin resistance gene (mcr) on the plasmid responsible for horizontal spread was absent in the isolate.
Conclusion
Colistin resistance has been reported previously in Klebsiella pneumoniae and it is a rare occurrence in Escherichia coli in Indian setting. Although the isolate lack mcr mediated colistin resistance, emergence and spread of colistin resistant in gram-negative bacteria pose a threat.
Keywords: Colistin, Antibiotic resistance, Escherichia, Whole genome sequencing
Introduction
Colistimethate or ‘colistin’ is polymyxin antibiotic, effective against multidrug-resistant Gram-negative infections caused by Acinetobacter baumanii, Pseudomonas aeruginosa, Klebsiella pneumoniae and other members of Enterobacteriaceae.1 The primary site of action of colistin is lipopolysaccharide layer of the outer membrane of Gram-negative bacterial cell wall. It disrupts the cell membrane by binding anionic LPS molecule by displacing Mg++ and Ca++ ions, resulting in altered cell permeability.2 Indiscriminate use of colistin in clinical practice and veterinary medicine has resulted in emergence of resistance to colistin. Colistin-resistant Enterobacteriaceae of human as well as animal origin have been reported from several countries now. The resistance to colistin is mediated primarily by chromosomal mutations. However, in 2015, horizontal transfer of resistance due to plasmid-mediated mobile colistin resistance gene (mcr-1 gene) was reported in Enterobacteriaceae3. Colistin is considered an antibiotic of choice for multidrug resistance Gram-negative organisms such as carbapenem-resistant Enterobacteriaceae. This mcr-1-mediated resistance, especially of animal origin has been reported in several countries.4, 5, 6, 7 In Indian setting, the overall susceptibility to colistin among Enterobacteriaceae is 92%.8 Emergence and spread of colistin-resistant in Gram-negative bacteria pose a public health threat in resource-limited settings. Genomic analysis to investigate colistin-resistant mechanism in Enterobacteriaceae is an important aspect of resistance surveillance strategy. Here we describe a whole genome sequencing (WGS) of colistin-resistant strain of Escherichia coli, isolated from an individual with urinary tract infection.
A 46-year-old female was admitted with a history of fever and burning micturition of 2-day duration. Clinical history revealed that she underwent hysterectomy for fibroid uterus a week back and was managed with injection amoxycillin-clavulanate as per hospital antibiotic policy. Her postoperative period was uneventful, and she was discharged 4 days after surgery.
The individual was readmitted, and the physician presumptively initiated the treatment with injection ceftriaxone and gentamicin, after sending urine sample for culture and antibiotic sensitivity testing. Urine culture showed significant growth (>105 cfu/ml) of lactose-fermenting Gram-negative bacilli on CLED agar. The identification and antibiotic sensitivity testing of the isolate was performed on Vitek 2 (Bio-Merieux) automated bacterial identification and susceptibility system. The isolate (Lab ID: AFMC_UC19) was identified as E. coli. The isolate was resistant to amoxycillin/clavulanic acid (MIC≥32 mg/L), piperacillin/tazobactam (MIC≥128 mg/L), cefuroxime (MIC≥64 mg/L), ceftriaxone (MIC = 32 mg/L), amikacin (MIC≥64 mg/L), gentamicin (MIC≥16 mg/L), ciprofloxacin (MIC≥4 mg/L), trimethoprim/sulphamethoxazole (MIC≥320 mg/L) and colistin (MIC≥16 mg/L). The isolate was sensitive to cefoperazone/sulbactam (MIC≤8 mg/L), imipenem (MIC≤0.25 mg/L) and tigecycline (MIC≤0.5 mg/L). Colistin MIC was further confirmed by microbroth dilution method. She was managed with parenteral imipenem, and her follow-up urine culture on 5th and 7th day of therapy was sterile. Considering the uncommon nature of colistin resistance in the E. coli strain, WGS was planned. WGS is an important research tool to study the entire genome of an organism for various mutations. The objective for WGS in this article is to study the genes responsible for virulence and antimicrobial resistance with special attention to plasmid-mediated colistin resistance.
Materials and methods
WGS of E. coli strain AFMC_UC19 was performed at the molecular laboratory of the National Centre for Microbial Resource, National Centre for Cell Sciences, Pune. Briefly, growth of the organism in pure culture was obtained by subculture on blood agar. Total DNA extraction from a colony was performed with QIAmp DNA Mini Kit (Qiagen) as per manufacturer's instruction. WGS was performed using Illumina MiSeq platform using 2x250bp V2 chemistry by following the manufacturer's protocol (Illumina Inc. USA) at the National Centre for Microbial Resource, Pune. The 16S rRNA gene sequence of the strain AFMC_UC19 was retrieved from the whole genome sequence, which was searched against the EzTaxon database to reveal its taxonomic affiliation.9 Multiple Web-based bioinformatic tools were used for analyses of genome.
Results
An approximate 5.4 Mb of genome of the urinary isolate AFMC_UC19 was sequenced successfully. EzTaxon database analysis of the 16 S RNA of isolate showed 99.9% sequence similarity with the Escherichia fergusonii ATCC 35469T. Whole genome comparisons of study isolate by computing the average nucleotide identity (ANI) indicated more than 98.4% homology with the E. coli type strain. Thus, the final identification of E. coli was accepted. Also, multidrug-resistant E. fergusonii is rare in clinical settings. It is a pathogen of zoonotic importance, isolated commonly from poultry and livestock.10
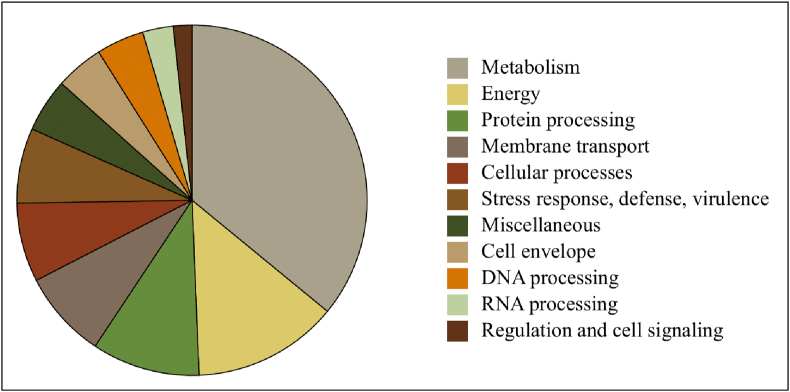
Further, annotation using PATRIC server (3.5.43) demonstrated the presence of 5547 coding DNA sequences of which 4735 were assigned to protein-coding genes and rest 812 as hypothetical proteins.11 The annotation detail is shown in Fig. 1 and Table 1. Categorizing at subsystem level revealed the dominance of cofactors, vitamins, prosthetic groups, protein synthesis and amino acids and derivatives followed by stress response, defence, and virulence and, energy and precursor metabolite generation (Fig. 2).
Fig. 1.
Annotation using PATRIC server (3.5.43) showing 5547 coding DNA sequences of Escherichia coli strain AFMC_UC19.
Table 1.
Genome sequencing statistics and functional features of the strainAFMC_UC19.
| AFMC_UC19 | |
|---|---|
| No. of pair-end reads | ∼3.4 million |
| Genome assembly | de novo |
| Genome annotation | PATRIC |
| Genome size | ∼5.4 Mb |
| Feature description | |
| No. of contigs | 71 |
| Protein-coding genes | 4735 |
| CDS | 5547 |
| tRNAs | 83 |
| Hypothetical proteins | 812 |
| GC content % | 50.71 |
Fig. 2.
Genome annotation for strains AFMC_UC19 using PATRIC server. The pie chart shows the abundance and distribution of the various subsystem categories.
To decipher the features of antimicrobial resistance and virulence factors, we carried out gene predictions using Resistance Gene Identifier, a Web tool by CARD and VFDB, respectively, which suggested presence of 88 genes conferring antibiotic resistance and 101 genes belonging to various classes of virulence factors (Fig. 3). Following the leads from antibiotic gene resistance, we noted the presence of complete colistin resistance pathway. The strain AFMC_UC19 shows persistence of all the genes, which were supported by significant homology. However, we did not find the localization of mobile colistin resistance gene (mcr) on the plasmid. Prophage sequences and features were identified by PHAge Search Tool (PHAST), which showed the presence of four intact prophages in the genome (Fig. 4) indicating the capability of horizontal transfer of antimicrobial resistance genes.12
Fig. 3.
Distribution of the AMR gene features accounting for the observed resistance in the strain AFMC_UC19.
Fig. 4.
PHASTviewer depicting the genomic constitutes for strain AFMC_UC19 representing the presence of various phage elements.
Discussion
Emergence of resistance to colistin and its global spread by plasmid-mediated mcr gene is worrisome in health-care setting. A recent systematic review indicated the presence of mcr genes in 47 countries across six continents with an average prevalence of 4.7%.13 Our strain AFMC_UC19 was resistant to beta-lactam antibiotics, third-generation cephalosporins, aminoglycosides, trimethoprim/sulphamethoxazole and colistin. The nature of colistin resistance was further elucidated by WGS. Genome sequence of colistin-resistant Klebsiella pneumonia was reported from India recently.14 Colistin-resistance E. coli case reports from hospital setting were also reported from the eastern India.15 Emergence of colistin resistance warrants strict antimicrobial stewardship practices to curb the menace of rising antimicrobial resistance.
In this study, we encountered an unusual colistin-resistant isolate of E. coli in a urinary specimen. We performed WGS to elucidate molecular mechanism of resistance. DNA sequencing technology provides more precise information about the identification and taxonomical classification of bacteria. EzBiocloud, a comprehensive database of 16S rRNA gene sequences and whole-genome sequence-based classification and identification software showed 99.9% sequence similarity with the E. fergusonii ATCC 35469T.9 However, ANI homology identified it to be E. coli, and the same was accepted by NCBI. Thus, taxonomic identification of such complex group of organisms represents a daunting task. E. fergusonii is closely related to E. coli and often misidentified by automated systems. E. fergusonii, a Gram-negative bacterium, is an emerging pathogen frequently isolated from wound and urinary specimen in India.16,17 Colistin resistance in E. coli and E. fergusonii is commonly reported from animal origin, such as poultry and pork meat. Emergence and spread of colistin resistance in Enterobacteriaceae especially from clinical specimen is a public health concern. Recently, Naveen et al performed WGS of 60 MDR E. coli isolates from individuals with blood stream infections from Southern India and found two colistin-resistant isolates expressing mcr-1.18 Our isolates lack mobile colistin resistance gene (mcr) on the plasmid, responsible for the horizontal transfer of polymyxin resistance. Colistin is an important antibiotic against carbapenemase-producing Enterobacteriaceae and considered as last resort against multidrug-resistant Gram-negative infections. There is lack of data on mcr-1-expressed colistin resistance from Indian setting. Although microbroth dilution method is recommended for detection of colistin resistance in clinical isolate, detection of mcr-1 by molecular methods in Enterobacteriaceae isolates with colistin MIC of 4 μg/ml or more is important to elucidate molecular mechanism.
To conclude, WGS is an important tool for taxonomic identification and molecular surveillance of drug-resistant genes. In the background of rising incidence of carbapenemase-producing Enterobacteriaceae, surveillance of antimicrobial resistance implementation of antimicrobial stewardship program and infection prevention measures is essential to restrict the spread of colistin resistance.
GenBank accession number
The draft genome sequence of multidrug-resistant E. coli AFMC_UC19 is available under BioProject Id PRJNA602785 with accession no. JAACYM000000000.
Disclosure of competing interest
The authors have none to declare.
References
- 1.Bialvaei A.Z., Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015;31(4):707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 2.Martis N., Leroy S., Blanc V. Colistin in multi-drug resistant Pseudomonas aeruginosa blood-stream infections: a narrative review for the clinician. J Infect. 2014;69(1):1–12. doi: 10.1016/j.jinf.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y.Y., Wang Y., Walsh T.R. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Gelbicova T., Kolackova I., Krutova M., Karpiskova R. The emergence of mcr-1-mediated colistin-resistant Escherichia coli and Klebsiella pneumoniae in domestic and imported Turkey meat in the Czech Republic 2017-2018. Folia Microbiol. 2020;65(1):211–216. doi: 10.1007/s12223-019-00709-z. [DOI] [PubMed] [Google Scholar]
- 5.Glover B., Wentzel J., Jenkins A., Van Vuuren M. The first report of Escherichia fergusonii isolated from non-human primates, in Africa. One health. 2017;3:70–75. doi: 10.1016/j.onehlt.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hmede Z., Sulaiman A.A.A., Jaafar H., Kassem Emergence of plasmid-borne colistin resistance gene mcr-1 in multidrug-resistant Escherichia coli isolated from irrigation water in Lebanon. Int J Antimicrob Agents. 2019;54(1):102–104. doi: 10.1016/j.ijantimicag.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Joshi P.R., Thummeepak R., Paudel S. Molecular characterization of colistin-resistant Escherichia coli isolated from chickens: first report from Nepal. Microb Drug Resist. 2019;25(6):846–854. doi: 10.1089/mdr.2018.0326. [DOI] [PubMed] [Google Scholar]
- 8.Walia K., Madhumathi J., Veeraraghavan B. Establishing antimicrobial resistance surveillance & research network in India: journey so far. Indian J Med Res. 2019;149(2):164–179. doi: 10.4103/ijmr.IJMR_226_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon S.H., Ha S.M., Kwon S. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forgetta V., Rempel H., Malouin F. Pathogenic and multidrug-resistant Escherichia fergusonii from broiler chicken. Poultry Sci. 2012;91(2):512–525. doi: 10.3382/ps.2011-01738. [DOI] [PubMed] [Google Scholar]
- 11.Wattam A.R., Davis J.J., Assaf R. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45(D1):D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbediwi M., Li Y., Paudyal N. Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018) Microorganisms. 2019;7(10) doi: 10.3390/microorganisms7100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul M., Narendrakumar L., Vasanthakumary A.R., Joseph I., Thomas S. Genome sequence of a multidrug-resistant Klebsiella pneumoniae ST78 with high colistin resistance isolated from a patient in India. J Glob Antimicrob Resist. 2019 Jun;17:187–188. doi: 10.1016/j.jgar.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Roy S., Das P., Das S. Detection of the emergence of mcr-1-mediated colistin-resistant Escherichia coli and Klebsiella pneumoniae through a hospital-based surveillance in an oncology center in eastern India. Infect Control Hosp Epidemiol. 2020;41(3):378–380. doi: 10.1017/ice.2019.363. [DOI] [PubMed] [Google Scholar]
- 16.Mahapatra A., Mahapatra S., Mahapatra A. Escherichia fergusonii: an emerging pathogen in South Orissa. Indian J Med Microbiol. 2005;23(3):204. doi: 10.4103/0255-0857.16598. [DOI] [PubMed] [Google Scholar]
- 17.Savini V., Catavitello C., Talia M. Multidrug-resistant Escherichia fergusonii: a case of acute cystitis. J Clin Microbiol. 2008;46(4):1551–1552. doi: 10.1128/JCM.01210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devanga Ragupathi N.K., Veeraraghavan B., Muthuirulandi Sethuvel D.P. First Indian report on genome-wide comparison of multidrug-resistant Escherichia coli from blood stream infections. PloS One. 2020;15(2) doi: 10.1371/journal.pone.0220428. [DOI] [PMC free article] [PubMed] [Google Scholar]