Abstract
The diversity of lichen photobionts is not fully known. We studied here the diversity of the photobionts associated with Cladonia, a sub-cosmopolitan genus ecologically important, whose photobionts belong to the green algae genus Asterochloris. The genetic diversity of Asterochloris was screened by using the ITS rDNA and actin type I regions in 223 specimens and 135 species of Cladonia collected all over the world. These data, added to those available in GenBank, were compiled in a dataset of altogether 545 Asterochloris sequences occurring in 172 species of Cladonia. A high diversity of Asterochloris associated with Cladonia was found. The commonest photobiont lineages associated with this genus are A. glomerata, A. italiana, and A. mediterranea. Analyses of partitioned variation were carried out in order to elucidate the relative influence on the photobiont genetic variation of the following factors: mycobiont identity, geographic distribution, climate, and mycobiont phylogeny. The mycobiont identity and climate were found to be the main drivers for the genetic variation of Asterochloris. The geographical distribution of the different Asterochloris lineages was described. Some lineages showed a clear dominance in one or several climatic regions. In addition, the specificity and the selectivity were studied for 18 species of Cladonia. Potentially specialist and generalist species of Cladonia were identified. A correlation was found between the sexual reproduction frequency of the host and the frequency of certain Asterochloris OTUs. Some Asterochloris lineages co-occur with higher frequency than randomly expected in the Cladonia species.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00248-020-01633-3.
Keywords: Asterochloris, Lichens , Specificity , Symbiosis , Trebouxiophyceae
Introduction
A lichen has been traditionally considered as a stable symbiotic association between a fungus (mycobiont) and at least a green alga or a Cyanobacterium (photobiont) [1]. However, a recent study considered a lichen to be a self-sustaining ecosystem constituted by a mycobiont, one or more photobionts, and a number of other microorganisms [2]. The photobionts transfer to the mycobiont, in form of polyols or glucose, the carbon they have fixed during photosynthesis. The mycobiont uses these compounds for its nutrition and for synthesizing secondary metabolites. For its part, the mycobiont provides the photobiont with adequate light, humidity, and gas exchange, which allows it to carry out the photosynthesis [3, 4]. This intimate relationship between the fungus and the alga is ecologically very successful, allowing lichens to inhabit the most extreme environments on the planet, where vascular plants have difficulties to grow. The adaptation of lichens to these environments depends, in part, on the photobiont and its photosynthetic ability under different conditions of light and temperature [5, 6]. Therefore, it is a very relevant task to study the diversity of photobionts, and likewise the mycobiont-photobiont association patterns, in order to understand the adaptations of lichens to environmental changes.
The diversity of the lichenizing fungi and that of the lichenizing algae are asymmetric. To date, nearly 19,000 lichenized species of fungi have been recognized [7], while the variety of algae and cyanobacteria acting as photobionts is much lower, around 120 species described, distributed in 40 genera [8]. Moreover, most authors agree that this diversity is underestimated [9–11]. Most of the green algae photobionts belong to the family Trebouxiaceae. Altogether 81 species have been described in this family [12], and these are either lichenizing or free-living [13]. Asterochloris is a genus of Trebouxiophyceae rather common as lichen photobiont [11, 14–16]. To date, 18 species of Asterochloris have been described [17, 18]. Asterochloris is mainly associated with species of the families Cladoniaceae [15] and Sterocaulaceae [19], although it can also associate with the families Verrucariaceae [20, 21], Psoraceae [17, 22], Lecideaceae [23], Parmeliaceae [24], and Thelotremataceae [25]. These lichens exhibit various growth forms and inhabit disparate environments, but most of them are terricolous. The taxonomy of the genus Asterochloris is very challenging, since the characters useful for distinguishing the species are those linked to the chloroplast morphology and the ultrastructure of the pyrenoid. In order to observe these traits, it is necessary to use confocal microscopy and transmission electron microscopy [17]. These difficulties, added to the necessity of isolating and culturing the specimens, have limited the studies on the diversity of lichenizing algae. However, the use of DNA sequences has allowed progress in the knowledge of the photobiont biodiversity and has made it possible to infer their phylogenetic relationships. Based on molecular studies, the results of several authors suggest that Asterochloris diversity is underestimated [15, 26, 27].
The genus Cladonia, comprising ca. 475 species, is one of the main host genera for Asterochloris. Up to 14 species of Asterochloris, along with some other phylogenetic lineages, have been found in symbiosis with species of Cladonia [17, 18, 28]. Cladonia has a sub-cosmopolitan distribution; many of its species are conspicuous, being part of the dominant vegetation in ecosystems such as the tundra, the boreal and antiboreal forests, bogs, temperate forests, various pioneer habitats (e.g., bank roads), tropical highlands, and even the sandy tropical lowlands of the Amazonia [29]. Though several studies have addressed the subject of the photobionts associated with the genus Cladonia, no exhaustive research has yet been carried out. Some of these works examined the associations mycobiont-photobiont in search of coevolutionary patterns, only to reject them and to find evidence of horizontal algal switching [30, 31]. The interactions between symbionts in lichens are described in terms of specificity and selectivity. Following the concepts adopted by Rambold et al. [32] and Yahr et al. [33], the term “specificity” refers to the range of compatible photobionts that can be associated with one species of mycobiont, while “selectivity” means the frequency of associations between the compatible symbionts. Some researches have focused on the specificity and selectivity of some species of Cladonia toward the photobiont and the population structure [34–37], finding different association patterns and showing that selectivity is dynamic and depends on the environmental conditions. The influence of the photobiont on the phenotype of lichen thalli has also been studied, but no correlation has been found [37]. The reproduction type has been identified as a key factor to explain the photobiont diversity in a small group of species of Cladonia [38]. Recently, the diversity of photobionts on Cladonia was examined in poorly explored regions, such as Nepal and India [26]. However, all these studies were based on a limited number of species (ca. 8% of all the described species of Cladonia). In order to reach a more complete view of the diversity of photobionts associated with Cladonia, a further, more extensive study of the photobionts of the genus Cladonia becomes necessary, including a wider taxon sampling and an extension of the geographic scope of the survey that embraces not much explored regions such as Africa, Asia, or Australasia.
The aims of the present study are (1) exploring the biodiversity of the photobionts associated with the genus of lichen-forming fungi Cladonia and (2) determining the factors that account for the diversity patterns of the photobionts, as well as for the specificity and selectivity of the mycobionts.
Material and Methods
Sampling
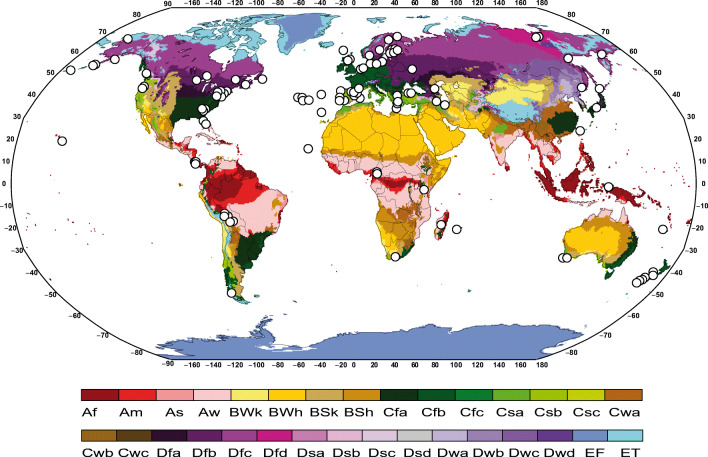
A total of 223 specimens of Cladonia, representing 135 species, were selected for studying the biodiversity of photobionts associated with this genus (supplementary material). The specimens are collected all over the world (Fig. 1), including specimens from 28 countries and 20 Köppen-Geiger bioclimatic regions [39]. The sampling represents the eleven major phylogenetic lineages of Cladonia [40]. The specimens are deposited at H and MACB herbaria (supplementary material). To complete the sampling, sequences from GenBank generated in the previous studies [15, 17, 18, 26–28, 30, 33–38, 41–43] were downloaded and included in our dataset. Only the sequences with associated species identification and adequate locality data were included (the latter for classifying the specimens in Köppen-Geiger’s ecoregions without ambiguity). The final ITS rDNA dataset included 545 sequences representing 172 Cladonia species and 24 ecoregions, while the actin type I dataset included 241 sequences representing 115 Cladonia species.
Fig. 1.
Geographical location of specimens newly sequenced in this study, represented by a dot. The colors of the map represent climatic classification of Köppen-Geiger. Af = equatorial rainforest, fully humid; Am = equatorial monsoon; Aw = equatorial savannah with dry Winter; BS = steppe climate; BW = desert climate; Cs = warm temperate climate with dry summer; Cw = warm temperate climate with dry winter; Cf = warm temperate climate, fully humid; Ds = snow climate with dry summer; Dw = snow climate with dry Winter; Df = snow climate, fully humid; ET = tundra climate; RF = frost climate
Amplification and Sequencing
In this study, Sanger sequencing technology has been used to determine the biodiversity of photobionts associated with Cladonia. Although some studies have showed that more than one photobiont can coexist in a lichen thallus [44–46], other studies have proved that most of thalli contain only a single photobiont and Sanger approach is suitable to study the diversity of photobionts [47].
Two loci were chosen to study the genetic diversity of Asterochloris, namely, ITS rDNA and a fragment of actin type I locus containing an intron. These loci were chosen according to previous results [15–17, 36] and the Asterochloris sequences available in GeneBank. Additionally, the locus rbcL was amplified in a selection of samples representing different lineages. The PCRs were carried out using Biotaq polymerase (Bioline), with 25 μl of final volume, 1 μl of each primer at 10 μM concentration, and 1 μl of DNA. The primers used were ITS1T/ITS4T or SSU-1780A/ITS4 [30, 48, 49] to amplify ITS rDNA; ActinF2 Astero/ActinR2 Astero [15] to amplify actin locus; and PRASF1/PRASR1 [50] to amplify rbcL. The PCRs were performed using an Eppendorf Mastercycler ep Gradient S thermal cycler with the following programs: 95 °C 2 min; 5 cycles of 30 s at 95 °C, 30 s at 54 °C, 60 s at 72 °C; 30 cycles of 30 s at 95 °C, 30 s at 48 °C, 60 s at 72 °C; 7 min at 72 °C for ITS rDNA; 95 °C 2 min; 30 cycles of 30 s at 95 °C, 30 s at 62 °C, 60 s at 72 °C; 10 min at 72 °C for actin; and 95 °C 2 min; 35 cyles of 45 s at 95 °C, 40 s at 52 °C, 90 s at 72 °C; 10 min at 72 °C for rbcL. PCR products were cleaned with Illustra ExoProStar TM 1-step (GE Healthcare). The sequencing was performed at Macrogen Spain service (www.macrogen.com) with the same primers used for the PCRs.
ITS rDNA sequences of mycobiont were generated previously by the authors [40, 51–53].
Alignments, OTUs Delimitation, and Phylogenetic Reconstructions
The sequences were assembled and edited using Sequencher 4.1.4 software (Gene Codes Corporation, Inc., Ann Arbor, MI, USA). The alignment for each region was conducted with MAFFT [54] with default parameters. Then, the alignments were improved manually in BIOEDIT 7.0 [55]. Gblock 0.91b [56] with the less stringent option was used to remove the ambiguous regions of actin alignment.
The diversity of Asterochloris is not well-known, and the species boundaries are not well-established [11, 17]. Therefore in this study, the sequences were clustered in operational taxonomic units (OTUs). Automatic Barcode Gap Discovery method (ABGD) was used to delimit the OTUs, following Leavitt et al. [57]. These analyses were conducted in the webserver (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) using Jukes-Cantor (JC69) model to calculate the genetic distances, Pmin = 0.001, Pmax = 0.01, step = 10, and Nb bins = 20, and several values of X were used, 0.5, 0.8, 1, and 1.5. This method as applied to ITS rDNA and actin datasets. Then, comparisons of the OTUs inferred with each value of X with both regions were done to assess the consistence of results.
Reference sequences of Asterochloris species [17, 18, 27, 28] were downloaded from GeneBank and included in our alignments. Maximum likelihood (ML) analyses were implemented using RAxML 7.0.3 [58] assuming the GTRGAMMA model for each alignment. The node support was estimated with rapid bootstrap algorithm, using 1000 pseudoreplicates. Congruence between the loci was tested manually, checking the clades with at least 75% bootstrap support. In order to reduce the computation time of concatenated analyses, a subset of 299 samples were selected for these analyses, removing sequences from the large OTUs and prioritizing samples with sequences for two and three loci. The concatenated dataset was analyzed by ML with the same conditions of that single-locus alignments and Bayesian inference. JModeltest [59] using the Akaike information criterion (AIC) was used to select the optimal substitution model for each locus. The models selected were: TrNef+I+G for actin, TrNef+G for ITS rDNA, and K80+I+G for rbcL. Bayesian analysis was run in MrBayes 3.2[60] in CIPRES portal [61] with three partitions and the substitution models selected by jModeltest. Two simultaneous runs with 20 000 000 generations, each starting with a random tree and employing four simultaneous chains, were executed. Every 1000th tree was saved into a file. The first 5, 000, 000 generations (i.e., the first 5000 trees) were deleted as the “burn-in” of the chain. The convergence of the chains was assessed with average standard deviation of split frequencies < 0.05 and plotting the likelihood versus generation number in Tracer v. 1.7 [62]. The different OTUs of Asterochloris were named using the reference sequences, and a number was assigned to the OTUs without species name.
Statistical Analyses
All the analyses were conducted in R 3.6.3 (http://www.r-project.org/). Redundancy (RDA) and partial redundancy analyses (pRDA) were done using the Vegan package [63] to determine the relative contribution of climate, geography, phylogeny of the mycobiont, and identity of the mycobiont on the genetic diversity of the photobiont. The genetic distances of photobionts were used to run a principal component analysis (PCA), and the three first components (explained 99% of the variance) were used as response matrix. Four explanatory matrices were used. Three of them were binary matrices containing the geographical origin (Europe, North America, South America, Africa, Asia, Australasia), the climatic region [39], and the phylogenetic clade of Cladonia [40]. The ITS rDNA sequences of the mycobiont were used to construct the fourth explanatory matrix. The genetic distances of the mycobiont were calculated under JC69 model, and the seven first components of a PCA (explained 95% of variance) were used as explanatory matrix. The variation explained by each variable group was estimated using adjusted R2, and the statistical significance was assessed using a permutation-based ANOVA test with 2000 permutations. Two different analyses were conducted using different dependent matrices, the ITS rDNA, and the actin type I locus alignments. Venn diagrams were generated in R, using the package Euler [64].
The specificity and selectivity were studied in a selection of Cladonia species for which more than four sequences of photobionts were available (Table 1). Given that numerous groups of Cladonia present taxonomical problems and the limits between species are not clear [40, 53], our analyses were restricted to include only those species of Cladonia whose borders are clear. Thus, all the following species were excluded: C. arbuscula, C. capitellata, C. cenotea, C. chlorophaea, C. coccifera, C. confusa, C. deformis, C. didyma, C. diversa, C. fimbriata, C. furcata, C. gracilis, C. grayi, C. macilenta, C. pleurota, C. pocillum, C. pyxidata, C. subsubulata, and C. veriticillata. In total, 150 ITS rDNA photobiont sequences were analyzed, associated with eighteen Cladonia species.
Table 1.
Genetic diversity of photobionts associated with Cladonia species, frequency of sexual reproduction, asexual reproduction type, and distribution
| Taxa | N | OTUs | H | Hd | π | Frequency of apothecia | Reproduction type | Distribution |
|---|---|---|---|---|---|---|---|---|
| C. coniocraea | 8 | 4 | 5 | 0.857 | 0.01948 | Frequent | Soredia | Subcosmopolitan |
| C. conista | 9 | 6 | 6 | 0.917 | 0.00990 | Rare | Soredia | Subcosmopolitan |
| C. corsicana | 4 | 2 | 2 | 0.500 | 0.00092 | Frequent | Thallus fragments | Mediterranea |
| C. cristatella | 4 | 2 | 2 | 0.500 | 0.00806 | Frequent | Thallus fragments | N America |
| C. evansii | 4 | 1 | 3 | 0.833 | 0.00202 | Rare | Thallus fragments | N & S America |
| C. foliacea | 39 | 4 | 4 | 0.282 | 0.00269 | Rare | Thallus fragments | W Eurasia, N Africa & Macaronesia |
| C. fruticulosa | 4 | 3 | 2 | 0.500 | 0.01042 | Rare | Soredia | Africa, Asia, Australasia |
| C. leporina | 4 | 3 | 3 | 0.833 | 0.01610 | Frequent | Thallus fragments | N & N America |
| C. portentosa | 4 | 1 | 2 | 0.667 | 0.00115 | Rare | Thallus fragments | W Europe, Macaronesia & N America |
| C. pachycladodes | 5 | 2 | 3 | 0.800 | 0.00322 | Rare | Thallus fragments | N America |
| C. ramulosa | 4 | 3 | 3 | 0.833 | 0.00700 | Frequent | Granulose | Europe |
| C. rangiferina | 10 | 4 | 5 | 0.756 | 0.00727 | Frequent | Thallus fragments | Eurasia, N America & S America |
| C. rangiformis | 18 | 4 | 5 | 0.484 | 0.00412 | Rare | Thallus fragments | W Eurasia, N Africa & Macaronesia |
| C. rei | 15 | 4 | 8 | 0.867 | 0.00938 | Frequent | Soredia | Circumpolar |
| C. strepsilis | 9 | 2 | 6 | 0.889 | 0.00306 | Rare | Thallus fragments | W Europe, E Asia, E N America & S America |
| C. subtenuis | 18 | 5 | 16 | 0.987 | 0.01757 | Rare | Thallus fragments | N & C America |
| C. subulata | 6 | 5 | 6 | 1.000 | 0.01663 | Rare | Soredia | Subcosmopolitan |
| C. uncialis | 4 | 2 | 3 | 0.833 | 0.0000016 | Rare | Thallus fragments | Circumpolar |
N number of sequences; H number of haplotypes; Hd haplotype diversity; π nucleotive diversity
Generalized linear models (GLM) with Poisson error distribution and log-link function were used to examine whether the frequency of OTUs was different among Cladonia species, climatic regions, geographic regions, type of asexual reproduction, and frequencies of sexual reproduction. The same categories used in the RDA analyses for geographic and climatic regions were used here. The type of asexual reproduction was categorized in soredia or other (the latter including thallus fragments, plates or schizidia). Based on our knowledge of species and on literature, e.g. [29], the frequency of sexual reproduction was split in two categories, frequent (for species with > 20% of specimens with apothecia) and rare (for species with < 20% of specimens with apothecia). The haplotypes, the haplotype diversity, and the nucleotide diversity (π) of the photobiont in each Cladonia species were calculated using DNAsp v6 [65]. T student analyses were used to compare the nucleotide diversity between sorediate and non-sorediate species and between species with frequent apothecia and with rare apothecia.
A bipartition network was used to visualize the association patterns between the 18 mycobionts and the OTUs of the photobiont. This network was constructed using the bipartite network analysis webservice (https://aaronecology.shinyapps.io/Network/).
The co-occurrence of Asterochloris OTUs within Cladonia species was tested statistically using co-occurrence analyses. The dataset contained 16 photobiont OTUs and 18 Cladonia species. This analysis constructs pair links of OTUs that co-occurred in the same Cladonia species. All possible pairwise Spearman’s rank correlations between OTUs were calculated. Only robust and statistically significant correlations (|ρ| > 0.65 and P value < 0.05) were selected. This analysis was implemented with Hmisc package [66] for R. One thousand random networks with the same number of edges and nodes, but assuming random connections, were generated using Erdös-Rényi model [67]. The networks properties, modularity, clustering coefficient, average degree, and graft density, were calculated. A GML network was generated using igraph package [68] for R and visualized with Gephi 0.9.2 [69].
Results
In total, 418 sequences of Asterochloris associated with Cladonia were generated (225 ITS rDNA, 162 actin type I, and 31 rbcL) and deposited in GenBank with accession numbers MW043487-MW043711 and MW073562-MW073754. Less than 1% of the sequences showed electropherograms with two peaks on some positions; for two samples, two different sequences of ITS rDNA were obtained (one for C. rappii, from Australia, and another for C. leprocephala, from Bolivia). In both cases, the two sequences from the same sample were grouped in the same OTU.
The ABGD analyses for ITS rDNA estimated 44 groups for values of maximal genetic distance of P = 0.002783–0.003594 for X = 0.5; P = 0.002154–0.003594 for X = 0.8; and P = 0.002154 for X = 1. Altogether, 154 groups were obtained using X = 1.5. The actin analyses generated 37 groups. They were generated with gap width X = 0.5 and maximal distance P = 0.005995; X = 0.8 and P = 0.005995; X = 1 and P = 0.003594; and X = 1.5 and P = 0.002154–0.004642. Some of the groups obtained with ITS were divided in several groups by the actin analyses. For example, the ITS OTU1 was divided in 3 OTUs in the actin analyses, and these 3 OTUs correspond with the species A. irregularis, A. glomerata, and A. pseudoirregularis. Therefore, the delimitation of the OTUs is based on the actin type I results, shown in Fig. 2. In total, 15 OTUs are represented by one only sequence (Fig. 2).
Fig. 2.


Phylogeny of Asterochloris based on ITS rDNA, actin type I, and rbcL. The 50% consensus majority tree of the Bayesian analysis. Bold branches were supported by ≥ 0.95 of posterior probability from Bayesian analysis and ≥ 75% of bootstrap values from ML analysis. Specimens sequenced in this study are in bold
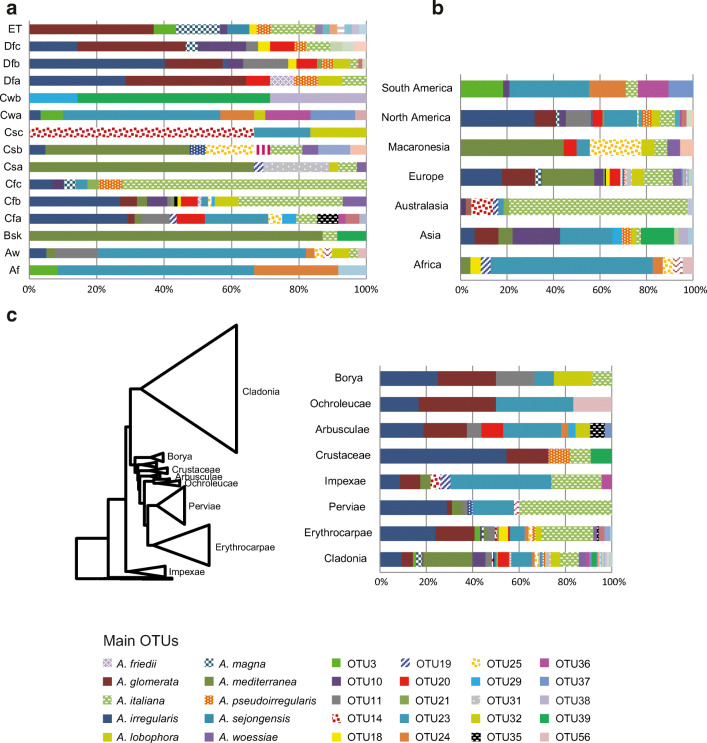
The ML analysis of the concatenated matrix ITS rDNA, actin type I, and rbcL generated a tree with – Lnl = 17756.02, while the Bayesian analysis generated a tree with an average value – LnL = 17886.61. The topologies of both trees are similar, whence only the tree for the Bayesian analysis is shown in Fig. 2. Twenty-six clades are well-supported, twelve of them represent described species and fourteen OTUs (Fig. 2). Most of the phylogenetic relationships among the clades were not supported. The commonest lineage of photobiont in Cladonia corresponds to Asterochloris glomerata (representing 16.5% of the photobionts). Other frequent photobionts were A. italiana (13.8%), A. mediterranea (12%), and an unnamed clade, the OTU23 (11.4%), that was earlier found [26, 30, 33, 70]. Many OTUs have a wide distribution and are found in specimens collected in different continents (Fig. 3). But different frequencies of OTUs in different geographical regions were found. Asterochloris italiana was the lineage most frequent in Australasia; OTU23 was the most frequent in Africa and South America; A. glomerata was the most frequent in North America; A. mediterranea was the most frequent in the Macaronesia; OTU23 and OTU10 were very frequent in Asia; four OTUs were found in similar frequency in Europe, A. glomerata, A. irregularis, A. mediterranea and OTU33. Few OTUs showed a restricted distribution, i.e., OTU14 (N = 4) in Australasia, OTU31(N = 7) in Europe, OTU35 (N = 3) in North America, and OTU39 (N = 7) in Asia.
Fig. 3.
Relative abundance of the different Asterochloris OTUs. a Frequency of different OTUs in the climatic regions. b Frequency of different OTUs in the continents. c Cartoon phylogeny of Cladonia and frequency of Asterochloris OTUs in species belonging to different clades
Some OTUs are clearly dominant in one or several climatic regions (Fig. 3a). For instance, OTU14 is dominant in subpolar oceanic climate (Cfc), OTU3 in subtropical highland climate (Cwb), and A. mediterranea in cold semi-arid climate (BsK), hot-summer Mediterranean climate (Csa), and warm-summer Mediterranean climate (Csb).
Figure 3c shows the OTUs associated with species belonging to different Cladonia lineages. In most of the Cladonia lineages, several OTUs occurred in similar proportion. The most diverse lineages were the clades Cladonia and Erythrocarpae (for clades, see [40]). But both gather together most of the specimens (53.1% and 23.1%, respectively).
The variation partitioning analyses explained between 0.37 and 0.67 of the genetic variation of the photobiont (Fig. 4). In the ITS rDNA dataset, the mycobiont explained the large proportion of the variation (0.635). The second factor that explained more variation was the climatic region (0.126). However, the effect of the climatic region alone is low (Fig. 4a). In the actin type I dataset, the factor that explained more variation was the climatic region (0.28) following by the identity of mycobiont (0.14). However, the proportion of variation explained by the climatic region and the identity of mycobiont are low after controlling the effect of the other factors (Fig. 4b). The phylogeny of the mycobiont was the factor that explained the smallest proportion of the variation in both analyses (0.03 in ITS rDNA and 0.05 in Actin type I).
Fig. 4.
Venn diagrams showing the variation partitioning of the genetic variation of photobiont explained by each group of explanatory variables (climatic region, geographical region, phylogeny of mycobiont, and identity of mycobiont) and the variation shared by groups of variables. The explained variation indicated is the adjusted R2 values. The significance was tested with 2000 permutations, *P < 0.05; **P < 0.01. a ITS rDNA. b Actin type I
Table 1 summarizes the genetic diversity of the photobionts found in different Cladonia species. The nucleotide diversity ranked from 0.0000016 in C. uncialis to 0.01948 in C. coniocraea. The haplotype diversity ranked from 0.282 in C. foliacea to 1.0 in C. subulata. The T student analyses indicated that there is not a significant difference in the nucleotide diversity between species with frequent and rare sexual reproduction (t = 1.1037, P value = 0.2860). However, significant differences were found between species with soredia and without soredia (t = 2.6075, P value = 0.0198). The nucleotide diversity was bigger in non-sorediate species.
The results of GLM analysis are shown in Table 2. The interaction between the photobiont OTUs and the frequency of apothecia was significant, indicating that the frequency of different OTUs is different among Cladonia species that differ in the frequency of sexual reproduction.
Table 2.
Summary of GLM analysis to assess the effect of Cladonia species, geographic region, climatic region, reproduction strategy on the frequency of photobiont OTUs, and probability of the associated Chi-square
| Df | Deviance | Resid.Df | Resd. Dev | P value | |
|---|---|---|---|---|---|
| NULL | 77 | 140.468 | |||
| OTU | 1 | 2.1811 | 76 | 138.286 | 0.1397147 |
| Mycobiont species | 1 | 0.0982 | 75 | 138.188 | 0.7540270 |
| Climate | 1 | 18.2673 | 74 | 119.921 | 1.920e-05 ** |
| Geography | 1 | 16.3660 | 73 | 103.555 | 5.221e-05 ** |
| Soredia | 1 | 11.0486 | 72 | 92.506 | 0.0008875 ** |
| Apothecia | 1 | 0.4931 | 71 | 92.013 | 0.4825596 |
| OTU:species | 1 | 0.2326 | 70 | 91.781 | 0.6295981 |
| OTU:climate | 1 | 3.3898 | 69 | 88.391 | 0.0656006 |
| OTU:geography | 1 | 0.4967 | 67 | 78.149 | 0.4809586 |
| OTU:soredia | 1 | 1.0753 | 64 | 73.695 | 0.2997566 |
| OTU:apothecia | 1 | 4.2523 | 60 | 67.448 | 0.0391970 * |
Df degrees of freedom. Only the two-term interaction in which the OTU is envolved is shown; all of them were not significant. *< 0.05, **< 0.01
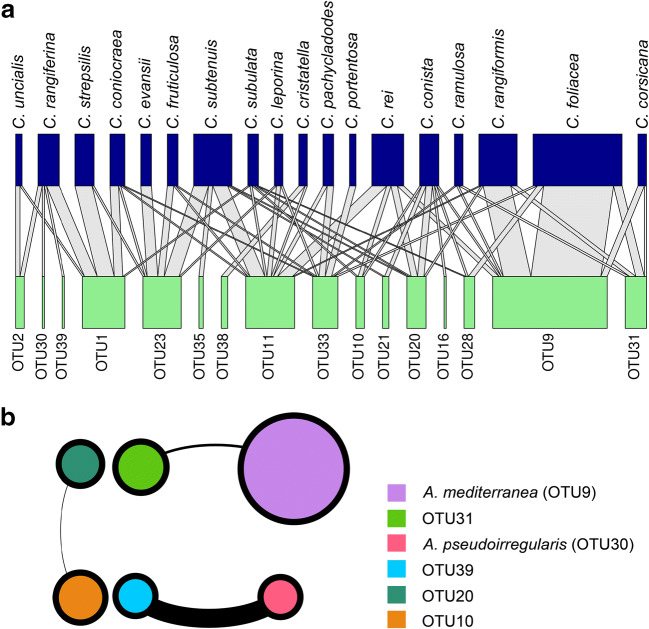
Figure 5a shows the association between photobiont OTUs and Cladonia species. The nestedness index was 26.97452. Most of the OTUs were associated with more than one Cladonia species; for instance, OTU1 (= A. glomerata) was found in 4 Cladonia species, OTU11 was found in 8 Cladonia species, and OTU20 was found in 4 Cladonia species. Most of Cladonia species are associated with more than one OTU of photobiont, but most of them have high selectivity (Fig. 5).
Fig. 5.
a Interactions network between Asterochloris OTUs and Cladonia species. The width of the lines is proportional to the frequency of the association. b Significant co-occurrence of Asterochloris OTUs in Cladonia. Each circle represents an OTU, and the size is proportional to the abundance.
Co-occurrence analyses are conducted to assess the correlations of photobiont OTUs in 18 Cladonia species (Fig. 5b). The network contained 6 nodes and 3 edges. Only three significant positive correlations were found and no negative correlation. The properties of the network were modularity = 0.666, clustering coefficient = NA, average degree = 1, and graft density = 0.2, while the values corresponding to the random networks were modularity = 0.205 ± 0.289, clustering coefficient = 0, average degree = 1, and graft density = 0.2. The modularity was higher than in the random networks, indicating that some OTUs interact more frequently with each other.
Discussion
Recent studies agree in that our knowledge on lichen photobiont diversity is still limited, and much more work using integrative approaches will be necessary in order to characterize that diversity [11, 57]. The present study compiles all the information currently available on the diversity of Asterochloris associated with Cladonia, along with new data coming from 223 specimens, contributing to achieve a more comprehensive and realistic view, as well as the interaction patterns of mycobiont-photobiont.
Biodiversity of Asterochloris Associated with Cladonia
A number of Asterochloris lineages associated with Cladonia have been found in this study (Fig. 2). These results agree with previous results that pointed out to a high diversity of Asterochloris associated with Cladonia [15, 17, 26]. The richness in species of Cladonia [7, 39], added to the cosmopolitan distribution of the genus, occupying disparate habitats, contributes indeed to the high diversity of photobionts. Some lineages are reported for the first time (OTU14, OTU19, OTU24, OUT37, CL190, and CL8), while others, already known, had never been found associated with Cladonia. This is the case of OTU3 that corresponds to a lineage recently reported of the genus Stereocaulon [71]. These results support that Asterochloris diversity is still under-sampled.
The most frequent lineage of Asterochloris in Cladonia was the one identified as A. glomerata. Previous studies had already found a high frequency of photobionts of this lineage [30, 38, 42], not only in Cladonia but also in other genera such as Stereocaulon [17, 71]. Other common lineages found are A. italiana, A. mediterranea, and OTU23. For most of them, the range of associated host species becomes widened (Fig. 2).
To date, the greatest diversity of Asterochloris had been found in the Holarctic region [17, 26, 71], and our results show a similar pattern. Europe is the region gathering the greatest number of lineages of Asterochloris associated with Cladonia (Fig. 4), but it is also the most sampled region. On the other hand, our results indicate that the climatic regions with the greatest diversity of Asterochloris are the tundra (ET), with long and cold winters and short, cold summers; the region Cfb, with a humid climate, short and dry summers; and the region Dfc, with a subarctic climate, and rainfall spread throughout the year. Similar results are reported by Steinová et al. [38], who found a great concentration of Asterochloris lineages in Central Europe, namely, in Austria and the Czech Republic, territories that fundamentally belong to these climates (Fig. 1). The warm regions AF, Csc and Cwb, were the least rich in Asterochloris lineages. This could be an indication of the Asterochloris diversity increasing with the latitude and the altitude. This agrees with the niche characterization of Asterochloris lineages proposed by Vančurová et al. [71], who found that most of the lineages appear in areas where the average annual temperature remains below 5 °C. This pattern is contrary to that found in most organisms, where diversity increases toward the tropic [72–74]. But the pattern agrees with the one found in other lichens, for example Protoparmelia, where no significant differences were found between the number of photobionts from tropical or boreal zones [75]. It would be worthwhile to note, however, that the new lineages of Asterochloris identified here come from poorly studied tropical regions as New Caledonia, South Africa, Costa Rica, and Bolivia; the diversity patterns currently known could change when territories such as Africa, Asia, and the Neotropic are better explored.
An accurate knowledge of species distribution is pivotal to understand their evolutionary history, abiotic tolerances, dispersion ability, and biotic interactions. The distribution ranges of lichen photobionts are still incomplete [26, 76, 77]. Our study reveals more accurately the distribution of the different lineages of Asterochloris. For example, OTU3 was only known for Faroe Island [71], but we have found it in Bolivia and Costa Rica. To date, it was thought that this lineage had a very restricted ecological niche, but our results indicate that it is tolerant to a wider range of environmental conditions. We corroborate that A. mediterranea is the more frequent lineage in the Mediterranean Basin [27], extending its distribution to Turkey, Iran, and Greece. The distribution of OTU23 is extended to Africa and New Zealand, previously found in India, North America, and South America [26, 30, 33, 41]. Asterochloris irregularis has been found in specimens restricted to cold areas of the Northern Hemisphere such as the Kamchatka Peninsula, Finland, or Canada. Prior studies had hypothesized that A. irregularis was not present in the tropical regions [70], and our results point in the same direction.
Drivers of Genetic Patterns of Asterochloris
On a large geographic scale, climate is considered the most decisive factor in determining the distribution of species [78]. Several researches have proved that climatic variables are relevant predictors to explain the genetic variation of photobionts [10, 31, 77, 79, 80]. In the present study, we find that the climate explains a considerable part of the genetic variation of the photobionts associated with Cladonia. In fact, different Asterochloris OTUs show a clear prevalence in certain climatic regions, despite the fact that numerous OTUs are shared among climatic regions (Fig. 3a). However, most of the variation explained by the climate is shared with other factors (Fig. 4). The identity of the mycobiont was the main driver of the photobiont’s genetic variation on the ITS rDNA analysis, which agrees with the results found in many other lichens [57, 71, 81, 82]. This is expected in lichens with dominant asexual reproduction [80]. In spite of many Cladonia species frequently developing apothecia, they usually produce few ascospores, and the reproduction is largely based on vegetative propagules [29]. Geographic region explained a small amount of the variance, which was expected since most of the lineages of Asterochloris have a broad geographic distribution [17]. Host phylogeny was the factor with the least influence on the photobiont’s genetic variation patterns, thus corroborating the lack of phylogenetic congruence between mycobiont and photobiont found in Cladonia [30, 31].
The RDA analyses based on the actin type I gene explained much less genetic variation than the ITS rDNA analysis. This is probably due to the fact that actin genetic variation of Asterochloris is much greater than that of ITS rDNA [17, 70]. As the patterns of both loci were also somewhat different (since the mycobiont was not the factor that explained most of the variance in actin type I), the ITS rDNA analysis was repeated for a subset of samples, the same ones included in the actin analysis. The ITS result is similar to that found for the actin (Table S2). The factor explaining the greatest amount of variance was the climate. This indicates that the two loci do not show different results, but that the differences in the patterns are due to the number of mycobiont species analyzed. When the sampling is reduced to a fewer mycobiont species, climate is the factor that most variation explains. This result emphasizes the importance of mycobiont-photobiont studies being carried out on a large scale, including most of the mycobiont species of the genera under study.
The results suggest that the diversity patterns of the photobionts associated with Cladonia are complex, with numerous factors intervening and acting jointly, as found in other genera [71, 82]. Biotic and abiotic factors shaped the distribution of Asterochloris diversity associated with the genus Cladonia across the landscape. However, the results of Table 2 indicate that other biotic factors such as the frequency of sexual reproduction are also relevant in the distribution of photobiont diversity at lower taxonomic scale.
Specificity and Selectivity of Cladonia species for the Photobiont
The association patterns of the mycobiont-photobiont are here analyzed for 18 species of Cladonia, either monophyletic or with a reviewed taxonomy. Cladonia is morphologically very variable, whose taxonomy at species level is one of the most complicated among macrolichens. Recent molecular studies have demonstrated that numerous species are polyphyletic and need a taxonomic revision [40]. The lack of clarity in the taxonomy of the mycobiont could show non-real association patterns, which has led us to limit our study.
In this study, we find mycobionts with different specificity, from specialist species to generalist ones. Cladonia evansii and C. portentosa associate with only one OTU and could be considered specialist species. Nevertheless, the number of specimens sampled of these species makes it difficult to establish unequivocally whether the species are specialists or not. Most of the species studied associate with more than one Asterochloris OTUs (Fig. 5, Table 1). The most generalist species are C. conista and C. subulata that associate with 6 and 5 OTUs, respectively. These species have also a medium or low selectivity. Different degrees of specificity and selectivity have been previously found in Cladonia [32, 33, 42]. Numerous studies have reported the ability of the mycobionts to establish associations with several photobionts much more frequently than the specificity toward one single lineage of photobiont, and the rarity of the reciprocal specificity between symbionts [83]. This is commonly considered an adaptive strategy of the mycobiont in order to tolerate a wider range of environmental conditions, selecting the photobiont best adapted to the local conditions or searching to extend the geographic range [33, 36, 80, 84].
In general, species with broad distributions are associated with a wide photobiont range [76, 85, 86], while the species with more restricted distributions are more specific [10, 77, 83, 87]. A high number of species of Cladonia have broad distribution ranges, embracing several continents and tolerating a vast range of environmental conditions [29]. Therefore, the association with several lineages of photobionts was expected in C. subulata, C. conista, and C. coniocraea, with a sub-cosmopolitan distribution [29], while C. corsicana and C. evansii, with a more restricted distribution [29, 88], were expected to be more specific. However, the differences in distribution range do not completely explain by themselves the differences found in specificity and selectivity, since some widely distributed species such as C. strepsilis associate with only two Asterochloris OTUs. The reproduction mode, sexual or asexual, has been strongly correlated with different specificity and selectivity of the mycobionts [76, 89]. Co-dispersion of the mycobiont and the photobiont occurs during asexual reproduction in lichens, and the species with predominant asexual reproduction tend to be more specific, while the species that reproduce sexually are expected to be generalist, since the mycobiont has to seek a new compatible photobiont. Evidences of the relevance of the reproduction strategy in the specificity and selectivity have been found in Cladonia [34, 38]. We did find differences in the frequency of OTUs between the species that commonly reproduce sexually and those who only rarely do (Table 2). For instance, OTUs 11, 23, and 33 are more frequently found in species where sexual reproduction is rare. Additionally, we find that the species having specialized asexual reproduction structures, such as soredia, have lower values of nucleotide diversity than those whose asexual reproduction is carried out by thallus fragments. These results support the hypothesis put forward by Steinová et al. [38], proposing that the specificity of the mycobiont toward the photobiont could be attributed to differences in size of the vegetative propagules and to the amount of them.
Alternative explanations of the differences in specificity and selectivity in Cladonia are the strategy of colonization and the morphological pattern of the thalli [34, 42]. Common habitats for many species of this genus are bare soils, rock banks, and postfire successional stages [29], of which these species are considered pioneer in the early stages. Previous studies found that these species had a lower selectivity [41], which would permit them to increase their fitness in these habitats. Most of the pioneer species studied here, C. conista, C. foliacea, C. rangiformis, C. subulata, and C. strepsilis (though they can live in other habitats too), showed a low specificity; besides, C. conista and C. subulata showed lesser selectivity.
Piercey-Normore [34] pointed out that the branching patterns of the species of Cladonia could be too an important factor to explain the association pattens with the photobionts, since the different thallus architecture influences the water contents and the gas exchange. Also in Parmeliaceae, some evidence of the influence of the growth forms on the selectivity of the mycobionts have been found, the fruticose genera being the most selective [57]. The species studied here have different morphology, from species whose thallus is basically formed by dominant squamulose primary thallus (C. corsicana, C. foliacea, and C. strepsilis) to species with richly branched thalli, lacking cortex (C. rangiferina, C. evansii, C. portentosa, and C. subtenuis) or corticate (C. rangiformis). But no clear pattern has been found, probably due to our data being insufficient. It will be necessary to examine more species of the different morphologies.
In summary, our results show that the specificity and selectivity in Cladonia may be influenced by a combination of factors, the reproduction mode (sexual or asexual), the type of asexual reproduction, the distribution of the mycobiont, and the habitat.
Our results suggest that some Asterochloris lineages co-occur not randomly in the species of Cladonia. Three clusters, formed by two OTUs each, are identified (Fig. 5). OTUs 9 and 31; 20 and 10; and 39 and 30 co-occur with higher frequency that randomly expected. Up to date, no test of this kind had been made, though some authors’ results seemed to suggest that some photobiont OTUs co-existed with high frequency [37]. Phylogenetically OTUs 9 and 31 are closely related (Fig. 2), and the co-occurrence can be due to habitat filtering. Closely related taxa tend to share certain traits that can facilitate their adaptation to a habitat. These OTUs appear with high frequency in the Mediterranean region and can be better adapted to withstand prolonged periods of water deficit. The two other clusters are formed by OTUs phylogenetically more distant, whereby it is expected that they differ in physiological and ecological adaptations and the mycobiont probably selects the photobiont best adapted to local conditions. The co-occurrence patterns can offer a new outlook on the interactions between the symbionts in lichens, helping us to answer ecological questions.
Supplementary Information
Specimens newly sequenced in this study. Locality, voucher specimen, mycobiont taxa, climatic region, and GenBank accession numbers (XLSX 30 kb)
(DOCX 15 kb)
Acknowledgments
We are grateful to Prof. Teuvo Ahti for providing collection data. We thank Olga Popova for her assistance in the lab.
Availability of Data
All DNA sequences have been submitted to GenBank, provided as supplementary material.
Authors’ Contribution
R P-B: conceived the study, conducted laboratory work, analyzed of data, and wrote the paper. S S: contributed to writing the paper.
Funding
This study was funded in part by Juan de la Cierva-Incorporación 2015-23526 and in part by FLF program, Royal Botanic Gardens, Kew, SYNTHESYS ES-TAF-922 and the European Union’s Seventh Framework Program under Grant No. PIEFGA-2013-62565.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Hawksworth DL. The variety of fungal-algal symbioses, their evolutionary significance, and the nature of lichens. Bot J Linn Soc. 1988;96(1):3–20. doi: 10.1111/j.1095-8339.1988.tb00623.x. [DOI] [Google Scholar]
- 2.Hawksworth DL, Grube M. Lichens redefined as complex ecosystems. New Phytol. 2020;227:1281–1283. doi: 10.1111/nph.16630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill DJ, Ahmadjian V. Relationship between carbohydrate movement and the symbiosis in lichens with green algae. Planta. 1972;103(3):267–277. doi: 10.1007/BF00386850. [DOI] [PubMed] [Google Scholar]
- 4.Honegger R. Lichen-forming fungi and their photobionts. In: Deising HB, editor. Plant relationships. 2. Berlin: Springer; 2009. pp. 307–333. [Google Scholar]
- 5.Lechowicz MJ, Adams MS. Ecology of Cladonia lichens. II. Comparative physiological ecology of C. mitis, C. rangiferina, and C. uncialis. Can J Bot. 1974;52(2):411–422. doi: 10.1139/b74-052. [DOI] [Google Scholar]
- 6.Casano LM, del Campo EM, García-Breijo FJ, Reig-Armiñana J, Gasulla F, Del Hoyo A, Guerá A, Barreno E. Two Trebouxia algae with different physiological performances are ever-present in lichen thalli of Ramalina farinacea. Coexistence versus competition? Environ Microbiol. 2011;13(3):806–818. doi: 10.1111/j.1462-2920.2010.02386.x. [DOI] [PubMed] [Google Scholar]
- 7.Lücking R, Hodkinson BP, Leavitt SD. The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota–approaching one thousand genera. Bryologist. 2017;119:361–416. doi: 10.1639/0007-2745-119.4.361. [DOI] [Google Scholar]
- 8.Honegger R. The symbiotic phenotype of lichen-forming ascomycetes and their endo-and epibionts. In: Hock B, editor. Fungal associations. The Mycota IX. Berlin: Springer; 2012. pp. 287–339. [Google Scholar]
- 9.Nelsen MP, Plata ER, Andrew CJ, Lücking R, Lumbsch HT. Phylogenetic diversity of Trentepohlialean algae associated with fungi. J Phycol. 2011;47(2):282–290. doi: 10.1111/j.1529-8817.2011.00962.x. [DOI] [PubMed] [Google Scholar]
- 10.Magain NJ, Miadlikowska B, Goffinet E, Sérusiaux E, Lutzoni F. Macroevolution of specificity in cyanolichens of the genus Peltigera section Polydactylon (Lecanoromycetes, Ascomycota) Syst Biol. 2017;66:74–99. doi: 10.1093/sysbio/syw065. [DOI] [PubMed] [Google Scholar]
- 11.Muggia L, Leavitt S, Barreno E. The hidden diversity of lichenized Trebouxiophyceae (Chlorophyta) Phycologia. 2018;57:503–524. doi: 10.2216/17-134.1. [DOI] [Google Scholar]
- 12.Guiry MD, Guiry GM (2020) AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 04 May 2020
- 13.Fučíková K, Lewis PO, Lewis LA. Widespread desert affiliation of trebouxiophycean algae (Trebouxiophyceae, C hlorophyta) including discovery of three new desert genera. Phycol Res. 2014;62(4):294–305. doi: 10.1111/pre.12062. [DOI] [Google Scholar]
- 14.Friedl T, Büdel B. Photobionts. In: Nash III, editor. Lichen biology. New York: Cambridge University Press; 2008. pp. 9–26. [Google Scholar]
- 15.Škaloud P, Peksa O. Evolutionary inferences based on ITS rDNA and actin sequences reveal extensive diversity of the common lichen alga Asterochloris (Trebouxiophyceae, Chlorophyta) Mol Phyl Evol. 2010;54:36–46. doi: 10.1016/j.ympev.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 16.Peksa O, Škaloud P. Do photobionts influence the ecology of lichens? A case study of environmental preferences in symbiotic green alga Asterochloris (Trebouxiophyceae) Mol Ecol. 2011;20(18):3936–3948. doi: 10.1111/j.1365-294X.2011.05168.x. [DOI] [PubMed] [Google Scholar]
- 17.Škaloud P, Steinová J, Řídká T, Vančurová L, Peksa O. Assembling the challenging puzzle of algal biodiversity: species delimitation within the genus Asterochloris (Trebouxiophyceae, Chlorophyta) J Phycol. 2015;51(3):507–527. doi: 10.1111/jpy.12295. [DOI] [PubMed] [Google Scholar]
- 18.Kim JI, Kim YJ, Nam SW, So JE, Hong SG, Choi H-G, Shin W. Taxonomic study of three new Antarctic Asterochloris (Trebouxio-phyceae) based on morphological and molecular data. Algae. 2020;35:17–32. doi: 10.4490/algae.2020.35.2.23. [DOI] [Google Scholar]
- 19.Nelsen MP, Gargas A. Dissociation and horizontal transmission of codispersing lichen symbionts in the genus Lepraria (Lecanorales: Stereocaulaceae) New Phytol. 2008;177(1):264–275. doi: 10.1111/j.1469-8137.2007.02241.x. [DOI] [PubMed] [Google Scholar]
- 20.Beck A, Kasalicky T, Rambold G. Myco-photobiontal selection in a Mediterranean cryptogam community with Fulgensia fulgida. New Phytol. 2002;153(2):317–326. doi: 10.1046/j.0028-646X.2001.00315.x. [DOI] [Google Scholar]
- 21.Thüs H, Muggia L, Pérez-Ortega S, Favero-Longo SE, Joneson S, O’Brien H, Nelsen MP, DuqueThüs R, Grube M, Friedl T, Brodie J, Andrew CJ, Lücking R, Lutzoni F, Gueidan C. Revisiting photobiont diversity in the lichen family Verrucariaceae (Ascomycota) Eur J Phycol. 2011;46(4):399–415. doi: 10.1080/09670262.2011.629788. [DOI] [Google Scholar]
- 22.Ruprecht U, Brunauer G, Türk R. High photobiont diversity in the common European soil crust lichen Psora decipiens. Biodivers Conserv. 2014;23(7):1771–1785. doi: 10.1007/s10531-014-0662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaper T, Ott S. Photobiont selectivity and interspecific interactions in lichen communities. I. Culture experiments with the mycobiont Fulgensia bracteata. Plant Biol. 2003;5(04):441–450. doi: 10.1007/s00300-018-2430-2. [DOI] [Google Scholar]
- 24.Friedl T (1989) Systematik und Biologie von Trebouxia (Microthamniales, Chlorophyta) als Phycobiont der Parmeliaceae (lichenisierte Ascomyceten). Dissertation, Universitat Bayreuth, Bayreuth
- 25.Friedl T. Thallus development and phycobionts of the parasitic lichen Diploschostes muscorum. Lichenologist. 1987;19:183–191. doi: 10.1017/S002428298700015X. [DOI] [Google Scholar]
- 26.Řidká T, Peksa O, Rai H, Upreti DK, Skaloud P. Photobiont diversity in Indian Cladonia lichens, with special emphasis on the geographical patterns. In: Rai H, Upreti DK, editors. Terricolous Lichens in India, Volume 1: Diversity Patterns and Distribution Ekology. New York: Springer; 2014. pp. 53–71. [Google Scholar]
- 27.Moya P, Škaloud P, Chiva S, García-Breijo J, Reig-Armiñana J, Vančurová L, Barreno E. Molecular phylogeny and ultrastructure of the lichen microalga Asterochloris mediterranea sp. nov. from Mediterranean and Canary Islands ecosystems. J Syst Evol Micr. 2015;65:1838–1854. doi: 10.1099/ijs.0.000185. [DOI] [PubMed] [Google Scholar]
- 28.Kim JI, Nam SW, So JE, Hong SG, Choi H-G, Shin W. Asterochloris sejongensis sp. nov. (Trebouxiophyceae, Chlorophyta) from King George Island, Antarctica. Phytotaxa. 2017;295:60–70. doi: 10.11646/phytotaxa.295.1.5. [DOI] [Google Scholar]
- 29.Ahti T. Cladoniaceae. Fl Neotrop Monogr. 2000;78:1–363. [Google Scholar]
- 30.Piercey-Normore MD, DePriest PT. Algal switching among lichen symbionts. Am J Bot. 2001;88:1490–1498. doi: 10.2307/3558457. [DOI] [PubMed] [Google Scholar]
- 31.Beiggi S, Piercey-Normore MD. Evolution of ITS ribosomal RNA secondary structures in fungal and algal symbionts of selected species of Cladonia sect. Cladonia (Cladoniaceae, Ascomycotina) J Mol Evol. 2007;64(5):528–542. doi: 10.1007/s00239-006-0115-x. [DOI] [PubMed] [Google Scholar]
- 32.Rambold G, Friedl T, Beck A. Photobionts in lichens: possible indicators of phylogenetic relationships? Bryologist. 1998;101:392–397. doi: 10.2307/3244177. [DOI] [Google Scholar]
- 33.Yahr R, Vilgalys R, DePriest PT. Strong fungal specificity and selectivity for algal symbionts in Florida scrub Cladonia lichens. Mol Ecol. 2004;13:3367–3378. doi: 10.1111/j.1365-294X.2004.02350.x. [DOI] [PubMed] [Google Scholar]
- 34.Piercey-Normore MD. Selection of algal genotypes by three species of lichen fungi in the genus Cladonia. Can J Bot. 2004;82(7):947–961. doi: 10.1139/b04-084. [DOI] [Google Scholar]
- 35.Robertson J, Piercey-Normore MD. Gene flow in symbionts of Cladonia arbuscula. Lichenologist. 2007;39(1):69–82. doi: 10.1017/S0024282906005809. [DOI] [Google Scholar]
- 36.Yahr R, Vilgalys R, DePriest PT. Geographic variation in algal partners of Cladonia subtenuis (Cladoniaceae) highlights the dynamic nature of a lichen symbiosis. New Phytol. 2006;171:847–860. doi: 10.1111/j.1469-8137.2006.01792.x. [DOI] [PubMed] [Google Scholar]
- 37.Kotelko R, Piercey-Normore MD. Cladonia pyxidata and C. pocillum genetic evidence to regard them as conspecific. Mycologia. 2010;102(3):534–545. doi: 10.3852/09-030. [DOI] [PubMed] [Google Scholar]
- 38.Steinová J, Škaloud P, Yahr R, Bestová H, Muggia L. Reproductive and dispersal strategies shape the diversity of mycobiont-photobiont association in Cladonia lichens. Mol Phylogenet Evol. 2019;134:226–237. doi: 10.1016/j.ympev.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Kottek MJ, Grieser C, Beck B, Rudolf, Rubel F. World map of Köppen-Geiger climate classification updated. Meteorol Z. 2006;15:259–263. doi: 10.1127/0941-2948/2006/0130. [DOI] [Google Scholar]
- 40.Stenroos S, Pino-Bodas R, Hyvönen J, Lumbsch HT, Ahti T. Phylogeny of the family Cladoniaceae (Lecanoromycetes, Ascomycota) based on sequences of multiple loci. Cladistics. 2019;35(4):351–384. doi: 10.1111/cla.12363. [DOI] [PubMed] [Google Scholar]
- 41.Cordeiro LM, Reis RA, Cruz LM, Stocker-Wörgötter E, Grube M, Iacomini M. Molecular studies of photobionts of selected lichens from the coastal vegetation of Brazil. FEMS Microbiol Ecol. 2005;54:381–390. doi: 10.1016/j.femsec.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Bačkor M, Peksa O, Škaloud P, Bačkorová M. Photobiont diversity in lichens from metal-rich substrata based on ITS rDNA sequences. Ecotox Environ Safe. 2010;73:603–612. doi: 10.1016/j.ecoenv.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Singh G, Kukwa M, Dal Grande F, Łubek A, Otte J, Schmitt I. A glimpse into genetic diversity and symbiont interaction patterns in lichen communities from areas with different disturbance histories in Białowieża forest, Poland. Microorganisms. 2019;7(9):335. doi: 10.3390/microorganisms7090335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piercey-Normore MD. The lichen-forming ascomycete Evernia mesomorpha associates with multiple genotypes of Trebouxia jamesii. New Phytol. 2006;169(2):331–344. doi: 10.1111/j.1469-8137.2005.01576.x. [DOI] [PubMed] [Google Scholar]
- 45.Dal Grande F, Alors D, Divakar PK, Bálint M, Crespo A, Schmitt I. Insights into intrathalline genetic diversity of the cosmopolitan lichen symbiotic green alga Trebouxia decolorans Ahmadjian using microsatellite markers. Mol Phylogenet Evol. 2014;72:54–60. doi: 10.1016/j.ympev.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Moya P, Molins A, Martinez-Alberola F, Muggia L, Barreno E. Unexpected associated microalgal diversity in the lichen Ramalina farinacea is uncovered by pyrosequencing analyses. PLoS One. 2017;12(4):e0175091. doi: 10.1371/journal.pone.0175091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul F, Otte J, Schmitt I, Dal Grande F. Comparing Sanger sequencing and high-throughput metabarcoding for inferring photobiont diversity in lichens. Sci Rep. 2018;8(1):1–7. doi: 10.1038/s41598-018-26947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kroken S, Taylor JW. Phylogenetic species, reproductive mode, and specificity of the green alga Trebouxia forming lichens with the fungal genus Letharia. Bryologist. 2000;103:645–660. doi: 10.1639/0007-2745(2000)103[0645:psrmas]2.0.co;2. [DOI] [Google Scholar]
- 49.White TJ, Bruns T, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 50.Sherwood AR, Garbary DJ, Sheath RG. Assessing the phylogenetic position of the Prasiolales (Chlorophyta) using rbcL and 18S rRNA gene sequence data. Phycologia. 2000;39:139–146. doi: 10.2216/i0031-8884-39-2-139.1. [DOI] [Google Scholar]
- 51.Pino-Bodas R, Martin MP, Burgaz AR. Insight into the Cladonia convoluta-C. foliacea (Cladoniaceae, Ascomycota) complex and related species, revealed through morphological, biochemical and phylogenetic analyses. Syst Biodivers. 2010;8(4):575–586. doi: 10.1080/14772000.2010.532834. [DOI] [Google Scholar]
- 52.Pino-Bodas R, Burgaz AR, Martín MP. Elucidating the taxonomic rank of Cladonia subulata versus C. rei (Cladoniaceae) Mycotaxon. 2010;113(1):311–326. doi: 10.5248/113.311. [DOI] [Google Scholar]
- 53.Pino-Bodas R, Martín MP, Burgaz AR, Lumbsch HT. Species delimitation in Cladonia (Ascomycota): a challenge to the DNA barcoding philosophy. Mol Ecol Resour. 2013;13:1058–1068. doi: 10.1111/1755-0998.12086. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics. 2017;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 56.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 57.Laevitt SD, Kraichak E, Nelsen MP, Altermann S, Divakar PK, Alors D, Esslinger TL, Crespo A, Lumbsch HT. Fungal specificity and selectivity for algae play a major role in determining lichen partnerships across diverse ecogeographic regions in the lichen-forming family Parmeliaceae (Ascomycota) Mol Ecol. 2015;24:3779–3797. doi: 10.1111/mec.13271. [DOI] [PubMed] [Google Scholar]
- 58.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 59.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 60.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA, pp 1–8. 10.1109/gce.2010.5676129
- 62.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67(5):901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. (2012) Vegan: community ecology package. R package version 2.0 ± 3. Available at: http://CRAN.R-project.org/package=vegan
- 64.Larsson J (2020) Eulerr: area-proportional Euler and Venn diagrams with ellipses. R package version 6.1.0, https://cran.r-project.org/package=eulerr
- 65.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 66.Harrell JR, Frank E, Harrell JR, Maintainer FE (2019). Package ‘Hmisc’. CRAN2018, pp 235–6. https://cran.rproject.org/web/packages/Hmisc/Hmisc.pdf
- 67.Erdös P, Rényi A. On random graphs. Publ Math-Debrecen. 1959;6:290–297. [Google Scholar]
- 68.Csardi G, Nepusz T. The igraph software package for complex network research. Inter Journal, Complex Systems. 2006;1695(5):1–9. [Google Scholar]
- 69.Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. In: Third international ICWSM Conference, pp 361–362. https://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154
- 70.Nelsen MP, Gargas A. Actin type I introns offer potential for increasing phylogenetic resolution in Asterochloris (Chlorophyta: Trebouxiophyceae) Lichenologist. 2006;38(5):435–440. doi: 10.1017/s0024282906005779. [DOI] [Google Scholar]
- 71.Vančurová L, Muggia L, Peksa O, Řídká T, Škaloud P. The complexity of symbiotic interactions influences the ecological amplitude of the host: a case study in Stereocaulon (lichenized Ascomycota) Mol Ecol. 2018;27(14):3016–3033. doi: 10.1111/mec.14764. [DOI] [PubMed] [Google Scholar]
- 72.Gaston KJ. Global patterns in biodiversity. Nature. 2000;405(6783):220–227. doi: 10.1038/35012228. [DOI] [PubMed] [Google Scholar]
- 73.Stevens GC. The latitudinal gradient in geographical range: how so many species co-exist in the tropics. Am Nat. 1989;133:240–256. doi: 10.1038/35012228. [DOI] [Google Scholar]
- 74.Hillebrand H. On the generality of the latitudinal diversity gradient. Am Nat. 2004;163:192–211. doi: 10.1086/381004. [DOI] [PubMed] [Google Scholar]
- 75.Singh G, Dal Grande F, Divakar PK, Otte J, Crespo A, Schmitt I. Fungal–algal association patterns in lichen symbiosis linked to macroclimate. New Phytol. 2017;214(1):317–329. doi: 10.1111/nph.14366. [DOI] [PubMed] [Google Scholar]
- 76.Blaha J, Baloch E, Grube M. High photobiont diversity associated with the euryoecious lichen-forming ascomycete Lecanora rupicola (Lecanoraceae, Ascomycota) Biol J Linn Soc. 2006;88(2):283–293. doi: 10.1111/j.1095-8312.2006.00640.x. [DOI] [Google Scholar]
- 77.Leavitt SD, Kraichak E, Vondrak J, Nelsen MP, Sohrabi M, Perez-Ortega S, St Clair L, Lumbsch HT. Cryptic diversity and symbiont interactions in rock-posy lichens. Mol Phylogenet Evol. 2016;99:261–274. doi: 10.1016/j.ympev.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 78.Currie DJ, Mittelbach GG, Cornell HV, Field R, Guégan JF, Hawkins BA, Kaufman DM, Kerr JT, Oberdorff T, O’Brien E, Turner JRG. Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol lett. 2004;7(12):1121–1134. doi: 10.1111/j.1461-0248.2004.00671.x. [DOI] [Google Scholar]
- 79.Nelsen MP, Gargas A. Symbiont flexibility in Thamnolia vermicularis (Pertusariales: Icmadophilaceae) Bryologist. 2009;112(2):404–417. doi: 10.1639/0007-2745-112.2.404. [DOI] [Google Scholar]
- 80.Fernandez-Mendoza F, Domaschke S, Garcıa MA, Jordan P, Martın MP, Printzen C. Population structure of mycobionts and photobionts of the widespread lichen Cetraria aculeata. Mol Ecol. 2011;20:1208–1232. doi: 10.1111/j.1365-294x.2010.04993.x. [DOI] [PubMed] [Google Scholar]
- 81.Buckley HL, Rafat A, Ridden JD, Cruickshank RH, Ridgway HJ, Paterson AM. Phylogenetic congruence of lichenised fungi and algae is affected by spatial scale and taxonomic diversity. Peer J. 2014;2:e573. doi: 10.7717/peerj.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jüriado I, Kaasalainen U, Jylhä M, Rikkinen J. Relationships between mycobiont identity, photobiont specificity and ecological preferences in the lichen genus Peltigera (Ascomycota) in Estonia (northeastern Europe) Fungal Ecol. 2019;39:45–54. doi: 10.1016/j.funeco.2018.11.005. [DOI] [Google Scholar]
- 83.Otálora MA, Martínez I, O’Brien H, Molina MC, Aragón G, Lutzoni F. Multiple origins of high reciprocal symbiotic specificity at an intercontinental spatial scale among gelatinous lichens (Collemataceae, Lecanoromycetes) Mol Phylogenet Evol. 2010;56(3):1089–1095. doi: 10.1016/j.ympev.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 84.Muggia L, Vancurova L, Škaloud P, Peksa O, Wedin M, Grube M. The symbiotic playground of lichen thalli–a highly flexible photobiont association in rock-inhabiting lichens. FEMS Microb Ecol. 2013;85(2):313–323. doi: 10.1111/1574-6941.12120. [DOI] [PubMed] [Google Scholar]
- 85.Ruprecht U, Brunauer G, Printzen C. Genetic diversity of photobionts in Antarctic lecideoid lichens from an ecological view point. Lichenologist. 2012;44(5):661–678. doi: 10.1017/s0024282912000291. [DOI] [Google Scholar]
- 86.Muggia L, Pérez-Ortega S, Kopun T, Zellnig G, Grube M. Photobiont selectivity leads to ecological tolerance and evolutionary divergence in a polymorphic complex of lichenized fungi. Ann Bot. 2014;114(3):463–475. doi: 10.1093/aob/mcu146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Otálora MA, Salvador C, Martínez I, Aragón G. Does the reproductive strategy affect the transmission and genetic diversity of bionts in cyanolichens? A case study using two closely related species. Microb Ecol. 2013;65(2):517–530. doi: 10.1007/s00248-012-0136-5. [DOI] [PubMed] [Google Scholar]
- 88.Burgaz AR, Ahti T, Pino-Bodas R. Mediterranean Cladoniaceae. Madrid: Spanish Lichen Society; 2020. [Google Scholar]
- 89.Cao S, Zhang F, Liu C, Hao Z, Tian Y, Zhu L, Zhou Q. Distribution patterns of haplotypes for symbionts from Umbilicaria esculenta and U. muehlenbergii reflect the importance of reproductive strategy in shaping population genetic structure. BMC Microbiol. 2015;15(1):212. doi: 10.1186/s12866-015-0527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specimens newly sequenced in this study. Locality, voucher specimen, mycobiont taxa, climatic region, and GenBank accession numbers (XLSX 30 kb)
(DOCX 15 kb)
Data Availability Statement
All DNA sequences have been submitted to GenBank, provided as supplementary material.