Abstract
Background
Intratumor heterogeneity (ITH) is described as the presence of various clones within one tumor, each with their own unique features in terms of morphology, inflammation, genetics or transcriptomics. Heterogeneity provides the fuel for drug resistance; therefore, an accurate assessment of tumor heterogeneity is essential for the development of effective therapies. The purpose of this study was to dissect morphologic and molecular ITH in colorectal adenocarcinoma.
Materials and methods
A series of 120 V600EBRAF-mutated (V600EBRAFmt) consecutive metastatic colorectal adenocarcinomas was assessed for morphologic heterogeneity. The two heterogeneous components of each specimen underwent a histopathological, immunohistochemical and molecular characterization to evaluate: histologic variant, grading, tumor-infiltrating lymphocytes (TILs), mismatch repair proteins' expression, KRAS/BRAF/NRAS mutations, microsatellite instability (MSI) status and consensus molecular subtype (CMS).
Results
Thirty-one out of 120 (25.8%) V600EBRAFmt primary colorectal adenocarcinomas presented a heterogeneous morphology. Among these, eight cases had adequate material for molecular profiling. Five out of the eight (62.5%) cases resulted instable at MSI testing. The majority (62.5%) of the samples showed a CMS4 phenotype based on gene expression profiling. Heterogeneity in CMS classification was observed in four out of eight cases. One out of eight cases presented significant heterogeneity in the number of TILs between the two components of the tumor.
Conclusions
Although the distribution of the immune infiltrate appears relatively conserved among heterogeneous areas of the same tumor, changes in gene expression profile and CMS occur in 50% of V600EBRAFmt adenocarcinoma cases in our small series and might contribute to variability in response to anticancer therapy and clinical outcomes. Assessment of morphological and molecular ITH is needed to improve colorectal cancer classification and to tailor anticancer treatments and should be included in the pathology report.
Key words: V600EBRAF-mutated colorectal cancer, morphologic heterogeneity, gene expression profiling, consensus molecular subtypes
Highlights
-
•
Morphologic ITH is linked to transcriptomic heterogeneity in 50% of BRAFmt colorectal cancers.
-
•
Morphologic ITH might contribute to variability in response to anticancer therapy and clinical outcomes.
-
•
Assessment of morphological and molecular ITH may improve colorectal cancer classification and tailor treatments.
Introduction
Intratumor heterogeneity (ITH) is an intrinsic feature of many cancers and is due to the presence of different subclones within one tumor, each with their own unique features at the level of morphology, inflammation, genetics or transcriptomics.1,2 Heterogeneity provides the fuel for drug resistance; therefore, an accurate assessment of ITH is essential for the development of effective therapies.3
Being a highly heterogeneous disease, colorectal cancers (CRCs) differ at various molecular levels; these differences result in significant variability in patients' prognosis and response to therapy.4 Based on its morphologic features, colorectal adenocarcinoma can be categorized into different histological subtypes, with the most frequent being: not otherwise specified (40%-60%), micropapillary (10%-20%) and mucinous (5%-20%).5 Nonetheless, in the era of personalized medicine, the investigation of tumor heterogeneity goes beyond the mere phenotype and requires an extensive matched morpho-molecular evaluation.6 Somatic mutations in KRAS/NRAS and BRAF and defective mismatch repair/microsatellite instability (MMRd/MSI) are prognostic and predictive biomarkers currently tested in the clinic to guide therapeutic decisions.7, 8, 9, 10
Gene expression-based subtyping is becoming widely accepted as a relevant source of disease stratification, as transcriptomics represents the level of high-throughput molecular data that is best linked to cellular or tumor phenotype and clinical behavior.11
By using multiple microarray or RNA-sequencing datasets of primary CRC samples, the Colorectal Cancer Subtyping Consortium identified four gene expression consensus molecular subtypes (CMS), CMS1 (immune), CMS2 (canonical), CMS3 (metabolic) and CMS4 (mesenchymal), which were later found to be independent prognostic factors.12 CMS distribution varies according to the molecular landscape of CRC. Previous studies have identified a high prevalence of CMS1 among V600EBRAF-mutated (V600EBRAFmt) CRCs, followed by CMS4.12
About 8%-15% of CRCs harbor the p.V600E somatic mutation in the BRAF gene (BRAFmt), which is almost always mutually exclusive with the RAS gene mutation.13 The presence of BRAF mutation is a well-established negative prognostic biomarker in metastatic CRC (mCRC)14,15 and patients bearing this alteration appear to be resistant to therapeutic regimens based on a single tyrosine-kinase inhibitor. However, the BEACON CRC trial8 established a new standard of care in BRAFmt progressive patients, consisting of the combination of the BRAF inhibitor encorafenib plus the anti-epidermal growth factor receptor monoclonal antibody cetuximab.
BRAFmt mCRCs are characterized by a heterogeneous clinical, therapeutic and biological landscape,16,17 and, so far, no key molecular biomarker has demonstrated to be clinically useful in the prognostic stratification of these tumors.18 Therefore, dissecting morphologic and molecular ITH of BRAFmt mCRCs is a step forward towards the optimization of targeted and combined therapies.
Materials and methods
Cases
A series of 120 mono-institutional formalin-fixed and paraffin-embedded (FFPE) V600EBRAFmt primary metastatic colorectal adenocarcinoma samples was collected between January 2005 and December 2020. The study was carried out within the frame of the ‘BRAF BeCool’ study,19 which received local ethics committees' approval (Veneto Institute of Oncology, code 2017/34).
Two experienced pathologists (MF and MS) jointly evaluated cases according to the morphologic World Health Organization 2019 criteria.5 Out of 120 BRAFmt primary colorectal adenocarcinomas, a subset of 31 (25.8%) presented a mixed histotype (i.e. the coexistence of two different histologic variants representing at least the 30% of the primary tumor).
A subset of eight cases, which displayed morphologic heterogeneity and for which enough material was available for molecular profiling, was subsequently evaluated to assess: tumor-infiltrating lymphocyte (TIL) score, CMS by using NanoString nCounter® platform (NanoString Technologies, Seattle, WA),20 CMS by using the immunohistochemistry (IHC)-based classifier and MSI status. The TIL score and CMS by using NanoString nCounter® platform were evaluated separately for the two morphologically heterogeneous areas of each case (named areas A and B in each case), while the CMS by using the IHC-based mini-classifier and MSI status were evaluated on the entire tumor area.
Tumor-infiltrating lymphocytes
As previously reported,21 the density of TILs was defined as the mean value of five random counts at high-power fields (×40) of tumor-enriched areas composed of >60% of neoplastic cells. In paucicellular tumors, such as mucinous adenocarcinomas, the fields with highest cancer cell density were assessed. Only tumor epithelium-infiltrating lymphocytes were counted. Based on previous data,21 the presence of TILs was dichotomized by using a cut-off of 2.0: tumors showing an average number of TILs <2.0 were defined as ‘low number of TILs’, while ≥2.0 TILs were defined as ‘high number of TILs’. All samples were jointly evaluated by two gastrointestinal pathologists who were unaware of any clinical information.
Consensus molecular subtypes by NanoString nCounter® platform
Two experienced pathologists carefully marked two representative areas for each of the two morphologically heterogeneous components of every CRC sample, to ensure that each area contained >60% of neoplastic cells. Five consecutive 5-μm-thick sections from each FFPE sample were obtained.
The previously marked areas were manually (i.e. scalpel blade-assisted) microdissected from adjacent tissue. The RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Invitrogen, Thermo Fisher Scientific, Waltham, MA) was used to isolate nucleic acids from the dissected material, according to the manufacturer's instructions. The concentration and the purity of RNA sample were evaluated by NanoDrop ND-100 Spectrophotometer (Thermo Fisher Scientific).
To assess the CRC subtypes by gene expression profiling, 38 published CRCAssigner subtype-specific genes (CRCA-38) were evaluated using the NanoString nCounter® platform according to a previously validated custom CRC subtype-based gene expression analysis assay.20 Although CMS and CRCA-38 are different classifiers, CRCAssigner was considered as a surrogate for CMS based on Guinney et al. in this study.12
MSI status
MSI was carried out by adopting the Titano MSI test (Diatech Pharmacogenetics, Jesi, Italy). Briefly, the extracted DNA of the tumor and the corresponding normal mucosa were analyzed with the Titano MSI kit following the manufacturer's instructions. The Titano MSI kit allows the determination of MSI status in CRC samples by multiplex amplification with fluorescent primers and subsequent DNA fragment analysis on an automated sequencer. Starting from 20 ng of the extracted DNA, this tool is able to detect variation in the number of microsatellite loci of 10 different molecular targets (BAT25, BAT26, D2S123, D17S250, D5S346, BAT40, D18S58, NR21, NR24 and TGFβRII) by comparing peak profiles generated from the capillary electrophoresis run of the tumor and the corresponding normal tissue samples for each patient.
Consensus molecular subtypes by IHC-based classifier
The CMS was assigned by assessing four IHC markers (FRMD6, ZEB1, HTR2B, CDX2) in combination with pan-cytokeratin (KER) to normalize the results. The primary CRC samples were then categorized into the three CMS classes (CMS1, CMS2/3 or CMS4) using the online classification tool (https://crcclassifier.shinyapps.io/appTesting). DNA MMR proteins' status was first used to define samples belonging to CMS1.22
Nuclear immunostaining for MLH1, PMS2, MSH2 and MSH6 was evaluated following the Italian Group of Gastrointestinal Pathologists (GIPAD-SIAPeC) criteria to identify MMR deficiency (MMRd) and MMR proficiency.
Results
Out of 120 V600EBRAFmt primary colorectal adenocarcinomas, a subset of 31 (25.8%) presented a heterogeneous morphology with the coexistence of two different histologic variants within the same tumor. From these 31 cases, 8 presented adequate material for molecular profiling (Table 1 and Figure 1). Overall, the mean age of the patients was 62.9 years (median 63.9 years; range 28-85 years); the male-to-female ratio was 1.1. Among the 31 heterogeneous cases, the mean age of the patients was 68.1 years (median 69.0 years; range 41-85 years); the male-to-female ratio was 0.82. Among the 89 non-heterogeneous cases, the mean age of the patients was 61.7 years (median 69.0 years; range 28-85 years); the male-to-female ratio was 1.23.
Table 1.
Evaluation of histopathological and molecular features in eight cases of morphologically heterogeneous V600EBRAFmt CRCs
| Case | Area | Histologic variant | CMS (NanoString) | TILs [low (L), high (H)] | CMS (IHC) | MSI status |
|---|---|---|---|---|---|---|
| #1 | A | Signet ring | CMS3 | 1.4/L | CMS2/3 | MSS |
| B | NOS HG | CMS3 | 1.6/L | |||
| #2 | A | Mucinous | CMS1 | 1.4/L | CMS4 | MSS |
| B | NOS LG | CMS4 | 0.4/L | |||
| #3 | A | Signet ring | CMS4 | 0.6/L | CMS4 | MSS |
| B | NOS HG | CMS4 | 0.8/L | |||
| #4 | A | Undifferentiated | CMS1 | 5.6/H | CMS1 | MSI |
| B | Signet ring | CMS4 | 1.0/L | |||
| #5 | A | Medullary | CMS4 | 6.6/H | CMS1 | MSI |
| B | NOS LG | CMS4 | 3.8/H | |||
| #6 | A | NOS LG | CMS3 | 3.0/H | CMS1 | MSI |
| B | Signet ring | CMS4 | 3.0/H | |||
| #7 | A | Mucinous | CMS4 | 6.0/H | CMS1 | MSI |
| B | NOS HG | CMS4 | 8.0/H | |||
| #8 | A | Mucinous | CMS1 | 5.2/H | CMS1 | MSI |
| B | NOS HG | CMS4 | 9.8/H |
CMS, consensus molecular subtype; CRC, colorectal cancer; HG, high grade; IHC, immunohistochemistry; LG, low grade; MSI, microsatellite instability; MSS, microsatellite stable; NOS, not otherwise specified; V600EBRAFmt, V600EBRAF-mutated.
Figure 1.
Representative histopathological and molecular characteristics of cases #2, 4 and 6.
CMS, consensus molecular subtype; HPF, high-power field; MMR, mismatch repair; MSI, microsatellite instability; MSS, microsatellite stable; NOS LG, not otherwise specified low grade; TILs, tumor-infiltrating lymphocytes.
As regards the tumor localization, 8 cases were localized in the rectum, 26 in the left colon and 86 in the right colon. Among the 31 heterogeneous cases, 7 were localized in the left colon and 24 in the right colon. Among the 89 non-heterogeneous cases, 8 cases were localized in the rectum, 19 in the left colon and 62 in the right colon. When evaluating the presentation of metastases, 39 cases were metachronous and 81 were synchronous. Among the 31 heterogeneous cases, 15 had metachronous metastases, while 16 had synchronous metastases. Among the 89 non-heterogeneous cases, 24 had metachronous metastases, while 65 had synchronous metastases.
Out of the 120 V600EBRAFmt CRCs, 36 (30.0%) were MSI, with 17 (54.8%) MSI cases among the heterogeneous CRCs and 19 (21.3%) MSI cases among the non-heterogeneous CRCs (P < 0.001).
The median overall survival since the diagnosis of metastatic disease was 20.0 months (mean 23.4 ± 16.9 months) for non-heterogeneous versus 18.8 months (mean 26.1 ± 21.3 months) for heterogeneous cases. Overall, 20 patients were treated with immunotherapy and 19 with BRAF-directed therapy. Among the heterogeneous cases, nine received immunotherapy and five received the BRAF inhibitor. Among the non-heterogeneous cases, 11 were treated with immunotherapy. Five out of the eight (63%) cases resulted instable at MSI testing. The median overall survival since the diagnosis of metastatic disease for the eight cases was 20.0 months [mean 23.4 ± 16.9 months; 8.8 months for the microsatellite stable (MSS) cases and 36.8 months for the MSI group]. Best response after first-line therapy was stable disease in three cases and progressive disease in five.
To further characterize the phenotypic heterogeneity, the two components were microdissected to extract total RNA. NanoString analysis revealed that, among the eight selected cases, four were characterized by a heterogeneous gene expression profile, which overlapped the morphologic heterogeneity. In fact, in cases 2, 4 and 8, area A and area B were classified as CMS1 and CMS4, respectively, whereas in case 2, area A and area B were classified as CMS3 and CMS4, respectively. On the contrary, within cases 3, 5 and 7, both areas belonged to CMS4, while in case 1 both areas belonged to CMS3 (Table 1). Of note, seven out of eight samples presented at least a component characterized by a CMS4 subtype.
Following dichotomization of the TIL score (low versus high), seven cases out of eight showed consistency in the number of TILs between the morphologically heterogeneous components. In cases 1, 2 and 3, both areas exhibited a low (L) number of TILs (TILs < 2), while in cases 5, 6, 7 and 8, both areas displayed a high (H) number of TILs (TILs ≥ 2). The cases with a homogenous low number of TILs (cases 1-3) were all MSS, whereas the case with a mixed number of TILs (case 4) and the cases with a homogenous high number of TILs (cases 5-8) were all classified as MSI. Among the eight cases, TIL score differences between area A and B were defined as max/min of 0.2 (cases 1, 3)/4.6 (cases 4, 8), with an average of 1.9. The MSI cases displayed an average number TILs of 5.2, while the MSS cases exhibited an average number of TILs of 1.0.
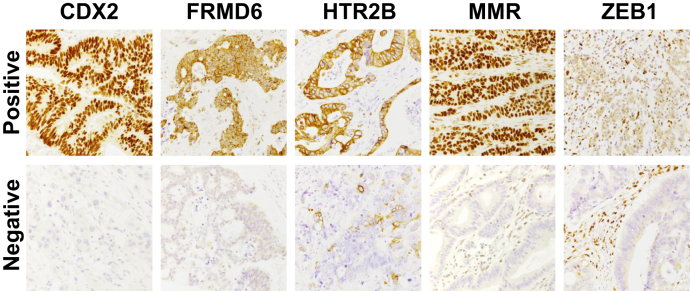
Using the IHC-based classifier (Figure 2), cases 4-8 were classified as CMS1, cases 2 and 3 as CMS4 and case 1 as CMS2/3. No significant intratumoral heterogeneity was observed between the two components, but two MMRd cases showed focal nuclear positivity for ZEB1. Cases 5, 6 and 7 showed a lack of consistency between the NanoString and immunohistochemical CMS classification. This is mainly due to the fact that by the IHC classifier all the MMRd tumors are classified as CMS1.
Figure 2.
Representative immunohistochemical positive and negative staining of the biomarkers used in the IHC classifier.
IHC, immunohistochemistry; MMR, mismatch repair.
Discussion
Despite the implementation of intensified chemotherapeutic regimens and targeted, patients with BRAFmt CRCs are often characterized by heterogeneous clinical outcomes and still present the poorer overall survival rates regardless of their stage at diagnosis compared to BRAF wild-type cases. Tumor heterogeneity may underlie poor clinical outcomes because diverse subclones can show distinct metastatic and drug resistance potential. We decided to characterize by gene expression a subset of phenotypically heterogeneous BRAFmt CRCs, in order to gather some preliminary data on the putative role of the mixed morphology to additionally stratify BRAFmt tumors.
Although recent studies found correlation between genomic heterogeneity and clinical prognosis in CRC patients,23 the prognostic role of morphologic heterogeneity has not been assessed yet.
According to the findings of a study by De Smedt et al.,24 50% of MSI colorectal tumors and 10% of MSS tumors revealed a mixed histology, and the fraction of cases presenting this feature among the V600EBRAFmt CRCs analyzed was comparable since the study comprised both MSI and MSS cases.
We found a statistically significant association between MSI and the presence of morphologic heterogeneity. Among the BRAFmt CRCs of our series, the higher incidence of morphological heterogeneous MSI cases may be explained by the mutator phenotype of MSI tumors, which results in several molecular clones with possible distinct morphological features. Furthermore, the MSI tumors often display a more poorly differentiated histology, with mucinous, signet cell and medullary differentiation and a higher density of TILs, as observed among our series of cases.
The distribution of the CMS across the morphologically heterogeneous V600EBRAFmt cases in our series was found to be significantly different from the one across V600EBRAFmt colorectal adenocarcinomas previously reported in literature. Indeed, the NanoString surrogate CMS classifier labeled the majority of samples as CMS4 (63%) and only a smaller fraction was labeled as CMS1 (19%). In particular, seven out of eight samples presented at least a CMS4 component. On the contrary, Barras and colleagues16 reported that the vast majority of BRAFmt patients (70%) were classified into CMS1 whereas only a few were found in CMS2 (2%), CMS3 (5%) and CMS4 (17%). Another interesting point is that, while MSI CRCs should be classified as CMS1, only two components of the five MSI cases were labeled as such, with the rest being CMS4 and CMS3. CMS4 being associated with the worst overall survival and relapse-free survival,12 it might be speculated that morphologic heterogeneity might predispose patients to inferior clinical outcomes.
While previous studies have documented the association between morphologic and genetic heterogeneity in relatively small CRC cohorts,25, 26, 27 no correlative analysis between morphology and gene expression has been reported so far. Here, for the first time, we have described a possible correlation between morphological and gene expression heterogeneity, indicating the presence of subpopulation with distinct morphologic and biological features within the primary tumor. Furthermore, transcriptomics being mostly linked to tumor behavior and clinical phenotype, this finding emphasizes the prognostic role of the different histologic variants.
Regarding the immune infiltrate, which has proven to be a positive prognostic marker for survival in patients with CRCs,28 morphologic heterogeneity does not seem to be immunogenic itself nor does it appear to be correlated to a heterogeneous distribution of TILs. Only one case presented a heterogeneous TIL status between the two components. This could be related to the fact that four out of five MSI cases were characterized by a diffuse elevated TIL score and all the three MSS cases presented a low number of infiltrating lymphocytes in both components.
Of note, for the purpose of this study, the IHC-based CMS classifier did not prove to be an adequate technique. In fact, the results show a lack of consistency between IHC and the NanoString classification, as all the MSI samples were classified as CMS1, by definition, using the IHC panel.
The relatively small sample size and the lack of a control cohort of BRAF wild-type tumors represent limitations of the study. A significant proportion of BRAF mutant cases in our series were microsatellite unstable and this is not unexpected given the frequent co-occurrence of BRAF mutations and DNA repair deficiencies in CRC.6 Our findings will need validation in larger and multicentric cohorts.
Nevertheless, we gathered preliminary data that seem to be consistent with the previous works that described ITH as a dynamic phenomenon that is observed at multiple levels. High levels of ITH could eventually predispose patients to inferior clinical outcomes and fuel resistance to targeted and immune therapy; however, further research on the matter is needed. Hence, an extensive morpho-molecular evaluation of ITH is required and should be integrated in the pathology report for a more effective disease stratification.
Acknowledgments
Funding
This work was partly supported by a grant from the Italian Health Ministry and Veneto Region [grant number NET-2016-02363853]. The funding agencies had no role in the design and performance of the study. NV is supported by Cancer Research UK [grant numbers A18052, A22909]; the National Institute for Health Research Biomedical Research Centre at The Royal Marsden NHS Foundation Trust and The Institute of Cancer Research [grant numbers A62, A100, A101, A159]; the European Union FP7 [grant number CIG 334261]; and the Katherine and Douglas Longden Chair in Oncology at Imperial College London (no grant number).
Disclosure
NV received honoraria from Merck Serono, Pfizer, Bayer, Eli-Lilly and Menarini Silicon Biosystems. FL had roles as consultant or advisor for Roche, Bayer, Amgen and Genentech. SL had roles as consultant or advisor for Amgen, Bayer, Merck Serono and Lilly; she received research funding from Amgen and Merck Serono and is part of speakers' bureau of Lilly and BMS. VZ received honoraria and had roles as consultant or advisor for Bristol-Myers Squibb, Bayer, Roche, Pfizer, Janssen, Novartis, Astellas and Servier; he had roles as consultant or advisor for Celgene and Merck. MF received research funding from Astellas Pharma, Macrophage Pharma and QED Therapeutics and had roles as consultant or advisor for Astellas Pharma, Roche, GSK-Tesaro and Diaceutics. All other authors have declared no conflicts of interest.
References
- 1.Sottoriva A., Kang H., Ma Z. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turajlic S., Sottoriva A., Graham T., Swanton C. Resolving genetic heterogeneity in cancer [published correction appears in Nat Rev Genet. 2020 Jan;21(1):65] Nat Rev Genet. 2019;20(7):404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 3.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 4.Sagaert X., Vanstapel A., Verbeek S. Tumor heterogeneity in colorectal cancer: what do we know so far? Pathobiology. 2018;85(1-2):72–84. doi: 10.1159/000486721. [DOI] [PubMed] [Google Scholar]
- 5.WHO Classification of Tumours Editorial Board . 5th ed. IARC; Lyon, France: 2019. Digestive System Tumours. [Google Scholar]
- 6.Fassan M. Molecular diagnostics in pathology: time for a next-generation pathologist? Arch Pathol Lab Med. 2018;142(3):313–320. doi: 10.5858/arpa.2017-0269-RA. [DOI] [PubMed] [Google Scholar]
- 7.Karapetis C.S., Khambata-Ford S., Jonker D.J. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 8.Kopetz S., Grothey A., Yaeger R. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med. 2019;381(17):1632–1643. doi: 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 9.André T., Shiu K.K., Kim T.W. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 10.Angerilli V., Galuppini F., Pagni F., Fusco N., Malapelle U., Fassan M. The role of the pathologist in the next-generation era of tumor molecular characterization. Diagnostics (Basel) 2021;11(2):339. doi: 10.3390/diagnostics11020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadanandam A., Lyssiotis C.A., Homicsko K. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guinney J., Dienstmann R., Wang X. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morkel M., Riemer P., Bläker H., Sers C. Similar but different: distinct roles for KRAS and BRAF oncogenes in colorectal cancer development and therapy resistance. Oncotarget. 2015;6(25):20785–20800. doi: 10.18632/oncotarget.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanelli G.N., Dal Pozzo C.A., Depetris I. The heterogeneous clinical and pathological landscapes of metastatic Braf-mutated colorectal cancer. Cancer Cell Int. 2020;20:30. doi: 10.1186/s12935-020-1117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fassan M., Milione M., Maddalena G. Synaptophysin expression in V600EBRAF-mutated advanced colorectal cancers identifies a new subgroup of tumours with worse prognosis. Eur J Cancer. 2021;146:145–154. doi: 10.1016/j.ejca.2021.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Barras D., Missiaglia E., Wirapati P. BRAF V600E mutant colorectal cancer subtypes based on gene expression. Clin Cancer Res. 2017;23(1):104–115. doi: 10.1158/1078-0432.CCR-16-0140. [DOI] [PubMed] [Google Scholar]
- 17.Schirripa M., Biason P., Lonardi S. Class 1, 2, and 3 BRAF-mutated metastatic colorectal cancer: a detailed clinical, pathologic, and molecular characterization. Clin Cancer Res. 2019;25(13):3954–3961. doi: 10.1158/1078-0432.CCR-19-0311. [DOI] [PubMed] [Google Scholar]
- 18.Loupakis F., Biason P., Prete A.A. CK7 and consensus molecular subtypes as major prognosticators in V600EBRAF mutated metastatic colorectal cancer. Br J Cancer. 2019;121(7):593–599. doi: 10.1038/s41416-019-0560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loupakis F., Intini R., Cremolini C. A validated prognostic classifier for V600EBRAF-mutated metastatic colorectal cancer: the ‘BRAF BeCool’ study. Eur J Cancer. 2019;118:121–130. doi: 10.1016/j.ejca.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Ragulan C., Eason K., Fontana E. Analytical validation of multiplex biomarker assay to stratify colorectal cancer into molecular subtypes. Sci Rep. 2019;9(1):7665. doi: 10.1038/s41598-019-43492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams D.S., Mouradov D., Jorissen R.N. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2019;68(3):465–474. doi: 10.1136/gutjnl-2017-315664. [DOI] [PubMed] [Google Scholar]
- 22.Ten Hoorn S., Trinh A., de Jong J., Koens L., Vermeulen L. Classification of colorectal cancer in molecular subtypes by immunohistochemistry. Methods Mol Biol. 2018;1765:179–191. doi: 10.1007/978-1-4939-7765-9_11. [DOI] [PubMed] [Google Scholar]
- 23.Oh B.Y., Shin H.T., Yun J.W. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci Rep. 2019;9(1):4542. doi: 10.1038/s41598-019-41098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Smedt L., Lemahieu J., Palmans S. Microsatellite instable vs stable colon carcinomas: analysis of tumour heterogeneity, inflammation and angiogenesis. Br J Cancer. 2015;113(3):500–509. doi: 10.1038/bjc.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehall V.L., Wynter C.V., Walsh M.D. Morphological and molecular heterogeneity within nonmicrosatellite instability-high colorectal cancer [published correction appears in Cancer Res 2002 Dec 1;62(23):7132] Cancer Res. 2002;62(21):6011–6014. [PubMed] [Google Scholar]
- 26.Bösmüller H., Kranewitter W., Webersinke G., Rumpold H., Hackl M., Fend F. Morphological and molecular heterogeneity in colorectal neoplasms with K-RAS mutation. A report of two cases. Pathol Res Pract. 2011;207(6):399–402. doi: 10.1016/j.prp.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Büttner J., Jöhrens K., Klauschen F. Intratumoral morphological heterogeneity can be an indicator of genetic heterogeneity in colorectal cancer. Exp Mol Pathol. 2018;104(1):76–81. doi: 10.1016/j.yexmp.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Idos G.E., Kwok J., Bonthala N., Kysh L., Gruber S.B., Qu C. The prognostic implications of tumor infiltrating lymphocytes in colorectal cancer: a systematic review and meta-analysis. Sci Rep. 2020;10(1):3360. doi: 10.1038/s41598-020-60255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]