Summary
The primary cilium is an essential organizing center for signal transduction, and ciliary defects cause congenital disorders known collectively as ciliopathies.1–3 Primary cilia form by two pathways that are employed in a cell type- and tissue-specific manner: an extracellular pathway in which the cilium grows out from the cell surface and an intracellular pathway in which the nascent cilium first forms inside the cell.4–8 After exposure to the external environment, cilia formed via the intracellular pathway may have distinct functional properties, as they often remain recessed within a ciliary pocket.9,10 However, the precise mechanism of intracellular ciliogenesis and its relatedness to extracellular ciliogenesis remain poorly understood. Here we show that Rab34, a poorly characterized GTPase recently linked to cilia11–13, is a selective mediator of intracellular ciliogenesis. We find that Rab34 is required for formation of the ciliary vesicle at the mother centriole and that Rab34 marks the ciliary sheath, a unique sub-domain of assembling intracellular cilia. Rab34 activity is modulated by divergent residues within its GTPase domain, and ciliogenesis requires GTP binding and turnover by Rab34. Because Rab34 is found on assembly intermediates that are unique to intracellular ciliogenesis, we tested its role in the extracellular pathway used by polarized MDCK cells. Consistent with Rab34 acting specifically in the intracellular pathway, MDCK cells ciliate independently of Rab34 and its paralog Rab36. Together, these findings establish that different modes of ciliogenesis have distinct molecular requirements and reveal Rab34 as a new GTPase mediator of ciliary membrane biogenesis.
Keywords: cilia, ciliogenesis, ciliopathy, centriole, GTPase, membrane, ciliary vesicle, Rab
eTOC Blurb:
Different cell types form primary cilia through different assembly pathways. Ganga et al. show that the GTPase Rab34 is needed for ciliary membrane formation in the intracellular ciliogenesis pathway but not the extracellular pathway. During intracellular ciliogenesis, Rab34 is recruited to nascent cilia and localizes to the ciliary sheath.
Graphical Abstract

Results
Rab34 is required for ciliogenesis
We and others recently identified Rab34 as a hit in genome-wide screens for regulators of cilium-dependent Hh signaling.11,14 Our CRISPR screen was based on an NIH-3T3 reporter cell line in which a blastidicin resistance gene is induced in response to Hh pathway activation. Notably, Rab34 was a top-scoring hit, with all ten Rab34-targeting sgRNAs causing decreased resistance to blasticidin14 (Figure 1A). We used two of these sgRNAs to generate Rab34 mutant NIH-3T3 cell pools and observed strongly reduced ciliogenesis, as assessed by ARL13B or acetylated tubulin staining (Figures 1B–C and S1A). We also knocked out RAB34 in RPE1 cells, a widely used model for ciliogenesis. We used two RAB34-targeting sgRNAs and generated clonal and polyclonal cell lines with frame-shift mutations and/or strongly reduced levels of Rab34 protein (Figure S1B). As in NIH-3T3 cells, ciliogenesis was greatly reduced in RPE1 RAB34 mutants (Figures 1D and S1C–D). Additionally, expression of human RAB34 rescued ciliogenesis in Rab34 mutant NIH-3T3 cells, confirming the specificity of the mutant phenotype (Figure 1G).
Figure 1. The nucleotide cycle of atypical GTPase Rab34 is required for ciliogenesis in NIH-3T3 and RPE1 cells.

A) Results of a genome-wide CRISPR screen for Hedgehog (Hh) signaling14 in which signaling-deficient mutants become hypersensitive to blasticidin. The sgRNAs targeting Rab34 are highlighted. B) Cilia (marked by ARL13B) and centrioles (marked by γ-tubulin) were stained in NIH-3T3 cells transduced with the indicated sgRNAs following 24 h serum starvation. Scale bar: 5 μm. C) Quantification of ciliogenesis in NIH-3T3 cells shown in (B); bars indicate means and dots show values from >120 cells analyzed in each of N=3 independent experiments. D) Quantification of ciliogenesis in RPE1 cells transduced with the indicated sgRNAs and stained as in (B) following 48 h serum starvation; bars indicate means and dots show values from >130 cells analyzed in each of N=3 independent experiments. See also Figure S1C. E) Illustration of the Rab34 nucleotide cycle including Guanine nucleotide Exchange Factors (GEFs), GTPase Activating Proteins (GAPs), and effectors that support its role in ciliogenesis. Mutants of interest and their nucleotide state are shown (predicted nucleotide states are denoted with asterisks; see also panel I for observed nucleotide states). F) Alignment of key regions of human Rab34 with other human Rab GTPases and KRAS. Conserved residues T66 and Q111 and divergent residues L61 and S166 are indicated. G) NIH-3T3 cells transduced with the indicated sgRNAs were transfected with GFP-FKBP or the indicated GFP-Rab34 constructs. Ciliogenesis was assessed as in C; bars indicate means and dots show values from >40 cells analyzed in each of N=3 independent experiments. H) Ciliogenesis was assessed in RPE1 stable cell lines expressing the indicated GFP-Rab34 variants under control of a doxycycline-inducible promoter. Where indicated, doxycycline was included for 24 h preceding serum starvation and during serum starvation. Bars indicate means and dots show values from N=4 independent experiments. I) Analysis of GTP and GDP bound to GFP-Rab34 following metabolic labeling in HEK-293T cells, anti-GFP immunoprecipitation, thin layer chromatography of bound nucleotides, and autoradiography. Values shown represent mean ± s.e.m. from N=3 independent experiments. See also Figure S1.
Rab34 GTP binding and hydrolysis are necessary for ciliogenesis
Rab GTPases cycle between an inactive GDP-bound state and an active GTP-bound state, and mutations at conserved residues can be used to alter this nucleotide cycle15–19 (Figure 1E). In the case of Rab34, a Q111L mutation is predicted to block GTP hydrolysis and lock Rab34 in the GTP-bound state, while a T66N mutation is expected to impair GTP binding (Figure 1E–F). We transfected Rab34 mutant NIH-3T3 cells with GFP-tagged Rab34 constructs and found that Rab34-WT and Rab34-Q111L efficiently rescued the ciliogenesis defect while Rab34-T66N did not. Thus, GTP-binding by Rab34 is essential for ciliogenesis (Figure 1G).
While analyzing the sequence of Rab34, we noted two residues—Leu61 and Ser166—that are conserved within Rab34 orthologs but diverge from the amino acids typically seen in Rab GTPases (Figures 1F and S1E). These residues are likely to be functionally important, as Leu61 lies in the P-loop and corresponds to Gly12 of KRAS, where G12D is a common oncogenic mutation.20 Ser166 is typically an asparagine in the NKxD motif, and mutations at this position impair nucleotide binding.21,22 We therefore examined ciliogenesis in Rab34 mutant cells expressing Rab34-S166N (reverting Ser166 to Asn) and Rab34-L61D (mimicking KRAS-G12D). Notably, both mutants failed to rescue ciliogenesis in Rab34 mutant cells (Figure 1G) and appeared to reduce ciliogenesis in wildtype cells (not shown). To further examine this dominant-negative effect, we generated RPE1 cell lines with doxycycline-inducible GFP-Rab34-WT, L61D, and S166N. While cells over-expressing wildtype Rab34 assembled cilia efficiently, induction of Rab34-L61D or Rab34-S166N caused a severe ciliogenesis defect (Figure 1H).
To determine how the S166N and L61D mutations affect the nucleotide cycle of Rab34, we analyzed Rab34-bound nucleotides in HEK293T cells that were transfected with GFP-tagged Rab34 variants and metabolically labeled with 32P-phosphate. Consistent with an earlier report23, wildtype Rab34 was mostly GDP-bound. However, Rab34-S166N and Rab34-L61D both bound more GTP, suggesting that these are GTP-locked mutants (Figure 1I). By contrast, the Q111L mutant canonically predicted to be GTP-locked was similar to wildtype, possibly because such Rab mutants can remain susceptible to GAP-stimulated GTP hydrolysis.24 Increased GTP binding by Rab34-S166N and Rab34-L61D is further supported by co-immunoprecipitation experiments in which these mutants exhibit increased binding to RILPL1, a putative effector that recognizes the GTP state25,26, and decreased binding to Rab-GDI, a binding partner selective for the GDP state18 (Figure S1F). Lastly, we examined how these mutations affect the GTPase activity of purified recombinant Rab34. Notably, Rab34-L61D exhibited a ~100-fold lower kcat than wildtype Rab34 and a ~100-fold tighter Km for GTP, while GTPase activity for Rab34-S166N was below our limit of detection (Figure S1G). These findings support our cell-based studies and indicate that GTP binding and hydrolysis by Rab34 are required for ciliogenesis.
Rab34 mediates ciliary vesicle formation
The intracellular ciliogenesis pathway used by NIH-3T3 and RPE1 cells involves a number of sequential steps and requires dynamic recruitment of proteins including MYO5A, EHD1, and Rab8 (Figures 2A and S2A). One of the earliest events is transport of pre-ciliary vesicles to the mother centriole by MYO5A, where they are captured at the distal appendages to give rise to MYO5A-labeled Distal Appendage Vesicles (DAVs).7,8,27 These events appear to occur normally in RAB34 mutant cells, as distal appendages marked by CEP164 are present, and MYO5A is recruited to the mother centriole (Figure 2B–C). After DAVs are captured at the mother centriole, they fuse into a larger ciliary vesicle through the action of EHD family ATPases.6,7 Notably, RAB34 knockout cells exhibited a significant defect in recruitment of EHD1-mScarlet-I to the mother centriole (Figure 2D). A similar defect was observed for GTPases Rab8 and ARL13B (Figures 2E and S1C), which are recruited after EHD1 to promote ciliary membrane growth.28,29 To directly observe the membrane remodeling events that occur during intracellular ciliogenesis, we used Focused Ion Beam Scanning Electron Microscopy (FIB-SEM). Strikingly, 3D reconstructions from RAB34 mutant cells consistently revealed multiple small vesicles docked to the distal appendages without any evident ciliary vesicle (seen for 7 RAB34 knockout cells analyzed; for 2 additional cells, details were obscured by neighboring organelles; Figures 2G and S2C). Thus, DAVs are recruited normally in RAB34 mutant cells, but fusion of DAVs to form a ciliary vesicle is defective, a phenotype similar to that seen upon EHD1 knockdown.7
Figure 2. Rab34 is required for formation of the ciliary vesicle from distal appendage vesicles.
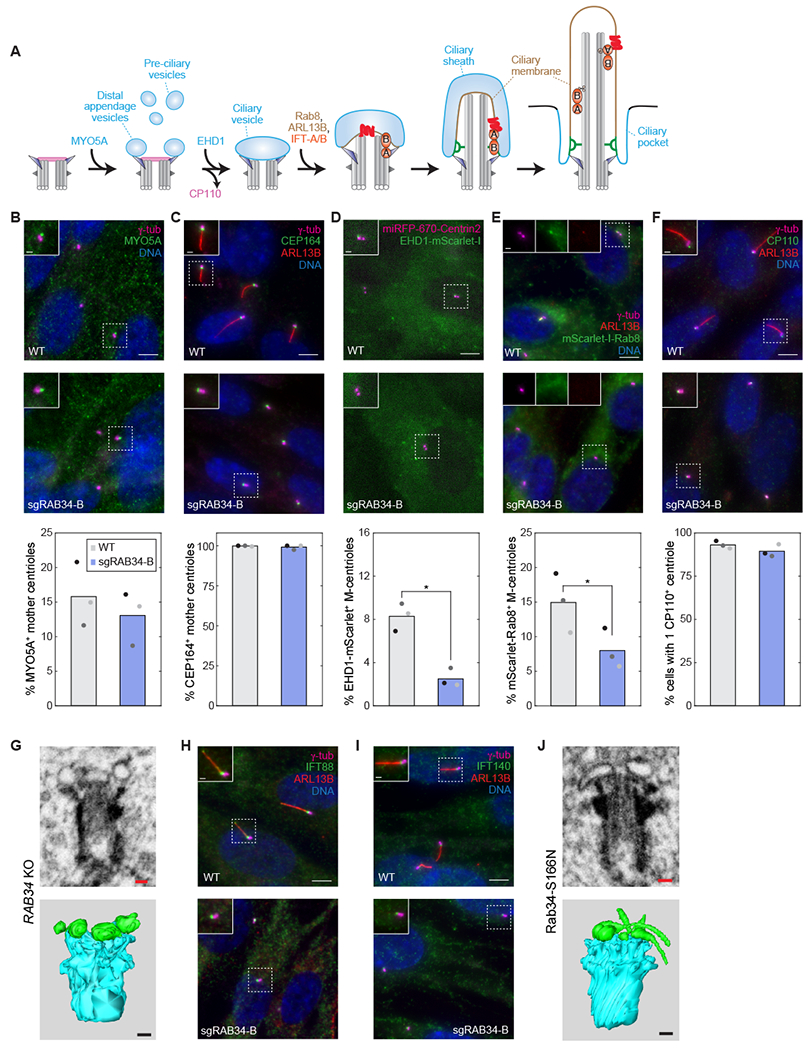
A) Schematic illustration of the intracellular ciliogenesis pathway, highlighting the stepwise formation of a ciliary vesicle associated with the mother centriole’s distal appendages, the growth of this membrane and the axonemal microtubules to yield a nascent intracellular cilium, and the subsequent fusion of the ciliary sheath with the plasma membrane that exposes the cilium to the external environment. IFT-complexes are indicated in orange and a ciliary membrane protein in red. B-F) The indicated markers were examined in RPE1 WT and RAB34 sgRNA-B cells. Distal appendage protein CEP164 and pre-ciliary vesicle trafficking motor MYO5A were immunostained after 48 h and 2 h serum starvation, respectively. Cells stably expressing EHD1-mScarlet-I and miRFP-670-Centrin2 were imaged live 4 h after serum starvation, and cells expressing mScarlet-I-Rab8, marking assembling cilia, were stained 24 h after serum starvation. For dynamic recruitment of MYO5A, EHD1 and Rab8, timepoints were chosen to match when these events occur during ciliogenesis; otherwise, cells were analyzed 48 h after serum starvation when mature cilia have formed. At bottom, the percentage of cells exhibiting localization of the indicated makers to the mother centriole (M-centriole) or cilium (nascent or mature) is shown. Bars indicate means and dots show values from N=3 independent experiments. P-values determined by two-sided paired t-test are 8.18x10−3 and 2.19x10−2 for EHD1 and Rab8, respectively; all other comparisons were not significant (P > 0.1). Scale bars: 5 μm (insets: 1 μm). G) Volumetric analysis of centrioles and associated membranes by focused ion beam scanning electron microscopy (FIB-SEM) was conducted in RAB34 knockout RPE1 cells (sgRAB34-B pool). 7x7x7nm voxels were acquired and individual slices as well as 3D reconstructions are shown (centrioles are segmented in cyan and vesicles in green). Scale bars: 100 nm. See also Figure S2C. H–I) IFT-B component IFT88 and IFT-A component IFT140 were examined after 48 h serum starvation in RPE1 WT and RAB34 sgRNA-B cells. Scale bars: 5 μm (insets: 1 μm). J) Volumetric analysis of centrioles and associated membranes by FIB-SEM was conducted as in (G) for doxycycline-induced RPE1 cells expressing GFP-Rab34-S166N. Scale bars: 100 nm. See also Figure S2.
In addition to ciliary membrane formation, ciliogenesis requires removal of capping protein CP110 from the mother centriole and recruitment of the intraflagellar transport (IFT) complexes IFT-A and IFT-B (Figure 2A).30 In RAB34 mutant cells, CP110 was removed normally, and IFT88 (IFT-B) and IFT140 (IFT-A) were still present at mother centrioles (Figures 2F, 2H–I, and S2A–B).
We next evaluated cells expressing dominant-negative Rab34 and found that, as in RAB34 knockout cells, Rab34-S166N did not inhibit CEP164 localization, CP110 removal, or IFT complex recruitment (Figure S2D–G). FIB-SEM reconstructions showed that cells expressing Rab34-S166N also arrested ciliogenesis with DAVs docked to the mother centriole (5 of 6 cells examined). However, the DAVs displayed a striking elongated or tubular morphology (Figures 2J and S2C). This finding strongly suggests that Rab34 acts directly on DAVs and that Rab34-S166N alters membrane fusion or remodeling at the mother centriole.
The ability to inducibly express Rab34-S166N allowed us to examine the effect of disrupting Rab34 function after cilium assembly. Interestingly, progressive loss of cilia was observed after induction of Rab34-S166N but not Rab34-WT (Figure S2H–I). Rab34 is therefore also needed to maintain cilia in cultured cells. Moreover, treating cells with the actin poison cytochalasin D (cytoD) rescued this defect. Thus, the ability of cytoD to stabilize cilia31 bypasses the need for Rab34 in cilium homeostasis (Figure S2J).
Rab34 dynamically localizes to the mother centriole and cilium during ciliogenesis
Because Rab GTPases are recruited to the membrane structures they regulate, we next examined Rab34 localization. Notably, staining with two antibodies revealed colocalization of Rab34 with ciliary markers in ~10% of ciliated RPE1 cells (Figures 3A and 3D). We also commonly observed a punctum of Rab34 at the mother centriole in unciliated cells, and similar localizations were observed in NIH-3T3 cells (Figure 3A–B). In serum-fed, sub-confluent RPE1 cells, ~12% of mother centrioles were Rab34-positive, while ~35% of mother centrioles became Rab34-positive early during ciliogenesis (Figures S3A and 3C). At this stage (4 h after serum starvation), only ~10% of mother centrioles were marked by ARL13B, suggesting that Rab34 is recruited before ARL13B (Figure 3C). By contrast, at later timepoints, the fraction of cilia with Rab34 decreased (Figure 3D). Thus, Rab34 may act early during ciliogenesis and be selectively present on nascent cilia.
Figure 3. Rab34 dynamically localizes to the mother centriole and cilium during ciliogenesis.
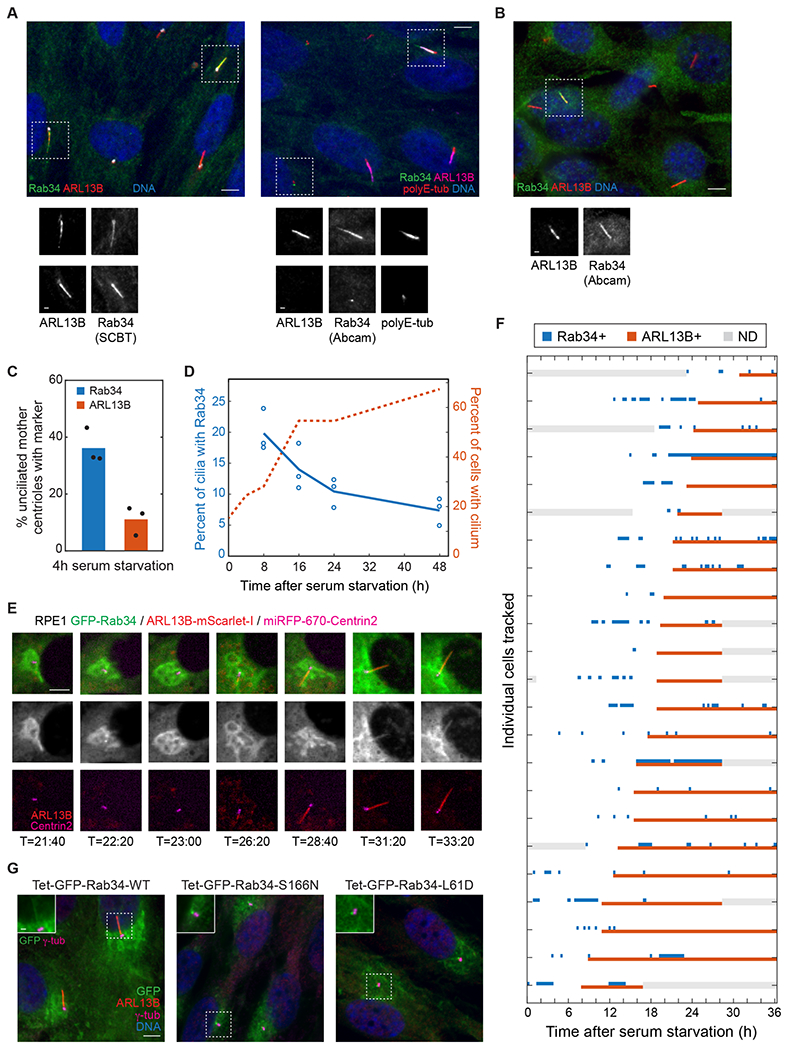
A) RPE1 cells were stained for the indicated ciliary and centriolar markers as well as for Rab34 using antibodies from Santa Cruz Biotechnology (SCBT) or Abcam. Scale bars: 5 μm (insets: 1 μm). B) NIH-3T3 cells were stained with the indicated antibodies as in (A). C) The percent of unciliated centrioles that have recruited puncta of Rab34 or ARL13B was determined 4 h after serum starvation. Bars indicate means and dots show values from each of N=3 independent experiments. D) The percentage of cilia (marked by ARL13B) that are positive for Rab34 is shown for the indicated times after serum starvation (left-hand Y-axis; the earliest timepoints are omitted due to the low level of ciliation). The percentage of cells with ARL13B-marked cilia is plotted on the right-hand Y-axis. Circles indicate values from individual experiments, and lines show mean from N=3 independent experiments. E) Time-lapse imaging of RPE1 cell line expressing GFP-Rab34, ARL13B-mScarlet-I, and miRFP-670-Centrin2 was performed, and select images of a representative cell are shown at the indicated times after serum starvation. Scale bar: 5 μm. F) Localization of GFP-Rab34 and ARL13B-mScarlet-I was assessed for 23 ciliating cells tracked for up to 36 h after serum starvation. Timepoints when GFP-Rab34 or ARL13B-mScarlet-I were detected at the mother centriole or cilium are indicated by blue and red boxes, respectively; gray boxes indicate when localization could not be determined (ND, e.g. due to cell leaving field of view). Cells are sorted by time of ARL13B arrival and cilium extension. G) Localization of GFP-tagged Rab34 was assessed in RPE1 cells stably expressing wildtype Rab34 or the indicated variants. Doxycycline was added prior to and during 48 h serum starvation. Scale bar: 5 μm (inset: 1 μm). See also Figure S3.
To further understand Rab34 localization dynamics, we carried out live-cell imaging in cells stably expressing ARL13B-mScarlet-I, miRFP-670-Centrin2, and GFP-Rab34. By directly monitoring cilium assembly, we observed, first, that all ciliating cells dynamically recruited Rab34 to the mother centriole during ciliogenesis (Figure 3E–F). Second, Rab34 consistently arrived at the mother centriole before ARL13B. Third, most cells exhibited dynamic ciliary localization of Rab34 after an ARL13B-positive cilium formed. Overall, the Rab34 localization dynamics we observed are consistent with a key role for Rab34 in ciliary membrane formation.
We also examined the localization of dominant-negative GFP-Rab34-S166N and GFP-Rab34-L61D and found that these proteins localized as a bright punctum at the mother centriole in ~90% of cells (Figures 3G and S3B). Given that these cells also exhibit distended DAVs (Figure 2J), it is likely that Rab34 is present on DAVs and that GTP-locked mutants become trapped on these structures, impairing DAV fusion.
Rab34 localization and function are specific to intracellular ciliogenesis
Because Rab34 is found on a subset of cilia that declines during the course of ciliogenesis (Figure 3D), we next asked whether Rab34 localizes to nascent intracellular cilia. To distinguish nascent intracellular (‘inside’) cilia from mature extracellular (‘outside’) cilia, we used the In/Out assay.32 Specifically, we used an anti-GFP Nanobody (Nb)33 to stain pHluorin-Smo in surface-exposed cilia of live cells, followed by fixation and anti-Rab34 immunostaining. Strikingly, even though most cilia were extracellular after 48 h of serum-starvation, all Rab34-positive cilia were negative for GFP Nb staining and therefore intracellular (Figure 4A–B). Furthermore, Rab34 localized to the majority of inside cilia (Figure S3C), indicating that Rab34 is a GTPase marker of intracellular cilia. We note that similar results were also obtained with an antibody-based In/Out assay protocol (Figure S3D–E). Thus, Rab34 localizes to a small fraction of cilia because it is found selectively on nascent intracellular cilia.
Figure 4. Rab34 localizes to the ciliary sheath of intracellular cilia and is dispensable for extracellular ciliogenesis in MDCK cells.
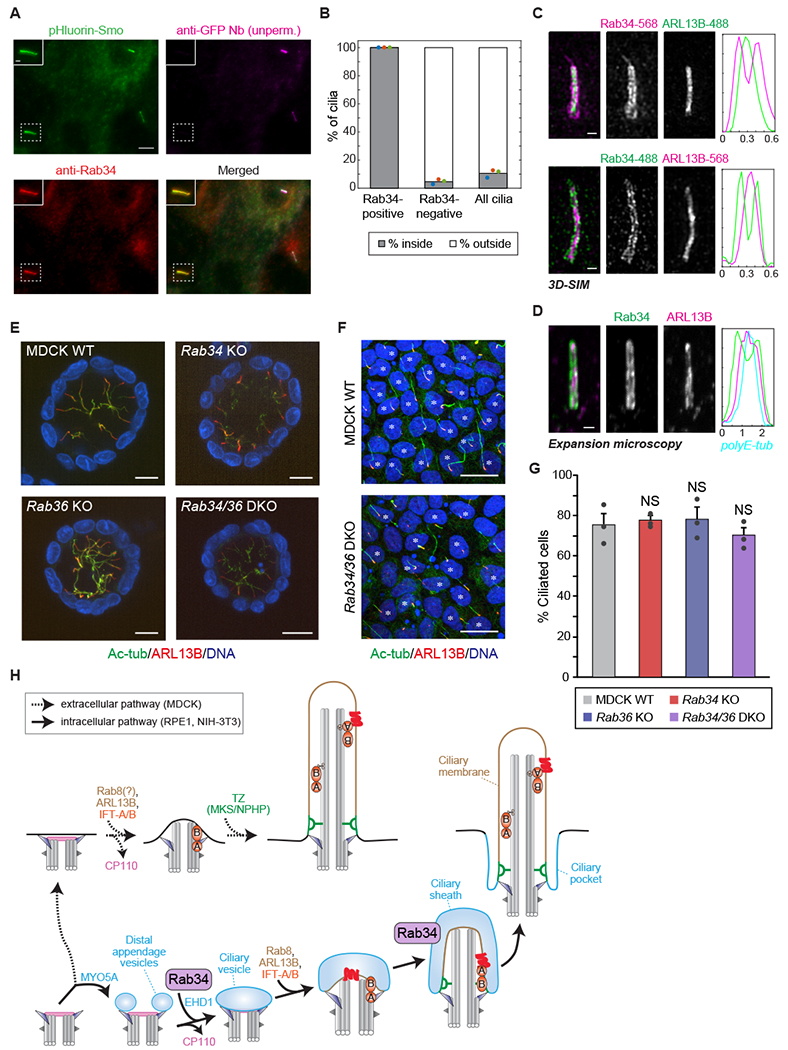
A) RPE1 cells expressing the pHluorin-Smo In/Out reporter were stained prior to permeabilization with anti-GFP nanobody (Nb) to reveal intracellular versus extracellular cilia followed by permeabilization and staining for Rab34 (Abcam antibody). Inset shows an intracellular (inside) cilium that is positive for Rab34. Scale bar: 5 μm (inset: 1 μm). B) Quantification of the fraction of cilia that are inside versus outside is shown for the cilia classes indicated. Dots indicate values from individual experiments, and bars indicate means from N=3 independent experiments encompassing 110 Rab34-positive cilia and 1,651 Rab34-negative cilia. See also Figure S3C. C) 3D-SIM (structured illumination microscopy) was performed on RPE1 cells stained for Rab34 and ARL13B. Graphs at right show normalized fluorescence intensity along line-profiles perpendicular to the longitudinal axis of the cilium. Scale bars: 100 nm. See also Figure S3F. D) Expansion microscopy of cells stained for Rab34 and ARL13B. Graph at right shows normalized fluorescence intensity along line-profile across the cilium, including for axonemal polyE-tubulin. Scale bar: 2 μm (after ~4-fold expansion). See also Figure S3G. E) Representative images of 3D cysts of parental, Rab34 knockout (KO), Rab36 KO, and Rab34/36 double-knockout (DKO) MDCK cells. The cells were fixed after 7-day culture in collagen gel and then stained for acetylated tubulin (Ac-tub; green), ARL13B (red), and DAPI (blue). Scale bars: 10 μm. F) Representative images of 2D monolayers of parental and Rab34/36 DKO MDCK cells. The cells were fixed 5 days after seeding and then stained as in (E). Scale bars: 10 μm. Asterisks indicate ciliated cells. G) The percentage of ciliated cells in 2D monolayers of parental, Rab34 KO, Rab36 KO, and Rab34/36 DKO MDCK cells is shown. Dots indicate values from individual experiments, bars indicate means from N=3 independent experiments, and error bars indicate standard error. Differences that are not significantly different are marked “NS” (Tukey-Kramer test; P > 0.60). H) Model for Rab34 function, in which it mediates ciliary membrane formation in the intracellular ciliogenesis pathway (bottom, solid arrows) but not the extracellular ciliogenesis pathway (top, dashed arrows). Rab34 is present on DAVs and the ciliary sheath (blue) but not the ciliary membrane (tan). See also Figures S3 and S4.
Intracellular cilia possess two adjacent membrane domains: the ciliary membrane and the surrounding ciliary sheath (Figure 2A). To determine which of these membranes harbors Rab34, we used 3D structured illumination microscopy (3D-SIM). Notably, Rab34 fluorescence consistently surrounded the ARL13B-labeled ciliary membrane (Figures 4C and S3F), as reported for ciliary sheath proteins such as EHD1 and MYO5A.8 We obtained similar results using expansion microscopy, in which samples are embedded in a swellable polymer to allow ~4-fold isotropic expansion.34,35 In expanded samples, Rab34 fluorescence surrounded ARL13B, with polyE-tubulin marking the center of the cilium (Figures 4D and S3G). These results establish that Rab34 localizes to the ciliary sheath and identify Rab34 as the first GTPase enriched on the ciliary sheath. Notably, although the sheath is a precursor to the ciliary pocket, Rab34 was largely absent from the ciliary pocket in RPE1 cells, as 25 of 26 EHDl-positive ciliary pockets lacked Rab34 staining (Figure S3H).
Because Rab34 localizes to structures unique to intracellular ciliogenesis, we hypothesized that Rab34 might be dispensable for extracellular ciliogenesis. As a model of extracellular ciliogenesis, we used the epithelial Madin-Darby Canine Kidney II (MDCK) cell line. During cilium assembly in MDCK cells, the mother centriole migrates to the apical surface before ciliary membrane formation, and intracellular intermediates are not readily detected by the In/Out assay.10,36,37 We used CRISPR to target Rab34 and its paralog Rab36 in MDCK cells and confirmed knockout by Western blot and DNA sequencing38 (Figure S4A). Consistent with our hypothesis, we did not observe any ciliogenesis defect in Rab34 mutant MDCK cells grown in 3D cysts or polarized 2D monolayers, nor did we observe any change in cilium length (Figures 4E, 4G and, S4B). Rab34/Rab36 double-knockout cells also exhibited normal cilia, ruling out the possibility that Rab34 and its paralog Rab36 function redundantly in MDCK ciliogenesis (Figure 4E–G). Finally, while Rab34 is expressed in MDCK cells, we did not observe Rab34 localization at cilia or centrioles in ciliating MDCK cells (Figures S4A and S4C). Thus, extracellular ciliogenesis in MDCK cells is independent of Rab34 and Rab36.
Discussion
Here, we found that Rab34 is a key mediator of ciliary membrane formation in the intracellular ciliogenesis pathway (Figure 4H). While Rab34 was recently suggested to promote early stages of ciliary membrane formation and to localize to some cilia12,13, we now show that Rab34 mediates ciliary vesicle formation and is specifically found on the ciliary sheath of intracellular cilia. Rab34 is also dynamically recruited to the mother centriole during ciliogenesis, and the persistent localization of Rab34-S166N at this site suggests that Rab34 is present on DAVs (Figure 3C–G). Rab34 may therefore establish the membrane identity of DAVs and the ciliary sheath. Because the ciliary sheath faces the cytoplasm, Rab34 may also promote vesicular trafficking to the nascent cilium and/or fusion of the ciliary sheath with the plasma membrane. Additionally, Rab34 may have a role in membrane trafficking to mature cilia given the impairment of cilium maintenance seen upon Rab34-S166N expression. A clearer picture of Rab34’s roles beyond DAV fusion awaits the development of tools to rapidly inactivate Rab34 during ciliogenesis. How Rab34’s localization is dynamically regulated will also be an important area for future study.
Rab34 is an atypical GTPase with key residues that differ from other family members. These divergent residues have critical roles, as Rab34-S166N and Rab34-L61D are GTP-locked dominant-negative mutants that block ciliogenesis (Figures 1H–I and S1F–G). Further insight into the Rab34 enzymatic cycle and the impact of these mutations awaits the identification of GEFs and GAPs that regulate Rab34. Similarly, Rab34’s effectors that mediate ciliogenesis remain unknown. We anticipate that the GTP-locked Rab34 mutants identified here will be valuable tools to uncover Rab34 effectors and regulators.
Our data indicate that cells using the extracellular pathway can form cilia independently of Rab34 (Figure 4E–G). It remains unclear if other proteins such as EHD1 and MYO5A also have selective roles in intracellular ciliogenesis. Both proteins promote ciliogenesis in RPE1 and IMCD3 cells, and Ehd gene knockdown in zebrafish impacts cilia in some epithelial tissues. However, mice lacking these proteins exhibit no (or variable) ciliary defects, and their roles have not yet been examined in MDCK cells.7,8,39–43 A pathway-specific role for Rab34 is nonetheless consistent with the fact that, unlike many genes needed for ciliogenesis, Rab34 is not conserved in all ciliated organisms but is restricted to metazoans.44 Indeed, one possibility is that Rab34 is selectively present in organisms that employ the intracellular ciliogenesis pathway.
Lastly, our findings provide a framework for understanding the physiologic roles of Rab34 and the consequences of its inactivation. Homozygous Rab34 mutant mice die perinatally and exhibit ciliopathy features such as polydactyly and craniofacial malformations.12,45 However, these phenotypes are milder than the early embryonic lethality seen for mutants that cause universal cilium loss46 and are instead consistent with disruption of a subset of ciliated tissues. For example, neural tube patterning is apparently normal in some Rab34 mutant embryos, which is consistent with these epithelial cells lacking a pronounced ciliary pocket and likely ciliating via the extracellular pathway.47–49 Conversely, limb buds of Rab34 mutant mice exhibit polydactyly and have fewer cilia.12 It is noteworthy that cells of the limb bud mesenchyme have a pronounced ciliary pocket and likely ciliate via the intracellular pathway.50 Cerebellar granule neuron progenitors also likely ciliate via the intracellular pathway51, and the limb bud and cerebellum are both tissues where Rab34 is induced by Hh signaling.52,53 Although the functional significance of Rab34 induction is unclear, one possibility is that Rab34 promotes cilium re-formation after mitogenic Hh signaling, as recently reported for Atohl.54 Finally, while patients with RAB34 mutations have not yet been reported, understanding the clinical manifestations of RAB34 disruption is likely to provide new insights into tissue-specific modes of cilium assembly and into the phenotypic variability seen in ciliopathies.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Breslow (david.breslow@yale.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact.
Data and code availability
Raw data not included in this article are available from the corresponding author on request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mammalian cell lines
NIH-3T3, RPE1-hTERT (RPE1), and HEK-293T cell lines were obtained from ATCC; MDCK-II cells were obtained from Katsuhiko Mikoshiba (ShanghaiTech University). Cultured cells were maintained in a humidified 37°C incubator with 5% CO2. NIH-3T3, HEK-293T, and MDCK-II cells were cultured in DMEM, high glucose (Gibco) supplemented with 10% fetal bovine serum (FBS; Sigma Aldrich), 100 units/ml Penicillin, 100 μg/ml Streptomycin, 2 mM Glutamine, and 1 mM sodium pyruvate (Gibco). RPE1 cells were cultured in DMEM/F-12 medium (Gibco) supplemented with 10% FBS, 100 units/ml of Penicillin, 100 μg/ml Streptomycin, and 2 mM Glutamine. To induce ciliogenesis, NIH-3T3 cells were starved in 0.5% FBS-containing medium for 24 h, and RPE-1 cells were starved in 0.2% FBS-containing medium for 48 h. For analysis of ciliogenesis of MDCK cells grown in two-dimensional (2D) monolayers, cells were cultured until confluence was achieved (5 days after seeding). Three-dimensional (3D) MDCK cysts were formed by culturing cells in collagen I gel (KOKEN #IPC-50), as described previously.38 Where specified, cells were treated with 100 nM cytochalasin D (Sigma Aldrich) or with 1 μg/ml doxycycline (Fisher Scientific). Cells were verified to be mycoplasma-negative using MycoAlert Plus detection kit (Lonza).
E. Coli
BL21(DE3) cells were used for recombinant protein expression and grown in LB medium.
METHOD DETAILS
DNA cloning
Gibson assembly, Gateway cloning, and standard molecular biology were used for DNA cloning. Human RAB34 cDNA was obtained from GE Healthcare (MHS6278-202807876) and cloned into Gateway pENTR plasmid pDONR221 (Invitrogen); RAB34 mutants were prepared by site-directed mutagenesis of pENTR-Rab34. Plasmids for tetracycline-inducible expression of LAP-Rab34 (LAP consists of GFP, TEV protease site, and S-tag) were constructed by modification of pCW-Cas9 (Addgene #50661, gift from Eric Lander and David Sabatini). Stable expression of fluorescently tagged ciliary and centriolar proteins was achieved by modification of pCW-Cas9 using cDNAs for the PGK promoter, ARL13B (Addgene #40879, gift from Tamara Caspary), EHD1 (gift from Chris Westlake), Rab8 (gift from Maxence Nachury), and Centrin2 (gift from Tim Stearns), miRFP-670 (Addgene #79987, gift from Vladislav Verkhusha), and mScarlet-I (Addgene #85044, gift from Dorus Gadella). Plasmids for CRISPR-based knockout were constructed using pMCB320 (Addgene #89359, gift from Michael Bassik) and pMJ179 (Addgene #89556, gift from Jonathan Weissman); see Key Resources Table for sgRNA sequences. Plasmids for transient transfection of RAB34 were made by Gateway cloning using pEF5-FRT-LAP-DEST55,56; for bacterial protein expression, RAB34 variants were cloned into pGEX-6P1 (GE Healthcare).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-ARL13B | Antibodies Inc. / NeuroMab | Cat#75-287; RRID: AB_2341543 |
| Rabbit anti-ARL13B | Proteintech | Cat#17711-1-AP; RRID: AB_2060867 |
| Rabbit anti-CP110 | Proteintech | Cat#12780-1-AP; RRID: AB_10638480 |
| Rabbit anti-CEP164 | Sigma Aldrich | Cat#HPA037606; RRID: AB_10672498 |
| Rabbit anti-GDI2 | Proteintech | Cat#10116-1-AP: RRID: AB_2279073 |
| Chicken anti-GFP | Invitrogen | Cat#A10262; RRID: AB_2534023 |
| Rabbit anti-GFP | Invitrogen | Cat#A11122; RRID: AB_221569 |
| Rabbit anti-IFT88 | Proteintech | Cat#13967-1-AP; RRID: AB_2121979 |
| Rabbit anti-IFT140 | Proteintech | Cat#17460-1-AP; RRID: AB_2295648 |
| Rabbit anti-MYO5A | Cell Signaling Technology | Cat#3402; RRID: AB_2148475 |
| Rabbit anti-Rab34 | Abcam | Cat#ab73383; RRID: AB_1952423 |
| Mouse anti-Rab34 | Santa Cruz Biotechnology | Cat#sc-376710; RRID: AB_11150504 |
| Rabbit anti-Rab34 | Proteintech | Cat#27435-1-AP; RRID: AB_2880870 |
| Rabbit anti-Rab34 | Fukuda lab13 | N/A |
| Rabbit anti-Rab36 | Fukuda lab26 | N/A |
| Rabbit anti-RILPL1 | Sigma Aldrich | Cat#HPA041314; RRID: AB_10795587 |
| Mouse anti-acetylated tubulin | Sigma Aldrich | Cat#T6793; RRID: AB_477585 |
| Mouse anti-gamma tubulin | Sigma Aldrich | Cat#T5326; RRID: AB_532292 |
| Mouse anti-glutamylated tubulin | Adipogen | Cat#AG-20B-0020; RRID: AB_2490210 |
| Donkey anti-Rabbit (Alexa Fluor 488, Cy3, Cy5) | Jackson Immunoresearch | Cat#711-[165/545/605]-152 |
| Goat anti-Mouse IgG1 (Alexa Fluor 488, Cy3, Cy5) | Jackson Immunoresearch | Cat#115-[165/545/605]-205 |
| Goat anti-Mouse IgG2a (Alexa Fluor 488, Cy3, Cy5) | Jackson Immunoresearch | Cat#115-[165/545/605]-206 |
| Goat anti-Mouse IgG2b-Cy3 | Jackson Immunoresearch | Cat#115-165-207 |
| Donkey anti-Rabbit-Alexa Fluor 568 | Abcam | Cat#ab175470; RRID: AB_2783823 |
| Goat anti-Mouse IgG1 CF-770 | Biotium | Catalog#20254; RRID: AB_10853480 |
| Goat anti-Mouse IgG2a-Alexa Fluor 568 | Invitrogen | Cat#A-21134; RRID: AB_2535773 |
| Goat anti-Chicken-Alexa Fluor 488 | Invitrogen | Cat#A-11039; RRID: AB_2534096 |
| Goat anti-Mouse-Alexa Fluor Plus 555 | Invitrogen | Cat#A-32727; RRID: AB_2633276 |
| Goat anti-Rabbit-Alexa Fluor 488 | Invitrogen | Cat#A-11034; RRID: AB_2576217 |
| Bacterial and Virus Strains | ||
| BL21 DE3 | Lucigen | Cat#60401 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protein-A-HRP | Invitrogen | Cat#101023 |
| Rab34 (WT, L61D, S166N) | This paper | |
| Anti-GFP Nanobody Alexa Fluor 647 | Breslow lab55 | N/A |
| GFP Trap | Chromotek | Cat#gta-20 |
| Collagen I | KOKEN | Cat#IPC-50 |
| Cytochalasin D | Sigma Aldrich | Cat#C8273 |
| Dialyzed fetal bovine serum | Gibco | Cat#A3382001 |
| Fluoromount-G mounting medium | Electron Microscopy Sciences | Cat#17984-25 |
| HRV3C protease | Pierce | Cat#88946 |
| Lenti-X Concentrator | Takara Biosystems | Cat#631231 |
| 32P orthophosphate | Perkin Elmer | Cat# NEX053H002MC |
| PEI Max (polyethyleneimine) | Polysciences | Cat#24765-1 |
| ProLong Diamond mounting medium | Thermo Fisher | Cat#P36965 |
| Sodium acrylate | Combi-Blocks | Cat#QC-1489 |
| X-tremeGENE 9 | Sigma Aldrich | Cat#6365779001 |
| Experimental Models: Cell Lines | ||
| Mouse: NIH-3T3 [Shh-BlastR;Cas9] cell line | Breslow lab14 | N/A |
| Human: RPE1-hTERT cell line | ATCC | Cat#CRL-4000; RRID: CVCL_4388 |
| Dog: MDCK II cell line | Katsuhiko Mikoshiba (ShanghaiTech Univ.) | Cat#RCB5148 (RIKEN BRC) |
| Dog: MDCK Rab34 KO, Rab36 KO, Rab34/Rab36 DKO cell lines | Fukuda lab38 | Cat#RCB5133, 35, 46 (RIKEN BRC) |
| Human: HEK293T cell line | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
| Oligonucleotides | ||
| Primer: ATCGTTCCATCTCGAAGTCC, Reverse primer for human RAB34 amplification and sequencing | This study | N/A |
| Primer: GGGTGTCTTAGAATATAGGAC, Forward primer for human RAB34 amplification and sequencing | This study | N/A |
| sgRNA: TTGAACCTCTCCTGCCCAG, sgRNA-A for human RAB34 | This study | N/A |
| sgRNA: ATCTCCAAGGTCATTGTGG, sgRNA-B for human RAB34 | This study | N/A |
| sgRNA: GAATTACAAGGCTACCAT, sgRNA-A for mouse Rab34 | This study | N/A |
| sgRNA: TGGCTCACTGATGCCCTCA, sgRNA-B for mouse Rab34 | This study | N/A |
| Recombinant DNA | ||
| pGro7 (GroEL/ES) | Yong Xiong, Yale Univ. | N/A |
| pGEX-6P-1-Rab34 (WT, L61D, and S166N) | This study | N/A |
| RAB34 (human) cDNA | GE Healthcare | Cat# MHS6278-202807876 |
| pCW-Tet-LAP-Rab34 | This Study | |
| pCW-Cas9 | Addgene | Cat#50661; RRID: Addgene_50661 |
| Lenti-Cas9-Blast | Addgene | Cat#52962; RRID: Addgene_52962 |
| pEF5-FRT-LAP-DEST | Breslow lab55,56 | N/A |
| pHR-Pgk-Cas9-BFP | Breslow lab14 | N/A |
| pDONR-221 | Invitrogen | Cat#12536017 |
| pCW-Pgk-ARL13B-mScarlet-I_Pgk-miRFP-670-Centrin2 | This study | N/A |
| pCW-Pgk-EHD1-mScarlet-I_Pgk-miRFP-670-Centrin2 | This study | N/A |
| pCW-Pgk- mScarlet-I-Rab8a_Pgk-NeoR | This study | N/A |
| pMCB320-sgRNA_PuroR-T2A-mCherry | Addgene | Cat#89359; RRID: Addgene_89359 |
| pMJ179-sgRNA_PuroR-T2A-BFP | Addgene | Cat#89556; RRID: Addgene_89556 |
| pMD2.G | Addgene | Cat#12259; RRID: Addgene_12259 |
| pRSV-Rev | Addgene | Cat#12253; RRID: Addgene_12253 |
| pMDLg/RRE | Addgene | Cat#12251; RRID: Addgene_12251 |
| pCMV-ΔR-8.91 | Bob Weinberg | N/A |
| pCMV-VSVG | Addgene | Cat#8454; RRID: Addgene_8454 |
| Software and Algorithms | ||
| Nikon Elements AR | Nikon Instruments | RRID: SCR_014329 |
| Fiji (ImageJ) | NIH | RRID: SCR_002285 |
| SmartSEM | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/int/products/microscope-software/smartsem.html |
| Atlas engine 5 | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/int/products/microscope-software/atlas.html |
| DragonFly | Object Research Systems | https://www.theobjects.com/dragonfly/index.html |
| IMOD | See 57,58 | RRID: SCR_003297 |
| ImageQuant | GE Healthcare | RRID: SCR_014246 |
| Softworx | GE Healthcare | RRID: SCR_019157 |
| Matlab | The Mathworks | RRID: SCR_001622 |
Lentivirus production and cell line generation
VSVG-pseudotyped lentiviral particles were produced by transfection of HEK293T cells with a lentiviral vector and packaging plasmids (pMD2.G, pRSV-Rev, pMDLg/RRE for sgRNAs; pCMV-ΔR-8.91 and pCMV-VSVG for protein-coding constructs). Following transfection using polyethyleneimine (Polysciences #24765-1), virus-containing supernatant was collected 48 h later and filtered through a 0.45 μm polyethersulfone filter (VWR 28145-505). For protein-coding constructs, lentiviral particles were concentrated 10-fold using Lenti-X Concentrator (Takara Biosystems).
Cells were transduced by addition of viral supernatants diluted to an appropriate titer in medium containing 4 mg/ml polybrene (Sigma Aldrich H9268). Following 24 h incubation at 37°C, virus-containing medium was removed, and cells were passaged and selected, as appropriate, using 2.0-8.0 μg/ml puromycin or 300 μg/ml G418 or 8.0 μg/ml blasticidin (InvivoGen). Alternatively, transduced cells were isolated by FACS using a FACSAria III sorter (Becton Dickinson). Unless otherwise indicated, polyclonal pools of transduced cells were used in subsequent experiments.
For CRISPR-based mutagenesis of NIH-3T3 cells, a cell line stably expressing Cas9 was used (3T3-[Shh-BlastR];Cas9).14 For mutagenesis of RPE1 cells, a cell line stably expressing Cas9 was generated by infection with lentiCas9-Blast (Addgene #52962, gift from Feng Zhang) and selection with blasticidin. Cas9-expressing cells were transduced with sgRNA constructs followed by antibiotic selection or FACS-based sorting of clones. Mutant alleles in knockout clones were assessed by sequencing of genomic DNA extracted using the QIAamp DNA Mini kit (Qiagen). The target locus was amplified using flanking primers (see Key Resources Table) and subjected to Sanger sequencing. Generation of Rab34, Rab36 and Rab34/Rab36 knockout MDCK cells was described previously.38
Staining for immunofluorescence microscopy
Staining for immunofluorescence microscopy was performed as described previously 14. Briefly, cells were seeded on acid-washed 12-mm #1.5 coverslips (Fisher Scientific). Cells were either fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 10 min or −20°C methanol for 10 min or 4% paraformaldehyde for 10 min followed by methanol or 10% TCA for 10 min (for MDCK cysts). Coverslips were permeabilized for 10 min in PBS containing 0.1 % Triton X-100, washed with PBS, and blocked for 20 min in PBS supplemented with 5% normal donkey serum (Jackson Immunoresearch) and 3% Bovine Serum Albumin (BSA). Coverslips were then incubated with appropriate primary antibodies diluted in PBS with 3% BSA at room temperature for 1 h, washed five times, incubated with secondary antibodies for 30-60 min at room temperature, washed again, stained with Hoechst 33258 dye, and mounted on glass slides in Fluoromount-G mounting medium (Electron Microscopy Sciences). MDCK cysts were transferred in PBS to glass-bottom dishes for imaging. Primary and secondary antibodies used are listed in the Key Resources Table.
In/Out Assay
In/Out Assay with anti-GFP antibody was carried out as described previously 32. Briefly, coverslips were fixed in 4% paraformaldehyde for 10 min and blocked in PBS containing 5% normal donkey serum and 3% BSA for 20 min. After blocking, unpermeabilized coverslips were incubated with rabbit anti-GFP antibody for 30 min at room temperature. After washing, coverslips were fixed again in 4% paraformaldehyde, permeabilized with PBS containing 0.1% Triton X-100, blocked again, and incubated with anti-Rab34 antibody (SCBT) for 1 h at room temperature. Coverslips were then washed, stained with secondary antibodies, and mounted on glass slides as described above.
The In/Out Assay with anti-GFP nanobody (Nb) was based on a modified version of the above protocol. Coverslips were placed on ice, washed with cold HKM-E1 buffer (20 mM HEPES, pH 7.4, 115 mM KOAc, 1 mM MgCl2, and 1 mM EGTA), and then incubated for 10 min on ice in HKM-E1 buffer supplemented with 0.1% BSA and with 70 nM Alexa Fluor 647-labeled anti-GFP Nb 33,55. After incubation with GFP Nb, coverslips were washed three times with HKM-E1 buffer, fixed in 4% paraformaldehyde, and shifted to room temperature. Coverslips were then permeabilized with PBS containing 0.1% Triton X-100 for 10 min, blocked, stained with anti-Rab34 antibody (Abcam), and processed as described above.
Sample preparation for expansion microscopy
Cells were seeded on 25mm #1.5 coverslips (Electron Microscopy Sciences) and prepared as in Sahabandu et al.35 Briefly, cells were fixed in 4% formaldehyde for 1 h, incubated in 30% acrylamide/4% formaldehyde at 37°C for 16 h, washed 3x 10min in PBS and then cooled on ice water. Pre-cooled gelling solution (20% Acrylamide, 0.04% bis-acrylamide, 7% sodium acrylate. 0.5% APS and 0.5% TEMED) was then added, and polymerization was allowed to proceed for 30 min on ice and 30-60 min at room temperature. 4-mm gel punches were taken using a biopsy punch (Integra Miltex , 33-34-P/25) and then denatured in SDS solution (50 mM Tris pH 9.0, 200 mM SDS, 200 mM NaCl) for 1 h at 90°C. Gel punches were allowed to cool, and SDS solution was washed out with PBS (9x 20 min washes) followed by incubation overnight in PBS at 4°C.
Denatured punches were then blocked in immunofluorescence buffer (1% BSA, 0.05% Tween-20 in PBS) for 2 h and incubated overnight at 4°C with primary antibodies in immunofluorescence buffer (see Key Resources Table). Punches were washed in PBS (6x 10 min), then incubated with secondary antibodies and Hoechst 33258 overnight at 4°C. Finally, stained punches were washed in water (12x 10 min), then expanded overnight in deionized water at 4°C. Gels were transferred to 35mm glass-bottom imaging dishes (Mattek P35G-1.5-14-C), immobilized using low-melt agarose, and imaged as described below. We estimate from the final gel size and the size of expanded nuclei that gels expanded ~4-fold, as previously reported.35
Fluorescence microscopy
Coverslips with fixed NIH-3T3 and RPE1 cells were imaged using a Nikon Eclipse Ti-2 widefield microscope equipped with a CMOS camera (Photometrics Prime BSI or Hamamatsu Orca-Fusion), a 60× PlanApo oil objective (NA 1.40; Nikon Instruments), and an LED light source (Lumencor Spectra X or SOLA-V-NIR). Fixed samples were mounted in Fluoromount-G and images acquired at room temperature or 37°C using Nikon Elements software. MDCK cells cultured as 2D monolayers were imaged on an Olympus FV1000 confocal microscope using a 60× UPLSAPO oil objective (NA 1.35) controlled with Fluoview software (Olympus). MDCK cells cultured as 3D cysts were imaged on an Olympus IX83 microscope equipped with a Dragonfly200 spinning disk unit (Andor), a CMOS camera (Andor Zyla 4.2 Plus), a 60× UPLSAPO silicone oil objective (NA 1.30), and Fusion software (Andor). Fixed 2D MDCK cells were mounted in ProLong Diamond (Thermo Fisher) and images were acquired at room temperature.
Live-cell imaging and expansion microscopy imaging were performed on a Nikon Eclipse Ti-2 equipped a with Yokogawa W1 spinning disk unit, CMOS camera (Photometrics Prime BSI), 40× PlanApo silicone oil objective (NA 1.25), and laser combiner (Nikon Instruments LUN-F XL 405/488/561/640). For live-cell imaging, 42,000 cells per well were seeded on a μ-slide 8 well plate (Ibidi 80826), and after 24 h cells were imaged in phenol-red-free DMEM/F-12 medium with 10-1000 ng/ml doxycycline and 0.2% serum at 37°C using Nikon Elements software. A Perfect Focus System was used to maintain focus, and a stage-top incubator (Tokai Hit) was used to maintain temperature and 5% CO2. This system was used for all live-cell imaging, except for imaging of Ehd1-mScarlet-I, which was performed on the widefield Eclipse Ti-2 used for fixed-cell imaging (using an Okolab enclosure to maintain cells at 37°C and 5% CO2). For expansion microscopy, samples were imaged at room temperature and the supplemental 1.5x magnification tube lens of the Eclipse Ti-2 was used with the 40× PlanApo silicone oil objective (NA 1.25).
Structured illumination microscopy (SIM) imaging was performed on a Deltavision OMX v3 microscope (Applied Precision) equipped with a U-PLANAPO 60× oil objective (NA 1.42; Olympus), CoolSNAP HQ2 CCD cameras (Photometrics), and solid-state lasers at 488, 561, and 642 nm (Coherent and MPB communications). Samples were illuminated by a coherent scrambled laser light source that had passed through a diffraction grating to generate the structured illumination by interference of light orders in the image plane to create a 3D sinusoidal pattern, with lateral stripes approximately 270 nm apart. The pattern was shifted laterally through five phases and through three angular rotations of 60° for each z section (separated by 125 nm). Cells were stained with antibodies to Rab34 (Abcam) and ARL13B (NeuroMab), mounted in Prolong Diamond medium (Invitrogen P36965), and imaged at room temperature. Raw images were processed and deconvolved using Softworx software (Applied Precision). Channels acquired on separate cameras were then aligned in x, y, z, and rotationally using the SoftWorx alignment tool and Tetraspeck beads (Invitrogen T7279).
Focused ion beam scanning electron microscopy (FIB-SEM)
Cells cultured on 12mm glass coverslips were fixed in 4% PFA for 20 min, washed in PBS, and further fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, 2% OsO4 and 1.5% K4Fe(CN)6 (Sigma-Aldrich). Fixed cells were then stained en bloc in 2% aqueous uranyl acetate, dehydrated, and embedded in Embed 812. Unless otherwise noted, all FIB-SEM reagents are from Electron Microscopy Sciences.
After removing the glass coverslip, the embedded monolayer of cultured cells was glued with EMS water-based conductive graphene carbon paint onto the sample mounting aluminum stub, with the coverslip-facing side of the cells exposed to the air. A 20-25 nm-thick conductive platinum coating was applied to the sample surface with a sputter coater (Ted Pella, Inc.), and the sample was then imaged in a Crossbeam 550 FIB-SEM workstation (Carl Zeiss Microscopy) operated with SmartSEM (Carl Zeiss Microscopy) or Atlas engine 5 (Fibics Incorporated, Ottawa, Canada) software. For image acquisition, a target area was first selected with the SE2 detector (Electron High Tension], 5kV; electron beam current, 2nA) at working distance of 10 nm, then the stage was inclined (to 54°) and lifted (to a working distance of 5 nm) as the eucentric height and coincident point were adjusted. The targeted cells’ surface (50x30 μm) was deposited with a 0.5-1 μm-thick protective platinum pad by using 30kV:3nA FIB. Autotune marks and 3D tracking marks were milled into the platinum pad, and carbon deposited by 30kV:50pA FIB, followed by deposition of another 0.5-1 μm-thick upper protective carbon pad. 30kV:30nA FIB was used for the coarse trench (20 μm deep), 30kV:3nA FIB was used for the fine trench, and 30kV:300pA FIB was used for milling the imaging surface during the 3D tracking. The dwell time for SEM imaging (Electron High Tension, 1.5 kV; electron beam current, 2 nA) with EsB detector was set at 2-3 μs with line averaging by 2. The filtering grid voltage of the EsB detector was set at 1000 V, and the collector voltage was set at 300 V. The imaging resolution was set at 7 nm/pixel at X, Y axis, slicing by 7 nm along Z axis, yielding final data sets with isotropic resolution at 7 nm/voxel. After the FIB-SEM imaging, the data sets were aligned and exported with Atlas 5 software, then further cropped and rotated with DragonFly software (Object Research Systems Inc.).
Image analysis
Widefield fluorescence microscopy images were analyzed using Fiji/ImageJ. Ciliogenesis and quantification of protein markers present at cilia or centrioles was performed by manual counting. For SIM microscopy, images that were deconvolved using Softworx software (see above) were further aligned using Tetraspeck bead images and the Fiji Descriptor-based registration (2D) plugin. Intensity values for line traces across SIM images were determined in Fiji/Image using the ‘Plot Profile’ tool and plotted using Matlab (Mathworks, Inc.).
FIB-SEM datasets were processed using IMOD software.57,58 Rotated sub-volumes were extracted using the ‘Rubber band’ tool. Centrioles and associated membranes were segmented using the ‘Sculp’ drawing tool and rendered with the Surface and Cap Meshing options.
Transfection, immunoprecipitation, SDS-PAGE, and Western blotting
HEK293T cells were transfected with LAP-Rab34 variants using X-tremeGENE 9 (Sigma Aldrich, 6365779001). After 48 h, the cells were collected, lysed on ice in Co-IP buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5 mM DTT, protease inhibitors), and centrifuged at 20,000 x g for 15 min. LAP-tagged proteins were captured on GFP nanobody beads (Chromotek #gta-20). After washing, the beads were resuspended in 2X NuPAGE LDS buffer (Invitrogen) and denatured at 95°C for 5 min to elute captured proteins. For analysis of Rab34 levels in whole-cell lysate, cells were lysed in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 2% NP40, 0.25% sodium deoxycholate, 0.5 mM DTT, protease inhibitors) on ice for 10 min and centrifuged at 20,000 x g for 10 min. The supernatant was collected, and equal amounts of protein were loaded, as determined by Bio-Rad Protein Assay.
Protein samples were separated on NuPAGE 4-12% Bis-Tris gels (Invitrogen) run in MOPS buffer (50 mM Tris, 50 mM MOPS, 3.5 mM SDS, 1 mM EDTA) and transferred to a PVDF membrane (Millipore Sigma #IPVH00010). The membranes were blocked in 1:1 PBS/SeaBlock (Thermo Fisher #37527) and incubated with primary antibodies overnight at 4°C (see Key Resources Table). After washing, the membranes were incubated with HRP-conjugated secondary antibodies or Protein A-HRP for 30 min at room temperature. The blots were developed using Clarity Western ECL Substrate (Bio-Rad) and imaged on a ChemiDoc Touch imager (Bio-Rad).
Analysis of GTP/GDP binding in cells
HEK293T cells were transfected with LAP-Rab34 variants using X-tremeGENE 9. After 48 h, the cells were incubated with 0.25 mCi of 32P orthophosphate (Perkin Elmer, NEX053H002MC) for 4 h in DMEM containing dialyzed FBS (Gibco A3382001). The labeled cells were washed with ice-cold PBS and lysed on ice in Co-IP buffer supplemented with 10 mM MgCl2. The lysate was clarified by centrifugation at 20,000 x g for 15 min at 4°C, and LAP-tagged proteins were captured on GFP nanobody beads. After washing, bound nucleotides were dissociated by incubation in Elution buffer (0.2% SDS, 4 mM EDTA, 2 mM DTT, 1 mM GDP, 1 mM GTP) for 6 min at 68°C. The eluate was then spotted on a PEI-cellulose TLC plate (Sigma Z122882) and developed in a chamber equilibrated with 0.75 M KH2PO4. The dried plate was exposed to a phosphorimaging screen for 24 h, imaged on a Typhoon FLA 9500 (GE Healthcare), and quantified using ImageQuant software (GE Healthcare).
Recombinant protein expression and purification
pGEX-Rab34 plasmids were transformed into BL21 DE3 competent cells expressing GroEL/ES (from pGro7 plasmid; gift from Yong Xiong, Yale University) and selected with 100 μg/mL ampicillin and 25 μg/mL chloramphenicol. Transformed cells were inoculated into a 100 mL culture of LB with ampicillin and chloramphenicol and grown at 37°C for 16 h. This culture was diluted to OD600=0.02-0.06 and expanded to 4L, followed by growth at 37°C until OD600=0.6. The cultures were then cooled in an ice water bath for 15 min. 2g/L arabinose was added to induce pGro7 expression and the cultures were transferred to an 18°C incubator. After 30 min, 200 μM IPTG was added to induce the expression of GST-Rab34. The cultures were grown at 18°C overnight; cultures were then collected by centrifugation, and the cells were flash frozen.
Cell pellets were suspended in lysis buffer (10 mM Tris pH 7.3, 1 mM DTT, 150 mM NaCl, 5 mM MgCl2, 0.001% Triton X-100, 1mM PMSF, 0.2 mM GDP, Roche cOmplete EDTA-free protease inhibitors) and lysed with a microfluidizer. Cell debris was removed through a high-speed (20,000 x g) clarification spin. The resulting supernatant was incubated with Glutathione Sepharose resin (GE Healthcare, 17-0756-05) for 2 h in lysis buffer. Bound Rab34 was eluted with elution buffer (10 mM Tris pH 8.2, 10 mM glutathione, 1 mM DTT, 70 mM NaCl, 5 mM MgCl2). Eluted Rab34 was then diluted 1:5 with Buffer A (10 mM Tris pH 8.2, 1 mM DTT, 70 mM NaCl, 5 mM MgCl2), loaded onto an anion exchange column (GE Healthcare HiTrap Q, 17-5156-01), washed with 10 column volumes of Buffer A, and eluted with a linear gradient of 0% to 100% Buffer B (10 mM Tris pH 8.2, 1 mM DTT, 0.5 M NaCl, 5 mM MgCl2) over 20 column volumes. Rab34-containing fractions were dialyzed overnight into storage buffer (50 mM Tris pH 7.5, 1 mM DTT, 150 mM NaCl, 5 mM MgCl2, 0.2 mM GDP) and simultaneously treated with HRV3C Protease (Pierce 88946) to cleave the GST tag. Dialyzed Rab34 was then concentrated to approximately 70 μM with an Amicon concentrator (Millipore, Z740199). Free GST tag, uncleaved GST-Rab34, and HRV3C were removed via a second incubation with Glutathione Sepharose resin. Lastly, glycerol was added to a final concentration of 40% (v/v) and the purified Rab34 was stored at −20°C. All steps were conducted at 4°C unless noted otherwise.
Analysis of GTPase activity in vitro
All Rab34 used in steady-state assays was treated with AG 1-X8 anion exchange resin (Bio-Rad, 1401441) immediately before use to remove bound GDP. Steady-state GTPase activity was measured with a modified version of the standard NADH-based assay59,60, in which the final concentrations of coupling reaction components were adjusted as follows: pyruvate kinase (Sigma Aldrich, P9136) – 400 units/mL; lactate dehydrogenase (BBI Enzymes, LDHP2FS) – 40 units/mL; phosphoenolpyruvate (Sigma Aldrich, 10108294) – 10 mM. Rab34 GTPase activity was assayed in Rab34 assay buffer (50 mM Tris pH 7.7, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT) at 25°C. Absorbance at 340 nm was measured on an Olis HP 8452 Diode Array spectrophotometer.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical tests were carried out using t-test (two-sided, paired) or Tukey-Kramer test, as indicated in Figure Legends. Figure legends also indicate the number of independent replicates performed and the number of cells analyzed for each condition of each replicate (N). All graphs show the mean and SEM unless otherwise noted.
Supplementary Material
Highlights:
Rab34 is required for ciliary vesicle formation during ciliogenesis
Rab34 localizes to nascent ciliary membranes and marks the ciliary sheath
GTP binding and hydrolysis by Rab34 are required for intracellular ciliogenesis
Rab34 is dispensable for the extracellular ciliogenesis pathway used by MDCK cells
Acknowledgments
We acknowledge David Mick and members of the D.K.B. lab for advice and discussion; Derek Toomre (Yale University) for the In/Out reporter construct and cell line; Jadranka Loncarek (National Cancer Institute) for advice on expansion microscopy; Chris Westlake (National Cancer Institute) for cDNA encoding EHD1; Tamara Caspary (Emory University) for ARL13B cDNA; Tim Stearns (Stanford University) for Centrin2 cDNA; and Yong Xiong (Yale University) for pGro7 plasmid. This work was supported by funding from the National Institutes of Health (R00HD082280 and R35GM137956 to D.K.B.; R35GM136656 to E.M.D.L.C.), the Charles H. Hood Foundation (to D.K.B.), the Alfred P. Sloan Foundation (FG-2018-10333 to D.K.B.), a Yale Anderson Endowed Fellowship (A.K.G.), the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grant-in-Aid for Young Scientists 20K15739 to Y.H.; Grant-in-Aid for Scientific Research(B) 19H03220 to M.F.), the Japan Science and Technology Agency (CREST grant JPMJCR17H4 to M.F.), the Japan Society for the Promotion of Science (to M.E.O.), and NSF major instrumentation grant 1725480. S.D.G. was supported by R35GM136656-S1. We acknowledge Felix Rivera-Molina, Derek Toomre, and the Yale CINEMA microscopy facility for assistance with SIM imaging; Yumei Wu, Xinran Liu, and the Yale Center for Cellular and Molecular Imaging for FIB-SEM sample processing and data collection; Ken Nelson and the MCDB FACS facility for assistance with cell sorting; Shirin Bahmanyar, Jing Yan, and Nadya Dimitrova for microscope use; Anna Pyle for phosphorimager use; Brandon Toyama for graphical assistance; and Kazuyasu Shoji for assistance with Western blotting.
Footnotes
Declaration of interests
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Braun DA, and Hildebrandt F (2017). Ciliopathies. Cold Spring Harb Perspect Biol 9, a028191. 10.1101/cshperspect.a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, and Christensen ST (2019). Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol 15, 199–219. 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiter JF, and Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorokin S (1962). Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol 15, 363–377. 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorokin SP (1968). Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 3, 207–230. [DOI] [PubMed] [Google Scholar]
- 6.Insinna C, Lu Q, Teixeira I, Harned A, Semler EM, Stauffer J, Magidson V, Tiwari A, Kenworthy AK, Narayan K, and Westlake CJ (2019). Investigation of F-BAR domain PACSIN proteins uncovers membrane tubulation function in cilia assembly and transport. Nat Commun 10, 428. 10.1038/s41467-018-08192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS, et al. (2015). Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol 17, 228–240. 10.1038/ncb3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CT, Chen HY, and Tang TK (2018). Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat Cell Biol 20, 175–185. 10.1038/s41556-017-0018-7. [DOI] [PubMed] [Google Scholar]
- 9.Mazo G, Soplop N, Wang WJ, Uryu K, and Tsou MF (2016). Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Dev Cell 39, 424–437. 10.1016/j.devcel.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghossoub R, Molla-Herman A, Bastin P, and Benmerah A (2011). The ciliary pocket:a once-forgotten membrane domain at the base of cilia. Biol Cell 103, 131–144. 10.1042/BC20100128. [DOI] [PubMed] [Google Scholar]
- 11.Pusapati GV, Kong JH, Patel BB, Krishnan A, Sagner A, Kinnebrew M, Briscoe J, Aravind L, and Rohatgi R (2018). CRISPR Screens Uncover Genes that Regulate Target Cell Sensitivity to the Morphogen Sonic Hedgehog. Dev Cell 44, 113–129 e8. 10.1016/j.devcel.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu S, Liu Y, Meng Q, and Wang B (2018). Rab34 small GTPase is required for Hedgehog signaling and an early step of ciliary vesicle formation in mouse. J Cell Sci 131, jcs213710. 10.1242/jcs.213710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oguchi ME, Okuyama K, Homma Y, and Fukuda M (2020). A comprehensive analysis of Rab GTPases reveals a role for Rab34 in serum starvation-induced primary ciliogenesis. J Biol Chem 295, 12674–12685. 10.1074/jbc.RA119.012233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breslow DK, Hoogendoorn S, Kopp AR, Morgens DW, Vu BK, Kennedy MC, Han K, Li A, Hess GT, Bassik MC, et al. (2018). A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat Genet 50, 460–471. 10.1038/s41588-018-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeffer SR (2017). Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol Biol Cell 28, 712–715. 10.1091/mbc.E16-10-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wandinger-Ness A, and Zerial M (2014). Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol 6, a022616. 10.1101/cshperspect.a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenmark H (2009). Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10, 513–525. 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 18.Muller MP, and Goody RS (2018). Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases 9, 5–21. 10.1080/21541248.2016.1276999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MT, Mishra A, and Lambright DG (2009). Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic 10, 1377–1389. 10.1111/j.1600-0854.2009.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prior IA, Lewis PD, and Mattos C (2012). A comprehensive survey of Ras mutations in cancer. Cancer Res 72, 2457–2467. 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, and Balch WE (1992). GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol 119, 749–761. 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walworth NC, Goud B, Kabcenell AK, and Novick PJ (1989). Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J 8, 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun P, Yamamoto H, Suetsugu S, Miki H, Takenawa T, and Endo T (2003). Small GTPase Rah/Rab34 is associated with membrane ruffles and macropinosomes and promotes macropinosome formation. J Biol Chem 278, 4063–4071. 10.1074/jbc.M208699200. [DOI] [PubMed] [Google Scholar]
- 24.Langemeyer L, Nunes Bastos R, Cai Y, Itzen A, Reinisch KM, and Barr FA (2014). Diversity and plasticity in Rab GTPase nucleotide release mechanism has consequences for Rab activation and inactivation. Elife 3, e01623. 10.7554/eLife.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuda M, Kanno E, Ishibashi K, and Itoh T (2008). Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics 7, 1031–1042. 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Matsui T, Ohbayashi N, and Fukuda M (2012). The Rab interacting lysosomal protein (RILP) homology domain functions as a novel effector domain for small GTPase Rab36:Rab36 regulates retrograde melanosome transport in melanocytes. J Biol Chem 287, 28619–28631. 10.1074/jbc.M112.370544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, and Pereira G (2012). Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol 199, 1083–1101. 10.1083/jcb.201202126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernabe-Rubio M, and Alonso MA (2017). Routes and machinery of primary cilium biogenesis. Cell Mol Life Sci 74, 4077–4095. 10.1007/s00018-017-2570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nachury MV, and Mick DU (2019). Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol 20, 389–405. 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breslow DK, and Holland AJ (2019). Mechanism and Regulation of Centriole and Cilium Biogenesis. Annu Rev Biochem 88, 691–724. 10.1146/annurev-biochem-013118-111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirvis M, Stearns T, and James Nelson W (2018). Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem J 475, 2329–2353. 10.1042/BCJ20170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kukic I, Rivera-Molina F, and Toomre D (2016). The IN/OUT assay: a new tool to study ciliogenesis. Cilia 5, 23. 10.1186/s13630-016-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. (2010). Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17, 133–138. 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 34.Tillberg PW, and Chen F (2019). Expansion Microscopy: Scalable and Convenient Super-Resolution Microscopy. Annu Rev Cell Dev Biol 35, 683–701. 10.1146/annurev-cellbio-100818-125320. [DOI] [PubMed] [Google Scholar]
- 35.Sahabandu N, Kong D, Magidson V, Nanjundappa R, Sullenberger C, Mahjoub MR, and Loncarek J (2019). Expansion microscopy for the analysis of centrioles and cilia. J Microsc 276, 145–159. 10.1111/jmi.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernabe-Rubio M, Andres G, Casares-Arias J, Fernandez-Barrera J, Rangel L, Reglero-Real N, Gershlick DC, Fernandez JJ, Millan J, Correas I, et al. (2016). Novel role for the midbody in primary ciliogenesis by polarized epithelial cells. J Cell Biol 214, 259–273. 10.1083/jcb.201601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jewett CE, Soh AWJ, Lin CH, Lu Q, Lencer E, Westlake CJ, Pearson CG, and Prekeris R (2021). RAB19 Directs Cortical Remodeling and Membrane Growth for Primary Ciliogenesis. Dev Cell 56, 325–340 e8. 10.1016/j.devcel.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Homma Y, Kinoshita R, Kuchitsu Y, Wawro PS, Marubashi S, Oguchi ME, Ishida M, Fujita N, and Fukuda M (2019). Comprehensive knockout analysis of the Rab family GTPases in epithelial cells. J Cell Biol 218, 2035–2050. 10.1083/jcb.201810134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohli P, Hohne M, Jungst C, Bertsch S, Ebert LK, Schauss AC, Benzing T, Rinschen MM, and Schermer B (2017). The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep 18, 1521–1535. 10.15252/embr.201643846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer JA, Seperack PK, Strobel MC, Copeland NG, and Jenkins NA (1991). Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature 349, 709–713. 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 41.Bhattacharyya S, Rainey MA, Arya P, Mohapatra BC, Mushtaq I, Dutta S, George M, Storck MD, McComb RD, Muirhead D, et al. (2016). Endocytic recycling protein EHD1 regulates primary cilia morphogenesis and SHH signaling during neural tube development. Sci Rep 6, 20727. 10.1038/srep20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rapaport D, Auerbach W, Naslavsky N, Pasmanik-Chor M, Galperin E, Fein A, Caplan S, Joyner AL, and Horowitz M (2006). Recycling to the plasma membrane is delayed in EHD1 knockout mice. Traffic 7, 52–60. 10.1111/j.1600-0854.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- 43.Rainey MA, George M, Ying G, Akakura R, Burgess DJ, Siefker E, Bargar T, Doglio L, Crawford SE, Todd GL, et al. (2010). The endocytic recycling regulator EHD1 is essential for spermatogenesis and male fertility in mice. BMC Dev Biol 10, 37. 10.1186/1471-213X-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diekmann Y, Seixas E, Gouw M, Tavares-Cadete F, Seabra MC, and Pereira-Leal JB (2011). Thousands of Rab GTPases for the cell biologist. PLoS Comput Biol 7, e1002217. 10.1371/journal.pcbi.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. (2016). High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514. 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, and Anderson KV (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87. 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 47.Liem KF Jr., Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, and Anderson KV (2012). The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol 197, 789–800. 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He M, Subramanian R, Bangs F, Omelchenko T, Liem KF Jr., Kapoor TM, and Anderson KV (2014). The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol 16, 663–672. 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goetz SC, Liem KF Jr., and Anderson KV (2012). The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151, 847–858. 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fonte VG, Searls RL, Hilfer SR The relationship of cilia with cell division and differentiation J. Cell Biol, 49 (1971), pp. 226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguilar A, Meunier A, Strehl L, Martinovic J, Bonniere M, Attie-Bitach T, Encha-Razavi F, and Spassky N (2012). Analysis of human samples reveals impaired SHH-dependent cerebellar development in Joubert syndrome/Meckel syndrome. Proc Natl Acad Sci U S A 109, 16951–16956. 10.1073/pnas.1201408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, and McMahon AP (2007). Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977–1989. 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- 53.Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA, McMahon AP, Wong WH, and Scott MP (2010). Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc Natl Acad Sci U S A 107, 9736–9741. 10.1073/pnas.1004602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang CH, Zanini M, Shirvani H, Cheng JS, Yu H, Feng CH, Mercier AL, Hung SY, Forget A, Wang CH, et al. (2019). Atoh1 Controls Primary Cilia Formation to Allow for SHH-Triggered Granule Neuron Progenitor Proliferation. Dev Cell 48, 184–199 e5. 10.1016/j.devcel.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 55.Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, and Nachury MV (2013). An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol 203, 129–147. 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye F, Breslow DK, Koslover EF, Spakowitz AJ, Nelson WJ, and Nachury MV (2013). Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. Elife 2, e00654. 10.7554/eLife.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mastronarde DN, and Held SR (2017). Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol 197, 102–113. 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76. 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 59.Ingerman E, and Nunnari J (2005). A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol 404, 611–619. 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
- 60.Bradley MJ, and De La Cruz EM (2012). Analyzing ATP utilization by DEAD-Box RNA helicases using kinetic and equilibrium methods. Methods Enzymol 511, 29–63. 10.1016/B978-0-12-396546-2.00002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data not included in this article are available from the corresponding author on request.


