Abstract
Objectives:
To examine the association of major depressive disorder (MDD) and selective serotonin reuptake inhibitor (SSRI) use with gut microbiome in older adolescents and younger adults.
Methods:
Fifteen to 20-year-old participants within a month of starting an SSRI and unmedicated controls were enrolled in a longitudinal study. They underwent a diagnostic evaluation comprising self-completed and rater-administered questionnaires and clinical interview. They also provided a stool sample, which was stored at −80°C until DNA extraction. Microbial DNA was extracted with the MoBio PowerSoil kit, and the V4 region of the 16S rRNA was amplified and sequenced. Raw sequence data was processed with the LotuS pipeline. Only samples with no antibiotic exposure in the last 6 months and with >1000 quality filtered reads were included in the analysis.
Results:
160 participants (57.5% female, mean age 20.0±1.9 years, 29% taking SSRIs) were enrolled, comprising 110 MDD patients (60% in acute episode), 27 healthy controls, and 23 psychiatric controls. No significant group differences were observed in bacterial richness or alpha and beta diversity. Differential abundance analysis of bacterial taxa found no significant group differences at the phylum and genus levels. Neither being in a major depressive episode vs. remission nor using SSRIs was associated with differential bacterial composition.
Conclusions:
In this sizeable sample of older adolescents, neither MDD nor SSRI use was associated with differences in gut bacterial microbiome. In this age group, the bi-directional interaction between the gut bacteria and brain may be more nuanced than in adults, requiring further investigation.
1. Introduction
Major depressive disorder (MDD) is a prevalent and recurrent condition, associated with significant functional impairment and representing a serious public health concern (Judd, 1997; Moussavi et al., 2007; Zheng et al., 2016). While past efforts to study MDD have focused on neurological, behavioral, and genetic factors underlying this condition, the role that the gut microbiome plays in the development of MDD has gained attention in recent years (Jiang et al., 2015).
The human gut microbiome, sometimes referred to as the second genome, comprises almost 100 trillion bacteria with non-redundant genes many-fold the number of the host genes (Gill et al., 2006; Li et al., 2016). In fact, a growing number of studies have established that the gut microbiome influences brain development, impacting central nervous system functions and behavior (Clarke et al., 2013; Collins et al., 2012; Diaz Heijtz et al., 2011; Li et al., 2016; K.-A. M. Neufeld et al., 2011; K. M. Neufeld et al., 2011; Sudo et al., 2004). Moreover, using advanced high-throughput sequencing technology, studies have sought to examine its role in MDD (Chung et al., 2019; Jiang et al., 2015; Lin et al., 2017; Naseribafrouei et al., 2014; Valles-Colomer et al., 2019), which may lead to the development of novel therapeutic options. In fact, fecal transplantation studies have shown that a “depressive” phenotype may be transferred from patients with MDD to germ-free or antibiotic-treated mice (Kelly et al., 2016; Zheng et al., 2016). Clinical studies in MDD, however, have neither yielded consistent findings nor involved children and adolescents. Given that early-onset MDD is associated with a more chronic course and treatment resistance and that it may implicate different pathophysiological mechanisms compared to MDD of adult onset, examining the younger age group is critical (Birmaher et al., 2007, 1996; Curtin et al., 2016). Moreover, the extent to which selective serotonin reuptake inhibitors (SSRIs), the most-widely used antidepressant class, might alter the gut microbiome is unclear. Thus, we investigated the association between the gut microbiome, MDD, and SSRI treatment in older adolescents and young adults.
2. Methods
2.1. Participants
Between 09/2010 and 12/2014, 15 to 20-year-old participants were recruited for a longitudinal observational study examining the skeletal effects of SSRIs (Calarge et al., 2017b). Participants were enrolled from outpatient settings, either unmedicated or within a month of starting an SSRI. Treatment with psychotropics, other than SSRIs, during the two years prior to study entry led to exclusion, with the exception of benzodiazepines, trazodone, α2-agonists, or psychostimulants. The presence of an eating disorder, substance dependence, pregnancy, significant medical or surgical history, the chronic use of medications potentially affecting bone metabolism, or plans to move out of state within a year also led to exclusion (Calarge et al., 2017b). The Institutional Review Board at the University of Iowa approved the study, and informed consent and assent were obtained.
After completing the intake visit, the participants were contacted monthly and returned for an in-person visit every four months for up to two years. At every contact, they were queried about their medical history and medication use. Treatment adherence was based on self-report and pharmacy records.
At each in-person visit, the Inventory of Depressive Symptomatology (IDS) (Rush et al., 1996) was completed by trained research staff and the Beck Depression Inventory (BDI-II) (Beck, 1967) and Beck Anxiety Inventory (BAI) (Beck et al., 1988) were completed by the participants. In addition, height and weight were measured following standard procedures (Calarge et al., 2014).
Clinical diagnoses, based on the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV-TR) (Association, 2000) incorporated information from the medical record, rating scales, and an unstructured interview by a child and adolescent psychiatrist (Calarge et al., 2017a). Moreover, the Longitudinal Interval Follow-up Evaluation modified for use with Adolescents (A-LIFE) (Calarge et al., 2017b; Keller et al., 1987) was utilized to rate the severity of depressive symptoms and whether participants met criteria for a major depressive episode (MDE), each week during the study period. Alcohol use was also recorded.
2.2. Sample collection
Starting in 08/2011, study participants were encouraged to provide a stool sample either during the research visit (53%) or at home. Stool samples collected fresh were transported to the lab to be aliquoted and frozen within 15–30 min. Otherwise, samples were placed on dry ice and picked up within 24 h (if the sample was provided at home). Samples were excluded from the analysis if antibiotic use occurred in the 6 months prior to sample collection. All samples were stored at −80°C until DNA extraction.
2.3. Sequencing and data analysis
2.3.1. DNA extraction and 16S rRNA sequencing
Genomic DNA (gDNA) was extracted from the stool samples using the MoBio PowerSoil kit (Qiagen, Hilden, Germany) following manufacturer recommendations. Bacterial 16S rRNA gene was PCR amplified for the V4 region using the NEXTFlex® V4 Amplicon-Seq Kit 2.0 (Bio Scientific, Austin, TX) as previously described (Luna et al., 2017). Paired-end (2×250) sequencing was performed on Illumina Miseq (Illumina, San Diego, CA) platform using v2 chemistry kit at the Texas Children‟s Microbiome Center (TCMC).
2.3.2. Sequence processing and analysis
Raw sequence data were processed with the LotuS pipeline (v1.462) (Hildebrand et al., 2014), as previously described (Thapa et al., 2020). Resulting sequences were then clustered into operational taxonomic units (OTUs) at 97% identity against the SILVA database (v123) using UPARSE (USEARCH v8.0.1623). Vsearch (v1.9.7) was used to identify and remove chimeric sequences. Representative sequences were taxonomically assigned by the RDP classifier (v2.2) with the threshold of 0.8 using the SILVA database (v123) as a reference. OTUs failing to classify as bacteria at the kingdom level were removed prior to further analysis as were archaeal and cyanobacterial OTUs (Pammi et al., 2020; Thapa et al., 2020). In addition, low-abundance OTUs (i.e. OTUs not observed more than 3 times in at least 20% of the samples) were also removed from the dataset. Only samples with greater than 1000 quality-filtered reads were included in the analysis.
Relative abundance of bacteria at various taxonomic levels was calculated based on the available demographic and clinical features using the biom file generated with QIIME 1.9.1 (Caporaso et al., 2010). Bacterial diversity (alpha and beta) metrics were computed from the biom file in R-Studio using PhyloSeq (McMurdie and Holmes, 2013). Alpha diversity was estimated using various measures (Observed OTUs, Chao1, ACE, phylogenetic diversity and Shannon index) in the rarefied data set (data was rarefied to the lowest sequencing depth to reduce bias due to varying sequencing depth). Principal coordinates analysis (PCoA) plots of both non-phylogenetic-based (Bray-Curtis dissimilarity index) and phylogenetic-based (unweighted and weighted UniFrac distances) metrics were used to compare beta diversity between the samples. In addition, principal component analysis (PCA) ordination of the Aitchison distance (Aitchison, 1982), a compositionally aware distance metric, following centered log-ratio (CLR) transformation of the read counts (Gloor et al., 2017) was used to compare beta diversity between samples. The Aitchison distance was calculated using the R-package „microbiome‟ (Lahti and Shetty, 2019).
2.3.3. Statistical analysis
As described elsewhere (Coryell et al., 2019), group-based trajectory modeling (GBTM) used weekly A-LIFE-based MDD symptom ratings to find clusters of individuals following similar patterns of depressive symptoms change over time (Nagin and Odgers, 2010). Four trajectories were identified, with individuals being assigned to the trajectory where the GBTM-determined probability of membership was highest. The GBTM analysis was conducted using PROC TRAJ in SAS 9.4 (Jones et al., 2001).
The statistical analysis of metagenomic profiles (STAMP) (v2.1.3) was used to analyze the bacterial community profiles. Relative abundance between groups was compared using the Wilcoxon rank sum test (two groups) and Kruskal-Wallis test (more than two groups) using the Phyloseq (v1.32.0) R-package. ANOVA-like differential expression version 2 (ALDEx2), a compositionally aware differential abundance analysis tool, was also used for differential abundance analysis of bacterial taxa (Gloor et al., 2017).
Alpha diversity comparisons among multiple groups was done using a Kruskal-Wallis test, while Wilcoxon rank sum test was used for comparing two groups. Comparisons of beta diversity metrics among and within groups were performed using a permutational multivariate analysis of variance (PERMANOVA), as implemented in the „Adonis‟ function in vegan R-package (v2.5.6). Adjustments for multiple comparisons testing were made using the Benjamini-Hochberg (BH) correction to maintain the false-discovery rate (FDR) at 0.05.
3. Results
3.1. Demographic and clinical characteristics
One hundred seventy three participants provided stool samples, of whom 13 were excluded either due to low reads (n=11) or missing data (n=2). The demographic and clinical characteristics of the 160 participants contributing to this study are summarized in Table 1. Psychiatric controls comprised 10 participants with a primary diagnosis of generalized anxiety disorder, 6 with social anxiety disorder, 5 with attention deficit hyperactivity disorder, 1 with simple phobia, and 1 with cannabis use disorder. None had a history of MDD.
Table 1:
Demographic and clinical characteristics of the participants.
| MDD (n=110) |
Healthy Controls (n=27) |
Psych Controls (n=23) |
P Value | |
|---|---|---|---|---|
| Age, mean (sd) | 19.5 (0.4) | 20.3 (0.2) | 19.1 (0.4) | <0.006 |
| Female, n (%) | 72 (65) | 10 (37) | 10 (43) | <0.010 |
| White Race, n (%) | 89 (86) | 24 (89) | 20 (87) | >0.90 |
| Hispanic, n (%) | 9 (9) | 2 (7) | 3 (9) | >0.90 |
| Vaginal delivery, n (%) | 76 (78) | 21 (91) | 17 (74) | >0.30 |
| Overweight/Obese, n (%) | 26 (24) | 9 (33) | 8 (35) | >0.50 |
| Antibiotic use, n (%) | 8 (7) | 2 (7) | 1 (4) | >0.90 |
| Alcohol Use, n (%) | 86 (83) | 17 (63) | 16 (70) | <0.060 |
| MDD Status, n (%) | <0.0001 | |||
| Taking SSRI, n (%) | 43 (39) | 0 | 3 (13) | <0.0001 |
| Trajectory Group, n (%) | <0.0001 |
MDD: major depressive disorder, Psych Controls: psychiatric controls.
Missing data: Race (6), Ethnicity (6), Vaginal Delivery (16), Trajectory Group (7), Alcohol Use (6)
“Antibiotic Use” relates to use within 6 to 12 months prior to study entry, given that use within the past 6 months led to exclusion.
Trajectory group 1) participants with no depressive symptoms, 2) participants with low-level depressive symptoms, 3) participants in episode, remitting during the course of the study, and 4) participants persisting in episode during the course of the study.
sd= standard deviation
3.2. Sequence statistics
After removing non-bacterial reads and the taxa observed ≤ 3 times in at least 20% of the samples, a total of 2,239,476 sequences, belonging to 163 unique OTUs, were obtained from 160 stool samples sequenced for 16S V4 rRNA. On average, there were 13,997 (± 3,851 standard deviation) quality sequences per sample (Supplemental Table 1). Rarefaction curves at a depth from 1,000 to 23,000 sequences suggested sufficient sequencing depth to capture the majority of the bacterial community, as shown by the representative curves of observed OTUs reaching a plateau (Supplemental Figure 1).
3.3. Microbiome diversity and composition by participants group
For diversity analyses, the data were normalized by rarefying to a depth of 1,053 reads. We found no significant differences in bacterial richness (number of observed OTUs) or alpha diversity (Shannon index) between individuals with MDD, psychiatric disorders other than MDD, and healthy controls (Figure 1). Similar results were obtained with other measures of alpha diversity (Supplemental Figure 2). However, diversity (Shannon index, Figure 1) and phylogenetic diversity (Supplemental Figure 2) tended to be lower in participants with MDD and psychiatric controls compared to healthy controls.
Figure 1:
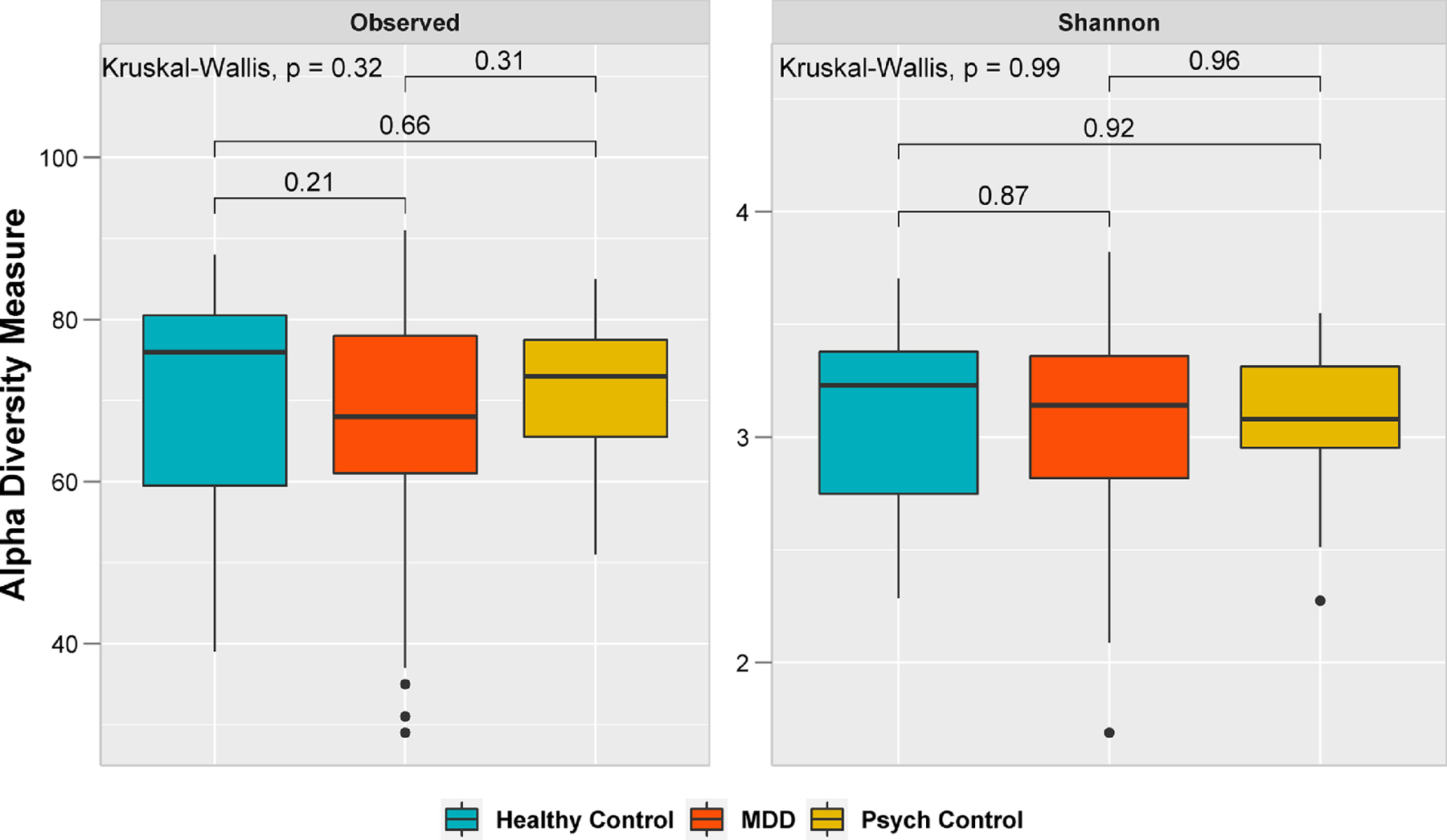
Alpha diversity of bacterial microbiome by group. Statistical analysis was performed using a global Kruskal-Wallis test between the three groups and a Wilcoxon rank sum test between the two groups. Healthy Controls (n=27); participants with major depressive disorder (MDD, n=110); and participants with psychiatric disorders other than MDD (Psych Control, n=23).
Two-dimensional PCoA plots of Bray-Curtis dissimilarity index and UniFrac distance (unweighted and weighted) metrics revealed no significant differences in gut microbiome composition between participants with MDD, psychiatric disorders other than MDD, and healthy controls (all p values > 0.05, Figure 2). Beta diversity ordination based on the Aitchison distance using CLR transformed data showed similar results (Supplemental Figure 3).
Figure 2:
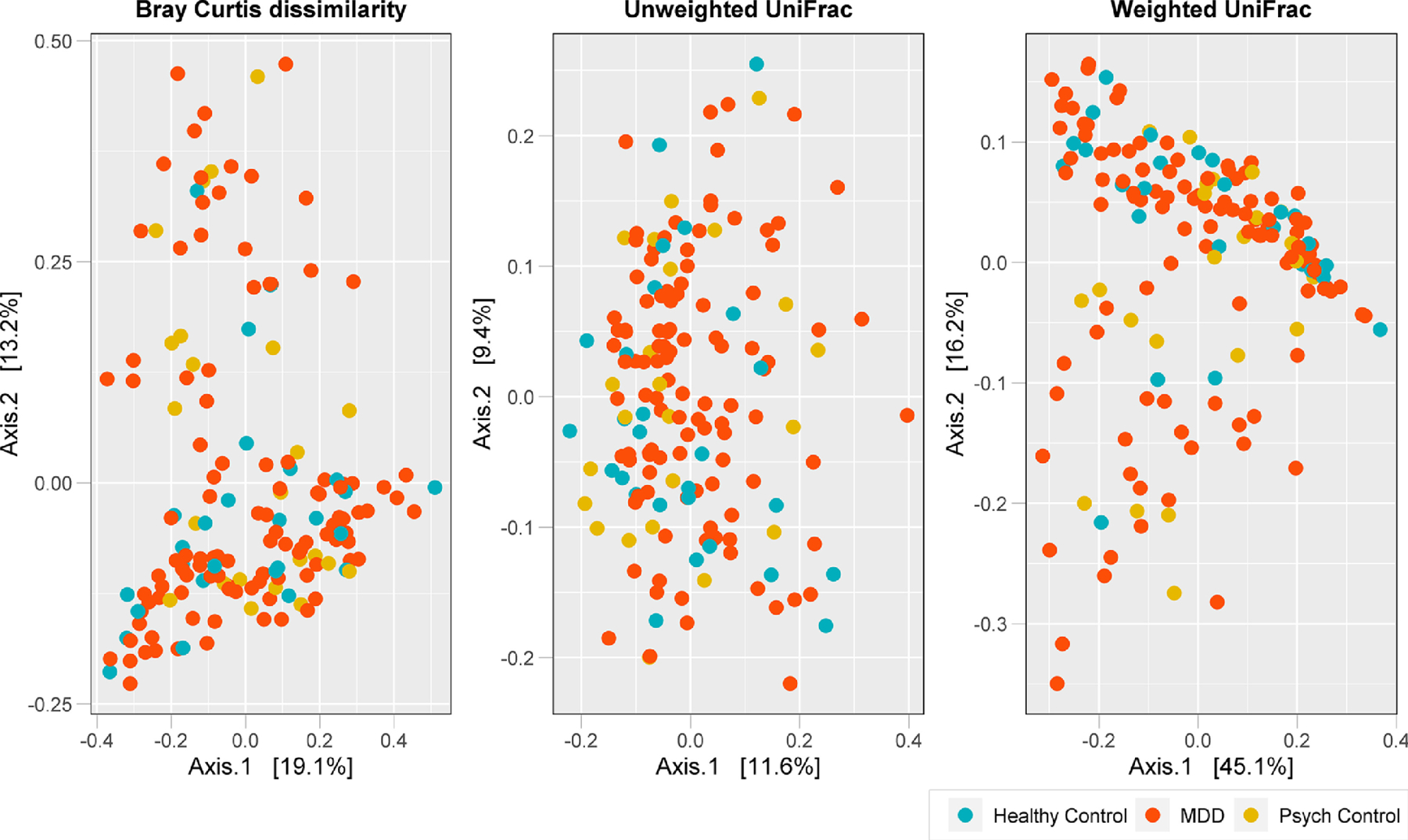
Beta diversity of bacterial microbiome by group. Two-dimensional principal coordinate analysis (PCoA) plots of Bray-Curtis dissimilarity index, unweighted and weighted UniFrac distances in healthy controls (n=27) and participants with major depressive disorder (MDD, n=110) or with psychiatric disorders other than MDD (psych controls, n=23). Axes represent the first two principal coordinates of the PCoA, and each point on the plot represents the bacterial microbiome of an individual stool sample (blue = healthy controls, red = MDD, orange = psych controls).
Differential abundance analysis of bacterial taxa revealed no significant group differences (BH-corrected Kruskal Wallis test p >0.05) at either the phylum or genus levels (Figure 3). Similar results were obtained with ALDEx2. However, relative abundance of phylum Actinobacteria was generally higher in both healthy controls (mean relative abundance= 4%) and psychiatric controls (4%), compared to MDD (2%) group. Overall, Firmicutes and Bacteroidetes were the major phyla, accounting for 85% or higher abundance in all three groups.
Figure 3:
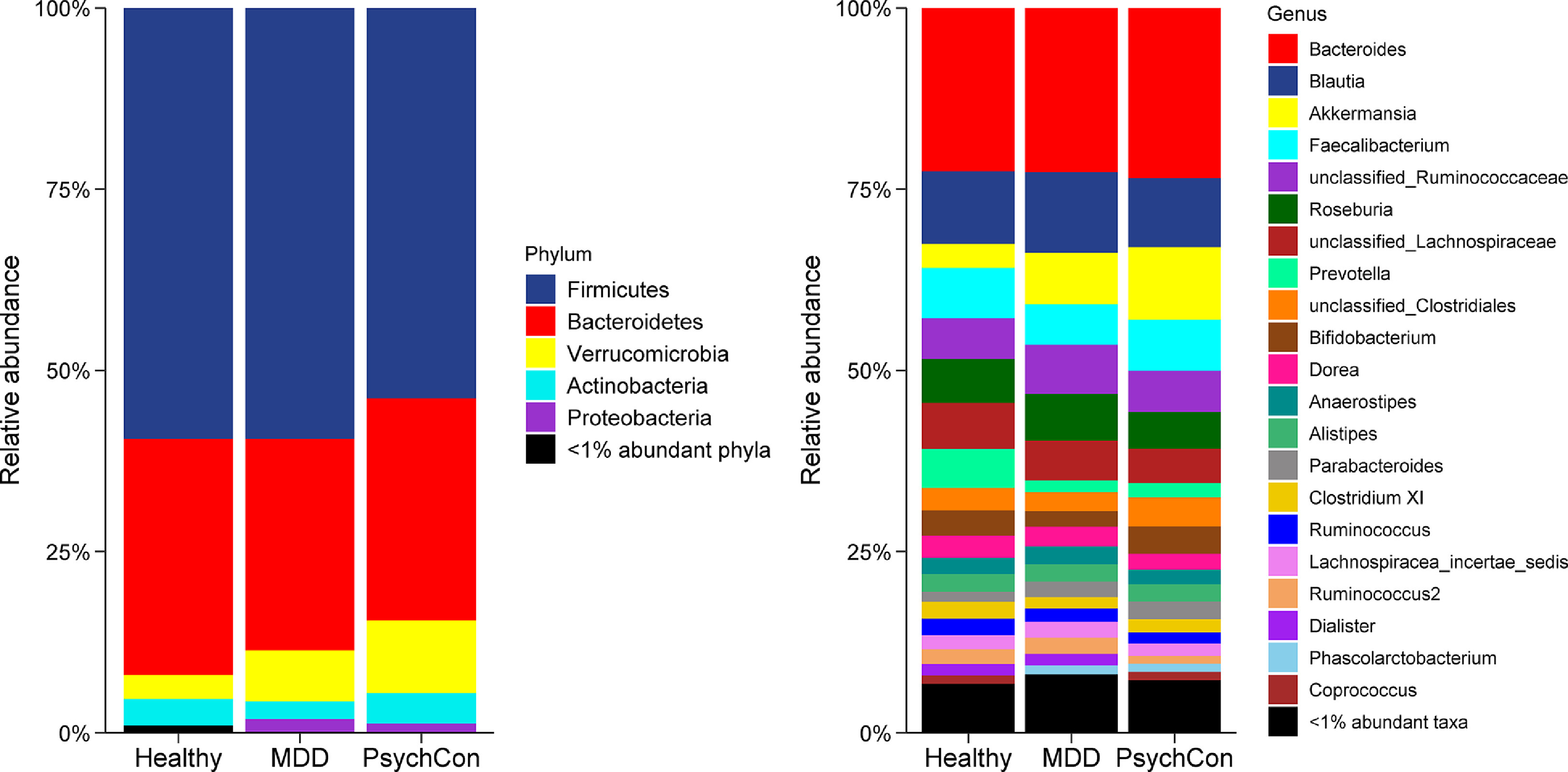
Comparison of bacterial relative abundance at the phylum and genus levels by participant groups. Healthy controls (n=27), major depressive disorder (MDD) (n=110) and psychiatric disorders other than MDD (PsychCon) (n=23). To optimize visualization, taxa with less than 1% abundance are grouped together as “<1% abundant phyla/taxa”.
At the genus level, mean relative abundance of Akkermansia was increased in MDD (7%) and psychiatric controls (10%) in comparison to the healthy controls (3%). In contrast, Faecalibacterium had a relatively similar abundance among healthy controls (7%), psychiatric controls (7%) and MDD group (6%), as did the genus Bacteroides (23%).
3.4. Effect of demographics and clinical factors on the gut microbiome composition
The effects of demographics and other factors on the gut microbiome composition as measured by PERMANOVA test of Bray-Curtis dissimilarity index is shown in Table 2.
Table 2:
Effects of demographics and other factors on Bray-Curtis dissimilarity index using PERMANOVA.
| Variable | F-value | R2 | p-value |
|---|---|---|---|
| Ethnicity | 1.33 | 0.01 | 0.19 |
| Race | 0.82 | 0.02 | 0.85 |
| Sex | 2.8 | 0.02 | 0.003 |
| Delivery mode | 0.99 | 0.007 | 0.43 |
| Overweight/Obese | 0.95 | 0.006 | 0.51 |
| Fresh Sample Aliquoting | 1.3 | 0.02 | 0.12 |
The effect of sex distribution on the microbiome composition is shown in Figure 4. PERMANOVA analysis of Bray-Curtis dissimilarity index revealed that the overall gut microbiome composition was significantly different between male and female participants differed (Table 2). Notably, the dispersion analysis (betadisper test at 1000 permutations) revealed no significant differences between male and female participants (i.e. equal sample variability between groups), suggesting that the microbiome composition is different between males and females.
Figure 4:
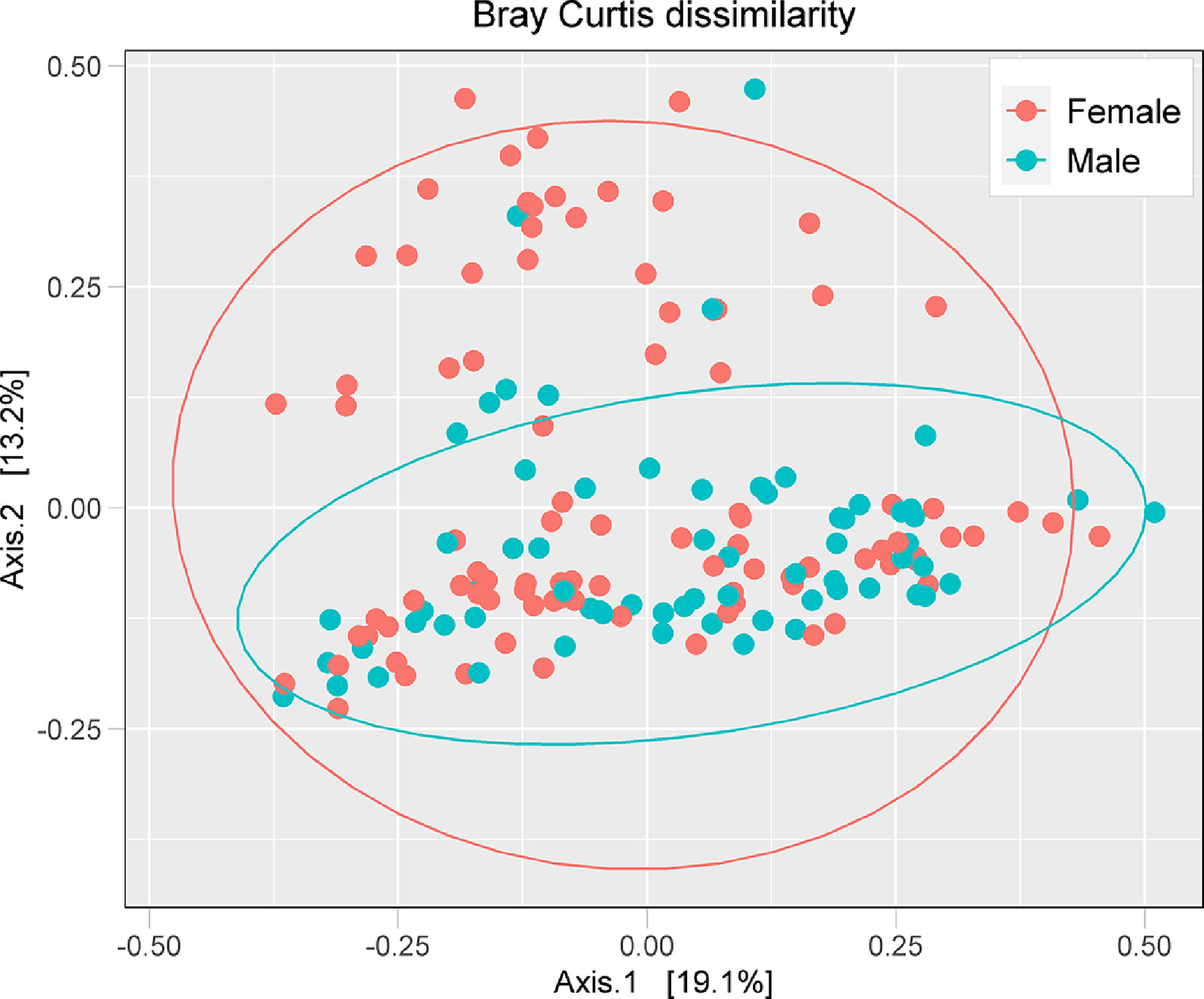
PCoA plot of Bray-Curtis distance metrics by sex distribution of all the participants.
Several variables reflecting psychiatric status or MDD severity were investigated to examine their association with gut microbiome composition (Table 3). Neither MDD status (i.e., being in episode or in remission, or never having had MDD), nor SSRI use (an index of clinically significant MDD), nor the interaction between them was significantly associated with a differential gut microbiome composition. Moreover, the GBTM analysis identified four trajectory groups, comprising participants with no depressive symptoms (i.e., never depressed), participants with low-level depressive symptoms (i.e., subclinical), participants who were in episode upon enrollment but remitted during the course of the study, and 4) participants persisting in episode throughout the course of the study (i.e., chronic depression). Again, these trajectories were not associated with differential gut bacterial composition, neither was alcohol use.
Table 3:
Effects of depression status, alcohol consumption and medications on Bray-Curtis dissimilarity using PERMANOVA test.
| Variable | F-value | R2 | p-value |
|---|---|---|---|
| Depression Status | 0.92 | 0.011 | 0.51 |
| Psychiatric Status | 0.93 | 0.017 | 0.57 |
| MDD Status | 0.91 | 0.008 | 0.52 |
| Taking SSRI | 0.97 | 0.006 | 0.42 |
| Depression Status by Taking SSRI | 0.95 | 0.024 | 0.54 |
| MDD Trajectory Group | 0.81 | 0.023 | 0.79 |
| Antibiotics use >6 months prior to study | 0.71 | 0.004 | 0.76 |
| Alcohol use | 1.12 | 0.007 | 0.34 |
SSRI: selective serotonin reuptake inhibitor. MDD: major depressive disorder
Depression Status included participants who were never depressed (n==50), in episode (n=66), and in remission (n=44).
Psychiatric Status included participants who were healthy controls (n=27), psychiatric controls (n=23), in episode (n=66), and in remission (n=44).
MDD status was restricted to participants who were in episode (n=66) or in remission (n=44).
Trajectory group 1) participants with no depressive symptoms (n=68), 2) participants with low-level depressive symptoms (n=17), 3) participants in episode, remitting during the course of the study (n=47), and 4) participants persisting in episode during the course of the study (n=21).
4. Discussion
To our knowledge, this is the first study to examine gut bacterial composition in adolescents with MDD, finding no significant association either with having ever had a MDD or with being in a current major depressive episode or with the trajectory of depressive symptoms over a 2-year follow-up period. Moreover, we also found no association with ongoing treatment with SSRIs.
Several studies have examined gut bacterial composition in adults with MDD, reaching disparate conclusions. Consistent with our findings, most studies failed to find differences in α-diversity between participants with versus those without MDD (Chung et al., 2019; Kelly et al., 2016; Lin et al., 2017; Naseribafrouei et al., 2014; Zheng et al., 2016). The study by Jiang et al. (Jiang et al., 2015) stands out, given that α-diversity was significantly different between participants in an ongoing depressive episode and healthy controls. This was not the case for treatment responders. In contrast, findings related to β-diversity are more inconsistent (Chung et al., 2019; Kelly et al., 2016; Lin et al., 2017; Naseribafrouei et al., 2014; Zheng et al., 2016).
Even in those studies that reported no differences in bacterial diversity, a wide range of differences were found at various taxonomic levels. For instance, Chung et al. (Chung et al., 2019) found the gut microbiome of MDD patients enriched with Firmicutes and Actinobacteria at the phylum level and with Bifidobacterium and Blatutia at the genus level but depleted of Prevotella compared to healthy controls. In contrast, Jiang et al. (Jiang et al., 2015) found that MDD patients had fewer Faecaelibacterium than controls and significantly more Alistipes and Enterobacteriaecae. Moreover, the prevalence of Faecalibacterium was inversely correlated with depressive symptom severity. In Naseribafrouei et al. (Naseribafrouei et al., 2014), the MDD group had increased relative abundance of Bacteroidales and decreased abundance of Lachnospiraceae compared to non-depressed controls. In Lin et al. (Lin et al., 2017) study, MDD patients have increased levels of Firmicutes and decreased levels of Bacteroidetes at the phylum level and increased levels of Clostridium XI, Klebsiella, Prevotella, and Streptococcus at the genus level. In Kelly et al. (Kelly et al., 2016), MDD was associated with increases in Thermoanaerobacteriaceae and relative depletion in Prevotellaceae at the family level, and increases in Anaerofilum, Eggerthella, Gelria, Holdemania, Turicibacter, Paraprevotella and relative depletion in Dialister and Prevotella at the genus level. Similarly, Zheng et al. (Zheng et al., 2016) reported significant alterations in the relative prevalence of Actinobacteria, Bacteroidetes, and Firmicutes between patients with MDD and healthy controls. More recently, in a significantly larger study, Valles-Colomer et al. (Valles-Colomer et al., 2019) found that Coprococcus and Dialister were relatively depleted in patients with MDD. In summary, even when specific taxonomic differences have been identified, only a few could be replicated (Valles-Colomer et al., 2019). Such inconsistency in the findings may be due to several factors including sample size, sex composition, ascertainment method (inpatient/outpatient), MDD chronicity, medication treatment, and comorbidity with other medical (i.e. obesity) or psychiatric conditions (i.e. alcohol, cigarette, or other substance use disorders). The findings may also be due to a type II error, given the relatively small sample size in most studies. In fact, compared to most studies (Chung et al., 2019; Jiang et al., 2015; Kelly et al., 2016; Lin et al., 2017; Liu et al., 2016; Naseribafrouei et al., 2014; Zheng et al., 2016), our study included a larger number of participants, in general, and those with MDD, in particular. Another unique feature of our study includes monitoring the participants for up to two years, which allowed for characterizing their symptom trajectory. We also included participants with other psychiatric conditions to determine the specificity of any potential finding to MDD and examined a variety of potential confounding factors, including antibiotic use. The latter is a significant improvement over other studies, which either did not mention prior antibiotic use (Zheng et al., 2016) or used a more liberal inclusion criterion, excluding only participants with antibiotic exposure in the prior 1 to 2 months (Chung et al., 2019; Jiang et al., 2015; Lin et al., 2017; Liu et al., 2016).
One factor that is unique to our study is the age of our patient population. The potential impact that MDD may have on the gut microbiome has had a shorter timeframe to manifest in this younger group of individuals. In fact, it may be that comorbid conditions that alter the gut microbiome have not had a long enough time to exert their effect. For example, although a substantial number of our participants, particularly those with MDD, used alcohol, this did not seem associated with alterations in the gut microbiome as others have found (Gorky and Schwaber, 2016; Hillemacher et al., 2018; Leclercq et al., 2014; Qamar et al., 2019; Vujkovic-Cvijin et al., 2020). Alcohol use disorder led to exclusion only in some studies examining gut microbiome composition in adults with MDD (Jiang et al., 2015; Kelly et al., 2016). Consistent with other published reports (Naseribafrouei et al., 2014; Zheng et al., 2016), overweight and obesity in our participants were also not associated with a specific gut microbiome signature.
Olanzapine-induced weight gain in rats is mediated by alteration in the gut microbiome composition (Davey et al., 2013, 2012). Consistent with these preclinical findings, our group has shown that risperidone treatment in children and adolescents similarly alters the Bacteroidetes to Firmicutes ratio in the gut microbiome (Bahr et al., 2015). However, neither in that study, nor in this much larger one, were SSRIs associated with significant gut microbiome changes. This is somewhat surprising given that the gut produces more than 95% of the body’s serotonin and the tight interactions the gut has with its microbiota.
While this is one of the larger studies seeking to characterize gut bacterial composition in patients with MDD, accounting for various confounding variables, some limitations must be acknowledged. First, our participants were enrolled from outpatient settings, with some of those with MDD not receiving medication treatment, suggesting perhaps a lower severity of the condition. Second, while this is the first study conducted in adolescents, it may be that a longer illness duration is needed to appreciably alter the gut microbiome. This is, however, unlikely, given that various factors, including diet, result in rapid changes in gut bacterial composition. We also did not investigate the effect of probiotic use as a potential confounding variable. Probiotics may improve depressive symptoms severity both in patients with MDD and in healthy controls (Huang et al., 2016; Kuo and Chung, 2019). Fourth, the sample size was substantially different between participants with MDD and those without, potentially limiting statistical power. Fifth, our analysis did not include metagenomic approaches, which may have exposed group differences in spite of the apparent absence of any specific associations of gut bacterial composition with MDD. Finally, the parent study collected the Block Adult Food Frequency Questionnaire (Calarge et al., 2017b), which characterizes the diet of participants over the prior year but fails to capture the granularity needed to examine how the diet in the day(s) preceding the visit may have affected gut microbiome composition.
5. Conclusions
In summary, this study contributes to the growing literature examining the role of the gut microbiome in psychiatric pathology, showing that neither MDD nor SSRI use was associated with differences in gut bacterial composition in older adolescents. This raises the possibility of a more nuanced microbiota-host interaction, whereby functional microbiome differences may be implicated in MDD. Additionally, the propensity of the host to respond to stress with increased gut permeability could also moderate the effect of the gut microbiome on the brain. In fact, we have previously reported increased gut permeability in unmedicated adolescents with MDD, compared to healthy controls (Calarge et al., 2019). Moreover, we found preliminary evidence linking this to activation of the sympathetic nervous system and innate immunity. As such, further research is needed to better delineate how the gut microbiome is implicated in psychopathology, in general, and in MDD, more specifically.
Supplementary Material
Highlights.
The role of the microbiome-gut-brain axis in psychopathology is gaining attention
Adolescents with and without major depression showed no gut microbiome differences.
Also, no significant effect of selective serotonin reuptake inhibitors was found.
Acknowledgements:
The authors thank the participants and their families, as well as the research team members who enrolled the participants and collected the data.
Role of Funding Source:
This study was funded, in part, by the National Institute of Mental Health (R01MH090072) and the National Center for Research Resources (2UL1TR000442-06). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors have no conflict of interest to report.
References
- Aitchison J, 1982. The Statistical Analysis of Compositional Data. Journal of the Royal Statistical Society. Series B (Methodological) 44, 139–77. [Google Scholar]
- Association, A.P., 2000. Diagnostic and Statistical Manual of Mental Disorders, 4th Edition, Text Revision, 4th Edition. ed. American Psychiatric Association, Washington, DC. [Google Scholar]
- Bahr SM, Tyler BC, Wooldridge N, Butcher BD, Burns TL, Teesch LM, Oltman CL, Azcarate-Peril MA, Kirby JR, Calarge CA, 2015. Use of the second-generation antipsychotic, risperidone, and secondary weight gain are associated with an altered gut microbiota in children. Transl Psychiatry 5, e652. 10.1038/tp.2015.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AT, 1967. Depression: Clinical, Experimental, and Theoretical Aspects, No additional printings listed Edition. ed. Harper and Row. [Google Scholar]
- Beck AT, Epstein N, Brown G, Steer RA, 1988. An inventory for measuring clinical anxiety: psychometric properties. J Consult Clin Psychol 56, 893–897. 10.1037//0022-006x.56.6.893 [DOI] [PubMed] [Google Scholar]
- Birmaher B, Brent D, AACAP Work Group on Quality Issues, Bernet W, Bukstein O, Walter H, Benson RS, Chrisman A, Farchione T, Greenhill L, Hamilton J, Keable H, Kinlan J, Schoettle U, Stock S, Ptakowski KK, Medicus J, 2007. Practice parameter for the assessment and treatment of children and adolescents with depressive disorders. J Am Acad Child Adolesc Psychiatry 46, 1503–1526. 10.1097/chi.0b013e318145ae1c [DOI] [PubMed] [Google Scholar]
- Birmaher B, Ryan ND, Williamson DE, Brent DA, Kaufman J, Dahl RE, Perel J, Nelson B, 1996. Childhood and adolescent depression: a review of the past 10 years. Part I. J Am Acad Child Adolesc Psychiatry 35, 1427–1439. 10.1097/00004583-199611000-00011 [DOI] [PubMed] [Google Scholar]
- Calarge CA, Butcher BD, Burns TL, Coryell WH, Schlechte JA, Zemel BS, 2014. Major depressive disorder and bone mass in adolescents and young adults. J. Bone Miner. Res 29, 2230–2237. 10.1002/jbmr.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarge CA, Devaraj S, Shulman RJ, 2019. Gut permeability and depressive symptom severity in unmedicated adolescents. J Affect Disord 246, 586–594. 10.1016/j.jad.2018.12.077 [DOI] [PubMed] [Google Scholar]
- Calarge CA, Mills JA, Janz KF, Burns TL, Coryell WH, Zemel BS, 2017a. Body Composition in Adolescents During Treatment With Selective Serotonin Reuptake Inhibitors. Pediatrics 140. 10.1542/peds.2016-3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarge CA, Mills JA, Janz KF, Burns TL, Schlechte JA, Coryell WH, Zemel BS, 2017b. The Effect of Depression, Generalized Anxiety, and Selective Serotonin Reuptake Inhibitors on Change in Bone Metabolism in Adolescents and Emerging Adults. J Bone Miner Res 32, 2367–2374. 10.1002/jbmr.3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R, 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y-CE, Chen H-C, Chou H-CL, Chen I-M, Lee M-S, Chuang L-C, Liu Y-W, Lu M-L, Chen C-H, Wu C-S, Huang M-C, Liao S-C, Ni Y-H, Lai M-S, Shih W-L, Kuo P-H, 2019. Exploration of microbiota targets for major depressive disorder and mood related traits. J Psychiatr Res 111, 74–82. 10.1016/j.jpsychires.2019.01.016 [DOI] [PubMed] [Google Scholar]
- Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF, 2013. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 18, 666–673. 10.1038/mp.2012.77 [DOI] [PubMed] [Google Scholar]
- Collins SM, Surette M, Bercik P, 2012. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol 10, 735–742. 10.1038/nrmicro2876 [DOI] [PubMed] [Google Scholar]
- Coryell W, Mills J, Dindo L, Calarge CA, 2019. Predictors of depressive symptom trajectories in a prospective follow-up of late adolescents. Psychol Med 1–6. 10.1017/S0033291719002551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SC, Warner M, Hedegaard H, 2016. Increase in Suicide in the United States, 1999–2014. NCHS Data Brief 1–8. [PubMed] [Google Scholar]
- Davey KJ, Cotter PD, O’Sullivan O, Crispie F, Dinan TG, Cryan JF, O’Mahony SM, 2013. Antipsychotics and the gut microbiome: olanzapine-induced metabolic dysfunction is attenuated by antibiotic administration in the rat. Translational Psychiatry 3, e309–e309. 10.1038/tp.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey KJ, O’Mahony SM, Schellekens H, O’Sullivan O, Bienenstock J, Cotter PD, Dinan TG, Cryan JF, 2012. Gender-dependent consequences of chronic olanzapine in the rat: effects on body weight, inflammatory, metabolic and microbiota parameters. Psychopharmacology 221, 155–169. 10.1007/s00213-011-2555-2 [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S, 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A 108, 3047–3052. 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE, 2006. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 312, 1355–1359. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ, 2017. Microbiome Datasets Are Compositional: And This Is Not Optional. Front Microbiol 8. 10.3389/fmicb.2017.02224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorky J, Schwaber J, 2016. The role of the gut-brain axis in alcohol use disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 65, 234–241. 10.1016/j.pnpbp.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Tadeo R, Voigt AY, Bork P, Raes J, 2014. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome 2, 30. 10.1186/2049-2618-2-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillemacher T, Bachmann O, Kahl KG, Frieling H, 2018. Alcohol, microbiome, and their effect on psychiatric disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 85, 105–115. 10.1016/j.pnpbp.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Huang R, Wang K, Hu J, 2016. Effect of Probiotics on Depression: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 8. 10.3390/nu8080483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, Li L, Ruan B, 2015. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun 48, 186–194. 10.1016/j.bbi.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Jones BL, Nagin DS, Roeder K, 2001. A SAS Procedure Based on Mixture Models for Estimating Developmental Trajectories. Sociological Methods & Research 29, 374–393. [Google Scholar]
- Judd LL, 1997. The clinical course of unipolar major depressive disorders. Arch. Gen. Psychiatry 54, 989–991. 10.1001/archpsyc.1997.01830230015002 [DOI] [PubMed] [Google Scholar]
- Keller MB, Lavori PW, Friedman B, Nielsen E, Endicott J, McDonald-Scott P, Andreasen NC, 1987. The Longitudinal Interval Follow-up Evaluation. A comprehensive method for assessing outcome in prospective longitudinal studies. Arch. Gen. Psychiatry 44, 540–548. 10.1001/archpsyc.1987.01800180050009 [DOI] [PubMed] [Google Scholar]
- Kelly JR, Borre Y, O’ Brien C, Patterson E, El Aidy S, Deane J, Kennedy PJ, Beers S, Scott K, Moloney G, Hoban AE, Scott L, Fitzgerald P, Ross P, Stanton C, Clarke G, Cryan JF, Dinan TG, 2016. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res 82, 109–118. 10.1016/j.jpsychires.2016.07.019 [DOI] [PubMed] [Google Scholar]
- Kuo P-H, Chung Y-CE, 2019. Moody microbiome: Challenges and chances. J. Formos. Med. Assoc 118 Suppl 1, S42–S54. 10.1016/j.jfma.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Lahti L, Shetty S, 2019. Introduction to the microbiome R package (Version 1.10.0) [WWW Document] URL https://microbiome.github.io/tutorials/ (accessed 12.14.20).
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, Windey K, Tremaroli V, Bäckhed F, Verbeke K, de Timary P, Delzenne NM, 2014. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. U.S.A 111, E4485–4493. 10.1073/pnas.1415174111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Su Q, Xie B, Duan L, Zhao W, Hu D, Wu R, Liu H, 2016. Gut microbes in correlation with mood: case study in a closed experimental human life support system. Neurogastroenterol. Motil 28, 1233–1240. 10.1111/nmo.12822 [DOI] [PubMed] [Google Scholar]
- Lin P, Ding B, Feng C, Yin S, Zhang T, Qi X, Lv H, Guo X, Dong K, Zhu Y, Li Q, 2017. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J Affect Disord 207, 300–304. 10.1016/j.jad.2016.09.051 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang L, Wang, Xiaoqi, Wang Z, Zhang J, Jiang R, Wang, Xiangqun, Wang K, Liu Z, Xia Z, Xu Z, Nie Y, Lv X, Wu X, Zhu H, Duan L, 2016. Similar Fecal Microbiota Signatures in Patients With Diarrhea-Predominant Irritable Bowel Syndrome and Patients With Depression. Clin. Gastroenterol. Hepatol 14, 1602–1611.e5. 10.1016/j.cgh.2016.05.033 [DOI] [PubMed] [Google Scholar]
- Luna RA, Oezguen N, Balderas M, Venkatachalam A, Runge JK, Versalovic J, Veenstra-VanderWeele J, Anderson GM, Savidge T, Williams KC, 2017. Distinct Microbiome-Neuroimmune Signatures Correlate With Functional Abdominal Pain in Children With Autism Spectrum Disorder. Cellular and Molecular Gastroenterology and Hepatology 3, 218–230. 10.1016/j.jcmgh.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S, 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi S, Chatterji S, Verdes E, Tandon A, Patel V, Ustun B, 2007. Depression, chronic diseases, and decrements in health: results from the World Health Surveys. The Lancet 370, 851–858. 10.1016/S0140-6736(07)61415-9 [DOI] [PubMed] [Google Scholar]
- Nagin DS, Odgers CL, 2010. Group-based trajectory modeling in clinical research. Annu Rev Clin Psychol 6, 109–138. 10.1146/annurev.clinpsy.121208.131413 [DOI] [PubMed] [Google Scholar]
- Naseribafrouei A, Hestad K, Avershina E, Sekelja M, Linløkken A, Wilson R, Rudi K, 2014. Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil 26, 1155–1162. 10.1111/nmo.12378 [DOI] [PubMed] [Google Scholar]
- Neufeld K-AM, Kang N, Bienenstock J, Foster JA, 2011. Effects of intestinal microbiota on anxiety-like behavior. Commun Integr Biol 4, 492–494. 10.4161/cib.4.4.15702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld KM, Kang N, Bienenstock J, Foster JA, 2011. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil 23, 255–264, e119. 10.1111/j.1365-2982.2010.01620.x [DOI] [PubMed] [Google Scholar]
- Pammi M, Thapa S, Balderas M, Runge JK, Venkatachalam A, Luna RA, 2020. Microbiome signatures in neonatal central line associated bloodstream infections. PLoS One 15, e0227967. 10.1371/journal.pone.0227967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar N, Castano D, Patt C, Chu T, Cottrell J, Chang SL, 2019. Meta-analysis of alcohol induced gut dysbiosis and the resulting behavioral impact. Behav. Brain Res 376, 112196. 10.1016/j.bbr.2019.112196 [DOI] [PubMed] [Google Scholar]
- Rush AJ, Gullion CM, Basco MR, Jarrett RB, Trivedi MH, 1996. The Inventory of Depressive Symptomatology (IDS): psychometric properties. Psychol Med 26, 477–486. 10.1017/s0033291700035558 [DOI] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Kubo C, Koga Y, 2004. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol 558, 263–275. 10.1113/jphysiol.2004.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa S, Runge JK, Venkatachalam A, Denne C, Luna RA, Anon JB, 2020. The Nasopharyngeal and Gut Microbiota in Children in a Pediatric Otolaryngology Practice. Pediatr Infect Dis J 39, e226–e233. 10.1097/INF.0000000000002703 [DOI] [PubMed] [Google Scholar]
- Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J, 2019. The neuroactive potential of the human gut microbiota in quality of life and depression. Nature Microbiology 4, 623–632. 10.1038/s41564-018-0337-x [DOI] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y, 2020. Host variables confound gut microbiota studies of human disease. Nature 587, 448–454. 10.1038/s41586-020-2881-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, Zeng L, Chen J, Fan S, Du X, Zhang X, Yang D, Yang Y, Meng H, Li W, Melgiri ND, Licinio J, Wei H, Xie P, 2016. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 21, 786–796. 10.1038/mp.2016.44 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


