Abstract
COVID-19 is an infectious and pathogenic viral disease caused by SARS-CoV-2 that leads to septic shock, coagulation dysfunction, and acute respiratory distress syndrome. The spreading rate of SARS-CoV-2 is higher than MERS-CoV and SARS-CoV. The receptor-binding domain (RBD) of the Spike-protein (S-protein) interacts with the human cells through the host angiotensin-converting enzyme 2 (ACE2) receptor. However, the molecular mechanism of pathological mutations of S-protein is still unclear. In this perspective, we investigated the impact of mutations in the S-protein and their interaction with the ACE2 receptor for SAR-CoV-2 viral infection. We examined the stability of pathological nonsynonymous mutations in the S-protein, and the binding behavior of the ACE2 receptor with the S-protein upon nonsynonymous mutations using the molecular docking and MM_GBSA approaches. Using the extensive bioinformatics pipeline, we screened the destabilizing (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) and stabilizing (H49Y, S50L, N501Y, D614G, A845V, and P1143L) nonsynonymous mutations in the S-protein. The docking and binding free energy (ddG) scores revealed that the stabilizing nonsynonymous mutations show increased interaction between the S-protein and the ACE2 receptor compared to native and destabilizing S-proteins and that they may have been responsible for the virulent high level. Further, the molecular dynamics simulation (MDS) approach reveals the structural transition of mutants (N501Y and D614G) S-protein. These insights might help researchers to understand the pathological mechanisms of the S-protein and provide clues regarding mutations in viral infection and disease propagation. Further, it helps researchers to develop an efficient treatment approach against this SARS-CoV-2 pandemic.
Keywords: SARS-CoV-2, Spike protein, ACE2 receptor, Stability, Nonsynonymous mutations, Binding affinity
1. Introduction
The first case of SARS-Cov-2 was reported in Dec 2019 in Wuhan, China. It was primarily called 2019-nCoV and later designated as SARS-Cov-2 because of its taxonomic and genomics relationship with SARS-CoV [1,2]. COVID-19, the exceedingly infectious and pathogenic viral disease that leads to septic shock, coagulation dysfunction, and acute respiratory distress syndrome, is caused by SARS-CoV-2 [3]. The transmission rate of SARS-CoV-2 is higher than MERS-CoV and SARS-CoV [4]. SARS-CoV-2 is a single-stranded RNA genome and has 29,930 nucleotides. It has 14 open reading frames (ORFs) that code for 29 proteins, including four crucial structural proteins: spike (S) proteins, membrane (M), nucleocapsid (N), and envelope (E), as well as nine supporting proteins and 16 non-structural proteins [5,6]. A recent study reported that mutations within the S-protein intercede the viral section that can tweak viral pathogenesis [7].
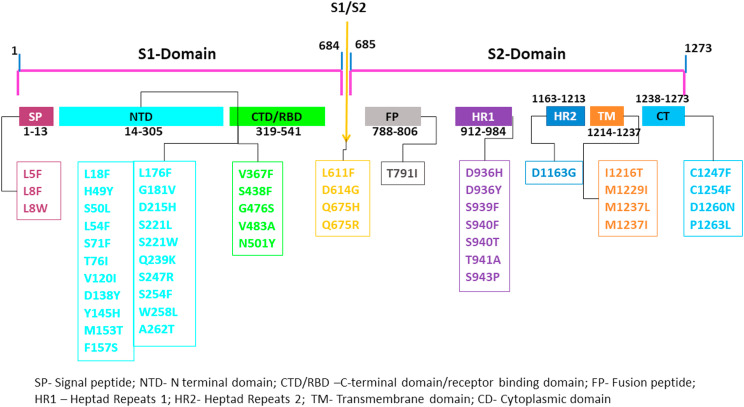
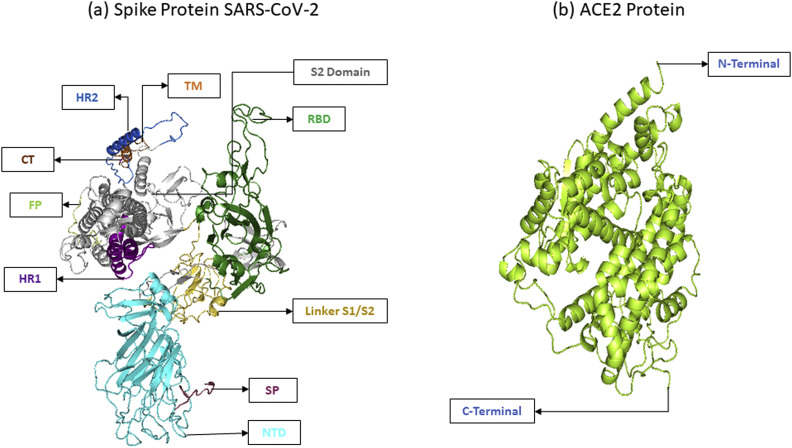
The S-protein is practically separated into two segments and known as S1 and S2. The S1 segment of the S-protein binds with the host angiotensin-converting enzyme2 (ACE2) receptor. The S2 region, which is isolated from the S1–S2 linker by the protease cleavage sites, aids in virus-host cell fusion [8]. The S1 segment of the S-protein contains a signal peptide (SP), N-terminal domain, and C-terminal domain. The C-terminal domain of the S1 segment contains a receptor-binding motif (RBM) and the receptor-binding domain (RBD), whereas the S2 segment of S-protein contains a heptad repeat (HR1&HR2), transmembrane domain (TM), fusion peptide (FP), and a small cytoplasmic domain (CP). The above features were combined, making the S-protein the critical target for developing antibodies, vaccines, and drugs [8,9]. Furthermore, nonsynonymous mutations found in the Spike-gene may have a vital role in the pathogen's host range and pathogenicity [10].
Moreover, discovering a complete set of nonsynonymous mutations in the S-protein of SARS-CoV-2 and its effect on human ACE2 affinity is required to develop therapeutic remedies. Hence, investigating the S- protein and its progression can improve our insights into host receptor interaction changes and pathogenic levels. We believe that Spike's essential significance, both in terms of antibody target and viral infection, is critical for developing an “early warning” pipeline to assess the pandemic's progression. The GISAID (A global initiative on sharing avian flu data) consortium has collected so far ~10,000 units of viral genome sequences and made them widely accessible to the research society. This effort allows scientists to examine the data to realize genome diversity [11,12], to postulate targetable targets for drug repurposing [13,14], and to build prevention approaches [15]. Mercatelli and Giorgi [16] completed the significant comparative analysis by inspecting more than 10,000 complete SARS-CoV-2 genomes. They reported every nonsynonymous mutation, further underlining the uprising of sub-clads, and genomic high mutation spots and arranged them genetically and geographically. These findings are mainly helpful for designing and measuring program efficacy for restricting the spread of SARS-Cov-2 on a regional basis [16].
Other recent studies have identified 1815 nonsynonymous mutations that belong to 1176 genomes from 29 countries [17,18]. These mutations fall into 62 distinct types, with 29 different amino acid substitutions present in the S1 segment, 28 in the S2 segment, and 5 at the S1–S2 junction of the S-protein. Mutation D614G was the most abundant mutation and showed the highly infectious A2 subtypes of SARS-CoV-2 [17,18]. RBD consists of 223 amino acid lengths in the S1-region of the S-protein that connects SARS-like coronaviruses to various hosts by directly binding to cellular ACE2 [19]. Shijulal and Umashankar et al. found six distinct types of mutations namely, V367F, P384L, S438F, K439N, G476S, and V483A in RBD domain of the S-protein. They further analyzed and identified that only ~2% of the RBD mutations were nonsynonymous reported from the total S-protein [17,18]. Recently, the mutation N501Y was observed in the RBD domain, which has spread rapidly in the UK and other countries [[20], [21], [22], [23], [24]]. The mutation N501Y has augmented the many discussions and questions, but only a small amount of data relating to it is currently available [21]. The mutation D614G, which originated either in China or Europe, and started to spread swiftly first in Europe and then throughout the world, is the focus of the current pandemic in a number of countries [[25], [26], [27]].
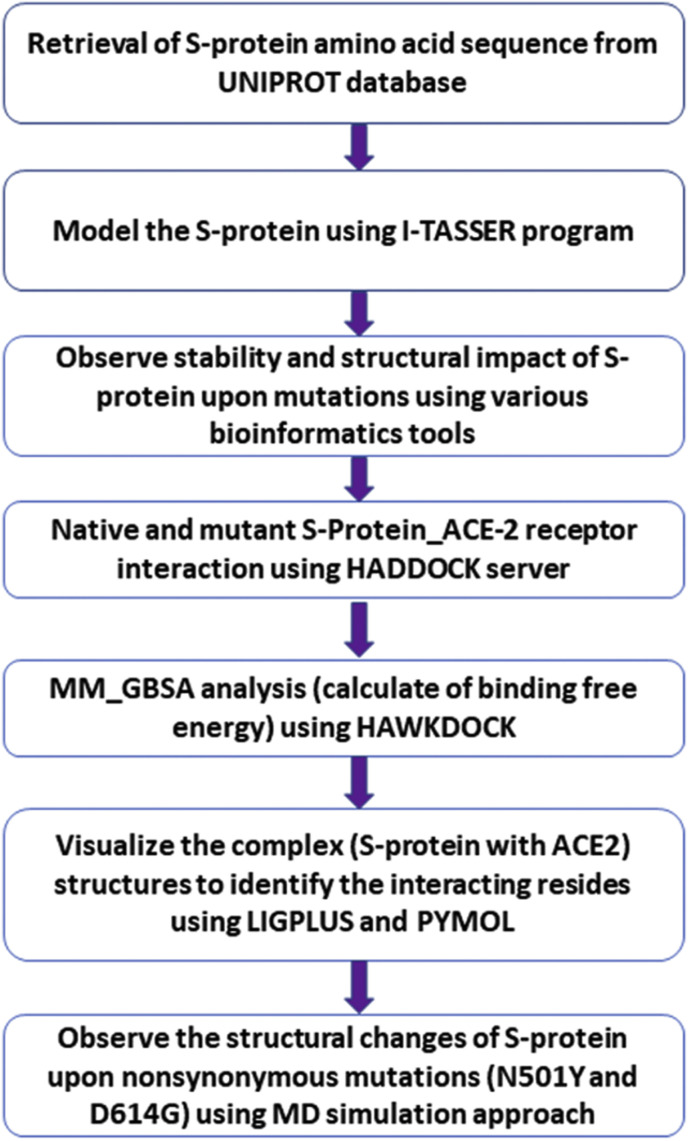
In biological studies, in-silico mutational investigations have proven to be a promising alternative to wet-bench techniques [28,29]. Further in-silico studies should play a vital role in developing information on this new virus to implement procedures that suppress its occurrence. We hypothesize that nonsynonymous mutations in the S-protein alter the stability of its structure and interaction with the ACE2 receptor. Therefore, in this study utilizing comprehensive bioinformatics, we detected the highly significant nonsynonymous mutations in the S-protein based on the stability of the S-protein. Further, we predicted the binding behavior of the S-protein upon nonsynonymous mutations with the ACE2 receptor using the molecular docking and MM_GBSA approaches. MD simulation approach revealed the structural changes of mutant (N501Y and D614G) proteins. Fig. 1 depicts the general workflow used in this work. This could help researchers in better understanding the pathogenic mechanism of S-protein and provide insights into the role of mutations in viral infection and disease propagation. Further, it helps researchers develop an efficient treatment approach against this pandemic SARS-CoV-2.
Fig. 1.
The workflow applied in this investigation.
2. Materials & methods
2.1. Dataset
The human S-protein of the SARS-CoV-2 sequence (length: 1273 amino acids) was obtained in FASTA format from the UNIPROT (ID: P0DTC2) database [30]. The 62 nonsynonymous mutations information of S-protein was collected from the recent articles [17,18,21] and displayed in Fig. 2 . The amino acid sequence was further used to construct the three dimensional (3D) protein conformation of native and mutant S-protein. The ACE2 receptor structure (PDB ID: 1R42_A) [31], was collected from the Protein Data Bank (PDB) database [32].
Fig. 2.
S-protein domains and their nonsynonymous mutations list.
2.2. Modeling the native and mutant S-protein
The available PDB structures of S-proteins show many missing residues in the 3-D structure. Some of the collected SNP residue positions do not present in the existed PDB structures. To fix this issue, we have used the previous research studies as an example and implemented them in this study to construct the three-dimensional (3-D) conformation of the native S-protein [[33], [34], [35], [36], [37], [38]]. Hence, we used the I-TASSER server [39] to model the native S-protein. It is a threading-based structure prediction program which used to generate the three-dimensional structure of proteins from their amino acid sequence. Some recent studies opted for the threading approach to model the protein to check the interaction with other biological molecules [[33], [34], [35], [36], [37], [38]]. I-TASSER produces great quality model predictions of three-dimensional structures from amino acid sequences. It is a very accurate and practical approach. We used the PDB ID: 6XR8_A [40] as a template, and it has shown 100% sequence coverage and similarity in the S-protein query sequence. We acquired the most acceptable modeled structure from I-TASSER, based on the high c-score. The predicted model of native S-protein was further refined by the molecular dynamics simulation (MDS) approach using the GROMACS [41] package. The OPLS-AA force field [42] was used for the refinement. Our MD simulations were carried out according to a procedure that has previously been published [[43], [44], [45]]. The pre-equilibrated protein was applied for MD simulations till 12 nanoseconds (ns). The RMSD value was calculated to examine the protein's structural alteration. To further inspect the effect of nonsynonymous mutations on the S-protein, we generated the nonsynonymous mutations in the predicted S-protein model. Moreover, we used the SwissPDB viewer tool [46] program and performed an energy minimization to create the perfect mutant protein structures. Lastly, the PROCHECK [47] and PROSA [48] tools were applied to assess the reliability of native and mutant S-protein.
2.3. Prediction of native and mutant S-protein stability
2.3.1. Sequence-based
The S-protein amino acid sequence was used as an input for estimating protein stability upon nonsynonymous mutations. i-Stable Server [49] was applied to predict the stability alterations of S-protein upon nonsynonymous mutations. It gives results from I-Mutant2.0 [50] and MUpro [51] programs, predicting the meta results. The DDG scores from I-Mutant 2.0 and Conf score from MUpro and i-Stable are considered to predict protein stability. First, I-Mutant-2.0, the DDG score less than 0 is predicted as decreasing stability, whereas DDG scores greater than 0 is predicted as increasing stability. Second, with MUpro, we obtained the Conf Score in the range of −1 to 1. A DDG score higher than 0 predicted increased stability, and a score lower than 0 predicted decreased stability. Last, the i-Stable conf score ranges between 0 and 1, where the higher value exposes higher confidence.
2.3.2. Structure-based
We used the modeled native and mutant S-protein structures as input to estimate how the protein's stability would vary as a result of nonsynonymous mutations.
-
i.
CUPSAT
CUPSAT (Cologne University Protein Stability Analysis Tool) [52] was used to analyze fluctuations in the stability of S-protein upon mutation. We uploaded the model structure of the S-protein as input and selected the location of the amino acid residue to be mutated. The stability of native and mutant S-proteins is predicted by calculating the difference in the free energy score (DDG score). Further, it provides information about nonsynonymous mutations, secondary structures, torsion angles, and solvent accessibility affected by the mutation.
-
ii.
SDM2
The Site-directed mutator 2 (SDM2) Server was employed to compute the effect of nonsynonymous mutations on the stability of proteins [53]. It is a knowledge‐based tool and applies ESSTs tables to analyze the alterations in protein stability upon mutation. Further, it computes the stability changes score between the native and mutant proteins. We uploaded the 3-D model structures of native and mutant S-proteins and point variant information as an input file.
-
ii.
DUET [54] is an integrated computational method used for calculating the impact of nonsynonymous mutations on the stability of the protein. We uploaded the 3-D model structures of native and mutant S-proteins as an input file.
2.4. S-protein and ACE2 docking by HADDOCK
The native and mutant S-proteins were docked with the ACE2 receptor molecule using HADDOCK v2. 4 [55]. The modeled native and mutant S-proteins and ACE2 protein (PDB ID: 1R42_A) molecules were used. We collected the interacting (binding) residues between the S-protein and ACE-2 receptor from recent studies [56]. The collected binding residues in S-protein together with the binding residues in the partner ACE-2 receptor were used as active residues, and the neighboring ones were used as passive residues. The default parameters we applied in our previous studies were also considered for the docking studies [[57], [58], [59], [60]]. The HADDOCK scoring function was executed based on the weighted aggregate of the various energy terms de-solvation (Desolv) and restraints energy, van der Waals (vdw), electrostatic (Elec), and buried surface area (BSA).
2.5. MM-GBSA calculation by HAWKDOCK
The MM-GBSA free energy decomposition analysis implemented in the S-protein and ACE2 receptor molecule and the binding affinity was estimated by the HawkDock server [61]. HawkDock considers a relatively more minor protein as a flexible receptor and a bigger protein as an inert receptor. The HawkRank scoring function and the ATTRACT docking algorithm with MM/GBSA free energy decomposition analysis were used to calculate the binding free energy between the protein complexes. We applied the haddock output complex structures (native and mutant S-proteins and ACE2 receptor) as an input to compute the binding free energy. Lastly, the best ten models of interacting proteins were re-ranked by MM/GBSA calculation [[62], [63], [64]]. All protein-protein interactions were represented diagrammatically using the LigPlot program [65] and PYMOL software [66].
2.6. Native and mutant (N501Y & D614G) S-proteins MD simulation
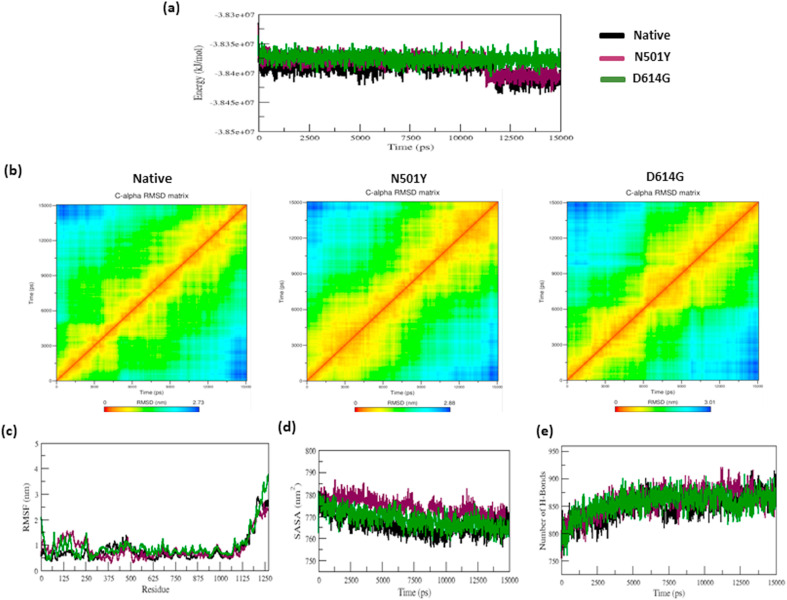
The native and most prominent stabilizing mutants (N501Y and D614G) of the S-protein were utilized as input for the molecular dynamics simulations. The initial setup of the MD simulation was prepared by following our previously published protocol [[43], [44], [45]]. The CHARMM36 m force field [67] was implemented in this simulation. The minimization, equilibration, and MD simulation procedures were performed per our previously published protocol [[43], [44], [45]]. Lastly, the MD simulation was carried out for 15 ns. XMGRACE [68] tool used to analyze the trajectories. We analyzed the energy plot, RMSD, RMSF, the solvent-accessible surface area (SASA), Principle component analysis (PCA) [69], and the number of hydrogen bonds (NH-bonds), and we made a comparison between native, stabilizing mutants (N501Y and D614G) to examine the structural behavior of the S-protein.
3. Results
3.1. Predicting the 3-D structures of native and mutant S-proteins
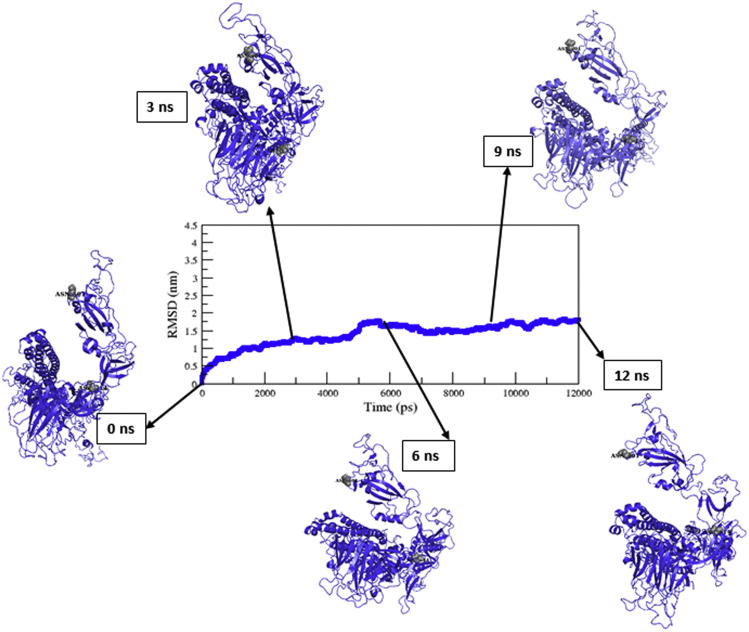
Observing the mutational effect on the S-protein and its interaction with the ACE2 receptor is very important for predicting the 3D conformation of the S-protein. Hence, we have used the I-TASSER online server to predict the 3D structure of the S-protein. The PDB ID: 6XR8_A was used as a template for predicting the 3D structure of the native S-protein. Further refinement of predicted model protein, MDS for 12ns performed to evaluate the stability of model protein for subsequent studies. The RMSD of the native S-protein is converged after 7 ns (Fig. 3 ). Further, the average structure of the native S-protein was shown at regular intervals. The best favorable structure was selected at the 12ns MDS, and subsequently used to build the mutant structures.
Fig. 3.
The backbone RMSD of the native S-protein versus time at 300 K. The average structure of native S-protein has shown in different timescale.
Further, the Swiss PDB viewer tool was used to build the mutant structures of the S-protein. Then, PROCHECK and PROSA software used to calculate the consistency of predicted model structures. The native S-protein showed that 99.7% favored and allowed region and z-score of −6.6. Mutant structures showed in the range of 96.4%–99.6% in favored residues and allowed region and z-score values in the range of 3.25–6.54. These predicted scores corroborate the high confidence level. Therefore, the native and mutant modeled S-protein structures were utilized for SNP and protein-protein docking analysis. The modeled structure of the native S-protein and ACE2 receptor was shown in Fig. 4 .
Fig. 4.
(a) Modeled Spike (S) - protein, (b) ACE2 protein (PDB ID: 1R42_A). This figure was prepared by PYMOL.
3.2. Protein stability analysis
To identify the effect of 62 nonsynonymous mutations of different domains (SP, NTD, CTD/RBD, Linker, S1/S2, FP, S2, HR1, HR2, TM, and CT domain) of S-protein, we utilized a broad approach of numerous coherent sequences and structure-based online servers. First, we used the sequence-based i-Stable Server to verify the stability of the S-protein (whether increased or decreased stability) upon mutations. The i-Stable tool collates I‐Mutant 2.0 SEQ, MUpro, and i-Stable to evaluate the stability. I-Mutant 2.0 SEQ predicted 49 of 62 nonsynonymous mutations with decreased stability, and 13 nonsynonymous mutations with increased stability (Table 1 ). On the other hand, Mupro predicted 39 and 23 nonsynonymous mutations as decrease stability and increase stability (Table 1). As a meta result, i-stable predicted, out of 62 nonsynonymous mutations, 40 nonsynonymous mutations (L5F, L8V, L8W, L18F, L54F, T76I, V120I, D138Y, Y145H, M153T, F157S, L176F, G181V, D215H, A262T, V367F, G476S, V483A, L611F, Q675H, A706V, T791I, P809S, A845S, A846V, A879S, V1040F, P1162L, D936H, S939F, T941A, D1163G, I1216T, M1229I, M1237L, M1237I, C1247F, C1254F, D1260 N, and P1263L) with decreased the stability and 22 nonsynonymous mutations (H49Y, S50L, S71F, S221L, S221W, Q239K, S247R, S254F, W258L, S438F, N501Y, D614G, Q675R, A831V, D839Y, A845V, A852V, P1143L, D936Y, S940F, S940T, and S943P) with increased stability (Table 1).
Table 1.
The sequence-based classification of nonsynonymous mutations of SPIKE protein (S-protein) as increasing or decreasing in stability by applying iStable Server.
|
Domain |
Mutation | I-Mutant2.0 SEQ |
Mupro |
Meta Result (iStable) |
|||
|---|---|---|---|---|---|---|---|
| DDG | Prediction | Score | Prediction | Score | Prediction | ||
| SP | L5F | −0.63 | Decrease | −0.72 | Decrease | 0.71 | Decrease |
| L8V | −2.82 | Decrease | −0.45 | Decrease | 0.83 | Decrease | |
| L8W | −1.20 | Decrease | −0.62 | Decrease | 0.82 | Decrease | |
|
NTD |
L18F | −0.82 | Decrease | −1.00 | Decrease | 0.86 | Decrease |
| H49Y | 0.69 | Increase | 0.98 | Increase | 0.84 | Increase | |
| S50L | −0.04 | Decrease | 1 | Increase | 0.52 | Increase | |
| L54F | −0.88 | Decrease | −1 | Decrease | 0.82 | Decrease | |
| S71F | 0.41 | Increase | 0.02 | Increase | 0.82 | Increase | |
| T76I | −0.93 | Decrease | −1 | Decrease | 0.60 | Decrease | |
| V120I | −0.57 | Decrease | −0.69 | Decrease | 0.78 | Decrease | |
| D138Y | 0.08 | Decrease | −0.67 | Decrease | 0.57 | Decrease | |
| Y145H | −1.43 | Decrease | −1 | Decrease | 0.82 | Decrease | |
| M153T | −1.07 | Decrease | −0.67 | Decrease | 0.84 | Decrease | |
| F157S | −1.94 | Decrease | −1 | Decrease | 0.89 | Decrease | |
| L176F | −0.88 | Decrease | −1 | Decrease | 0.81 | Decrease | |
| G181V | −0.48 | Decrease | −0.15 | Decrease | 0.81 | Decrease | |
| D215H | −0.97 | Decrease | −1 | Decrease | 0.69 | Decrease | |
| S221L | −0.25 | Decrease | 1 | Increase | 0.51 | Increase | |
| S221W | 0.14 | Increase | 0.89 | Increase | 0.77 | Increase | |
| Q239K | −0.56 | Increase | −0.01 | Decrease | 0.51 | Increase | |
| S247R | 0.01 | Increase | 0.50 | Increase | 0.78 | Increase | |
| S254F | 0.21 | Increase | 0.44 | Increase | 0.82 | Increase | |
| W258L | −0.84 | Decrease | 0.11 | Increase | 0.62 | Increase | |
| A262T | −0.73 | Decrease | −0.83 | Decrease | 0.77 | Decrease | |
|
CTD/RBD |
V367F | −2.07 | Decrease | −0.19 | Decrease | 0.79 | Decrease |
| S438F | 0.00 | Increase | 0.75 | Increase | 0.82 | Increase | |
| G476S | −1.45 | Decrease | −0.45 | Decrease | 0.82 | Decrease | |
| V483A | −1.46 | Decrease | −0.51 | Decrease | 0.75 | Decrease | |
| N501Y | 0.18 | Increase | 0.39 | Increase | 0.51 | Increase | |
|
Linker (S1/S2) |
L611F | −0.72 | Decrease | −1 | Decrease | 0.82 | Decrease |
| D614G | −1.10 | Decrease | 0.19 | Increase | 0.68 | Increase | |
| Q675H | −0.64 | Decrease | −0.06 | Decrease | 0.80 | Decrease | |
| Q675R | −0.06 | Decrease | 0.50 | Increase | 0.59 | Increase | |
| A706V | −0.26 | Decrease | 0.16 | Increase | 0.52 | Decrease | |
| FP | T791I | −0.35 | Decrease | −1 | Decrease | 0.66 | Decrease |
|
S2 |
P809S | −1.59 | Decrease | −1 | Decrease | 0.79 | Decrease |
| A831V | −0.06 | Decrease | 0.85 | Increase | 0.56 | Increase | |
| D839Y | 0.04 | Increase | 0.31 | Increase | 0.80 | Increase | |
| A845S | −0.77 | Decrease | −1 | Decrease | 0.88 | Decrease | |
| A845V | −0.20 | Decrease | 0.09 | Increase | 0.51 | Increase | |
| A846V | −0.18 | Decrease | 0.89 | Increase | 0.54 | Decrease | |
| A852V | −0.14 | Decrease | 0.72 | Increase | 0.58 | Increase | |
| A879S | −0.62 | Decrease | −0.50 | Decrease | 0.86 | Decrease | |
| V1040F | −1.72 | Decrease | −0.92 | Decrease | 0.83 | Decrease | |
| P1143L | −0.64 | Decrease | 0.16 | Increase | 0.55 | Increase | |
| P1162L | −0.94 | Decrease | −0.39 | Decrease | 0.81 | Decrease | |
|
HR1 |
D936H | −0.94 | Decrease | −0.61 | Decrease | 0.76 | Decrease |
| D936Y | −0.58 | Decrease | 0.09 | Increase | 0.69 | Increase | |
| S939F | −0.09 | Increase | −0.24 | Decrease | 0.52 | Decrease | |
| S940F | 0.09 | Increase | 0.99 | Increase | 0.75 | Increase | |
| S940T | −0.07 | Increase | 0.55 | Increase | 0.81 | Increase | |
| T941A | −1.04 | Decrease | −0.51 | Decrease | 0.72 | Decrease | |
| S943P | −0.21 | Increase | 0.47 | Increase | 0.81 | Increase | |
| HR2 | D1163G | −1.78 | Decrease | −0.83 | Decrease | 0.75 | Decrease |
|
TM |
I1216T | −1.96 | Decrease | −1 | Decrease | 0.86 | Decrease |
| M1229I | −0.85 | Decrease | −1 | Decrease | 0.71 | Decrease | |
| M1237L | −0.91 | Decrease | −0.25 | Decrease | 0.82 | Decrease | |
| M1237I | −0.75 | Decrease | −0.63 | Decrease | 0.74 | Decrease | |
|
CT |
C1247F | 0.01 | Decrease | −0.42 | Decrease | 0.69 | Decrease |
| C1254F | −0.13 | Decrease | −0.87 | Decrease | 0.76 | Decrease | |
| D1260 N | −0.94 | Decrease | −0.57 | Decrease | 0.85 | Decrease | |
| P1263L | −0.66 | Decrease | −0.41 | Decrease | 0.80 | Decrease | |
Further, we have collated the CUPSAT, SDM 2.0, and DUET structure-based online servers to predict S-protein stability upon nonsynonymous mutations. CUPSAT predicted that out of 62 nonsynonymous mutations, 36 and 26 nonsynonymous mutations were destabilizing and stabilizing (Table 2 ). The SDM 2.0 server depicted 28 nonsynonymous mutations with increased stability, and 34 nonsynonymous mutations with decreased stability (Table 2). While the DUET server predicted 46 and 16 nonsynonymous mutations as destabilizing and stabilizing (Table 2). In combination, structure-based online servers predicted 17 nonsynonymous mutations (L8V, L8W, L18F, S71F, Y145H, M153T, F157S, S221L, S221W, S247R, G476S, L611F, A831V, A852V, A879S, C1247F, and C1254F) with decreased stability, and 7 nonsynonymous mutations (H49Y, S50L, D215H, N501Y, D614G, A845V, and P1143L) with increased stability in the S-protein (Table 2). Together, both sequence and structure-based online servers predicted 11 nonsynonymous mutations (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) exhibiting decreased stability and 6 nonsynonymous mutations (H49Y, S50L, N501Y, D614G, A845V, and P1143L) exhibiting increased stability in the S-protein upon mutations (Table 1, Table 2). These nonsynonymous mutations were separated based on each domain (SP, NTD, CTD/RBD, Linker, S2, and TM) of the S-protein. Further, we examined binding behavior of nonsynonymous mutations in S-protein with the ACE2 receptor using molecular docking and MM_GBSA studies.
Table 2.
The structure-based classification of nonsynonymous mutations of SPIKE protein (S-protein) as increasing or decreasing in stability using CUPSAT, SDM 2.0, DUET Server.
| Domain | Mutation | CUPSAT |
SDM 2.0 |
DUET |
|||
|---|---|---|---|---|---|---|---|
| Predicted DDG (kcal/mol) | Prediction | DDG | Prediction | DDG | Prediction | ||
| SP | L5F | −0.29 | Destabilizing | 0.2 | Increased stability | −0.611 | Destabilizing |
| L8V | −3.18 | Destabilizing | −2.69 | Reduced stability | −2.13 | Destabilizing | |
| L8W | −8.17 | Destabilizing | −1.11 | Reduced stability | −2.003 | Destabilizing | |
| NTD | L18F | −2.64 | Destabilizing | −1.11 | Reduced stability | −1.72 | Destabilizing |
| H49Y | 0.96 | Stabilizing | 0.78 | Increased stability | 1.293 | Stabilizing | |
| S50L | 4.18 | Stabilizing | 1.2 | Increased stability | 0.734 | Stabilizing | |
| L54F | 4.41 | Stabilizing | −0.08 | Reduced stability | −0.866 | Destabilizing | |
| S71F | −0.53 | Destabilizing | −0.66 | Reduced stability | −1.772 | Destabilizing | |
| T76I | 1.1 | Stabilizing | 1.1 | Increased stability | −0.06 | Destabilizing | |
| V120I | 0.89 | Stabilizing | −0.65 | Reduced stability | −0.029 | Destabilizing | |
| D138Y | 1.71 | Stabilizing | −0.21 | Reduced stability | −1.505 | Destabilizing | |
| Y145H | −1.6 | Destabilizing | −0.29 | Reduced stability | −1.08 | Destabilizing | |
| M153T | −0.58 | Destabilizing | −0.09 | Reduced stability | −0.733 | Destabilizing | |
| F157S | −4.01 | Destabilizing | −0.83 | Reduced stability | −0.977 | Destabilizing | |
| L176F | −3.11 | Destabilizing | 0.2 | Increased stability | −0.537 | Destabilizing | |
| G181V | 3.27 | Stabilizing | −1.05 | Reduced stability | −0.466 | Destabilizing | |
| D215H | 1.17 | Stabilizing | 0.85 | Increased stability | 1.087 | Stabilizing | |
| S221L | −1.23 | Destabilizing | −0.08 | Reduced stability | 0.102 | Stabilizing | |
| S221W | −3.44 | Destabilizing | −0.74 | Reduced stability | −0.793 | Destabilizing | |
| Q239K | 6.79 | Stabilizing | −1.01 | Reduced stability | −1.223 | Destabilizing | |
| S247R | −3.25 | Destabilizing | −1.01 | Reduced stability | −1.155 | Destabilizing | |
| S254F | 0.11 | Stabilizing | −0.73 | Reduced stability | −1.638 | Destabilizing | |
| W258L | 0.71 | Stabilizing | −1.07 | Reduced stability | −1.932 | Destabilizing | |
| A262T | 1.51 | Stabilizing | 0.13 | Increased stability | −0.865 | Destabilizing | |
| CTD/RBD | V367F | −0.11 | Destabilizing | 0.14 | Increased stability | −0.636 | Destabilizing |
| S438F | 3.64 | Stabilizing | −0.38 | Reduced stability | −0.485 | Destabilizing | |
| G476S | −0.8 | Destabilizing | −4.37 | Reduced stability | −1.112 | Destabilizing | |
| V483A | −0.13 | Destabilizing | 0.12 | Increased stability | −0.317 | Destabilizing | |
| N501Y | 0.07 | Stabilizing | 0.91 | Increased stability | 0.207 | Stabilizing | |
| Linker | L611F | −1.01 | Destabilizing | −0.59 | Reduced stability | −1.308 | Destabilizing |
| D614G | 0.3 | Stabilizing | 0.87 | Increased stability | 0.173 | Stabilizing | |
| (S1/S2) | Q675H | −3.05 | Destabilizing | 0.33 | Increased stability | −0.71 | Destabilizing |
| Q675R | −0.87 | Destabilizing | 0.12 | Increased stability | 0.106 | Stabilizing | |
| A706V | 0.17 | Stabilizing | −0.12 | Reduced stability | −0.104 | Destabilizing | |
| FP | T791I | −2.58 | Destabilizing | 0.07 | Increased stability | 0.257 | Stabilizing |
| S2 | P809S | 2.16 | Stabilizing | −0.63 | Reduced stability | −0.236 | Destabilizing |
| A831V | −1.46 | Destabilizing | −0.84 | Reduced stability | −0.127 | Destabilizing | |
| D839Y | −0.98 | Destabilizing | 0.67 | Increased stability | −0.362 | Destabilizing | |
| A845S | 0.77 | Stabilizing | −0.14 | Reduced stability | −0.328 | Destabilizing | |
| A845V | 0.36 | Stabilizing | 1.16 | Increased stability | 0.119 | Stabilizing | |
| A846V | 1.47 | Stabilizing | −0.12 | Reduced stability | −0.018 | Destabilizing | |
| A852V | −0.5 | Destabilizing | −0.84 | Reduced stability | −0.55 | Destabilizing | |
| A879S | −1.52 | Destabilizing | −2.18 | Reduced stability | −1.393 | Destabilizing | |
| V1040F | −1.11 | Destabilizing | 0.14 | Increased stability | −0.781 | Destabilizing | |
| P1143L | 0.45 | Stabilizing | 1.45 | Increased stability | 0.358 | Stabilizing | |
| P1162L | 0.23 | Stabilizing | −0.58 | Reduced stability | −0.085 | Destabilizing | |
| HR1 | D936H | −0.91 | Destabilizing | 0.13 | Increased stability | −0.684 | Destabilizing |
| D936Y | −1.69 | Destabilizing | 0.77 | Increased stability | −0.434 | Destabilizing | |
| S939F | −0.37 | Destabilizing | 0.54 | Increased stability | −1.037 | Destabilizing | |
| S940F | −0.36 | Destabilizing | 0.54 | Increased stability | −0.89 | Destabilizing | |
| S940T | 0.06 | Stabilizing | −0.14 | Reduced stability | 0.342 | Stabilizing | |
| T941A | −1.07 | Destabilizing | 1.1 | Increased stability | −0.594 | Destabilizing | |
| S943P | 0.75 | Stabilizing | −0.48 | Reduced stability | −0.138 | Destabilizing | |
| HR2 | D1163G | −1.93 | Destabilizing | 0.87 | Increased stability | 0.411 | Stabilizing |
| TM | I1216T | 1.46 | Stabilizing | −1.06 | Reduced stability | −0.86 | Destabilizing |
| M1229I | −0.46 | Destabilizing | 0.11 | Increased stability | −0.18 | Destabilizing | |
| M1237L | −1.1 | Destabilizing | 0.54 | Increased stability | 0.498 | Stabilizing | |
| M1237I | −1.75 | Destabilizing | 0.11 | Increased stability | 0.439 | Stabilizing | |
| CT | C1247F | −1.53 | Destabilizing | −0.2 | Reduced stability | −0.369 | Destabilizing |
| C1254F | −1.24 | Destabilizing | −0.2 | Reduced stability | −0.424 | Destabilizing | |
| D1260 N | 0.27 | Stabilizing | −0.11 | Reduced stability | 0.616 | Stabilizing | |
| P1263L | −1.83 | Destabilizing | 2.24 | Increased stability | 0.394 | Stabilizing | |
3.3. Docking the S-protein and its predicted mutations with the ACE2 receptor molecule
The HADDOCK tool and HawkDock Server was used to examine the binding energy between the native and mutant S-proteins [destabilizing (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) and stabilizing (H49Y, S50L, N501Y, D614G, A845V, and P1143L)] with the ACE2 receptor. The HADDOCK score must be computed in order to understand the interaction between biological partners. During docking, each structure is assigned a HADDOCK score, which allows the structures to be classified. The score is a sum of the intermolecular AIR energies, buried surface area (BSA) Electrostatic, van der Waals, and desolvation (Dsolv) energies [[70], [71], [72]].
The Haddock scores of the native S-protein-ACE2 complex, and destabilizing S-proteins of S-protein-ACE2 (L8V-ACE2, L8W-ACE2, L18F-ACE2, Y145H-ACE2, M153T-ACE2, F157S-ACE2, G476S-ACE2, L611F-ACE2, A879S-ACE2, C1247F-ACE2, and C1254F-ACE2) complexes were −123.5 ± 13.2, −118.8 ± 6.1, −120.8 ± 16.0, −116.0 ± 3.2, −119.0 ± 9.9, −115.5 ± 1.2, −118.4 ± 2.8, −119.2 ± 6.5, −117.0 ± 14.4, −116.8 ± 12.7, −117.1 ± 4.0, and −113.4 ± 8.0, respectively, shown in Table 3 a. Whereas the HADDOCK score of stabilizing nonsynonymous mutations of S-protein -ACE2 complexes (H49Y-ACE2, S50L-ACE2, N501Y-ACE2, D614G-ACE2, A845V-ACE2, and P1143L-ACE2) were −136.5 ± 6.7, −131.3 ± 10.3, −136.3 ± 6.3, −128.2 ± 12.0, −127.7 ± 4.7, and −125.9 ± 17.1, respectively, are shown in Table 3 b.
Table 3a.
Docking analysis of native and destabilizing nonsynonymous mutants of S-protein with ACE2 receptor using HADDOCK.
| Protein Type | Domain | Haddock Score | Van der waal energy (Kcal mol−1) | Electrostatic energy (Kcal mol−1) | Desolvation energy (Kcal mol−1) | Restraints violation energy (Kcal mol−1) | Buried surface area (A °2) |
|---|---|---|---|---|---|---|---|
| Native-ACE2 | – | −123.5 ± 13.2 | −51.8 ± 3.3 | −374.5 ± 80.4 | −10.3 ± 3.7 | 134.0 ± 27.6 | 2099.9 ± 186.4 |
| L8V-ACE2 | SP | −118.8 ± 6.1 | −44.1 ± 4.6 | −345.6 ± 18.0 | −12.8 ± 5.0 | 73.0 ± 3.6 | 2012.7 ± 146.2 |
| L8W-ACE2 | −120.8 ± 16.0 | −56.9 ± 8.9 | −320.2 ± 29.3 | −10.5 ± 2.8 | 106.3 ± 36.1 | 2082.0 ± 84.8 | |
| L18F-ACE2 | NTD | −116.0 ± 3.2 | −54.4 ± 11.4 | −319.1 ± 51.9 | −8.6 ± 3.3 | 108.3 ± 57.1 | 2004.5 ± 81.5 |
| Y145H-ACE2 | −119.0 ± 9.9 | −57.7 ± 4.0 | −294.7 ± 21.8 | −13.3 ± 2.8 | 108.7 ± 28.4 | 1986.3 ± 52.0 | |
| M153T-ACE2 | −115.5 ± 1.2 | −47.7 ± 9.6 | −343.4 ± 77.9 | −7.1 ± 6.1 | 80.0 ± 29.1 | 1946.9 ± 59.7 | |
| F157S-ACE2 | −118.4 ± 2.8 | −56.9 ± 1.5 | −282.9 ± 27.0 | −13.4 ± 4.6 | 85.0 ± 36.9 | 1954.0 ± 25.2 | |
| G476S-ACE2 | CTD/RBD | −119.2 ± 6.5 | −50.7 ± 4.5 | −375.0 ± 33.8 | −4.9 ± 0.9 | 113.9 ± 60.6 | 1910.9 ± 123.7 |
| L611F-ACE2 | Linker | −117.0 ± 14.4 | −66.2 ± 6.1 | −242.3 ± 39.5 | −7.9 ± 2.6 | 55.4 ± 16.2 | 2019.9 ± 243.6 |
| A879S-ACE2 | S2 | −116.8 ± 12.7 | −48.8 ± 12.0 | −339.1 ± 44.8 | −6.9 ± 5.7 | 66.8 ± 22.6 | 2001.8 ± 193.5 |
| C1247F-ACE2 | TM | −117.1 ± 4.0 | −51.5 ± 6.4 | −315.3 ± 19.5 | −13.3 ± 3.3 | 108.1 ± 28.7 | 1977.5 ± 72.2 |
| C1254F-ACE2 | −113.4 ± 8.0 | −65.8 ± 9.7 | −216.8 ± 48.0 | −11.6 ± 5.3 | 73.5 ± 35.5 | 2098.8 ± 135.2 |
Table 3b.
Docking analysis of native and nonsynonymous stabilizing mutants of S-protein with ACE2 receptor using HADDOCK.
| Protein Type | Domain | Haddock Score | Van der waal energy (Kcal mol−1) | Electrostatic energy (Kcal mol−1) | Desolvation energy (Kcal mol−1) | Restraints violation energy (Kcal mol−1) | Buried surface area (A °2) |
|---|---|---|---|---|---|---|---|
| Native-ACE2 | – | −123.5 ± 13.2 | −51.8 ± 3.3 | −374.5 ± 80.4 | −10.3 ± 3.7 | 134.0 ± 27.6 | 2099.9 ± 186.4 |
| H49Y-ACE2 | NTD | −136.5 ± 6.7 | −50.8 ± 5.5 | −450.8 ± 19.9 | −6.4 ± 4.2 | 108.8 ± 37.7 | 2278.1 ± 100.4 |
| S50L-ACE2 | −131.3 ± 10.3 | −55.0 ± 2.2 | −381.8 ± 44.3 | −10.8 ± 2.7 | 108.0 ± 22.7 | 2232.6 ± 166.2 | |
| N501Y-ACE2 | RBD | −136.3 ± 6.3 | −61.5 ± 4.9 | −406.1 ± 23.8 | −2.6 ± 2.4 | 90.8 ± 30.6 | 2128.1 ± 109.5 |
| D614G-ACE2 | Linker | −128.2 ± 12.0 | −45.8 ± 7.5 | −427.7 ± 26.5 | −6.9 ± 4.1 | 99.4 ± 43.2 | 2135.1 ± 87.8 |
| A845V-ACE2 | S2 | −127.7 ± 4.7 | −43.9 ± 6.4 | −434.2 ± 47.7 | −9.0 ± 5.2 | 120.7 ± 19.4 | 2213.1 ± 90.2 |
| P1143L-ACE2 | −125.9 ± 17.1 | −47.2 ± 7.5 | −398.4 ± 82.8 | −9.5 ± 3.2 | 105.2 ± 47.0 | 2119.3 ± 118.2 |
The buried surface area (BSA) is applied to calculate the surface of the protein. The native-complex displays a BSA value of 2099.9 ± 186.4, while the BSA values of the destabilizing nonsynonymous mutations of S-protein – ACE2 complexes are in between the range of 1910.9 ± 123.7 and 2098.8 ± 135.2 (Table 3a), and the stabilizing nonsynonymous mutations of S-protein- ACE2 complexes exhibit a higher BSA score between the range of 2119.3 ± 118.2, and 2278.9 ± 100.4 (Table 3b), compared to the native complex. We applied the MM_GBSA approach to evaluate the binding free energy (ddG) between the native and mutants S-protein and ACE2 receptor molecules to verify this further. The MM_GBSA score (ddg) of native complex and destabilizing mutation complexes were −60.8, −45.35, −48.39, −28.55, −52.57, −32.55, −36.89, −47.3, −30.7, −48.06, −54.27, and −40.78, respectively, as shown in Table 4 a. Whereas the MM_GBSA score of stabilizing nonsynonymous mutations of S-protein (H49Y, S50L, N501Y, D614G, A845V, and P1143L) and ACE2 receptor complexes were −73.06, −80.89, −66.53, −79.81, −63.05, and −68.62, respectively (Table 4 b). Further, the interaction between the native and mutant S-protein and ACE2 receptor molecules is shown in Fig. 5a, Fig. 5b a and b.
Table 4a.
MM-GBSA binding free energy (ddG) score using HawkDock, number of Hydrogen bonds and binding site residues of native and destabilizing nonsynonymous mutants of S-protein and ACE2 receptor.
| Protein Type | Domain | MM-GBSA Binding free energy (kcal/mol) | No of HBONDS | Binding site residues of S protein | Binding site residues of ACE2 protein |
|---|---|---|---|---|---|
| Native-ACE2 | – | −60.8 | 13 | Arg403, GLU406, LYS417, VAL445, TYR453, GLU484, GLN493, SER494, GLY496, THR500, ASN501, TYR505 | GLU23, ASP30, LYS31, HIS34, GLU35, ASP38, GLN42, LYS74, GLU75, GLN76 |
| L8V-ACE2 | SP | −45.35 | 9 | LYS417, TYR449, TYR453, LEU455, GLU484, TYR489, THR500, ASN501 | SER19, GLN24, LYS31, GLU35, GLN42, LYS68, GLU75, TYR83, LYS353 |
| L8W-ACE2 | −48.39 | 10 | ARG403, ASP405, LYS417, GLY446, TYR453, GLY476, ASN487, GLN498, ASN501, GLY502 | LYS31, GLU35, ASP38, GLN42, ASN61, LYS68, GLU75, GLN325 | |
| L18F-ACE2 | NTD | −28.55 | 11 | LYS417, ARG457, LYS458, GLN474, GLU484, CYS488, TYR489, GLN493, SER494, THR500, GLY502 | SER19, LYS31, GLU35, ASP38, GLN42, LYS68, GLU75, MET82, LYS353 |
| Y145H-ACE2 | −52.57 | 11 | TYR449, TYR453, GLU484, GLY485, ARG457, TYR489, GLN493, TYR505 | THR27, HIS34, GLU35, ASP38, LYS68, GLU75, GLN76, MET82, LYS353 | |
| M153T-ACE2 | −32.55 | 9 | ARG403, TYR453, LEU455, GLU484, CYS488, GLN493, SER494 | HIS34, GLU35, ASP38, LYS68, GLU75, GLN76, LYS353 | |
| F157S-ACE2 | −36.89 | 8 | GLY446, TYR453, LEU455, GLU484, CYS488, GLN493, SER494, TYR505 | GLN24, LYS31, HIS34, GLU35, ASP38, LYS68, GLU75 | |
| G476S-ACE2 | CTD/RBD | −47.3 | 9 | GLU406, LYS417, TYR449, TYR453, GLU484, THR500, ASN501, GLY502 | THR27, GLU35, ASP38, TYR41, LYS353, ASP355, ARG559 |
| L611F-ACE2 | Linker | −30.7 | 9 | ARG403, GLU406, LYS417, TYR453, GLN474, ASN487, TYR489, THR500, TYR505 | SER19, GLN24, ASP30, ASP38, GLN42, ASN49, LYS453 |
| A879S-ACE2 | S2 | −48.06 | 9 | ARG403, LYS417, TYR453, GLU484, ASN501, GLY502, TYR505 | GLN24, LYS31, GLU35, GLU75, MET82, TYR83, LYS353 |
| C1247F-ACE2 | TM | −54.27 | 13 | ARG403, GLY446, TYR449, TYR453, GLU484, GLY485, CYS488, GLN493, SER494, THR500, ASN501 | GLN24, LYS31, HIS34, GLU35, ASP38, GLU75, GLN76, THR78, GLN81, TYR83, LYS353 |
| C1254F-ACE2 | −40.78 | 8 | ARG403, GLU406, ARG408, LYS417, TYR449, ASN487, ASN501, VAL503 | GLN24, GLN42, ASN49, GLN325, ASN330, LYS353, ASP355 |
Table 4b.
MM-GBSA binding free energy (ddG) score using HawkDock, number of Hydrogen bonds and binding site residues of native and stabilizing nonsynonymous mutants of S-protein and ACE2 receptor.
| Protein Type | Domain | MM-GBSA Binding free energy (kcal/mol) | No of HBONDS | Binding site residues of S protein | Binding site residues of ACE2 protein |
|---|---|---|---|---|---|
| Native-ACE2 | – | −60.8 | 13 | Arg403, GLU406, LYS417, VAL445, TYR453, GLU484, GLN493, SER494, GLY496, THR500, ASN501, TYR505 | GLU23, ASP30, LYS31, HIS34, GLU35, ASP38, GLN42, LYS74, GLU75, GLN76 |
| H49Y-ACE2 | SP | −73.06 | 17 | ARG403, GLU406, LYS417, VAL445, GLY446, TYR453, GLU484, GLN493, SER494, GLY496, GLN498, THR500, ASN501, GLY504 | ASP30, LYS31, HIS34, GLU35, ASP38, GLN42, LYS68, LYS74, GLU75, GLN76 |
| S50L-ACE2 | −80.89 | 15 | ARG403, GLU406, VAL445, GLY446, TYR 453, GLU484, TYR489, GLN493, SER494, GLY496, GLN498, THR500, ASN501 | ASP30, LYS31, HIS34, GLU35, ASP38, GLN42, LYS68, LYS74, GLU75, GLN76, MET82 | |
| N501Y-ACE2 | RBD | −66.53 | 13 | ARG403, ASP405, LYS417, TYR421, TYR453, LEU455, ALA475, GLU484, PHE486, GLN493, GLY502 | ASP30, LYS31, GLU35, ASP38, GLN42, TRP48, ASN61, LYS68, GLU75, GLN325, ASN330, LYS353 |
| D614G-ACE2 | Linker | −79.81 | 16 | ARG403, GLU406, LYS417, TYR421, TYR453, GLU484, GLN493, GLY496, GLN498, THR500, ASN501 | SER19, GLU23, ASP30, LYS31, HIS34, GLU35, ASP38, LYS74, GLU75, GLN76, LYS353 |
| A845V-ACE2 | S2 | −63.05 | 13 | ARG403, GLU406, LYS417, GLU484, GLN493, SER494, GLY496, THR500, ASN501, TYR505 | ASP30, LYS31, HIS34, GLU35, ASP38, LYS68, LYS74, GLU75, GLN76 |
| P1143L-ACE2 | -68.62 | 15 | ARG403, GLU406, LYS417, TYR449, TYR453, GLU484, GLN493, SER494, GLY496, GLN498, THR500, ASN501 | ASP30, LYS31, HIS34, GLU35, ASP38, LYS68, LYS74, GLU75, GLN76, LYS353 |
Fig. 5a.
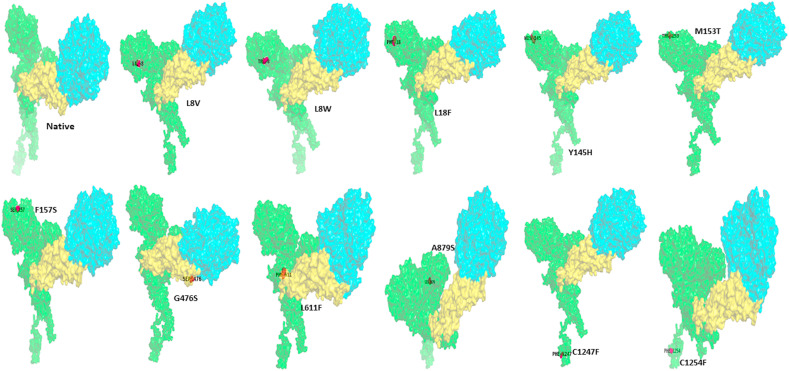
Native and destabilizing nonsynonymous mutations (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) of S-protein interaction with ACE2 receptor. All the proteins were represented in the surface style. The color coding represents the S-protein in green color, the RBD domain of S-protein in yellow color, and ACE2 protein in cyan color. The image was prepared by PYMOL.
Fig. 5b.
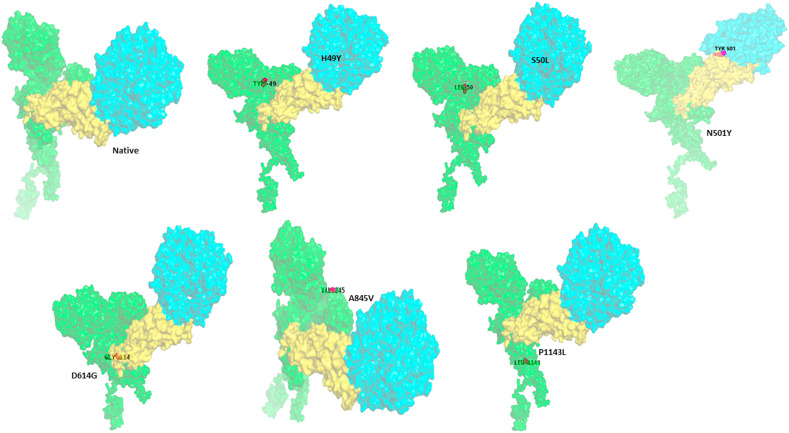
Native and nonsynonymous stabilizing mutations (H49Y, S50L, N501Y, D614G, A845V, and P1143L) of S-protein interaction with ACE2 receptor. All the proteins were represented in the surface style. The color coding represents the S-protein in green color, the RBD domain of S-protein in yellow color, and ACE2 protein in cyan color. The image was prepared by PYMOL.
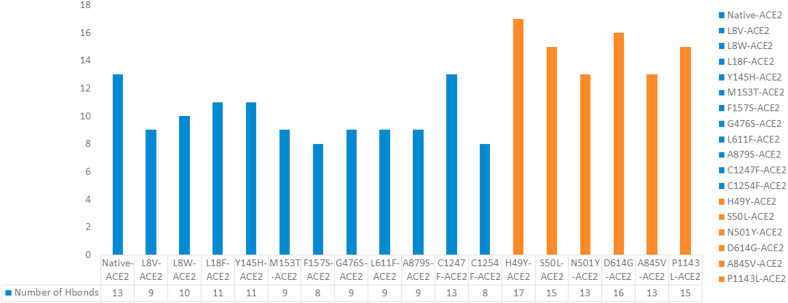
Therefore, the no. Of H-bonds was calculated for the native, destabilizing, and stabilizing nonsynonymous mutations of S-protein-ACE2 complexes, and the values are depicted in Fig. 6 and Table 4a, Table 4ba–b. The native S-protein -ACE2 receptor complex displays a total of 13 hydrogen bonds. The destabilizing nonsynonymous mutations (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) of S-protein and ACE2 complexes exhibits a lesser number of H-bonds compared to the native S-protein _ACE2 complex (Fig. 6).
Fig. 6.
The no. H-bonds formed between the native and nonsynonymous mutants spike receptor and ACE2 receptor.
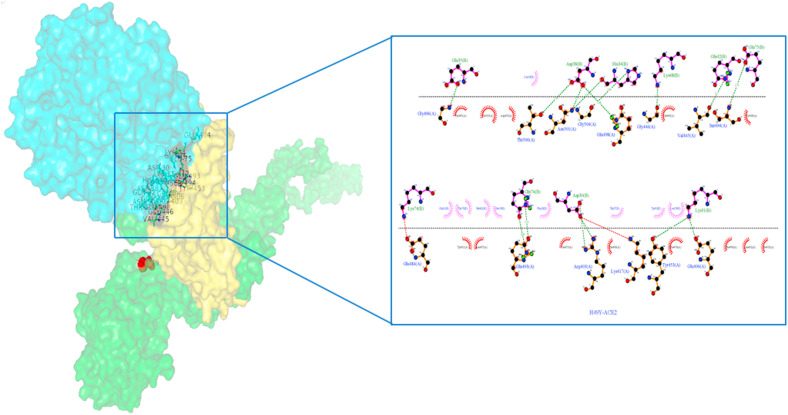
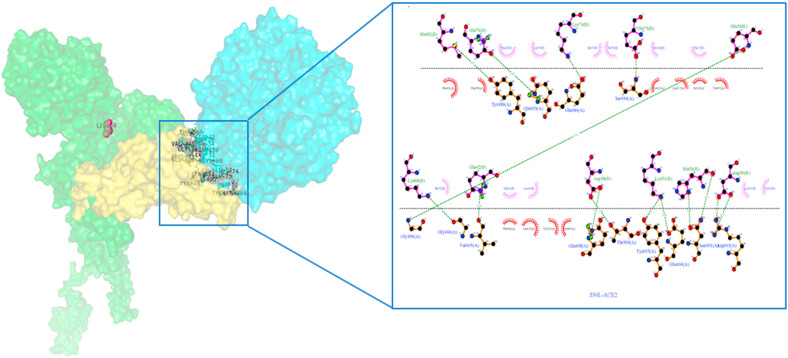
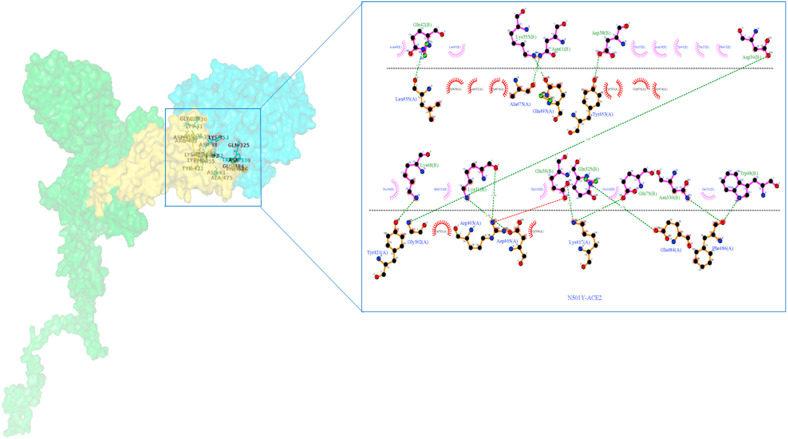
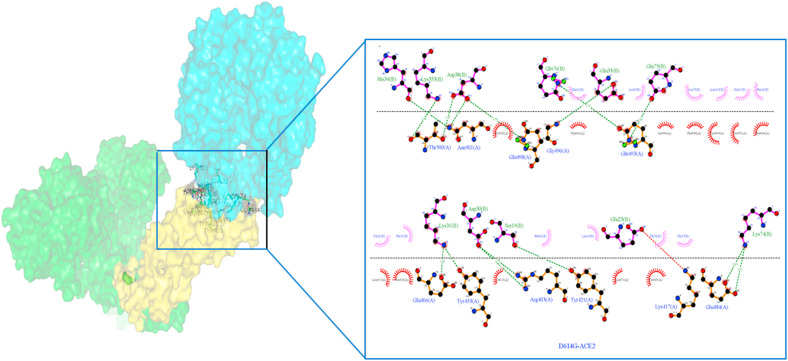
The H-bond interactions between the native complex and destabilizing nonsynonymous mutations are shown in Fig. 7 a and Fig. S1, respectively. The four stabilizing nonsynonymous mutations (H49Y, S50L, D614G, and P1143L) of S-protein_ACE2 complexes show a greater number of h-bonds and other nonsynonymous mutations (N501Y andA845V) shows the same number of h-bonds compared to the native complex. (Fig. 6). The hydrogen bond interactions between the stabilizing nonsynonymous mutations (H49Y, S50L, N501Y, and D614G) of the S-protein with the ACE2 receptor are depicted in Fig. 7b, Fig. 7c, Fig. 7d, Fig. 7e b–e . The other two stabilization nonsynonymous mutations are depicted in Fig. S2. The essential binding site residues of native and mutant S-protein and ACE2 receptor complexes are indicated in Table 4a, Table 4ba–b.
Fig. 7a.
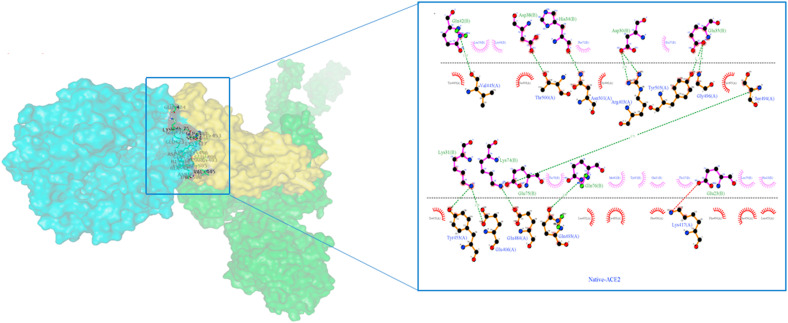
Native S-protein residue interaction with ACE-2 receptor prepared by Ligplot. The S-protein is represented by a brown color, while the ACE2 protein is represented by a purple color. The green dashed lines represent hydrogen bonding interactions.
Fig. 7b.
H49Y nonsynonymous mutant S-protein residue interaction with ACE-2 receptor prepared by Ligplot. The S-protein is represented by a brown color, while the ACE2 protein is represented by a purple color. The green dashed lines represent hydrogen bonding interactions.
Fig. 7c.
S50L nonsynonymous mutant S-protein residue interaction with ACE-2 receptor prepared by Ligplot. The S-protein is represented by a brown color, while the ACE2 protein is represented by a purple color. The green dashed lines represent hydrogen bonding interactions.
Fig. 7d.
N501Y nonsynonymous mutant S-protein residue interaction with ACE-2 receptor prepared by Ligplot. The S-protein is represented by a brown color, while the ACE2 protein is represented by a purple color. The green dashed lines represent hydrogen bonding interactions.
Fig. 7e.
D614G nonsynonymous Mutant S-protein residue interaction with ACE-2 receptor prepared by Ligplot. The S-protein is represented by a brown color, while the ACE2 protein is represented by a purple color. The green dashed lines represent hydrogen bonding interactions.
3.4. MD simulation of native and mutants (N501Y and D614G) S-proteins
The recent studies reported that N501Y and d614G stabilized nonsynonymous mutations are the most promising in S-protein [[73], [74], [75], [76]]. Our analysis confirmed that both the nonsynonymous mutations (N501Y and d614G) stabilized and showed better interaction with ACE2. These results motivated us to observe the structural changes of native and these two advantageous nonsynonymous mutations (N501Y and d614G) at the atomic level. Hence, we implemented the MD simulation approach to investigate how the structural transition in the mutant (N501Y and d614G) S- proteins influencing the interaction with ACE2. We analyzed the total energy, Root mean square deviation (RMSD), Root mean square fluctuation (RMSF), Solvent accessible surface area (SASA), and Hydrogen bond (H-bonds) analysis to investigate the differences in structural variations between the native and mutant (N501Y and D614G) S-proteins. The average RMSD, RMSF, SASA, and H-bond values of the native and mutant (N501Y and D614G) structures are listed in Table 5 .
Table 5.
The average values of RMSD matrix, RMSF, SASA, covariance value and NH-bonds value of native and nonsynonymous mutant (N501Y & D614G) spike proteins.
|
Type of Protein |
Parameters |
||||
|---|---|---|---|---|---|
| RMSD (nm) | RMSF | SASA (nm2) | Covariance value (nm2) | NH-bond | |
| Native | 2.73 | 0.81 ± 0.50 | 765.55 ± 4.12 | 1149.32 | 849.64 ± 23.80 |
| N501Y | 2.88 | 0.87 ± 0.46 | 773.86 ± 4.04 | 1221.26 | 859.02 ± 23.70 |
| D614G | 3.01 | 0.95 ± 0.55 | 768.55 ± 3.84 | 1551.80 | 854.31 ± 21.09 |
To investigate the convergence of the native and mutant (N501Y and D614G) protein systems, the total energy was measured and displayed in Fig. 8 a. The native and mutant (N501Y and D614G) S-protein systems show the convergence from the beginning to the end of the simulation. To investigate the stability of the native and mutant (N501Y and D614G) protein system, the RMSD matrix for all Cα-atoms from the initial structure was measured (Fig. 8b). Fig. 8b shows that the RMSD value of the native and mutant structures (N501Y and D614G) is 2.73 nm, 2.88 nm, and 3.01 nm, respectively (Table 5). Furthermore, the RMSF value analysis revealed a significant difference in the fluctuation of residues between the native and mutant (N501Y and D614G) S-proteins (Fig. 8c).
Fig. 8.
a–e: The total energy, RMSD matrix, RMSF, SASA, and NH-bonds of native and nonsynonymous mutants (N501Y and D614G) of the S-protein versus time at 300 K.
The Mutants N501Y and D614G structures have a greater degree of fluctuation in the residue of 501 and 614 along with neighboring residues than native S-protein throughout the simulation and shown in Fig. 8c. By analyzing the solvent-accessible surface area (SASA) Plots, we determined the geometry and surface of native and mutant (N501Y and D614G) S- proteins. The changes of SASA of native and mutant (N501Y and D614G) S-protein over time are depicted in Fig. 8d. In Fig. 8d, from the beginning to the end of the simulation, the N501Y and D614G mutants have higher fluctuation in the SASA values compared to native structure (Fig. 8d).
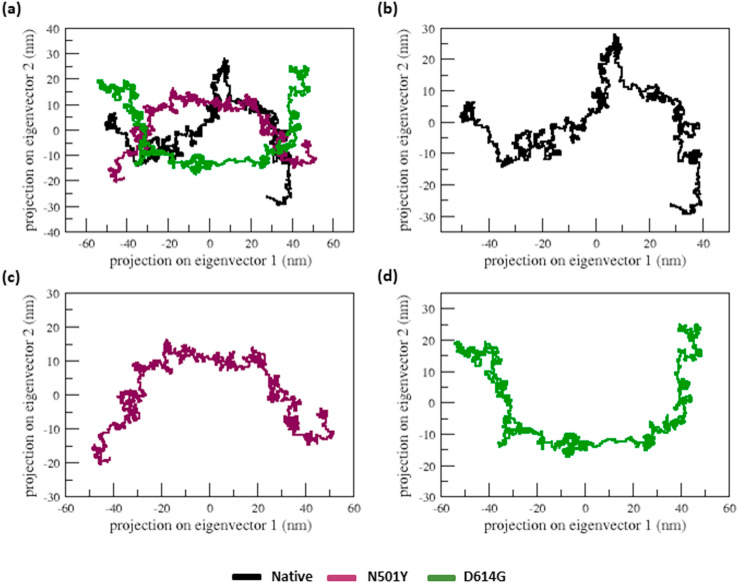
The protein folding, stability, and function are all dependent on the hydrogen bond. To better understand the stability of native and mutant (N501Y and D614G) S-proteins, we measured the intramolecular H-bond concerning time (Fig. 8e). From the beginning to the end of the simulation, both the mutants show higher numbers of h-bonds than the native structure (Fig. 8e). Further, we performed PCA analysis to attain the motion of S- protein upon mutation. The projection of the first two eigenvectors in the PCA plot (Fig. 9 ) shows that the structures of the mutants cover a broader region of phase space in both PC1 and PC2 planes than the native S-protein, again indicating the expansion in the structures. The covariance value of native and mutant structures is listed in Table 5. This further confirms the overall increased flexibility of the mutants (N501Y and D614G) compared to the native S-protein at 300 K. Overall, the mutant structures (N501Y and D614G) exhibited more motion and flexibility than the native S-protein.
Fig. 9.
The projection of native and nonsynonymous mutant spike proteins motion in dimensional space through the initial two significant eigenvectors at 300 K. (a) The native is depicted in black, the N501Y nonsynonymous mutant is depicted in green, and the D614G nonsynonymous mutant is depicted in yellow. Each trajectory is also illustrated independently in (b), (c), and (d) for clarification.
4. Discussion
Nonsynonymous mutations in the S-protein may influence the stability of its structure and interaction with the ACE2 receptor, which provides clues for viral infection and disease spread. Identifying the mutational effect on the S-protein and its interaction with the ACE2 receptor is very important for predicting the 3D conformation of the S-protein. Unfortunately, the entire length of the S-protein structure is not available in the protein data bank. We have used the earlier proved studies [[33], [34], [35], [36], [37], [38]] to fix this issue. Hence, we have preferred the I-TASSER online server to predict the 3D structure of the S-protein. Further refinement of predicted model S-protein, the MDS performed to assess the stability of model structure for further studies. Fig. 3 shows that the RMSD of native S-protein is converged after 7 ns and produces stable conformations. Further, the average structure of the native s-protein was shown at regular intervals (Fig. 3). The most favorable structure of native S-protein was selected at the 12 ns MD simulation, which was then used to build the mutant structures and validated with PROCHECEK and PROSA programs. The sequence and structure-based in-silico tools predict destabilizing and stabilizing nonsynonymous mutations in S-protein associated with COVD-19 infection. The sequence-based approach predicted 40 and 22 nonsynonymous mutations with decreased stability and increased stability. The structure-based approach predicted 36 and 26 nonsynonymous mutations as destabilizing and stabilizing, respectively. Combination of both sequence and structure-based online servers predicted 11 (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) nonsynonymous mutations with decreased stability and six nonsynonymous mutations (H49Y, S50L, N501Y, D614G, A845V, and P1143L) with increased stability in the S-protein upon mutations (Table 1, Table 2). We considered these 11 destabilizing and six stabilizing nonsynonymous mutations for further docking analysis to investigate the interaction behavior with the ACE2 receptor.
The screened nonsynonymous mutations in the S-protein may affect the stability of its structure and interaction with the ACE2 receptor, which provides clues for SARS-CoV-2 infection. Corroborating this further, we employed molecular docking and Molecular Mechanics, and energy merged with Generalized Born and Surface Area (MM/GBSA) computation to evaluate the affinity between the native and mutants (Destabilizing and stabilizing) of S-protein with the ACE-2 receptor. The results of the examination of the top complex are illustrated in Table 3a, Table 3ba–b & Fig. 5a, Fig. 5ba and b. The native complex (native S-protein_ACE2) shows more negative values in the HADDOCK score, which means there is higher binding interaction between the molecules (Table 3a & Fig. 7a). On the other hand, the destabilizing nonsynonymous mutations of S-protein and ACE2 complexes (L8V-ACE2, L8W-ACE2, L18F-ACE2, Y145H-ACE2, M153T-ACE2, F157S-ACE2, G476S-ACE2, L611F-ACE2, A879S-ACE2, C1247F-ACE2, and C1254F-ACE2) show less negative value than the native complex and indicate a lower binding interaction with biological partners (Table 3a & Fig. S1). On the other hand, the stabilizing nonsynonymous mutations of S-protein-ACE2 complexes have greater negative values as a HADDOCK score than the native S-protein-ACE2 complex and illustrate a better interaction between the biological partners (Table 3b Fig. 7b, Fig. 7c, Fig. 7d, Fig. 7eb–e, & Fig. S2). A more excellent BSA value facilitates an immediacy between the two molecules. The BSA, restraint and desolvation energy significantly associate with the HADDOCK docking score [[70], [71], [72]]. The S-protein loses its interaction with the ACE2 receptor upon destabilizing nonsynonymous mutations (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F), whereas the stabilizing nonsynonymous mutations (H49Y, S50L, N501Y, D614G, A845V, and P1143L) in the S-protein shows increased interaction with the ACE2 receptor (Table 3a, Table 3ba–b).
We applied the MM_GBSA approach to evaluate the binding free energy (ddG) between the native and mutants S-protein and ACE2 receptor molecules to verify this further. From Table 4a, it is thus again clear that the destabilizing nonsynonymous mutations show less binding free energy than the native complex, and they lose their interaction with the ACE2 receptor. The stabilizing nonsynonymous mutations (H49Y, S50L, N501Y, D614G, A845V, and P1143L) of S-protein and ACE2 complexes show more binding free energy native complex (Table 4b). Hydrogen bonds are the essential interactions in biological identification processes and vital for establishing the binding specificity [[57], [58], [59], [60]]. The intermolecular hydrogen bonds can provide favorable binding energy [77,78]. This principle is the destabilizing nonsynonymous mutations (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) have lost their binding affinity between the S-protein and the ACE2 receptor molecule, and this affects their function. The stabilizing nonsynonymous mutations (H49Y, S50L, N501Y, D614G, A845V, and P1143L) of the S-protein have a better binding affinity with ACE2 receptor complexes with the native complex (Table 4b). It is confirmed that all six stabilizing nonsynonymous mutations show better affinity with the ACE2 receptor compared to the native S-protein.
Further, MDS was performed using several parameters like total energy RMSD matrix, RMSF, SASA, and NH-bond to evaluate the conformational transitions of native and mutant S-proteins. The total energy plot shows the convergence for the native and mutants S-protein system during the simulation and generates stable conformation, thus providing an appropriate foundation for further analysis (Fig. 8a). In the RMSD matrix plot, the mutant (N501Y and D614G) structures showed higher deviation values, whereas the native structure showed lower deviation values (Fig. 8b). It defines that the N501Y and D614 nonsynonymous mutations have structural transitions on the conformational geometry of S-protein. The RMSF values were calculated for native and mutant S-proteins to determine the dynamic behavior of residues (Fig. 8c). During simulation, higher flexibility was observed in both the mutant structures compared to native S-protein. It furthers confirms that mutant protein residues undergo a structural transition as compared to native S-protein. The variation in SASA of the native and mutant S-proteins with time is shown in Fig. 8d. Mutant (N501Y and D614G) S-protein structures showed higher values of SASA with time, whereas native structure shows lower values of SASA with time. It further confirms that mutant S-protein residues were flexible throughout the simulation and attained the extended conformation compared to native S-protein (Table 5, & Fig. 8b–d).
For further reinforcement of our results, we performed the PCA analysis better to understand the flexibility behavior of native and mutant S-protein. From, Fig. 9, we observed that the mutation induces the structural transitions and causes the loss of the native conformation of the native S-protein, making it more flexible. As a result, the mutant S-protein structures (N501Y and D614G) had a larger fluctuation in RMSD, RMSF, SASA, and PCA, indicating that the native S-protein is undergoing a major structural shift, which could be the reason for better ACE2 receptor interaction. The number of H-bond (Fig. 8e) analyses confirms it. Due to the structural transitions in mutants S-protein, yield more hydrogen bonds than native S-protein, which could be the reason for better interaction with ACE2 receptors. Docking results well support this. The Mutant (N501Y and D614G) -ACE2 complex structures show similar or more h-bonds than other destabilizing nonsynonymous mutations (Fig. 6 & Table 4a, Table 4ba–b).
Overall, the docking results confirm that the ACE2 receptor interacts with the residues of the RBD domain of the S-protein. The destabilizing nonsynonymous mutations alter the binding site residues of the RBD domain of S-protein and decrease the interaction with the ACE2 receptor. However, the stabilizing nonsynonymous mutations increase the interaction with ACE2. The MD simulation results confirm that the two advantageous stabilizing nonsynonymous mutations (N501Y and D614G) undergo the structural transitions and might be the reason for enhancing the interaction with ACE2. This confirms and provides evidence that was stabilizing nonsynonymous mutations have a high affinity with ACE2 and high virulent levels compared to native and destabilizing nonsynonymous mutations of the S-protein. There is some experimental evidence to corroborate our findings, which proves the better association between ACE2 and S-protein upon stabilizing nonsynonymous mutations [[79], [80], [81], [82], [83], [84]]. Many recent studies reported that mainly the stabilizing nonsynonymous mutations N501Y and D614G have a better affinity with the ACE2 receptor [[73], [74], [75], [76],[79], [80], [81], [82], [83], [84]]. This result will help experimental scientists understand the mechanism of the S-protein and its interaction with the ACE2 receptor upon nonsynonymous mutations.
5. Conclusion
Using the vast bioinformatics approaches, we segregated and screened the destabilizing and stabilizing nonsynonymous mutations in the Spike protein. Further, we examined the interaction between the S-protein and the ACE2 receptor. The docking and free binding energy (ddG) scores exhibited destabilizing nonsynonymous mutations (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) show lower interaction with the ACE2 receptor compared to native S-protein. On the other hand, the stabilizing nonsynonymous mutations (H49Y, S50L, N501Y, D614G, A845V, and P1143L) show greater interaction with the ACE2 receptor than native and destabilizing nonsynonymous mutations of the S-protein. Thus, the MD simulation of stabilizing nonsynonymous mutations (N501Y and D614G) undergo structural changes and might influence interaction with ACE2. The outcome of this study can support the other scientists, enabling them to comprehend the virulent level of nonsynonymous mutations in the Spike protein and provide clues regarding a mutation's role in infection and disease spread, contributing to the development of effective therapy against SARS-CoV-2.
Declaration of competing interest
We have no conflicts of interest to disclose.
Acknowledgment
We would like to acknowledge the Bioinformatics Laboratory at College of Applied Medical Sciences in Jubail, Imam Abdulrahman Bin Faisal University, Jubail, 35816, Saudi Arabia, for their computing facility to carry out this work. The authors would also like to take this opportunity to thank the management of the Vellore Institute of Technology, India, for providing the necessary facilities and encouragement to carry out this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104654.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV) Euro Surveill. 2020;25:2000058. doi: 10.2807/1560-7917.ES.2020.25.4.2000058. December 2019 to January 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . World Health Organization; 2020. Coronavirus Disease 2019 (COVID-19): Situation Report; p. 52.https://apps.who.int/iris/handle/10665/331476 [Google Scholar]
- 3.Yang P., Wang X. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 2020;17:555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microb. Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu A., Peng Y., Huang B., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon D.E., Jang G.M., Bouhaddou M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shang J., Wan Y., Luo C., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q., Zhang Y., Wu L., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du L., He Y., Zhou Y., et al. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112:3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen K.G., Rambaut A., Lipkin W.I. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Z., Xiao Y., Kang L., et al. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin. Infect. Dis. 2020;4:536. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C., Liu Y., Yang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y., Hou Y., Shen J., et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao S., Chen H. Modeling the epidemic dynamics and control of COVID-19 outbreak in China. Quant. Biol. 2020;8:11–19. doi: 10.1007/s40484-020-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson-Sathi S., Umasankar P.K., Sreekumar E., et al. Structural and functional implications of spike protein mutational landscape in SARS-CoV-2. BioRxiv. 2020:1–11. doi: 10.1101/2020.05.02.071811. [DOI] [Google Scholar]
- 18.Nelson-Sathi S., Easwaran S., Nair R.R., et al. Structural and functional implications of non-synonymous mutations in the spike protein of 2,954 SARS-CoV-2 genomes. BioRxiv. 2020:1–11. doi: 10.1101/2020.05.02.071811. [DOI] [Google Scholar]
- 19.Zhou P., Yang X., Wang X., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortuso F., Mercatelli D., Guzzi P.H., Giorgi F.M. Structural genetics of circulating variants affecting the SARS-CoV-2 spike/human ACE2 complex. J. Biomol. Struct. Dyn. 2021:1–11. doi: 10.1080/07391102.2021.1886175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fratev F. The SARS-CoV-2 S1 spike protein mutation N501Y alters the protein interactions with both hACE2 and human derived antibody: a Free energy of perturbation study. BioRxiv. 2020:1–8. doi: 10.1101/2020.12.23.424283. [DOI] [PubMed] [Google Scholar]
- 22.Kupferschmidt K. Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear. Science. 2020;80 doi: 10.1126/science.abg2626. [DOI] [Google Scholar]
- 23.Rambaut A., Loman N., Pybus O., et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations: COVID-19 genomics UK consortium. 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-ukdefined-by-a-novel-set-of-spike-mutations/563
- 24.Hadfield J., Megill C., Bell S.M., et al. Next strain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiyotani K., Toyoshima Y., Nemoto K., et al. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isabel S., Graña-Miraglia L., Gutierrez J.M., et al. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sneha P., Doss C.G.P. Gliptins in managing Diabetes-Reviewing computational strategy. Life Sci. 2016;166:108–120. doi: 10.1016/j.lfs.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Sneha P., Doss C.G.P. Molecular dynamics: new frontier in personalized medicine. Adv. Protein Chem. Struct. Biol. 2016;102:181–224. doi: 10.1016/bs.apcsb.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Consortium UniProt, UniProt A hub for protein information. Nucleic Acids Res. 2014;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towler P., Staker B., Prasad S.G., et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman H.M., Battistuz T., Bhat T.N., et al. The protein data bank. Acta Crystallogr. D. 2002;58:899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- 33.Barros E.P., Casalino L., Gaieb Z., et al. The flexibility of ACE2 in the context of SARS-CoV-2 infection. Biophys. J. 2021;120:1072–1084. doi: 10.1016/j.bpj.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2, Comput. Biol. Med. 2021;104555 doi: 10.1016/j.compbiomed.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., Zheng W., Cheng M., et al. Functions of essential genes and a scale-free protein interaction network revealed by structure-based function and interaction prediction for a minimal genome. J. Proteome Res. 2021;20:1178–1189. doi: 10.1021/acs.jproteome.0c00359. https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pourseif M.M., Parvizpour S., Jafari B., et al. A domain-based vaccine construct against SARS-CoV-2, the causative agent of COVID-19 pandemic: development of self-amplifying mRNA and peptide vaccines. Bioimpacts: BI. 2021;11:65. doi: 10.34172/bi.2021.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadat S.M., Aghadadeghi M.R., Yousefi M., et al. Bioinformatics analysis of SARS-CoV-2 to approach an effective vaccine candidate against COVID-19. Mol. Biotechnol. 2021;63:389–409. doi: 10.1007/s12033-021-00303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frota N.F., de Sousa Rebouças A., Fuzo C.A., Lourenzoni M.R. Alemtuzumab scFv fragments and CD52 interaction study through molecular dynamics simulation and binding free energy. J. Mol. Graph. Model. 2021;107949 doi: 10.1016/j.jmgm.2021.107949. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Y., Zhang J., Xiao T. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abraham M.J., Van Der Spoel D., Lindahl E., Hess B. Royal Institute of Technology and Uppsala University; 2014. GROMACS User Manual Version 5.0. 4. [Google Scholar]
- 42.Sambasivarao S.V., Acevedo O. Development of OPLS-AA force field parameters for 68 unique ionic liquids. J. Chem. Theor. Comput. 2009;5:1038–1050. doi: 10.1021/ct900009a. [DOI] [PubMed] [Google Scholar]
- 43.Kamaraj B., Purohit R. R. Computational screening of disease-associated mutations in OCA2 gene. Cell Biochem. Biophys. 2014;68:97–109. doi: 10.1007/s12013-013-9697-2. [DOI] [PubMed] [Google Scholar]
- 44.Kamaraj B., Purohit R. Silico screening and molecular dynamics simulation of disease-associated nsSNP in TYRP1 gene and its structural consequences in OCA3. BioMed Res. Int. 2013;2013:1–13. doi: 10.1155/2013/697051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balu K., Purohit R. Mutational analysis of TYR gene and its structural consequences in OCA1A. Gene. 2013;513:184–195. doi: 10.1016/j.gene.2012.09.128. [DOI] [PubMed] [Google Scholar]
- 46.Kaplan W., Littlejohn T.G. Swiss-PDB viewer (deep view) Briefings Bioinf. 2001;2:195–197. doi: 10.1093/bib/2.2.195. [DOI] [PubMed] [Google Scholar]
- 47.Laskowski R.A., Rullmannn J.A., MacArthur M.W. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 48.Wiederstein M., Sippl M.J., ProSA-web Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C.W., Lin J., Chu Y.W. iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinf. 2013;14:1–14. doi: 10.1186/1471-2105-14-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capriotti E., Fariselli P., Casadio R. I-Mutant2. 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng J., Randall A., Baldi P. Prediction of protein stability changes for single‐site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 52.Parthiban V., Gromiha M.M., Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandurangan A.P., Ochoa-Montaño B., Ascher D.B., Blundell T.L. SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017;45:W229–W235. doi: 10.1093/nar/gkx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pires D.E., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M. The HADDOCK2. 2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Chowdhury R., Boorla V.S., Maranas C.D. Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity. Comput. Struct. Biotechnol. J. 2020;18:2573–2582. doi: 10.1016/j.csbj.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamaraj B., Al-Subaie A.M., Ahmad F., Surapaneni K.M., Alsamman K. Effect of novel leukemia mutations (K75E & E222K) on interferon regulatory factor 1 and its interaction with DNA: insights from molecular dynamics simulations and docking studies. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1784790. [DOI] [PubMed] [Google Scholar]
- 58.Gopalakrishnan C., Al‐Subaie A.M., Yeh H.Y., et al. Prioritization of SNPs in y+ LAT‐1 culpable of Lysinuric protein intolerance and their mutational impacts using protein‐protein docking and molecular dynamics simulation studies. J. Cell. Biochem. 2019;120:18496–18508. doi: 10.1002/jcb.29172. [DOI] [PubMed] [Google Scholar]
- 59.Kamaraj B., Bogaerts A. Structure and function of p53-DNA complexes with inactivation and rescue mutations: a molecular dynamics simulation study. PloS One. 2015;10 doi: 10.1371/journal.pone.0134638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balu K., Rajendran V., Sethumadhavan R., Purohit R. Investigation of binding phenomenon of NSP3 and p130Cas mutants and their effect on cell signalling. Cell Biochem. Biophys. 2013;67:623–633. doi: 10.1007/s12013-013-9551-6. [DOI] [PubMed] [Google Scholar]
- 61.Weng G., Wang E., Wang Z. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47:W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun H., Li Y., Tian S., et al. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014;16:16719–16729. doi: 10.1039/C4CP01388C. [DOI] [PubMed] [Google Scholar]
- 64.Chen F., Liu H., Sun H., et al. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein–protein binding free energies and re-rank binding poses generated by protein–protein docking. Phys. Chem. Chem. Phys. 2016;18:22129–22139. doi: 10.1039/C6CP03670H. [DOI] [PubMed] [Google Scholar]
- 65.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 66.DeLano W.L. The PyMOL molecular graphics system. 2002. http://www.pymol.Org
- 67.Huang J., Rauscher S., Nawrocki G. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nature. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turner P.J. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology; Beaverton, OR: 2005. XMGRACE, Version 5.1. 19. [Google Scholar]
- 69.Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 70.Halperin I., Ma B., Wolfson H., Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 71.Janin J., Henrick K., Moult J., et al. CAPRI: a critical assessment of predicted interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 72.McDonald I.K., Thornton J.M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 73.Ozono S., Zhang Y., Ode H., et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan A., Zia T., Suleman M., et al. Higher infectivity of the SARS‐CoV‐2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;Mar 23:10.1002/jcp.30367. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Korber B., Fischer W.M., Gnanakaran S. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daniloski Z., Jordan T.X., Ilmain J.K., et al. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife. 2021;10 doi: 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartlett P.A., Marlowe C.K. Evaluation of intrinsic binding energy from a hydrogen bonding group in an enzyme inhibitor. Science. 1987;235:569–571. doi: 10.1126/science.3810155. [DOI] [PubMed] [Google Scholar]
- 78.Gao J., Mammen M., Whitesides G.M. Evaluating electrostatic contributions to binding with the use of protein charge ladders. Science. 1996;272:535–537. doi: 10.1126/science.272.5261.535. [DOI] [PubMed] [Google Scholar]
- 79.Jafary F., Jafari S., Ganjalikhany M.R. In silico investigation of critical binding pattern in SARS-CoV-2 spike protein with angiotensin-converting enzyme 2. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-021-86380-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laurini E., Marson D., Aulic S., et al. Computational mutagenesis at the SARS-CoV-2 spike protein/angiotensin-converting enzyme 2 binding interface: comparison with experimental evidence. ACS Nano. 2021;15:6929–6948. doi: 10.1021/acsnano.0c10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ali F., Kasry A., Amin M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med. Drug Discov. 2021;10:100086. doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luan B., Wang H., Huynh T. Enhanced binding of the N501Y‐mutated SARS‐CoV‐2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595:1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saha P., Majumder R., Srivastava A.K. Mutations in spike protein of SARS-CoV-2 modulate receptor binding. membrane fusion and immunogenicity: an Insight into viral tropism and pathogenesis of COVID-19. 2020;ChemRxiv. Cambridge: Cambridge Open Engage; doi: 10.26434/chemrxiv.12320567.v1. [DOI] [Google Scholar]
- 84.Bhattarai N., Baral P., Gerstman B.S., Chapagain P.P. Structural and dynamical differences in the spike protein RBD in the SARS-CoV-2 variants B. 1.1. 7 and B. 1.351. J. Phys. Chem. B. 2021;125(26):7101–7107. doi: 10.1021/acs.jpcb.1c01626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.