SUMMARY
Clinical oncology is experiencing rapid growth in data that are collected to enhance cancer care. With recent advances in the field of Artificial Intelligence (AI), there is now a computational basis to integrate and synthesize this growing body of multi-dimensional data, deduce patterns, and predict outcomes to improve shared patient and clinician decision-making. While there is high potential, significant challenges remain. In this perspective, we propose a pathway of clinical, cancer care touchpoints for narrow-task AI applications and review a selection of applications. We describe the challenges faced in the clinical translation of AI and propose solutions. We also suggest paths forward in weaving AI into individualized patient care, with an emphasis on clinical validity, utility, and usability. By illuminating these issues in the context of current AI applications for clinical oncology, we hope to help advance meaningful investigations that will ultimately translate to real-world clinical use.
INTRODUCTION
Over the last decade, there has been a resurgence of interest for artificial intelligence (AI) applications in medicine. This is driven by the advent of deep learning algorithms, computing hardware advances, and the exponential growth of data that are being generated and used for clinical decision making (Esteva et al., 2019; Kann et al., 2020a; LeCun et al., 2015). Oncology is particularly poised for transformative changes brought on by AI, given the proven advantages of individualized care and recognition that tumors and their response rates differ vastly from person to person (Marusyk et al., 2012; Schilsky, 2010). In oncology, much like other medical fields, the overarching goal is to increase quantity and quality of life, which, from a practical standpoint, entails choosing the management strategy that optimizes cancer control and minimizes toxicity.
As multidimensional data is increasingly being generated in routine care, AI can support clinicians to form an individualized view of a patient along their care pathway and ultimately guide clinical decisions. These decisions rely on the incorporation of disparate, complex datastreams, including clinical presentation, patient history, tumor pathology and genomics, as well as medical imaging, and marrying these data to the findings of an ever-growing body of scientific literature. Furthermore, these datastreams are in a constant state of flux over the course of a patient’s trajectory. With the emergence of AI, specifically deep learning (LeCun et al., 2015), there is now a computational basis to integrate and synthesize these data, to predict where the patient’s care path is headed, and ultimately improve management decisions.
While there is much reason to be hopeful, numerous challenges remain to the successful integration of AI in clinical oncology. In analyzing these challenges, it is critical to view the promise, success, and failure of AI not only in generalities, but on a clinical case-by-case basis. Not every cancer problem is a nail to AI’s hammer; its value is not universal, but inextricably linked to the clinical use case (Maddox et al., 2019). The current evidence suggests that clinical translation of the vast majority of published, high-performing AI algorithms remains in a nascent stage (Nagendran et al., 2020). Furthermore, we posit that the imminent value of AI in clinical oncology is in the aggregation of narrow task-specific, clinically validated and meaningful applications at clinical “touchpoints” along the cancer care pathway, rather than general, all-purpose AI for end-to-end decision-making. As the global cancer incidence increases and the financial toxicity of cancer care is increasingly recognized, many societies are moving towards value-based care systems (Porter, 2009; Yousuf Zafar, 2016). With development of these systems, there will be increasing incentive for the adoption of data-driven tools - potentially powered by AI - that can lead to reduced patient morbidity, mortality, and healthcare costs (Kuznar, 2015).
Here, we will describe the key concepts of AI in clinical oncology and review a selection of AI applications in oncology from the lens of a patient moving through clinical touchpoints along the cancer care path. We will therein describe the challenges faced in the clinical translation of AI and propose solutions, and finally suggest paths forward in weaving AI into individualized patient cancer care. By illuminating these issues in the context of current AI applications for clinical oncology, we hope to provide concepts to help drive meaningful investigations that will ultimately translate to real-world clinical use.
Artificial Intelligence: from shallow to deep learning
The concept of AI, formalized in the 1950’s, was originally defined as the ability of a machine to perform a task normally associated with human performance (Russell and Haller, 2003). Within this field, the concept of machine learning was born, which refers to an algorithm’s ability to learn data and perform tasks without explicit programming (Samuel, 1959). Machine learning research has led to development and use of a number of “shallow” learning algorithms, including earlier generalized linear models like logistic regression, Bayesian algorithms, decision-trees, and ensemble methods (Bhattacharyya et al., 2019; Richens et al., 2020). In the simplest of these models, such as logistic regression, input variables are assumed to be independent of one another, and individual weights are learned for each variable to determine a decision boundary that optimally separates classes of labelled data. More advanced shallow learning algorithms, such as random forests, allow for the characterization and weighting of input variable combinations and relationships, thus learning decision boundaries that can fit more complex data.
Deep learning is a newer subset of machine learning, which has the ability to learn patterns from raw, unstructured input data by incorporating layered neural networks (LeCun et al., 2015). In supervised learning, which represents the most common form within medical AI, a neural network will generate a prediction from this input data and compare it to a “ground truth” annotation. This discrepancy between prediction and ground truth is encapsulated in a loss function which is then propagated back through the neural network, and over numerous cycles, the model is optimized to minimize this loss function.
For the purpose of clinical application, we can view AI as a spectrum of algorithms, the utility of which are inextricably linked to the characteristics of the task under investigation. Thorough understanding of the data stream is necessary to choose, develop, and optimize an algorithm. In general, deep learning networks offer nearly limitless flexibility in input, output, architectural and parameter design, and thus are able to fit vast quantities of heterogeneous and unstructured data never before possible (Esteva et al., 2017). Specifically, deep learning has a high propensity to learn non-linear and high-dimensional relationships in multi-modal data including time series data, pixel-by-pixel imaging data, unstructured text data, audio/video data, or biometric data. Data with significant spatial and temporal heterogeneity are particularly well-suited for DLNNs (Zhong et al., 2019). On the other hand, this power comes at the expense of limited interpretability and a proclivity for overfitting data if not trained on a large enough dataset (Zhu et al., 2015). While traditional machine learning and statistical modeling can perform quite well at certain predictive tasks, they generally struggle to fit unprocessed, unstructured, and high dimensional data compared to deep learning. Therefore, despite its limitations, deep learning has opened the door to big data analysis in oncology and promises to advance clinical oncology, so long as certain pitfalls in development and implementation can be overcome.
Cancer care as a mathematical optimization problem
To appreciate the promise surrounding AI applications for clinical oncology, it is essential to incorporate a mathematical lens to the patient care path through cancer risk prediction, screening, diagnosis and treatment. From the AI perspective, the patient path is an optimization problem, wherein heterogeneous data streams converge as inputs into a mathematical scaffold (i.e. machine learning algorithms) (Figure 1). This scaffold is iteratively adjusted during training until the desired output can be reliably predicted and an action can be taken. In this setting, an ever-growing list of inputs include patient clinical presentation, past medical history, genomics, imaging, and biometrics, and can be roughly subdivided as tumor, host, or environmental factors. The complexity of the algorithms is often driven by the quantity, heterogeneity, and dimensionality of such data. Outputs are centered, most broadly, on increasing survival and/or quality of life, but are often evaluated by necessity as a series of more granular surrogate endpoints.
Figure 1:
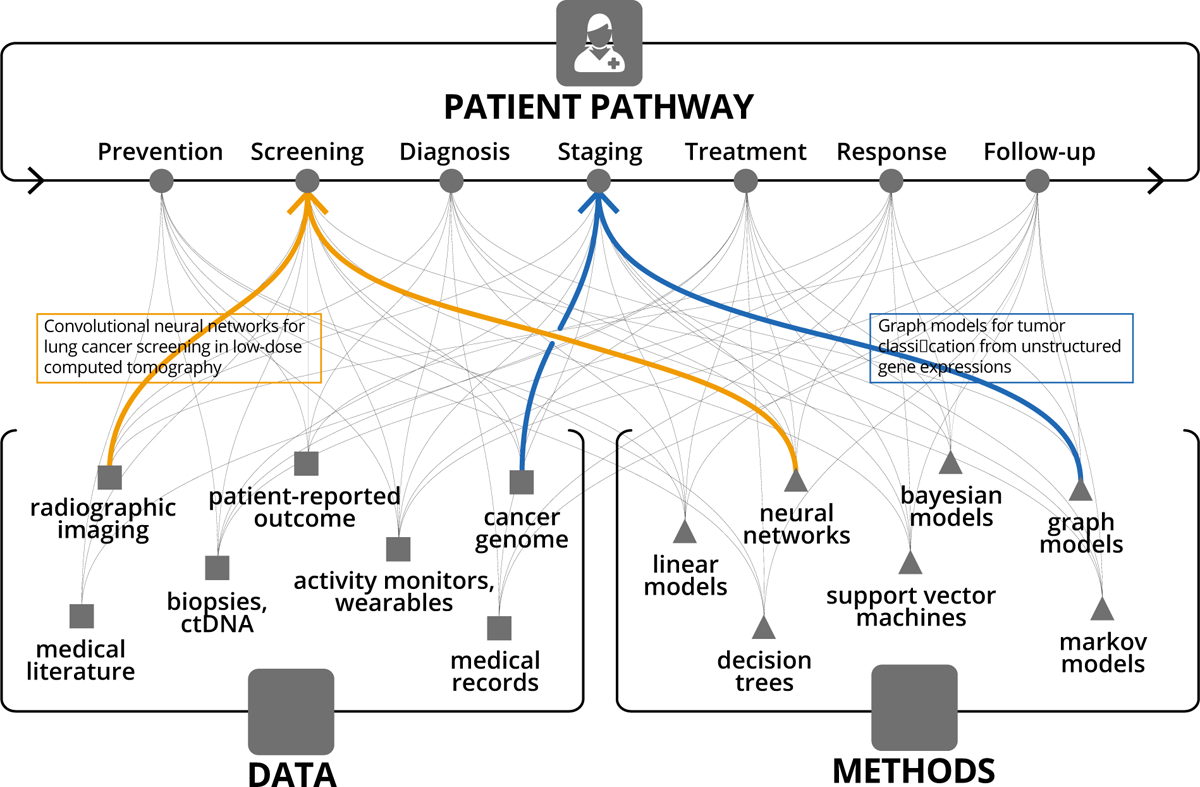
Narrow task-specific AI applications addressing a specific touchpoint along the patient pathway, and utilizing a specific data type and AI method.
Datastreams for clinical oncology
The arc of research in oncology, increasing data generation, and advances in computational technology have collectively resulted in a frameshift from low-dimensional to increasingly high-dimensional patient data representation. Earlier data and computational limitations often necessitated reducing unstructured patient data (e.g. medical images and biopsies) into a set of human-digestible discrete measures of disease extent. One notable example of such simplification lies within cancer staging systems, most prominently the AJCC TNM classification (Amin et al., 2017). In 1977, with only three inputs commonly available - tumor size, nodal involvement, and presence of metastasis - the first edition AJCC TNM staging became standard of care for risk-stratification and decision-management in oncology. Over the subsequent decades, with the incorporation of other discrete data points, predictive nomograms could be generated using simple linear models, which have found practical use in certain situations (Bari et al., 2010; Creutzberg et al., 2015; Mittendorf et al., 2012; Stephenson et al., 2007). More recently, improved methods to extract and analyze existing data coupled with new data streams and a growing understanding of inter- and intra-tumoral heterogeneity, have all led to the development of increasingly complex and specific stratification models. Key examples of novel data streams introduced over the past two decades are the Electronic Health Record, The Cancer Genome Atlas (Weinstein et al., 2013), The Cancer Imaging Archive (Clark et al., 2013), and the Project GENIE initiative (AACR Project GENIE Consortium, 2017). Key examples of advanced risk-stratification and prediction models are the prostate cancer Decipher score (Erho et al., 2013) and breast cancer OncotypeDx score (Paik et al., 2004), which utilize discrete genomic data and shallow machine learning algorithms to form clinically validated predictive models. Useful oncology datastreams, roughly following historical order of availability, include: clinical presentation, tumor stage, histopathology, qualitative imaging, tumor genomics, patient genomics, quantitative imaging, liquid biopsies, electronic medical record mining, wearable devices, and digital behavior (Figure 1). Furthermore, as a patient moves along the cancer care pathway, the number of influxing, intra-patient datastreams grows. With each step through the pathway, new data is generated out of the pathway with the potential to be reincorporated at a later time back into the pathway (Figure 2).
Figure 2:
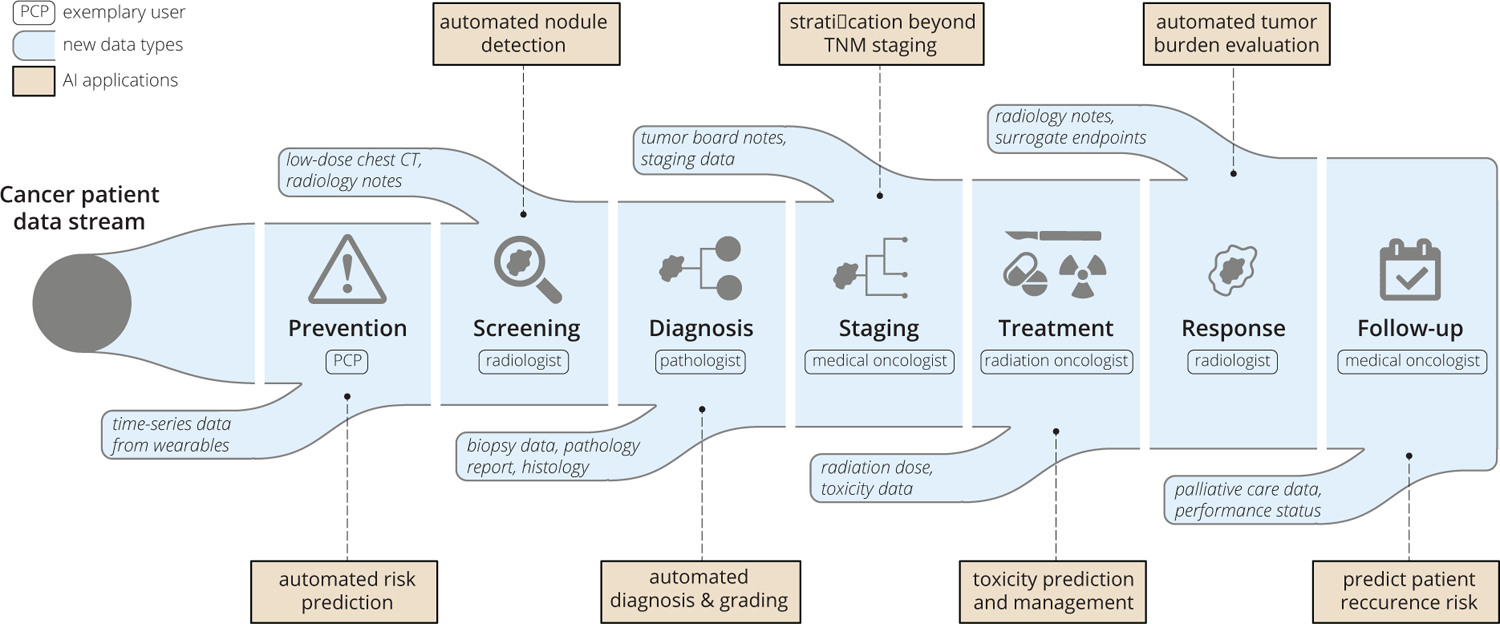
An example cancer patient pathway converges with an ever-increasing data stream. Potential AI applications and exemplary clinical users at each touchpoint are also illustrated.
As our biological knowledge base and datastreams grow in clinical oncology, machine learning algorithms can be deployed to learn patterns that apply to more and more precise patient groups and generate predictions to guide treatment for the next, “unseen” patient. As we assimilate more data, optimal cancer care, i.e. the care that results in the best survival and quality of life for a patient, inevitably becomes precision care, assuming we have the necessary tools to fully utilize the data. Here, at this intersection of data complexity and precision care in clinical oncology, is where the promise of AI has been so tantalizing, though as of yet, unfulfilled.
AI Applications and Touchpoints along the Clinical Oncology Care Path
We propose that AI development for clinical oncology should be approached from patient and clinician perspectives across the following cancer care touchpoints: Risk Prediction, Screening, Diagnosis, Prognosis, Initial Treatment, Response Assessment, Subsequent Treatment, and Follow-up (Figure 2). The clinical touchpoint pathway shares features with the “cancer continuum,” (Chambers et al., 2018) though it consists of more granular, patient and clinician decision-oriented points of contact for AI to add clinical benefit. Each of these touchpoints involves a critical series of decisions for oncologists and patients to make and yields a use-case for AI to provide an incremental benefit. Furthermore, touchpoint details will vary by cancer subtype. Within these touchpoints, ideal AI use-cases are ones with significant unmet need and large available datasets. In the context of supervised machine learning, these datasets require robust and accurate annotation to form a reliable “ground-truth” on which the AI system can train.
Narrow tasks with high reliability
As clinical oncology datastreams increase in complexity, the tools needed to discern patterns from these data are necessarily more complex, as well. Amidst this flood of heterogeneous intra-patient data, there is a relative dearth of inter-patient data which is needed to train large scale models. Therefore, to accumulate the training data required for generalizable models, it will likely be more fruitful to target and evaluate individual AI models towards specific datastreams at a particular touchpoint along the care pathway.
It is tempting to think that, given the increasing data streams that encompass multiple patient characteristics and outcomes, one could develop a unifying, dynamic model to synthesize and drive precision oncology, developing a “virtual-guide” of sorts for the oncologist and patient (Topol, 2019). Analogies are often made to transformative technologies, such as self-driving cars and social media recommendations that leverage powerful neural networks on top of streams composed of billions of incoming data points, to predict real-time outcomes and continually improve performance. While in theory, this strategy could one day be deployed in a clinical setting, there are vast differences between these domains that question whether or not we should or even could pursue this strategy currently. One of the most glaring differences between the healthcare and technology domains, in terms of AI application, is the striking difference in data quality and quantity. While there has been a sea change in the collection of data within the healthcare field over the past decade, driven by the adoption of the Electronic Health Record, datasets still remain virtually siloed, intensely regulated, and, particularly in cancer care, much too small to leverage the most powerful AI algorithms available (Bi et al., 2019; Kelly et al., 2019). One of the most high-profile of these endeavors, IBM’s Watson Oncology project, has attempted to develop a broad prediction machine to guide cancer care, but has been limited by suboptimal concordance with human oncologists’ recommendations and subsequent distrust (Gyawali, 2018; Lee et al., 2018; Somashekhar et al., 2017).
As our biological perspective has evolved, we now know that cancer is made up of thousands of distinct entities that will follow different trajectories, each with different treatment strategies (Dagogo-Jack and Shaw, 2018; Polyak, 2011). In computational model development, there is thought to be a bare minimum number of data samples required for each model input feature (Mitsa, 2019). As we seek to make recommendations more and more bespoke, it becomes more challenging to accrue the quantity of training data necessary to leverage complex algorithms. Fortunately, this data gap in healthcare is well-recognized, and a number of initiatives have been proposed to streamline and unify data collection (Wilkinson et al., 2016). However, given the innately heterogeneous, fragmented, and private nature of healthcare data, we in the oncology field may never achieve a level of data robustness enjoyed by other technology sectors. Therefore, strategies are necessary to mitigate the data problem, such as proper algorithm selection, model architecture improvements, data preprocessing, and data augmentation techniques. Above all, thoughtful selection of narrow use cases across cancer care touchpoints is paramount in order to yield clinical impact.
Once rigorously tested, these narrow-AIs could then be aggregated over the course of a patient’s care to provide a measurable, clinical benefit. This sort of AI-driven dimensionality reduction of a patient’s feature space allows for optimizing the development process and exporting of quality AI applications in the present environment of siloed data, expertise, and infrastructure. As of writing, there are approximately 20 FDA-approved AI applications targeted specifically for clinical oncology, and each of these performs a narrow task, utilizing a single data stream at a specific cancer care touchpoint (Benjamens et al., 2020; Hamamoto et al., 2020; Topol, 2019) (Table 1). We hypothesize that the future of AI in oncology will continue to consist of an aggregation of rigorously evaluated, narrow-task models, each one providing small, incremental benefits for patient quantity and quality of life. In the next sections, we will review select AI applications that have excelled with this narrow-task approach.
Table 1.
FDA approvals to date for deep-learning applications in clinical oncology
| Name | Data type | Task | FDA summary | Year | |
|---|---|---|---|---|---|
| Thoracic/liver | |||||
| 1 | Arterys Oncology DL | CT, MRI | segmentation of lung nodules and liver lesions, automated reporting | https://www.accessdata.fda.gov/cdrh_docs/pdf17/K173542.pdf | 2017 |
| 2 | Siemens AI-Rad Companion (Pulmonary) | CT | segmentation of lesions of the lung, liver, and lymph nodes | https://www.accessdata.fda.gov/cdrh_docs/pdf18/K183271.pdf | 2019 |
| 3 | Riverain ClearRead CT | CT | detection of pulmonary nodules in asymptomatic population | https://www.accessdata.fda.gov/cdrh_docs/pdf16/k161201.pdf | 2016 |
| 4 | Siemens syngo.CT Lung CAD | CT | detection of solid pulmonary nodules, alerts to overlooked regions | https://www.accessdata.fda.gov/cdrh_docs/pdf19/K193216.pdf | 2020 |
| 5 | GE Hepatic VCAR | CT | liver lesion segmentation and measurement | https://www.accessdata.fda.gov/cdrh_docs/pdf19/K193281.pdf | 2020 |
| 6 | Coreline AView LCS | CT | characterization of nodule type, location, measurements, and Lung-RADS category | https://www.accessdata.fda.gov/cdrh_docs/pdf20/K201710.pdf | 2020 |
| 7 | MeVis Veolity | CT | detection of solid pulmonary nodules, alerts to overlooked regions | https://www.accessdata.fda.gov/cdrh_docs/pdf20/K201501.pdf | 2021 |
| 8 | Philips Lung Nodule Assessment and Comparison Option (LNA) | CT | characterization of nodule type, location, and measurements | https://www.accessdata.fda.gov/cdrh_docs/pdf16/K162484.pdf | 2017 |
| 9 | NinesMeasure | CT | characterization of nodule type, location, and measurements | https://www.accessdata.fda.gov/cdrh_docs/pdf20/K202990.pdf | 2021 |
| Breast | |||||
| 10 | iCAD ProFound AI | 3D DBT mammography | detection of soft tissue densities and calcifications | https://www.accessdata.fda.gov/cdrh_docs/pdf19/K191994.pdf | 2019 |
| 11 | cmTriage | 2D FFDM | triage and passive notification | https://www.accessdata.fda.gov/cdrh_docs/pdf18/K183285.pdf | 2019 |
| 12 | Screenpoint Transpara | FFDM | detection of suspicious soft tissue lesions and calcifications | https://www.accessdata.fda.gov/cdrh_docs/pdf19/K192287.pdf | 2019 |
| 13 | Zebra Medical Vision HealthMammo | 2D FFDM | triage and passive notification | https://www.accessdata.fda.gov/cdrh_docs/pdf20/K200905.pdf | 2020 |
| 14 | Koios DS for Breast | ultrasonography | classification of lesion shape, orientation, and BI-RADS category | https://www.accessdata.fda.gov/cdrh_docs/pdf19/K190442.pdf | 2019 |
| 15 | Hologic Genius AI Detection | DBT mammography | detection of suspicious soft tissue lesions and calcifications | https://www.accessdata.fda.gov/cdrh_docs/pdf20/K201019.pdf | 2020 |
| 16 | Therapixel MammoScreen | FFDM | detection of suspicious findings and level of suspicion | https://www.accessdata.fda.gov/cdrh_docs/pdf19/K192854.pdf | 2020 |
| 17 | QuantX | MRI | image registration, automated segmentation, and analysis of user-selected regions of interest | https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN170022.pdf | 2020 |
| 18 | ClearView cCAD | ultrasonography | classification of shape and orientation of user-defined regions, and BI-RADS category | https://www.accessdata.fda.gov/cdrh_docs/pdf16/K161959.pdf | 2016 |
| Prostate | |||||
| 19 | Quantib Prostate | MRI | semi-automatic segmentation of anatomic structures, volume computations, automated PI-RADS category | https://www.accessdata.fda.gov/cdrh_docs/pdf20/K202501.pdf | 2020 |
| 20 | GE PROView | MRI | prediction of PI-RADS category | https://www.accessdata.fda.gov/cdrh_docs/pdf19/K193306.pdf | 2020 |
| Central nervous system | |||||
| 21 | Cortechs NeuroQuant | MRI | automated segmentation and volumetric quantification of brain lesions | https://www.accessdata.fda.gov/cdrh_docs/pdf17/K170981.pdf | 2017 |
DBT, digital breast tomosynthesis; FFDM, full-field digital mammography; BI-RADS, Breast Imaging Reporting and Data System; PI-RADS, Prostate Imaging Reporting and Data System.
Narrow-task AI examples across the clinical oncology touchpoints
T1. Risk Prediction and Prevention.
Given the burden to people and healthcare systems of cancer diagnosis and management, there is a significant opportunity for AI to help predict an individual’s risk of developing cancer, and thereby target screening and early interventions effectively and efficiently. In a mathematical sense, the patient’s entire personal history up until diagnosis makes up a vast and extremely heterogeneous datastream to be evaluated, positioning deep learning to have an impact. This is evidenced by the steady development of tools that leverage computational modeling to refine cancer risk. In the past few years, several DL algorithms have been investigated to further tailor risk prediction beyond traditional models. Some of these algorithms utilize novel datastreams that were not available until recently: satellite imagery (Bibault et al., 2020), internet search history (White and Horvitz, 2017), and wearable devices (Beg et al., 2017). Others maximize the utility of pre-existing datastreams, including patient genomics, routine imaging, unstructured health record data, and deeper family history to improve predictions (Ming et al., 2020).
T2. Screening.
Cancer screening involves the input and evaluation of data at a distinct time-point to determine whether or not additional diagnostic testing and procedures are warranted. Datastreams can be in the form of serum markers, medical imaging, or visual or endoscopic examination. Each of these modalities provides opportunities for the integration of AI to improve prediction of cancer. For serum markers, such as prostate specific antigen (PSA), early research suggests that machine learning algorithms modeling PSA at different timepoints, in conjunction with other serum markers, may be able to better predict presence of prostate cancer than PSA alone (Nitta et al., 2019). Perhaps more than in any other application, AI has found high impact use in medical imaging screening. Narrow-task models have been developed to localize lesions and predict risk of malignancy on lung cancer CT (Ardila et al., 2019) and breast cancer mammography (McKinney et al., 2020), with applications that have been shown to perform on par, or sometimes better than expert diagnosticians (Salim et al., 2020). In these applications, raw pixel data of the image is utilized as input into a deep learning convolutional neural network that is trained based on radiologist-labelled ground-truth outputs. Importantly, while the algorithms demonstrate impressive results in terms of area under the curve, sensitivity and specificity, they do not evaluate direct clinical endpoints, such as cancer mortality, healthcare costs, or quality of life. Outside of medical imaging, AI has found utility in screening endoscopy for colorectal carcinoma, with an application that guides biopsy site selection (Guo et al., 2020; Zhou et al., 2020). Furthermore, there are opportunities to improve diagnostic yield for other malignancies for which screening has been traditionally difficult and unproven. This could be accomplished by AI improving analysis of pre-existing datastreams, such as abdominal CT or MRI imaging, or via its ability to integrate multi-modal datastreams, like EHR and genomic data. While currently the United States Preventive Services Task Force (USPSTF,2021) recommends against screening for many cancers, there are a number of ongoing investigations to determine if incorporation of AI into screening criteria and technology may allow screening to be utilized in a wider array of disease sites, such as pancreatic cancer.
T3. Diagnosis.
Diagnosing involves the exclusion of other benign disease processes and the characterization of cancer by primary site, histopathology, and increasingly, genomic classification. Diagnosis represents an AI touchpoint for these three domains by analyzing their respective datastreams: including clinical exam and medical imaging (i.e. Radiomics), digital pathology, and genomic sequencing. A key study that revealed the promise of deep learning for cancer diagnosis showed that convolutional neural networks could achieve dermatologist level accuracy in the classification of skin cancers utilizing digital photographs (Esteva et al., 2017). Other promising areas of investigation in this realm include non-invasive brain tumor diagnosis (Chang et al., 2018) and prostate cancer Gleason grading (Schelb et al., 2019) via MRI, automated histopathologic diagnosis for breast cancer (Ehteshami Bejnordi et al., 2017) and prostate cancer (Nagpal et al., 2020), and utilization of radiographic and histopathologic data to predict underlying genomic classification (Lu et al., 2018). Thus far, the Screening and Diagnosis touchpoints account for nearly all FDA-approved AI applications for clinical oncology, with three algorithms focusing on mammography and three focusing on CT-based lesion diagnosis (Benjamens et al., 2020).
T4. Risk Stratification and Prognosis.
Historically, risk-stratification consisted of TNM staging, though increasingly additional datastreams such as genomics, advanced imaging, and serum markers have allowed for more precise risk stratification. Given the vast heterogeneity in cancer risk, risk-stratification presents a highly attractive use case for AI. Over the past two decades, genomic classifiers, developed with machine learning, have been integrated into risk-stratification for a number of malignancies. Classifiers such as OncotypeDx for breast cancer, a logistic regression based classifier, and the Decipher score, a random forest-based classifier, have demonstrated the ability to improve prognostication (Spratt et al., 2017) and guide treatment (Sparano et al., 2018). The Decipher score genomic classifier is based on 22 genomic expression markers input into a random forest model that was trained to predict metastasis after prostatectomy for patients with prostate cancer at a single institution(Erho et al., 2013). This classifier has been subsequently validated in several external settings, and is now undergoing investigation in several randomized control trials (NCT04513717; NCT02783950). Deep learning strategies have been explored to integrate multi-omic data sources into risk-stratification models utilizing combinations of diagnostic imaging (Kann et al., 2020b), EHR data (Beg et al., 2017; Manz et al., 2020), and genomic information (Qiu et al., 2020). Furthermore, there is the potential for deep learning to better risk-stratify patients based on large population databases, such as the Surveillance, Epidemiology, and End Results Program, by learning non-linear relationships between database variables, though preliminary efforts require validation (She et al., 2020).
T5. Initial Treatment Strategy.
The formulation of initial treatment strategy is arguably the most pivotal touchpoint for AI in the cancer pathway, as it directly influences patient management. The last two decades have seen exponential growth in the number and complexity of initial treatment options for common cancers (Kann et al., 2020a). A common predicament for initial treatment is what combination of systemic therapy, radiotherapy, and surgery is optimal for a given patient. Machine learning methods utilizing genomic (Scott et al., 2017) and radiomic data (Lou et al., 2019) have been investigated to predict radiation sensitivity. While immunotherapy has been adopted in an increasing number of disease settings, it remains difficult to predict response based on currently available biomarkers, and machine learning algorithms with radiomic input have demonstrated the ability to improve response prediction (Sun et al., 2018). Furthermore, deep learning has demonstrated the ability to analyze multi-modal datastreams within the genomic realm: a recent analysis demonstrated that integration of tumor mutational burden, copy number alteration, and microsatellite instability code can help predict response to immunotherapy (Xie et al., 2020). AI is also enabling more accurate “evidence-based treatment”. Natural language processing and powerful language models can help analyze published scientific works and utilize existing oncology literature e.g. extracting medical oncology concepts from EHR and linking these to a literature corpus (Simon et al., 2019).
T6. Response Assessment.
Assessment of response to treatment generally includes radiographic and clinical assessments. Quantitative response assessment criterias like RECIST and RANO have long been established as reproducible ways to assess response to therapy, though in the age of targeted immunotherapies, validity has been questioned (Villaruz and Socinski, 2013). As targeted therapeutics and immunotherapies have entered the clinic, however, it has become clear that response assessment via RECIST is inadequate, due to phenomena such as pseudoprogression (Gerwing et al., 2019). Detailed response assessment is often a time intensive process that requires a high degree of human expertise and experience, not to mention high intra- and inter-reader variability. Additionally, despite periodic review and revision of these criterion, they remain inapt at capturing edge cases, such as variable lesion response, in the case of patients receiving immunotherapy. Deep learning has demonstrated potential for automated response assessment, including automated RANO assessment (Kickingereder et al., 2019) and RECIST response in patients undergoing immunotherapy (Arbour et al., 2020).
T7. Subsequent Treatment Strategy.
When approaching AI algorithm development for subsequent treatment strategy, there are a number of specific considerations that generate complexity as compared to from initial treatment strategy. Firstly, there are additional datastreams to consider, such as prior treatments, treatment-related toxicity, restaging imaging, and often multiple tissue specimens. Given the heterogeneity in datastreams and the shrinking patient populations from which to build these models, subsequent treatment strategy is a challenging space for evidence-based decision-making, and in turn, for reliable AI applications. Algorithms that utilize longitudinal follow-up information may help here. In one example, AI has demonstrated the ability to synthesize serial CT follow-up imaging for lung cancer patients post-chemoradiation, and demonstrated the ability to predict later recurrence (Xu et al., 2019). An intervention such as this could guide selection for patients to undergo consolidative treatments like surgery or immunotherapy.
T8. Follow-up.
Another underexplored area for AI oncologic applications is development of tools to guide precision follow-up. Diagnostic and screening algorithms may often be transferable to the follow-up setting, but will require retraining and validation for the task of interest. Similar to T7, the effect of prior cancer treatment on the datastream will often shift things significantly. For example, radiomic features extracted from the same tumor, pre- and post-treatment, show significant discrepancies (van Dijk et al., 2019). These “delta” features could be used to predict patient recurrence risk and late toxicity, helping to tailor follow-up plans (Chang et al., 2019). Appropriately triaging patients for escalated follow-up and attention can promote decreased morbidity and more efficient healthcare resource utilization; AI leveraging EHR data has demonstrated the ability to accomplish this, by selecting patients at high risk for acute care visit while undergoing cancer therapy and assigning them to an escalated preventative care strategy (Hong et al., 2020). In cases where patients have untreatable relapse, end-of-life care becomes an extremely important and challenging process. AI has shown potential here as well, as a way to triage patients at high risk of mortality and nudge physicians to converse with patients regarding their values, wishes, and quality of life options (Ramchandran et al., 2013).
Challenges for clinical translation: beyond performance validation
While tremendous strides have been made in the development of oncologic AI, as evidenced by the surge in publications and published datasets in recent years, there remains a large gap between evidence for AI performance and evidence for clinical impact. While there have been thousands of published studies of deep learning algorithm performance (Kann et al., 2019), a recent systematic review, found only nine prospective trials, and two published randomized clinical trials of deep learning in medical imaging (Nagendran et al., 2020).
As alluded to above, perhaps the defining barrier to development of clinical AI applications in oncology, and healthcare overall, is data limitation, both in quality and quantity. The problems with data curation, aggregation, transparency, bias, and reliability have been well-described (Norgeot et al., 2020; Thompson et al., 2018). Additionally, the lack of AI model interpretability, trust, reproducibility, and generalizability have received ample, and well-justified attention (Beam et al., 2020). While all of these challenges must be overcome for successful AI development, here we will introduce several concepts specific to clinical translation of models that have already succeeded in preliminary stages of development and validation: clinical validity, utility, and usability (Figure 3). Incorporation of these concepts into model design and evaluation is easy to overlook, yet is critical to move clinical AI beyond the research and development stage into real-world cancer care.
Figure 3:
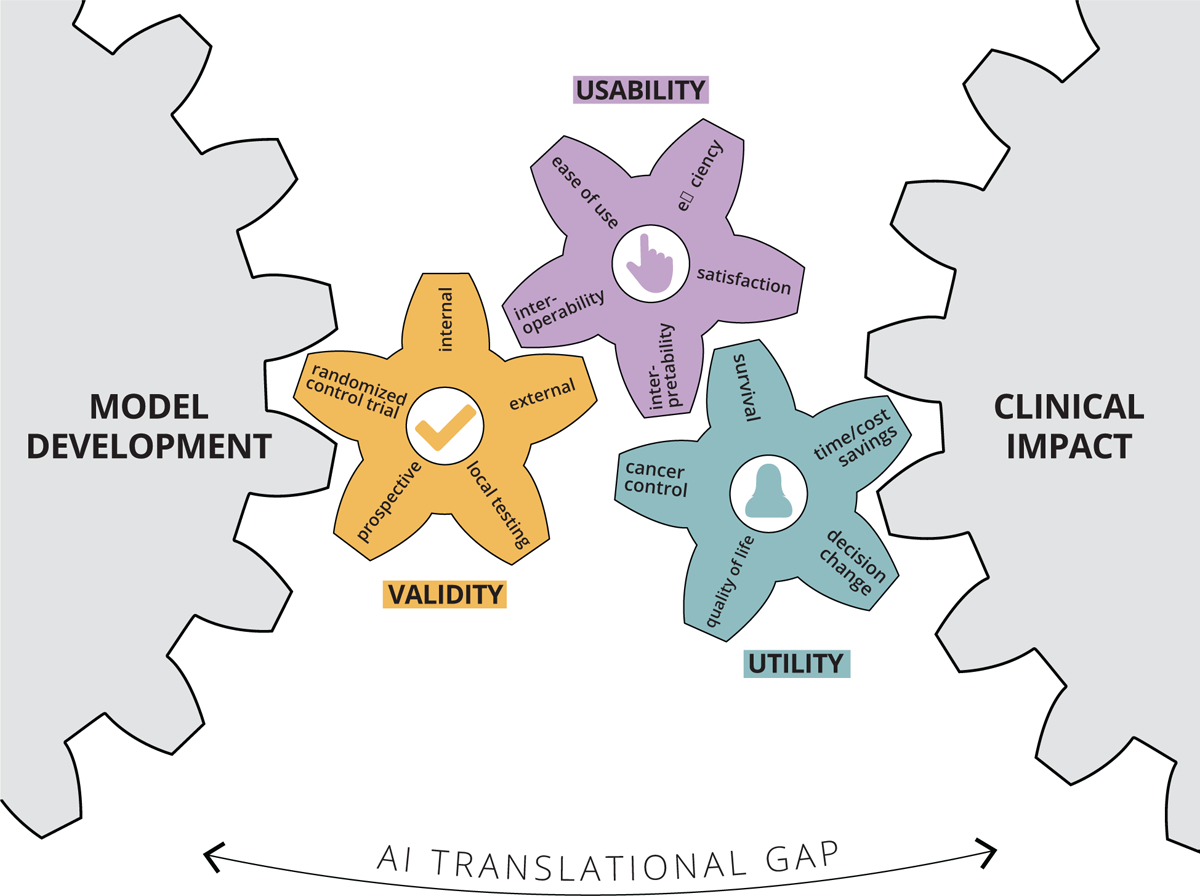
Bridging the AI translational gap between initial model development and routine clinical cancer care by emphasizing and demonstrating three essential concepts: clinical validity, utility, and usability.
To demonstrate clinical validity, a model is often evaluated in the following general sequence: internal validation, external validation, prospective testing, and local testing in the real-world population of interest (Park et al., 2020). Recently developed guidelines such as FAIR data, CONSORT/SPIRTAI, and the (in development) TRIPOD-AI checklists should be followed to ensure reproducibility, transparency, and methodologic rigor (Liu et al., 2019). These guidelines are an important step forward in standardizing AI model development pathways and establishing a basis to determine AI study methodological rigor. While the vast majority of AI published reports include an internal, blinded test set, far fewer utilize an external validation set, and an even smaller proportion employ prospective testing and benchmark comparisons with human experts (Kim et al., 2019). Given the lack of hypothesize-driven feature selection in most AI models, performance in real-world scenarios can vary dramatically if the test data distribution varies from the training data (Moreno-Torres et al., 2012). For this reason, multiple external validation sets are of utmost importance. Beyond this, it is often difficult to predict how a model will perform on edge cases - those that were under-represented in training data (Oakden-Rayner et al., 2020). In the practice of oncology, detection of rare findings can be critical to safe cancer care, and thus must be taken into account to demonstrate a model is clinically valid. One way to mitigate the risk of model failure in real-world use, is to conduct trial, run-in periods of “silent” prospective testing in the scenario of interest (Kang et al., 2020). If a model performs well in the run-in period, there is some assurance that it will be safe to utilize, though its performance on extremely rare cases may be still difficult to presume.
Demonstrating clinical utility, requires clinical validity as a prerequisite, but goes beyond performance validation to the testing of clinically meaningful endpoints. High performance on commonly used endpoints, such as area under the receiver operating characteristic curve, sensitivity, or specificity, may suffice for certain diagnostic applications, but real-world impact will require validation of clinical endpoints as appropriate for each touchpoint along the care pathway. In the case of oncology, this includes overall survival, disease control, toxicity reduction, quality of life improvement, and decrease of healthcare resource utilization. Testing of these endpoints should be ideally performed in the setting of a randomized trial. The gold standard would be randomizing patients to the AI intervention and directly comparing clinical endpoints. A few of these trials have been completed, with one notable example involving testing accuracy for polyp detection rate on colonoscopy (Wang et al., 2019). In this study, the primary outcome was adenoma detection rate. Despite demonstrating the superiority of the AI systems, downstream clinical benefit in terms of quality of life or survival requires yet further investigation. Another approach to AI clinical trials is to apply a validated model to all patients for risk-stratification, and then to apply randomized interventions. This was pursued successfully in a trial that utilized EHR data to predict patients at high-risk for emergency department (ED) visits during radiotherapy (Hong et al., 2020). High-risk patients were then randomized to usual care, or extra preventative provider visits. It was found that high-risk patients randomized to extra visits had significantly fewer ED and hospital admissions, while low-risk patients had uniformly low rates of ED and hospital admissions without extra care. While providing a lower level of clinical utility evidence than a true randomized trial, this type of study strategy is attractive and practical for AI-based risk-prediction models, which make up a large proportion of AI models in development. Randomized clinical trials are notoriously difficult and time-consuming to execute, and AI interventions have unique characteristics that make such undertakings even more daunting. Notably, AI models are able to adapt to new data and improve over time; how would one take this into account in a traditional randomized trial? While we need AI to embrace randomized trials to truly prove clinical utility, it may be time to recognize that a re-imagining of the traditional randomized clinical trial may be necessary to appropriately study the benefits of AI applications (Haring, 2019).
Beyond validation of clinically meaningful endpoints, demonstrating clinical usability involves study of the AI model in a real-world setting, where it interfaces with clinical practitioners and patients. Evaluation of effects of the model on time task, user satisfaction, and acceptance of AI recommendations should be performed (Kumar et al., 2020). A mechanism of feedback should be integrated into the design of the platform to identify weak points and opportunities for improved interface (Cutillo et al., 2020). Additionally, interoperability between systems at the facility-to-facility, intra-facility, and point-of-care levels are crucial to streamline workflow (He et al., 2019). Usability issues are also specific to the datastreams being analyzed. New datastreams such as mobile health data and wearable activity monitors each present unique challenges to usability and adoption (Beg et al., 2017). A key component of promoting usability is interpretability of the AI algorithm. As data streams become more dimensional, it is increasingly difficult to discern a biological or clinical rationale supporting an algorithm’s predictions. This “black-box” effect may be acceptable in certain consumer electronics industries, but due to the consequential and medicolegal nature of healthcare decision-making, lack of interpretability poses a tremendous barrier to clinical use (Doshi-Velez and Kim, 2017; Wang et al., 2020). Fortunately, there is a growing research field dedicated to investigation of interpretability issues, and several techniques, such as saliency maps, hidden-states analysis, variable importance metrics, and feature visualizations can illuminate some aspects of AI prediction rationale (Guo et al., 2019; Olah et al., 2018). Beyond this, an appreciation of advances in Human Factors research and collaboration with appropriate experts can help streamline the adoption of otherwise clinically validated algorithms. Finally, translating algorithms into clinical usable solutions requires robust information technology support services that may require dedicated investment from clinical institutions and departments.
Another key concept related to clinical usability is addressing the challenges that emerge when multiple AI models are deployed sequentially or simultaneously at a given touchpoint or series of touchpoints. Orchestration of these situations, which are expected to become more common, require attention to end-user responsibilities, interoperability, access, and training. As a patient moves through the oncology care path, they interact (directly or indirectly) with many different care providers who may be the primary users of a given AI application (Figure 2). These users may have a primarily diagnostic or therapeutic role (or both). From a simplified perspective, the primary diagnosticians of the cancer care path are pathologists and radiologists, while the therapeutic clinicians tend to be medical, radiation, and surgical oncologists. Multidisciplinary touchpoints along the pathway, e.g. tumor boards, represent opportunities to collate and orchestrate disparate AI applications. In addition to physicians, there are numerous advanced practice providers such as nurses and physician assistants, as well as therapists, social workers, and medical students, who may be users of a specific AI application. If, for example, a patient receives a CT scan with an AI-generated prediction of malignancy, and this prediction is subsequently utilized as input for another algorithm to recommend surgery as treatment, who is the “designated user” primarily responsible for utilizing and disseminating that information? A further issue, which logically follows, is who is legally liable for decisions based on the use of the model. Specific solutions have not yet been developed to address these issues, and are, unfortunately, likely to arise on an ad hoc, case by case basis. This clinical orchestration of AI models merits further resources, investigation, and guidelines aimed at medical AI developers and cancer care providers to navigate these complex issues.
Despite the vanishingly few FDA-approved AI applications for oncologic indications, with numerous applications in the pipeline, there is high interest in streamlining ways to bridge the gap between development and clinical translation. Accordingly, the FDA is in the process of devising AI and machine learning-specific guidelines for approved clinical use. The recently released action plan incorporates the above clinical concepts and sets the stage for further defining a framework for safe AI translation to the clinic (FDA, 2021).
Conclusions
Increasing datastreams and advances in computational algorithms have positioned artificial intelligence to improve clinical oncology via rigorously evaluated, narrow-task applications interacting at specific touchpoints along the cancer care path. While there are a number of promising artificial intelligence applications for clinical oncology in development, substantial challenges remain to bridge the gap to clinical translation. The most successful models have leveraged large-scale, robustly annotated datasets for narrow tasks at specific cancer care touchpoints. Further development of artificial intelligence applications for cancer care should emphasize clinical validity, utility, and usability. Successful incorporation of these concepts will require bringing a patient-provide, clinical decision-centric focus to model development and evaluation.
ACKNOWLEDGEMENTS
The authors acknowledge financial support from NIH (HA: NIH-USA U24CA194354, NIH-USA U01CA190234, NIH-USA U01CA209414, and NIH-USA R35CA22052; BK: NIHK08:DE030216), the European Union - European Research Council (HA: 866504), as well as the Radiological Society of North America (BK: RSCH2017).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interests
H.A. is a shareholder of and receives consulting fees from Onc.Ai and BMS, outside submitted work. A.H. is a shareholder of and receives consulting fees from Altis Labs, outside submitted work.
REFERENCES
- AACR Project GENIE Consortium (2017). AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 7, 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, and Winchester DP (2017). The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin 67, 93–99. [DOI] [PubMed] [Google Scholar]
- Arbour KC, Luu AT, Luo J, Rizvi H, Plodkowski AJ, Sakhi M, Huang KB, Digumarthy SR, Ginsberg MS, Girshman J, et al. (2020). Deep Learning to Estimate RECIST in Patients with NSCLC Treated with PD-1 Blockade. Cancer Discov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, et al. (2019). End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med 25, 954–961. [DOI] [PubMed] [Google Scholar]
- Bari A, Marcheselli L, Sacchi S, Marcheselli R, Pozzi S, Ferri P, Balleari E, Musto P, Neri S, Aloe Spiriti MA, et al. (2010). Prognostic models for diffuse large B-cell lymphoma in the rituximab era: a never-ending story. Ann. Oncol 21, 1486–1491. [DOI] [PubMed] [Google Scholar]
- Beam AL, Manrai AK, and Ghassemi M (2020). Challenges to the Reproducibility of Machine Learning Models in Health Care. JAMA 323, 305–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg MS, Gupta A, Stewart T, and Rethorst CD (2017). Promise of Wearable Physical Activity Monitors in Oncology Practice. Journal of Oncology Practice 13, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamens S, Dhunnoo P, and Meskó B (2020). The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. NPJ Digit Med 3, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R, Ha MJ, Liu Q, Akbani R, Liang H, and Baladandayuthapani V (2019). Personalized Network Modeling of the Pan-Cancer Patient and Cell Line Interactome. [DOI] [PMC free article] [PubMed]
- Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, et al. (2019). Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin 69, 127–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibault J-E, Bassenne M, Ren H, and Xing L (2020). Deep Learning Prediction of Cancer Prevalence from Satellite Imagery. Cancers 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers DA, Vinson CA, and Norton WE (2018). Advancing the Science of Implementation across the Cancer Continuum (Oxford University Press; ). [Google Scholar]
- Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, Kavouridis VK, Senders JT, Boaro A, Beers A, et al. (2018). Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res 24, 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Lafata K, Sun W, Wang C, Chang Z, Kirkpatrick JP, and Yin F-F (2019). An investigation of machine learning methods in delta-radiomics feature analysis. PLOS ONE 14, e0226348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, et al. (2013). The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutzberg CL, van Stiphout RGPM, Nout RA, Lutgens LCHW, Jürgenliemk-Schulz IM, Jobsen JJ, Smit VTHBM, and Lambin P (2015). Nomograms for Prediction of Outcome With or Without Adjuvant Radiation Therapy for Patients With Endometrial Cancer: A Pooled Analysis of PORTEC-1 and PORTEC-2 Trials. International Journal of Radiation Oncology*Biology*Physics 91, 530–539. [DOI] [PubMed] [Google Scholar]
- Cutillo CM, MI in Healthcare Workshop Working Group, Sharma KR, Foschini L, Kundu S, Mackintosh M, and Mandl KD (2020). Machine intelligence in healthcare—perspectives on trustworthiness, explainability, usability, and transparency. Npj Digital Medicine 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagogo-Jack I, and Shaw AT (2018). Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol 15, 81–94. [DOI] [PubMed] [Google Scholar]
- van Dijk LV, Langendijk JA, Zhai T-T, Vedelaar TA, Noordzij W, Steenbakkers RJHM, and Sijtsema NM (2019). Delta-radiomics features during radiotherapy improve the prediction of late xerostomia. Sci. Rep 9, 12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi-Velez F, and Kim B (2017). Towards A Rigorous Science of Interpretable Machine Learning.
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak JAWM, the CAMELYON16 Consortium, Hermsen M, Manson QF, et al. (2017). Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 318, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, Bergstralh EJ, Kollmeyer T, Fink S, Haddad Z, et al. (2013). Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 8, e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, and Thrun S (2017). Corrigendum: Dermatologist-level classification of skin cancer with deep neural networks. Nature 546, 686. [DOI] [PubMed] [Google Scholar]
- Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, and Dean J (2019). A guide to deep learning in healthcare. Nat. Med 25, 24–29. [DOI] [PubMed] [Google Scholar]
- FDA (2021). Artificial Intelligence/Machine Learning (AI/ML)-Based.:Jf/<X Software as a Medical Device (SaMD) Action Plan.
- Gerwing M, Herrmann K, Helfen A, Schliemann C, Berdel WE, Eisenblätter M, and Wildgruber M (2019). The beginning of the end for conventional RECIST — novel therapies require novel imaging approaches. Nature Reviews Clinical Oncology 16, 442–458. [DOI] [PubMed] [Google Scholar]
- Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, et al. (2020). Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest. Endosc 91, 41–51. [DOI] [PubMed] [Google Scholar]
- Guo T, Lin T, and Antulov-Fantulin N (2019). Exploring Interpretable LSTM Neural Networks over Multi-Variable Data.
- Gyawali B (2018). Does global oncology need artificial intelligence? Lancet Oncol. 19, 599–600. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Suvarna K, Yamada M, Kobayashi K, Shinkai N, Miyake M, Takahashi M, Jinnai S, Shimoyama R, Sakai A, et al. (2020). Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring A (2019). In the age of machine learning randomized controlled trials are unethical.
- He J, Baxter SL, Xu J, Xu J, Zhou X, and Zhang K (2019). The practical implementation of artificial intelligence technologies in medicine. Nat. Med 25, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JC, Eclov NCW, Dalal NH, Thomas SM, Stephens SJ, Malicki M, Shields S, Cobb A, Mowery YM, Niedzwiecki D, et al. (2020). System for High-Intensity Evaluation During Radiation Therapy (SHIELD-RT): A Prospective Randomized Study of Machine Learning–Directed Clinical Evaluations During Radiation and Chemoradiation. Journal of Clinical Oncology 38, 3652–3661. [DOI] [PubMed] [Google Scholar]
- Kang J, Morin O, and Hong JC (2020). Closing the Gap Between Machine Learning and Clinical Cancer Care—First Steps Into a Larger World. JAMA Oncol 6, 1731–1732. [DOI] [PubMed] [Google Scholar]
- Kann BH, Thompson R, Thomas CR Jr, Dicker A, and Aneja S (2019). Artificial Intelligence in Oncology: Current Applications and Future Directions. Oncology 33, 46–53. [PubMed] [Google Scholar]
- Kann BH, Johnson SB, Aerts HJWL, Mak RH, and Nguyen PL (2020a). Changes in Length and Complexity of Clinical Practice Guidelines in Oncology, 1996–2019. JAMA Netw Open 3, e200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann BH, Hicks DF, Payabvash S, Mahajan A, Du J, Gupta V, Park HS, Yu JB, Yarbrough WG, Burtness BA, et al. (2020b). Multi-Institutional Validation of Deep Learning for Pretreatment Identification of Extranodal Extension in Head and Neck Squamous Cell Carcinoma. J. Clin. Oncol 38, 1304–1311. [DOI] [PubMed] [Google Scholar]
- Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, and King D (2019). Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, Brugnara G, Schell M, Kessler T, Foltyn M, et al. (2019). Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 20, 728–740. [DOI] [PubMed] [Google Scholar]
- Kim DW, Jang HY, Kim KW, Shin Y, and Park SH (2019). Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers. Korean J. Radiol 20, 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Chiang J, Hom J, Shieh L, Aikens R, Baiocchi M, Morales D, Saini D, Musen M, Altman R, et al. (2020). Usability of a Machine-Learning Clinical Order Recommender System Interface for Clinical Decision Support and Physician Workflow. medRxiv. [Google Scholar]
- Kuznar W (2015). The Push Toward Value-Based Payment for Oncology. Am Health Drug Benefits 8, 34. [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, and Hinton G (2015). Deep learning. Nature 521, 436–444. [DOI] [PubMed] [Google Scholar]
- Lee W-S, Ahn SM, Chung J-W, Kim KO, Kwon KA, Kim Y, Sym S, Shin D, Park I, Lee U, et al. (2018). Assessing Concordance With Watson for Oncology, a Cognitive Computing Decision Support System for Colon Cancer Treatment in Korea. JCO Clin Cancer Inform 2, 1–8. [DOI] [PubMed] [Google Scholar]
- Liu X, Faes L, Calvert MJ, Denniston AK, and CONSORT/SPIRIT-AI Extension Group (2019). Extension of the CONSORT and SPIRIT statements. Lancet 394, 1225. [DOI] [PubMed] [Google Scholar]
- Lou B, Doken S, Zhuang T, Wingerter D, Gidwani M, Mistry N, Ladic L, Kamen A, and Abazeed ME (2019). An image-based deep learning framework for individualising radiotherapy dose: a retrospective analysis of outcome prediction. The Lancet Digital Health 1, e136–e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C-F, Hsu F-T, Hsieh KL-C, Kao Y-CJ, Cheng S-J, Hsu JB-K, Tsai P-H, Chen R-J, Huang C-C, Yen Y, et al. (2018). Machine Learning–Based Radiomics for Molecular Subtyping of Gliomas. Clin. Cancer Res 24, 4429–4436. [DOI] [PubMed] [Google Scholar]
- Maddox TM, Rumsfeld JS, and Payne PRO (2019). Questions for Artificial Intelligence in Health Care. JAMA 321, 31–32. [DOI] [PubMed] [Google Scholar]
- Manz CR, Chen J, Liu M, Chivers C, Regli SH, Braun J, Draugelis M, Hanson CW, Shulman LN, Schuchter LM, et al. (2020). Validation of a Machine Learning Algorithm to Predict 180-Day Mortality for Outpatients With Cancer. JAMA Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, and Polyak K (2012). Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334. [DOI] [PubMed] [Google Scholar]
- McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, Back T, Chesus M, Corrado GS, Darzi A, et al. (2020). International evaluation of an AI system for breast cancer screening. Nature 577, 89–94. [DOI] [PubMed] [Google Scholar]
- Ming C, Viassolo V, Probst-Hensch N, Dinov ID, Chappuis PO, and Katapodi MC (2020). Machine learning-based lifetime breast cancer risk reclassification compared with the BOADICEA model: impact on screening recommendations. Br. J. Cancer 123, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsa T (2019). How Do You Know You Have Enough Training Data?
- Mittendorf EA, Hunt KK, Boughey JC, Bassett R, Degnim AC, Harrell R, Yi M, Meric-Bernstam F, Ross MI, Babiera GV, et al. (2012). Incorporation of Sentinel Lymph Node Metastasis Size Into a Nomogram Predicting Nonsentinel Lymph Node Involvement in Breast Cancer Patients With a Positive Sentinel Lymph Node. Annals of Surgery 255, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Torres JG, Raeder T, Alaiz-Rodríguez R, Chawla NV, and Herrera F (2012). A unifying view on dataset shift in classification. Pattern Recognit. 45, 521–530. [Google Scholar]
- Nagendran M, Chen Y, Lovejoy CA, Gordon AC, Komorowski M, Harvey H, Topol EJ, Ioannidis JPA, Collins GS, and Maruthappu M (2020). Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ 368, m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal K, Foote D, Tan F, Liu Y, Chen P-HC, Steiner DF, Manoj N, Olson N, Smith JL, Mohtashamian A, et al. (2020). Development and Validation of a Deep Learning Algorithm for Gleason Grading of Prostate Cancer From Biopsy Specimens. JAMA Oncol 6, 1372–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta S, Tsutsumi M, Sakka S, Endo T, Hashimoto K, Hasegawa M, Hayashi T, Kawai K, and Nishiyama H (2019). Machine learning methods can more efficiently predict prostate cancer compared with prostate-specific antigen density and prostate-specific antigen velocity. Prostate Int 7, 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgeot B, Quer G, Beaulieu-Jones BK, Torkamani A, Dias R, Gianfrancesco M, Arnaout R, Kohane IS, Saria S, Topol E, et al. (2020). Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat. Med 26, 1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakden-Rayner L, Dunnmon J, Carneiro G, and Ré C (2020). Hidden Stratification Causes Clinically Meaningful Failures in Machine Learning for Medical Imaging. Proc ACM Conf Health Inference Learn (2020) 2020, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah C, Satyanarayan A, Johnson I, Carter S, Schubert L, Ye K, and Mordvintsev A (2018). The building blocks of interpretability. Distill 3. [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. (2004). A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med 351, 2817–2826. [DOI] [PubMed] [Google Scholar]
- Park Y, Jackson GP, Foreman MA, Gruen D, Hu J, and Das AK (2020). Evaluating artificial intelligence in medicine: phases of clinical research. JAMIA Open 3, 326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K (2011). Heterogeneity in breast cancer. J. Clin. Invest 121, 3786–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME (2009). A strategy for health care reform--toward a value-based system. N. Engl. J. Med 361, 109–112. [DOI] [PubMed] [Google Scholar]
- Qiu YL, Zheng H, Devos A, Selby H, and Gevaert O (2020). A meta-learning approach for genomic survival analysis. Nat. Commun 11, 6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandran KJ, Shega JW, Von Roenn J, Schumacher M, Szmuilowicz E, Rademaker A, Weitner BB, Loftus PD, Chu IM, and Weitzman S (2013). A predictive model to identify hospitalized cancer patients at risk for 30-day mortality based on admission criteria via the electronic medical record. Cancer 119, 2074–2080. [DOI] [PubMed] [Google Scholar]
- Richens JG, Lee CM, and Johri S (2020). Improving the accuracy of medical diagnosis with causal machine learning. Nat. Commun 11, 3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell I, and Haller S (2003). Introduction: Tools and Techniques of Artificial Intelligence. International Journal of Pattern Recognition and Artificial Intelligence 17, 685–687. [Google Scholar]
- Salim M, Wåhlin E, Dembrower K, Azavedo E, Foukakis T, Liu Y, Smith K, Eklund M, and Strand F (2020). External Evaluation of 3 Commercial Artificial Intelligence Algorithms for Independent Assessment of Screening Mammograms. JAMA Oncol 6, 1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel AL (1959). Some Studies in Machine Learning Using the Game of Checkers. IBM J. Res. Dev 3, 210–229. [Google Scholar]
- Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S, Kuder TA, Stenzinger A, Hohenfellner M, Schlemmer H-P, et al. (2019). Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology 293, 607–617. [DOI] [PubMed] [Google Scholar]
- Schilsky RL (2010). Personalized medicine in oncology: the future is now. Nat. Rev. Drug Discov 9, 363–366. [DOI] [PubMed] [Google Scholar]
- Scott JG, Harrison LB, and Torres-Roca JF (2017). Genomic biomarkers for precision radiation medicine – Authors’ reply. The Lancet Oncology 18, e239. [DOI] [PubMed] [Google Scholar]
- She Y, Jin Z, Wu J, Deng J, Zhang L, Su H, Jiang G, Liu H, Xie D, Cao N, et al. (2020). Development and Validation of a Deep Learning Model for Non-Small Cell Lung Cancer Survival. JAMA Netw Open 3, e205842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G, DiNardo CD, Takahashi K, Cascone T, Powers C, Stevens R, Allen J, Antonoff MB, Gomez D, Keane P, et al. (2019). Applying artificial intelligence to address the knowledge gaps in cancer care. Oncologist 24, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somashekhar SP, Kumarc R, Rauthan A, Arun KR, Patil P, and Ramya YE (2017). Abstract S6–07: Double blinded validation study to assess performance of IBM artificial intelligence platform, Watson for oncology in comparison with Manipal multidisciplinary tumour board – First study of 638 breast cancer cases. Cancer Res. 77, S6–S07–S6–S07. [Google Scholar]
- Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE Jr, Dees EC, Goetz MP, Olson JA Jr, et al. (2018). Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med 379, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt DE, Yousefi K, Deheshi S, Ross AE, Den RB, Schaeffer EM, Trock BJ, Zhang J, Glass AG, Dicker AP, et al. (2017). Individual Patient-Level Meta-Analysis of the Performance of the Decipher Genomic Classifier in High-Risk Men After Prostatectomy to Predict Development of Metastatic Disease. J. Clin. Oncol 35, 1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson AJ, Scardino PT, Kattan MW, Pisansky TM, Slawin KM, Klein EA, Anscher MS, Michalski JM, Sandler HM, Lin DW, et al. (2007). Predicting the outcome of salvage radiation therapy for recurrent prostate cancer after radical prostatectomy. J. Clin. Oncol 25, 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, Verlingue L, Brandao D, Lancia A, Ammari S, et al. (2018). A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 19, 1180–1191. [DOI] [PubMed] [Google Scholar]
- Thompson RF, Valdes G, Fuller CD, Carpenter CM, Morin O, Aneja S, Lindsay WD, Aerts HJWL, Agrimson B, Deville C Jr, et al. (2018). Artificial intelligence in radiation oncology: A specialty-wide disruptive transformation? Radiother. Oncol 129, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol EJ (2019). High-performance medicine: the convergence of human and artificial intelligence. Nat. Med 25, 44–56. [DOI] [PubMed] [Google Scholar]
- Villaruz LC, and Socinski MA (2013). The clinical viewpoint: definitions, limitations of RECIST, practical considerations of measurement. Clin. Cancer Res 19, 2629–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Kaushal R, and Khullar D (2020). Should Health Care Demand Interpretable Artificial Intelligence or Accept “Black Box” Medicine? Ann. Intern. Med 172, 59–60. [DOI] [PubMed] [Google Scholar]
- Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, et al. (2019). Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut 68, 1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Network CGAR, et al. (2013). The cancer genome atlas pan-cancer analysis project. Nat. Genet 45, 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RW, and Horvitz E (2017). Evaluation of the Feasibility of Screening Patients for Early Signs of Lung Carcinoma in Web Search Logs. JAMA Oncol 3, 398–401. [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Duffy AG, Brar G, Fioravanti S, Mabry-Hrones D, Walker M, Bonilla CM, Wood BJ, Citrin DE, Gil Ramirez EM, et al. (2020). Immune Checkpoint Blockade in Combination with Stereotactic Body Radiotherapy in Patients with Metastatic Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res 26, 2318–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, Mak RH, and Aerts HJWL (2019). Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin. Cancer Res 25, 3266–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf Zafar S (2016). Financial Toxicity of Cancer Care: It’s Time to Intervene. J. Natl. Cancer Inst 108. [DOI] [PubMed] [Google Scholar]
- Zhong G, Ling X, and Wang L (2019). From shallow feature learning to deep learning: Benefits from the width and depth of deep architectures. Wiley Interdiscip. Rev. Data Min. Knowl. Discov 9, e1255. [Google Scholar]
- Zhou D, Tian F, Tian X, Sun L, Huang X, Zhao F, Zhou N, Chen Z, Zhang Q, Yang M, et al. (2020). Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat. Commun 11, 2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Vondrick C, Fowlkes C, and Ramanan D (2015). Do We Need More Training Data?
- (2021). USPSTF: A and B Recommendations. https://www.uspreventiveservicestaskforce.org/uspstf/recommendation-topics/uspstf-and-b-recommendations.


