Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a treatment-refractory malignancy in urgent need of a molecular framework for guiding therapeutic strategies. Bulk transcriptomic efforts over the past decade have yielded two broad consensus subtypes: classical-pancreatic/epithelial versus basal-like/squamous/quasi-mesenchymal. While this binary classification enables prognostic stratification, it does not currently inform the administration of treatments uniquely sensitive to either subtype. Furthermore, bulk mRNA studies are challenged by distinguishing contributions from the neoplastic compartment versus other cell types in the microenvironment, which is accentuated in PDAC given that neoplastic cellularity can be low. The application of single-cell transcriptomics to pancreatic tumors has generally lagged behind other cancer types due in part to the difficulty of extracting high-quality RNA from enzymatically-degradative tissue, but emerging studies have and will continue to shed light on intra-tumoral heterogeneity, malignant-stromal interactions, and subtle transcriptional programs previously obscured at the bulk level. In conjunction with insights provided by single-cell/nucleus dissociative techniques, spatially resolved technologies should also facilitate the contextualization of gene programs and inferred cell-cell interactions within the tumor architecture. Finally, given that patients often receive neoadjuvant chemotherapy and/or chemoradiotherapy even in resectable disease, deciphering the gene programs enriched in or induced by cytotoxic therapy will be crucial for developing insights into complementary treatments aimed at eradicating residual cancer cells. Taken together, single-cell and spatial technologies provide an unprecedented opportunity to refine the foundations laid by prior bulk molecular studies and significantly augment precision oncology efforts in pancreatic cancer.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) has a 5-year survival rate of only 9% and is projected to become the second highest cause of cancer deaths in the US by 2030 (1,2). Standard management often involves multi-agent chemotherapy in the form of FOLFIRINOX or gemcitabine/nab-paclitaxel, but the administration of either regimen is not generally guided by molecular characteristics of individual tumors (3,4). The rare examples of precision oncology in pancreatic cancer involve patients with germline BRCA1/BRCA2 mutations who may benefit from platinum-based agents and poly(adenosine diphosphate–ribose) polymerase (PARP) inhibition, and those with high microsatellite instability (MSI-high) or defective DNA mismatch repair (dMMR) who may respond to immune checkpoint inhibitors, but these subgroups account for less than 4% and 1% of all PDAC patients, respectively (5-9) (Table 1). As such, there remains a paramount need to refine the molecular characterization of PDAC to stratify patients based on potentially targetable vulnerabilities and efficacious therapeutic strategies.
Table 1.
Associations between specific biomarkers/aberrations and potential treatment sensitivities in pancreatic ductal adenocarcinoma.
| Biomarker Type | Aberration | Potential Treatment Sensitivities | References |
|---|---|---|---|
| Genomic | Unstable genomes (>200 structural variation events) | Platinum-based agents, PARP inhibitor | Waddell et al, Nature 2015 (6) Golan et al, N Engl J Med 2019 (9) |
| Genomic | BRCA mutational signature; BRCA1/2, PALB2 mutations | Platinum-based agents, PARP inhibitor | Waddell et al, Nature 2015 (6) Golan et al, N Engl J Med 2019 (9) Raphael et al, Cancer Cell 2017 (19) Golan et al, Br J Cancer 2014 (21) |
| Gene expression | Basal-like/quasi-mesenchymal/squamous subtype | Gemcitabine, immune checkpoint blockade* | Collisson et al, Nat Med 2011 (25) Chan-Seng-Yue et al, Nat Genet 2020 (29) Hwang & Jagadeesh et al, bioRxiv 2020 (40) Porter et al, Proc Natl Acad Sci 2019 (65) |
| Gene expression | Classical-like epithelial subtype | FOLFIRINOX, Vitamin D, CD40 agonist* | Chan-Seng-Yue et al, Nat Genet 2020 (29) Hwang & Jagadeesh et al, bioRxiv 2020 (40) Porter et al, Proc Natl Acad Sci 2019 (65) |
| Cellular markers | ACTA2+ / LRRC15+ cancer associated myofibroblasts (myCAFs) | Losartan (TGF-β modulator)* | Elyada et al, Cancer Discov 2019 (44) Xu et al, Am J Pathol 2010 (50) Von Ahrens et al, J Hematol Oncol 2017 (51) Öhlund et al, J Exp Med 2017 (52) Ligorio et al, Cell 2019 (53) Dominguez et al, Cancer Discov 2020 (58) |
Denotes lack of strong supporting evidence.
Although a clinically informative taxonomy has yet to be established for pancreatic cancer, there have been numerous attempts at molecular subtyping to date. In this review, we will first briefly highlight the groundwork laid by bulk genomic and transcriptomic characterizations of human PDAC, which have been more extensively reviewed elsewhere (10-12). We then focus most of the review on insights gleaned from single-cell studies of human specimens. Although applications of single-cell technologies to pancreatic cancer have generally lagged behind other neoplasms due in part to the difficulty of extracting high-quality RNA from enzymatically-degradative tissue (13,14), such studies are now rapidly emerging and will continue to elucidate intra-tumoral heterogeneity and subtle transcriptional programs once obscured at the bulk level. Insights revealed from enhanced cellular resolution are particularly valuable in this disease given the relatively low neoplastic cellularity of PDAC tumors (as low as <5% of total cells in the tumor specimen) and pathological contributions from the tumor microenvironment (TME). In tandem with biology gleaned through dissociative techniques, spatially-resolved technologies should also facilitate the contextualization of expression programs and cellular interactions within the tumor architecture. Taken together, the collective insights from these approaches have the potential to inspire future directions for scientific investigation and spawn treatment strategies aimed at modulating multiple cell type compartments within the TME.
Profiling genomic aberrations in pancreatic cancer
It has been well-established from both clinical biopsies and in vivo models that four of the most common genetic mutations in pancreatic cancer are KRAS, TP53, SMAD4, and CDKN2A (15-18). Unfortunately, with the exception of some recent trials (e.g., NCT04330664), mutant KRAS has thus far been largely recalcitrant to drug targeting, and the latter three most commonly mutated genes are typically affected by loss-of-function mutations that are difficult to revert therapeutically. As such, several large cohort studies of primary tumor specimens were carried out over the past decade to discover additional mutations enriched in pancreatic cancer (18). To date, the subpopulation of PDAC patients for whom mutational signatures have guided treatment and conferred overall survival (OS) benefits are those with forms of homologous recombination deficiency (HRD) such as BRCA1 and BRCA2 mutations receiving platinum agents, though they constitute less than 5% of all patients (9,19-21) (Table 1). This appears to be the case for most individual genetic alterations in pancreatic cancer, which appear in only a minor subset of the patient population (6). Given this, notable efforts over the past decade have shifted towards subcategorizing PDAC based on transcriptomic information, which may be more conducive to the generation of broader, clinically-meaningful subtypes.
Molecular subtyping using bulk transcriptomics
The ability to capture mRNA in a relatively unbiased manner across the entire transcriptome has yielded gene expression based subtyping of various cancers, which in many cases, has already generated prognostic and therapeutic insights (22-24). Unlike mutationally-driven subtyping that is intrinsically limited by the prevalence of any particular genomic aberration within the patient population, transcriptome-based subtypes have tunable granularity as they are driven instead by expression signatures of multiple genes; the optimal extent of subcategorization is therefore one that is precise enough to inform subgroup-specific treatments but also broad enough to inspire therapeutic development and clinical deployment.
Bulk transcriptomic analyses over the past decade have resulted in a general distinction between two consensus subtypes: classical/epithelial and basal-like/squamous/quasi-mesenchymal (Table 2). The classical phenotype is characterized by robust expression of GATA6-driven endodermal programs and genes involved in epithelial differentiation, which was orthogonally confirmed through histological detection of abundantly secreted mucins (25,26). Putative variants of this subtype, such as immunogenic-progenitor and aberrantly differentiated endocrine exocrine (ADEX) (27), have also been proposed therein, but are now thought to describe contributions from immune infiltrate in the TME, or acinar and endocrine contamination, respectively (28). As a whole, the classical/epithelial subtype is thought to be the ‘default’ pathway in PDAC pathogenesis given GATA6 expression in adjacent normal tissue and its higher prevalence in the patient population (29). Basal-like tumors, in contrast, are characterized by an abundance of laminins and keratins, and exhibit expression patterns consistent with previously-described basal-like subtypes in breast and bladder cancer (30,31). This rarer subtype is thought to be correlated with high tumor grade, some level of KRAS-independence, and poorer outcomes. Indeed, basal-like tumors are associated with approximately 8 months shorter median survival in the localized disease setting (26). As with its classical counterpart, however, a recurring challenge in characterizing this subtype at the bulk level has been differentiating between neoplastic-intrinsic and TME-derived contributions. For example, an early study by Collisson et al. in 2011 suggested that basal-like tumors exhibit mesenchymal-like features (25), though retrospective comparisons have raised the possibility that these observations may have been due to an admixture of the basal-like subtype and stromal contamination (26), a challenge that is frequently accentuated in analyses of PDAC due to pervasive desmoplastic stroma.
Table 2.
Summary of key bulk transcriptomic studies of pancreatic cancer that have yielded the classical/epithelial vs. basal-like/quasi-mesenchymal/squamous consensus subtypes.
| Study | Technique(s) | Sample Size | Key Findings |
|---|---|---|---|
| Collisson et al, Nature Medicine 2011 (25) | Microarray | 27 microdissected 36 PDAC tumors, previously published |
Non-negative matrix factorization and consensus clustering yields three subtypes: classical, quasi-mesenchymal, and exocrine-like |
| Moffitt et al, Nature Genetics 2015 (26) | Microarray | 145 primary 61 metastatic 17 cell lines |
Identification of classical and basal-like subtype, with the latter exhibiting poorer survival |
| Bailey et al, Nature 2016 (27) | RNA-seq Microarray | 96 RNA-seq 232 microarray |
Unsupervised clustering of RNA-seq data from 96 tumors with at least 40% epithelial content yielded four subtypes: squamous, pancreatic progenitor, immunogenic, aberrantly differentiated endocrine exocrine (ADEX); additionally examined 232 tumors with microarray data (median cellularity 30%) |
| Raphael et al, Cancer Cell 2017 (19) | RNA-seq | 150 | Reconciliation of previously identified subtypes; found that high-purity tumors can consistently be classified into either basal-like/squamous or classical/progenitor |
| Puleo et al, Gastroenterology 2020 (28) | Microarray | 309 | Validated basal-like vs. classical distinction found from prior studies. Incorporated consideration of microenvironment, which yielded five subtypes: pure basal-like, stroma activated, desmoplastic, pure classical, and immune classical |
Bulk transcriptomic subtypes and response to therapy
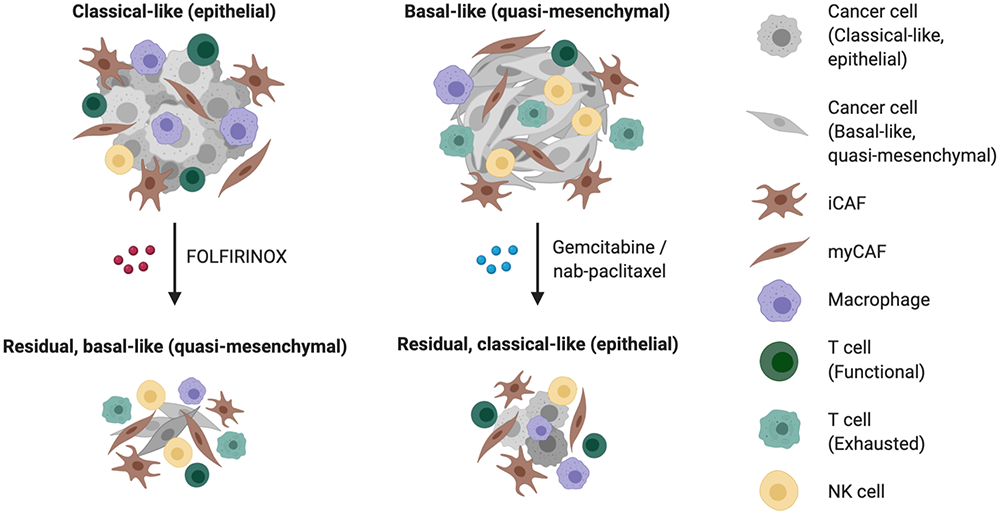
Beyond prognostication, subtype determination currently does not influence the administration of chemotherapy for patients, but preliminary pre-clinical and clinical data suggest that there may be differential responses of basal-like and classical/epithelial tumors to FOLFIRINOX vs. gemcitabine/nab-paclitaxel (Table 1; Figure 1). In 2011, Collisson et al. showed that cell lines classified as quasi-mesenchymal were more sensitive to gemcitabine relative to their classical counterparts (25). Furthermore, among the 12 patients who had basal-like tumors in the COMPASS cohort, seven were responders (stable disease or partial response) while five were non-responders (progressive disease) (32). Of note, nearly half of patients in the former group received gemcitabine/nab-paclitaxel instead of modified FOLFIRINOX while no patients in the latter group received gemcitabine/nab-paclitaxel, though larger sample sizes are needed to substantiate this potential heightened sensitivity of basal-like tumors to gemcitabine-based regimens (Table 1; Figure 1). Forthcoming randomized controlled trials such as the PASS-01 study may provide further insights into these outstanding questions by integrating molecular profiling (e.g., GATA6) into comparisons of modified FOLFIRINOX vs. gemcitabine/nab-paclitaxel (NCT04469556).
Figure 1.
Schematic diagram of potential transcriptomic subtype-dependent response to therapy, associations among different cell types, and treatment-induced plasticity.
Challenges of bulk transcriptomic profiling
Although mRNA-based subtyping efforts to date have improved our understanding and classification of PDAC, they are intrinsically challenged by the mixing of mRNA derived from unknown proportions of malignant and non-malignant cell types, providing a low-resolution ensemble readout. Prior studies recognized this limitation and, as a result, frequently performed microdissections or screened for high epithelial content prior to bulk processing. Nonetheless, the limitations of bulk transcriptomics were evident in the conception of malignant subtypes now believed to result from stromal contamination. This strongly indicates the need to dissect this disease using emerging single-cell technologies that can compartmentalize the various cell types within the TME and differentiate neoplastic from non-neoplastic programs. If combined with spatially-resolved analyses, single-cell RNA-seq (scRNA-seq) can further reveal how various subtypes co-localize with one another across cell types. Indeed, an outstanding question in the field is how neoplastic subtypes differentially associate with cancer-associated fibroblast (CAF) subtypes, for instance. We thus dedicate the remainder of this review to emerging research that leverages single-cell transcriptomics and spatial technologies to illuminate the intra- and inter-tumoral heterogeneity of PDAC at unprecedented resolution.
Single cell and spatial technologies redefine the molecular diversity of pancreatic cancer
The maturation of scRNA-seq in recent years has enabled the discovery of previously-unknown cell types and improved characterizations of transitory cell states (33-36). In human cancers, these approaches have already been applied to sites such as the brain, breast, skin, and colon, among many others (33,35,37-39), but have lagged behind in the pancreas by several years. However, technological improvements have led to emerging single cell studies in PDAC that shed light on intra-tumoral heterogeneity, malignant-stromal interactions, treatment-induced alterations, and gene expression programs in rarer cell types or subtypes (29,40-47) (Table 3). In this section, we discuss the various techniques that have been used as well as the insights specifically derived from the malignant, CAF, and immune compartments.
Table 3.
Summary of select single-cell/nucleus RNA-seq studies of pancreatic ductal adenocarcinoma.
| Study | Technique(s) | Specimens | Number of Cells | Key Findings |
|---|---|---|---|---|
| Bernard et al, Clinical Cancer Research 2018 (42) | Drop-seq | 2 Low Grade-IPMN 2 High Grade-IPMN 2 primary PDAC |
3,343 | - Found epithelial and stromal heterogeneity in progression from precursor lesions to invasive carcinoma - Pro-inflammatory immune infiltrate seen in LG-IPMNs was progressively depleted during neoplastic progression, accompanied by infiltration of myeloid-derived suppressor cells and myofibroblasts |
| Peng et al, Cell Research 2019 (43) | 10x 3’ v2 | 24 primary PDAC 11 normal |
41,986 | - Subset of ductal cells with specific proliferative features associated with inactivation programs in T cells -Pseudotime trajectory showed EPCAM highly expressed throughout transition from abnormal ductal to malignant while other genes such as MUC1 increased with progression |
| Elyada et al, Cancer Discovery 2019 (44) | 10x 3’ v2 | 6 primary PDAC 2 normal |
21,200 | - Identified previously validated and new subtypes of fibroblasts: myofibroblastic CAFs (myCAFs), inflammatory CAFs (iCAFs), and antigen-presenting CAFs (apCAFs) but required enrichment first - Interconversion between CAF subtypes demonstrated in culture |
| Moncada et al, Nature Biotechnology 2020 (45) | inDrop, ST | 2 primary PDAC | 3,659 | - Integrated scRNA-seq with spatial transcriptomics using multimodal intersection analysis (MIA) -Co-localization of inflammatory CAFs and cancer cells with a stress-response gene module |
| Chan-Seng-Yue et al, Nature Genetics 2020 (29) | 10x 3’ v2 | 13 primary PDAC 2 metastatic |
31,195 | - Basal-like and classical-like programs co-exist intratumorally - Subtypes can be further subdivided into 5 groups: basal-like A, basal-like B, hybrid, classical-A, classical-B |
| Lin et al, Genome Medicine 2020 (47) | 10x 3’ v2 | 10 primary PDAC 6 metastatic biopsies |
14,926 | - Cell type-specific marker expression for EMT+ neoplastic cells, activated CAFs, and endothelial cells are associated with survival - Significant heterogeneity between primary and metastatic lesions |
| Dominguez et al, Cancer Discovery 2020 (58) | 10x 3’ v2 | 20 primary mouse pancreata (normal, KPP) | 13,454 | - LRRC15+ CAFs reprogrammed by TGF-β surround tumor islets but are absent from normal pancreas - Examination of immunotherapy-based clinical trials from six cancer types show associations between high levels of LRRC15+ CAF and poor response to anti–PD-L1 |
| Juiz & Elkaoutari et al, The FASEB Journal 2020 (46) | SPLiT-seq | 6 primary PDAC (classical tumors) by fine needle aspiration | 8,934 | - Single cell RNA-seq of biopsy-derived pancreatic cancer organoids from ‘classical-like’ tumors each exhibit at least one cluster corresponding to basal-like phenotype |
| Raghavan, Winter, Navia, & Williams et al, BioRxiv 2020 (41) | Seq-Well | 23 metastatic PDAC needle biopsies | 23,042 | - Identified continuum of basal-like and classical-like malignant cells including hybrid cells - Matched organoids exhibit bias against growth of basal-like cells in standard organoid media, but modification of culture conditions can rescue basal-like phenotype |
| Hwang & Jagadeesh et al, BioRxiv 2020 (40) | 10x 3’ v2/v3 (snRNA-seq) | 26 primary PDAC, untreated and neoadjuvant chemotherapy and radiotherapy | 138,547 | - Single nucleus RNA-seq captures various cell types in expected proportions and enables processing of banked frozen specimens dating back years - Enrichment of basal-like over classical-like programs in the post-neoadjuvant chemoradiotherapy context - Malignant cells expressing basal-like programs spatially associate with greater immune infiltration and increased lymphocytic content compared to those expressing classical-like programs |
Malignant Cells
In 2018, Bernard et al. used Drop-seq (36) to analyze 3343 cells from two PDAC, two low grade intraductal papillary mucinous neoplasms (IPMN), and two high-grade IPMNs to dissect the epithelial cell programs involved in the progression of nascent lesions (42). They found numerous similarities in signaling programs between invasive PDAC and high-grade IPMNs, and conversely, fewer between invasive cancer and low-grade lesions. By capturing other cell types in the microenvironment, they also revealed the acquisition of heterogeneous alterations not only in the epithelium but also in non-neoplastic compartments throughout tumor progression. In spite of these insights, no modification to the existing molecular taxonomy was proposed perhaps in part due to the low numbers of cells and specimens examined.
A larger-scale effort was carried forth in 2019 by Peng et al. (43) in which 41,986 single-cells were analyzed from 24 primary tumors and 11 ‘normal’ control pancreata using the commercially-available 10× Genomics Chromium Single Cell Gene Expression (3’ v2) platform, revealing 10 distinct clusters in expression space that corresponded to specific known cell types. The epithelial cells, in particular, were separated into type 1 and 2 ductal cells, with the former approximating moderately atypical ductal cells and the latter malignant cells with pronounced chromosomal copy number variations (CNVs). A series of differential gene expression analyses showed that the type 1 ductal cells were enriched for cell adhesion, migration, and inflammatory response but still maintained perfunctory pancreatic roles, while type 2 ductal cells were enriched for malignant phenotypes such as proliferation and hypoxia. When superimposed onto previous Bailey et al. subtypes (27), the classical-progenitor and basal-like/squamous subtypes scored highly in the ‘malignant’ type 2 ductal cells as expected, though they were also present in other non-ductal cells, underscoring the cell type admixing present in these bulk expression subtypes. Using a subset of markers from the malignant ductal cells (type 2), the authors clustered The Cancer Genome Atlas (TCGA) patients into three distinct groups based on bulk RNA-seq data with differing survival and immune composition associations such as an inverse relationship between high levels of proliferative ductal markers and cytotoxic T cell markers, marking the first attempt at de novo patient-level categorization using single-cell insights. The authors also performed pseudotime trajectory analysis and found that EPCAM maintained high expression across the transition from abnormal ductal to malignant while other genes known to be involved with tumor progression, such as MUC1, gradually increased during PDAC progression (43).
In addition to performing bulk RNA-seq on 248 PDAC specimens, the COMPASS/PanCuRx investigators also conducted scRNA-seq on a limited subset of 15 samples (13 primary, 2 metastatic) using 10× Genomics Chromium Single Cell Gene Expression (3’ v2), which yielded 31,195 cells for downstream analysis (29). Although the single-cell data was not used to perform a de novo analysis, perhaps owing in part to an insufficient number of cells, the data was helpful for validating the reclassification derived from non-negative matrix factorization of the bulk RNA-seq data (Basal-like A, Basal-like B, Hybrid, Classical A, Classical B). The intra-tumoral co-existence of basal-like and classical programs in different malignant cells was demonstrated (29,46), and other studies have also shown that cells generally exist along a continuum of basal-like and classical-like states, with certain cells exhibiting characteristics of both (41). Such findings may have profound therapeutic ramifications and raise the question of whether combinations of agents aimed at both basal-like and classical cells may be advantageous for tumors harboring characteristics of both subtypes. Further investigation into the defining signatures and dependencies of ‘hybrid cells’ should also be integrated into therapeutic considerations.
The loss of spatial relationships that comes with dissociation into a single-cell suspension is a notable limitation when asking questions about the heterogeneity of cell types and cell-cell interactions within the tumor architecture. To address this challenge, Moncada and colleagues integrated single-cell RNA-seq with spatial transcriptomics (ST) on bisected PDAC samples (45). As a proof-of-principle, they demonstrated their multimodal intersection analysis (MIA) technique on two specimens. The ST component had a resolution of 20-70 cells and captured 1-2 orders of magnitude fewer unique genes and transcripts compared to scRNA-seq, which meant that reliable cell type identification in space using such an approach required a robust scRNA-seq atlas. Through their method, the authors were able to demonstrate that subpopulations of neoplastic cells, ductal cells, macrophages, and other immune cells had distinct spatial associations with one another; among these, neoplastic cells exhibiting a stress response gene module were shown to co-localize with a subset of CAFs called inflammatory CAFs (iCAFs), which we discuss in greater detail in subsequent sections.
Cancer Associated Fibroblasts
While transcriptional programs intrinsic to malignant cells are generally the focus of subtyping efforts, many groups have also recognized the prominent role of the stroma in pancreatic cancer and have sought to characterize their cell-type constituents at the bulk and single-cell level. In particular, cancer associated fibroblasts (CAFs) are an abundant member of the tumor stroma (48) and have received considerable interest given their capacity to facilitate tumor growth and metastasis, interfere with drug delivery (44,49,50), and augment desmoplasia and immune suppression (51).
Single-cell analyses of CAFs have not only validated the existence of known subtypes but also refined markers and revealed rarer subpopulations that were obscured by bulk analyses. Elyada et al., for instance, confirmed the presence of tumor-adjacent myofibroblastic CAFs (myCAFs) with elevated ACTA2 expression and iCAFs within regions of high desmoplasia expressing various cytokines and chemokines such as IL6 (44,52). In doing so, they discovered novel markers for each subtype, such as TAGLN, MYL9, TPM1, TPM2, MMP11, POSTN, and HOPX for myCAFs, and CFD, LMNA, and DPT for iCAFs. More broadly, they were also able to reclassify general CAF markers as subtype-specific ones (e.g. PDGFRA for iCAFs) and identify additional hallmarks of CAFs (e.g., PDPN, DCN) in pancreatic cancer. Finally, using scRNA-seq to analyze mouse models of pancreatic cancer in parallel with patient samples, Elyada and colleagues found a novel antigen-presenting CAF (apCAF) subpopulation that upregulates MHC class II genes and interacts with CD4+ T cells (44). Interestingly, this apCAF subpopulation lacks costimulatory molecules necessary for T cell activation and is therefore purported to be an immunosuppressive decoy that unfavorably skews the ratio of CD8+ to regulatory T cells. Of note, however, the authors observed that these CAFs are plastic and have the ability to interconvert among the various phenotypes (44), which may present an opportunity to modulate the TME for either endogenous recognition by the host immune system or facilitate immunotherapies that have shown limited success in PDAC to date.
Although myCAFs are generally well-demarcated across single-cell studies examining fibroblasts, some have not been able to identify iCAFs or apCAFs from de novo unsupervised clustering (40,47). Lin et al., for instance, reported three distinct fibroblast clusters but found that two of them exhibited quiescent/normal and smooth muscle signatures (RSG5, NOTCH3, CSRP2) instead of iCAFs and apCAFs (47). It may therefore be helpful for future work to examine whether iCAFs or apCAFs exist as smaller subpopulations within one or more of these clusters, and to decipher the gene expression patterns that obscure their identification in unsupervised clustering analyses.
In addition to classifying the CAF subpopulations, it has also been of great interest to learn of the cell types they co-enrich with, their spatiotemporal orientations, as well as their differential compatibilities with various therapeutic modalities. Toward this end, Bernard et al. noted in their single-cell study that myCAFs were well-represented in high-grade IPMNs and may therefore result from or contribute to pre-invasive dysplastic processes. Conversely, iCAFs were only found in invasive cancer specimens and notably absent from non-invasive IPMNs, suggesting that this CAF sub-population may not arise until more mature stages of oncogenic transformation (42). This is consistent with the notion that the TME becomes increasingly immune-suppressed throughout tumor development. Our group has also undertaken efforts to examine interactions between stromal and cancer cells through RNA in situ hybridization (RNA-ISH) with the stromal marker, SPARC, and the neoplastic markers MKI67 (proliferation, PRO) and FN1 (epithelial-mesenchymal transition, EMT). In this study (53), we found that distinct neoplastic gland types (PRO vs. EMT vs. both) were differentially associated with stromal abundance. Specifically, PRO+EMT+ glands were enriched in high-stroma tumors, EMT+ in medium-stroma tumors, and PRO+ in low-stroma tumors. More broadly, these results revealed that the CAF-rich stroma in PDAC has a large influence on the overall tumor architecture and the heterogeneity that lies therein.
Immune Cells
While dissociating viable malignant cells and CAFs from PDAC tissues is challenging, immune cells are readily detached, achieving non-enriched viable cell fractions of approximately 90% (44). Therefore, several scRNA-seq studies to date have assessed the immune microenvironment of PDAC at the single-cell level. Elyada and colleagues identified two major immune cell clusters representing the myeloid and lymphoid lineages (44). Within the myeloid cluster, sub-clustering revealed six distinct populations that were identified as resident macrophages, alternatively activated (M2-like) macrophages, classic monocytes, conventional type 1 dendritic cells (cDC1), and two types of Langerhans-like dendritic cells. Within the lymphoid cluster, sub-clustering identified five discrete cell types: CD8+ T cells, CD4+ T cells, regulatory T cells (Tregs), proliferating Tregs, and natural killer (NK) cells. In addition, several groups have examined features of the tumor immune microenvironment (TIME) during the tumorigenic process, relative to malignant cell transcriptional state, and in primary versus metastatic deposits.
Low-grade IPMNs are enriched for cytotoxic T lymphocytes and CD4+ effector T cells compared to high-grade IPMNs and PDACs, suggesting an inflammatory reaction early in pancreatic tumorigenesis that becomes suppressed over time (42). In contrast, PDACs are enriched for granulocytic myeloid-derived suppressor cells (MDSCs) compared to precursor IPMNs (42). Once invasive cancer has developed, cancer cell-intrinsic transcriptional features appear to have a role in modulating the TIME. For example, Peng and colleagues used their single-cell malignant ductal markers to cluster 178 pancreatic cancer patients from TCGA based on bulk RNA-seq data (43). Among the three PDAC patient clusters identified, cluster 3 featured proliferation markers and was associated with worse survival compared with patients in clusters 1 and 2. Differential gene expression analysis of cluster 3 versus clusters 1 and 2 revealed an enrichment of cell cycle, DNA replication, and DNA repair pathways and a depletion in several immune/T cell activation gene sets (43). In both the TCGA and scRNA-seq cohorts, there was an inverse correlation between high expression of proliferative ductal markers and low expression of T cell activation markers (43). Immunohistochemistry demonstrated that low Ki67+ ductal cells were spatially associated with high T cell infiltration and vice versa, indicating that dysregulated ductal cell proliferation and the local immune response are closely linked and pointing to a potential therapeutic strategy combining cell-cycle inhibitors and immunotherapy (54).
The TIMEs of primary tumors and metastatic lesions are distinct in numerous ways, including organ-specific differences. Using scRNA-seq, Lin and colleagues extracted immune cells from primary tumor resections and metastatic biopsies (mostly liver) (47). Clustering tumor-infiltrating lymphocytes across primary tumors and metastases yielded two mixed clusters, indicating similar functional states among lymphocytes in these two contexts. The first cluster exhibited high levels of activation/exhaustion markers (e.g., PDCD1, TIGIT, CTLA4, HAVCR2, LAG3) while the second cluster was characteristic of naïve, antigen-inexperienced T cells (47). In contrast, macrophages from primary tumors and metastases clustered separately with the former enriched in genes associated with extracellular matrix production and would healing processes (M2-like polarization) while the latter expressed MHC II-related genes associated with antigen-presentation (47). These findings warrant further exploration, though intrinsic distinctions between pancreas-resident and liver-resident macrophages may contribute to the observed differences.
The tradeoff for the depth of phenotypic characterization offered by scRNA-seq is the loss of tissue architecture, which hinders inferences of intercellular interactions. A complementary approach with single-cell spatial resolution but much lower molecular resolution involves multiplexed immunolabelling or RNA in situ hybridization and imaging. For example, Carstens and colleagues developed an eight-plex immunofluorescence (IF) assay (α-smooth muscle actin, collagen I, cytokeratin 8, CD3, CD8, CD4, Foxp3, DAPI) and performed multispectral imaging with spectral unmixing for simultaneous assessment of all markers on tissue microarrays derived from 132 PDAC specimens (55). They found that high levels of total T cell, CD8+ cytotoxic T cell, and CD4+ effector T cell infiltration were all independently associated with improved survival in a multivariate Cox regression analysis (55). Focusing on a 20 μm radius around each CK8+ cancer cell, only high infiltration of cytotoxic CD8+ T cells was significantly associated with improved patient survival. Notably, αSMA levels did not correlate with T-cell infiltration and collagen I deposition was actually positively correlated with T-cell infiltration, indicating that desmoplastic stroma does not appear to have a net hinderance on lymphocyte infiltration in this context (49,56). Recently, a similar multiplexed IF approach was used to investigate myeloid cells (CD14, CD15, CD33, ARG1, HLA-DR) and macrophages (CD68, CD86, CD163, CD206, IRF5) in tissue microarrays assembled from 305 primary resection specimens (57). Using four polarization markers to calculate a continuous M1-M2 macrophage polarization index (M1: CD86, IRF1; M2: CD163, CD206), Vayrynen and colleagues discovered that M1-polarized macrophages were located significantly closer to cancer cells than M2-polarized macrophages (57). Furthermore, reduced survival was associated with high density of CD15+ARG1+ immunosuppressive granulocytic cells and M2-polarized macrophages, as well as closer proximity of M2-polarized macrophages to cancer cells. Finally, the authors found that myeloid cell densities were associated with alterations in PDAC driver genes (e.g., higher CD15+ granulocytic cell density associated with TP53 alterations, lower CD14+ monocytic cell density associated with SMAD4 inactivation), which provides further evidence that cancer cell-intrinsic factors influence the PDAC TIME and are important considerations in future efforts at therapeutic immunomodulation (57).
Emerging evidence and future directions
Early single-cell studies have demonstrated promise and have overcome prior challenges posed by stromal admixing with neoplastic cells. In addition, the cumulative availability of novel datasets that now constitute profiling from dozens of patients and hundreds of thousands of cells (40,41,43) provides an exceptional opportunity to confirm cell-type-specific findings derived from orthogonal settings or models (58). Nonetheless, there remain several outstanding issues that still need to be resolved. First, single-cell studies have not yet resulted in a molecular taxonomy that anchors a substantive clinical framework. Next, the requirement of scRNA-seq for freshly dissociated tissue limits data quality due to suboptimal dissociation and lower RNA quality—as well as hampers the potential for multi-institutional collaboration. This results in relatively low stromal capture of primary PDAC tumors, leading to a non-representative distribution of the cell types acquired (44). Conversely, extracting single nuclei from difficult to dissociate tissues such as benign pancreas and neuronal specimens has been demonstrated, and comparisons between the mRNA transcripts derived from nuclei and the cytoplasmic compartment have shown similar abundance patterns (14,59-63). As such, one alternative strategy would be to use single-nucleus RNA-seq (snRNA-seq) for PDAC, which has shown compatibility with frozen specimens and bypasses some of the challenges of balancing viability, dissociation, and RNA integrity seen with scRNA-seq (40). Indeed, snRNA-seq in primary PDAC captures a histologically representative distribution of the major cell types (40). It should be noted, however, that many patients with PDAC present with metastatic disease, and as such, there may be additional insights that can be gleaned from examination of non-primary lesions. Towards this end, a recent study used Seq-Well to profile 23,042 single cells from mostly metastatic liver lesions, where the challenges of dissociating fibrotic tissue for scRNA-seq are thought to be less accentuated (41).
Emerging clinical evidence suggests that neoadjuvant chemotherapy and/or radiotherapy may improve clinical outcomes, even in resectable disease (64). Nevertheless, there is still much room for improvement. Prior transcriptomic studies of PDAC, both bulk and single-cell, have focused on untreated disease but given the increasing prevalence of neoadjuvant therapies and second-line treatments after recurrence or metastatic spread, the need to study the disease after molecular alteration by the selection pressure of treatment has never been higher. Our group recently found in patient-derived cell lines that ex vivo treatment with FOLFIRINOX induces a shift towards a more basal-like phenotype (Figure 1), whereas treatment with Vitamin D augments the baseline basal-like or classical/epithelial state of the cell line (65). Separately, in human primary resection specimens exposed to FOLFIRINOX and radiotherapy with capecitabine, an enrichment of basal-like over classical/epithelial-like signatures was observed in post-treatment malignant cells, though it remains unclear whether these observations were induced by treatment or resultant from the depletion of more vulnerable cell subtypes/states, given the absence of matched specimens (40). Additional mRNA profiling in the context of randomized controlled clinical trials with matched pre- and post-treatment specimens would further elucidate the significance of expression patterns observed after therapy.
The loss of spatial information inherent to dissociated single-cell methods is problematic when an understanding of interacting cell partners is required to fully dissect the complex tumor ecosystem. As such, Moncada and colleagues have made important technological advances in combining scRNA-seq with spatial transcriptomics (45), revealing specific associations between neoplastic programs and CAF subtypes. However, it should also be noted there are orthogonal methods compatible with archival formalin-fixed paraffin-embedded (FFPE) sections and higher transcript capture rates that can be leveraged. For example, digital spatial RNA profiling on FFPE sections has revealed that basal-like cancer cells spatially associate with lymphocytic infiltration relative to classical-like cancer cells (40), and conversely, that classical-like neoplastic cells appear to associate more strongly with a myeloid-rich microenvironment. These results mirror those seen in breast cancer, in which the triple-negative subtype that features basal-like characteristics exhibits greater lymphocytic infiltration and responsiveness to immune checkpoint inhibition (66). One outstanding question in this regard, however, is how the various CAF subpopulations associate with both malignant subtypes and immune cell types. One might posit that classical-like/macrophage microniches also engage with immunosuppressive iCAFs, for instance, though this requires further validation. Interestingly, a 2013 study by Mitsunaga et al. found that high serum levels of IL-6 and IL-1β, markers of iCAFs, associated with poor response to gemcitabine in patients with advanced PDAC (67), substantiating the notion that classical-like/iCAF/macrophage co-enrichment may be an orchestrated, multi-cellular module conferring resistance to gemcitabine-based chemotherapies. However, it should also be noted that a prior study by Somerville et al. (68) showed that pancreatic cancer cells expressing the squamous/basal-like associated p63ΔN isoform could induce iCAF-like states in pancreatic stellate cells through conditioned media experiments and orthotopic transplantation into mouse pancreata.
Ultimately, the value of the biology uncovered using single-cell and -nucleus approaches and the molecular subtyping of PDAC lies in the applicability to patients. Through cell type-specific biology, spatial relationships among different cells, and temporal dynamics during tumor evolution and response to therapy, we should be well equipped to design rational therapeutic strategies to overcome the treatment-refractory nature of this disease. While precision oncology in pancreatic cancer remains in its nascent stages compared to other common cancers, there is hope from the recent momentum and results described in this review that clinical trials for PDAC will soon be stratified by meaningful molecular subtypes and discerning biomarkers. The era of managing PDAC patients with a one-size-fits-all approach is nearing its conclusion.
Acknowledgments
Financial Support
This work was supported in part by the UCSF Dean’s Yearlong Fellowship (J.A.G.), Chinese American Medical Society (J.A.G.), American Society for Clinical Oncology/Conquer Cancer Foundation Young Investigator Award (W.L.H.), Hopper-Belmont Foundation Inspiration Award (W.L.H), and American Cancer Society/Massachusetts General Hospital Institutional Research Grant (W.L.H.) . W.L.H. is an Andrew L. Warshaw, M.D. Institute for Pancreatic Cancer Research Fellow. D.T.T. has research funding from the NIH (U01CA228963, R01CA240924, R01CA235412), The Robert L. Fine Cancer Research Foundation, an SU2C-NSF-Lustgarten Foundation Pancreatic Cancer Convergence Research Team grant, and a Stand Up To Cancer-Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (grant number: SU2C-AACR-DT26-17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. SU2C-AACR-DT26-17 grant is administered by the American Association for Cancer Research, the scientific partner of SU2C.
Footnotes
Disclosures
D.T.T. has received consulting fees from ROME Therapeutics, Foundation Medicine, Inc., NanoString Technologies, EMD Millipore Sigma, Pfizer, and Third Rock Ventures that are not related to this work. D.T.T. is a founder and has equity in ROME Therapeutics, PanTher Therapeutics and TellBio, Inc., which is not related to this work. D.T.T. receives research support from ACD-Biotechne, PureTech Health LLC, and Ribon Therapeutics, which was not used in this work. D.T.T.’s interests were reviewed and are managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. All other authors declare no competing interests.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014; 74:2913–21. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. N Engl J Med 2011; 364:1817–25. [DOI] [PubMed] [Google Scholar]
- 4.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013; 369:1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu ZI, Shia J, Stadler ZK, Varghese AM, Capanu M, Salo-Mullen E, et al. Evaluating mismatch repair deficiency in pancreatic adenocarcinoma: Challenges and recommendations. Clin Cancer Res 2018; 24:1326–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupinacci RM, Goloudina A, Buhard O, Bachet JB, Maréchal R, Demetter P, et al. Prevalence of Microsatellite Instability in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology 2018; 154:1061–1065. [DOI] [PubMed] [Google Scholar]
- 8.Sohal DPS, Kennedy EB, Khorana A, Copur MS, Crane CH, Garrido-Laguna I, et al. Metastatic pancreatic cancer: ASCO clinical practice guideline update. J Clin Oncol 2018; 36:2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019; 381:317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2019; 16:207–20. [DOI] [PubMed] [Google Scholar]
- 11.Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer 2016; 16:553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2006; 20:1218–49. [DOI] [PubMed] [Google Scholar]
- 13.Jun E, Oh J, Lee S, Jun HR, Seo EH, Jang JY, et al. Method Optimization for Extracting High-Quality RNA From the Human Pancreas Tissue. Transl Oncol 2018; 11:800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosti L, Hang Y, Trefzer T, Steiger K, Ten FW, Lukassen S, et al. Single nucleus RNA sequencing maps acinar cell states in a human pancreas cell atlas. Gastroenterology 2019; 0016-5085:35399–3. [DOI] [PubMed] [Google Scholar]
- 15.Waters AM, Der CJ. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med 2018; 8:a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009; 324:1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005; 7:469–83. [DOI] [PubMed] [Google Scholar]
- 18.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012; 491:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raphael BJ, Hruban RH, Aguirre AJ, Moffitt RA, Yeh JJ, Stewart C, et al. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017; 32:185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golan T, Kanji ZS, Epelbaum R, Devaud N, Dagan E, Holter S, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer 2014; 111:1132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guinney J, Dienstmann R, Wang X, De Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015; 21:1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mooi JK, Wirapati P, Asher R, Lee CK, Savas P, Price TJ, et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: Molecular analysis of the AGITG MAX clinical trial. Ann Oncol 2018; 29:2240–2246. [DOI] [PubMed] [Google Scholar]
- 24.Brenton JD, Carey LA, Ahmed A, Caldas C. Molecular classification and molecular forecasting of breast cancer: Ready for clinical application? J Clin Oncol 2005; 23:7350–60. [DOI] [PubMed] [Google Scholar]
- 25.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011; 17:500–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015; 47:1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531:47–52. [DOI] [PubMed] [Google Scholar]
- 28.Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology 2018; 155:1999–2013. [DOI] [PubMed] [Google Scholar]
- 29.Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O’Kane GM, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet 2020; 52:231–240. [DOI] [PubMed] [Google Scholar]
- 30.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV., et al. Signatures of mutational processes in human cancer. Nature 2013; 500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res 2015; 5:2929–43. [PMC free article] [PubMed] [Google Scholar]
- 32.Aung KL, Fischer SE, Denroche RE, Jang GH, Dodd A, Creighton S, et al. Genomics-driven precision medicine for advanced pancreatic cancer: Early results from the COMPASS trial. Clin Cancer Res 2018; 16:207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014; 344:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018; 560:319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018; 175:984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017; 171:1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018; 556:457–462. [DOI] [PubMed] [Google Scholar]
- 39.Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 2019; 177:1330–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang WL, Jagadeesh KA, Guo JA, Hoffman HI, Yadollahpour P, Mohan R, et al. Single-nucleus and spatial transcriptomics of archival pancreatic cancer reveals multi-compartment reprogramming after neoadjuvant treatment. bioRxiv 2020. [Google Scholar]
- 41.Raghavan S, Winter PS, Navia AW, Williams HL, Kalekar RL, Galvez-Reyes J, et al. Transcriptional subtype-specific microenvironmental crosstalk and tumor cell plasticity in metastatic pancreatic cancer. bioRxiv 2020. [Google Scholar]
- 42.Bernard V, Semaan A, Huang J, San Lucas FA, Mulu FC, Stephens BM, et al. Single Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clin Cancer Res 2018; 25:2194–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019; 29:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov 2019; 9:1102–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol 2020; 38:333–342. [DOI] [PubMed] [Google Scholar]
- 46.Juiz N, Elkaoutari A, Bigonnet M, Gayet O, Roques J, Nicolle R, et al. Basal-like and classical cells coexist in pancreatic cancer revealed by single-cell analysis on biopsy-derived pancreatic cancer organoids from the classical subtype. FASEB J 2020; 34:12214–12228. [DOI] [PubMed] [Google Scholar]
- 47.Lin W, Noel P, Borazanci EH, Lee J, Amini A, Han IW, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med 2020; 12:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin A V., et al. Pancreatic cancer. Nat Rev Dis Prim 2016; 2:16022. [DOI] [PubMed] [Google Scholar]
- 49.Ene-Obong A, Clear AJ, Watt J, Wang J, Fatah R, Riches JC, et al. Activated pancreatic stellate cells sequester CD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology 2013; 145:1121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Z, Vonlaufen A, Phillips PA, Fiala-Beer E, Zhang X, Yang L, et al. Role of pancreatic stellate cells in pancreatic cancer metastasis. Am J Pathol 2010; 177:2585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol 2017; 10:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017; 214:579–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell 2019; 178:160–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goel S, Decristo MJ, Watt AC, Brinjones H, Sceneay J, Li BB, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017; 548:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carstens JL, De Sampaio PC, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun 2017; 8:15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feig C, Jones JO, Kraman M, Wells RJB, Deonarine A, Chan DS, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci 2013;110:20212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Väyrynen JP, Lau MC, Haruki K, Väyrynen SA, Dias Costa A, Borowsky J, et al. Prognostic Significance of Immune Cell Populations Identified by Machine Learning in Colorectal Cancer Using Routine Hematoxylin and Eosin-Stained Sections. Clin Cancer Res 2020; 26:4326–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dominguez CX, Müller S, Keerthivasan S, Koeppen H, Hung J, Gierke S, et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov 2020; 10:232–253. [DOI] [PubMed] [Google Scholar]
- 59.Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 2016; 353:925–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 2017; 14:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barthelson RA, Lambert GM, Vanier C, Lynch RM, Galbraith DW. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics 2007; 8:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdelmoez MN, Iida K, Oguchi Y, Nishikii H, Yokokawa R, Kotera H, et al. SINC-seq: Correlation of transient gene expressions between nucleus and cytoplasm reflects single-cell physiology. Genome Biol 2018; 19:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Slyper M, Porter CBM, Ashenberg O, Waldman J, Drokhlyansky E, Wakiro I, et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med 2020; 26:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Versteijne E, Suker M, Groothuis K, Akkermans-Vogelaar JM, Besselink MG, Bonsing BA, et al. Preoperative Chemoradiotherapy Versus Immediate Surgery for Resectable and Borderline Resectable Pancreatic Cancer: Results of the Dutch Randomized Phase III PREOPANC Trial. J Clin Oncol 2020; 38:1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Porter RL, Magnus NKC, Thapar V, Morris R, Szabolcs A, Neyaz A, et al. Epithelial to mesenchymal plasticity and differential response to therapies in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci 2019; 116:26835–26845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nanda R, Chow LQM, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: Phase Ib keynote-012 study. J Clin Oncol 2016; 34:2460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitsunaga S, Ikeda M, Shimizu S, Ohno I, Furuse J, Inagaki M, et al. Serum levels of IL-6 and IL-1β can predict the efficacy of gemcitabine in patients with advanced pancreatic cancer. Br J Cancer 2013; 108:2063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Somerville TDD, Biffi G, Daßler-Plenker J, Hur SK, He XY, Vance KE, et al. Squamous trans-differentiation of pancreatic cancer cells promotes stromal inflammation. Elife 2020;9:e53381. [DOI] [PMC free article] [PubMed] [Google Scholar]