Abstract
LTR retrotransposons (LTR-RTs) are ubiquitous and represent the dominant repeat element in plant genomes, playing important roles in functional variation, genome plasticity and evolution. With the advent of new sequencing technologies, a growing number of whole-genome sequences have been made publicly available, making it possible to carry out systematic analyses of LTR-RTs. However, a comprehensive and unified annotation of LTR-RTs in plant groups is still lacking. Here, we constructed a plant intact LTR-RTs dataset, which is designed to classify and annotate intact LTR-RTs with a standardized procedure. The dataset currently comprises a total of 2,593,685 intact LTR-RTs from genomes of 300 plant species representing 93 families of 46 orders. The dataset is accompanied by sequence, diverse structural and functional annotation, age determination and classification information associated with the LTR-RTs. This dataset will contribute valuable resources for investigating the evolutionary dynamics and functional implications of LTR-RTs in plant genomes.
Subject terms: Mobile elements, Data mining
| Measurement(s) | LTR_retrotransposon • genome |
| Technology Type(s) | bioinformatics method • digital curation |
| Factor Type(s) | Species |
| Sample Characteristic - Organism | Rhodophyta • Chlorophyta • Bryophyta • Tracheophyta |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.14684877
Background & Summary
Transposable elements (TEs) are mobile DNA sequences that can move, propagate, and integrate into new positions in the host genomes, and which are ubiquitous in nearly all living organisms1,2. All TEs manage to increase their copy number via transposition processes. Depending on the mechanism used for transposition, TEs can be divided into two classes: Class I retrotransposons, which commonly transpose through ‘copy-and-paste’ mechanism of a transcribed RNA intermediate and Class II DNA transposons that move via a ‘cut-and-paste’ mechanism that mobilizes the DNA directly3. TEs are often considered as “junk DNA” because of their continuous amplification and potential impairment on the host gene function4. However, recently, numerous studies clearly indicated that TEs play a major role in reshaping genome structure through chromosomal rearrangements, gene capture, movement, and exon shuffling2,5–7, in creating mutagenic and regulatory variation through their insertion within or near genes8,9, and in creating additional genetic diversity underlying species adaptation and evolution10,11. Hence, knowledge of their impact on the structure, function and evolution of plant genomes is a priority in the field of genomics.
Among class I elements, the long terminal repeats retrotransposons (LTR-RTs), have been observed to be the most abundant TE component of plant genomes12,13, contributing up to 70% of the plant genome size, as reported in maize14, wheat15, or sugar pine16. Moreover, these elements have been considered to be the major source for the observed extensive genome variation of flowering plants, along with polyploidization17. In addition, epigenetic silencing of LTR-RTs can affect their impact on major fitness-related traits, including flowering time, a key process of plant life cycle11,13. TE-genome wide association studies (TE-GWAS) uncovered that the insertion of LTR-RTs is associated with grain width in rice and fruit weight in tomato18,19. LTR-RTs also show unique patterns of development or environment regulation. For instance, maize transcripts Opie-1 element20, barley BARE-121, and soybean SIRE-122 have been detected primarily in roots, leaves, and seedlings, respectively. Therefore, understanding the molecular causes of genome evolution is of utmost importance, so that the mechanisms regulating LTR-RTs are established, as well as the importance of their transcription to host biology is also better known.
An autonomous LTR-RT that bears all features essential for retrotransposition is composed of two nearly identical LTR sequences which are flanked by target site duplications (TSDs) of usually 4–6 bp13,23. In some species, small palindromic motifs at the 5′ and 3′ end of the LTRs are observed24. The internal region contains open reading frame, Gag-Pol2,25. Gag, a gene that encodes a polyprotein comprising subcomponents of the virus-like particle (VLP) is involved in the maturation and packaging of retrotransposon RNA, Pol products that encode protease (PR), reverse transcriptase (RT), RNase H (RH), and integrase (INT) that are involved in the synthesis of retrotransposon DNA and integration into the host genome13. Based on the order of RT and INT in POL, LTR-RTs are classified into Gypsy and Copia superfamilies26, which are further divided into an enormous number of lineages according to phylogenetic analysis of the polyprotein domains. Usually, plants’ Copia retrotransposons are sub-classified into Ivana (Sirevirus/Oryco), Osser (hemivirus), Bianca and SIRE27–30, while Gypsy retrotransposons are grouped into CRM, Galadriel, Reina, Tcn1, Tekay (Del/Del1), Athila, Phygy and Tat (Metavirus). Gypsy lineages are further grouped into different branches according to the presence of a chromodomain, grouping together CRM, Galadriel, Reina, Tcn1, and Tekay (Del/Del1) lineages into the Chromovirus branch27,28. Moreover, previous studies have found that plant Tcn1 sequences representatives share high similarity to that of Cryptococcus neoformans, which may be the result of a horizontal transfer from fungi, which have not been deeply studied31,32. So, to better understand the hierarchical classification and complicated pattern of evolution, compiling a multi-species, comprehensive large-scale LTR-RT dataset is of great necessity.
With the advent of modern sequencing technologies and the availability of genomic resources for many organisms, different TEs databases have become available. These databases can be divided into two main types of focus: 1) analysis and classification of TE based on their phylogenetics (per lineage and protein domain), such as GyDB28 and REXdb27 and 2) identification and characterization of TEs in specific species, such as GrTEdb33 and DPTEdb34. However, there is no database for systematic and unified processing of LTR-RTs of plants, including Rhodophyta, Chlorophyta, Bryophytes, Pteridophyta, Gymnosperm, and Angiosperm. To make better use of and compare LTR-RTs in plants, it is necessary to establish a dataset containing these plants phylum to annotate LTR-RTs comprehensively and uniformly.
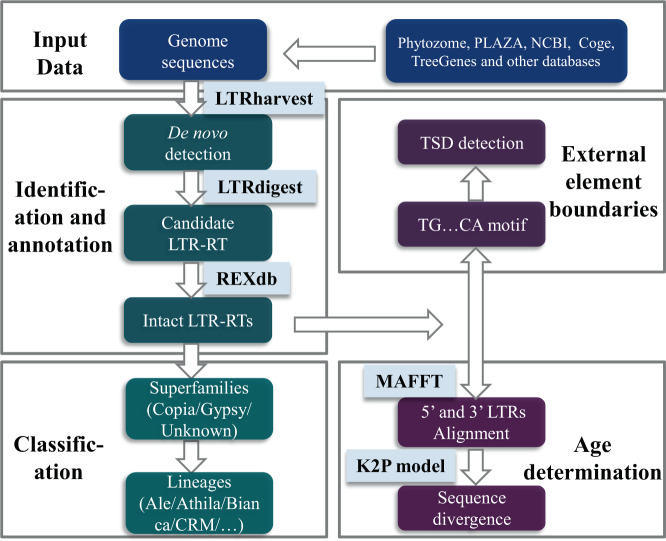
The LTR-RTs dataset presented here has been established following the schematic shown in Fig. 1. In this framework, a comprehensive annotated dataset of a total of 2,593,685 intact LTR-RTs from 300 plant genomes is presented. This dataset contributes to broadening the availability of information useful for the classification of LTR-RTs by: 1) identifying all intact LTR-RTs from diverse whole plant genomes; 2) accomplishing the functional annotation (coding domains including GAG, AP, INT, RT and RH) and classification of intact LTR-RTs; and 3) determining the age distribution of intact LTR-RTs with Kimura two-parameter model. Further details of dataset generation and contents are also provided. The dataset released in this study covers a wide breadth of highly complex plants and is expected to provide a useful resource of LTR-RTs.
Fig. 1.
Schematic diagram illustrating the overall process of the intact LTR-RTs characterization in plant genomes. The top section shows the data sources of plant genomes, and the following four different modules represent different analyses.
Methods
Genomic data collection
A total of 301 plants genome assemblies were collected from multiple comprehensive databases such as Phytozome (v12, https://phytozome.jgi.doe.gov/pz/portal.html)35, PLAZA (https://bioinformatics.psb.ugent.be/plaza/), NCBI GenBank (https://www.ncbi.nlm.nih.gov/genome/), CoGe (https://genomevolution.org), TreeGenes (https://treegenesdb.org/Drupal) and other individual genome databases. In this study, the collected genomic data represent 93 families of 46 orders. Our taxon sampling includes 2 Rhodophyta, 5 Chlorophyta, 3 Bryophytes, 4 Pteridophyta, 10 Gymnosperm, and 277 Angiosperm species. Detailed information (species, genus, family, order names, links to the published genome articles and URLs for the species genome assemblies) is provided in Supplementary Table 1.
Identification of LTR-RTs
All 301 plant genomes were searched for the de novo detection of LTR-RTs using LTRharvest36 and LTRdigest37 programs. We required that an LTR-RT is separated by 1 to 15 kb from other candidates and flanked by a pair of putative LTRs ranging from 100 to 3,000 bp with similarity > 80%. We obtained 12,829,207 candidate LTR-RTs from the 301 plant genomes, except for Genlisea aurea, a carnivorous plant with an unusually small genome size of 63.6 Mb, one of the smallest known among all higher plants. The genome of G. aurea was investigated for LTR-RT content using the default settings in RepeatMasker v4.0.738 with the RepBase version 20170127 library39, and we found a few fragmented LTR-RTs but potentially no full-length intact LTR-RTs, which is consistent with a previous study40. Further, all the internal sequences of candidate LTR-RTs were annotated by aligning the Gag-Pol protein sequences to the reference library REXdb (http://repeatexplorer.org/?page_id=918)27. Alignment was performed, using LAST v983 (http://last.cbrc.jp)41, with the following parameters: “-L 10 -m 70 -P BL80 -e 80”. Those LTR-RTs containing alignments with the domains of “GAG” (Capsid protein), “AP” (Aspartic proteinase), “INT” (Integrase), “RT” (Reverse transcriptase), and “RH” (RNase H) were considered as intact LTR-RTs. Finally, the resulting dataset consisted of 2,593,685 intact LTR-RTs from 300 plant genomes.
Reconstruction of LTR-RTs superfamilies and lineages
Depending on the order and similarity of protein domains in the Pol gene, the identified intact LTR-RTs were mainly classified into Copia and Gypsy superfamilies. We found some unclassified elements (8,682) because there were multiple Gag-Pol protein sequences that occurred inside the LTR-RTs. The Copia and Gypsy sequences were further grouped into 18 lineages based on their phylogenetic relationships and structural features of the elements within the REXdb database27.
TGCA and TSD detection
In plants, LTRs are typically flanked by 2-bp palindromic motifs, commonly 5′-TG…CA-3′, with some rare exceptions and TSD is a small exact repeat that may occur at the insertion site. They normally show a high sequence identity but may have acquired mutational variation over evolutionary processes. The two nearly identical LTR sequences of LTR-RTs were flanked by TSDs of usually 4–6 bp. We determined LTR ends (TG at the 5′ end of 5′ LTR and CA at the 3′ end of 3′ LTR) and then searched for how often the next 4, 5 and 6 bp can be used to identify their direct orientation precisely flanking each side of the LTR ends.
Age determination of LTR-RTs with Kimura distance-based calculation
To assess the evolutionary role of LTR-RTs, it is important to estimate when LTR-RT integration into the genome took place. The insertion of an LTR-RT creates a pair of LTRs with identical sequences at the two breakpoints, and subsequent accumulation of mutations between the pair of LTRs of one LTR-RT can be used as a measure of the elapsed time after the insertion. Here, we used nucleotide sequence divergence of a pair of LTRs as a proxy for LTR-RT’s insertion age. MAFFT42 with default parameters was used to align the 5′ and 3′ LTRs of each intact LTR-RT. Sequence divergence was then calculated using Kimura two-parameter (K2P) model43. Insertion times can be converted into million years given a lineage-specific synonymous substitution rate per site per year.
Data Records
The dataset containing the intact LTR-RTs information from 300 plant genomes resulting in 2,593,685 intact LTR-RTs with diverse structural, functional annotation, age determination, and classification information is available from the Figshare Repository44. The organization of the data collection is illustrated in Fig. 2. The top-level folder contains six sub-folders containing the intact LTR-RT data from Rhodophyta, Chlorophyta, Bryophytes, Pteridophyta, Gymnosperm, and Angiosperm and each sub-folder is further subdivided according to the plant order, family, and genus assignment.
Fig. 2.
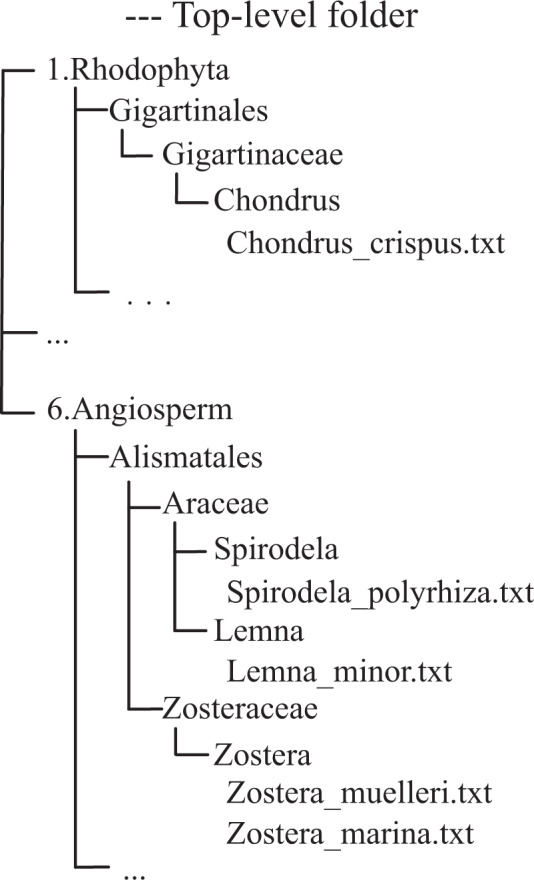
Illustration of the data structure.
File format
All data are stored in plain text (txt) format. The file is named as “X.txt”, where “X” is a species’ scientific name. Each text file includes the structure information and divergence of intact LTR-RTs detected for a specific plant genome. Table 1 summarizes the keys for the metadata.
Table 1.
Description of metadata keys for the plain text (.txt) files.
| Key | Type | Description |
|---|---|---|
| Species | string | Species name |
| LTR_ID | string | ID of intact LTR-RTs |
| Chromosome | string | Chromosome of intact LTR-RTs |
| Start | int | Start position of domain in intact LTR-RTs |
| End | int | End position of domain in intact LTR-RTs |
| Domain | string | Type of domain in intact LTR-RTs |
| Length(bp) | int | Length of intact LTR-RTs |
| Superfamilies | string | Type of superfamilies |
| Lineages | string | Type of Lineages |
| Divergence | float | Sequence divergence of intact LTR-RTs |
Graphical representation of the dataset
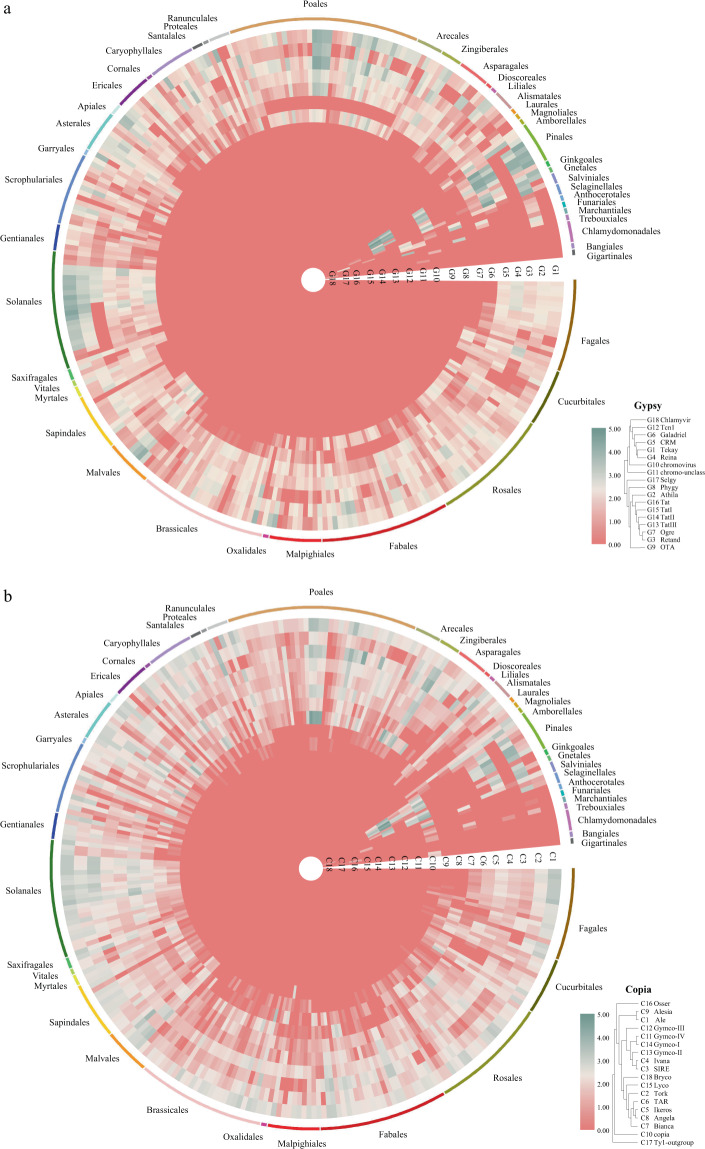
Figure 3 presents the distribution of intact LTR-RT lineages of the studied 300 plant genomes.
Fig. 3.
Intact LTR-RT (Gypsy and Copia) occupation of plant genomes. Resolved intact LTR-RT lineages were identified in 300 plant genomes of diverse systematic assignment. The presence of intact LTR-RT lineages is shown as heatmap determined by the log-transformed (log10) value of the intact LTR-RT copy number. The realized phylogenetic relationship of LTR-RT lineages24 is shown in the bottom right corner. (a) Gypsy superfamily. (b) Copia superfamily.
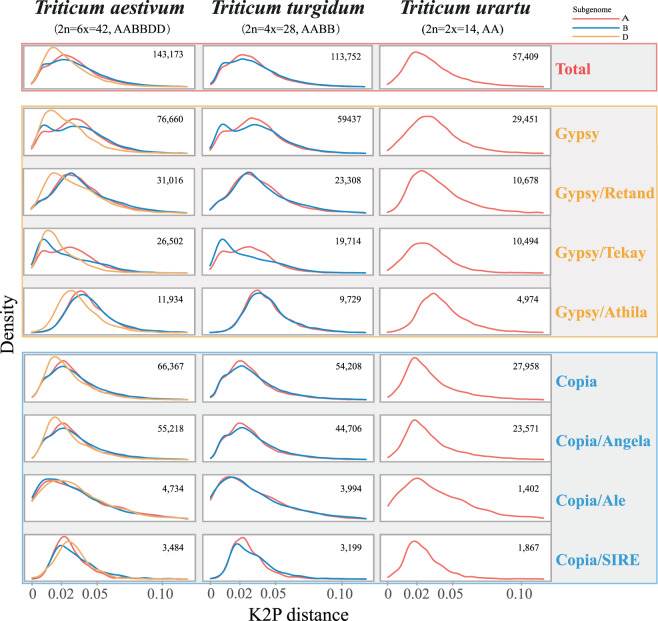
We further analyzed sequence divergence measured by K2P distance and intact LTR-RT activity pattern of representative wheat (Triticum) species, one of the most important cereal grain crops (Fig. 4). Differences in historical proliferation dynamics were shown among different LTR-RT subfamilies of different subgenomes in different plants with different ploidy.
Fig. 4.
Density map of age distribution of intact LTR-RTs in representative Triticum species. For each species, intact LTR-RTs were grouped in both superfamilies and lineages (only the first few dominant lineages are shown here). The proportion of intact LTR-RTs of each specific age bin is shown, and subgenomes (A, B and D) from three Triticum species are colored red, blue and yellow, respectively.
Technical Validation
To validate the dataset, we compared the intact LTR-RTs annotation acquired by sequence similarity and the de novo (used in this study) method. We chose rice (Oryza sativa. ssp. japonica) as a representative species for quality control as its genome is intensively examined and well annotated. A manually curated LTR-RTs library including 897 elements of rice was prepared in a previous study45. This library included known LTR-RT elements like RIRE1 (named as Angela in our dataset), RIRE2 (Retand), RIRE3 (Tekay), CRR (CRM) and Truncator (Tekay). Next, we annotated 897 LTR-RT sequences against the REXdb database27 using LAST software41. Among them, 247 sequences possessed complete Gag-Pol protein sequences, which were considered as intact LTR-RTs. These candidate intact LTR-RTs sequences were then mapped to the Oryza sativa. ssp. japonica genome (Nip-BRI) using RepeatMasker software38 with default parameters. Finally, we acquired in total the 3,002 intact LTR-RTs, of which 2,332 elements were consistent with our results (2,941 elements) obtained by de novo method. This comparison confirmed the reliability of our dataset.
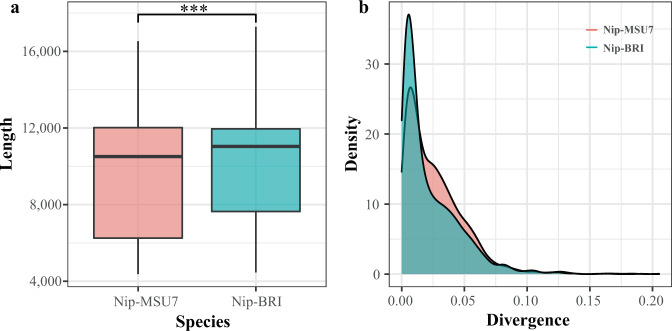
The differences in LTR-RT content, length and age structure in Oryza sativa. ssp. japonica may be influenced by assembly quality. A total of 2,941 LTR-RTs was detected in O. sativa. ssp. japonica (Nip-BRI)46, an updated assembly from long-reads sequencing, compared with 2,636 in Nip-MSU747, a short-read based assembly, and also, no LTR-RT with multiple Gag-Pol was identified in the updated assembly (Table 2). Wilcoxon test showed that the LTR-RT length identified in the Nip-BRI was significantly longer than that in the Nip-MSU7 (Fig. 5a). We further found that the insertion time of an LTR-RT estimated assembly by sequence divergence of the two LTRs in the Nip-BRI was significantly younger than that in the Nip-MSU7 (Wilcoxon test, p < 2.22e-16), indicating that many recently inserted LTR-RTs were unidentified in the Nip-MSU7 genome generated by short-reads sequencing (Fig. 5b). These findings suggest that high-quality assembled genomes obtained by long-read sequencing technology are critical to the identification and classification of LTR-RTs.
Table 2.
Comparison of LTR-RT annotated in two Oryza sativa genome assemblies.
| Key | Nip-BRI | Nip-MSU7 |
|---|---|---|
| Assembly size/Mb | 380.70 | 373.25 |
| Contig N50/Mb | 17 | 7.7 |
| Number of Gap | 18 | 905 |
| Number of Intact LTR-RTs | 2,941 | 2,636 |
| Length/Mb | 29,589,668 | 25,529,468 |
| Percent/% | 7.78 | 6.84 |
| Number of Intact LTR-RTs with multiple Gag-Pol | 0 | 2 |
Fig. 5.
Comparison of LTR-RT length and insertion time identified in two rice genome assemblies. (a) Difference of LTR-RT length between Nip-BRI and Nip-MSU7. *** shows a P-value of less than 0.001. (b) Insertion time of LTR-RT estimated by sequence divergence of the two LTRs in Nip-BRI and Nip-MSU7.
Several databases describing TE reference sequences have been published. The Repbase Update contains consensus sequences of LTR-RT superfamilies and lineages39, but lacks information on internal structure. The Gypsy database (GyDB) compiles LTR-RTs and Retroviridae-like elements28, but the metadata of Gypsy/Copia lineages is not comprehensive. REXdb divides Copia and Gypsy retrotransposons into 16 and 14 lineages, respectively, based on the conserved polyprotein domains27, but is derived from a relatively small sampling of sequences from 80 species. In the current study, we compile a dataset of LTR-RTs in plants to further enable comparative and evolutionary studies in plants. The dataset is dedicated to the identification and classification of intact LTR-RTs in 300 plant genomes using comprehensive and unified annotation approaches. Furthermore, it provides information on age distribution of intact LTR-RTs with Kimura two-parameter model.
Usage Notes
We envision many possible uses for this dataset, especially for the study of the origin, amplification, functional impact, and evolutionary dynamics of LTR-RTs among species and to encourage its use for evaluating the impact of LTR-RTs on host genomes, and to analyze the potential interaction between LTR-RTs and protein-coding genes such as:
Solo LTRs and truncated LTR-RTs detection. Unpaired LTRs (solo LTRs and truncated LTR-RTs) could further be determined based on the information of intact LTR-RTs48, since the ratios between intact LTR-RTs and solo LTRs have been used to estimate purge rates (removal rate). Removal rate could help to further research on why different plant genomes have distinct removal rate and understand molecular mechanisms of DNA amplification and removal.
LRT-RT insertion and associated factors. LTR-RT insertion times can be used to reveal the dynamics of LTR-RTs and their impact on genome evolution. For available sequence divergences of each species, insertion times could be converted into million years when given a lineage-specific synonymous substitution rate per site per year. When an LTR-RT’s proliferation time is determined, it could further be associated with historical processes, like environmental changes, mating transition, historical hybridization, and polyploidization/diploidization, so as to reveal the potential biological mechanism.
LTR-RT’s expression and its functional impact. Quantitative expression of LTR-RTs could be performed by RNA sequencing or RT-qPCR in plant tissues. In addition, RNA-seq data can be used to dissect the effect of LTR-RT insertions and analyze the expression from the targeted genomic region. Furthermore, R packages, like TEtranscripts, could be used to analyze TEs, including LTR-RTs in differential expression analysis of RNA-seq data49. The analysis of LTR-RTs expression could help understanding how these elements affect cell function to preserve specific tissues physiology and homeostasis in the plant.
LTR-RT’s involvement in gene regulation. DNA methylation and hydroxymethylation could be measured to understand the genome-wide epigenetic regulation of LTR-RTs. Additionally, several transcription factors were found to have their binding sites frequently located within various types of TEs, particularly LTR-RTs for ChIP-seq data, potentially leading to cell-specific gene regulation50,51. LTR-RT changes in adjacent gene regulation could further infer whether the contribution to plant fitness is positive, neutral, or negative.
LTR-RTs derived gene duplication. Genes can be duplicated through an RNA intermediate in a process mediated by retrotransposons as functional retrocopies or retrogenes, and they are mostly flanked by LTR-RTs in plants. Our dataset could help identify retrogenes and related duplicates, thus can help further investigate their contribution to species-specific phenotypic variation. For example, Sun is involved in the morphological variation of the tomato fruit52.
Lateral transfer of LTR-RTs. Horizontal transfers (HTs) usually represent the transmission of genetic material between reproductively isolated species and could allow TEs to escape their original host by transposing into a new organism, ensuring their survival. However, although HTs are common in plants, studies of horizontal TE transfers (HTTs) remain scarce because of limited taxa sampling53. Our dataset is valuable for further study of HTTs based on a larger taxon sampling covering most major plant orders.
LTR-RTs and genome size variation. In flowering plants, changes in copy number of retrotransposons appear to be the main factor responsible for genome size differences between species, in addition to polyploidy. It is found that the maize genome is 3–4 times as large as the sorghum genome, which is mainly caused by the extensive proliferation of retrotransposons (especially LTR-RTs) after the divergence of the two species54. Differences in the activity of retrotransposon regulation mechanisms (the proliferation of LTR-RTs) or their deletion generation (removal rate of LTR-RTs mentioned above) between species could explain current genome size variation. The present dataset brings a starting point for further systematic investigation of LTR-RT’s roles in genome size variation.
Supplementary information
Acknowledgements
This research was supported by National Natural Science Foundation of China (31670664), Project of Construction of World Class Universities in Beijing Forestry University (2019XKJS0308) and the Fundamental Research Funds for the Central Universities (2018BLCB08) to J.F.M.
Author contributions
J.F.M. conceived and designed the study; S.S.Z. collected and analyzed the data; X.M.Y., K.F.Z., H.L., J.X., S.N., K.H.J., S.Q.J., W.Z. and Y.J.Z. provided valuable assistance to data analysis; S.S.Z. and J.F.M. wrote the manuscript; I.P., Y.A.E. and T.L.W. provided suggestions and revised the manuscript.
Code availability
To prepare this dataset, we used LTRharvest and LTRdigest from genometools version 1.5.10 software55 and REXdb database (http://repeatexplorer.org/)27. The sources for the 301 plant genomes can be downloaded through the link provided in Supplementary Table 1 and scripts for intact LTR-RTs annotation are available at GitHub link (https://github.com/sszhou9/intact-LTR-RTs).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-021-00968-x.
References
- 1.Tenaillon MI, Hollister JD, Gaut BS. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010;15:471–478. doi: 10.1016/j.tplants.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Bennetzen JL, Wang H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. plant Biol. 2014;65:505–530. doi: 10.1146/annurev-arplant-050213-035811. [DOI] [PubMed] [Google Scholar]
- 3.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 5.Vitte C, Fustier MA, Alix K, Tenaillon MI. The bright side of transposons in crop evolution. Brief. Funct. Genomics. 2014;13:276–295. doi: 10.1093/bfgp/elu002. [DOI] [PubMed] [Google Scholar]
- 6.Sharma A, Wolfgruber TK, Presting GG. Tandem repeats derived from centromeric retrotransposons. BMC Genomics. 2013;14:142. doi: 10.1186/1471-2164-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 2000;42:251–269. doi: 10.1023/A:1006344508454. [DOI] [PubMed] [Google Scholar]
- 8.Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noshay, J. M. et al. Cis-regulatory elements within TEs can influence expression of nearby maize genes. Preprint at https://www.biorxiv.org/content/10.1101/2020.05.20.107169v1 (2020).
- 10.Babushok DV, Ostertag EM, Kazazian HH., Jr. Current topics in genome evolution: molecular mechanisms of new gene formation. Cell Mol. Life Sci. 2007;64:542–554. doi: 10.1007/s00018-006-6453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quadrana L, et al. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat. Commun. 2019;10:3421. doi: 10.1038/s41467-019-11385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandbastien MA. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta. 2015;1849:403–416. doi: 10.1016/j.bbagrm.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Kumar A, Bennetzen JL. Plant retrotransposons. Annu. Rev. Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 14.Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 15.Sabot F, et al. Updating of transposable element annotations from large wheat genomic sequences reveals diverse activities and gene associations. Mol. Genet. Genomics. 2005;274:119–130. doi: 10.1007/s00438-005-0012-9. [DOI] [PubMed] [Google Scholar]
- 16.Stevens KA, et al. Sequence of the sugar pine megagenome. Genetics. 2016;204:1613–1626. doi: 10.1534/genetics.116.193227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitte C, Panaud O, Quesneville H. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics. 2007;8:218–218. doi: 10.1186/1471-2164-8-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akakpo R, Carpentier MC, Ie Hsing Y, Panaud O. The impact of transposable elements on the structure, evolution and function of the rice genome. New Phytol. 2020;226:44–49. doi: 10.1111/nph.16356. [DOI] [PubMed] [Google Scholar]
- 19.Alseekh S, Scossa F, Fernie AR. Mobile transposable elements shape plant genome diversity. Trends Plant Sci. 2020;25:1062–106. doi: 10.1016/j.tplants.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butelli E, et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24:1242. doi: 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao D, Ferguson AA, Jiang N. What makes up plant genomes: the vanishing line between transposable elements and genes. Biochim Biophys Acta. 2016;1859:366–380. doi: 10.1016/j.bbagrm.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Gao X, Havecker ER, Baranov PV, Atkins JF, Voytas DF. Translational recoding signals between gag and pol in diverse LTR retrotransposons. RNA. 2003;9:1422–1430. doi: 10.1261/rna.5105503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann P, Novák P, Hoštáková N, Macas J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile. DNA. 2019;10:1. doi: 10.1186/s13100-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llorens C, et al. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 2010;39:D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wicker T, Keller B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007;17:1072–1081. doi: 10.1101/gr.6214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King AMQ, et al. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch. Virol. 2018;163:2601–2631. doi: 10.1007/s00705-018-3847-1. [DOI] [PubMed] [Google Scholar]
- 31.Goodwin TJ, Poulter RT. The diversity of retrotransposons in the yeast Cryptococcus neoformans. Yeast. 2001;18:865–880. doi: 10.1002/yea.733. [DOI] [PubMed] [Google Scholar]
- 32.Novikova O, Smyshlyaev G, Blinov A. Evolutionary genomics revealed interkingdom distribution of Tcn1-like chromodomain-containing Gypsy LTR retrotransposons among fungi and plants. BMC Genomics. 2010;11:231. doi: 10.1186/1471-2164-11-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Z, et al. GrTEdb: the first web-based database of transposable elements in cotton (Gossypium raimondii) Database. 2017;2017:bax013. doi: 10.1093/database/bax013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li SF, et al. DPTEdb, an integrative database of transposable elements in dioecious plants. Database. 2016;2016:baw078. doi: 10.1093/database/baw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2011;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinbiss S, Willhoeft U, Gremme G, Kurtz S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009;37:7002–7013. doi: 10.1093/nar/gkp759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit, A. H. R. & Green, P. RepeatMasker Open-4.0, http://www.repeatmasker.org (2013-2015).
- 39.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leushkin EV, et al. The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences. BMC Genomics. 2013;14:476. doi: 10.1186/1471-2164-14-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 44.Zhou SS, 2021. A comprehensive annotation dataset of intact LTR retrotransposons of 300 plant genomes. figshare. [DOI] [PMC free article] [PubMed]
- 45.Ou S, Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q, et al. N6-methyladenine DNA methylation in japonica and indica rice genomes and its association with gene expression, plant development, and stress responses. Mol Plant. 2018;11:1492–1508. doi: 10.1016/j.molp.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu CQ, et al. Genome sequence of Malania oleifera, a tree with great value for nervonic acid production. GigaScience. 2019;8:giy164. doi: 10.1093/gigascience/giy164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015;31:3593–3599. doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 51.de Souza FS, Franchini LF, Rubinstein M. Exaptation of transposable elements into novel cis-regulatory elements: is the evidence always strong? Mol. Biol. Evol. 2013;30:1239–1251. doi: 10.1093/molbev/mst045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 53.El Baidouri M, et al. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 2014;24:831–838. doi: 10.1101/gr.164400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanmiguel P, Bennetzen JL. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 1998;82:37–44. doi: 10.1006/anbo.1998.0746. [DOI] [Google Scholar]
- 55.Gremme G, Steinbiss S, Kurtz S. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013;10:645–656. doi: 10.1109/TCBB.2013.68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Zhou SS, 2021. A comprehensive annotation dataset of intact LTR retrotransposons of 300 plant genomes. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
To prepare this dataset, we used LTRharvest and LTRdigest from genometools version 1.5.10 software55 and REXdb database (http://repeatexplorer.org/)27. The sources for the 301 plant genomes can be downloaded through the link provided in Supplementary Table 1 and scripts for intact LTR-RTs annotation are available at GitHub link (https://github.com/sszhou9/intact-LTR-RTs).