Summary
• Two natural auxins, phenylacetic acid (PAA) and indole-3-acetic acid (IAA), play crucial roles in plant growth and development. One route of IAA biosynthesis uses the glucosinolate intermediate indole-3-acetaldoxime (IAOx) as a precursor, which is thought to occur only in glucosinolate-producing plants in Brassicales. A recent study showed that overproducing phenylacetaldoxime (PAOx) in Arabidopsis increases PAA production. However, it remains unknown whether this increased PAA results from hydrolysis of PAOx-derived benzyl glucosinolate or, like IAOx-derived IAA, is directly converted from PAOx. If glucosinolate hydrolysis is not required, aldoxime-derived auxin biosynthesis may occur beyond Brassicales.
• To better understand aldxoime-derived auxin biosynthesis, we conducted isotope labeled aldoxime feeding assay using an Arabidopsis glucosinolate-deficient mutant sur1 and maize, and transcriptomics analysis.
• Our study demonstrates that the conversion of PAOx to PAA does not require glucosinolates in Arabidopsis. Furthermore, maize produces PAA and IAA from PAOx and IAOx respectively indicating that aldoxime-derived auxin biosynthesis also occurs in maize.
• Considering that aldoxime production occurs widely in the plant kingdom, aldoxime-derived auxin biosynthesis is likely more widespread than originally believed. A genome-wide transcriptomics study using PAOx-overproduction plants identified complex metabolic networks among IAA, PAA, phenylpropanoid, and tryptophan metabolism.
Keywords: auxin, phenylacetic acid (PAA), indole-3-acetic acid (IAA), aldoxime, phenylpropanoid, Arabidopsis thaliana
Introduction
Auxins are plant hormones that are vital to plant growth and development, and play significant roles in a variety of other biological processes such as plant defense and stress responses (Zhao, 2010; Kidd et al., 2011; Korver et al., 2018). Plants naturally produce several auxins, including indole-3-acetic acid (IAA), phenylacetic acid (PAA), 4-chloroindole-3-acetic acid (4-Cl-IAA), and indole-3-butyric acid (IBA) (Simon and Petrášek, 2011). Among them, tryptophan-derived IAA has the greatest biological activity and is the first identified and most well-studied auxin (Haagen Smit and Went, 1935). The phenylalanine-derived auxin, PAA, has lower biological activity than IAA but accumulates to higher levels in some tissues (Schneider and Wightman, 1986; Ludwig-Muller and Cohen, 2002; Sugawara et al., 2015). Like many auxins, PAA can promote growth; however, it can’t undergo polar auxin transport (Morris and Johnson, 1987; Sugawara et al., 2015; Aoi et al., 2020a).
The YUCCA pathway is a major route of IAA biosynthesis (Kasahara, 2016). In this pathway, tryptophan is converted to indole-3-pyruvate by Tryptophan Aminotransferase of Arabidopsis (TAA) family aminotransferases (Stepanova et al., 2008; Tao et al., 2008; Wang et al., 2020). Then, YUCCA family flavin-containing monooxygenases (YUC) convert indole-3-pyruvate to IAA (Cheng et al., 2007; Cao et al., 2019). The YUCCA pathway may also act as a route of PAA biosynthesis, as several TAA and YUC enzymes can use phenylalanine and phenylpyruvate as substrates, respectively (Sugawara et al., 2015; Cook et al., 2016). In addition to the YUCCA pathway, several other routes of IAA biosynthesis, such as an indole-3-acetaldoxime (IAOx)-dependent route, have been proposed (Zhao, 2002; Sugawara et al., 2009; Wang et al., 2015). IAOx is a tryptophan-derived metabolite that is a precursor of Brassicales-specific defense compounds such as indole glucosinolates and camalexin (Mikkelsen et al., 2000; Glawischnig et al., 2004). In Arabidopsis, tryptophan is converted to IAOx by the action of two redundant cytochrome P450 monooxygenases, CYP79B2 (At4g39950) and CYP79B3 (At5g05260) (Zhao, 2002). CYP79B2 overexpression lines in Arabidopsis and Camelina sativa contain increased levels of IAA, whereas cyp79b2 cyp79b3 double mutants (hereafter called b2b3) are deficient in high-temperature-dependent IAA production (Zhao, 2002; Zhang et al., 2020). Furthermore, IAOx consumption mutants such as ref5 and sur1 accumulate IAA due to the redirection of IAOx to IAA (Mikkelsen et al., 2004; Kim et al., 2015).
IAOx metabolism impacts plant growth not only via IAOx-derived IAA production but also through modulating phenylpropanoid metabolism (Kim et al., 2015; Kim et al., 2020). The accumulation of IAOx or its derivatives increased the degradation of phenylalanine ammonia lyase (PAL), the entry point to the phenylpropanoid pathway in Arabidopsis and other Brassicales plants such as Camelina sativa (Zhang et al., 2020; Kim et al., 2020).
Plants also produce the phenylalanine-derived aldoxime phenylacetaldoxime (PAOx), which is a widely distributed metabolite within the plant kingdom (Irmisch et al., 2013; Luck et al., 2016; Luck et al., 2017; Sørensen et al., 2018; Thodberg et al., 2020). In Arabidopsis, CYP79A2 converts phenylalanine to PAOx which serves as a precursor for the defense compound benzyl glucosinolate (Wittstock and Halkier, 2000). A recent study showed that overexpression of CYP79A2 increases PAA and its conjugates (Aoi et al., 2020a). It has been suggested that glucosinolates are turned over in vivo (Petersen et al., 2002; Wittstock and Burow, 2010; Jeschke et al., 2019). Thus, it is possible that the increased PAA in CYP79A2 overexpression plants results either from direct conversion of PAOx to PAA similar to the IAOx-derived IAA production, or indirectly via benzyl glucosinolate which can be hydrolyzed in benzyl cyanide, a possible PAA precursor (Urbancsok et al., 2018; Günther et al., 2018). If aldoxime-derived auxin production does not require glucosinolate hydrolysis, it may be relevant beyond Brassicales.
In this study we examined glucosinolate dependency in PAOx-derived PAA production, if it occurs in non-Brassicacea species such as maize, and if enhanced PAOx production represses phenylpropanoid production as shown for IAOx. Our genetic study and stable isotope labeling assay demonstrate the conversion of PAOx to PAA in Arabidopsis glucosinolate-deficient plants. Furthermore, we show that maize can produce both IAA and PAA from their respective aldoximes. Our transcriptomics study using Arabidopsis plants with elevated PAOx production revealed complex metabolic interactions among PAOx, phenylpropanoids, and tryptophan-derived metabolites.
Materials and Methods
Growth Conditions and Genetic Material
Arabidopsis thaliana Col-0 and Zea mays B73 were used as wild-type plants. Plants were grown at 22°C ± 1°C with 16-h light/8-h dark photoperiod. For seedings grown on Murashige and Skoog (MS) growth medium plates, seeds were sterilized with 20% (v/v) bleach containing 0.005% triton X-100 (Sigma-Aldrich, MO) for 10 min. After being washed with water multiple times, the seeds were cold-treated at 4°C for 3 days before being planted on MS media containing 2% sucrose and 0.8% agar. For soil-grown plants, seeds were directly planted on soil after 3 days of cold treatment at 4°C.
The T-DNA insertion mutant sur1 (SAIL_1164_G12) was obtained from the ABRC (Ohio State University, Columbus, OH). Genotyping of sur1 was done using primers P367 and P368 (Table S1). The ref2 mutant, ref2-1, was genotyped by following a previously defined method (Hemm et al., 2003). cyp79b2 cyp79b3 (b2b3) plants and their genotyping method were published previously (Zhao, 2002).
Plasmid Construction and Transgenic Plant Generation
To generate AtCYP79A2 and ZmCYP79A61 overexpression constructs, the AtCYP79A2 and GRMZM2G138248 open reading frames were synthesized within the pUC57 vector (Genewiz, NJ). The synthesized entry vectors were subsequently recombined with the destination vector, pCC0995 (Kim et al., 2015), in which expression of the open reading frames is driven by the 35S promoter to generate the 35S:AtCYP79A2 and 35S:ZmCYP79A61 constructs. The 35S:AtCYP79A2 and 35S:ZmCYP79A61 constructs were introduced into wild type (Col-0), or ref2-1 plants via Agrobacterium tumefaciens (GV3101)-mediated transformation by following a method shown in Zhang et al. (Zhang et al., 2020). More than ten T1 plants were screened by application of 0.2% Basta (Rely 280, BASF, NJ). Several single-insertion homozygous T3 lines were established based on Basta resistance.
HPLC Analysis of Soluble Metabolites
For phenylpropanoid and glucosinolate analysis, whole rosette leaves of soil-grown 2-week-old plants were extracted with 50% methanol (v/v) at 65°C for 1 hour at a tissue concentration of 200 mg/mL. Samples were centrifuged at 10,000 g for 10 min, and the supernatant was collected for HPLC analysis. 10 μL of extract was analyzed on an UltiMate 3000 HPLC system (ThermoFisher Scientific, MA) equipped with an autosampler cooled to 10°C and a diode array detector (DAD). The compounds were separated on an Acclaim™ RSLC120 C18 column (100mm×3mm; 2.2μm) (ThermoFisher Scientific, MA), with a mobile phase consisting of solvent A (0.1% formic acid (v/v) in water) and solvent B (acetonitrile) with a linear gradient program (14-18% solvent B over 10 min) to separate metabolites. The flow rate was 0.5 mL/min and the column temperature was 40°C. The content of sinapoylmalate was quantified based on the peak area at 328 nm, and sinapic acid equivalents (Sigma, MO, D7927). Glucosinolate contents were quantified based on the peak area at 220 nm and authentic standards were used for quantification. Soluble metabolite analyses were conducted with two to four biological replicates.
Confirmation of Benzyl Glucosinolate using LC-ESI-MS and LC-ESI-MS/MS
Methanol extracts were cleaned using disposable SPE columns (Strata™-X 33 μm Polymeric, 8B-S100-TAK, Phenomenex Inc.) following the manufacturer’s protocol. A Jupiter® 4 μm Proteo 90 Å, LC Column 50 x 1 mm (Phenomenex, US) was used for LC-HRMS analysis. Data of LC-HRMS and MS/MS analysis was obtained using a ThermoFisher Q Exactive Focus mass spectrometer equipped with electrospray probe on Universal Ion Max API source. Full scan mass spectra and targeted MS/MS spectra were extracted from raw files using Xcalibur™ 2.1 (ThermoFisher Scientific, MA).
Solvent A (0.1% formic acid (v/v) in water) and solvent B (acetonitrile) were used as mobile phases with a linear gradient program (10-40% solvent B over 11 min) to separate chemicals by the above reverse phase HPLC column at a flow rate of 0.2 mL/min. MS1 signals were acquired under the Full Scan mode of the Orbitrap, in which a mass range of m/z 50-2000 was covered and data were collected in the negative ion mode. Fragmentation was introduced by HCD technique with optimized collision energy ranging from 18 to 30 eV. For each sample, the parent ion was selected as the precursor ion for MS/MS analysis. Other settings for the Orbitrap scan include resolution at 15000 and AGC target at 5×105. Two independent LC-MS analyses were conducted to confirm benzyl glucosinolate.
Auxin and Aldoxime Quantification
Auxin and aldoxime extraction were performed using methods adapted from Novák et al 2012 (Novák et al., 2012). Briefly, 30-100 mg fresh weight samples were extracted in 1 mL cold sodium phosphate buffer (50 mM, pH 7.0) containing 0.1% diethyldithiocarbamic acid sodium salt. For auxin quantification, 100 ng per sample of [13C6]-IAA and [13C6]-PAA were added as internal standards to the buffer solution. Plant samples were incubated at 4°C with continuous shaking for 40 min and then centrifuged at 13,000g at 4°C for 15 min. After adjusting the pH to 2.7, samples were purified by solid-phase extraction using Oasis™ HLB columns (Waters, MA, WAT094225). The final elutes with 80% methanol were evaporated to dryness in vacuo and stored at −20°C until LC/MS analysis.
All samples were resuspended in water and analyzed with Vanquish Horizon ultra-high performance liquid chromatography (UHPLC) installed with an Eclipse Plus C18 column (2.1× 50 mm, 1.8 μm) (Agilent) and mass analysis was performed using a TSQ Altis Triple Quadrupole (Thermo Scientific) MS/MS system with ion funnel. MRM parameters of the standards (precursor m/z, fragment m/z, radio frequency (RF) lens, and collision energy) of each compound was optimized on the machine using direct infusion of the authentic standards. PAA, [13C6]-PAA, D5-PAA, IAA, [13C6]-IAA, and D5-IAA were purchased from Cambridge Isotope laboratories, and IAOx, PAOx, D5-IAOx and D5-PAOx were synthesized as stated below.
For IAA quantification, the mass spectrometer was operated in positive ionization mode at ion spray voltage 4800V. Formic acid (0.1%) in water and 100% acetonitrile were employed as mobile phases A and B respectively with a gradient program (0-95% solvent B over 4 min) at a flow rate of 0.4 ml/min. The sheath gas, aux gas, and sweep gas were set at 50, 9, 1 (arb unit), respectively. Ion transfer tube and vaporizer temperatures were set at 325°C and 350°C, respectively. For MRM monitoring, both Q1 and Q3 resolutions were set at 0.7 FWHM with CID gas at 1.5 mTorr. Te scan cycle time was 0.8 s. MRM for IAA and aldoximes was used to monitor parent ion→ product ion reactions for each analyte as follows: m/z 175.983 → 130.071 (CE, 18V) for IAA; m/z 182.091→ 136 (CE, 18V) for [13C6]-IAA; m/z 181.102→134.083 (CE, 19V) for D5-IAA.
For PAA quantification, samples were analyzed in negative ionization mode, ion spray voltage −4500V. 100% water and 100% acetonitrile were used as mobile phases A and B, respectively. The gradient profile and the mass setting were same as stated previously. MRM for PAA was used to monitor parent ion→ product ion reactions as follows: m/z 135.045→ 91 (CE, −6V) for PAA; m/z 141.065→ 97 (CE, −7V) for [13C6]-PAA; and m/z 140.077→ 96 (CE, −7V) for D5-PAA. Auxin analysis was conducted with two to four biological replicates.
Feeding Assay
Detached leaves of 2-week-old Arabidopsis b2b3 mutants, leaf segments of the first leaf from 12-day-old maize plants, or 2-week-old whole Arabidopsis sur1 seedlings were placed in solutions containing 0.005% triton X-100 with or without 30 μM of unlabeled or deuterium-labeled aldoximes. After 24 hours, samples were removed from the solutions, weighed, frozen and stored at −80°C until extraction. Feeding assays were conducted with two to four biological replicates.
Aldoxime Synthesis
Aldoxime synthesis was done using procedures modified from Sugawara et al. (Sugawara et al., 2009). For the LiAlH4 reduction in PAOx formation, PAA (80.0 mg, 0.567 mmol) was dissolved in 10 mL of dry THF (0.06 M) was added to a flame dried Schlenk under N2. LiAlH4 (50.1 mg, 1.32 mmol) was added to the reaction mixture portion-wise at room temperature over 1 minute. The reaction mixture was left to stir for 1 hour at room temperature and then quenched with 2 M HCl at 0 °C. The ice bath was removed after the reaction was quenched (indicated by no more vigorous bubbling), and then 20 mL of an aqueous, saturated solution of Rochelle’s salt was added to the reaction flask. The solution continued to stir for an additional 2 hours at room temperature. The crude mixture was transferred to a separatory funnel and the aqueous layer was extracted three times with EtOAc. The organic layer was then washed with brine, dried with Na2SO4, and the solvent was evaporated in vacuo to afford 2-phenylethanol. For the LiAlH4 reduction in IAOx formation, IAA (100mg, 0.57 mmol) was added to a flame dried Schlenk under N2. LiAlH4 (50.1 mg, 1.32 mmol) was added to the reaction mixture portion-wise at room temperature over 1 minute. The reaction mixture was left to stir for 1 hour at room temperature and then quenched with 2 M HCl at 0 °C. The ice bath was removed after the reaction was quenched (indicated by no more vigorous bubbling), and then the whole mixture was extracted three times with EtOAc. The organic layer was then washed with brine, dried with Na2SO4, and the solvent was evaporated in vacuo to afford indole-3-ethanol.
For Parikh-Doering oxidation, the crude materials from the previous step (69.7 mg, 0.548 mmol) were dissolved in the 1:4 DMSO:DCE (0.1 M) solvent mixture and were added to a flame dried Schlenk flask under N2. Et3N (382 μL, 2.74 mmol) was then added and the reaction mixture was cooled to 0 °C in an ice bath, followed by the addition of SO3•pyr (261.6 mg, 1.64 mmol). The reaction continued to stir at 0 °C for 30 minutes. The crude reaction mixture was transferred to a separatory funnel followed by the addition of 20 mL of deionized H2O. The organic layer was washed with deionized water a total of three times in order to remove DMSO. Then, the aqueous layer was extracted three times with DCM. The organic layer was washed with brine, dried with Na2SO4, and the solvent was evaporated to afford crude indole-3-acetaldehyde (yellow-brown oil) or 2-phenylacetaldehyde (yellow oil).
For oxime formation, the crude materials from the previous step were dissolved in 5 mL of EtOH (0.1 M) in a round bottom flask. Hydroxylamine hydrochloride (76.2 mg, 1.10 mmol) and sodium acetate (89.9 mg, 1.10 mmol) were added to the reaction flask. The reaction mixture was left to stir at room temperature overnight. The crude mixture was filtered with EtOAc through a pad of silica gel and concentrated in vacuo. The crude material was then subjected to silica gel flash column chromatography (20% EtOAc: Hexanes) to afford a mixture (38.6 mg, 50% yield) of the E and Z isomers of the oxime products (IAOx and PAOx, respectively). The Z isomer was obtained in pure crystalline form after recrystallization from hexanes and CHCl3. Deuterium-labeled aldoximes D5-IAOx and D5-PAOx were synthesized from [indole-D5]-IAA and [phenyl-D5]-PAA as described above. Characterization Data [indole-D5]-IAA: 1H NMR (600 MHz, CD3OD) δ (ppm): 6.81 (t, J = 5.3 Hz, 1H), 3.80 (d, J = 5.3 Hz, 2H). The indole N-H and oxime O-H signals are not present in the spectrum taken in CD3OD due to rapid exchange with CD3OD.13C{1H} NMR (151 MHz, CD3OD) δ (ppm): 152.1, 138.0, 128.5, 123.6, 123.4, 123.2, 122.2, 122.0, 121.8, 119.4, 119.2, 119.1, 119.0, 118.9, 118.8, 118.6, 112.1, 112.0, 111.9, 111.7, 110.8, 22.3. [phenyl-D5]-PAA: 1H NMR (600 MHz, CDCl3) δ (ppm): 7.37 (bs, 1H), 6.90 (t, J = 5.4 Hz, 1H), 3.74 (d, J = 5.5 Hz, 2H).13C{1H} NMR (151 MHz, CDCl3) δ (ppm): 151.0, 136.8, 128.9, 128.9, 126.8, 31.8.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from whole areal parts of two-week old plants unless otherwise specified using the Trizol method as per the manufacturer’s protocol. (ThermoFisher Scientific, MA, 15596026). DNAse treatment, cDNA synthesis, and qRT-PCR analysis were conducted by following a method shown in a previous report (Zhang et al., 2020). 750 ng RNA was used for first-strand cDNA synthesis using reverse transcription kit (ThermoFisher Scientific, MA, 4368814) and qPCR reactions were performed in a 10 μL volume mix using SYBR Green components (ThermoFisher Scientific, MA, A25742). The expression was normalized to the reference gene Tubulin3 (At5g62700). A list of the primers used in this study is shown in Supplementary Table S1. qRT-PCR analysis was conducted with three biological replicates.
RNA Sequencing and Differential Expression Analysis
Whole rosettes of 2-week-old ox-2 and wild type were harvested in triplicate. Total RNA was extracted with Trizol as stated above. Paired-end, unstranded, 150-bp sequencing of these total RNA samples was performed by an Illumina NovaSeq 6000 machine. These RNA-seq data have been deposited in NCBI’s Sequencing Read Archive (SRA) under bioproject number PRJNA682862. Raw reads were trimmed and filtered using trimmomatic v0.39. Trimmed reads were aligned to the TAIR10 genome build using HISAT2 v2.2.0. Read counts were generated with the featureCounts function from subread v2.0.0 with fractional counting enabled for overlapping and multimapping reads. Differential expression analysis was completed using edgeR v3.30.3. To filter out genes expressed at low levels, a counts-million-cutoff (CPM) was calculated for each group of samples as 10/L, where L was the minimum library size in millions. Only genes which were expressed at a level above this threshold in all the samples in either group were retained. After normalizing for library size, a negative binomial model was fit to contrast expression in mutant samples to expression in wild type. p-values were calculated using empirical Bayes quasi-likelihood F-tests. Genes with a p-value smaller than 0.05 were considered differentially expressed (Fig S8A, see later). RNA-seq data for ref2 and ref5 mutants, and the associated wild type samples, were downloaded from GEO accession GSE99581.
Hypocotyl length measurement
Seven-days-old wild-type, ref2, and CYP79A2 overexpression lines were photographed. Using imageJ (ver1.52), hypocotyl length was measured. The data presented are from six to eight individual plants. Statistical significant was calculated by ANOVA with post-hoc Tukey’s test.
Results
Elevated PAOx production in Arabidopsis leads to benzyl glucosinolate and PAA accumulation.
To assess any impact of increased PAOx production on PAA and benzyl glucosinolate levels (Fig 1A), we engineered Arabidopsis wild type to constitutively express CYP79A2 and further studied two lines overexpressing CYP79A2 (Fig 1B–C). These lines were named ox-1 and ox-2 respectively. They displayed increased hypocotyl length, a characteristic high auxin morphology (Fig 1D)(Kim et al., 2007), and accumulated 6- to 10- fold more PAA than wild type (Fig 1E), which is consistent with a previous report (Aoi et al., 2020a). In non-stressed Arabidopsis (ecotype Col-0), the expression of CYP79A2 is very low in most organs (Fig S1A), so that its derived compound, benzyl glucosinolate, was below detectible levels in leaves of wild type (Fig 1F). Our HPLC analysis identified new peaks in both ox-1 and ox-2 which wild type does not accumulate. Our LC-MS analysis identified that the new peak is benzyl glucosinolate, which was further confirmed with its authentic standard (Fig 1F, Fig S2) suggesting conversion of PAOx to benzyl glucosinolate in these overexpression lines.
Figure 1.
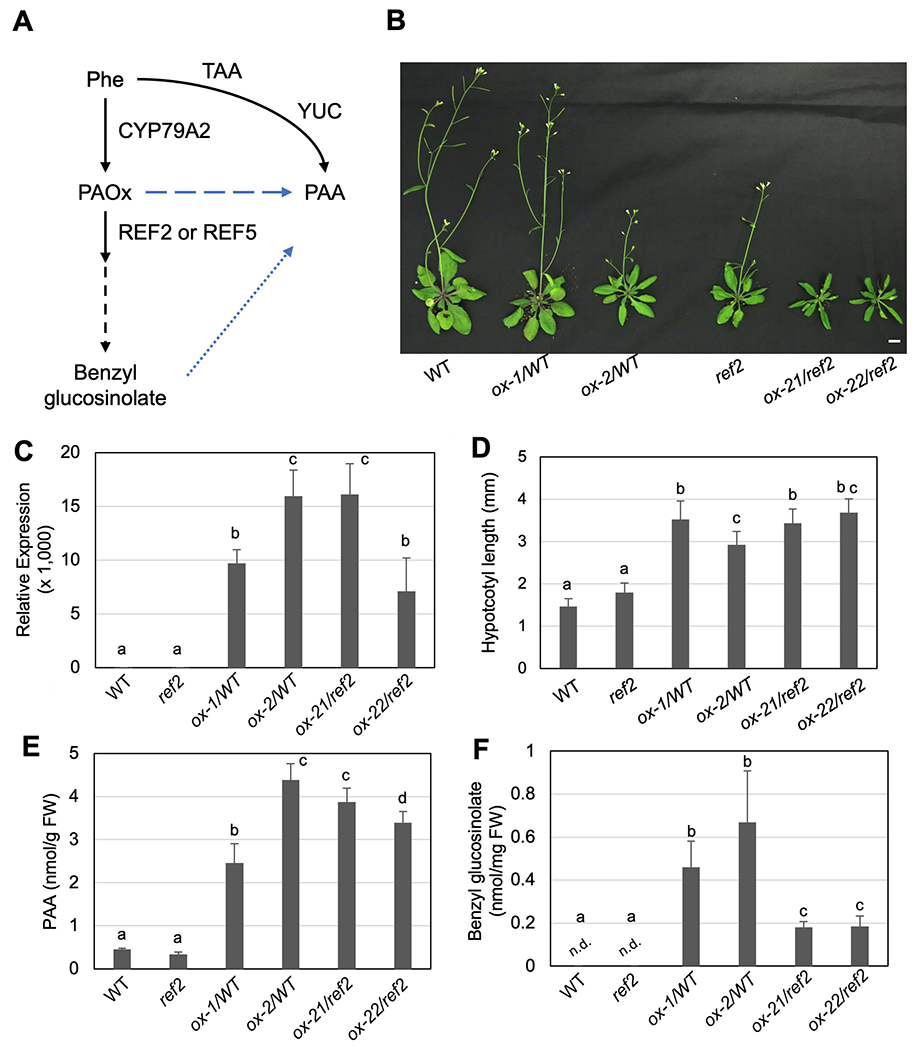
Increased PAOx production results in the accumulation of benzyl glucosinolate and PAA.
A) A schematic diagram of PAOx metabolism and PAA biosynthesis in Arabidopsis thaliana. Proposed routes of PAOx-derived PAA biosynthesis are shown with the dashed blue line. B) Representative 4-week-old CYP79A2 overexpression lines in the wild-type and ref2 genetic backgrounds compared to their controls. Scale bar = 1 cm. C) Relative expression of CYP79A2 in wild type, ref2, and CYP79A2 overexpression lines in the wild-type (ox-1, ox-2) and ref2 (ox-21, ox-22) genetic backgrounds (N=3). D) Hypocotyl length of seven-days-old wild-type, ref2, and CYP79A2 overexpression lines (N=8). E) Free PAA content of wild-type, ref2, and CYP79A2 overexpression lines (N=3). F) Benzyl glucosinolate content of wild-type, ref2, and CYP79A2 overexpression lines (N=4). 2-week-old whole aerial parts were extracted to measure relative amount of benzyl glucosinolate. Data represent mean ± SD. The means were compared by one-way ANOVA, and statistically significant differences (P < 0.05) were identified by Tukey’s test and indicated by letters to represent difference among groups.
It was shown that a benzyl glucosinolate hydrolysis product, benzyl cyanide, can be converted to PAA by nitrilases (Fig 1A) (Urbancsok et al., 2018; Günther et al., 2018). Thus, it is possible that the increased PAA in ox-1 and ox-2 results from either benzyl glucosinolate catabolism or conversion of PAOx to PAA bypassing benzyl glucosinolate (Fig 1A). To better understand the contribution of benzyl glucosinolate to PAA production, we generated CYP79A2 overexpression lines in ref2-1, a null allele for CYP83A1 (REF2) (Hemm et al., 2003). REF2 functions redundantly with CYP83B1 (REF5) in the committed step of benzyl glucosinolate biosynthesis by converting PAOx to phenylacetothiohydroximate (Fig 1A) (Naur et al., 2003). Due to the severe growth defects of REF5 null (Kim et al., 2015), which made them unsuitable for genetic study, we tested our hypothesis with ref2-1 (hereafter called ref2). Although REF5 and REF2 function redundantly, we thought that disruption of REF2 may impact on the flux toward benzyl glucosinolate production since they have slightly different substrate specificity (Naur et al., 2003). Two CYP79A2 overexpressing lines in the ref2 background accumulated 15,000- and 7,000- fold increase in CYP79A2 expression (Fig 1C). These lines were named ox-21/ref2 and ox-22/ref2 respectively. This increase in expression was equivalent to that of the wild type overexpressors ox-1/WT and ox-2/WT and led to the production of benzyl glucosinolate (Fig 1F). The CYP79A2 overexpressors in the ref2 background produced significantly less benzyl glucosinolates than those in the wild-type background (Fig 1F), yet the increases in PAA production and hypocotyl length were comparable (Fig 1D–E) suggesting a possibility that PAOx-derived PAA production may occur by bypassing benzyl glucosinolate hydrolysis.
Biosynthesis of PAA from PAOx does not require benzyl glucosinolate production.
To test if PAA production requires benzyl glucosinolate, we analyzed the production of PAA from PAOx in the glucosinolate-deficient mutant sur1, a null allele of the C-S lyase SUR1 (AT2g20610) which converts S-alkylthiohydroximates made by REF5 or REF2 to thiohydroximates (Fig 2A) (Mikkelsen et al., 2004). SUR1 requires for the production of all glucosinolates including indole and benzyl glucosinolates, and disruption of SUR1 causes high levels of IAA due to redirection of IAOx to IAA, leading to severe growth defects (Fig 2A–B) (Boerjan et al., 1995). Since CYP79A2 expression is negligible in non-stress condition (Fig 1C, Fig S1), accumulation of PAOx or its derivatives in sur1 are not expected. To test if our feeding system worked, IAOx, PAOx, or water were fed to the b2b3 mutant which fails to produce IAOx or its derivatives such as indole glucosinolates. As shown in Fig 2C, b2b3 plants fed with IAOx accumulated indole glucosinolates but water-fed controls did not. Similarly, benzyl glucosinolate accumulated in b2b3 plants fed with PAOx but not in water-fed controls (Fig 2D). These data demonstrate that aldoximes can be successfully taken up by Arabidopsis and incorporated into their respective pathways. As expected, sur1 failed to accumulate both indole and benzyl glucosinolates even after IAOx and PAOx feeding (Fig 2C–D). However, PAOx supplementation led to an increase in PAA levels in the sur1 mutant when compared to water-fed controls (Fig 2E), suggesting that PAOx-derived PAA production can occur without glucosinolate hydrolysis in Arabidopsis.
Figure 2.
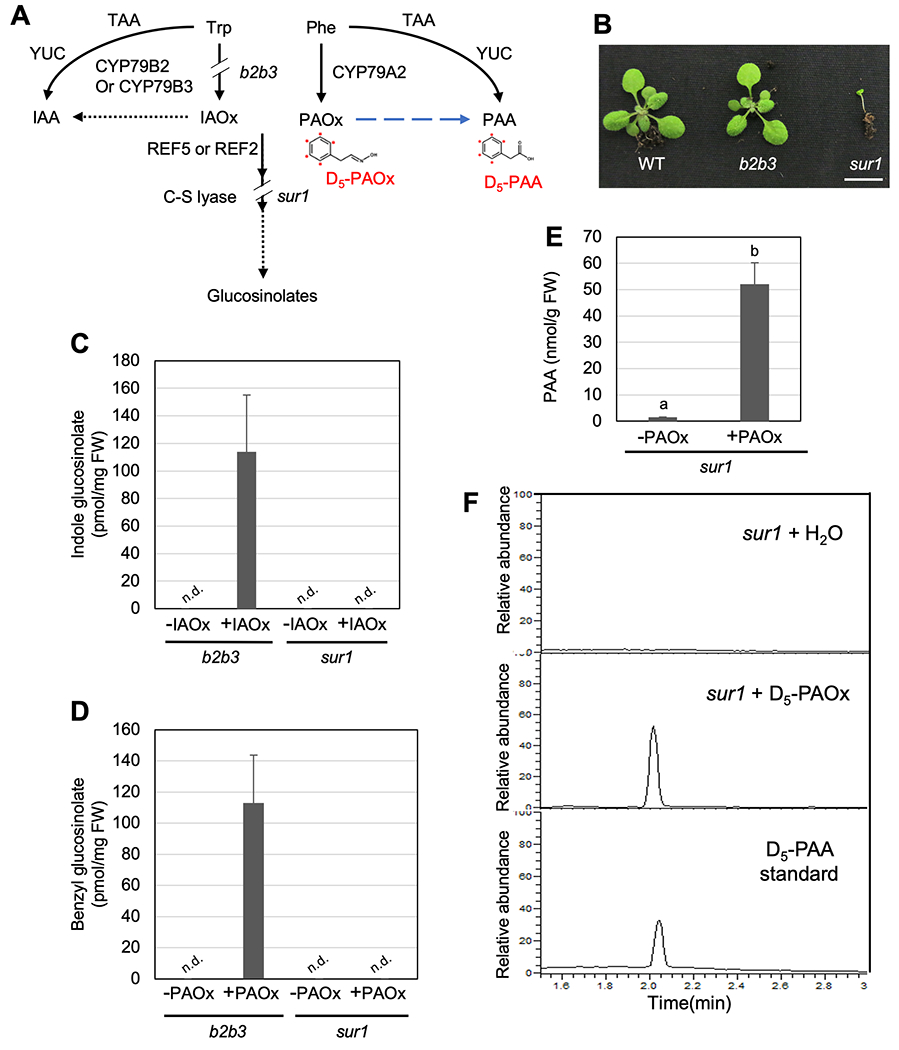
PAOx can be converted to PAA without benzyl glucosinolate hydrolysis.
A) A schematic diagram of PAOx and IAOx metabolism in Arabidopsis. The point of disruption in b2b3 and sur1 is marked in the schematic. The chemical structures shown represent deuterium labeled compound D5-PAOx and D5-PAA (the input and expected output of the labeled PAOx feeding assays done with sur1), with the red dots denoting the location of deuterium atoms. B) Representative 3-week-old wild type, cyp79b2 cyp79b3 double mutant (b2b3), and sur1 mutant. Scale bar = 1 cm. C) Indole-3-ylmethyl glucosinolate level in b2b3 and sur1 plants fed with water or IAOx for 24 hours (N=2 for sur1 and N=4 for b2b3). D) Benzyl glucosinolate level in b2b3 and sur1 plants fed with water or PAOx for 24 hours (N=2 for sur1 and N=4 for b2b3). C-E) Data represent mean ± SD. The means were compared by one-way ANOVA, and statistically significant differences (P < 0.05) were identified by Tukey’s test and indicated by letters to represent difference among groups. E) Free PAA content of water-fed and PAOx-fed 2-week-old sur1 seedlings (N=4). F) LC-MS chromatograms for the D5-PAA standard (bottom) and endogenous D5-PAA in sur1 seedlings after feeding D5-PAOx (middle) or water (top).
There are two possibilities for PAOx-mediated PAA accumulation: 1) PAOx or its derivative serves as a direct precursor of PAA, or 2) PAOx accumulation induces other routes of PAA biosynthesis, such as the YUCCA pathway. To determine if PAOx functions as a precursor for PAA biosynthesis, we fed sur1 mutants with deuterium-phenyl ring labeled PAOx (D5-PAOx) (Fig 2A). Our LC-MS analysis identified that sur1 fed with D5-PAOx accumulated D5-PAA, which was absent in water-fed sur1 (Fig 2F, Fig S3) demonstrating that PAOx can be converted to PAA in Arabidopsis in a manner reminiscent of the IAOx-derived IAA pathway (Sugawara et al., 2009). These data show that PAOx-derived PAA production in Arabidopsis can indeed occur directly without the need for glucosinolate-derived intermediates.
Maize can convert IAOx and PAOx to IAA and PAA, respectively.
Given that aldoxime-dependent auxin biosynthesis does not require Brassicales-specific glucosinolates as an intermediate, this pathway may also occur in non-glucosinolate-producing species. Several plant species possess CYP79 enzymes capable of generating IAOx, PAOx, or both. A recent in vitro study has shown that the maize enzyme ZmCYP79A61 (GRMZM2G138248) can convert tryptophan and phenylalanine to IAOx and PAOx respectively (Irmisch et al., 2015). Consistently, we found that Arabidopsis transgenic lines overexpressing ZmCYP79A61 contained increased IAA and PAA contents and displayed characteristic high-auxin morphological changes such as narrow, curled down leaves, elongated hypocotyls and elongated petioles, which are often observed in other high auxin mutants (Fig S4, S5).
Since ZmCYP79A61 produces aldoximes, and its overexpression can increase auxins in Arabidopsis, we hypothesize that maize can engage in aldoxime-dependent auxin biosynthesis. To test this, we performed unlabeled or labeled aldoxime feeding assays with maize. As shown in Fig 3, maize leaves fed with IAOx or PAOx increased IAA or PAA significantly compared to water-fed maize. Additionally, when maize leaves were fed with D5-IAOx or D5-PAOx, they produced D5-IAA or D5-PAA respectively whereas water-fed maize did not produce D5-IAA or D5-PAA (Fig 3C, D, Fig S6, S7). These results suggest that, like Arabidopsis, maize can utilize both IAOx and PAOx to produce IAA and PAA respectively.
Figure 3.
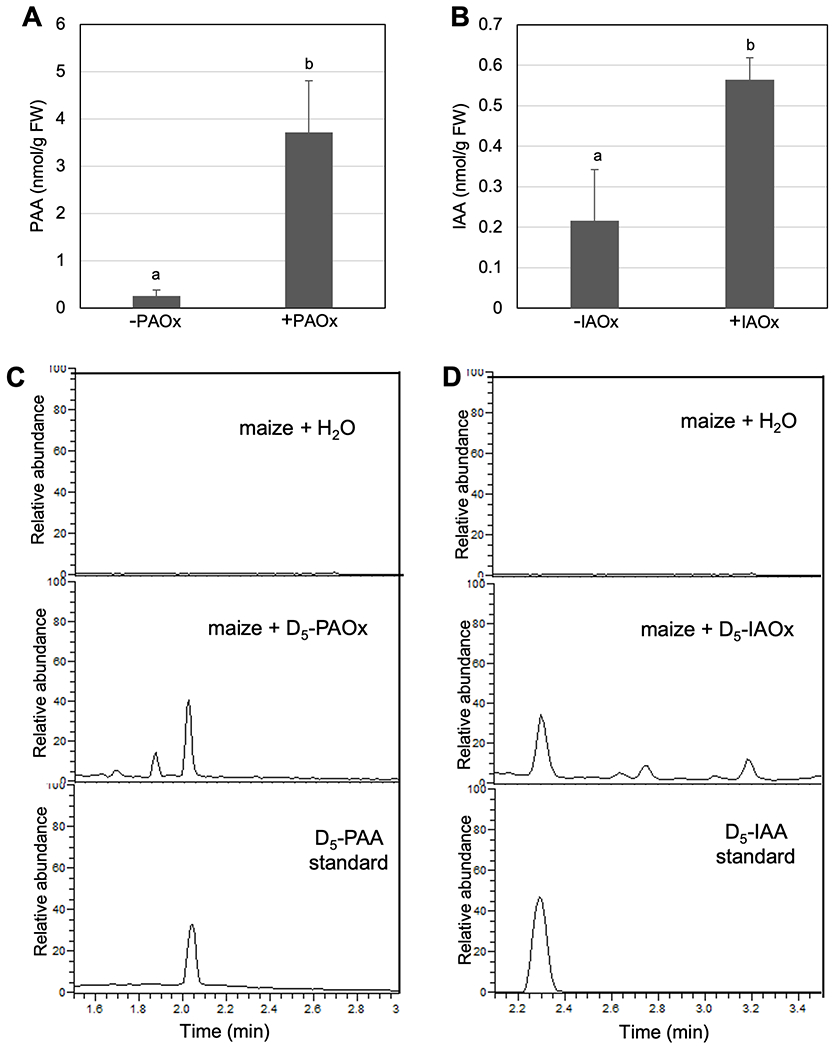
Maize converts IAOx and PAOx to IAA and PAA, respectively.
A) Free PAA content of maize leaves fed with water or PAOx (N=3). B) Free IAA content of maize leavs fed with water or IAOx (N=2). Data represent mean ± SD. First leaves from 12-day-old maize plants were incubated in either water or water containing 30 μM of aldoximes for 24 hours. C) LC-MS chromatograms for the D5-PAA standard (bottom) and endogenous D5-PAA in maize leaves after feeding with water (top) or D5-PAOx (middle). D) LC-MS chromatograms for the D5-IAA standard (bottom) and endogenous D5-IAA in maize leaves after feeding with water (top) or D5-IAOx (middle). The means were compared by one-way ANOVA, and statistically significant differences (P < 0.05) were identified by Tukey’s test and indicated by letters to represent difference among groups.
Enhanced PAOx production suppresses phenylpropanoid production.
Previous studies have demonstrated that enhanced IAOx metabolism, either by overexpressing the IAOx production enzyme CYP79B2 or by disruption of the IAOx consumption enzyme REF5, represses phenylpropanoid production in Arabidopsis and Camelina sativa (Kim et al., 2015; Zhang et al., 2020; Kim et al., 2020). In addition, disruption in REF2, which can catalyze aliphatic aldoximes, also repress phenylpropanoid production (Hemm et al., 2003). One cause for the repressed phenylpropanoid biosynthesis in ref5 and ref2 is the increased expression of a set of F-box genes (KFBs) that function in the ubiquitination of the PAL enzyme for degradation (Zhang et al., 2013; Kim et al., 2020). The induction of these KFB genes (KFB1/20/39/50) ultimately reduces flux through the phenylpropanoid pathway and results in a reduction in the accumulation of various phenylpropanoids, such as sinapoylmalate. Knocking out all four KFBs partially rescued phenylpropanoid production in ref5 and ref2 mutants, indicating that post-translational modification of PALs through transcriptional activation of KFBs is one mechanism underlying the crosstalk between aldoxime metabolism and phenylpropanoid biosynthesis (Fig 4A) (Kim et al., 2020).
Figure 4.
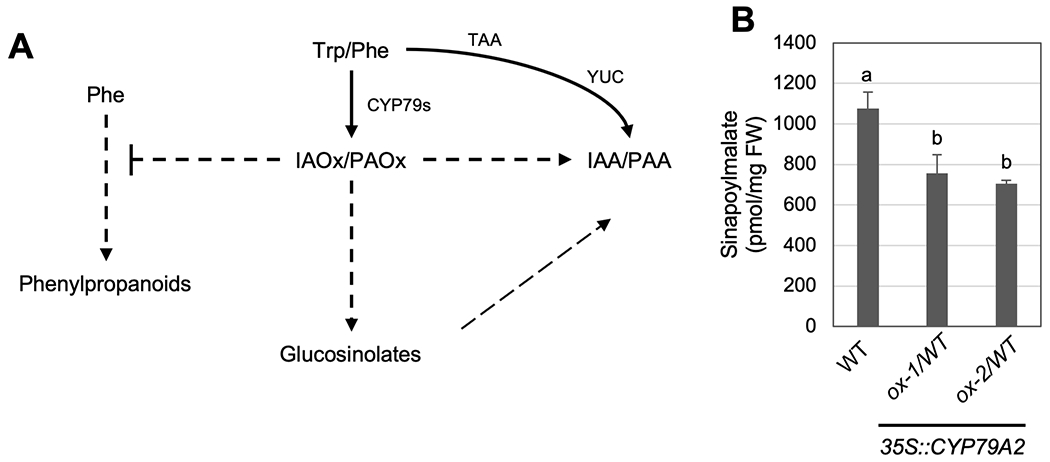
PAOx metabolism is linked to phenylpropanoid production.
A) A schematic of the metabolic network of IAOx and PAOx in Arabidopsis, as well as the major route of IAA and PAA biosynthesis. B) The level of sinapoylmalate in wild type and CYP79A2 overexpression (ox-1, ox-2) (N=4). Data represent mean ± SD. The means were compared by one-way ANOVA, and statistically significant differences (P < 0.05) were identified by Tukey’s test and indicated by letters to represent difference among groups.
Despite these findings, little has been done to determine the effect of PAOx accumulation on phenylpropanoid biosynthesis. We therefore analyzed the metabolite profile of CYP79A2 overexpressing lines to determine if elevated PAOx production impacts phenylpropanoid biosynthesis. We found that our CYP79A2 overexpression lines accumulate lower levels of sinapoylmalate (Fig 4B) similar to that observed in ref5 and ref2 mutants (Hemm et al., 2003; Kim et al., 2015; Kim et al., 2020).
Transcriptomics analysis shows that enhanced PAOx production affects multiple metabolic pathways through transcriptional reprogramming.
To understand the impact of increased PAOx production on global gene expression, we investigated differentially expressed genes in CYP79A2 overexpression line ox-2/WT compared to wild type. 1339 genes were up-regulated and 1024 genes were down-regulated in ox-2/WT compared to wild-type plants (Fig 5A, B). Gene ontology (GO) term analysis of up-regulated genes showed an enrichment of genes involved in response to abiotic stresses such as temperature change and water deprivation, and response to hormones (Table S2). In particular, genes functioning in auxin-activated signaling pathways such as GRETCHEN HANGEN3 (GH3), small auxin up-regulated RNA (SAUR), and IAA genes were enriched, which is consistent with increased PAA content in ox-2/WT (Table S2, Fig 5C). GH3 genes encode auxin-amido synthetases (Staswick et al., 2005; Korasick et al., 2013) and the accumulation of PAA amino acid conjugates has been previously shown upon PAA accumulation in Arabidopsis (Aoi et al., 2020a). This suggests that the upregulation of these genes is likely an autoregulatory mechanism attempting to re-balance PAA overproduction in these lines. Interestingly, GO term analysis of the down-regulated genes in ox-2/WT revealed an enrichment of genes functioning in tryptophan related pathways (Table S3). Several tryptophan biosynthesis genes including ASA1, ASB1, TRP1, TSB1, TSB2, and TSA1, and genes involved in the YUCCA pathway such as YUC2, YUC8 and TAR2 were down-regulated (Fig 5C). In addition, CYP79B2 and CYP79B3 were substantially down-regulated, whereas aliphatic aldoxime biosynthesis genes (CYP79F1 and CYP79F2) were less impacted (Fig 5C, Fig S8). The down-regulation of tryptophan biosynthesis, the YUCCA pathway, and CYP79B2/B3 genes is likely to result in the repression of IAA production. These findings agree with the previous observation that increased PAA accumulation reduced IAA content and vice versa (Aoi et al., 2020a). Our data indicate that the balance between IAA and PAA homeostasis is associated with transcriptional regulation of not only IAA biosynthetic genes but also those of tryptophan (Fig 5C). Also, down-regulated CYP79B2 and CYP79B3 can explain reduced indole glucosinolate in Arabidopsis transgenic lines with increased benzyl glucosinolate (Fig 1D, Fig S9).
Figure 5.
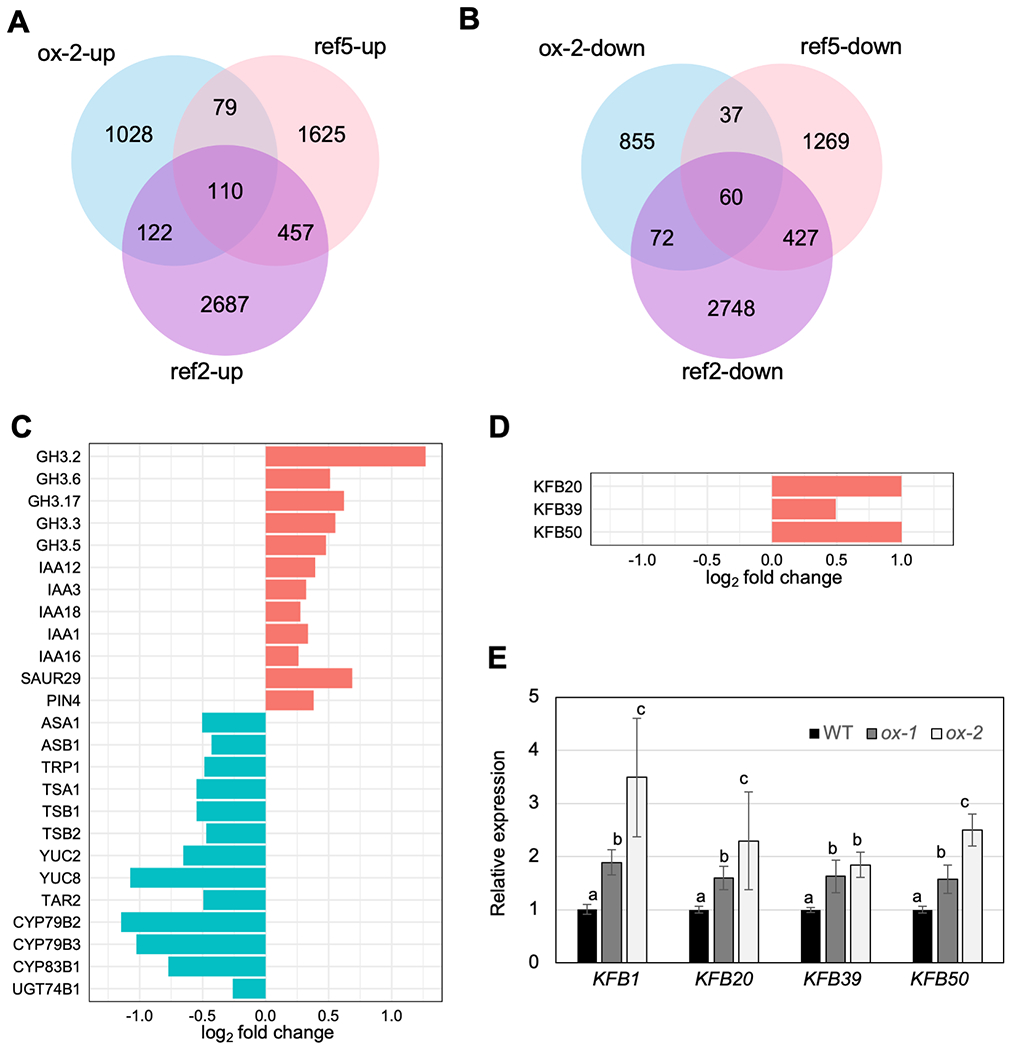
Increased PAOx metabolism alters the expression of genes related to auxin response, tryptophan metabolism, and phenylpropanoid biosynthesis.
A) The number of genes with significantly increased expression in CYP79A2 overexpression line ox-2 (ox-2-up), ref2 (ref2-up), and ref5 (ref5-up), compared to wild type. B) The number of genes with significantly decreased expression in CYP79A2 overexpression line ox-2 (ox-2-down), ref2 (ref2-down), and ref5 (ref5-down), compared to wild type. C) The expression levels (log2 fold change) of select genes differentially regulated in ox-2 compared to wild type. D) The expression levels of the KFB genes that are upregulated in ref2, ref5, and ox-2 compared to wild type. E) Relative expression, determined by qRT-PCR, of KFB genes in ox-1 and ox-2 compared to wild type (N=3). 3rd to 6th rosette leaves from two-week-old plants were harvested for RNA extraction. Data represent mean ± SE. The means were compared by one-way ANOVA, and statistically significant differences (P < 0.05) were identified by Tukey’s test and indicated by letters to represent difference among groups.
Although ref5, ref2 and CYP79A2 overexpression lines are associated with different type of aldoximes (IAOx for ref5, mainly aliphatic aldoximes for ref2, and PAOx for CYP79A2 overexpression lines), all show repression of phenylpropanoid biosynthesis (Fig 4) (Hemm et al., 2003; Kim et al., 2015). Thus, we hypothesized that there is a common mechanism underlying this crosstalk. To identify differentially expressed genes (DEGs) in high aldoxime mutants, we compared transcriptome data of ox-2/WT and published RNAseq data for ref5 and ref2 (Kim et al., 2020). 110 genes are up-regulated and 60 genes are down-regulated in all high aldoxime plants (Fig 5A, B, Table S4, S5). GO term analysis of the down-regulated genes did not give any enriched GO category. However, GO term analysis of the up-regulated genes in all high aldoxime plants identified a GO term category of three genes involved in regulation of phenylpropanoid metabolic process (Table S6). They are the KFB genes KFB20, KFB39 and KFB50, all of which function in PAL degradation (Fig 5D) (Zhang et al., 2013; Zhang et al., 2015). We verified that all four KFBs that function redundantly in PAL degradation were up-regulated in both ox-1/WT and ox-2/WT in a manner similar to IAOx- or aliphatic aldoxime-based induction (Fig 5E) (Zhang et al., 2020; Kim et al., 2020). This suggests that IAOx and PAOx share all three of the known outcomes of aldoxime metabolism in Brassicales species; production of defense compounds, auxin biosynthesis, and repression of the phenylpropanoid pathway (Fig 5C).
Discussion
PAOx-dependent production of PAA can occur in plants independently of benzyl glucosinolate formation.
It has been shown that PAA can be synthesized via the two-step YUCCA pathway using phenylpyruvate as an intermediate (Dai et al., 2013; Sugawara et al., 2015; Cook et al., 2016). Consistently, we observed that yuc6-1D overexpressing YUC6 increased both IAA and PAA (Fig S5) (Kim et al., 2007). However, analysis of specific mutants of the YUCCA pathway’s genes, such as de18 of maize, tar2-1 of pea, and yuc1yuc2yuc6 of Arabidopsis (Cook et al., 2016; Cook and Ross, 2016), suggests that an alternative pathway or enzymes for PAA biosynthesis may also exist. Indeed, activation of arogenate dehydratase (ADT) increases phenylpyruvate and PAA in Arabidopsis (Aoi et al., 2020b). Here, we showed that PAA can indeed be synthesized from PAOx as when the metaobolic flux through PAOx was enhanced via the overexpression of Arabidopsis or maize PAOx-producing enzymes (Fig 1, Fig S4). Our data also revealed that PAOx-dependent PAA biosynthesis is active even when glucosinolate accumualtion is disrupted, as PAOx-fed sur1 failed to produce benzyl glucosinolate but had increased PAA content (Fig 2D, E). The conversion of labeled PAOx to labeled PAA in sur1 and in maize further further confirms that benzyl glucosinolate hydrolysis is not vital for PAOx-derived PAA biosynthesis (Fig 2F). The identity of the enzymes involved in conversion of PAOx to PAA and any intermediates in the pathway remain unknown, and further study is needed to find these enzymes and assess whether they also function to convert IAOx to IAA. Given that benzyl cyanide, a hydrolysis product of benzyl glucosinolate, can be converted to PAA via the action of nitrilase enzymes (Ludwig-Muller and Cohen, 2002; Urbancsok et al., 2018; Günther et al., 2018), it is still possible that benzyl glucosinolate catabolism also contributes to PAA production in Arabidopsis.
Aldoxime-derived auxin biosynthesis in plants.
It has been thought that the IAOx-derived IAA biosynthetic pathway is limited to glucosinolate-producing species in Brassicales. However, our results ultimately suggest that aldoxime-dependent auxin biosynthesis, rather than being a family-specifc pathway, is instead a widely distributed route of auxin biosynthesis. As ZmCYP79A61 can fit into the biochemical pathway present in Arabidopsis to produce auxins from aldoximes, and maize can convert labeled aldoximes into auxin in vivo, it is likely that the aldoxime derived auxin biosynthetic route is present in maize. These data strongly support the idea that aldoxime-derived auxin biosynthesis may occur widely in plants, however, loss-of-function studies in other species are needed to definitively confirm this. It has long been known that CYP79 enzymes in various species produce aldoximes from aliphatic and aromatic amino acids (Towers et al., 1964; Andersen et al., 2000; Luck et al., 2017; Sørensen et al., 2018). For instance, CYP79D enzymes from poplar (Populus trichocarpa and Populus nigra) produce IAOx and PAOx (Irmisch et al., 2013); CYP79D73 in plumeria (Plumeria rubra) converts phenylalanine into PAOx (Dhandapani et al., 2019); CYP79D in coca (Erythroxylum coca) and its related African wild species (Erythroxylum fischeri) produce PAOx and IAOx (Luck et al., 2016); CYP79A118 from the gymnosperm European yew (Taxus baccata) produces IAOx and PAOx (Luck et al., 2017); and Citrus unshiu, Carica papaya, and Calanthe sylvatica also produce PAOx (Sakurai et al., 1979; Olafsdottir, 2002). Besides CYP79 enzymes, flavin containing monooxygenase (FMO) can convert phenylalanine to PAOx through two-step reactions in the fern species Phlebedium aureum and Pteridium aquilinum (Thodberg et al., 2020). Whether the aldoxime-derived auxin production occurs in these species remains unexplored.
Our data indicate that maize can produce IAA and PAA from respective aldoximes. It was previously reported that 3-day-old maize coleoptiles did not accumulate IAOx (Sugawara et al., 2009). However, maize leaves treated with simulated hervibory display increases in PAOx, PAA, and IAA (Irmisch et al., 2015). It is possible that aldoxime-derived auxin biosynthesis may be active only under certain stress conditions in maize. Indeed, most characterized CYP79 genes are induced by stresses or the stress-related hormone jasmonic acid (Irmisch et al., 2013; Irmisch et al., 2015; Luck et al., 2016). In Arabidopsis, the IAOx-dependent IAA biosynthesis pathway is by all accounts considered a minor or stress-responsive route of IAA production. Arabidopsis CYP79B2 expression increases under high temperature and the IAOx-deficient mutant b2b3 shows growth defects under suboptimal growth temperatures (Zhao, 2002; Franklin et al., 2011). In maize, four CYP79 homologs have been reported (Irmisch et al., 2015). These four CYP79 homologs (GRMZM2G138248 (ZmCYP79A61), GRMZM2G178351, GRMZM2G105185, GRMZM2G011156) display similar tissue specific expression although ZmCYP79A61 is highly expressed compared to the other three homologs (Fig S10)(Stelpflug et al., 2016; Hoopes et al., 2019). Biochemical activities of the CYP79 homologs and biological roles of aldoxime metabolism and aldoxime-derived auxin production in maize are still unknown and await further investigation.
Arabidopsis thaliana has a single copy of CYP79A2 encoding a PAOx production enzyme and its expression is low in most organs under optimal growth conditions (Fig S1). Consistently, benzyl glucosinolate does not accumulate in most parts of Arabidopsis ecotype Col-0 except in its seeds, whereas IAOx-derived indole glucosinolates are abundant in leaves and roots (Kliebenstein et al., 2001). This implies that PAOx-derived PAA biosynthesis using CYP79A2 is mostly inactive, at least in Arabidopsis Col-0, unlike the IAOx-IAA pathway. It is possible that the PAOx-PAA pathway is active only in specific tissues or cell types or its activation requires specific stressors in Arabidopsis. It is worth noting that Arabidopsis CYP79A2 is expressed exclusively in chalazal endosperm where both of the PAOx utilizing enzymes REF5 and REF2 are scarcely expressed (Fig S1B) (Belmonte et al., 2013), which may result in the activation of PAOx-dependant PAA production. Further biochemical and genetic study is needed to understand the role of aldoxime-derived auxin biosynthesis in plants.
Aldoxime metabolism affects multiple aspects of plant growth and metabolism.
Studies of aldoxime metabolism have uncovered the roles of these metabolites in plant defense. The tryptophan- and phenylalanine-derived aldoximes can all act as precursors for diverse defens compouds (Halkier, 2016; Sørensen et al., 2018; Blažević et al., 2020). The initial discovery of IAOx-dependent IAA biosynthesis uncovered a role for IAOx in growth regulation, which is unique in its use of a defense-related compound precursor as a key intemediate in the biosynthesis of a phytohormone that governs growth and development. Our findings have expanded upon this initial discovery, identifying this aldoxime-dependent pathway as a biosynthetic route for PAA.
It was also found that various aldoximes and their derivatives could limit the biosynthesis of phenylpropanoids (Hemm et al., 2003; Kristensen et al., 2005; Kim et al., 2015; Kim et al., 2020). This repression occurs partially through the increased expression of a set of KFB genes whose proteins target the PAL enzyme of phenylpropanoid biosynthesis for degradation (Kim et al., 2020); the identity of additional components of this crosstalk remians unknown. This crosstalk links aldoxime metabolism to plant growth because accelerated PAL degradation has a negative impact on the entire phenylpropanoid pathway including the biosynthesis of lignin, a key structural polymer and major component of plant cell walls (Bonawitz and Chapple, 2010; Zhang et al., 2013). Our work has shown that PAOx accumulation has a negative impact on phenylpropanoid biosynthesis through transcriptional activation of the genes functioning in post-translational modification of PAL similar to other aldoximes (Fig 4). It was proposed that there are additional step(s) of the phenylpropanoid pathway affected by increased aldoxime besides PALs because the disruption of KFBs partially restored phenylpropanoid production in high aldoxime plants (Kim et al., 2020). Since both our RNAseq analysis and the previous transcriptomics study did not identify any altered expression of phenylpropanoid biosythesis genes, it is likely that the crosstalk occurs via mechanisms other than transcriptional regulation of its biosynthesis genes. It will be interesting to determine if this aldoxime-phenylpropanoid crosstalk also exists in other aldoxime producing species.
A previous study has shown the presence of a balance between IAA and PAA contents (Aoi et al., 2020a). Increased PAA production reduced IAA content while overproduction of IAA reduced the level of PAA. Our study revealed the repression of a series of tryptophan and IAA biosynthesis genes in plants with increased PAA content (Fig 5C), suggesting that these are the mechanism(s) used to maintain the balance of these important natural auxins. This transcriptional repression of tryptophan and its derivatives’ biosynthesis genes may explain reduction of indole glucosinolates in CYP79A2 overexpression lines (Fig S9A).
The increased PAA in CYP79A2 overexpression lines may affect plant growth and development (Fig 1D). It has been shown that PAA is more active in roots than in aerial parts (Aoi et al., 2020a), but under our growth conditions, plants with elevated PAA (ox-1 and ox-2) displayed mild phenotypes in aerial parts such as elongated hypocotyls, whereas the roots of ox-1 and ox-2 were indistinguishable from wild-type roots (Fig S11).
In summary, this study has demonstrated that PAA biosynthesis from PAOx can bypass benzyl glucosinolate hydrolysis in Arabidopsis. We also showed that maize can produce auxins from aldoximes, which has been long believed to occur only in Brassicales. Given that IAOx and/or PAOx production enzymes are found in diverse species, it is necessary to revisit these aldoxime-producing species to asess auxin production from aldoximes. These aldoximes are well-known as precursors of defense compounds but our data indicate that aldoxime metabolism appears to be linked to plant growth as well through the altered production of auxin and phenylpropanoids, which are crucial for plant growth and development. Further studies will address molecular mechanism(s) underlying aldoxime-derived modulation of plant growth and defense simultaneously, the role of aldoxime-derived auxin biosynthesis in maize, and what kinds of compounds are derived from aldoximes in maize and other non-Brassicales.
Supplementary Material
Acknowledgments
This work was supported by the United States Department of Agriculture (USDA)-National Institute of Food and Agriculture Hatch (005681) and a startup fund from the Horticultural Sciences Department and Institute of Food and Agricultural Sciences at the University of Florida to J.K. The United States Department of Agriculture (USDA)-Agricultural Research Service Project number 6036-11210-001-00D and the USDA- National Institute of Food and Agriculture-Specialty Crop Research Initiative grant 2018-51181-28419 to A.K.B. This work was supported, in part, by the NIH R35GM128742 to Y.D. We thank ICBR Proteomics & Mass Spectrometry Facility at UF with LTS operation.
Data availability
Accessions:
AtCYP79A2 (At5g05260)
AtCYP79B2 (At4g39950)
AtCYP79B3 (At2g22330)
AtCYP83A1/REF2 (At4g13770)
AtCYP83B1/REF5 (At4g31500)
SUR1 (AT2g20610)
ZmCYP79A61 (GRMZM2G138248)
References
- Andersen MD, Busk PK, Svendsen I, and Møller BL (2000). Cytochromes P-450 from Cassava (Manihot esculentaCrantz) Catalyzing the First Steps in the Biosynthesis of the Cyanogenic Glucosides Linamarin and Lotaustralin: CLONING, FUNCTIONAL EXPRESSION IN PICHIA PASTORIS, AND SUBSTRATE SPECIFICITY OF THE ISOLATED RECOMBINANT ENZYMES*. Journal of Biological Chemistry 275:1966–1975. [DOI] [PubMed] [Google Scholar]
- Aoi Y, Tanaka K, Cook SD, Hayashi K-I, and Kasahara H (2020a). GH3 Auxin-Amido Synthetases Alter the Ratio of Indole-3-Acetic Acid and Phenylacetic Acid in Arabidopsis. Plant and Cell Physiology 61:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Oikawa A, Sasaki R, Huang J, Hayashi K, and Kasahara H (2020b). Arogenate dehydratases can modulate the levels of phenylacetic acid in Arabidopsis. Biochemical and Biophysical Research Communications 524:83–88. [DOI] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. PNAS 110:E435–E444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blažević I, Montaut S, Burčul F, Olsen CE, Burow M, Rollin P, and Agerbirk N (2020). Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry 169:112100. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Onckelen HV, Montagu MV, and Inzé D (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. The Plant Cell 7:1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, and Chapple C (2010). The Genetics of Lignin Biosynthesis: Connecting Genotype to Phenotype. Annu. Rev. Genet. 44:337–363. [DOI] [PubMed] [Google Scholar]
- Cao X, Yang H, Shang C, Ma S, Liu L, and Cheng J (2019). The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. International Journal of Molecular Sciences 20:6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, and Zhao Y (2007). Auxin Synthesized by the YUCCA Flavin Monooxygenases Is Essential for Embryogenesis and Leaf Formation in Arabidopsis. Plant Cell 19:2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SD, and Ross JJ (2016). The auxins, IAA and PAA, are synthesized by similar steps catalyzed by different enzymes. Plant Signal Behav 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SD, Nichols DS, Smith J, Chourey PS, McAdam EL, Quittenden L, and Ross JJ (2016). Auxin Biosynthesis: Are the Indole-3-Acetic Acid and Phenylacetic Acid Biosynthesis Pathways Mirror Images? Plant Physiol. 171:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Mashiguchi K, Chen Q, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, and Zhao Y (2013). The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 288:1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhandapani S, Jin J, Sridhar V, Chua N-H, and Jang I-C (2019). CYP79D73 participates in biosynthesis of floral scent compound 2-phenylethanol in Plumeria rubra. Plant Physiology 180:171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011). PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences 108:20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, and Halkier BA (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proceedings of the National Academy of Sciences 101:8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther J, Irmisch S, Lackus ND, Reichelt M, Gershenzon J, and Köllner TG (2018). The nitrilase PtNIT1 catabolizes herbivore-induced nitriles in Populus trichocarpa. BMC Plant Biology 18:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagen Smit AJ, and Went FW (1935). A physiological analysis of the growth substance. Proc. R. Acad. Amsterdam 38:852–857. [Google Scholar]
- Halkier BA (2016). General Introduction to Glucosinolates. Advances in Botanical Research 80:1–14. [Google Scholar]
- Hemm MR, Ruegger MO, and Chapple C (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15:179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes GM, Hamilton JP, Wood JC, Esteban E, Pasha A, Vaillancourt B, Provart NJ, and Buell CR (2019). An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J 97:1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, McCormick AC, Boeckler GA, Schmidt A, Reichelt M, Schneider B, Block K, Schnitzler J-P, Gershenzon J, Unsicker SB, et al. (2013). Two Herbivore-Induced Cytochrome P450 Enzymes CYP79D6 and CYP79D7 Catalyze the Formation of Volatile Aldoximes Involved in Poplar Defense. The Plant Cell 25:4737–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, Zeltner P, Handrick V, Gershenzon J, and Köllner TG (2015). The maize cytochrome P450 CYP79A61 produces phenylacetaldoxime and indole-3-acetaldoxime in heterologous systems and might contribute to plant defense and auxin formation. BMC Plant Biol 15:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke V, Weber K, Moore SS, and Burow M (2019). Coordination of Glucosinolate Biosynthesis and Turnover Under Different Nutrient Conditions. Front. Plant Sci. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H (2016). Current aspects of auxin biosynthesis in plants. Bioscience, Biotechnology, and Biochemistry 80:34–42. [DOI] [PubMed] [Google Scholar]
- Kidd BN, Kadoo NY, Dombrecht B, Tekeoglu M, Gardiner DM, Thatcher LF, Aitken EAB, Schenk PM, Manners JM, and Kazan K (2011). Auxin Signaling and Transport Promote Susceptibility to the Root-Infecting Fungal Pathogen Fusarium oxysporum in Arabidopsis. MPMI 24:733–748. [DOI] [PubMed] [Google Scholar]
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, et al. (2007). yucca6, a Dominant Mutation in Arabidopsis, Affects Auxin Accumulation and Auxin-Related Phenotypes. Plant Physiology 145:722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Dolan WL, Anderson NA, and Chapple C (2015). Indole Glucosinolate Biosynthesis Limits Phenylpropanoid Accumulation in Arabidopsis thaliana. Plant Cell 27:1529–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Zhang X, Pascuzzi PE, Liu C, and Chapple C (2020). Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol 225:154–168. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, and Mitchell-Olds T (2001). Genetic Control of Natural Variation in Arabidopsis Glucosinolate Accumulation. Plant Physiology 126:811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, and Strader LC (2013). Auxin biosynthesis and storage forms. J Exp Bot 64:2541–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver RA, Koevoets IT, and Testerink C (2018). Out of Shape During Stress: A Key Role for Auxin. Trends Plant Sci 23:783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen C, Morant M, Olsen CE, Ekstrøm CT, Galbraith DW, Møller BL, and Bak S (2005). Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. PNAS 102:1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck K, Jirschitzka J, Irmisch S, Huber M, Gershenzon J, and Köllner TG (2016). CYP79D enzymes contribute to jasmonic acid-induced formation of aldoximes and other nitrogenous volatiles in two Erythroxylum species. BMC Plant Biology 16:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck K, Jia Q, Huber M, Handrick V, Wong GK-S, Nelson DR, Chen F, Gershenzon J, and Köllner TG (2017). CYP79 P450 monooxygenases in gymnosperms: CYP79A118 is associated with the formation of taxiphyllin in Taxus baccata. Plant Mol Biol 95:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Muller J, and Cohen JD (2002). Identification and quantification of three active auxins in different tissues of Tropaeolum majus. Physiol Plant 115:320–329. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, and Halkier BA (2000). Cytochrome P450 CYP79B2 from Arabidopsis Catalyzes the Conversion of Tryptophan to Indole-3-acetaldoxime, a Precursor of Indole Glucosinolates and Indole-3-acetic Acid. J. Biol. Chem. 275:33712–33717. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Naur P, and Halkier BA (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37:770–777. [DOI] [PubMed] [Google Scholar]
- Morris DA, and Johnson CF (1987). Regulation of auxin transport in pea (Pisum sativum L.) by phenylacetic acid: inhibition of polar auxin transport in intact plants and stem segments. Planta 172:408–416. [DOI] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, and Halkier BA (2003). CYP83A1 and CYP83B1, Two Nonredundant Cytochrome P450 Enzymes Metabolizing Oximes in the Biosynthesis of Glucosinolates in Arabidopsis. Plant Physiology 133:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, and Ljung K (2012). Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. The Plant Journal 72:523–536. [DOI] [PubMed] [Google Scholar]
- Olafsdottir E (2002). Cyanogenesis in glucosinolate-producing plants: Carica papaya and Carica quercifolia. Phytochemistry 60:269–273. [DOI] [PubMed] [Google Scholar]
- Petersen B, Chen S, Hansen C, Olsen C, and Halkier B (2002). Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214:562–571. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Toyoda T, Muraki S, and Yoshida T (1979). Odorous Constituents of the Absolute from Flower of Citrus unshiu Marcovitch. Agricultural and Biological Chemistry 43:195–197. [Google Scholar]
- Schneider EA, and Wightman F (1986). Auxins of non-flowering plants. I. Occurrence of 3-indoleacetic acid and phenylacetic acid in vegetative and fertile fronds of the ostrich fern (Matteucia struthiopteris). Physiol Plant 68:396–402. [Google Scholar]
- Simon S, and Petrášek J (2011). Why plants need more than one type of auxin. Plant Sci. 180:454–460. [DOI] [PubMed] [Google Scholar]
- Sørensen M, Neilson EHJ, and Møller BL (2018). Oximes: Unrecognized Chameleons in General and Specialized Plant Metabolism. Molecular Plant 11:95–117. [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, and Suza W (2005). Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. The Plant Cell 17:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, and Kaeppler SM (2016). An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome 9. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie D-Y, Doležal K, Schlereth A, Jürgens G, and Alonso JM (2008). TAA1-Mediated Auxin Biosynthesis Is Essential for Hormone Crosstalk and Plant Development. Cell 133:177–191. [DOI] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, and Kasahara H (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. PNAS 106:5430–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Mashiguchi K, Tanaka K, Hishiyama S, Sakai T, Hanada K, Kinoshita-Tsujimura K, Yu H, Dai X, Takebayashi Y, et al. (2015). Distinct Characteristics of Indole-3-Acetic Acid and Phenylacetic Acid, Two Common Auxins in Plants. Plant Cell Physiol 56:1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer J-L, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thodberg S, Sørensen M, Bellucci M, Crocoll C, Bendtsen AK, Nelson DR, Motawia MS, Møller BL, and Neilson EHJ (2020). A flavin-dependent monooxygenase catalyzes the initial step in cyanogenic glycoside synthesis in ferns. Commun Biol 3:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GHN, McInnes AG, and Neish AC (1964). The absolute configurations of the phenolic cyanogenetic glucosides taxiphyllin and dhurrin. Tetrahedron 20:71–77. [Google Scholar]
- Urbancsok J, Bones AM, and Kissen R (2018). Benzyl Cyanide Leads to Auxin-Like Effects Through the Action of Nitrilases in Arabidopsis thaliana. Front. Plant Sci. 9:1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, and Li J (2015). Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci U S A 112:4821–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Qin G, Cao M, Chen R, He Y, Yang L, Zeng Z, Yu Y, Gu Y, Xing W, et al. (2020). A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nature Communications 11:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, and Burow M (2010). Glucosinolate Breakdown in Arabidopsis: Mechanism, Regulation and Biological Significance. Arabidopsis Book 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, and Halkier BA (2000). Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. Catalyzes the Conversion of l-Phenylalanine to Phenylacetaldoxime in the Biosynthesis of Benzylglucosinolate. J. Biol. Chem. 275:14659–14666. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gou M, and Liu C-J (2013). Arabidopsis Kelch Repeat F-Box Proteins Regulate Phenylpropanoid Biosynthesis via Controlling the Turnover of Phenylalanine Ammonia-Lyase. The Plant Cell 25:4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gou M, Guo C, Yang H, and Liu C-J (2015). Down-Regulation of Kelch Domain-Containing F-Box Protein in Arabidopsis Enhances the Production of (Poly)phenols and Tolerance to Ultraviolet Radiation. Plant Physiology 167:337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Song YH, Dai R, Lee TG, and Kim J (2020). Aldoxime Metabolism Is Linked to Phenylpropanoid Production in Camelina sativa. Frontiers in Plant Science 11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes & Development 16:3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y (2010). Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant Biol. 61:49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accessions:
AtCYP79A2 (At5g05260)
AtCYP79B2 (At4g39950)
AtCYP79B3 (At2g22330)
AtCYP83A1/REF2 (At4g13770)
AtCYP83B1/REF5 (At4g31500)
SUR1 (AT2g20610)
ZmCYP79A61 (GRMZM2G138248)


