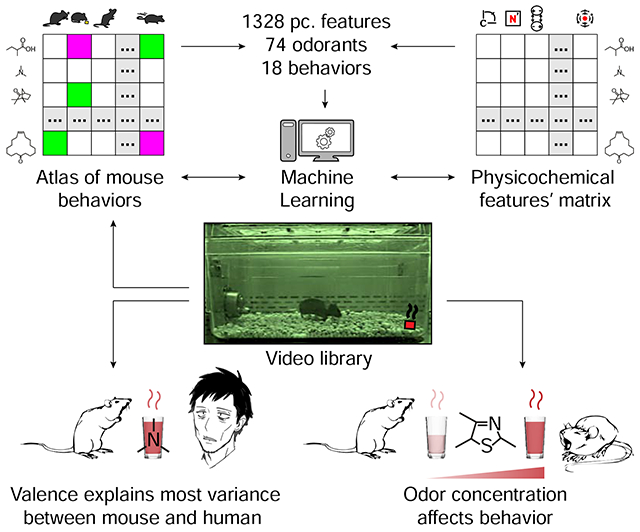
SUMMARY
Odor perception in non-humans is poorly understood. Here, we generated the most comprehensive mouse olfactory ethological atlas to date, consisting of behavioral responses to a diverse panel of 73 odorants, including 12 at multiple concentrations. These data revealed that mouse behavior is incredibly diverse and changes in response to odorant identity and concentration. Using only behavioral responses observed in other mice, we could predict which of two odorants was presented to a held-out mouse 82% of the time. Considering all 73 possible odorants, we could uniquely identify the target odorant from behavior on the first try 20% of the time, and 46% within five attempts. While mouse behavior is difficult to predict from human perception, they share three fundamental properties: First, odor valence parameters explained the highest variance of olfactory perception. Second, physicochemical properties of odorants can be used to predict the olfactory percept. Third, odorant concentration quantitatively and qualitatively impacts olfactory perception. These results increase our understanding of mouse olfactory behavior, how it compares to human odor perception, and provide a template for future comparative studies of olfactory percepts among species.
Keywords: Odor, behavior, olfactory, perception, machine learning, chemoinformatics
eTOC BLURB
Manoel et al. score 18 behaviors in 525 mice across 98 odorant conditions to generate a murine olfactory ethological atlas, which they use to deconstruct mouse olfactory behavior, predict it from odorant structure, evaluate its major factors, and compare it to human perception.
Graphical Abstract
INTRODUCTION
How sensory cues translate into perceptual objects or complex behaviors remains a major unanswered question in neuroscience. Odor transduction in the nose leads to odor perception and to changes in behavior or physiology (e.g., aggression, feeding) that are key for survival and reproduction, making the olfactory system an attractive model to address this question1,2.
Many studies have used mice to elucidate molecular, cellular, and neural processes underlying mammalian olfaction1. The proliferation of annotated genomes and high-throughput sequencing technologies have yielded new clues into the functional logic and the evolutionary dynamics of mammalian olfaction more broadly3–5. However, our understanding of olfactory perception derives from large psychophysical datasets in humans combined with chemoinformatic, statistical, and machine learning tools6–11. These studies have yielded three key findings regarding human olfactory perception. First, the first principal component of human olfactory perception is highly associated (r=0.79) with a single dimension – odor valence6,7,12. Second, the human olfactory perceptual ratings for most odorous molecules can be predicted from chemical structure with surprising accuracy (r=0.3-0.7), a value limited primarily by rater reliability8. Third, odorant concentration can qualitatively alter perceived odor intensity and character9,13. Do these principles also apply to olfactory perception in non-humans?
Characterizing olfactory perception in an animal relies on accurately quantifying multiple behaviors in response to large numbers of odorants, ideally at various concentrations. This assumes that visible mouse behaviors either encode for mouse perception or at least report something informative about the meaning of the stimulus. For example, mice may exhibit differential behavioral responses depending on odor valence (attractive vs aversive), odor novelty, or implications of the presence of an odor for a broader behavioral strategy (e.g. exploration of surroundings). They may also use behavior to communicate odor information to conspecifics. Despite recent efforts14–25, a systematic characterization of various mouse behaviors in response to a large panel of diverse odorants and several concentrations is still lacking. This prevents a systematic understanding of mouse olfactory behavior and how it relates to perception in humans and other species and limits our ability to study the neural computations underlying the transformation of odor stimuli at the nose to odor objects in the brain.
Here, we generated and investigated a mouse atlas of odor-guided behaviors in response to a diverse panel of odorants, at different concentrations, to deconstruct olfactory behavior in mice and compare it to human perception.
RESULTS
The olfactory-ethological atlas
We recently generated a mouse behavioral video library and quantified the cumulative duration of olfactory investigation over a three-minute-long assay (3’dOI) to a panel of 73 odorants at 85 mM and the odorless control water (H2O)25. These stimuli include 61 general odorants, seven pheromones (IPT, IAA, BFA, 2HO, TMA, AST, FAR), and five kairomones (TMT, PEA, 2PT, BZL, QUI)2,26–30. Odorants were classified as eliciting avoidance or approach if the 3’dOI was significantly lower or higher than H2O, respectively25. Here, we enhance the utility of this video library by scoring 17 additional behavioral parameters from 410 mice exposed to 73 odorants and H2O (Methods, Figures 1A–D). Each mouse was exposed to only one odorant (n=5-10 per odorant) and scored on 18 distinct behavioral parameters, chosen for discernibility, quantifiability, and reliability and as surrogate measures of odor ‘valence’, exploration, or stress14,15,20,23,25,31–34.
Figure 1. An olfactory-ethological atlas and the primary axis of olfactory perception in mouse.
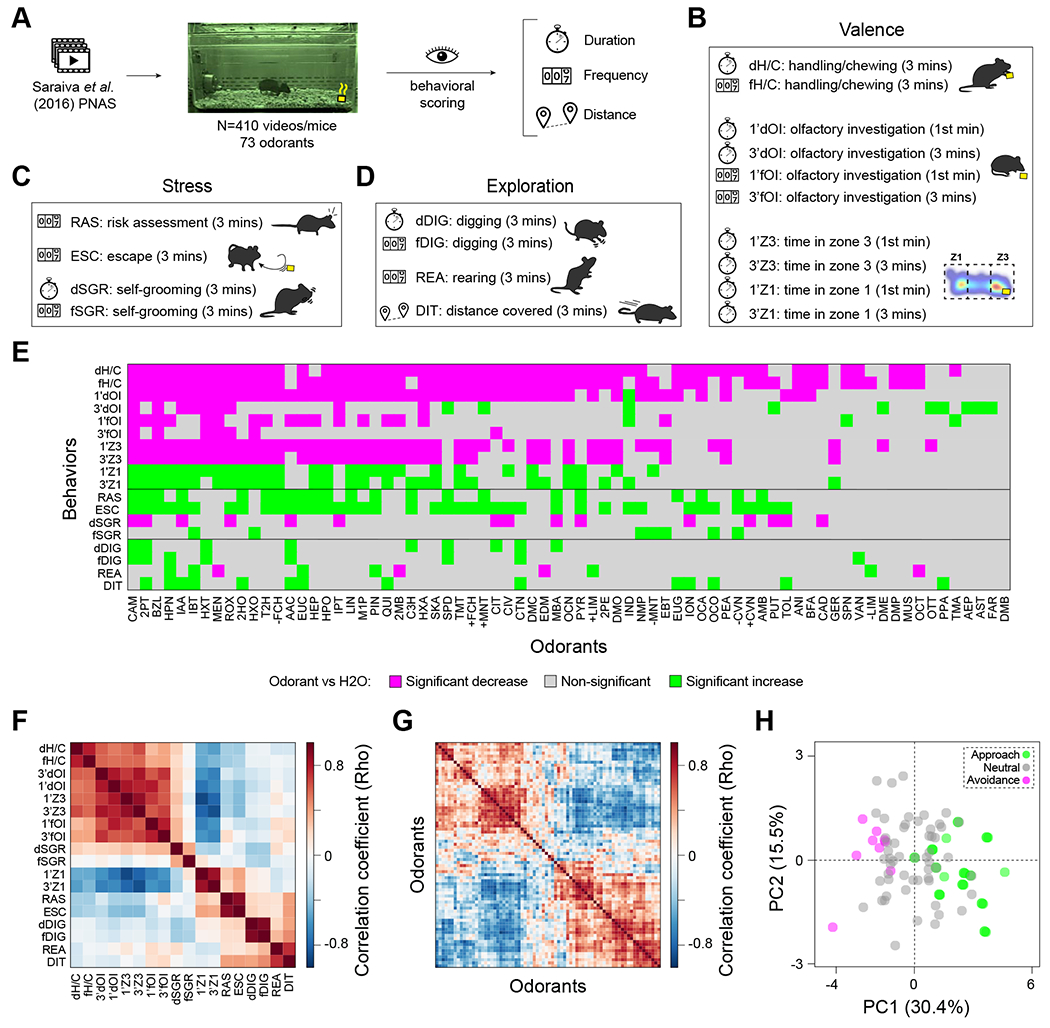
(A) Videos were scored for 18 behavioral parameters, grouped in three broad categories: (B) valence, (C) stress, (D) exploration. (E) A graphical display summarizing the combinatorial behavioral patterns for the 73 odorants tested. Behaviors showing significant (one-way ANOVA, BKY multiple comparisons correction, n = 5-10 per odorant) increases, decreases and non-significant responses compared to H2O are indicated in green, magenta, and grey squares, respectively. (F) Correlation across odorants between each of the 18 behaviors (n = 3-7 per odorant). (G) Correlation across behaviors between each of the 73 odorants and H2O (n = 3-7 per odorant). In both F and G, odorants and behaviors are ordered to illustrate clustering. Two major clusters of odorants stand out. (H) Principal Component Analysis of the 18 behaviors for H2O and the 73 tested odorants (n = 3-7 per odorant). Circles are colored to indicate avoidance (magenta), neutral (grey), or approach (green) odorants. See also Figure S1, and Data S1.
From this new dataset totaling 7606 individual data points (Figure S1A, Data S1), corresponding to individual-odorant-behavior triads, we calculated the across-individual average value for each of 1314 odorant-behavior pairs: 470 (35.8%) are significantly different (p<0.05, one-way ANOVA, BKY multiple-comparisons correction) from H2O, with 293 decreases and 177 increases of a behavioral parameter (Figure 1E, Data S1). Of the 72 odorants eliciting at least one significant behavioral change, 57 (78%) exhibited unique patterns of significant increases and decreases among the 18 behaviors, indicating that odor-guided behavioral patterns are incredibly diverse.
Valence is the primary axis of olfactory perception in mouse
Pleasantness, a surrogate for odor valence, is the primary axis of human olfactory perception7,12,35. However, whether this is conserved in mouse remains unknown. We examined the structure of the behavioral response matrix (behaviors x odorants) to address this question. From this matrix, we computed two correlation matrices (between behaviors, Figure 1F, and between odorants, Figure 1G) and ordered each to match a hierarchical clustering. We identified two self-similar groups (upper left and lower right patches in Figures 1F, G). Principal components analysis (PCA) showed that principal component 1 (PC1) explains 30.4% of the data variance (Figures 1H, S1B). We observed only a weak relationship between molecular structure and PC1 or PC2 (Figure S1C), suggesting that odor valence is not a trivial consequence of molecular structure. Nevertheless, PC1 cleanly separates approached from avoided odorants, which we interpret as a valence axis (Figure 1H). This result is robust to alternative subsamples of behaviors in which valence, exploration, and stress have equal representation; valence-related behaviors always comprise the top-5 highest-weighted behaviors in PC1.
Discriminability of odorants using behaviors
Odorants appear to lie in stereotypical locations in a behavioral space defined by PC1 and PC2 of the response matrix (Figure 1H). We next asked how well odorants can be distinguished using the original 18-dimensional space. We computed D’, a measure of discriminability between two signals, for all odorant pairs. Higher D’ values indicate that behavior more easily discriminates between two odorants; a value of 1.0 occurs when the mean behavioral difference between odorants equals the behavioral variability across mice exposed to the same odorant; it is thus also a measure of effect size.
D’ for each behavior (across odorant pairs) ranged between 0.6 and 1.0; by contrast, shuffling odorant labels between mice resulted in lower D’ values (0.5-0.6) (Figure 2A). Every single behavior exhibited higher D’ in the original than in the shuffled data (Figure 2B).
Figure 2. Discriminability of odorants using behaviors and out-of-sample prediction of odor identity.
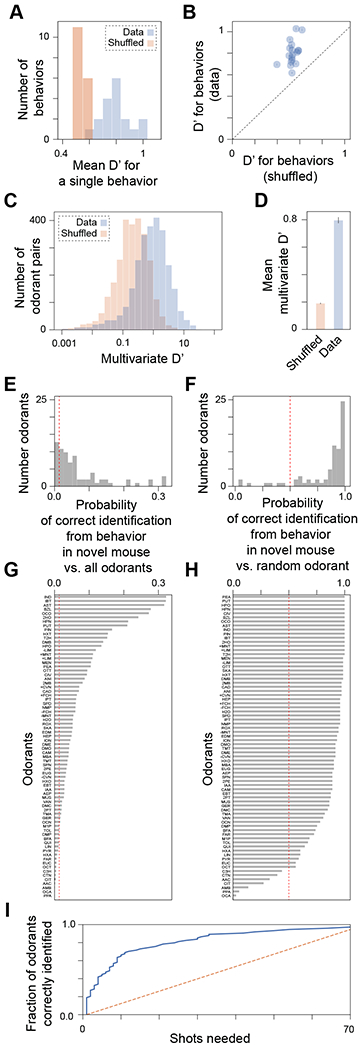
(A) D’, a measure of discriminability between two odorants, is greater for the data than for a shuffling (across mice) of the data. (B) Same data as in A, except shown for each behavior (circle) vs. its corresponding shuffle. Error bars (inside circles) represent SEM taken over all odorant pairs. (C) Using all behaviors simultaneously, the multivariate measure D’ is computed. (D) Multivariate D’ is ~4x larger for the real data than for shuffled data. (E-H) A linear discriminant analysis classifier was trained on all odorants, using all but one mouse for each odorant. Predictive performance was evaluated for the remaining mice (one odorant each). (E) A histogram of the probability that the correct odorant (out of 74 possibilities) is identified from a new mouse’s behavior. The dashed red line reflects chance performance. (F) Mean performance for each odorant; higher values mean the odorant is easier to uniquely identify from behavior. (G) Similar to E, but for classification of the true odorant against a random alternative odorant. Chance is now 50%, as reflected by the dashed red line. (H) Mean performance for each odorant in G. 1.0 means that behavior was always sufficient to identify the odorant vs. any specific alternative odorant. (I) Number of shots (guesses) that the classifier needs to determine the correct odorant (out of 74 possibilities) from novel mouse behavior. This value (solid blue line) is shown for all 74 odorants, ranked from fewest to greatest number of shots required. The orange dashed line represents chance performance. See also Figures S2, and S3.
Because the behavioral code may be combinatorial, we also computed a multivariate D’ which uses all behaviors simultaneously. If most behavioral variability is mouse-specific and not odorant-specific, D’ would be much less than 1. However, we found that multivariate D’ – while heterogeneous across odorant pairs (Figure 2C) –was on average 4x larger for the real data (mean D’=0.8) than for shuffled data (Figure 2D). Thus, this behavioral ensemble can represent an odorant-specific behavioral code. D’ can alternatively be expressed in terms of the area (AUC) under the receiver-operator characteristic (ROC) curve quantifying a sensitivity vs. specificity tradeoff; under a Gaussian assumption a D’=0.8 corresponds to an AUC=0.66 for a typical pair of odorants.
D’ can overestimate discriminability when the number of replicates (individuals) per stimulus is small (so D’ for shuffled data remains >0). Instead, we can ask how accurately a predictive model can select the correct odorant from out-of-sample observations of behavior, i.e., a mouse the model has not observed. We trained a linear discriminant classifier on all odorants, but withheld one mouse per odorant for cross-validation testing. We asked the classifier to perform two tasks: first, to predict the correct odorant (out of 74 possibilities) given a new observation of behavior, and second, to predict the correct odorant (out of two choices: the correct odorant and one other chosen at random). In the first task, most odorants could be predicted at above chance (1/74) levels (Figures 2E,F). Some could even be predicted correctly >30% of the time (IND, IBT, AST). In the second task, 67/74 odorants could be identified from behavior at above chance (½) levels, and 32/74 could be identified >95% of the time in the same comparison (Figures 2G,H). If we gave the classifier additional “shots on goal” in the first task, allowing 5 shots made 46% of odorants identifiable from the ensemble (Figure 2I). Thus, odor-evoked behavior for most odorants was stereotypical enough to help identify which odorant was presented to a novel mouse.
Reconstructing a low-dimensional space of mouse olfactory behavior
We recorded 18 distinct behaviors but many of these behaviors are correlated (Figure 1F). Thus, the underlying dimensionality of the olfactory behavioral space may be <<18. PCA indicated that 90% of the variance was explained by ten dimensions (Figure S1B). However, this is likely to be an overestimate for two reasons: first, at least some of this variance is noise, driven by within-odorant, across-mouse behavioral variability; second, PCA does not produce a natural decomposition of data in many applications36. A particular concern here is that PCA might represent valence as a single dimension, while the underlying concepts “aversive” and “approach” could be distinct perceptual categories which just happen to produce behaviors of opposite sign. To overcome the first concern, we asked how many dimensions are required to optimally represent each odorant’s behavioral phenotype. To address the second we used non-negative matrix factorization (NMF)36, a decomposition technique known for producing compact, intuitive, parts-based representations in diverse domains, including olfactory perception37. Specifically, we computed an NMF decomposition of behavior and asked for what number of factors the intraclass correlation coefficient (ICC) was maximized when projecting data from novel mice onto these factors. Here the ICC measures the fraction of behavioral variance explained by odorant identity. Theory suggests that a low-factor NMF decomposition might denoise behavioral data by identifying and discarding noisy, irrelevant dimensions, thus increasing ICC. However, with too few factors, odorant-associated structure could be lost, reducing ICC. The number of factors that maximizes ICC thus reflects the most efficient decomposition of olfactory behavior.
We found that ICC was greatest for a 2-factor space (ICC=0.57, Figure 3A), substantially higher than for the original 18-factor space (ICC=0.34) or a 1-factor space (ICC=0.35). A 2-factor space was thus optimal for explaining mouse behavioral variance in terms of odorant identity. By contrast, shuffling odorant labels across mice produced a consistently low value (ICC=0.2) for all choices of factor number. To interpret these two factors, we examined the factor weights for each behavior (Figure 3B). The highest-weighted factors corresponded to valence-related behaviors (handling/catching – H/C, olfactory investigation – OI, zone assays – Z1/Z3), followed by the exploratory (rearing – REA, digging – DIG, distance covered – DIT) and stress-related behaviors (risk assessment – RAS, escape – ESC). The weights for each factor were mostly orthogonal (Figure S2A, r=−0.24, p=0.35, Fisher Z transformation test) but the positions of odorants along each factor were anti-correlated (Figure S2B, r=−0.79, p<0.0001), indicating that these two factors are still largely capturing a single behavioral category and its opposite (i.e. approach and aversion). This is consistent with PCA – the valence axis is primary – but goes a step further by separating out an attractive from an aversive factor and showing that these factors (but no others) are shared identifiably in cross-animal comparisons. This provides a parts-based understanding of the fundamental units of olfactory behavior in mice.
Figure 3. Optimal reconstruction of mouse behavior and the alignment between mouse and human behavioral spaces.
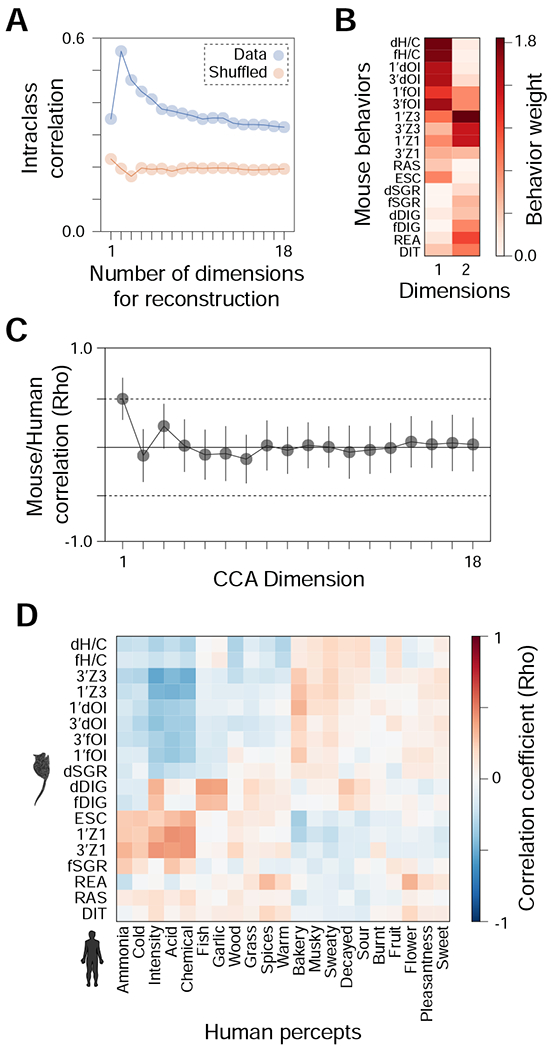
(A-B) The optimal reconstruction of mouse behavior requires few dimensions. (A) Non-negative matrix factorization (NMF) is used to learn a low-dimensional (≤ the number of measured behaviors) representation of the behavioral data. The intra-class correlation coefficient (ICC), reflecting the behavioral agreement within (vs. across) odorants is shown, as a function of the number of factor used. Lower numbers of factors effectively denoise the data. Results for the data are shown in blue, results for the data with shuffled odorant labels are shown in orange. Eighteen factors would reflect independent contributions of each behavior. ICC is maximized for a 2-factor representation of behavior. (B) Contributions of behaviors to the resulting 2 factors. (C) Alignment of mouse and human behavioral spaces. Canonical correlation analysis co-aligns mouse behavioral features and human-provided descriptors for the same odorants. Canonical dimensions were computed using all but one odorant, and the remaining odorant was used to evaluate the correlation (Pearson) between mouse behavior and human percepts. Error bars represent standard deviation across held-out odorants. P-values were computed by comparing to shuffled data. (D) Comparing mouse behaviors and human percepts. Correlation matrix heatmap comparing the 18 mouse behavioral parameters with 21 human odor descriptors for all 73 odorants tested here (odor descriptors for 22 odorants taken from9 and 51 predicted using the DREAM model8). Rows and columns are sorted to maximize clustering in the heatmap, revealing ~2 major clusters of perception/behavior. See also Figures S2, S3, and Table S1.
Mouse vs. Human
Comparative studies of odor valence in mice and humans are scarce and have yielded conflicting results17,38. Indeed, whether mice and humans share a common olfactory perceptual space is still unknown. To answer this question, we first compared our mouse behavioral dataset to the human-rated intensity and pleasantness reported in a previous study9. For the 23 overlapping odorants between both studies, we found no significant correlation among the mouse and human-rated parameters (Figure S3A), consistent with our previous study38. We then extended this analysis to include 19 additional human semantic descriptors (e.g., fishy, sweet)9, but found only seven significant correlations (out of a possible 475) encompassing five mouse behaviors and four human semantic descriptors (Figure S3A).
To identify shared structure between mouse olfactory behavior and human olfactory perception, 23 odorants may be insufficient. To make use of a larger sample we used the data and machine-learning algorithms from the DREAM Olfaction Prediction Challenge8 to obtain predictions of human-rated intensity, pleasantness, and 19 semantic descriptors for the remaining 51 odorants tested in our mouse experiments (see STAR Methods). Rather than identify simple correlations, we asked whether mouse behavioral space and human perceptual space could be projected onto a common basis. If so, they might simply be two views of a common mammalian olfactory perceptual space. We used canonical correlation analysis (CCA) to obtain this basis using all but one odorant, and then asked whether this basis could identify shared inter-species structure using the remaining (out-of-sample) odorant. Our results (Figure 3C) identified a single dimension for this hypothesized shared structure (Pearson’s r=0.50, p=0.02, shuffle test), with additional dimensions failing to capture any additional shared structure. This means that a one standard deviation change in the optimal linear combination of mouse behaviors is associated with a ½ standard deviation change in an optimal linear combination of human percepts for novel odorants. What human percepts comprise this shared dimension? The factor weights are shown in Table S1. No single human percept dominates this dimension, and individually each of them is not statistically significant (p>0.1). Similarly, all human percepts have a Pearson correlation r<0.5 vs. either of the first two principal components of mouse behavioral space in Fig. 1 (Figure S3B) or the first two NMF factors obtained from the same space in Fig. 3 (Figure S3C). The correlation between specific human percepts and mouse behaviors is shown in Figures 3D, and S3A. We conclude that the shared dimension is multifactorial: it cannot be easily reduced to a single percept or behavior.
Predicting behavior from chemical structure
Prior studies identified relationships between hydrocarbon chain length or chemical functional group of aliphatic odorants, and their induced neural responses or perceptual similarity39–41. For the twelve aliphatic odorants in our dataset, we found no apparent association between chemical functional groups and valence (Figure 4A) but observed a strong rank correlation between hydrocarbon chain length and 3’dOI (rs=0.795, P=0.0034; Figure 4B, Data S2). We observed similar significant relationships for three additional valence-related olfactory investigation parameters (1’dOI, 1’fOI, and 3’fOI) but not for other behavioral parameters. However, a broader understanding of odor perception requires a more complete description of the stimulus7,42. Indeed, robust correlations have been identified between large sets of physicochemical descriptors and multiple olfactory perceptual qualities in human, as well as simple mouse behavioral measures like investigation time8,9,17,43.
Figure 4. Predicting mouse olfactory-driven behaviors.
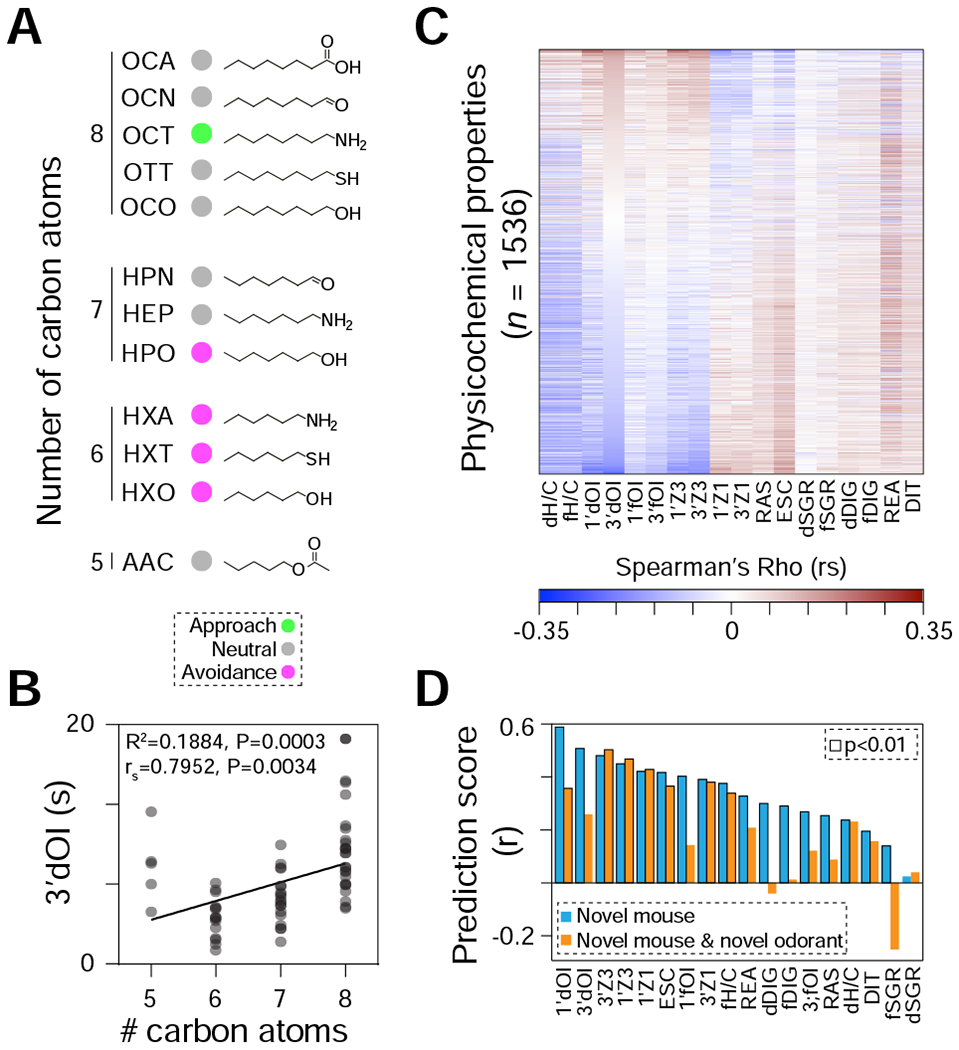
(A) Twelve n-aliphatic odorants in our dataset include molecules with varying lengths and different functional groups: amyl acetate (AAC), hexanethiol (HXT), hexylamine (HXA), hexanol (HXO), heptanol (HPO), heptanal (HPN), heptylamine (HEP), octanoic acid (OCA), octanol (OCO), octanal (OCN), octylamine (OCT), and octanethiol (OTT). Circles are colored to indicate aversive (magenta), neutral (grey), or approached (green) odorants. (B) Correlation plot between the numbers of carbon atoms and the total duration of olfactory investigation (3’dOI), for the 12 odorants displayed in panel A (n = 5-9 mice per odorant). The R2 value for the linear regression, Spearman’s correlation coefficient (rs), and associated p-values are indicated in the top left corner of the graph. (C) Heatmap depicting the rs between 1536 physicochemical descriptors and the 18 mouse behavioral parameters. The physicochemical descriptors are sorted in descending order of the rs for 3’dOI. (D) Performance of the support vector regression model on the 18 behavioral parameters. Blue bars are for models trained on all but one mouse and tested on that mouse. Orange bars are for models trained on all but one odorant and tested on novel mice for that odorant. Black borders around the bars indicate significant associations (P < 0.01. See also Figure S3 and Data S2.
To test whether mouse behaviors are related to distinct physicochemical descriptors, we retrieved 4,870 physicochemical Dragon descriptors for all odorants. After removing descriptors with near-zero variance or missing data, we calculated the correlations between the remaining subset of 1,536 physicochemical descriptors and the 18 behavioral parameters for all 410 individual mice (Figure 4C, Data S2). Of the possible 27,648 interactions, 29.5% (or 8,141) resulted in significant (p<0.05) interactions, with the durations of self-grooming (dSGR) and handling/catching (dH/C) eliciting the minimum (16) and maximum (968) significant interactions, respectively.
Physicochemical descriptors can be used to reverse-engineer perceptual descriptors of odorants in humans8,43,44. Can mouse olfactory-driven behaviors also be predicted by the chemical structure of odorants? To answer this question, we used an unsupervised machine-learning approach (see STAR Methods) evaluated on either held-out mice or held-out odorants. 17/18 behaviors could be predicted above chance (p<0.01) using physicochemical features on held-out mice, with predictive accuracy varying greatly from 1’dOI (r=0.59, p<1e-40) to dSGR (r=0.02, p=0.31). Since within-odorant behavior across mice is highly correlated, a stricter test is to predict behavior for all mice on a held-out odorant. There 7/18 behaviors could be predicted (p<0.01; or 9/18, p<0.05) from physicochemical descriptors, with the four zone parameters (indicating approach vs. avoidance) having the strongest predictability (r=0.38-0.50, p<0.001) (Figure 4D).
Behavioral effects of odorant concentration
In humans, the quality, valence, and intensity of odorants can change with concentration11,13,45. For example, humans perceive (R)-1-p-menthen-8-thiol as a pleasant grapefruit odor at low concentrations, but as an unpleasant sulfur odor at high concentrations. In mouse, the valence25,31 and intensity46 of odorants can also be concentration-dependent, but whether concentration affects other odor-guided behaviors remains largely unexplored. To address this question, we scored 123 videos from mice exposed to a subset of 12 odorants at two additional descending concentrations (850 μM and 8.5 μM) (Figure S5A, Data S3). At the highest concentration tested (85 mM), odorants have diverse valence, with some eliciting avoidance (IBT, IAA, TMT), no change (PEA, 2HO, DMP, VAN, SKA, AMB), or approach (IND, TMA, PUT) (Figure 5A). However, at lower concentrations all odorants were neutral or elicited approach (Figure 5A)25. Scoring the 17 other behaviors for each concentration yielded 3474 individual data points (Data S3). Of the 12×3×18=648 odorant-concentration-behavior pairwise comparisons, 117 (18.1%) significantly differed from H2O (Figures 5A, and S5A, Data S3). 3’dOI elicited the highest (23) and dSGR the lowest (1) number of significant behavioral changes (Figure 5A, Data S3), consistent with odor valence driving the largest fraction of behavioral variation. Of the 12 odorants tested, 10 elicited significant behavioral changes for all concentrations tested, while the remaining 2 (DMP and PEA) did so for only 2 out of 3 concentrations. Across all odorants, 85 mM elicited the highest number of combined significant behavioral changes (60), followed by 8.5 μM (30) and 850 μM (27). We identified 28/36 unique combinations of significant behavioral changes: 12 for 85 mM, 8 for 850 μM, and 8 for 8.5 μM (Figure 5A).
Figure 5. The effect of odorant concentration on behavior.
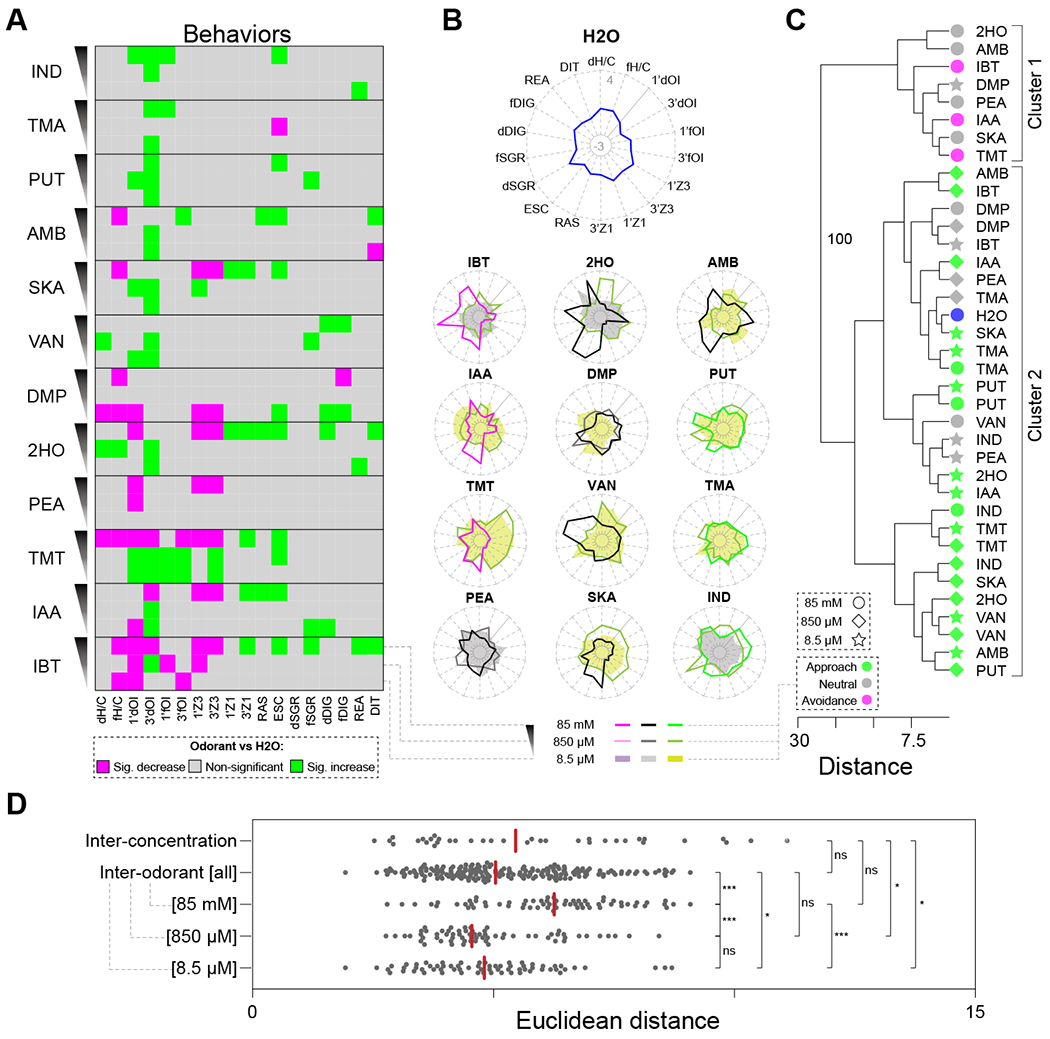
(A) A graphical display summarizing the behavioral codes for the 12 odorants tested at three different concentrations. Behaviors showing significant (one-way ANOVA, BKY multiple comparisons correction, n = 5-9 mice per odorant) increases, decreases, and non-significant responses compared to H2O are indicated in green, magenta, and grey squares, respectively. (B) The behavioral profiles (radar plots, z-scores) for H2O (top center) and the 12 odorants tested at three concentrations (85 mM, 850 μM and 8.5 μM) are shown. The behavioral profiles for the odorants classified at 85 mM as aversive, neutral, and approached are shown in different shades of magenta, grey/black, and green, respectively. The corresponding descending concentrations are depicted in lighter hues of the same color, irrespective of their valence at 850 μM and 8.5 μM. (C) Hierarchical clustering analysis of the behavioral profiles for the 12 odorants tested at the three different concentrations supported the existence of 2 clusters. Bootstrap values56 are shown only for strongly supported nodes, i.e., nodes displaying bootstrap values >70 (total 100 replications). (D) Plot comparing the Euclidean distances calculated for all pairwise inter-concentration comparisons (restricted to different concentrations within each odorant), inter-odorant comparisons (restricted to different odorants at the same concentration), and subsets of inter-odorant comparisons for each concentration separately (85mM, 850μM, and 8.5μM). Asterisks indicate significant differences (one-way ANOVA, BKY multiple comparisons correction): ns, non-significant; *P < 0.05; ***P < 0.001. See also Figure S5, and Data S3.
We next estimated the role of concentration and odorant-concentration interactions in generating behavioral responses. ANOVA showed a main effect of either concentration or an interaction between odorant and concentration for nearly all behavioral parameters (p<0.005 for 16/18; p<10−8 for 13/18). The effect size for odorant identity (η2=0.27 ± 0.05) was slightly greater than for odorant concentration (η2=0.18 ± 0.04), but the interaction between odorant identity and concentration was stronger than either one alone (η2=0.43 ± 0.04).
Thus, even at low concentrations odorants impact mouse behavior and produce diverse behavioral responses. But how does mouse olfactory behavior change with concentration and how is odor identity preserved across concentrations?
To answer these questions, we compared the behavioral profiles across odorants and concentrations. The highest concentration (85 mM) yielded the most distinguishable behavioral profiles across odorants, and some of these profiles were conserved for the same odorants at 850 μM and 8.5 μM (Figure 5B). As concentration decreased, the scores for some behaviors associated with stress (RAS, ESC) and negative odor valence (3’Z1) typically decreased, while the ones associated with positive odor valence (e.g., dH/C, 1’dOI, 3’dOI, 3’Z3) increased.
Next, we performed a hierarchical clustering of odorants-concentrations pairs using the behavioral data. This analysis yielded two clusters (Figure 5C). Cluster 1 is composed mostly (7/8) of odorants presented at 85 mM (five neutral, and three elicit avoidance), which elicit on average 7.00 ± 0.85 (SEM) significant behavioral changes. By contrast, cluster 2 includes H2O, eight neutral, and 20 approached odorants. These odorants elicit on average only 2.18 ± 0.28 (SEM) significant behavioral changes. Moreover, in cluster 2, five odorants (TMA, DMP, PUT, TMT, and VAN) produce behaviors which, while diverse across odorants, were quite similar across the two lowest concentrations of the same odorant. However, all other odorants are not tightly clustered across their 3 concentrations, suggesting that the behavioral profile changes with concentration. PCA further supported these results (Figure S5B).
Finally, to compare the magnitude of changes in odorant identity vs. changes in odorant concentration, we calculated the Euclidean distances between all possible odorant-concentration pairs. The average distances for different odorant pairs of equal concentrations (i.e., inter-odorant), and for different concentration pairs of the same odorant (i.e., inter-concentration) are not significantly different (Figure 5D). However, the average inter-odorant distances were significantly higher for odorant pairs at the highest concentration (85 mM) compared to pairs at the two lower concentrations (850 μM and 8.5 μM). Odorant identity and concentration thus had a similar quantitative impact on mouse olfactory behavior, with higher odorant concentrations producing greater behavioral diversity.
DISCUSSION
Studies from the last 30 years have contributed to quantification, characterization, and prediction of human olfactory perception. This was enabled by physicochemical descriptions of odorants, novel statistical methods, and machine learning algorithms applied to large human olfactory psychophysical datasets6–11. In comparison, mouse olfactory perception is still poorly understood, mainly due to the lack of a large odor-guided behavioral dataset. Here, we conducted a large-scale study of olfactory-driven mouse behavior by generating the most comprehensive mouse olfactory ethological atlas to date. We scored 18 behaviors in 525 mice across 98 odorant conditions, generating 9765 data points encompassing 1764 odorant-behavior interactions.
In humans, odor character is quantified using behavioral methods such as free labeling, odor profiling, or pairwise similarity47. These methods show that, despite interindividual variation, humans have shared and reproducible olfactory percepts for most odorants. Similarly, we find that mouse olfactory perception can be quantified by measuring several behaviors, and that such behaviors demonstrate shared and reproducible olfactory perception across individuals. Many odorants were distinguishable from each other or from the ensemble using such quantified behaviors as observed in a novel mouse. Behavior in novel mice was best reconstructed using a low-dimensional space built from the behavior of other mice. Furthermore, a single dimension showed a modest correlation (r=0.5) between mouse behavior and human perception. Lastly, some mouse behaviors were at least partly predictable from chemical structure alone. Together, these data indicate that there is indeed a rich, canonical set of odor-evoked behaviors in mouse, and this analysis begins to provide insight into mouse olfactory perception.
We found unique behavioral patterns for most odorants and for different concentrations of the same odorant, highlighting the diversity of odor-guided behavioral responses. The olfactory system employs a combinatorial strategy to maximize the discriminability of distinct odorants at different concentrations39,40. This strategy involves hundreds to thousands of OSN/OR subtypes, present in the olfactory mucosa at different abundances3. Despite this peripheral diversity, it is hypothesized that behavioral/perceptual dimensionality is lower than it first appears6. Our data were consistent with a low-dimensional space for behavior. Approach-avoid behaviors, surrogate for mouse odor valence, were the most important behavioral dimension across our analyses. Analogously, rated pleasantness, a surrogate for odor valence, is the principal perceptual dimension in human odor descriptions7,12,35. Odor valence may thus play a central role in olfactory perception in mammals more generally. However, we offer only a lower bound on odor-evoked behavioral diversity, complexity, and dimensionality; alternative future techniques could capture additional information spanning additional dimensions. Furthermore, olfactory perception, resulting from a combinatorial code of peripheral neural activity, could be higher-dimensional still. The transformation from perception to behavior may consist of a projection to lower dimension, a computational transformation consistent with olfactory circuitry48.
Valence is the principal axis for both mouse behavior and human perception43, and indeed olfactory receptor responses in both mice and humans predict human-rated valence12. However this does not imply that specific valence percepts for odorants are shared across species. Indeed, the shared factor that we identified using CCA did not have a strong loading for human-rated pleasantness, or any other specific human percept, suggesting that (at least for this molecular panel), any shared perceptual/behavioral representation of specific odorants across species may have a more multifactorial origin.
In humans, the same odorant can be perceived as pleasant or unpleasant, depending on its concentration. We and others have shown that odorant concentration also impacts mouse olfactory preferences14,25,31,49,50. Here we extended the publicly available data on this topic significantly (18 behaviors, 12 odorants, 3 concentrations). We found that behavior within-odorant was not fixed across concentrations, and that higher concentrations elicited more diverse behavior across odorants. This is consistent with numerically greater OR and glomerular activation by higher odorant concentrations39, 41, 51, thus increasing potential combinatorial complexity and facilitating downstream pattern separation. Similar to our findings, human studies also show that similarities in odor quality between distinct molecules are inversely related to concentration45. Odor character is not consistent across concentrations and so it is not an intrinsic property of the molecule. Indeed, our analysis suggests that mouse behavior is a consequence of an interaction between molecular structure and concentration.
While we cannot exclude the possibility that lower concentrations are undetectably weak for some odorants, we observed significant (vs. odorless H2O control) behavioral changes for all odorants except PEA at the lowest concentration (and PEA activates OSNs at concentrations much lower than that) 52.
Molecular features of odorants are closely linked to psychophysical and behavioral measures of odor valence in human and mouse, respectively7,17,43. Predictive models based on such features can now accurately predict the human olfactory percept of many odorants8,53,54. We employed similar strategies and found that some mouse odor-guided behaviors can also be partly predicted by the physicochemical properties of odorants. The observation that specific physicochemical properties of odorants can predict some behavioral outputs in mice also suggests new possibilities for studying the functional organization of the mouse olfactory system. Subsets of physicochemical properties predictive of odor-guided behaviors could potentially be linked to the spatial organization of ORs/OSN subtypes in the nasal epithelium. Future large-scale experiments focused on connecting the zonal expression patterns for all mouse ORs to the physicochemical descriptors of their respective agonists will be critical to test this hypothesis.
Future studies using recently developed and fully-automated techniques, such as MoSeq and DeepLabCut24,55, in combination with behavioral recordings meeting or exceeding the volume and diversity of odorants and mice used here, have the potential to further elucidate the structure of mouse behavioral space and how it relates to odorant identity, valence, and character. We hope that the results presented here will motivate such future work, as we have shown the link between odorant and behavior is strong and shared across individuals.
In conclusion, our study provides a foundational quantitative database of odor-guided behaviors in the mouse that can be exploited in future studies to further deconstruct many aspects of mouse olfactory behavior and putative perception, and facilitate future comparative studies of olfactory percepts among different species.
STAR*METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and data should be directed to and will be fulfilled by the Lead Contacts, Luis R. Saraiva (saraivalmr@gmail.com) or Richard C. Gerkin (rgerkin@asu.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study, and the code generated during this study is available upon manuscript acceptance at http://github.com/rgerkin/manoel-2021.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
The animals used in the video library from our previous study25 were adult male C57Bl/6J mice (aged 8-14 weeks, The Jackson Laboratory). Each mouse was randomly assigned and exposed to only a single odorant, and thus the data for each odorant consisted of 5-10 mice each scored on 18 behaviors. The experiments performed in Saraiva et al 2016 were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee.
METHOD DETAILS
Behavioral scoring
We retrieved a video library from our previous study25, in which we subjected adult male mice to the olfactory preference test for a total duration of 3 minutes. In this study we analyzed 410 videos from mice exposed to an odorless control (i.e., water, or H2O) or one of the 73 odorants at a single concentration (at 85 mM), and 123 additional videos from mice exposed to two other descending concentrations (850 μM and 8.5 μM) for a subset of 12 odorants. The odorants tested in the videos include 61 general odorants and 12 ethologically relevant odorants (7 mouse pheromones, and 5 kairomones). All these compounds were chemically diverse and belong to multiple chemical structural classes (underlined, for more details see Saraiva et al 2016) as shown below, followed by their 3-letter abbreviation in parentheses: Alcohols: 2-phenylethanol (2PE); geraniol (GER); heptanol (HPO); hexanol (HXO); linalool (LIN); octanol (OCO); cis-3-hexenol (C3H). Aldehydes: benzaldehyde (BZL); citral (CIT); citronellal (CTN); heptanal (HPN); octanal (OCN); trans-2-hexenal (T2H). Amines: 2-methylbutylamine (2MB); 1-(2-aminoethyl)piperidine (AEP); aniline (ANI); 3-amino-s-triazole (AST); cadaverine (CAD); N,N-dimethylbutylamine (DMB); N,N-dimethylethylamine (DME); N,N-dimethyloctylamine (DMO); N,N-dimethylcyclohexylamine (DMC); heptylamine (HEP); hexylamine (HXA); isoamylamine (IAA); 2-methyl-1-pyrroline (M1P); N-methylpiperidine (NMP); octylamine (OCT); 2-phenylethylamine (PEA); trimethylamine (TMA); putrescine (PUT); pyrrolidine (PYR); spermidine (SPD); spermine (SPN); o-toluidine (TOL). Azines. 2,5-dimethylpirazine (DMP); 2-ethyl-3,5(6)-dimethylpirazine (EDM); indole (IND); quinoline (QUI); skatole (SKA). Camphors: (+/−)-camphor (CAM); (−)-fenchone (-FCH); (+)-fenchone (+FCH); eucalyptol (EUC). Carboxylic acids: 2-methylbutyric acid (MBA); octanoic acid (OCA); propionic acid (PPA). Esters, amyl acetate (AAC); ethyl butyrate (EBT). Ketones. 2-heptanone (2HO); alpha-ionone (ION). Musks: ambrettolide (AMB); civettone (CIV); muscone (MUS). Terpenes: beta-farnesene (BFA); (−)-carvone (-CVN); (+)-carvone (+CVN); farnesene mixed isomers (alpha+beta) (FAR); (−)-limonene (−LIM); (+)-limonene (+LIM); (+)-menthol (+MEN); (−)-menthone (-MNT); (+)-menthone (+MNT); alpha-pinene (PIN); rose oxide (ROX). Thiazoles. 2-isobutylthiazole (IBT); 2-isopropyl-4-5-dihydrothiazole (IPT); 2,5-dihydro-2,4,5-trimethylthiazoline (TMT). Thiols: 2-propylthietane (2PT); hexanethiol (HXT); octanethiol (OTT). Vanillin-like compounds: eugenol (EUG); vanillin (VAN).
For each video, in addition to the cumulative duration of olfactory investigation (3’dOI) reported in Saraiva et al 2016 for the 3 minutes-long assay, we scored 17 new behavioral parameters indicative of either valence, stress, and exploration. The videos were randomized and scored blind (to the odorant) using the following criteria:
- Olfactory investigation (OI) parameters: 1’dOI, 1’fOI, and 3’fOI represent the cumulative duration (d) or frequency (f) of olfactory investigation during the 1st minute (1’) or the full 3 minutes (3’) of the assay. We considered OI only if the nose of the mouse was overlapping, or in very close proximity (~0.5 cm) of the stimulus. For these parameters, the videos were randomized and scored by the experimenter.
- Zone (Z) parameters: 1’Z1, 1’Z3, 3’Z1, and 3’Z3 represent the cumulative time mice spent in either Zone 1 (Z1) or Zone 3 (Z3) of the cage during the 1st minute (1’) or the full 3 minutes (3’) of the assay. Here, the test cage was divided into three equal-sized zones, with Z1 representing the zone furthest away from the odor stimulus and Z3 the zone containing the stimulus. Time spent inside each zone (head of the animal had to be within the zone) was scored using the Ethovision XT software (version 11, Noldus Information Technology), and videos in which the mouse transported the piece of filter paper outside Z3 were not included. For these parameters, the videos were scored by a non-experimenter.
- Handling (H) or catching (C) parameters: dH/C and fH/C represent the cumulative duration (d) or frequency (f) where mice handled and/or caught the stimulus with its front paws during the 3 minutes of the assay. For these parameters, the videos were scored by a non-experimenter.
- Risk assessment (RAS): this parameter represents the number of episodes the mouse displays the flat-back/stretch-attend response, followed by a sniff in the direction of the stimulus, during the 3 minutes of the assay. For this parameter, the videos were scored by a non-experimenter.
- Escape (ESC): this parameter represents the number of episodes the mouse displays a quick and contactless approach towards the stimulus, followed by an even faster withdrawal/darting to the opposite end of the cage, during the 3 minutes of the assay. For this parameter, the videos were scored by a non-experimenter.
- Digging (DIG) parameters: dDIG and fDIG represent the cumulative duration (d) or frequency (f) that the mouse digs into the bedding with the forelimbs, often kicking it away with the hindlimbs, during the 3 minutes of the assay. For these parameters, the videos were scored by a non-experimenter.
- Distance (DIT): this parameter represents the total length the mouse walked/ran through during the three minutes duration of the video. Distance traveled was scored using the Ethovision XT software (version 11, Noldus Information Technology), and video tracking done using the center-point of the mouse. For this parameter, the videos were scored by a non-experimenter.
- Rearing (REA): this parameter represents the number of episodes the mouse stands on its hindlegs (rearing) anywhere in the cage, including when rearing against the walls during the 3 minutes of the assay. For this parameter, the videos were scored by a non-experimenter.
- Self-grooming (SGR) parameters: dSGR and fSGR represent the cumulative duration (d) or frequency (f) where the mice are self-grooming, defined by when the mouse was licking its fur, grooming itself with the forepaws, or scratching any part of its body with any limb. For these parameters, the videos were scored by a non-experimenter.
Machine Learning
We used Python 3.8 and the Pyrfume package (version 0.15) to calculate 1826 physicochemical descriptors for 74 molecules (Mordred, version 1.2). Descriptors with zero variance or >25% missing values were removed leaving 1328 descriptors, and the remaining missing values were iteratively imputed using (scikit-learn, version 0.23.2). We also used Pyrfume to calculate the Morgan fingerprint similarity (rdkit, version 2020.09.1) of the 74 molecules to a reference set of 9645 (mostly) odorous compounds. We used support vector regression with leave-one-out cross-validation (scikit-learn) to train two models, one based on Mordred descriptors and one on Morgan similarities, and their predictions were averaged before validation. We evaluated two kinds of predictions: (1) One where the models were trained on all but a single mouse, i.e. the training set included other instances of the same odorant presented to different mice, and (2) one where the models were trained on all but a single odorant, i.e. the training set excluded all mice presented with that odorant. For (1) evaluation was done per mouse and for (2) averaging across mice within odorant was performed first and training/testing was done on these averages. We reported the Pearson correlation r between the observed (out-of-sample) behavior and the predicted behavior; r was chosen to facilitate direct comparison to several predictive models in human olfactory perception that also use this measure8, 53, 54. Significance was assessed by converting r values to Z-scores using the Fisher transformation and using the standard error for the Fisher Z of 1/sqrt(n-3) in conjunction with the cumulative normal distribution. We did not correct our reported values for multiple comparisons, but a False Discovery Rate correction would have had minimal impact.
DREAM predictions
The DREAM model developed in Keller et al8 was applied to H2O and each of the 73 odorants used in the current study to generate predictions for each of 21 perceptual descriptors. Briefly, an isomeric, canonical SMILES string was generated for each odorant using rdkit (Python) and used to generate a rdkit mol object in which 3-dimensional coordinates of each atom position were estimated. In order to match the methods of Keller et al, Dragon 6.0 was used to compute features from these 3-dimensional structures. The DREAM model was re-trained from scratch on the 476 original molecules (from Keller et al) and used to predict 21 perceptual descriptors from the 74 odorants used in the current study.
Data Analysis
The number of mice tested for each odorant and the quantification of each behavioral measurement are described in the “Behavioral Scoring” section above. The raw behavioral scores and numbers of animals used can be consulted in Data S1. One-way ANOVA tests were performed using GraphPad Prism 8.0.0 software for each of the 18 behavioral parameters. P-values for the pairwise comparisons between H2O control and each of the tested odorant stimuli were computed with a two-stage Benjamini, Krieger and Yekutieli (BKY) multiple comparisons correction57 (Figures 1E, 5A, and Data S1).
Correlation matrices generated with pandas and scikit-learn were used to compute the eigenvalues and eigenvectors in the principal components analysis (Figures 1F,G). For Figure S1C, the odorant coloring was done by computing the first three principal components of the Morgan fingerprint similarity matrix (computed using rdkit and pyrfume), projecting the odorants onto this 3d-space, normalizing each dimension between 0 and 1, and setting the 53 values of each odorant according to these normalized values.
Custom code using numpy and scipy was used to compute multivariate D’ (Figure 2A–D) and scikit-learn was used to train the linear classifiers (Figure 2E–I). Scikit-learn was used to perform the NMF analysis (Fig. 3A–B) and custom code58 was used to implement the ridge-regularized CCA (Figure 3C).
Spearman’s correlation coefficients of the 1,536 physicochemical descriptors against the 18 behavioral parameters were computed using Graphpad Prism 8.0.0 software (Figure 4C).
The standardized data (z-scores) in Figure S4A were further analyzed through hierarchical clustering performed with PAlaeontological STatistics (version 4.06) using Euclidean distances with Ward’s method (Figure 5C). The z-score data matrix was also used for principal component analyses and performed using PAlaeontological STatistics (version 4.06) (Figure S4B).
Supplementary Material
Data S1. Microsoft Excel format file, related to Figure 1. Raw behavioral scores for the 410 mice exposed to odorants; P-values for the odorant vs. H2O comparison for each of the behaviors (one-way ANOVA, BKY multiple comparisons correction, see STAR Methods): ns, non-significant; *P < 0.05; ***P < 0.001. Behaviors showing significant increases and decreases are highlighted in green and magenta, respectively. Grey color indicates no significant statistical difference. See also Figures 1, and S1.
Data S2. Microsoft Excel format file, related to Figure 4. Linear regression and correlation stats for 3’dOI vs. the number of carbons; Spearman correlation coefficient (rs) matrix of the 18 behavioral parameters vs. 1536 physicochemical descriptors. See also Figure 4.
Data S3. Microsoft Excel format file, related to Figure 5. Raw behavioral scores for 36 odor conditions and H2O across 18 behavioral parameters; p-values of 36 odor conditions across 18 behavioral parameters vs. H2O control (one-way ANOVA, BKY multiple comparisons correction); Euclidean distances between 36 odor conditions and H2O, across 18 behavioral parameters. See also Figures 5, and S4.
KEY RESOURCES TABLE
| RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Mouse behavior video library | ||
| Behavior video library | Saraiva et al. 2016 | PMID: 27208093 |
| Software and algorithms | ||
| Code generated here | This manuscript | http://github.com/rgerkin/manoel-2021 |
| Ethovision XT | Noldus | https://www.noldus.com/ethovision-xt |
| Dragon 6.0 | Talete | http://www.talete.mi.it/ |
| Python 3 | Python | python.org |
| Pandas | AQR Capital Management | https://pandas.pydata.org/ |
| Scikit-Learn | scikit-learn | https://scikit-learn.org/stable/ |
| Pyrfume | The Pyrfume Project | http://pyrfume.org |
| Mordred | Mordred Descriptors | https://github.com/mordred-descriptor/mordred |
| Rdkit | Rdkit | http://www.rdkit.org |
| R Statistical Software 3.5.1 | R project for Statistical Computing | https://www.r-project.org/ |
| GraphPad Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| PAlaeontological STatistics | Paleontological Museum, University of Oslo | https://folk.uio.no/ohammer/past |
HIGHLIGHTS.
We produced a novel atlas of mouse odor-guided behavior for 73 molecules
Odorant structure and mouse behavior are mutually predictive in novel mice
Odor valence explains most behavioral variance and covariance with human perception
Concentration qualitatively and quantitatively impacts mouse olfactory behavior
ACKNOWLEDGMENTS
We would like to thank Dr. Darren W. Logan and the members of the Saraiva Lab for the constructive feedback, and Alex Williams for the ridge-regularized CCA code. This work was supported by Sidra Medicine (SDR400077), a member of Qatar Foundation, and the National Institute of Health (U19NS112953 and R01DC018455).
DECLARATION OF INTERESTS
J.D.M. receives research funding from Google Research, Procter & Gamble, was on the scientific advisory board of Aromyx, and received compensation for these activities. R.C.G. receives research funding from Google Research, The Taylor Corporation, and is on the advisory board of Climax Foods. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All other authors declare that they have no competing interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bear Daniel M., Lassance J-M, Hoekstra Hopi E., and Datta Sandeep R. (2016). The Evolving Neural and Genetic Architecture of Vertebrate Olfaction. Current Biology 26, R1039–R1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, and Liberles SD (2015). Aversion and attraction through olfaction. Curr Biol 25, R120–R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saraiva LR, Riveros-McKay F, Mezzavilla M, Abou-Moussa EH, Arayata CJ, Makhlouf M, Trimmer C, Ibarra-Soria X, Khan M, Van Gerven L, et al. (2019). A transcriptomic atlas of mammalian olfactory mucosae reveals an evolutionary influence on food odor detection in humans. Sci Adv 5, eaax0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saraiva LR, Ahuja G, Ivandic I, Syed AS, Marioni JC, Korsching SI, and Logan DW (2015). Molecular and neuronal homology between the olfactory systems of zebrafish and mouse. Scientific reports 5, 11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niimura Y (2012). Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr Genomics 13, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Secundo L, Snitz K, and Sobel N (2014). The perceptual logic of smell. Curr Opin Neurobiol 25, 107–115. [DOI] [PubMed] [Google Scholar]
- 7.Yeshurun Y, and Sobel N (2010). An odor is not worth a thousand words: from multidimensional odors to unidimensional odor objects. Annu Rev Psychol 61, 219–241, C211-215. [DOI] [PubMed] [Google Scholar]
- 8.Keller A, Gerkin RC, Guan Y, Dhurandhar A, Turu G, Szalai B, Mainland JD, Ihara Y, Yu CW, Wolfinger R, et al. (2017). Predicting human olfactory perception from chemical features of odor molecules. Science 355, 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller A, and Vosshall LB (2016). Olfactory perception of chemically diverse molecules. BMC neuroscience 17, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dravnieks A (1982). Odor quality: semantically generated multidimensional profiles are stable. Science 218, 799–801. [DOI] [PubMed] [Google Scholar]
- 11.Moskowitz HR, Dravnieks A, and Klarman LA (1976). Odor intensity and pleasantness for a diverse set of odorants. Perception & Psychophysics 19, 122–128. [Google Scholar]
- 12.Haddad R, Weiss T, Khan R, Nadler B, Mandairon N, Bensafi M, Schneidman E, and Sobel N (2010). Global features of neural activity in the olfactory system form a parallel code that predicts olfactory behavior and perception. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 9017–9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross-Isseroff R, and Lancet D (1988). Concentration-dependent changes of perceived odor quality. Chemical senses 13, 191–204. [Google Scholar]
- 14.Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. (2007). Innate versus learned odour processing in the mouse olfactory bulb. Nature 450, 503–508. [DOI] [PubMed] [Google Scholar]
- 15.Root CM, Denny CA, Hen R, and Axel R (2014). The participation of cortical amygdala in innate, odour-driven behaviour. Nature 515, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logan DW, Brunet LJ, Webb WR, Cutforth T, Ngai J, and Stowers L (2012). Learned recognition of maternal signature odors mediates the first suckling episode in mice. Curr Biol 22, 1998–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandairon N, Poncelet J, Bensafi M, and Didier A (2009). Humans and mice express similar olfactory preferences. PloS one 4, e4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, Datta SR, Spehr M, Fendt M, and Liberles SD (2011). Detection and avoidance of a carnivore odor by prey. Proceedings of the National Academy of Sciences of the United States of America 108, 11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno H, Shimada A, Suemitsu S, Murakami S, Kitamura N, Wani K, Matsumoto Y, Okamoto M, and Ishihara T (2019). Anti-depressive-like effect of 2-phenylethanol inhalation in mice. Biomed Pharmacother 111, 1499–1506. [DOI] [PubMed] [Google Scholar]
- 20.Papes F, Logan DW, and Stowers L (2010). The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell 141, 692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brechbuhl J, Moine F, Klaey M, Nenniger-Tosato M, Hurni N, Sporkert F, Giroud C, and Broillet MC (2013). Mouse alarm pheromone shares structural similarity with predator scents. Proceedings of the National Academy of Sciences of the United States of America 110, 4762–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondoh K, Lu Z, Ye X, Olson DP, Lowell BB, and Buck LB (2016). A specific area of olfactory cortex involved in stress hormone responses to predator odours. Nature 532, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito H, Nishizumi H, Suzuki S, Matsumoto H, Ieki N, Abe T, Kiyonari H, Morita M, Yokota H, Hirayama N, et al. (2017). Immobility responses are induced by photoactivation of single glomerular species responsive to fox odour TMT. Nature communications 8, 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP, and Datta SR (2015). Mapping Sub-Second Structure in Mouse Behavior. Neuron 88, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saraiva LR, Kondoh K, Ye X, Yoon KH, Hernandez M, and Buck LB (2016). Combinatorial effects of odorants on mouse behavior. Proceedings of the National Academy of Sciences of the United States of America 113, E3300–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortes-Marco L, Lanuza E, and Martinez-Garcia F (2013). Of pheromones and kairomones: what receptors mediate innate emotional responses? Anat Rec (Hoboken) 296, 1346–1363. [DOI] [PubMed] [Google Scholar]
- 27.Novotny MV (2003). Pheromones, binding proteins and receptor responses in rodents. Biochem Soc Trans 31, 117–122. [DOI] [PubMed] [Google Scholar]
- 28.Liberles SD (2014). Mammalian pheromones. Annu Rev Physiol 76, 151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Achiraman S, and Archunan G (2002). Characterization of urinary volatiles in Swiss male mice (Mus musculus): bioassay of identified compounds. J Biosci 27, 679–686. [DOI] [PubMed] [Google Scholar]
- 30.Apfelbach R, Soini HA, Vasilieva NY, and Novotny MV (2015). Behavioral responses of predator-naive dwarf hamsters (Phodopus campbelli) to odor cues of the European ferret fed with different prey species. Physiology & behavior 146, 57–66. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Korzan WJ, Ferrero DM, Chang RB, Roy DS, Buchi M, Lemon JK, Kaur AW, Stowers L, Fendt M, et al. (2013). Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr Biol 23, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuzesi T, Daviu N, Wamsteeker Cusulin JI, Bonin RP, and Bains JS (2016). Hypothalamic CRH neurons orchestrate complex behaviours after stress. Nature communications 7, 11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Gomez A, Bleymehl K, Stein B, Pyrski M, Birnbaumer L, Munger SD, Leinders-Zufall T, Zufall F, and Chamero P (2015). Innate Predator Odor Aversion Driven by Parallel Olfactory Subsystems that Converge in the Ventromedial Hypothalamus. Curr Biol 25, 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cichy A, Shah A, Dewan A, Kaye S, and Bozza T (2019). Genetic Depletion of Class I Odorant Receptors Impacts Perception of Carboxylic Acids. Curr Biol 29, 2687–2697 e2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crampin EJ, Haddad R, Carmel L, Sobel N, and Harel D (2008). Predicting the Receptive Range of Olfactory Receptors. PLoS computational biology 4, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DD, and Seung HS (1999). Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791. [DOI] [PubMed] [Google Scholar]
- 37.Castro JB, Ramanathan A, and Chennubhotla CS (2013). Categorical dimensions of human odor descriptor space revealed by non-negative matrix factorization. PloS one 8, e73289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manoel D, Makhlouf M, Scialdone A, and Saraiva LR (2019). Interspecific variation of olfactory preferences in flies, mice, and humans. Chemical senses 44, 7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malnic B, Hirono J, Sato T, and Buck LB (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 40.Nara K, Saraiva LR, Ye X, and Buck LB (2011). A large-scale analysis of odor coding in the olfactory epithelium. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 9179–9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson BA, and Leon M (2000). Modular representations of odorants in the glomerular layer of the rat olfactory bulb and the effects of stimulus concentration. J Comp Neurol 422, 496–509. [DOI] [PubMed] [Google Scholar]
- 42.Haddad R, Khan R, Takahashi YK, Mori K, Harel D, and Sobel N (2008). A metric for odorant comparison. Nat Methods 5, 425–429. [DOI] [PubMed] [Google Scholar]
- 43.Khan RM, Luk CH, Flinker A, Aggarwal A, Lapid H, Haddad R, and Sobel N (2007). Predicting odor pleasantness from odorant structure: pleasantness as a reflection of the physical world. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 10015–10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Licon CC, Bosc G, Sabri M, Mantel M, Fournel A, Bushdid C, Golebiowski J, Robardet C, Plantevit M, Kaytoue M, et al. (2019). Chemical features mining provides new descriptive structure-odor relationships. PLoS computational biology 15, e1006945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laing DG, Legha PK, Jinks AL, and Hutchinson I (2003). Relationship between molecular structure, concentration and odor qualities of oxygenated aliphatic molecules. Chemical senses 28, 57–69. [DOI] [PubMed] [Google Scholar]
- 46.Sirotin YB, Shusterman R, and Rinberg D (2015). Neural Coding of Perceived Odor Intensity. eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wise PM, Olsson MJ, and Cain WS (2000). Quantification of odor quality. Chemical senses 25, 429–443. [DOI] [PubMed] [Google Scholar]
- 48.Pashkovski SL, Iurilli G, Brann D, Chicharro D, Drummey K, Franks KM, Panzeri S, and Datta SR (2020). Structure and flexibility in cortical representations of odour space. Nature 583, 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato-Akuhara N, Horio N, Kato-Namba A, Yoshikawa K, Niimura Y, Ihara S, Shirasu M, and Touhara K (2016). Ligand Specificity and Evolution of Mammalian Musk Odor Receptors: Effect of Single Receptor Deletion on Odor Detection. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 4482–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horio N, Murata K, Yoshikawa K, Yoshihara Y, and Touhara K (2019). Contribution of individual olfactory receptors to odor-induced attractive or aversive behavior in mice. Nature communications 10, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fried HU, Fuss SH, and Korsching SI (2002). Selective imaging of presynaptic activity in the mouse olfactory bulb shows concentration and structure dependence of odor responses in identified glomeruli. Proceedings of the National Academy of Sciences of the United States of America 99, 3222–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Pacifico R, Cawley D, Feinstein P, and Bozza T (2013). Ultrasensitive detection of amines by a trace amine-associated receptor. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 3228–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kowalewski J, Huynh B, and Ray A (2021). A systems-wide understanding of the human olfactory percept chemical space. Chemical senses. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Lengeling B, Wei JN, Lee BK, Gerkin RC, Aspuru-Guzik A, and Wiltschko AB (2019). Machine Learning for Scent: Learning Generalizable Perceptual Representations of Small Molecules. p. arXiv:1910.10685. [Google Scholar]
- 55.Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, and Bethge M (2018). DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat Neurosci 21, 1281–1289. [DOI] [PubMed] [Google Scholar]
- 56.Jain AK, and Moreau JV (1987). Bootstrap technique in cluster analysis. Pattern Recognition 20, 547–568. [Google Scholar]
- 57.Benjamini Y, Krieger AM, and Yekutieli D (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507. [Google Scholar]
- 58.Williams A (2020). Ridge CCA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Microsoft Excel format file, related to Figure 1. Raw behavioral scores for the 410 mice exposed to odorants; P-values for the odorant vs. H2O comparison for each of the behaviors (one-way ANOVA, BKY multiple comparisons correction, see STAR Methods): ns, non-significant; *P < 0.05; ***P < 0.001. Behaviors showing significant increases and decreases are highlighted in green and magenta, respectively. Grey color indicates no significant statistical difference. See also Figures 1, and S1.
Data S2. Microsoft Excel format file, related to Figure 4. Linear regression and correlation stats for 3’dOI vs. the number of carbons; Spearman correlation coefficient (rs) matrix of the 18 behavioral parameters vs. 1536 physicochemical descriptors. See also Figure 4.
Data S3. Microsoft Excel format file, related to Figure 5. Raw behavioral scores for 36 odor conditions and H2O across 18 behavioral parameters; p-values of 36 odor conditions across 18 behavioral parameters vs. H2O control (one-way ANOVA, BKY multiple comparisons correction); Euclidean distances between 36 odor conditions and H2O, across 18 behavioral parameters. See also Figures 5, and S4.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study, and the code generated during this study is available upon manuscript acceptance at http://github.com/rgerkin/manoel-2021.


