Abstract
Under the current pandemic situation caused by the novel coronavirus SARS-CoV-2, wastewater monitoring has been increasingly investigated as a surveillance tool for community-wide disease prevalence. After a year into the pandemic, this review critically discusses the real progress made in the detection of SARS-CoV-2 using wastewater monitoring. The limitations and the key challenges faced in improving the detection methods are highlighted. As per the literature, the complex nature of the wastewater matrix poses problems in processing the samples and achieving high sensitivity at low loads of viral RNA using the current detection methods. Furthermore, in the absence of a gold standard analytical method for wastewater, the validation of the generated data for use in wastewater-based epidemiological modeling of the disease becomes practically difficult. However, research is advancing in adopting clinical methods to the wastewater by using appropriate processing controls, and recovery methods. Besides, the technological advances made by the industry including the development of PCR kits with improved detection limits, easy-to-use viral RNA concentration methods, ability to detect the coronavirus variants, and artificial intelligence and advanced data modeling for continuous and remote monitoring greatly help to debottleneck some of these problems. Currently, these technologies are limited to healthcare systems, however, their use for wastewater monitoring is expected to provide opportunities for wide-scale applications of wastewater-based epidemiology (WBE). Moreover, the data from wastewater monitoring act as the initial checkpoint for human health even before the appearance of symptoms, hence WBE needs more attention to manage current and future infectious transmissions.
Keywords: COVID-19, Wastewater-based epidemiology, Big data analytics, Digital droplet PCR, Artificial intelligence, Automated wastewater sampling
Graphical Abstract
1. Introduction
A year has passed since the emergence of the novel coronavirus was considered serious enough to lead to a ‘global lockdown state’ beginning from China and quickly spreading to the rest of the world. COVID-19 is an ongoing pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which belongs to the family of enveloped, single-stranded, positive-sense RNA viruses. The pandemic has severely impacted the public health system and the global economy while comprehensive knowledge of the virus remains limited [1]. The past year witnessed the most rapid advances ever made in epidemiological solutions ranging from clinical coronavirus testing to the discovery of potential vaccines in an attempt to control the pandemic. However, any epidemiological indicator is confronted with many biases and limitations and the same has been true for the COVID-19 pandemic as well [2]. It has been frequently reported that about 40% of infected individuals are asymptomatic and are therefore precluded from clinical testing data. Additionally, diagnostic testing has remained in high demand even with improvements in the mass production of diagnostic kits, thus raising continuous concerns of a shortage. The main cause for this has been the sudden emergence and re-emergence of COVID-19 positive cases in several countries due to hidden, community transmissions, imported cases from incoming travelers or inadvertent spread by asymptomatic yet infectious individuals. This has led to several cycles of lockdown-open-lockdown of cities and even entire countries in some cases, while a clear understanding of the full extent of the pandemic is still not achieved.
Since the beginning of the pandemic, vaccine development and availability have been touted as the magic bullet to control it. However, as already witnessed at the beginning of the year 2021, vaccine roll-out and distribution is a key impediment to manifest real progress even by the end of the year 2021 for most countries. Furthermore, the recent concerns over the finding of the new SARS-CoV-2 variants such as the UK variant and the efficacy of the already developed vaccines against it are additional challenges to the proposed vaccine solution [3]. In view of the above social, economic and technological challenges, a smarter, robust and more holistic approach is required for pandemic management that can complement the existing clinical approach while also facilitate targeting wider communities.
Wastewater is the first and foremost entry point for fecal and urine excretions of humans. Moreover, most of the wastewater effluent is released into surface water and recycled to produce drinking water. Hence, all contaminants including microorganisms that pass through human bodies will recycle back to humans if they are not detected and treated efficiently at their initial stages. To this, the experience from other viral diseases in the past has shown that wastewater-based epidemiology (WBE) is an effective surveillance tool to detect the presence of pathogens in entire communities and provide a strong indication of increasing or decreasing transmission [4]. Moreover, continuous monitoring of wastewater provides the early detection of virus mutations/variants in a community that helps to take the necessary precautions. It has been reported that both viable SARS-CoV-2 and viral RNA are shed in bodily excreta, such as saliva, sputum and feces, and thus eventually reach the wastewater [5], [6], [7], [8], [9]. Not surprisingly, the novel coronavirus has been detected in wastewater in several countries. Viral constituents are shed by both symptomatic and asymptomatic infected individuals which makes it more useful and reliable to find the presence of the virus by testing wastewater as compared to clinical testing which only usually informs about the symptomatic individuals unless in the case of mass testing of a population. Benefits from wastewater monitoring of COVID-19 have been described as (i) being a cost-effective method to determine transmission trends in entire communities, (ii) collection of data from communities that lack healthcare, and (iii) capability to provide near real-time information to the public health officials [10]. Furthermore, even in the optimistic scenario of large-scale vaccination controlling COVID-19 transmission in the future, wastewater testing of coronavirus would be an effective tool to quickly pick up any outbreaks so they could be squelched with clinical intervention before they spread, as was shown for the case of poliovirus infections in the past [11], [12]. Concerning COVID-19, limited data is available across few countries for wastewater monitoring, thereby making it a bit difficult to apply WBE for a diverse mass population on a global scale. However, studies have found a significant correlation between WBE and clinical data of COVID-19 [13], [14]. Considering this aspect, clinical data could be used to extrapolate to the WBE data which in turn can be used to take preliminary caution and prevent the spread of the pandemic.
Since March 2020, several review papers have been published that discussed the status of WBE for COVID-19 while mainly focussing on providing a summary of literature findings from countries wherein the novel coronavirus was detected in wastewater and hence described the potential benefits for wastewater monitoring [15], [16], [17], [18]. After a year into the pandemic, this review aims to critically analyze and discuss the real progress made in the detection of novel coronavirus using wastewater monitoring in the past year while highlighting the limitations of the methods used and the key challenges faced in improving the detection methods. It further describes the new technologies being developed for wastewater monitoring and the crucial role played by the industry in the advancement of the field of WBE, which is currently lacking in the published literature. Finally, some perspectives are provided on the wide-scale applications of wastewater monitoring as an effective tool for COVID-19 pandemic management. A systematic search with different keywords and combinations for each topic in this review has been conducted. In the case of SARS-CoV-2 detection section, “wastewater+COVID-19”, “COVID-19+PCR+wastewater”, “wastewater+COVID-19 detection+limitation”, for the section on the role of industry,– “industry+COVID-19 detection”, “industry+COVID wastewater”, “wastewater COVID-19 detection kits”, “COVID-19 dashboard”, “big data+COVID-19”, “wastewater surveillance+COVID+company” combination search terms were used to review the literature published since 2020.
2. Detection of SARS-CoV-2 in wastewater – current status
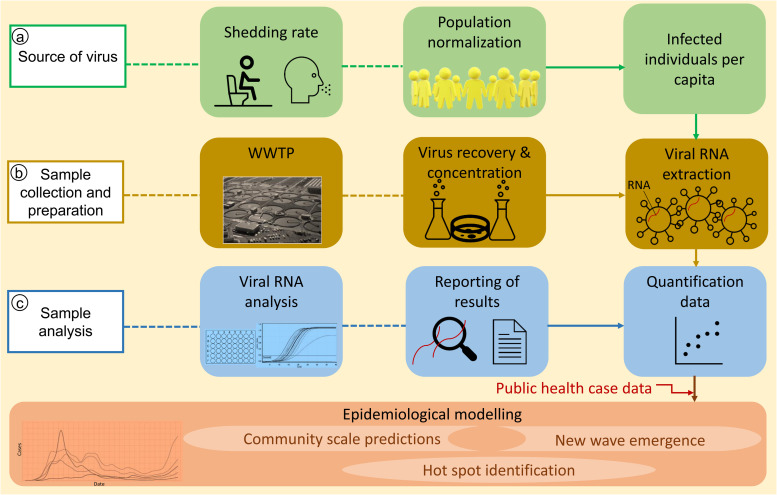
Since the COVID-19 pandemic, several studies have provided evidence regarding the presence of SARS-CoV-2 RNA in municipal wastewater across the globe [17], [18], [19], [20], [21], [22]. These studies were mainly focused on advancing our understanding of the epidemiology of SARS-CoV-2 RNA in wastewater by measuring the viral RNA [1]. Current research findings highlight the potential role of WBE in evaluating the virus spread, predominance in water sources, molecular epidemiology, the efficiency of wastewater treatment plants (WWTPs) and possible strategies for the eradication of the virus. A schematic of steps involved in WBE monitoring of COVID-19 for use in community surveillance is shown in Fig. 1.
Fig. 1.
Schematic of steps involved in WBE monitoring of COVID-19 for use in community surveillance.
To study WBE for COVID-19, two sampling methods have been predominantly used by the research community. Researchers collected and analysed both grab and composite type of wastewater samples to monitor viral RNA levels and compared the infection cases versus SARS-CoV-2 RNA concentrations detected [19]. Most of the studies used only three types of wastewaters, including untreated wastewater, secondary treated wastewater and tertiary wastewater samples to detect the viral RNA [19], [20], [21]. Among these, untreated wastewater was studied the most, followed by secondary treated and tertiary wastewater samples. Regardless of the wastewater sample collection type and detection method, untreated wastewater showed a higher detection frequency range of 22–100% compared to 0–20% detection range in secondary wastewater of many countries, including Japan, Spain, Australia, Italy, USA, and the Netherlands [14], [21], [22], [23]. In most of the countries, this detection frequency data was correlated with the COVID-19 cases and it was observed that the estimated number of infections and the prevalence of viral RNA copy numbers were significantly correlated [13]. A community study in Japan reported the increase in viral RNA detection in secondary wastewater samples with the rise of infection cases [24]. This was further proved by studies where SARS-CoV-2 RNA detection frequency was raised in wastewater after the number of confirmed cases reached 1–100 per million population [14]. However, the SARS-CoV-2 RNA concentrations do not directly represent COVID-19 case numbers. This is because in a single infected patient with COVID-19 there are 106–1011 viral RNAs in an mL of sputum or throat swab [25]. Hence, the detected viral RNAs in mL of sludge/wastewater will not represent the infectious cases. In addition, WBE gives both viable and non-viable counts of RNAs, hence most of the studies have considered the frequency calculations instead of viral count in WBE.
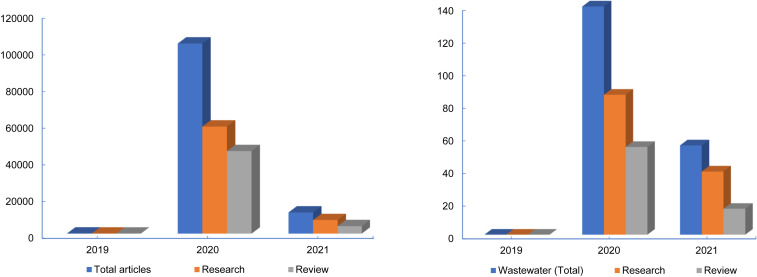
The increased viral levels detection in treated wastewater indicates the low removal efficiency of current municipal WWTPs under the conditions of increasing COVID-19 infection in the population and/or increasing viral RNA load in wastewater influent. Apart from low removal efficiency, these reports also implicate the lack of efficient sample processing methods and/or sensitive detection methods to detect low levels of viral RNA in complex wastewater samples. Fig. 2 presents the current research data from published literature related to COVID-19 since the beginning of the pandemic, based on the Scopus web. Out of all types of data that have been published, 57% was related to research and 43% was on all types of review articles (Fig. 2a). These figures directly represent the high intensity of research devoted towards critical reviewing of the existing [26] research data instead of generating new research. For wastewater, only 0.17% of articles were published that directly emphasizes the lack of new research developments towards wastewater analysis (Fig. 2b). Moreover, around 60% of data were generated from only 5 different countries including the USA, UK, China, Italy, and India. Though WBE is not a new concept, still, the deficiency of research and less tendency to advance the research in wastewater is notable even in the current advancing era of technologies due to the complexity of the wastewater matrix. Having technologies (analytical methods and industrial evolutions) for wastewater with similar efficiency as is available for clinical samples could allow better handling of the current and future infectious diseases due to their early detection. Fig. 2 depicts the need for intense and advanced research specific to wastewater to become more reliable on WBE and allow better control of future pandemics. Furthermore, the high infection rates, scientific advancements, population, and socio-economic factors might determine the research expansion in developing and developed countries.
Fig. 2.
Available published literature related to COVID-19 since the beginning of the pandemic (based on Scopus web).
The vast differences in the detection range of viral RNA among various countries and within a country might be affected by various parameters. The key parameters responsible for such a wide variety of detection frequencies include the infection rate of the community/country, detection methods, and wastewater sample matrix. Among these, only detection methods and sampling processing methods could be standardized until now and used worldwide to reduce the variation arising from the method used for coronavirus detection. Furthermore, as per the authors’ knowledge, very limited studies have reported the levels of SARS-CoV-2 RNA in wastewater sludge samples [27] as well as in wastewater samples from other stages of treatment processes such as screening, grit removal, primary treatment and tertiary treatment. Analysis of viral RNA across the WWTPs is important because quantitative data of viral RNA at different stages of treatment will provide information on the fate of viral RNA and the removal efficiency of each stage of treatment. Moreover, such data will determine the partitioning of viral load between wastewater and sludge phases. Similar to other organic and inorganic contaminants, viral RNA might retain in the wastewater sludge. Hence, the quantification of viral RNA in sludge provides the effect of environmental factors such as pH, total solids, suspended solids, and organic matrix on the partitioning of virus between sludge and wastewater. The generated data can be used to improve the precision of WBE thereby making it more reliable. The rising question on wastewater-based data of COVID-19 infections can be resolved one step further using sludge analysis. Further landfilling and/or fertilizer applications of municipal sludge might act as a primary point source contributing to the viral RNA load to surface water. Hence, strategic wastewater sampling and analysis are needed to identify the parameters necessary to improve the removal efficiency of WWTPs towards viral RNA.
2.1. Quantitative analysis of SARS-CoV-2 in wastewater
Prior to detecting/quantifying SARS-CoV-2, it will likely be necessary to perform virus concentration steps in wastewater samples to eliminate matrix interference and enrich the media with viral RNA. However, the wastewater samples' RNA viral loads are comparable to many enteric viruses’ concentrations [8]. Multiple reviews summarized the various viral concentration methods to extract the SARS-CoV-2 and its RNA from untreated and treated wastewater samples in different countries such as China, Australia, France, Italy, Israel, Japan, Spain, Netherlands, and the US [14], [22], [23], [28], [29], [30]. Compared to drinking water samples, wastewater has a complex matrix. It needs efficient sample processing and viral RNA extraction methods to eliminate matrix interferences and improve detection sensitivity to ensure WBE reliability. The extraction methods used for SARS-CoV-2 RNA in wastewater samples were initially derived for the non-enveloped viruses, such as adenovirus, norovirus, hepatitis A virus [31] and enterovirus. However, the SARS-CoV-2 has enveloped nucleic acid (RNA), and yet similar extraction methods were applied, thereby raising questions on the extraction efficiency towards specific SARS-CoV-2 RNA due to the significant differences in their structural and physical properties from other non-enveloped viruses. Compared to non-enveloped viruses, little scientific evidence is available to conclude the efficiency of the existing concentration methods for enveloped SARS-CoV-2 to date.
In the recent literature, researchers have reported mainly four different sample concentration processes for SARS-CoV-2 RNA in wastewater samples, including (i) adsorption-extraction/elution- (Electropositive or electronegative membranes) with/without pre-treatment options, (ii) ultra-centrifugal filter device methods, iii) polyethylene glycol (PEG 8000) precipitation and ultracentrifugation [32], [33], [34], [35]. Among these methods, adsorption-membrane-MgCl2 pre-treatment (electrostatic interactions) and ultra-centrifugal filtration methods (size exclusion) have shown a good recovery, around 54–60.5% and 56–65.7%, respectively and have been mostly used in the current research for extracting the SARS-CoV-2 RNA [22], [28] and Cross-assembly phage (crAssphage) [36]. Other sample processes such as acidification, centrifugation, precipitation and filtration have shown a low recovery of < 50%. As per research findings, adsorption processes can provide rapid and cost-effective recovery of SARS-CoV-2 RNA in wastewater. To validate or standardize these concentration methods, studies tested other enveloped viruses belonging to the same genus and low-pathogenic CoV strains such as the classical human CoVs, murine hepatitis virus (MHV) [28], F-specific RNA phages [37], Pseudomonas bacteriophage Φ6 [22] as a surrogate for biosafety reasons. These validations reported a mean recovery ranging from 18.2% to 73% [22], [28]. In addition to limited research, constricted choice of use of surrogates and/or sample process controls for SARS-CoV-2 RNA in wastewater samples urge advanced research in this field to assess their usefulness as suitable process controls [38]. Inter and intra-laboratory variability among the developed methods studied across Canada and the US suggest that reproducible results are possible through the same standard operating method between laboratories. In addition, matrix spikes can be used to correct the method recoveries to quantify the viral particles between methods to generate reproducible numbers. Even the inter-laboratory results require adequate quality control protocols with sufficient detail to track SARS-CoV-2 and harmonize the trends [39], [40].
Regarding the analysis method, the detection and quantification of SARS-CoV-2 primarily rely on reverse transcription-quantitative polymerase chain reaction (RT-qPCR) or nested RT-PCR techniques. Current RT-qPCR assays to detect and/or quantify CoV-2 RNA are mainly focused on targeting specific genes of SARS-CoV-2 such as RNA-dependent RNA polymerase (RdRp), envelope (E), ORF1a, ORF1b, ORFab, S protein and nucleocapsid (N) protein genes. This selection is mainly based on the clinical testing methods and later adopted for wastewater testing. Corman et al., reported an absolute limit of detection (LOD) of 3.8, 5.2, and 8.3 RNA copies per reaction for RdRp, E and N genes, respectively in clinical samples [20]. Other studies have reported that N gene-RT-qPCR assay also has a comparable LOD around ~5–6.3 RNA copies per reaction [20], [41], [42]. For each assay, different combinations of primers and probes have been usually tested to improve the specificity and sensitivity of the method. However, these methods and their LOD had been developed for clinical samples where a higher abundance of SARS-CoV-2 RNA is usually found, and a less complex matrix is involved. Unlike in clinical samples, once the SARS-CoV-2 enters the wastewater samples, it undergoes dilution and becomes less concentrated, and therefore, wastewater samples will likely need more sensitive techniques with lower LOD values. Apart from dilution, a low prevalence of COVID-19 infections in the population will also affect their wastewater samples' concentration levels depending on the infection rate. As shown in Table 1, a very limited number of studies have reported the LOD and limit of quantification (LOQ) values of SARS-CoV-2 RNA in wastewater samples. Unlike the clinical samples’ LODs, wastewater samples have shown higher LOD and high variability among the reported values with different types of measurement units used in various studies (Table 1). The LOD values of clinical samples and wastewater samples have a huge difference in the order of 101–103. The existing detection limits from the adopted clinic method indicate that LOD < 10 RNA copies per reaction in wastewater samples could be needed to efficiently screen SARS-CoV-2. Researchers are also trying to test an SYBR Green-based qPCR targeting spike (S) protein gene and digital RT-PCR specific to SARS-CoV-2 RNA to enable a more sensitive and accurate detection/quantification [26], [43], [44]. No LOD for these methods has been reported yet using wastewater samples though [10].
Table 1.
Summary of detection methods used for quantification of SARS-CoV-2 RNA in wastewater.
| Sample type and country location | Extraction method | PCR assays and primers used for detection | Limit of detectiona | Reference |
|---|---|---|---|---|
| Raw wastewater, Italy | Two phases (PEG dextran method) | Nested RT-PCR assays (ORF1ab, 332 bp fragment of ORF1ab) and one real time qPCR assay (RdRP gene) | > 500 genome units/reaction | [13] |
| Influent, secondary and tertiary treated effluent, Spain | Aluminum hydroxide adsorption-precipitation | TaqMan real-time RT-PCR (RT-qPCR) | 50 genome units/ reaction | [22] |
| Raw wastewater, Paris | Centrifugation | RT-qPCR primers | 103 genome units/L | [28] |
| Raw wastewater, Japan | – | RT-qPCR | 2000 genome units/L | [35], [36] |
| Raw wastewater, Montana, USA. | Filtration and centrifugation | RT-qPCR (N1 and N2) | 10 genome units/ reaction | [37] |
| Influent, secondary and tertiary treated effluent, Japan | Electronegative membrane-vortex and adsorption | RT-qPCR (N_sarbeco, NIID_2019-n, COV_N CDC-N1) | 4.0 × 103 – 8.2 × 104 copies/L (Influent) 1.4 × 102–2.5 × 103 copies/L (Secondary wastewater) | [19] |
| Influent, secondary and tertiary treated effluent, Louisiana, USA | Ultrafiltration and adsorption–elution method using an electronegative membrane | RT-qPCR | 1.7 × 102 - 10 × 103 - copies/L | [26] |
| Influent and tertiary treated effluent, Germany | Centrifugal ultrafiltration | RT-qPCR for M-gene | 200 genome units/ reaction | [27] |
| Influent and effluent wastewater, Australia | Adsorption–extraction with electronegative and Centrifugal ultrafiltration | RT-ddPCR (CDC N1) | 1000–4000 copies/L | [44] |
PCR: Polymerase chain reaction; RT-qPCR: Reverse transcription quantitative polymerase chain reaction; L: Liter; PEG: Polyethylene glycol.
Different measurement units for limit of detection due to different methods developed by various studies.
Similarly, attempts have also been made to track the recent and future genetic diversity in SARS-CoV-2 RNA. To this, the researchers have used two new nested RT-PCR assays targeting ORF1a and S protein genes [41] for higher specificity. Besides, a loop-mediated isothermal amplification (LAMP) technique was also developed to target more regions (orf1ab, S gene and N gene) of SARS-CoV-2 RNA using twenty-four primers and improve the sensitivity to 80 copies of viral RNA per mL [45]. In addition, a multiplex LAMP coupled with a nanoparticle- biosensor was developed and this assay is characterized by high analytical specificity (100%) and a LOD of 12 copies per reaction [30]. In general, most studies reported a higher and different LOD (depending on the wastewater type) as compared to the clinical samples. In one study, wastewater influent showed a LOD of 4.0 × 103 – 8.2 × 104 copies/L with 200 mL filtration volume while the secondary-treated wastewater had a LOD of 1.4 × 102 – 2.5 × 103 copies/L with 5000 mL filtration volume were two sample processing methods including electronegative membrane-vortex and membrane adsorption were used to extract RNA [32]. However, with such high LOD values, most of the wastewater samples of Yamanashi Prefecture, Japan, have shown negative results. Similarly, different LODs of 1.0 × 103 copies/L and 1.7 × 102 copies/L were observed for other extraction processes such as centrifugal filter and adsorption–elution method using an electronegative membrane, respectively [22]. Another assay, RdRp-qPCR, showed a LOD of 500, 200 viral genomic equivalents per reaction and 100–400 RNA copies/100 mL in untreated wastewater in Italy [29], Germany [23] and airline and cruise ship wastewater [46], respectively. Most wastewater sample studies have reported the quantification range 0.1–104 (min-max) of SARS-CoV-2 RNA copies. However, the reported detection methods were not developed as per the standardized procedure comprising a range of validation parameters, including LOD, the limit of quantification, reproducibility, accuracy and precision. In addition, the various measurement units for the virus such as copies/L or genome units/ reaction and high variability among the reported LODs make it impossible to compare the efficiency of the existing methods with respect to the analysis of wastewater samples. Hence, advanced research is needed to get a ‘gold standard’ analytical method for a complex matrix such as wastewater which is a potential source for WBE scaleup and accuracy. As per the authors’ analysis, so far, most of the studies on detection methods and analysis related to the COVID-19 monitoring of wastewater since 2019 have mostly attempted to make some improvements only by introducing small ‘tweaks’ in the existing methods. However, this has not led to the development of a validated method that could form a gold standard to be used globally for wastewater detection of COVID-19.
2.1.1. Challenges with current detection methods
In view of the wastewater matrix complexity, sample processing to concentrate and achieve highly sensitive assay at low loads of viral RNA are major problems with the current detection methods. Consequently, the main challenges faced in the current research on WBE monitoring of COVID-19 can be summarized as follows:
-
i)
Lack of reproducibility of the concentration methods in wastewater samples is a significant limitation in current protocols. Hence, pre-treatment processes should be improved to achieve a good recovery of > 80% so that the sensitivity of the method could be increased.
-
ii)
Larger volume of wastewater used for filtration to concentrate maximum viral quantity often requires dealing with more solids/co-concentration of inhibitors, especially for adsorption or filtration pre-treatment process. Hence, the volume of wastewater samples for extraction of the virus also needs to be optimized and standardized globally.
-
iii)
Finding the best surrogate to validate current pre-treatment methods has become another challenge. Identification of a surrogate that is stable during wastewater processing, has no toxicity, possesses similar structural features to SARS-CoV-2, is not affected by the wastewater matrix (e.g., solids content) and is not expected to be present in the wastewater is needed to validate the recovery percentage reported in the current methods.
-
iv)
A critical issue in applying PCR techniques in wastewater samples in PCR inhibition during the detection process. Hence, appropriate process controls are needed during the analysis to screen the recovery of RNA and/or inhibition across all analytical steps.
-
v)
Downstream RT-PCR assays are necessary for refined and validated assays having 100% specificity and sensitivity.
-
vi)
Targeting multiple genes on viral RNA and different combinations of primers and probes raises the specificity of the assay towards SARS-CoV-2 RNA. However, this aspect of research is currently limited to clinical samples. Hence, an extension of this application to wastewater samples is necessary to navigate easy identification and monitoring of genetic modifications of viral RNA in the future and generate accurate WBE data for further evaluation.
-
vii)
Another major challenge is that qPCR results do not specify the information on the viability of the virus. Hence, it is crucial to study the viability of SARS-CoV-2 in wastewater samples and their half-life to understand the virus transmission among different environmental compartments. However, urine and stool sample analysis provide a reliable viable count of viral particles [47].
Cumulative improvements in both concentration methods and assays will help epidemiologists, government health officials, and modelers evaluate frequencies of SARS-CoV-2 infection rates using WBE strategies to support community-level epidemic mitigation and risk assessment as well as virus genetic modifications. Therefore, cross-validated/ inter-laboratory validated methods are necessary to establish a standard quantitative method for viral RNA.
3. Industry interest in monitoring wastewater for COVID-19: development of new technologies
Early at the beginning of the pandemic, the use of WBE for COVID-19 gained momentum. This was witnessed by the announcement of plans for network-based environment surveillance projects by more than 50 countries including the US, Canada, Australia, the Netherlands and the UK [48], [49], [50], [51]. In a great number of cases, these plans involved industry partnerships to allow an extensive community coverage of tens and in some cases hundreds of WWTPs for wastewater testing of the novel coronavirus.
To address the challenges in WBE monitoring of COVID-19 as discussed in Section 2 above, different companies have participated in wastewater monitoring through different routes. These include (i) development of advanced RT-qPCR kits, (ii) providing services for sampling and its analysis for COVID-19, (iii) development of automated wastewater sampling instruments to facilitate continuous sampling, (iv) R&D in RNA extraction and concentration process, which is a major bottleneck in RT-qPCR, the current gold standard for testing, and (v) development of advanced modeling and data analytics platforms for greater accuracy in estimation of some COVID-19 cases from viral titers in a community ( Table 2). Considering the enormous amount of data being generated for COVID-19 detection, Big Data analytics plays an important role in providing a fast and almost real-time evaluation of complex datasets, thereby facilitating a quicker decision-making process. Artificial intelligence (AI) can be used along with big data for recognizing, explaining and predicting the patterns in, for example, new or sudden outbreaks, etc. [52], [53]. These digital technologies can also be further integrated with smart manufacturing and used to provide precise control systems in inspection, testing, quality assurance and quality control of products and equipment needed to meet the escalating demands during the ongoing pandemic [54], [55]. Thus, various kinds of technological advancements have been witnessed within a short span of the last year for the management of the COVID-19 pandemic.
Table 2.
List of companies and their roles in WBE monitoring of COVID-19.
| Company name | Country | Role in WBE monitoring of COVID-19 | University and/or Government engagement for WBE |
|---|---|---|---|
| Biobot Analytics | United States | Data analytics for development of advanced mathematical models to corelate virus concentration in wastewater with number of cases | Massachusetts Institute of Technology, Massachusetts Water Resources Authority (MWRA) |
| Kando | Israel | IoT sensors and AI algorithms for autonomous sampling, data analysis and live streaming via dashboards | Ben Gurion University, Technion – Israel Institute |
| AquaVitas, LLC | United States | Testing and data analytics for virus surveillance and online dashboards | Arizona State University, US Department of Health and Human Science (HHS), Centre for Disease Control and Prevention (CDC) |
| GoAigua | Spain | IoT sensors, AI/mL algorithms for sampling, data integration and analysis | The Institute of Agrochemistry and Food Technology (IATA), Spanish National Research Council (CSIC) |
| Pace Analytical | United States | Services for lab testing and analysis of wastewater samples | – |
| OSP Microcheck | Canada | Services for lab testing and analysis of wastewater samples; selling of qPCR kits to clients | – |
| Eurofins | Luxembourg | Services for lab testing and analysis of wastewater samples | – |
| LuminUltra | Canada | Development of testing equipment for on-site analysis of wastewater samples | Dalhousie University, Halifax Water |
| GT Molecular | United States | Development of testing method including testing of new UK variant | Colorado State University, Metropolitan State University, State of Colorado |
| CEC Analytics | Canada | Development of sampling equipment | – |
| Teledyne Isco | United States | Development of sampling equipment | – |
Most of the companies involved in wastewater surveillance of COVID-19 had been pioneers in the field of water and/or wastewater testing and analysis for tracking the presence of pollutants/contaminants, drugs, pharmaceuticals, microbes, etc. For example, Biobot Analytics, an MA-based company, and AquaVitas, LLC, a spin-off company from Arizona State University, pivoted from tracing drugs in wastewater to measure coronavirus when the pandemic began [56], [57]. For WBE of COVID-19 program design, the two most important actions are (i) to determine the location where a wastewater sampling system should be installed, including the selection and installation of wastewater samples at appropriate monitoring points, and (ii) the provision of qualified analytical laboratories which can detect the presence of coronavirus in the samples [18]. Finally, the data need to be processed in a meaningful manner to enable accurate analysis. Considering the above points, it has not been uncommon to see partnerships at the industry level too. E.g. Teledyne ISCO, a leader in the development of automated sampling instruments uses its Compact Portable sampler to measure the presence of virus in wastewater and sends the results to Biobot Analytics for processing and analysis of the results [58].
Wastewater analysis is being viewed by technology companies as a long-term opportunity for public health. To this end, Biobot Analytics, Kando and GoAigua are the companies at the forefront of wastewater COVID-19 analysis by using their advanced WBE technologies including sensors and real-time data analysis to identify and track COVID-19 levels in municipal wastewater systems. Biobot Analytics, predominantly a data company, has developed advanced mathematical models that can accurately predict the relationship between the amount of virus in the sewage and the associated number of reported cases [57]. As discussed in Section 2, the conventional methods of deriving the number of cases based on wastewater monitoring are to divide the total amount of virus in wastewater by the amount of virus shed per infected person. However, the major challenge in this method is its dependence on a virus shedding factor which is obtained from clinical studies. The latter is extremely difficult to reliably measure since the clinical studies are often limited to a small number of individuals and usually for hospitalized individuals at later stages of infection. Also, the exclusion of pre-symptomatic individuals, who also shed virus, from these studies, makes it unreliable for use in modeling studies. These problems have been addressed by Biobot during the development of their models for early detection of the coronavirus. The main advantage of Biobiot’s modeling approach is the fact that it is independent of clinical studies on virus shedding including differences among individuals in virus shedding and temporal differences within an individual [59]. This results in the high accuracy of predicted data from these models. Another key feature of Biobot’s approach is the continuous up-gradation of data models. This is required since the laboratory protocols are being constantly revised based on an evolving understanding of the virus in wastewater. Consequently, the model results are liable to change and/or be updated based on the reprocessing of previously obtained wastewater sample results using the most current data analysis methods. Thus, the variations in the laboratory processing are reflected in the range of result variability generated by the models to allow greater accuracy. Due to its high accuracy, Biobot’s data have been used for the safe reopening of schools in the City of Cambridge.
Another big data analytics company, Kando, based in Israel and the US, has combined its expertize in the Internet of Things (IoT) technology, AI and advanced data modeling to operate continuously and remotely for COVID-19 monitoring in wastewater using its solution, Clear Upstream [60]. Kando uses factors such as population density, social factors, population age and public transportation usage to develop algorithms that are then used to install IoT units at the right locations to optimize monitoring capabilities. The IoT autonomous sampler collects the samples triggered by predetermined wastewater chemical and hydraulic properties, thereby using AI to respond to network conditions in real-time. Similar to Biobot, the results are then fed to a model. Combined with the laboratory findings, Kando’s system can identify remnants of viral RNA and pinpoint a local outbreak with the ability to narrow down information about hotspots as well. The integration of a live map, online dashboards and text messages, provides continuous information on COVID-19. Likewise, GoAigua uses its BigData platform to synthesize automatic sampling results from sewage networks [61]. Similar to Kando, it identifies strategic sampling locations based on the morphology and topology of the sewer network and uses data integration and analytical tools combined with real-time maps to help public health agencies assess the spread of the coronavirus. It also provides a mobile-based app to operators wherein the monitoring information along with other sewage parameters such as pH, chlorine levels, etc. can be integrated to adjust the analysis results. Implementation of smart decision systems has been successful in allowing the city of Valencia, Spain, to anticipate outbreaks, thus providing evidence of its usefulness in tracking COVID-19. The system has also been adopted by the City of Burlington, Vermont, US, for sewage surveillance [62].
A common feature of the technology companies involved in wastewater COVID-19 monitoring is the rapid, live streaming of data to municipalities and public health agencies through a dashboard. This allows the latter to look at the data in a real-time manner and quickly identify the problems (spikes, hotspots, etc.), sources of events and take appropriate prompt actions to control the spread of the virus. The dashboard built by Biobot and AquaVitas, LLC in collaboration with Massachusetts Water Resources Authority (MWRA) and Arizona State University, Tempe, respectively, are good examples of such wide-scale surveillance projects [63], [64] (see Table 2). The use of smartphone-based applications to capture and quickly transfer the data has been significant in making this possible [65].
In addition to data analytics, companies have also been engaged in extensive R&D to provide innovative solutions for efficient sampling from sewage collection points. CEC Analytics has designed a compact composite sampler that can be deployed at previously inaccessible locations, thereby expanding the coverage of sampling points for large data collection and improving the accuracy of results [66]. Similarly, Teledyne Isco is using its cutting-edge technology for virus detection via its automatic water sample collectors [67]. Its compact portable sampler equipped with 24 1-L bottles allows daily sampling at multiple sites throughout the sewer network and measures the presence of the virus in parts per million quantities. It has been installed in the City of Cody and the Middle East. Another variant in the form of a fiberglass refrigerated sampler is designed to withstand the harshest of environments.
The heart of virus tracking lies in its accurate detection in wastewater samples. Research efforts have been directed in this direction too by the industry and rapid advances have been made for faster, efficient and accurate detection. Companies such as Pace Analytics, Eurofins, OSP Microcheck, LuminUltra, GT Molecular have been providing specialized equipment and services for testing wastewater samples for COVID-19. Pace Analytics provides commercial laboratory testing of wastewater samples using its qPCR technology while OSP Microcheck provides both services and easy-to-use qPCR kits to the clients [68], [69]. As discussed in Section 2.1.1, the conventional methods for wastewater monitoring require specialized equipment and skilled operators to pre-concentrate large volumes of wastewater prior to its analysis. This is one of the major challenges for COVID-19 testing of wastewater. This problem has been addressed by LuminUltra which has developed a wastewater test kit that can extract SARS-CoV-2 RNA directly from a 1 mL sample of raw wastewater using its magnetic binding bead technology. This obviates the need for pre-concentrating samples. LuminUltra’s rapid and portable GeneCount® qPCR device incorporating its innovative RNA extraction and concentration process can examine multiple samples on-site within 90 min [70]. Another attractive feature of this device is that it can be used without any special laboratory expertize which is generally required for other available wastewater testing methods. The company has filed a patent in October 2020 for this process which it claims to be more efficient, of lower cost than other methods and high capacity. In a further study conducted this year, the company performed a side-by-side comparison using their direct RNA extraction technology and electronegative membrane concentration method, a commonly used laboratory method, to detect SARS-CoV-2 genes N1 and N2 in wastewater samples. The Pearson correlation coefficient was used to report the results of both extraction methods and it showed no significant difference between the results for the N2 gene target (p-value < 0.01). However, a significant difference was seen for N1 (p-value = 0.15), which is being further investigated by the company. It was also reported that the SARS-CoV-2 concentrations obtained from the direct extraction method compared well with the reported clinical cases across the same time period. The company further demonstrated the ability of their developed surveillance method to detect trends in the community COVID-19 cases 7 days prior to cases being reported.
Similarly, GT Molecular has developed a digital droplet PCR (ddPCR) technique for coronavirus testing which has played a key role in wastewater monitoring in Colorado. The company has also launched a community-wide test for the highly contagious UK variant of SARS-CoV-2 [71]. With the greater emergence of variants of concern such as the UK, South African, Brazilian/Japanese, California variants, their fast detection is of paramount importance. The currently used method for variant detection is based on mass sequencing of a large population of individuals which is both expensive and time-consuming taking several days to give results. In this scenario, the prospect of their detection in wastewater is lucrative. This is what is being done by GT Molecular as it performs ‘genotyping’ of the sewer and can detect all important variants with one sample. Its customizable and highly sensitive PCR method can detect as little as 1–3 molecules of the target nucleic acid. The developed method can test the variant in 24 h using its ultra-sensitive ddPCR at a fraction of the cost required to test individuals in a population.
Not surprisingly, various advanced technologies in wastewater sampling technique, virus testing in wastewater samples, and data analysis, developed by the industry have been sought by municipalities in various countries for nationwide COVID-19 monitoring in wastewater. For example, since March 2020, Biobot Analytics has worked with about 400 WWTPs in 42 states in the US to accurately correlate the virus concentration in wastewater with clinical cases in communities. Similarly, AquaVitas, LLC, began with a sampling of up to 100 WWTPs in Phase I to scaling up to 340 plants in Phase II and covering about 30% of the US population. While an increasing number of programs have been launched and are successfully running using the continuously improving WBE technologies, widespread sewage testing for COVID-19 remains a pipedream. This is mainly due to the cost and equipment shortages that prevent the communities from tracking virus outbreaks using wastewater. It is expected that with further research and technological improvement, and greater impetus from governments in support of WBE, this bottleneck can also be addressed to allow its greater utility as a pandemic management tool complementing the existing clinical technologies.
4. Limitations and future scope
The current literature on COVID-19 has serious limitations to analyze and harmonize the global data. These include the lack of standard protocols for various steps including the sample collection strategies, sampling processing and extraction, validated methods of viral RNA concentration (sensitivity and detection limit), and definitive use of primer/probe design for PCR analysis in wastewater and surface water. Further, the reported data on COVID-19 in wastewater has been mainly focused on accuracy and recovery of method but does not account for the effect of other environmental factors contributing to the variability in the decay of viruses such as wastewater temperature (frozen and thawed), pH, suspended solids, and methods of wastewater treatment [72]. Furthermore, the process efficiency recovery is highly variable among the developed methods even after using appropriate process controls [73]. Compared to the clinical data that provide the infection rate based on the live virus, COVID-19 data in wastewater does not provide accurate data of infection rate because the developed method only measures the total virus particle irrespective of its live or dead state. In addition, although industrial revolutions (4.0 and 5.0) such as machine learning and AI have been increasingly developed to support global healthcare systems, these technologies are not yet applied to the wastewater/environmental samples at the same scale as the clinical samples, albeit some progress has been made in this direction [52], [53]. The uncontrolled mutation of the COVID-19 can be first detected using WBE by continuous wastewater monitoring before the appearance of symptoms. Hence, monitoring of wastewater contributes a potential approach to predict an upcoming situation across the globe/community especially where clinical surveillance capacity might be limited. Data of WBE provide an initial indication of human health which can be used to detect and manage infectious disease transmission in the future more intelligently.
5. Conclusions
Studies have been using WBE in wastewater as an effective and efficient tool to provide information on new wave emergence, identification of hot spots and community-scale prediction on COVID-19 transmission. Complementing the existing clinical practices along with public health care data can allow timely intervention in the efficient management of the COVID-19 situation. This review analyzed and discussed the actual progress made using WBE monitoring of COVID-19 since the beginning of the pandemic, the challenges encountered and the road ahead. Investigation of the available literature published in the last year in the wastewater domain showed the lack of standard methods for SARS-CoV-2 detection in wastewater. These problems are further aggravated by incoherent reporting of data from various literature reports which makes it difficult to compare the methods used and analyse and validate the generated data for its utility in epidemiological modeling of COVID-19 on a comprehensive scale.
Encountering some of these challenges, the tremendous efforts made by the industry in cooperation with government organizations in this short time for WBE monitoring of COVID-19 are worthy of appreciation. The advances made in the development of a new generation of PCR kits with enhanced detection limits, improved and easy viral RNA concentration methods which is a critical bottleneck in this process, and detection of SARS-CoV-2 variants with high sensitivity, thus obviating the need for large-scale sequencing, are some commendable examples. Such rapid advances provide confidence in the quick establishment of robust and standard protocols for viral analysis using wastewater in the nearest future. These are expected to debottleneck the existing challenges in WBE monitoring and pave the way for integrating wastewater infrastructure with healthcare systems in the future.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC-Discovery Grant 355254) and James and Joanne Love Chair in Environmental Engineering at York University.
Editor: Yang Liu
References
- 1.Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38(10):1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chand M. Public Health England. PHE; 2020. Investigation of Novel SARS-COV-2 Variant: Variant of Concern 202012/01 (PDF) [Google Scholar]
- 4.Saawarn B., Hait S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: current knowledge and future perspectives. J. Environ. Chem. Eng. 2021;9(1) doi: 10.1016/j.jece.2020.104870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung K.S., Hung I.F., Chan P.P., Lung K., Tso E., Liu R., Ng Y., Chu M.Y., Chung T.W., Tam A.R. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 8.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems. 2020;5(4) doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8(5) doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pöyry T., Stenvik M., Hovi T. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 1988;54(2):371–374. doi: 10.1128/aem.54.2.371-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berchenko Y., Manor Y., Freedman L.S., Kaliner E., Grotto I., Mendelson E., Huppert A. Estimation of polio infection prevalence from environmental surveillance data. Sci. Transl. Med. 2017;9(383) doi: 10.1126/scitranslmed.aaf6786. [DOI] [PubMed] [Google Scholar]
- 13.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 14.Wurtzer S., Marechal V., Mouchel J.-M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. MedRxiv. 2020 [Google Scholar]
- 15.La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods-A scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman A.I., Boehm A.B. Systematic review and meta-analysis of the persistence and disinfection of human coronaviruses and their viral surrogates in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(8):544–553. doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- 18.Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta. 2021;224 doi: 10.1016/j.talanta.2020.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020 [Google Scholar]
- 28.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. 2020.06.04.20122747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hata A., Honda R. Association of albuminuria with white matter hyperintensities volume on brain magnetic resonance imaging in elderly Japanese - The Hisayama study. Circ. J. 2020;84:935–942. doi: 10.1253/circj.CJ-19-1069. [DOI] [PubMed] [Google Scholar]
- 34.Mlejnkova H., Sovova K., Vasickova P., Ocenaskova V., Jasikova L., Juranova E. Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health. 2020;17(15):5508. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater cell reports. Medicine. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. MedRxiv. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 38.Bhatt A., Arora P., Prajapati S.K. Occurrence, fates and potential treatment approaches for removal of viruses from wastewater: a review with emphasis on SARS-CoV-2. J. Environ. Chem. Eng. 2020;8(5) doi: 10.1016/j.jece.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chik A.H.S., Glier M.B., Servos M., Mangat C.S., Pang X.-L., Qiu Y., D’Aoust P.M., Burnet J.-B., Delatolla R., Dorner S., Geng Q., Giesy J.P., McKay R.M., Mulvey M.R., Prystajecky N., Srikanthan N., Xie Y., Conant B., Hrudey S.E. Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: results and implications from a collaborative inter-laboratory study in Canada. J. Environ. Sci. 2021;107:218–229. doi: 10.1016/j.jes.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the US. Environ. Sci. Water Res. Technol. 2021;7(3):504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020 doi: 10.7883/yoken.JJID.2020.061. JJID. 2020.061. [DOI] [PubMed] [Google Scholar]
- 42.N. Coronavirus, Real-Time rRT-PCR Panel Primers and Probes, US Centers for Disease Control and Prevention, https://www. cdc. gov …, 2019.
- 43.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv. 2020 [Google Scholar]
- 44.Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X. ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. medRxiv. 2020 doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M. RT‐LAMP for rapid diagnosis of coronavirus SARS‐CoV‐2. Microb. Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020;27(5):taaa116. doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong H.W., Kim S.-M., Kim H.-S., Kim Y.-I., Kim J.H., Cho J.Y., Kim S.-h, Kang H., Kim S.-G., Park S.-J., Kim E.-H., Choi Y.K. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26(11):1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Wastewater Surveillance System (NWSS), 〈https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/wastewater-surveillance.html〉, (accessed 23 December 2020).
- 49.CWN, Canadian coalition on wastewater-related COVID-19 research. Web page maintained by Canadian Water Network, 〈http://cwn-rce.ca/wastewater-coalition/〉, (accessed 23 December 2020).
- 50.WaterRA, The ColoSSoS Project — Collaboration on Sewage Surveillance of SARS-COV-2, COVID-19 National Research Initiative., Water Research Australia (〈https://www.waterra.com.au/research/communities-of-interest/covid-19/〉; 〈https://www.waterra.com.au/research/open-rffs-and-rfps/2020/monitoring-covid-19-virus-presence-and-persistence-in-the-australian-sewage-networks/〉), (accessed 23 December 2020).
- 51.Naughton C.C., Roman F.A., Alvarado A.G.F., Tariqi A.Q., Deeming M.A., Bibby K., Bivins A., Rose J.B., Medema G., Ahmed W. Show us the data: global COVID-19 wastewater monitoring efforts, equity, and gaps. medRxiv. 2021 doi: 10.1093/femsmc/xtad003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Javaid M., Haleem A., Vaishya R., Bahl S., Suman R., Vaish A. Industry 4.0 technologies and their applications in fighting COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14(4):419–422. doi: 10.1016/j.dsx.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haleem A., Javaid M. Medical 4.0 and its role in healthcare during COVID-19 pandemic: a review. J. Ind. Integr. Manag. 2020;05(4):531–545. [Google Scholar]
- 54.Javaid M., Haleem A. Critical components of Industry 5.0 towards a successful adoption in the field of manufacturing. J. Ind. Integr. Manag. 2020;5(03):327–348. [Google Scholar]
- 55.Vaishya R., Haleem A., Vaish A., Javaid M. Emerging technologies to combat the COVID-19 pandemic. J. Clin. Exp. Hepatol. 2020;10(4):409–411. doi: 10.1016/j.jceh.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.AquaVitas, 〈https://aquavitas.com/hhs-project〉., (accessed 17 December 2020).
- 57.Biobot, 〈https://www.biobot.io/〉, (accessed 17 December 2020).
- 58.T. Isco, 〈https://www.teledyneisco.com/en-us/water-and-wastewater/partnering-with-biobot-analytics〉, (accessed 20 December 2020).
- 59.Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Medrxiv. 2020 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kando, 〈https://www.kando.eco/kando-covid-19〉, (accessed 17 December 2020).
- 61.GiAiugua, 〈https://www.idrica.com/case-studies/goaigua-sars-analytics-early-warning-system-detection-covid-19/〉, (accessed 17 December 2020).
- 62.〈https://www.waterworld.com/wastewater/press-release/14187697/burlington-vt-leverages-wastewater-epidemiology-program-to-stop-the-spread-of-covid19〉, (accessed 20 December 2020).
- 63.〈https://www.mwra.com/biobot/biobotdata.htm〉., (accessed 20 December 2020).
- 64.〈https://covid19.tempe.gov/〉., (accessed 20 December 2020).
- 65.Singh R.P., Javaid M., Haleem A., Suman R. Internet of things (IoT) applications to fight against COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14(4):521–524. doi: 10.1016/j.dsx.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.C. Analytics, 〈https://cecanalytics.com/covid-19-wastewater-testing/〉, (accessed 20 December 2020).
- 67.〈https://www.teledyneisco.com/en-us/waterandwastewater/Pages/COVID-19-Sampler-Selcetion.asp〉, (accessed 20 December 2020).
- 68.P. Analytical, 〈https://www.pacelabs.com/environmental-sciences/testing-services/specialty-services/covid-19-wastewater-testing.html〉, (accessed 17 December 2020).
- 69.Eurofins, 〈https://www.eurofinsus.com/environment-testing/testing-services/covid-19-testing-solutions/coronavirus-wastewater-testing/〉, (accessed 17 December 2020).
- 70.LuminUltra, 〈https://www.luminultra.com/covid-19-testing/wastewater-testing/〉 (accessed 17 December 2020).
- 71.GTMolecular, 〈https://gtmolecular.com/covid19-wastewater-test〉 (accessed 17 December 2020).
- 72.Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bivins A., North D., Wu Z., Shaffer M., Ahmed W., Bibby K.J. Within-day variability of SARS-CoV-2 RNA in municipal wastewater influent during periods of varying COVID-19 prevalence and positivity. medRxiv. 2021 [Google Scholar]