Abstract
Background:
While the detection of the AR-V7 in CTCs is associated with resistance to abiraterone or enzalutamide (abi/enza) in men with metastatic castration resistant prostate cancer (mCRPC), it only accounts for a minority of this resistance. Neuroendocrine (NE) differentiation or chromosomal instability (CIN) may be additional mechanisms that mediate resistance.
Methods:
PROPHECY was a multicenter prospective study of men with poor risk mCRPC starting abi/enza. A secondary objective was to assess Epic CTC CIN and NE phenotypes before abi/enza and at progression. The proportional hazards (PH) model was used to investigate the prognostic importance of CIN and NE in predicting progression free survival (PFS) and overall survival (OS) adjusting for CTC number (CellSearch), AR-V7, prior therapy, and clinical risk score. The PH model was utilized to validate this association of NE with OS in an external dataset of patients treated similarly at MSKCC.
Results:
We enrolled 118 men with mCRPC starting on abi/enza; 107 were evaluable on the Epic platform. Of these, 36.4% and 8.4% were CIN positive and NE positive, respectively. CIN and NE were independently associated with worse OS (HR 2.2, 95% CI 1.2-4.0 and HR 3.8, 95% CI 1.2-12.3, respectively) when treated with abi/enza. The prognostic significance of NE positivity for worse OS was confirmed in the MSKCC dataset (n=173, HR 5.7, 95% CI 2.6-12.7).
Conclusions:
A high chromosomal instability and neuroendocrine CTC phenotype is independently associated with worse survival in men with mCRPC treated with abi/enza, warranting further prospective controlled predictive studies to inform treatment decisions.
Keywords: CTC, biomarkers, prognostic, resistance
Introduction
The treatment paradigm for men with metastatic castration-resistant prostate cancer (mCRPC) has changed dramatically with the approval and availability or approval and widespread use of next generation androgen receptor (AR) antagonists such as enzalutamide or the androgen synthesis inhibitor abiraterone acetate (1–4). Unfortunately, approximately 10-20% of men have tumors that resist these agents at the outset, while virtually all men progress on therapy after a median of 1-2 years. CTC AR-V7 mRNA or nuclear AR-V7 protein detection has been shown to be associated independently with a poor prognosis among men with high-risk mCRPC who have progressed on abiraterone or enzalutamide, but detection only accounts for a minority of the cross-resistance to subsequent AR inhibitor therapy (5–9). Additional predictive biomarkers are needed to optimally select men at highest likelihood for a beneficial response to therapy.
Recognized now through molecular profiling efforts of mCRPC is that a proportion of patients harbor germline or somatic alterations in DNA repair genes such as BRCA1/BRCA2/ATM that leads to homologous recombination deficiency (HRD) and is detected in 20-25% of men with mCRPC (10). Studies have suggested that men with tumors with HRD have worse outcomes when treated with androgen receptor signaling inhibitors (ARSI) (11,12) while others have shown no difference in outcome relative to those without HRD (13). A hallmark of HRD is a high number of chromosomal breaks, which can be measured by the number of large-scale transitions (LSTs) on single cell whole genome sequencing, defined as breakages that lead to chromosomal gains or losses of greater than 10Mb termed chromosomal instability (CIN) (14–16). Epic Sciences (San Diego, CA) has developed and validated a CTC assay that reports CIN positivity based on a phenotypically defined CIN high score, which aims to predict the presence of 9 or more large scale transitions (pLST) in a CTC and has been shown to be associated with worse OS among men with mCRPC treated with androgen receptor signaling inhibitors and taxanes (17).
A second mechanism of AR therapy resistance is lineage plasticity that can result in a small cell or neuroendocrine (NE) like phenotype (18–21). These transitions can develop from treated adenocarcinoma and is likely found in upwards 20% of mCRPC compared to 1-2% at initial diagnosis (19,22,23). Outcomes are poor (24) despite treatment with platinum-based combinations such as carboplatin plus etoposide or cabazitaxel plus carboplatin (25,26) considered by some to be the de facto standard. Epic Sciences has also developed a CTC based assay that identifies cells that are primarily small, round, and have high nuclear to cytoplasmic ratios in order to detect men potentially harboring de novo or undergoing neuroendocrine or small-cell transition (27).
Detection of chromosomal instability (CIN) or neuroendocrine (NE) phenotype on a blood-based assay may improve diagnostic accuracy and impact therapeutic decision making. Here we sought to explore the relationship between the CIN biomarker and progression free survival and confirm the association with worse OS previously shown, and separately to relate the presence of the NE biomarker with clinical outcomes in the PROPHECY trial: an independent, multi-center, prospective and blinded validation study of men with high-risk mCRPC treated with abiraterone or enzalutamide.
Methods
Patients
The PROPHECY study was a prospective multicenter clinical trial investigating clinical outcomes among men with progressive mCRPC initiating standard-of-care enzalutamide or abiraterone. Study details have been described previously (9,28). See supplementary appendix for full eligibility criteria and definitions of high-risk disease, which required ≥2 poor prognosis clinical factors (29,30). All patients provided written informed consent. The study was approved by institutional review boards of all participating centers within the Department of Defense Prostate Cancer Clinical Trial Consortium (PCCTC)(31), and Duke University as lead coordinating center. All the authors vouch for the completeness and integrity of the data and for the fidelity of the study to the clinical protocol (available online). A second cohort was used to validate findings observed in the PROPHECY trial. Independently, blood samples from men with progressing mCRPC about to initiate either abiraterone or enzalutamide and treated at Memorial Sloan Kettering Cancer Center (MSKCC) between December 2012 and September 2016 and followed until July 2020. All patients provided informed consent for biospecimen collection as part of an IRB approved protocol and all had histologic confirmation of prostate cancer.
Epic Analysis of CTC Chromosomal Instability and Neuroendocrine Phenotypes
The PROPHECY study was designed initially to validate the ability of baseline AR-V7 status in CTCs to predict clinical outcomes with abiraterone/enzalutamide. CellSearch CTC enumeration was performed at these timepoints on all men and processed in a CAP/CLIA approved central laboratory at MSKCC (29,32). Peripheral blood was collected for CTC analysis from men at prespecified time points: baseline and at the time of clinical, radiographic, or biochemical progression. CTCs were sent to two blinded central laboratories, one of which was Epic Sciences (Epic; San Diego, Ca). Epic Sciences performed the approved CTC nuclear-specific AR-V7 protein assay and CTC heterogeneity evaluations as previously described (9). As an exploratory but prospectively defined objective, CTCs were also analyzed by Epic for chromosomal instability (CIN) phenotype and neuroendocrine (NE) phenotype as predicted by an algorithm based on CTC immunofluorescent staining (DAPI, CK, CD45, and AR) and cell phenotypic characteristics (cytoplasmic and nuclear characteristics). The CIN phenotype algorithm was developed previously to predict CTCs harboring CIN high score defined as the prediction of the presence of 9 or more large scale transitions (pLST) in a CTC (17) and the NE phenotype algorithm to identify men with tumors that have undergone a neuroendocrine transformation (27). Laboratory investigators were blinded to the clinical information and patient outcomes.
The CIN algorithm uses digital pathology features including CTC morphologic and immunofluorescent staining to make a binary prediction of whether an individual CTC has 9 or more LSTs. (Figure 1). Important features in the CIN classifier include among others cell size, nuclear entropy (texture metric), CK protein expression, and AR protein expression. CIN positivity was previously defined as having ≥3 CIN positive CTC/mL, a cut-off chosen for 100% specificity for identifying CTCs with 9 or more LSTs. The NE algorithm uses digital pathology features such as CTC morphologic and immunofluorescent staining to predict whether an individual CTC is NE positive (Figure 1). NE positive CTCs generally have lower AR expression, higher cytoplasmic circularity and small cell features including a high nuclear to cytoplasmic ratio. NE positivity was previously defined as having ≥3 NE positive CTC/mL.
Figure 1:

(A) CTC identification on Epic Sciences platform and (B) biomarker identification algorithm
The laboratory data was sent to the study statistician (SH) and unblinded after the database was locked. The primary clinical efficacy end point of the PROPHECY study was progression free survival (PFS), defined as the time from registration to clinical or radiographic progression, clinical progression, or death, whichever occurred first, and excluded PSA progression (9). Radiographic progression was determined using Prostate Cancer Working Group 2 soft tissue and bone scan criteria (33). Clinical progression was defined by death, pain, or other symptomatic progression; initiation of new systemic therapy; or a new skeletal event. Key secondary end points were confirmed 50% or greater prostate-specific antigen declines (PSA50) on therapy from baseline, radiographic response per RECIST version 1.1 (defined as partial or complete response), and overall survival (OS) defined from the date of study registration to date of death or last follow-up. Confirmed 50% decline in PSA required subsequent PSA measurement at least 2 weeks later. Separately from Epic CTC processing, CTCs were enumerated using the FDA-approved CellSearch platform (34) in a Clinical Laboratory Improvement Amendments (CLIA) approved laboratory at baseline and at progression on abiraterone/enzalutamide.
The MSKCC cohort was analyzed separately to validate the NE biomarker. Pretreatment CTCs from blood draws taken within 30 days of therapy initiation were analyzed and categorized as NE positive or negative (27). Overall survival was calculated as the date from initiation of therapy until date of death from any cause or date of last follow-up (July 2020).
Data Analysis
A key secondary analysis of the PROPHECY study was to correlate the CTC biomarkers (CIN and NE) with clinical outcomes, and thus no specific hypothesis testing was performed. The pre-specified analysis was to correlate each biomarker with PFS, OS, ≥50% PSA decline from baseline and objective response rate. The maximum rank statistical method based on asymptotic distribution was used to find cut-off points for CIN and NE corresponding to the largest discrepancy between the lower- and higher-risk groups (35). The primary analysis was based on the optimal cut-off points. Men who had biomarker levels above these cut-off points were considered positive status for the biomarker. The proportional hazards model was then used to test if these biomarkers were prognostic of PFS and OS adjusting for AR-V7 status, CellSearch CTC number (≥5), prior therapy, and established prognostic factors (risk score) (36). In addition, two exploratory analysis of the association of CIN/NE with clinical outcomes were performed. The first exploratory analysis analyzed the number biomarker positive CTCs/mL for NE and CIN as a continuous variable (log2(biomarker level+1)). The second exploratory analysis utilized the biomarker cut-offs for the CIN and NE assays from prior publications (17,27) that defined ≥3 positive CTCs/mL as biomarker positive in contrast to the optimized cut-offs. The Kaplan-Meier product-limit approach was used to estimate the PFS and OS distributions by each biomarker status. Investigators and laboratory personnel were blinded to the biomarker status and outcome data prior to statistical analysis. The clinical database was locked on February 4, 2020.
For the MSKCC cohort of men treated with an ARSI the Kaplan-Meier method was used to estimate the OS distribution. The proportional hazards model was utilized to assess the prognostic importance of the NE biomarker as a binary variable, using the optimal cut-offs from the PROPHECY study, as well as a continuous variable (log2(CTC NE+1) in univariate and multivariable analyses for OS. The proportional hazards models included NE biomarker status, prior therapy, pre-treatment clinical prognostic variables, and Epic CTCs ≥3/mL.
Results
Between May 2015 and January 2017, 118 men were enrolled in the PROPHECY trial from five academic medical centers. Of the 118 men enrolled, 11 were unevaluable for Epic CTC analysis leaving 107 eligible for inclusion in this analysis who were evaluable for CIN, NE, and AR-V7 by Epic Nuclear AR-V7 protein assay at baseline (Figure 1). See Supplemental Figure 1 for CONSORT diagram and Supplemental Figure 2 for example images of biomarker positive and negative CTCs. Table 1 summarizes patient characteristics from the PROPHECY study for the overall cohort and by CTC biomarker defined cohorts. Analysis of the association between CIN and NE with clinical outcomes was performed and determined that the optimal threshold for biomarker positivity was ≥1 CTC/mL for CIN and ≥2 CTC/mL for NE given CTC counts are whole integers. CIN or NE positivity, unless otherwise specified, is defined using these optimized thresholds.
Table 1:
Baseline characteristics of evaluable men from the PROPHECY study.
| Baseline characteristics | All Men (n=107) |
CIN(+) Men (≥1 CTCs/mL) (n=39) |
CIN (−) Men (<1 CTCs/mL) (n=68) |
NE(+) Men (≥2 CTCs/mL) (n=9) |
NE (−) Men (<2 CTCs/mL) (n=98) |
|---|---|---|---|---|---|
| Age, median years (range) | 73 (44-92) | 74 (48-92) | 72 (44-87) | 77 (58-87) | 72 (44-92) |
| Race: white/black/other (%) | 81/12/7 | 77/13/10 | 84/12/4 | 67/22/11 | 83/11/6 |
| Gleason Sum 8-10 (%) | 60 | 56 | 62 | 33 | 62 |
| Karnofsky Score ≥90 (%) | 73 | 56 | 82 | 33 | 77 |
| High Risk Features | |||||
| Hemoglobin <12 g/dl (%) | 39 | 49 | 34 | 89 | 35 |
| Elevated alkaline phosphatase (%) | 44 | 64 | 32 | 89 | 40 |
| Elevated serum LDH (%) | 36 | 74 | 13 | 100 | 30 |
| Prior abiraterone or enzalutamide (%) | 37 | 36 | 38 | 33 | 38 |
| Presence of liver or lung metastasis (%) | 29 | 31 | 28 | 33 | 29 |
| Presence of clinically significant pain requiring opiates (%) | 28 | 26 | 29 | 33 | 28 |
| CellSearchS CTC ≥ 5 cells per 7.5 mL (%) | 48 | 82 | 28 | 100 | 43 |
| Radiographic progression at entry(%) | 77 | 85 | 72 | 67 | 78 |
| PSA doubling time <3 months (%) | 64 | 67 | 62 | 78 | 62 |
| Prior docetaxel for mHSPC (%) | 19 | 21 | 18 | 11 | 19 |
| M1 stage at diagnosis (%) | 32 | 49 | 22 | 33 | 32 |
| >20 bone metastases (%) | 35 | 54 | 24 | 67 | 32 |
| Median Baseline PSA ng/ml (range) | 22 0.08-4195 |
42 0.32-4195 |
14 0.08-482 |
297 0.54-4195 |
18 0.08-1105 |
| Epic nuclear AR-V7 positive (%) | 10.3 | 23.1 | 2.9 | 33.3 | 8.2 |
CTC biomarkers at baseline
At baseline, 39 men (36.4%) were positive for the CTC CIN phenotype, 9 men (8.4%) for the CTC NE phenotype, and 11 men (10%) had nuclear AR-V7 positive CTCs by Epic (≥ nuclear localized AR-V7 positive CTC). Of the 39 CIN positive men, 9 were also positive for NE phenotype and 9 were positive by AR-V7 (Figure 2A and Supplemental Table 1). All 9 NE positive men were CIN positive, and 3/9 (33.3%) were AR-V7 positive. The number of men who were negative for CIN, NE and AR-V7 was 66/107 (61.7%). Both CIN positive and NE positive men had baseline characteristics suggestive of a higher burden of disease such as higher rates of anemia, higher baseline LDH, alkaline phosphatase, and PSA, higher CellSearch CTC burden, visceral metastases, and a greater number of bone metastasis (Table 1). AR N-terminal protein expression was common and overlapped with CTC CIN and NE phenotype detection. Biomarker positive CTC numbers are shown for each patient along with Epic CTC and CellSearch CTC quantification in Figure 2C. Men who were CIN biomarker positive had a median of 4.7 CIN positive CTCs/mL (range 1.1-880.7 CTCs/mL) while NE positive men had a median of 7.2 NE positive CTCs/mL (range 2.6-85.1) (Supplemental Table 2). On the single CTC level, most NE positive CTCs were also CIN positive (83%, 186/223) while a minority of CIN positive CTCs were also NE positive (8.3%, 186/2235). AR-V7 is performed on a separate set of slides from CIN or NE and thus cannot be correlated on a single CTC level. AR N-terminal overexpression, which is part of the digital pathology algorithm, was positive in 39% of NE positive CTCs and 48% of CIN positive CTCs.
Figure 2:

(A) CTC biomarker overlap, (B) CTC biomarker group incidence, (C) CTC enumeration by patient at baseline including CellSearch CTC/7.5mL (grey), AR-V7 (orange), AR-N-terminal positive (yellow), NE (purple), and CIN (cyan). Dark Blue bars show total number of Epic CTC/mL. Note that CellSearch CTC enumeration is provided per 7.5 mL whole blood, while Epic CTC enumeration is reported per mL whole blood, so enumeration results are not directly comparable, and (D) CTC Biomarkers groups at baseline and progression. Positive CIN defined as ≥1 CTCs/mL, negative CIN <1 CTC/mL, positive NE defined as ≥2 CTCs/mL and negative <2 CTC/mL.
CTC CIN and NE phenotypes and clinical outcomes
The median follow-up among surviving men was 31 months (range 3.4 – 42.3), 104/107 men have progressed on abiraterone or enzalutamide and 83/107 men have died. The Kaplan Meier curves and analysis for PFS and OS for each biomarker are shown in Figures 3A–3D and Table 2. CIN positive men experienced shorter median PFS and OS durations compared to CIN negative men, 3.7 months versus 7.5 months, (univariate hazard ratio (HR)= 2.3, 95% CI 1.5-3.6), and OS of 11.5 months versus 25.0 months (univariate HR 3.2, 95% CI 2.0-5.1) for positive vs. negative cases respectively. NE positivity was similarly associated with shorter PFS, 1.6 months versus 6.0 months (univariate HR 3.4, 95% CI 1.7-6.8) and OS of 5.7 months versus 20.5 months (univariate HR 5.5, 95% CI 2.7-11.2) for positive and negative cases.
Figures 3:



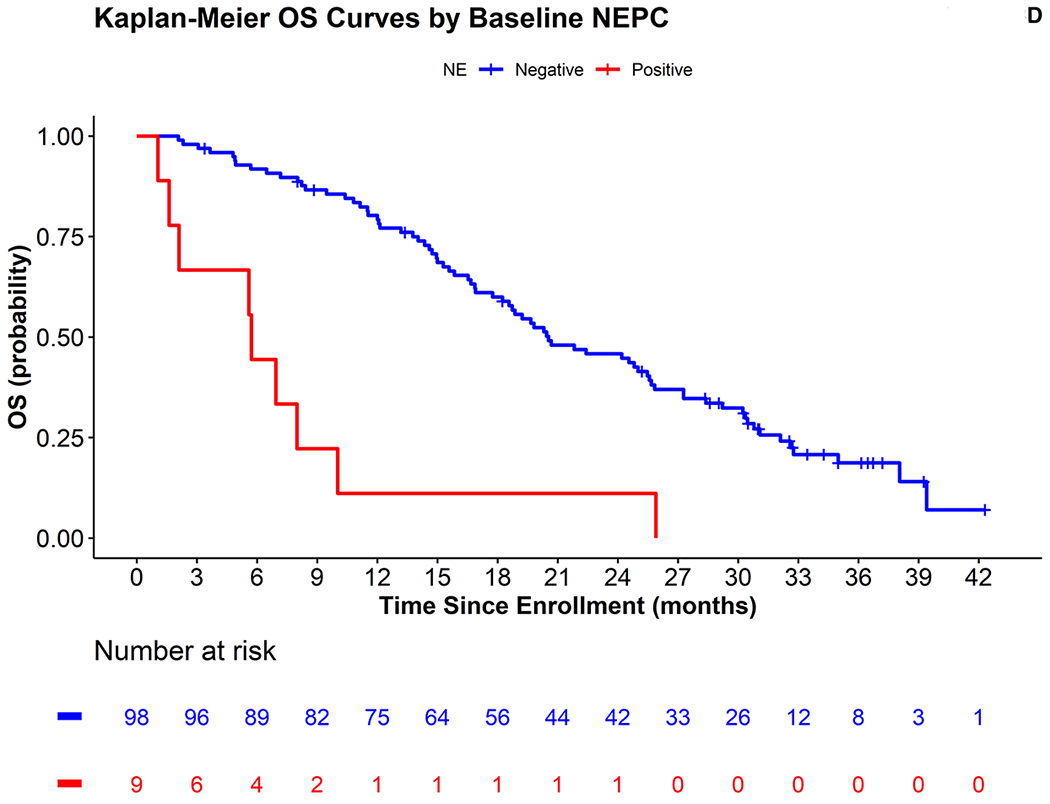

(A-D) Progression free survival (PFS) and overall survival (OS) curves for chromosomal instability (CIN) and neuroendocrine (NE) biomarkers. Positive CIN defined as ≥1 CTCs/mL, negative CIN <1 CTC/mL, positive NE defined as ≥2 CTCs/mL and negative <2 CTC/mL. (E) OS curves for MSKCC cohort treated with ARSI for external validation of NE biomarker.
Table 2:
Association of CIN and NE biomarkers with clinical outcomes from the PROPHECY study.
| Chromosomal Instability (CIN) Phenotype n=107 |
Neuroendocrine (NE) Phenotype n= 107 |
|||
|---|---|---|---|---|
| Biomarker Status | Positive (≥1 CTCs/mL) | Negative (<1 CTCs/mL) | Positive (≥2 CTCs/mL) | Negative (<2 CTCs/mL) |
| Number (%) | 39 (36.4) | 68 (63.6) | 9 (8.4) | 98 (91.6) |
| Median PFS (months; 95% CI) | 3.7 (3.0-6.0) | 7.5 (5.5-10.1) | 1.6 (1.5-NR) | 6.0 (5.3-7.8) |
| HR* (95% CI) | 2.3 (1.5-3.6) | 3.4 (1.7-6.8) | ||
| HR**(95% CI) | 1.8 (1.0-3.0) | 2.1 (0.6-7.5) | ||
| HR †(95% CI) | 1.5 (0.8-2.7) | 2.2 (0.7-7.2) | ||
| Median OS (months; 95% CI) | 11.5 (8.4-18.9) | 25.0 (19.8-32.1) | 5.7 (2.1-NR) | 20.5 (18.2-25.7) |
| HR* (95% CI) | 3.2 (2.0-5.1) | 5.5 (2.7-11.2) | ||
| HR**(95% CI) | 2.2 (1.2-4.0) | 3.8 (1.2-12.3) | ||
| HR†((95% CI) | 2.0 (1.1-3.8) | 3.0 (1.0-9.4) | ||
| % of men ≥ 50% confirmed PSA decline (95% CI)* | 18 (10-41) | 28 (25-54) | 12 (0.4-58) | 26 (24-46) |
| % of men with Best response (CR+PR, 95% CI)* | 7.7 (1.8-22.5) | 4.4 (1.0-12.9) | 11.1 (0.3-52.7) | 5.1 (1.8-12.1) |
univariate
adjusted for CellSearch CTCs (≥5), AR-V7 status, prior therapy and clinical risk score
model with CIN and NE and adjusted for CellSearch CTCs (continuous (log2(CTC+1)), AR-V7 status, prior therapy and clinical risk score. NR=not reached
The prognostic significance of CIN positivity was retained in multivariable analysis of PFS adjusting for AR-V7 status, CellSearch CTC number (≥5), prior therapy, and clinical prognostic factors for a shorter PFS (adjusted HR 1.8, 95% CI 1.0-3.0) while NE was not (adjusted HR 2.1, 95% CI 0.6-7.5), shown in Table 2. In contrast, the significance of both biomarkers was retained in the multivariable model of OS, (adjusted HR 2.2, 95% CI 1.2-4.0; adjusted HR 3.8 95% CI 1.2-12.3), respectively. Similar results were found when CIN and NE were included in the same model with CellSearch CTC number as a continuous covariate (log2(CTC+1) (Table 2). Further, in univariate analysis where the number of CIN or NE positive CTCs were modelled as continuous variables (log2(biomarker+1)), CIN and NE were associated with worse OS and PFS. However, in multivariable analysis, CIN as a continuous variable was associated with OS but not PFS. Conversely, NE as a continuous variable was associated with PFS but not OS (Supplemental Table 3). Using the threshold of positivity of ≥3 CTC/mL for CIN and NE positivity from prior publications, similar results were observed where both CIN positivity and NE positivity were associated with worse OS, but not worse PFS (Supplemental Table 4, Supplemental Figure 3, and Supplemental Figure 4A–F).
Confirmed ≥50% PSA declines and radiographic responses were also assessed as key secondary endpoints and the waterfall plots by biomarker status are shown in Figure 4A. A lower proportion of CIN positive men experienced a confirmed ≥50% PSA decline, 18% (95% CI 10-41%) compared to 28% (95% CI 25-54%) for CIN negative men (Table 2). Similarly, a smaller proportion of NE positive men experienced a confirmed PSA decline compared to NE negative men, 12% (95% CI 0.4-58) versus 26% (95% CI 24-46), respectively. Objective responses were similar between CIN positive and CIN negative as well as NE positive and NE negative men (Table 2 and Supplemental Table 5). A swim-lane plot is shown in Figure 4B with patients grouped by biomarker category using the three CTC biomarkers (AR-V7, CIN, and NE), demonstrating the shorter progression free and overall survival times in biomarker positive men.
Figure 4:


(A) PSA waterfall plot showing CTC biomarker categories (n=106). Asterix (*) indicates confirmed 50% PSA decline. One patient is excluded due to no PSA follow up measurements who was CIN+, NE+, and AR-V7 –. (B) Swim-lane plot showing CTC biomarker categories (n=107).
In exploratory analyses, the CTC biomarkers were categorized into three groups: any positive biomarker (AR-V7, NE, or CIN; n=41), triple biomarker negative and CellSearch CTC >0 (n=38), and triple biomarker negative and CTC count of 0 (n=23). The Kaplan-Meier PFS and OS curves for this analysis are shown in Supplemental Figure 5A–B. These analyses showed that men positive for any of the three biomarkers experienced worse outcomes with a median PFS of 3.5 months (95% CI=3.0-5.9) and OS of 11.5 (95% CI=8.4-18.9) months. Men who were negative for all three biomarkers with CTC >0 by CellSearch, had a median PFS and OS of 5.5 month (95% CI= 3.5-9.5) and 19.2 months (95% CI=16.7-30.2), respectively, while men who had no CTCs by Epic or Cellsearch and thus negative for all 3 CTC biomarkers had the best outcomes with a median PFS of 9.2 months (95% CI=6.7-24.3) months and OS of 30.4 months (95% CI=24.5-NR).
CTC NE biomarker external validation
An independent cohort of 173 men contributed pre-treatment CTC samples collected before starting therapy with an ARSI at MSKCC and were used for validation of the association between NE and OS. This cohort was utilized previously as part of the clinical validation of the CIN biomarker showing relationship between CIN and OS (17). Demographics for the MSKCC cohort are shown in Supplemental Table 6. As a group, the median PSA was 20.6 ng/mL (0.5-2010) and 57.8%, 27.7%, and 14.5% of samples were from patients starting an ARSI in the first, second, or third-line or greater setting, respectively (17). When defining NE and CIN by the biomarker cut-offs established from the PROPHECY study, the NE positivity (≥2 CTC/mL) rate was 7.4% (13/173), and all 13 (100%) of NE positive patients were also CIN positive (≥1 CTC/mL).
NE positive men had a worse OS when treated with an ARSI, HR 14.5 (95% CI 7.5-28.0) in univariate analysis (Supplemental Table 7), shown in Figure 3E, which was retained in the multivariable analysis, HR 5.7 (95% CI 2.6-12.7) when adjusting for treatment line (1, 2, 3+), lung or liver metastasis, PSA >20 ng/mg, LDH >250 U/L, hemoglobin >12 g/dL, alkaline phosphatase >140 U/L, and total CTC (Epic defined CK+, CD45-) ≥3/mL vs < 3/mL. This association with OS was similarly retained when the NE biomarker was considered as a continuous variable (log2 (CTC NE+1) in univariate (HR 1.7, 95% CI 1.4-2.0) and in the above multivariable model (HR 1.5, 95% CI 1.3-1.9) for OS (Supplemental Table 7).
CTC biomarkers at progression
In the PROPHECY trial CTCs were again collected at progression on abiraterone or enzalutamide and analyzed for multiple biomarkers; 68 men had CTCs measured at progression for analysis of whom 10 (15%) were missing AR-V7 status. At baseline and first progression, 74% (79/107) and 79% (46/58) men, respectively, had detectable CTCs on the Epic platform. Biomarker positive CTC numbers are shown for each patient at progression along with Epic CTC and CellSearch CTC quantification in Supplemental Figure 6 The incidence of CIN, NE, and AR-V7 biomarker positivity at progression was 55% (32/58), 14% (8/58), and 17% (10/58) (17.2%), respectively, all of which were increased from their incidence at baseline. The percentage of men with baseline and progression samples who were positive for any of the three CTC biomarkers increased from 38% at baseline to 57% at progression (Figure 2D). Of the 39 men positive for CIN at baseline 15 remained CIN positive, 5 became CIN negative, and 19 were not assessed at progression, whereas of the 9 men positive for NE at baseline, 2 converted to NE negative, and 7 were not assessed at progression. Of the 32 men positive for CIN at progression, 15 men were previously CIN positive while 17 men were previously CIN negative. All 8 NE positive men at progression were previously NE negative at baseline prior to treatment with abiraterone or enzalutamide. NE positive patients at baseline were more likely to not have biomarkers assessed at progression: 7/9 NE positive men compared to 41/98 NE negative men, possibly due to more rapid clinical progression. The prevalence of CIN, NE, and AR-V7 positivity among men with 3 or more CTCs/mL (CTCs defined by Epic) was found to be 34/38 (89.5%), 99/38 (23.7%), and 8/38 (21.1%) at baseline, respectively, and 24/27 (88.9%), 8/27 (29.6%), 7/27 (25.9%) at first progression, respectively.
Association of CIN and NE positive CTCs with CTC genomic alterations
In order to associate CTC genotype with CTC phenotype, we performed copy number variation (CNV) analysis on pooled CTCs by array comparative genomic hybridization (aCGH) and whole exome sequencing (WES) on a cohort of men from PROPHECY (n=12) with sufficient evaluable CTCs and CTC DNA (see Supplemental Methods). These results are very limited by the low sample size and the bias towards genomic data being available on patients with sufficient CTCs and CTC DNA and are presented only as hypothesis generating. For individual gene alteration analysis, we focused on genes found to be associated with aggressive prostate cancer (MYCN, PTEN, RB1, and TP53) and DNA repair genes (BRCA1, BRCA2, FANCA, ATM, PALB2). See Supplemental Figure 7A–E for mutation and copy number alteration frequency for each gene stratified by biomarker status as well as the total copy number burden for patients stratified by biomarker status. Both CIN and NE positive men had a higher average of whole genome copy number alterations, 3988 versus 2430 for CIN and 6589 versus 2845 for NE (Supplemental Figure 8).
CIN positive men (n=8) had a higher rate of pathogenic mutation or copy number loss compared to CIN negative men (n=4) in TP53 (4/8 vs 0/4), BRCA1 (6/8 vs 0/4), and BRCA2 (4/8 vs 1/4). The rates of copy number loss or mutation for CIN positive versus negative men was similar for PTEN (5/8 vs 2/4), RB1 (2/8 vs 1/4), FANCA (2/8 vs 2/4), ATM (1/8 vs 0/4), and PALB2 (0/8 vs 1/4). Notably MYCN gain was seen in 2/8 CIN positive men compared to 3/4 CIN negative men. NE positive men (n=2) had a higher rate of pathogenic mutation of copy number loss compared to NE negative men (n=10) in TP53 (2/2 vs 2/10) and RB1 (1/2 vs 2/10), but not PTEN (1/2 vs 6/10). MYCN gain was found in 1/2 NE positive men and 4/10 NE negative men. Higher rates of BRCA1 (2/2 vs 4/10) and FANCA (2/2 vs 2/10) mutation or copy number loss were also observed in NE positive compared to negative. Other genes assessed include BRCA2 (1/2 vs 4/10), ATM 1(/2 vs 0/8), and PALB2 (0/2 vs 1/10). Lastly, patients with available CTC whole genomic data were categorized by the previously described aggressive variant prostate cancer (AVPC) molecular criteria proposed by Aparicio et al (21) which defines the molecular AVPC subtype as harboring 2 or more alterations in PTEN, RB1, and TP53. AVPC molecular criteria were met for 4/8 CIN positive men and 1/4 CIN negative men as compared to 2/2 NE positive men and 3/10 NE negative men.
Discussion
The present study intended to associate two CTC biomarkers, chromosomal instability (CIN) and neuroendocrine (NE) phenotype, with overall and progression free survival in men with mCRPC and identify optimal thresholds for outcome discrimination for future predictive studies. These results from the PROPHECY study serve as an optimization cohort for the previously described CTC CIN biomarker (17). For the CTC NE biomarker, we present both the initial biomarker analysis and optimization from the PROPHECY study as well as separate external clinical validation cohort at MSKCC. We observed that both biomarkers are independently associated with worse overall survival among men with high-risk mCRPC in the PROPHECY study treated with abiraterone or enzalutamide.
After adjusting for AR-V7 status, CTC enumeration by CellSearch, prior therapy, and clinical risk score, CIN positivity (defined as ≥1 CIN positive CTC/mL) was associated with worse progression free survival following treatment with abiraterone or enzalutamide, though NE positivity (defined as ≥2 NE positive CTC/mL) was not. This could be due to the small NE positive sample and the high rates of early progression in this high-risk cohort of men treated with abiraterone or enzalutamide. Pre-treatment detection of CTC CIN or NE was not associated clearly with differential PSA declines or objective responses, and thus our data refutes our initial hypothesis that CIN and NE may be mediators of primary resistance to novel hormonal therapies. Rather, these biomarkers may identify men at high-risk of rapid progression through hormonal as well as subsequent non-hormonal treatments (37,38). PSA declines were uncommon for each biomarker positive category (18% for CIN and 12% for NE), though this is higher than the 0% confirmed PSA declines seen in men with Epic AR-V7 positive mCRPC treated with AR inhibitors. Together these data suggest a poor prognosis for both CIN and NE positive men but does not directly suggest that novel hormonal agents should be withheld in these agents. Rather, these men represent an identifiable subgroup with poor survival outcomes that could benefit from novel approaches such as PARP inhibitors, immunotherapy, or platinum-based chemotherapy combinations (25,26).
Cross-resistance between AR targeted therapies is an unmet medical of clinical significance given the earlier use of AR therapies in men with metastatic hormone sensitive prostate cancer (mHSPC) (39–43) and non-metastatic CRPC (44,45). However, while AR-V7 has been shown to have prognostic value in mCRPC for poor outcomes and predictive of a lack of sensitivity to AR targeted therapies in mCRPC, its clinical predictive utility is limited by the low prevalence of AR-V7 detection and that AR-V7 detection only accounts for a minority of this cross-resistance. Thus, additional actionable predictive biomarkers are needed to guide treatment choice. CIN attempts to predict men with CTCs harboring a high number of chromosomal breaks, which has been hypothesized be associated with homologous recombination deficiency such as mutations or deletions of BRCA1/2, ATM, or other DNA repair enzymes. Our preliminary and limited results showing BRCA1 and BRCA2 alterations in 6/8 and 4/8 CIN positive patients versus 0/4 and 1/4 CIN negative patients, respectively, need further validation. Whether CIN or other markers of a functional HRD phenotype in prostate cancer may complement genomic testing for selecting men for PARP inhibitor therapy will require prospective testing, given the enrichment for TP53 alterations in CIN positive patients as well. Additional studies are also warranted to investigate whether platinum agents alone or platinum agents in addition to a taxane such as cabazitaxel could also be a therapeutic strategy for these men (26).
The emergence of NEPC in tissue biopsies or clinically has been known to convey a poor prognosis and detecting this aggressive form of transformed prostate cancer on a blood-based assay is appealing (20,25). Our study had a lower than expected overall number of NE positive men at 8.4%, though this is consistent with the NE positivity rate from the separate MSKCC validation cohort at 7.4%. Among men enrolled in PROPHECY with sufficient evaluable CTCs, the prevalence of NE positivity was 23.7%, which is similar to contemporary biopsy prevalence studies of men with mCRPC (20). The PROPHECY trial specifically excluded men with known small cell/NEPC and included only men with prostate adenocarcinoma suitable for treatment with abiraterone or enzalutamide for mCRPC, reducing the overall number of men who would be expected to test NE positive. Systematic metastatic biopsies were not performed, and thus tissue correlation is not available. We have previously published one case of documented NEPC transformation at progression on enzalutamide from the PROPHECY study, and subsequently found this patient to be NE positive at baseline (46). Additionally, two thirds of men had not previously received an AR antagonist or abiraterone and thus have not been exposed to the therapeutic pressure that may drive NEPC development. Lastly, sufficient CTCs were required to detect NEPC, thus potentially leading to false negatives for those with NEPC and low number of CTCs. We did note an increase in biomarker rates at progression for both NE (8.4% increased to 13.8% at progression) and CIN (36.4% increased to 55.2%) at progression, using the newly defined cut-offs suggesting that these biomarkers may emerge in the setting of hormonal therapies. Of the men who were biomarker positive at progression, many were previously biomarker negative (8/8 NE positive men at progression and 17/32 CIN positive men at progression). A small group of men converted from biomarker positive at baseline to negative at progression (2/9 NE positive men at baseline and 5/39 CIN positive men at baseline). Collectively these results suggest biomarker positivity with either CIN or NE increases over time.
One interesting observation that we made was that all NE positive men at baseline were also CIN positive. This may either reflect an overlap of the digital pathology algorithm or suggest that NE men have a high number of chromosomal breaks and DNA repair machinery defects, suggesting a connection to platinum sensitivity. Malihi and Graf et al. recently performed a single CTC genome analysis that noted an association between a higher number of LSTs and aggressive variant prostate cancer both defined by clinical variables as well as an aggressive variant molecular signature (loss of at least two of PTEN, RB1, and TP53) (47), which may explain the overlap of CIN and NE in this study as many men with aggressive variant prostate cancer may harbor neuroendocrine features. Our limited genomic analysis of 12 patients do show that this aggressive variant molecular signature may be enriched among NE and CIN positive men (2/2 and 4/8, respectively) but this molecular signature was also observed in some NE and CIN negative men (3/10 and 1/4, respectively). Of the nine men who were NE positive, 3 were noted to be AR-V7 positive and AR N-terminal positivity was common in NE positive cases (Figure 2C). While NEPC is not considered AR dependent, it has been shown that increased AR expression but low AR activity is common in transformed NEPC/small cell prostate cancer (20) and also that AR-V7 can be detected by IHC in prostate cancer tissue among men after neoadjuvant hormonal therapy (48). We are unable to correlate AR-V7 with NE or CIN positivity on a single CTC level because AR-V7 is run on a separate set of slides from NE and CIN; however, we did find that 39% of NE positive CTCs and 48% of CIN positive CTCs were also positive for AR N-terminal overexpression on the single cell level. These results concur that while NEPC may be AR independent, AR expression can still be detected. Importantly, we show enrichment of both CTC CIN and NE immunophenotypes at progression on AR targeted therapies, suggesting the importance of these genetic phenotypic transitions or clonal selections in drug resistance. Only through specific AR variant and NE/CIN specific therapeutic clinical trials can the question of whether these are passenger or driver alterations be directly addressed.
Men with de novo or transformed small cell/NEPC are frequently treated with platinum-based chemotherapy (either etoposide/platinum or taxane/platinum), though making this diagnosis can be challenging as it frequently requires a biopsy. Additionally, given the known heterogeneity within individual men with mCRPC, the utility of a single site biopsy that is negative for NEPC is not known. A theoretical benefit of a blood-based assay is to aggregate this heterogeneity. Future clinical trials should assess whether men with mCRPC and are NE positive by a peripheral blood assay have improved outcomes with platinum-based chemotherapy. A limitation of this assay is the lack of tissue validation of small cell transformation for some of the NE positive patients. The response to abiraterone/enzalutamide among the NE positive patients suggests that these patients may have harbored tumor heterogeneity, wherein the AR positive clones responded to AR inhibition, which the AR independent NE clones eventually progressed. Future studies of CTC genomic and phenotypic evolution over time with tissue correlates are needed. Other promising methods of detecting transformation to NEPC include detection of ctDNA (49). Additionally, given the higher number of mutations found in NEPC and the improvements in survival with immunotherapy seen in small cell lung cancer, clinical trials of chemotherapy plus immune checkpoint inhibitor therapy should be considered (50,51). Future refinement of this assay may include incorporating staining for neuroendocrine markers such as synaptophysin, chromogranin, or CD56, higher magnification/resolution to better assess nuclear texture (i.e. salt-and-pepper chromatin) (52), as well as incorporation of CTC or cell free genomic analysis.
The strengths of our study include blinded application of previously defined CTC biomarker algorithms to a prospectively enrolled multicenter cohort of men with mCRPC treated with abiraterone or enzalutamide with a long follow-up. Laboratory personnel and investigators were blinded to the biomarker status and clinical outcomes of men prior to data analysis. An additional strength is that we further optimized the cut-offs for both the CIN and NE assay and found that a threshold of ≥1 CTC/mL for CIN and ≥2 CTC/mL for NE improved prognostic ability of each assay, though these new cut-offs will require future validation. Weaknesses include a relatively small sample size of biomarker positive men due to the low prevalence of CIN and NE and that these analyses were secondary analyses of the overall PROPHECY trial. In addition, treatments were not assigned prospectively based on testing so no commentary can be made on whether these assays are predictive. An additional weakness is our relative lack of genomic and mutation information on all men to correlate with these CTC biomarkers. However, our subset phenotype-genotype analysis suggests enrichment of CIN/NE phenotypes with TP53, BRCA1, and BRCA2 alterations. Future prospective and larger trials are clearly needed to define the clinical utility of a CTC phenotypic and genotypic classifier of primary and acquired AR therapy resistance, including AR-V7 and other AR variants, chromosomal instability, lineage plasticity and neuroendocrine transformation, and other mechanisms.
In conclusion, chromosomal instability and neuroendocrine phenotypes on a CTC based assay in high-risk mCRPC are independently associated with worse overall survival among men treated with abiraterone or enzalutamide. Further study of these two biomarkers for prognostic as well as predictive utility is warranted and ongoing.
Supplementary Material
Translational relevance.
Here we present further evidence to support the clinical validation of circulating tumor cell prognostic biomarkers: chromosomal instability (CIN) and neuroendocrine (NE) immunomorphology phenotypes. In this prospective study, we found that the CTC CIN phenotype was associated with shorter progression free and overall survival times in univariate and multivariable analysis among men with mCRPC treated with abiraterone or enzalutamide, after adjusting for CTC AR-V7 status, CTC enumeration by CellSearch, prior therapy, and clinical risk score. CTC NE phenotype was similarly associated with worse overall survival in multivariable analysis in two independent datasets and strongly associated with CTC CIN. These data support the utility of the defined CIN and CTC NE phenotype biomarkers to identify men with mCRPC and poor outcomes when considering androgen receptor targeting therapies, who should be considered for clinical trials investigating alternative therapeutic approaches directed toward the underlying chromosomal instability or neuroendocrine biology.
Acknowledgements
We wish to thank the Prostate Cancer Foundation and Movember for their financial support of this Global Treatment Sciences Challenge Award (A Armstrong, Duke), and the US Department of Defense Prostate Cancer Clinical Trial Consortium for infrastructural support for this multicenter study. A. Armstrong was supported by a Prostate Cancer Foundation grant, an NIH R01 (1R01CA233585-01) and the DCI P30 CA014236 as well as Duke Cancer Institute shared resources for biostatistics, Flow Cytometry, and Sequencing and Genomic Technologies. This work was partially funded by Department of Defense grants W81XWH-13-PCRP-CCA, W81XWH-17-2-0021 and W81XWH-14-2-0179 (D George, A Armstrong, Duke), W81XWH-5-1-0467 (S Halabi, Duke), W81XWH-18-1-0278 (S Halabi, Duke), W81XWH-14-2-0159 (D Nanus, Weill Cornell), W81XWH-15-2-0018 (R Szmulewitz, Chicago), W81XWH-15-2-0018 (H Scher, MSKCC), and W81XWH-16-PCRP-CCRSA (E Antonarakis, Johns Hopkins). D Danilla and H Scher were also supported in part by National Cancer Institute Grants P30CA008748 and the MSKCC Sidney Kimmel Center for Prostate and Urologic Cancers. J Zhao is supported by an ASCO Conquer Cancer Young Investigator Award, a K12 Paul Calabresi Career Development Award for Clinical Oncology, and a Prostate Foundation Young Investigator Award. This research was supported in part by National Cancer Institute (NCI) Grant NIH T32CA062948 and the CTSC UL1 TR002384-01 (Weill Cornell). We wish to thank the study coordinators at Weill Cornell, Duke University, Johns Hopkins, University of Chicago, and Memorial Sloan Kettering Cancer Center. We wish to acknowledge the dedication of our patients to provide blood samples at no clear benefit to them but for the benefits of all patients with prostate cancer.
Conflicts of interest
LCB has no conflicts of interest
SH sits on DSMB for Ferring and Sanofi and receives funding via ASCO as a consulting statistician on the TAPUR trial.
JDS is/was an employee of Epic Sciences during the writing, data collection, and analysis
QY has no conflicts of interest
JL has served as a paid consultant/advisor for Sun Pharma, Janssen, Tolero, and Sanofi; has received research funding to his institution from Orion, Mirati, Astellas, Sanofi, Constellation, Calibr, Pandomedx, Trovagene, and Gilead; and is a co-inventor of a technology that has been licensed to Tokai, Qiagen, and A&G.
DMN has served as a paid consultant/advisor for Roche, Genentech; has received research funding to his institution from Sanofi, Medivation, Astellas, Janssen, Amgen, Progenics, Dendreon, Lilly, Genentech, Newlink, BMS, Inovio, AstraZeneca, Immunomedics, Aveo, Rexahn, Atlab, Boehringer Ingelheim, Millennium, Bayer, Merck, Abbvie, Karyopharm, Endocyte, Clovis, Seattle Genetics, AAA/Novartis
PG is employed and has stock/ownership interest in Novartis; research funding from Sanofi; patent/royalties/intellectual property International patent application docket No. 1676.083WO1 “Identifying Taxane Sensitivity in Prostate Cancer Patients” (Giannakakou, Plymate, co-inventors)
RZS has served as a paid consultant/advisor for AstraZeneca, AbbVie, Exelixis, Merck, Amgen, Janssen Pharmaceuticals, Sanofi, Astellas Pharma, Pfizer; has received research funding to their institution from AbbVie, Astellas Pharma, Incyte, Macrogenics, Janssen Pharmaceuticals; travel/accommodation expenses from Corcept Therapeutics; and patent/royalties/intellectual property from a Patent licensed by University of Chicago of which co-inventor to Corcept Therapeutics for combination AR/GR inhibition in prostate cancer
DCD has research support from US Department of Defense, American Society of Clinical Oncology, Prostate Cancer Foundation, Stand Up 2 Cancer, Janssen Research & Development, Astellas, Medivation, Agensys, Genentech, CreaTV. Consultant for Angle LLT, Janssen Research & Development, Astellas, Agensys, Medivation.
ESB has no conflicts of interest
EAC has no conflicts of interest
JLZ has no conflicts of interest
PH has no conflicts of interest
MA has no conflicts of interest
AG is/was an employee of Epic Sciences during the writing, data collection, and analysis
AJ is/was an employee of Epic Sciences during the writing, data collection, and analysis
WRB has received paid honoraria/speakers bureau/travel and accommodation expenses for Bayer AG; received research funding (to institution) from Merck.
SG has no conflicts of interest
SGG is a paid consultant for Pfizer and receives research funding to the institution from Janssen Pharmaceuticals.
RW is/was an employee of Epic Sciences during the writing, data collection, and analysis
ESA is a paid consultant/advisor to Janssen, Astellas, Sanofi, Dendreon, Pfizer, Amgen, AstraZeneca, Bristol-Myers Squibb, Bayer, Clovis, and Merck; has received research funding (to his institution) from Janssen, Johnson & Johnson, Sanofi, Dendreon, Genentech, Novartis, Bristol Myers-Squibb, AstraZeneca, Clovis, and Merck; and is the co-inventor of an AR-V7 biomarker technology that has been licensed to Qiagen.
DJG has research support (to Duke) from Acerta, Astellas, BMS, Bayer, Calithera, Exelixis, Janssen, Myovant, Pfizer, Novartis, Sanofi Aventis. Consulting income from Vizuri health sciences, UroToday, Sanofi, Pfizer, Nektar, Myovant, Modra, Merck, Ipsen, Flatiron, Exelixis, Capio, EMD Serono, BMS, Bayer, Astrazeneca, Astellas.
HIS has research support related to this study to Institution EPIC and Menarini Silicon Biosystems, Menarini; Additional support outside of this study is: compensated board of directors member of Asterias Biotherapeutics: compensated consultant/advisor to Ambry Genetics Corp, Konica Minolta Inc, Pfizer Inc, WCG Oncology; uncompensated consultant/advisory to Amgen, Bayer, ESSA Pharma Inc, Janssen Research & Development, LLC, Janssen Biotech, Inc, Sanofi Aventis; received research funding (to his institution) from Epic Sciences, Illumina, Inc, Janssen, Menarini Silicon Biosystems, Prostate Cancer Foundation and ThermoFisher; non-financial support from Amgen, Asterias Biotherapeutics, Bayer, Clovis Oncology, ESSA Pharma Inc., Menarini Silicon Biosystems, Phosplatin, Pfizer Inc, Prostate Cancer Foundation, Sanofi Aventis and WCG Oncology.
AJA is a paid consultant with Pfizer, Astellas, Janssen, Bayer, Astrazeneca, and Merck and receives research funding (to his institution) from Pfizer, Astellas, Janssen, Bayer, Dendreon, Novartis, Genentech/Roche, Merck, BMS, Astrazeneca, Constellation, Beigene.
Footnotes
Prior presentation: Poster presented, in part, at the 2020 American Society of Clinical Oncology Genitourinary Symposium, San Francisco, CA February 2020
References
- 1.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013;368:138–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364:1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371:424–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187–97 [DOI] [PubMed] [Google Scholar]
- 5.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Zhu Y, et al. Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J Clin Oncol 2017;35:2149–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scher HI, Graf RP, Schreiber NA, Jayaram A, Winquist E, McLaughlin B, et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol 2018;4:1179–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2016;2:1441–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong AJ, Halabi S, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J Clin Oncol 2019;37:1120–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015;162:454. [DOI] [PubMed] [Google Scholar]
- 11.Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov 2018;8:444–57 [DOI] [PubMed] [Google Scholar]
- 12.Annala M, Struss WJ, Warner EW, Beja K, Vandekerkhove G, Wong A, et al. Treatment Outcomes and Tumor Loss of Heterozygosity in Germline DNA Repair-deficient Prostate Cancer. Eur Urol 2017;72:34–42 [DOI] [PubMed] [Google Scholar]
- 13.Antonarakis ES, Lu C, Luber B, Liang C, Wang H, Chen Y, et al. Germline DNA-repair Gene Mutations and Outcomes in Men with Metastatic Castration-resistant Prostate Cancer Receiving First-line Abiraterone and Enzalutamide. Eur Urol 2018;74:218–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 2019;571:576–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marquard AM, Eklund AC, Joshi T, Krzystanek M, Favero F, Wang ZC, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res 2015;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popova T, Manie E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012;72:5454–62 [DOI] [PubMed] [Google Scholar]
- 17.Schonhoft JD, Zhao JL, Jendrisak A, Carbone EA, Barnett ES, Hullings MA, et al. Morphology-Predicted Large-Scale Transition Number in Circulating Tumor Cells Identifies a Chromosomal Instability Biomarker Associated with Poor Outcome in Castration-Resistant Prostate Cancer. Cancer Res 2020;80:4892–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Han B, Huang J. Morphologic Spectrum of Neuroendocrine Tumors of the Prostate: An Updated Review. Arch Pathol Lab Med 2020;144:320–5 [DOI] [PubMed] [Google Scholar]
- 19.Aggarwal R, Zhang T, Small EJ, Armstrong AJ. Neuroendocrine prostate cancer: subtypes, biology, and clinical outcomes. J Natl Compr Canc Netw 2014;12:719–26 [DOI] [PubMed] [Google Scholar]
- 20.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol 2018;36:2492–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aparicio AM, Shen L, Tapia EL, Lu JF, Chen HC, Zhang J, et al. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin Cancer Res 2016;22:1520–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Coleman IM, Brown LG, True LD, Kollath L, Lucas JM, et al. SRRM4 Expression and the Loss of REST Activity May Promote the Emergence of the Neuroendocrine Phenotype in Castration-Resistant Prostate Cancer. Clin Cancer Res 2015;21:4698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017;32:474–89.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsaur I, Heidegger I, Kretschmer A, Borgmann H, Gandaglia G, Briganti A, et al. Aggressive variants of prostate cancer - Are we ready to apply specific treatment right now? Cancer Treat Rev 2019;75:20–6 [DOI] [PubMed] [Google Scholar]
- 25.Aparicio AM, Harzstark AL, Corn PG, Wen S, Araujo JC, Tu SM, et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res 2013;19:3621–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corn PG, Heath EI, Zurita A, Ramesh N, Xiao L, Sei E, et al. Cabazitaxel plus carboplatin for the treatment of men with metastatic castration-resistant prostate cancers: a randomised, open-label, phase 1-2 trial. Lancet Oncol 2019;20:1432–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beltran H, Jendrisak A, Landers M, Mosquera JM, Kossai M, Louw J, et al. The Initial Detection and Partial Characterization of Circulating Tumor Cells in Neuroendocrine Prostate Cancer. Clin Cancer Res 2016;22:1510–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armstrong AJ, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, Danila DC, et al. Prospective Multicenter Study of Circulating Tumor Cell AR-V7 and Taxane Versus Hormonal Treatment Outcomes in Metastatic Castration-Resistant Prostate Cancer. JCO Precis Oncol 2020;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. ClinCancer Res 2008;14:6302–9 [DOI] [PubMed] [Google Scholar]
- 30.Halabi S, Lin CY, Kelly WK, Fizazi KS, Moul JW, Kaplan EB, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol 2014;32:671–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris MJ, Basch EM, Wilding G, Hussain M, Carducci MA, Higano C, et al. Department of Defense prostate cancer clinical trials consortium: a new instrument for prostate cancer clinical research. Clinical genitourinary cancer 2009;7:51–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 2015;33:1348–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol 2008;26:1148–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heller G, McCormack R, Kheoh T, Molina A, Smith MR, Dreicer R, et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison With Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J Clin Oncol 2018;36:572–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Computational Statistics & Data Analysis 2003;43:121–37 [Google Scholar]
- 36.Halabi S, Lin CY, Small EJ, Armstrong AJ, Kaplan EB, Petrylak D, et al. Prognostic model predicting metastatic castration-resistant prostate cancer survival in men treated with second-line chemotherapy. J Natl Cancer Inst 2013;105:1729–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleppe A, Albregtsen F, Vlatkovic L, Pradhan M, Nielsen B, Hveem TS, et al. Chromatin organisation and cancer prognosis: a pan-cancer study. Lancet Oncol 2018;19:356–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A 2009;106:8671–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY, et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. New England Journal of Medicine 2017;377:352–60 [DOI] [PubMed] [Google Scholar]
- 40.Davis ID, Martin AJ, Stockler MR, Begbie S, Chi KN, Chowdhury S, et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N Engl J Med 2019;381:121–31 [DOI] [PubMed] [Google Scholar]
- 41.Chi KN, Agarwal N, Bjartell A, Chung BH, Pereira de Santana Gomes AJ, Given R, et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med 2019;381:13–24 [DOI] [PubMed] [Google Scholar]
- 42.James ND, de Bono JS, Spears MR, Clarke NW, Mason MD, Dearnaley DP, et al. Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N Engl J Med 2017;377:338–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armstrong AJ, Szmulewitz RZ, Petrylak DP, Holzbeierlein J, Villers A, Azad A, et al. ARCHES: A Randomized, Phase III Study of Androgen Deprivation Therapy With Enzalutamide or Placebo in Men With Metastatic Hormone-Sensitive Prostate Cancer. J Clin Oncol 2019;37:2974–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain M, Fizazi K, Saad F, Rathenborg P, Shore N, Ferreira U, et al. Enzalutamide in Men with Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med 2018;378:2465–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fizazi K, Shore N, Tammela TL, Ulys A, Vjaters E, Polyakov S, et al. Darolutamide in Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med 2019;380:1235–46 [DOI] [PubMed] [Google Scholar]
- 46.Gupta S, Hovelson DH, Kemeny G, Halabi S, Foo WC, Anand M, et al. Discordant and heterogeneous clinically relevant genomic alterations in circulating tumor cells vs plasma DNA from men with metastatic castration resistant prostate cancer. Genes Chromosomes Cancer 2020;59:225–39 [DOI] [PubMed] [Google Scholar]
- 47.Malihi PD, Graf RP, Rodriguez A, Ramesh N, Lee J, Sutton R, et al. Single-Cell Circulating Tumor Cell Analysis Reveals Genomic Instability as a Distinctive Feature of Aggressive Prostate Cancer. Clin Cancer Res 2020;26:4143–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao P, Zhu Y, Cheng L, Luo J. Detection of androgen receptor (AR) and AR-V7 in small cell prostate carcinoma: Diagnostic and therapeutic implications. Asian J Urol 2019;6:109–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beltran H, Romanel A, Conteduca V, Casiraghi N, Sigouros M, Franceschini GM, et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 2020;130:1653–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220–9 [DOI] [PubMed] [Google Scholar]
- 52.Scher HI, Jendrisak A, Gill A, Barnett E, Gopalan A, Zaidi S, et al. Circulating tumor cells (CTCs) with small-cell like pathology are prevalent in metastatic castration-resistant prostate cancer (mCRPC) and show selective pharmacodynamic reductions in patients treated with platinum but not ARSI or taxane. Journal of Clinical Oncology 2020;38:5572- [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


