Summary
Studies in experimental autoimmune encephalomyelitis (EAE), the animal model of multiple sclerosis, have shown potential links between diet components, microbiome composition, and modulation of immune responses. In this review, we reanalyze and discuss findings in an outbred marmoset EAE model in which a yogurt-based dietary supplement decreased disease frequency and severity. We show that although diet has detectable effects on the fecal microbiome, microbiome changes are more strongly associated with the EAE development. Using an ecological framework, we further show that the dominant factors influencing the gut microbiota were marmoset sibling pair and experimental time point. These findings emphasize challenges in assigning cause-and-effect relationships in studies of diet-microbiome-host interactions and differentiating the diet effects from other environmental, stochastic, and host-related factors. We advocate for animal experiments to be designed to allow causal inferences of the microbiota's role in pathology while considering the complex ecological processes that shape microbial communities.
Subject areas: Microbiology, Microbiome, Nutrition
Graphical abstract
Ecology; Microbiology; Microbiome; Nutrition;
Introduction
Adequate nutrition is an important factor for the normal development of the immune system, as well as for its correct function throughout life. Several nutrients, such as retinol, tocopherol, dietary fiber, and essential fatty acids, are capable of modulating the intensity of inflammatory processes (Uauy, 2007). In addition, carefully orchestrated nutrient delivery is required for the assembly and maintenance of the myelin sheath. Autoimmune and neurodegenerative diseases, such as multiple sclerosis (MS) and its animal model experimental autoimmune encephalomyelitis (EAE), can develop when the myelin sheath and axons are damaged by autoimmune inflammation (Dobbing, 1964). Long-chain polyunsaturated fatty acids, zinc, iron, choline, cholesterol, phospholipids, and sphingomyelin from diet sources have all been shown to play important roles in myelin production and repair, either as energy sources or as key components of the myelin sheath (Oshida et al., 2003; Saher et al., 2005; van Rensburg et al., 2006; Vallet et al., 2014; Skripuletz et al., 2015; Hadley et al., 2016; Berghoff et al., 2017).
Some effects of diet on MS development might be mediated via the gut microbiome, which has been implicated in the etiology of both MS (Chen et al., 2016; Jangi et al., 2016) and EAE (Berer et al., 2011; Chen et al., 2019a, 2019b; Stanisavljević et al., 2018). Diet is one of the main factors that determines the composition and metabolism of the gut microbiota, which benefits the host by defending against pathogens and modulating the immune system (Fan and Zhang, 2019; Hirschberg et al., 2019; Zapatera et al., 2015). Because diet impacts both host immune-metabolic functions and the gut microbiota and these three factors interact dynamically to impact intestinal innate and adaptive immune mechanisms, all three components of this triad (diet, the gut microbiome, and the immune system) and the interactions among them must be accounted for when evaluating patient outcomes in clinical interventions targeting autoimmune and inflammatory processes. The cause-and-effect relationships connecting diet to disease and whether the underlying mechanisms are microbiome-dependent or microbiome-independent are difficult to unravel in humans, thus providing strong rationale to conduct diet studies in animal models in combination with detailed monitoring of both host and microbiome responses. Animal studies further have the advantage that environmental factors, which have been shown to be the strongest determinant of gut microbiome composition in humans (Rothschild et al., 2018), can be more strictly controlled. However, the success of animal studies still depends on careful monitoring of ecological cues, such as cage and sibling effects, that can uniquely affect the course of disease in animal models.
It was recently discovered that a daily dietary supplement prepared from whole-milk yogurt decreased the incidence of MS-like disease (EAE) from 100% to 65% in a marmoset model of MS (Kap et al., 2018). This discovery was made by chance when, in 2014, the Biomedical Primate Research Centre (Rijswijk, The Netherlands) introduced a yogurt-based dietary supplement (YBS) to replace the previous water-based dietary supplement (WBS) in the diet of their pedigreed marmoset colony. This switch was made in an effort to reduce the risk of wasting marmoset syndrome, a protein-calorie deficiency that is clinically characterized by weight loss, chronic diarrhea, muscle atrophy, and anemia (Barnard et al., 1988), and has been described in several New World monkey species, including common marmosets (Callithrix jacchus) and cotton-top tamarins (Saguinus oedipus). It was unclear if or how the change to the YBS diet caused the incidence of EAE to decline in marmosets selected from the colony.
To address whether the change in dietary supplement caused the decreased incidence of EAE and whether these benefits were linked to gut microbiota composition, Kap et al. (2018) performed a controlled intervention in eight adult outbred marmoset dizygotic twin pairs. One member of each twin pair was reverted to the original WBS for eight weeks, while the other members were maintained on the YBS, which all animals had been receiving prior to selection into the experiment. Although the study was preliminary and small (with only 16 animals), the findings suggested that the YBS was associated with reduced disease incidence (six out of eight YBS-fed monkeys versus eight out of eight WBS-fed monkeys) and significantly reduced spinal cord demyelination (Kap et al., 2018). This work also assessed, for the first time, the gut microbiome of healthy marmosets in captivity and how diet and/or disease induction affected the natural marmoset microbiome composition.
Here, we begin by reviewing the evidence for a role of the gut microbiota in MS and EAE. We then provide the results from a re-analysis of the 16S rRNA sequencing data collected by Kap et al. (2018), which we performed in order to (1) re-evaluate the natural microbiome composition in healthy marmosets and (2) more thoroughly explore how diet and pathology influenced the diversity and composition of the marmoset gut microbiome in their experiment. Guided by our results and by other studies on the role of nutrition in neurogenerative diseases, we hypothesize how specific components of the YBS diet may have contributed to the decreased incidence of disease and spinal cord pathology in the marmosets via both “microbiome-dependent” and “microbiome-independent” effects. We then use the Kap et al. (2018) data as an example to discuss how experimental and statistical limitations, such as cage housing and genetic relatedness among study subjects, can make it challenging to accurately determine causal or correlational relationships among diet, the gut microbiome, and MS. We conclude by reviewing the lessons learned about human MS from animal models and providing concrete recommendations for future studies to disentangle the complexities of diet-microbiome-pathology interactions in human MS.
Evidence for a diet-microbiome-disease axis in MS/EAE
Mammals have evolved a mutualistic relationship with microbial communities in which gut microbes enhance their host's mucosal and systemic immune responses, protect against pathogen colonization, and aid in the maintenance of a healthy intestinal barrier, among other contributions (Kinross et al., 2011; Ogunrinola et al., 2020; Takiishi et al., 2017; Lei et al., 2015). A healthy gut microbiota helps maintain homeostasis by balancing pro- and anti-inflammatory responses and is generally characterized as being diverse, stable, and resilient (Shahi et al., 2017; Lozupone et al., 2012).
Different avenues of research have suggested that alterations or disruptions of the gut microbiota, often referred to as “dysbiosis,” may contribute to the development of MS. This hypothesis has been supported by epidemiological studies that have linked stressors that disrupt the gut microbiota early in life, such as antibiotics and caesarean sections, with an increased risk of MS (Dalla Costa et al., 2019; Maghzi et al., 2012; Norgaard et al., 2011). Studies in mouse models of MS have also clearly shown that the development of EAE is attenuated in germ-free mice and in mice treated with antibiotics (Ochoa-Reparaz et al., 2009; Lee et al., 2011; Yokote et al., 2008), although this research cannot be conclusively extrapolated to humans. It has therefore been suggested that distinct microbial signatures can determine the course of MS, specifically by modulating between the chronic progressive and remitting-relapsing variants of the disease (Gandy et al., 2019).
Are there consistent microbiome patterns in human MS?
In the last decade, several research groups have shown that specific bacterial taxa are depleted or enriched in the fecal microbiota of patients with MS relative to healthy controls. Excellent reviews on how the relative abundances of gut bacteria change in patients with MS have been published elsewhere (Freedman et al., 2018; Mirza et al., 2020; Ochoa-Reparaz et al., 2017; Pröbstel and Baranzini, 2018; Shahi et al., 2017), so a detailed description will not be provided here. However, attempts to identify consistent microbial associations with MS remain inconclusive because, with the exception of a few select species, results are not always consistent among studies (Mirza et al., 2020). For example, Chen et al. (2016) noted that while some research groups reported decreases in the Actinobacteria genera Adlercreutzia and Collinsella in patients with MS (Chen et al., 2016), others reported no difference in these genera (Jangi et al., 2016) and instead reported reductions in Slackia (Jangi et al., 2016) or increases in Bifidobacterium or Eggerthella (Miyake et al., 2015; Tremlett et al., 2016).
There are similar disagreements among studies about the roles of the Firmicutes, Bacteroidetes, and Proteobacteria, which are the main constituents of the human gut microbiome. The few consistent microbial signatures have mostly been found when comparing human remitting-relapsing MS to healthy controls and include decreases in Parabacteroides distasonis and Bacteroides, Prevotella, and Clostridium species (Chen et al., 2016; Miyake et al., 2015; Cekanaviciute et al., 2017) and increases in Akkermansia and Dorea species (Chen et al., 2016; Berer et al., 2017; Gandy et al., 2019); these patterns were confirmed in a recent systematic review of a small number of MS microbiome studies (Mirza et al., 2020).
Inconsistent results among studies might arise from differences in the population under investigation (e.g., differences in age, comorbidities, ethnicities, or geographic locations); the laboratory methods used (e.g., primers used for amplification of the 16S rRNA gene); the study design (e.g., studying one time point or temporal variations); the approach to data analyses (e.g., sample quality control criteria); the pipelines and software used for analysis (e.g., percent identity threshold, taxonomy database); or the statistical interpretation of results (e.g., potential for false-positive findings). There is also considerable variation among studies in terms of the sample size, sample collection technique, and disease-modifying treatment being tested, among other factors. In addition, even though some bacterial taxa have been consistently associated with MS across several studies, many of these associations are small or subtle (Janakiraman and Krishnamoorthy, 2018), and a clearly defined MS-associated gut microbial signature has not been identified to date.
Are the microbiome features altered in MS causal to disease?
Interestingly, even though animal models do not perfectly recapitulate all the aspects of human MS, some of the gut bacterial taxa associated with human MS have been assigned specific causal or protective roles in EAE, providing further evidence that they play a direct role in human MS (Berer et al., 2011; He et al., 2019; Lynch et al., 2019; J Ochoa-Repáraz et al., 2010; Walter et al., 2020). For example, Prevotella histolica was found to ameliorate EAE by inducing the proliferation of CD4+ FoxP3+ regulatory T cells in the gut and the periphery and repressing pro-inflammatory IFNγ and IL-17-producing CD4+ T cells in the central nervous system (CNS) (Shahi et al., 2019; Mangalam et al., 2017), while Parabacteroides distasonis was found to induce IL-10-producing CD4+ CD25+ regulatory T cells (Cekanaviciute et al., 2017). On the other hand, ex vivo studies using stimulated whole blood or peripheral blood mononuclear cells (PBMCs) from healthy subjects have shown that Dorea may play a pro-inflammatory role by inducing IFNγ and potentially degrading mucins (Schirmer et al., 2016).
For some bacterial taxa or animal models, such as the mucin degrader Akkermansia in the context of myelin oligodendrocyte peptide (MOG35-55)-induced EAE in C57BL/6 mice, conflicting results prevent the species from being definitively classified as pathogenic or protective. While one study on the mechanistic effect of cannabinoids found that Akkermansia increases with disease and decreases with drug treatment (a pathogenic role) (Al-Ghezi et al., 2019), another study found that this species increases in mice that receive a synthetic form of microRNA and subsequently increases CD4+ FoxP3+ regulatory T cells and ameliorates EAE (a protective role) (Liu et al., 2019). Other examples of bacteria (or their components or products) that have been identified as protective in animal models of EAE through modulation of FoxP3+ regulatory T cells include Salmonella typhimurium (Jun et al., 2012; Ochoa-Repáraz et al., 2007) and Bacteroides fragilis (Ochoa-Repáraz et al., 2010a; Ochoa-Repáraz et al., 2010b).
Broadly, the results from studies of the causal role of bacterial taxa in the development of MS in animal models, though also plagued by inconsistencies among studies, suggest that some bacterial taxa are mechanistically involved in the etiology or the pathology of EAE. This opens the possibility that dysbiosis in the gut microbiota might predispose a human to MS, a hypothesis that is further supported by the observation that human microbiota-associated mice developed disease after receiving feces from human patients with MS (Berer et al., 2017; Cekanaviciute et al., 2017).
How does the microbiota impact disease development?
Four primary mechanisms have been described by which the gut microbiome could impact disease development, either by increasing risk or by modulating the disease course. Firstly, gut bacteria can translocate to the bloodstream when intestinal permeability is low, leading to activation of the immune system. Both MS and EAE are characterized by immune-triggered damage of the blood-brain barrier (BBB), leading to leukocyte infiltration and concomitant demyelination and lesion development (Alvarez et al., 2011). Gut permeability is impaired in EAE mice at the onset of disease, which in turn exacerbates infiltration of pro-inflammatory Th1/Th17 cells and is associated with a decrease in numbers of regulatory T cells (Nouri et al., 2014). Secondly, bacteria could trigger disease by molecular mimicry. The gut microbiome collectively encodes millions of genes (Grice and Segre, 2012), and bacterial gene products with enough homology to the host's proteins could activate autoimmune responses, which has been shown to occur in neuromyelitis optica (Cree et al., 2016), a disease with similar features to MS.
Thirdly, some members of the gut microbiome are able to produce neurotransmitter-like molecules and metabolites that signal to the brain via the vagus nerve and the enteric nervous system (also known as the gut-brain axis) (Forsythe and Kunze, 2013; Cryan et al., 2019). Lastly, the gut microbiota and their products are able to directly impact the development of immune cells (Wesemann et al., 2013; Kim and Kim, 2017; Cekanaviciute et al., 2017), such as B cells and T cells, which have been implicated in the development of disease. For example, segmented filamentous bacteria, which are found in mice but not in humans, support the differentiation of IL-17-producing lymphocytes via induction of the transcription factor RORγt in immune cells (Wekerle, 2015). Indeed, the discovery that members of the microbiome are capable of modulating immune responses made researchers hypothesize that activation of autoimmune T cells involved in inflammatory diseases of the CNS takes place in the intestine (Wekerle, 2017). This mechanism would also explain how an altered microbiome could contribute to a predisposition to disease risk.
How might diet modulate microbiome-disease relationships?
Studies in both humans and animals have consistently demonstrated that the composition of the gut microbiome is inextricably linked to host diet (David et al., 2014; Wu et al., 2011), and it is therefore likely that diet may affect the microbiota to modulate immune responses in MS (Berer et al., 2018; Fan and Zhang, 2019; Hirschberg et al., 2019; Saresella et al., 2017). Diet is capable of influencing MS/EAE through gut microbiota-derived metabolites and diet-mediated compositional and functional changes in the gut microbiota, as well as directly through the anti-inflammatory properties of specific food components (Katz Sand, 2018; Janakiraman and Krishnamoorthy, 2018). In animals, studies have shown potential links between diet components and modulation of immune responses in EAE that could be of benefit in the prevention or treatment of MS/EAE (Berer et al., 2018; Sonner et al., 2019). A consistent theme of potential MS diets implicated in animal studies, including diets rich in dietary fibers and omega-3 fatty acids and low in red meat, is the provision of nutrients that interact in a beneficial way with the gut microbiota to increase anti-inflammatory immune responses (AlAmmar et al., 2019; Berer et al., 2018; Esposito et al., 2021). However, despite ongoing interest in how diet impacts disease progression in MS, either directly or via the microbiome, no official organizations have released generally accepted diet recommendations for the management, prevention, or treatment of MS.
Animal models offer several advantages relative to humans for the continued study of diet-microbiome-MS connections: (i) the environmental factors that shape the gut microbiota can be controlled; (ii) time scales are shorter, thus allowing longitudinal studies over time lines that span the entire life of the animal and disease development; (iii) mechanisms can be directly studied; and (iv) follow-up studies to address causality or determine causal components within the microbiota are tractable. However, even in animal models, the ecological drivers that shape microbiomes are often not sufficiently considered (Ubeda et al., 2012; McCoy et al., 2017; Martínez et al., 2018), and cause-and-effect relationships are difficult to unravel (Fischbach, 2018). With these challenges in mind, we reanalyzed 16S rRNA sequencing data from our previous study of a marmoset model of MS, which we refer to as Kap et al. (see Data S1). Our goal in doing so was to gain a more detailed nutritional and ecological perspective on the drivers that may have shaped the gut microbiome and the onset of pathology in the marmoset model used in our experiment.
The microbiome in healthy marmosets and how it compares to humans
In their experiment, Kap et al. studied eight pairs of outbred adult marmoset twins; for each pair, one twin was reverted to the WBS diet while one was maintained on the YBS diet. In addition to the dietary supplement, the marmosets received pelleted food and regular servings of fruit; WBS-fed monkeys also received “play-and-food enrichment” consisting of mealworms, Arabic gum, and raisins, whereas YBS-fed monkeys received Arabic gum, peanuts, and bread. Microbiome samples were collected from each marmoset at (1) “day −56,” the baseline time point (immediately before the diet adjustment); (2) “day −7,” seven weeks after the diet change; (3) “day 21,” three weeks after EAE was induced by injection of recombinant MOG1-155 in incomplete Freund adjuvant (IFA); and (4) “day 49,” seven weeks after disease induction (see Data S1).
Like Kap et al., we first characterized the gut microbiota in marmosets at the two time points prior to disease induction (days −56 and −7) to obtain a baseline evaluation of gut microbiota structure and composition in healthy marmosets. Our reanalysis confirmed several of Kap et al.’s original results, including the fact that Actinobacteria were the predominant phylum in the marmoset gut in this study (69.0% ± 17.3%; Figure S1 and Table S1) and that Proteobacteria occurred at low abundances (1.1% ± 1.0%). At lower taxonomic levels, eight families and eight genera comprised approximately 96% and 91%, respectively, of the gut microbiome of healthy marmosets (Table S1). Seventeen of the 74 operational taxonomic units (OTUs) detected in our re-analysis comprised approximately 85% of the gut communities (Table S1). As in Kap et al., the most dominant OTUs were Bifidobacterium callithrichos and Bifidobacterium aesculapii.
There were a small number of discrepancies between Kap et al.’s original conclusions and our re-analyzed data. For example, while Kap et al. found the Firmicutes to be twice as abundant as the Bacteroidetes (20.3% ± 6.0% vs. 10.9% ± 7.5%, respectively), we found the relative abundances of these two phyla to be relatively similar (13.7% ± 5.4% vs. 14.0% ± 9.8%, respectively; Figure S1). At the OTU level, Kap et al. (2018) assigned a large group of Coriobacteriaceae OTUs to Collinsella tanakaei, which was among the five most dominant species; we found C. tanakaei only comprised 0.06% ± 0.08% (min = 0.001%, max = 0.36%) of the sequences, though the family Coriobacteriaceae, to which this species belongs, was still among the most abundant families in our re-analyses. In fact, this dominant OTU appeared to belong to the newly described species Parolsenella catena (Sakamoto et al., 2018), which was not present in the databases at the time that the work described by Kap et al. was published. Other discrepancies may be attributed to the fact that Kap et al. used Paired-end assembler for Illumina sequences (PANDAseq) for read assembly and Quantitative Insights into Microbial Ecoloby (QIIME) for quality control and analysis, whereas here a pipeline that combines several other publicly available tools was used (Illumina-utils, USEARCH, and QIIME, among others; see Data S1). This moderate pipeline-based variation in results is expected (O'Rourke et al., 2020; Bokulich et al., 2018; Xue et al., 2018; Rajan et al., 2019; Barnes et al., 2020). Despite these discrepancies in the abundances of select taxa, we emphasize that our re-analysis identified the same suite of dominant taxa in the marmoset microbiome that were originally described by Kap et al., with Actinobacteria, Firmicutes and Bacteroidetes present at high (>10%) relative abundances.
Because animal models are used to identify mechanisms that induce disease in order to develop therapeutics targeted for humans, it is important to understand how the gut microbiome in the animal model compares to the human gut microbiome. In humans, the composition of the healthy infant gut microbiome resembles the adult gut microbiome by the age of around three years (Voreades et al., 2014). The marmosets used in this study, which were between one and two years, would be considered young adults by human standards. While Actinobacteria was the predominant phylum in the marmosets, Bacteroidetes and Firmicutes are the predominant phyla in humans (Claesson et al., 2011). At the genus level, Bifidobacterium comprises over 60% of the gut microbiome in marmosets, while this genus reaches abundances of 60%–99% only in breastfed newborn humans (Turroni et al., 2012; Arboleya et al., 2016) and then declines consistently upon the introduction of solid foods, reaching a stable abundance of less than 10% throughout adulthood that decreases to approximately 5% in the elderly (>65 years old) (Arboleya et al., 2016). However, despite any gross differences in taxon abundances, many bacterial taxa are still shared between marmosets and humans at lower taxonomic levels. These taxa would still employ similar mechanisms to metabolize food sources and have similar pathways for stimulating the immune system or otherwise affecting the host and could therefore affect disease development in either host.
Effects of diet, pathology, and time on the marmoset gut microbiome
Driven by Kap et al.’s interpretation that there were few diet-specific effects on the microbiota but rather “a close interplay between diet, microbiota, and the immune system,” we used a variety of mixed-effect modeling procedures and distance-based community analyses to more comprehensively explore how diet and pathology shaped the diversity and taxonomic composition of the marmoset gut microbiome in the experiment (see Data S1). All models included diet, pathology (symptomatic or asymptomatic), and experimental time point as predictors, thus ensuring that any results for one variable are controlled for variation in the other two. Models also included a random effect term to account for natural similarities within marmoset twin pairs. We also tested for interactions between diet and time and between diet and pathology, but because few interactions were significant, we only present the results for the three main experimental variables (diet, pathology, and time).
Microbiome diversity
Two main parameters were used to assess bacterial diversity in relation to diet and pathology: α-diversity, which considers the presence and abundance of taxa within a single habitat and time point (e.g., the gut of an individual marmoset), and β-diversity, which considers variation in species presence and abundance among habitats or time points (Tuomisto, 2010). Higher α-diversity is often associated with more stable ecosystems that are more resistant to environmental pressures, such as dietary changes or pathogen invasions, whereas higher β-diversity can indicate if microbiome composition is significantly different between two groups (e.g., between WBS-fed and YBS-fed marmosets or healthy and symptomatic marmosets). Kap et al. limited their discussion of α-diversity to the Simpson index, which considers both species richness (the number of different species in the ecosystem) and evenness (whether there are a few dominant species or all species are equally abundant) and is influenced by the dominant OTUs (Hill, 1973). A detailed analysis of β-diversity was not included in Kap's manuscript. To provide a more complete image of how diet and EAE development shape the microbiome, we therefore analyzed species richness and evenness separately and thoroughly tested for changes in β-diversity.
Alpha diversity
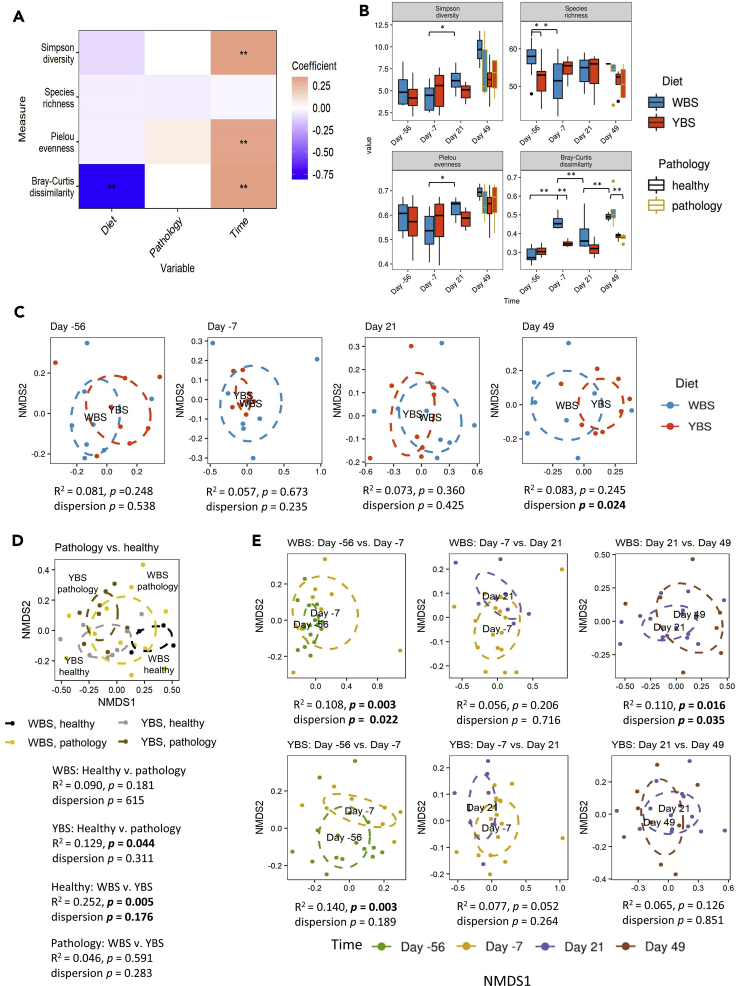
Mixed-effects models showed that Simpson diversity and Pielou evenness (a measure of species evenness) (Pielou, 1966) increased significantly over the course of the experiment in both diet groups; this increase was slightly larger in the WBS-fed marmosets (Figures 1A and 1B; Table S2). There were no significant associations between the development of pathology and any α-diversity measure. The observed temporal changes could be an effect of drift or stochastic changes over time due to inherent random processes. Pairwise statistical analyses of the α-diversity metrics showed that Simpson diversity started to increase in WBS-fed monkeys between days −7 and 21 (p = 0.022), presumably due to a corresponding increase in evenness (p = 0.010) following the induction of disease (Figure 1B). The observation of higher diversity in WBS-fed monkeys contradicts the conventionally positive relationship between microbiome diversity in animal health, as YBS-fed animals were on average healthier than WBS-fed animals, especially at the later time points. In addition, although WBS-fed marmosets had significantly higher species richness than the YBS-fed marmosets at the beginning of this experiment (p = 0.012), this difference was not observed at subsequent time points (Figure 1B). This increased richness was unexpected, as all marmosets were receiving the same diet at the first time point.
Figure 1.
Influence of diet, pathology, and time on microbiome diversity (related to Table S2)
(A) Heatmap of coefficients obtained from a mixed effect model analyses of α-diversity metrics and Bray Curtis distances. Double asterisks indicate coefficients for which the 95% confidence interval did not overlap zero.
(B) Boxplots showing various diversity metrics, including the Simpson index, species richness, Pielou evenness, and Bray-Curtis distances. An analysis of variance (ANOVA) followed by Tukey's post hoc test was used to identify significantly different diversity measures between diets at each time point and within diets relative to the previous time point. Lines represent minimum and maximum values. Significance was established at p < 0.05.
(C–E) Non-metric multidimensional scaling (NMDS) plots based on the Bray-Curtis distance comparing (C) diets at each time point, (D) healthy and pathology within each diet, and (E) time points within each diet. PERMANOVA was used to determine whether two groups were significantly different, with a significance threshold of p < 0.05.
Beta diversity
We used the Bray-Curtis distance, a β-diversity measure in which 0 means identical communities while a value of 1 means complete dissimilarity, to assess “inter-individual” (within-group) variation in microbiome composition or how different marmosets were from each other. Inter-individual Bray-Curtis dissimilarity was significantly lower in YBS-fed monkeys at each experimental time point but increased in both diet groups throughout the experiment (Figure 1B). Pairwise comparisons confirmed that inter-individual Bray-Curtis dissimilarity was significantly lower in YBS-fed monkeys at all time points except baseline and day 21 (day −7 p < 0.001, day 49 p = 0.004; Figure 1B). This finding suggests that the WBS diet increases inter-individual variation in the marmoset gut microbiota, potentially because the shift to the WBS diet represents a disturbance to the microbiota not experienced by the YBS treatment group.
Diet
We also used permutational multivariate analysis of variances (PERMANOVAs) based on the Bray-Curtis distance to evaluate whether microbial communities became recognizably different during the experiment due to the experimental interventions or outcomes described by Kap et al. (diet change, pathology development, and experimental time point). We visualized the results using non-metric multidimensional scaling (Figures 1C–1E). In broad agreement with Kap et al., we concluded that the overall effect of the dietary shift on the marmoset gut microbiome was small, as there were no significant compositional differences associated with diet over the course of the experiment (Figure 1C). Multivariate dispersion, which is another measure of inter-individual variation, was also higher in WBS-fed marmosets (i) after the diet switch, relative to the previous time point (p = 0.022), and (ii) at day 49, relative to both YBS-fed monkeys and the previous time point (p = 0.024 and p = 0.035, respectively), supporting our earlier conclusion that the WBS diet increased inter-individual variation.
Pathology
Other community-level changes indicated that the strongest associations were between microbiome composition and the development of pathology. YBS-fed monkeys exhibiting symptoms of EAE had significantly different gut microbiomes than healthy YBS-fed monkeys, whereas there was no significant difference in microbiome composition between healthy and symptomatic WBS-fed monkeys (Figure 1D). However, in the WBS group, microbial communities were significantly different between days 21 and 49 (R2 = 0.110 p = 0.016) (Figure 1E), when some animals began to exhibit symptoms. Kap et al. reported that all WBS-fed marmosets eventually developed pathology, though some did not exhibit symptoms until after the last experimental time point, and that disease onset occurred much later in YBS-fed marmosets. The shift in composition over time for the WBS-fed monkeys, combined with the shift associated with pathology in the YBS-fed monkeys, could therefore support a role for the microbiome in the development of pathology. Under this hypothesis, the YBS potentially promoted taxa that prevented or ameliorated disease development. Notably, symptomatic marmosets exhibited similar microbiome composition, regardless of their diet, suggesting that there may be a consistent microbial signature of EAE in this animal model (Figure 1D).
Time point
There were also significant differences in community composition between experimental time points (Figure 1E), as the microbial communities at day −7 were significantly different from the communities at baseline (day −56) for both the WBS group (R2 = 0.108, p = 0.003) and the YBS group (R2 = 0.140, p = 0.003). This was not expected for the YBS group, as all marmosets in this group were kept on the same diet for the duration of the experiment. However, twin siblings that were previously housed together were separated and re-paired with another monkey for the experiment, and it is possible that this change in the environment caused the observed microbiome change.
Taxon abundances
With respect to how diet and pathology influenced specific taxa, Kap et al. reported that twin siblings “displayed an essentially unaltered microbiota composition” between the first and second time points, despite the change in diets. The only exceptions were “a twofold increase of Collinsella tanakaei in the YBS group and a threefold decrease of Marvinbryantia formatexigens in the WBS group” (Kap et al., 2018). Kap et al. additionally reported that the effect of diet was more noticeable at the phylum level three weeks after disease induction, with the WBS group showing progressive decreases in Actinobacteria from days 21–49 and increases in Bacteroidetes and Firmicutes at day 49. At the species level, Kap et al. identified statistically significant increases in Bifidobacterium stellenboschense in both diet treatments from day −7 to day 21. At day 49, Bifidobacterium callitrichos, Bifidobacterium stellenboschense, and Bifidobacterium reuteri were significantly less abundant in the WBS group compared with the YBS group, whereas Collinsella tanakaei was significantly more abundant.
To determine (i) taxonomic signatures consistently associated with diet and pathology (Figure 2) and (ii) which taxa showed the most diet-related or pathology-related shifts in abundance over time (Figure 3), we used mixed-effect models predicting log-transformed taxon relative abundances and log-transformed “changes” in relative abundance, respectively (see Data S1).
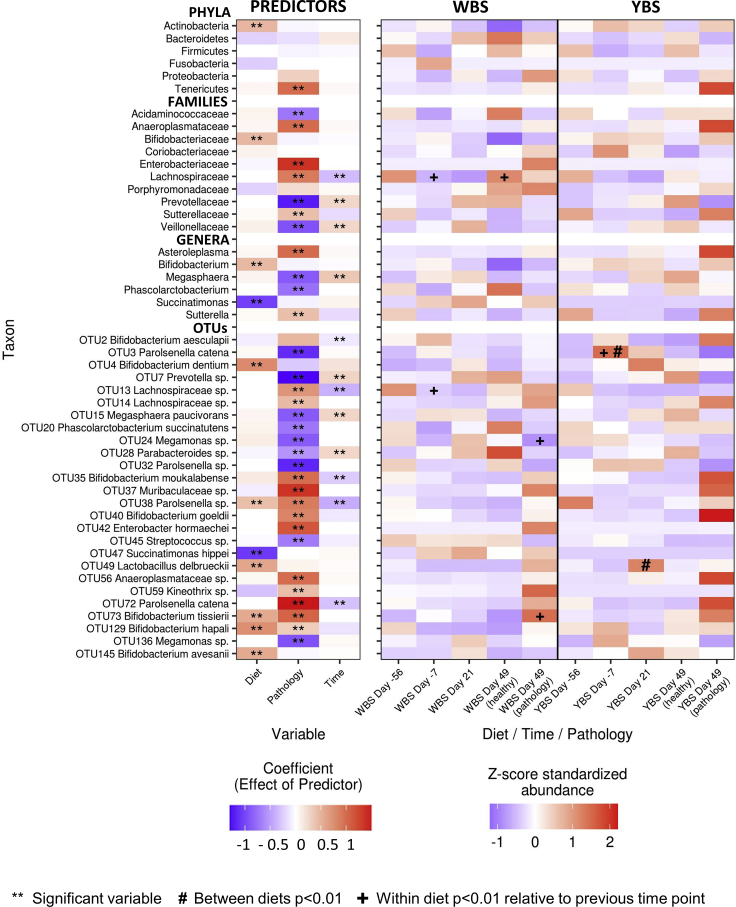
Figure 2.
Taxonomic signatures associated with diet, pathology, and time (related to Tables S3 and S4)
(Left) Heatmap shows the coefficients in mixed-effect models predicting the abundances of taxa present in the marmoset gut. Double asterisks indicate coefficients for which the 95% confidence interval did not overlap zero. (Right) Heatmap shows Z score standardized relative abundances for the same taxa. Student's t-test was used to identify significantly differentially abundant taxa between diets at the same time point (labeled with #) and within diets relative to the previous time point (labeled with +). Significance was established at p < 0.05 after the Benjamini-Hochberg correction for multiple comparisons. Only taxa with at least one coefficient > |0.5| are shown. WBS, water-based supplement; YBS, yogurt-based supplement.
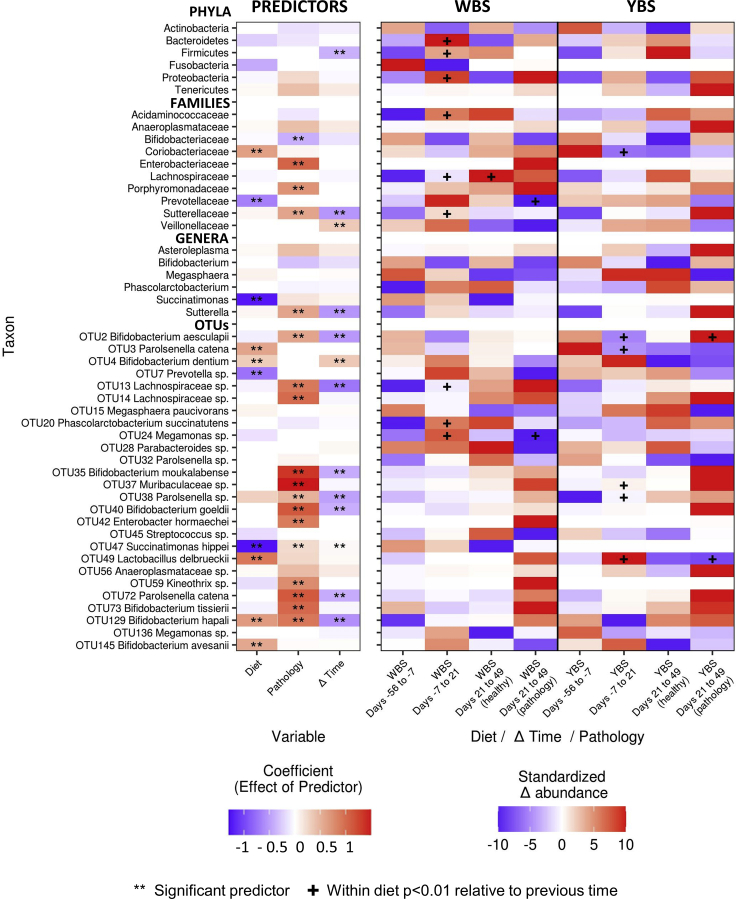
Figure 3.
Diet- and pathology-related shifts in taxon abundances (related to Tables S3 and S5)
(Left) Heatmap shows the coefficients of mixed-effect models predicting the change in taxon abundances between time points. Double asterisks indicate coefficients for which the 95% confidence interval did not overlap zero. (Right) Heatmap shows the change in relative abundance between time points for the same taxa. For visualization, changes in abundance were scaled between −10 and 0 for decreases in abundance and between 0 and 10 for increases in abundance. Student's t-test was used to identify significantly different changes in taxonomic abundances between diets at the same time point (labeled with #) and within diets relative to the previous time point (labeled with +). Significance was established at p < 0.05 after the Benjamini-Hochberg correction for multiple comparisons. All taxa included in Figure 3 are repeated here, regardless of coefficient values. WBS, water-based supplement; YBS, yogurt-based supplement.
Diet
In line with the results described by Kap et al., Actinobacteria was the sole phylum that was significantly associated with the YBS diet, and Bacteroidetes was associated with the WBS diet, although this relationship was not significant (Figure 2 and Table S3). Firmicutes was not associated with either diet. At the genus level, Bifidobacterium and Succinatimonas were the best indicators of YBS and WBS, respectively (Figure 2 and Table S3). Among OTUs, our reanalysis suggested that only a select number of low-abundance taxa were consistently associated with diet throughout the entire experiment: a small group of Bifidobacterium species (OTUs 4, 72, 129, and 145) and the yogurt species Lactobacillus delbrueckii were the best indicators of the YBS diet in mixed-effect models, while OTU47 Succinatimonas hippei was the best indicator of the WBS diet (Figure 2; Table S4). Another common yogurt bacterium, Streptococcus thermophilus, was not detected in our analysis despite being inconsistently associated with MS in other studies (Miyake et al., 2015; Dargahi et al., 2020; Tankou et al., 2018), and the single Streptococcus OTU (OTU45) was not significantly associated with diet. We could not make definitive conclusions about some of the diet-related species implicated by Kap et al., including C. tanakaei, M. formatexigens, and B. callitrichos, as these three species were undetected, detected at very low abundances, or not significantly associated with YBS, respectively, in our re-analysis.
With respect to our tests for how diet may have affected the magnitude of shifts in taxon abundances, we found that the rate at which OTU4 B. dentium and OTU49 Lactobacillus delbrueckii increased in abundance rose steadily from days −56 to 21 in YBS-fed monkeys, and the rate of change in OTU47 Succinatimonas increased over the same period in WBS-fed monkeys (Figure 3). Curiously, the YBS diet also induced larger shifts in OTU3 Parolsenella catena (phylum Actinobacteria, order Coriobacteriales). P. catena showed a threefold increase in YBS-fed marmosets between day −56 and −7 and then decreased progressively after disease induction, despite the fact that the marmosets assigned to this group kept receiving the same diet throughout the entire experiment (Figure 3).
Pathology
Importantly, the development of pathology led to much more pronounced differences in taxon abundances; these signatures of pathology were independent of diet and could be observed at all taxonomic levels (Figure 2). Although Tenericutes, represented primarily by the genus Asteroleplasma, comprised less than 0.5% of the marmoset microbiome, they were an order of magnitude more abundant in diseased marmosets relative to healthy marmosets in both the WBS- and YBS-fed groups (Figure 2 and Table S4). Sutterellaceae and Porphyromondaceae were also twofold more abundant in diseased marmosets, independently of diet (Figure 2 and Tables S3 and S4). The OTUs most strongly associated with pathology in mixed-effect models included OTU72 Parolsenella catena, OTU37 Muribaculaceae sp., OTU42 Enterobacter hormaechei, and OTU73 Bifidobacterium tissierii (Figure 2 and Table S4). Healthy marmosets tended to have higher abundances of OTU7 Prevotella sp., OTU32 Parolsenella sp., and OTU136 Megamonas sp. The higher-level taxa to which these OTUs belong, including Veillonellaceae and Prevotellaceae, were similarly associated with health.
We correspondingly found that the development of pathology was associated with larger shifts in taxon abundances and that these associations were generally stronger than those detected for diet. For example, the development of pathology was associated with larger increases in the abundances of OTU14 Lachnospiraceae sp., OTU72 Parolsenella catena, and OTU37 Muribaculaceae sp. in both diets; larger increases in OTU2 Bifidobacterium aesculapii and OTU35 Bifidobacterium moukalabense in the YBS-fed marmosets; and larger increases in OTU42 Enterobacter hormaechei in the WBS-fed group (Figure 3 and Table S5). The fact that pathology was consistently associated with a greater shift in taxon abundances (Figure 3), even though different taxa show distinct and taxon-specific associations with pathology (Figure 2), suggests that the development of pathology is broadly related to a larger disruption of the microbiome: taxa generally showed larger increases or decreases in abundance in response to pathology than they showed in response to other experimental changes. The spread of these taxonomic changes across all taxonomic levels underscores the magnitude of this overall pathology-related shift in community composition.
Time point
Experimental time point was included as a predictor in our models to control for both natural and stochastic changes in the microbiome that were not consistently associated with diet or pathology. Notably, several taxa exhibited these significant temporal shifts (Figures 2 and 3), which could reflect changes caused by introduction to new housing conditions at the beginning of the experiment or by the process of immunization with MOG/IFA halfway through the experiment. For many taxa, the “time” coefficients are opposite in sign to the corresponding “pathology” coefficients, suggesting that, in marmosets that developed symptoms of EAE, the changes in their microbiome were opposite to the pattern of changes that occurred naturally throughout the experiment. This result further emphasizes that the development of symptoms is associated with the emergence of new microbial signatures that are distinct from the microbiome of healthy individuals, independent of diet.
Additional trends
We additionally noted several important trends with respect to bacterial species that have either been linked to MS or are hypothesized to have a potential role in the marmoset model of MS (Table S3), although these trends were not significant in our re-analyzed data. First, the genus Bifidobacterium progressively decreased over time in the WBS group, while remaining relatively constant in the YBS group. Second, although the Prevotellaceae and Bacteroides increased with disease induction in both diet groups, the increases were twofold larger in the WBS group than in the YBS group. Third, several OTUs classified as Megasphaera species increased with disease induction in both groups, but while they decreased with disease onset in the WBS group, they remained relatively constant in the YBS group. These findings suggest that there are interactions between diet and pathology that could be associated with the microbiome but may have escaped detection due to the study's modest sample size. Further discussion of these potential interactions is provided in the following section, where we discuss the mechanisms by which diet could impact disease development.
In summary, our re-analysis of the data obtained by Kap et al. suggests that reverting marmosets to the WBS diet caused decreases in the abundances of species in the phylum Actinobacteria, especially of Bifidobacterium and Parolsenella spp., and increased inter-individual heterogeneity. However, relationships between pathology and specific microbial taxa were generally stronger and more numerous than the relationships with diet, and the microbiomes of symptomatic marmosets were similar regardless of diet, indicating that pathology constitutes a larger perturbation to the microbiome. Pathology was specifically associated with increased abundances of Tenericutes, Enterobacteriaceae spp., Lachnospiraceae spp., Parolsenella spp., and some Bifidobacterium spp. and caused larger shifts in taxon abundances at all taxonomic levels.
Possible mechanisms underlying diet-microbiome-host interactions in marmoset EAE
The associations between the fecal microbiota and each of diet and pathology in the marmoset model of MS suggest that the health benefits of the YBS diet may have been mediated by the gut microbiota. In this section, we discuss both “microbiome-dependent” and “microbiome-independent” mechanisms that might explain why the YBS diet attenuated EAE development in marmosets (Figure 4). Because these effects could be contingent on the nutrient composition of each supplement, the amounts of protein, carbohydrates, fats, and calories in each supplement are provided in Table 1.
Figure 4.
Potential beneficial diet components and diet-gut microbiome interactions in the marmoset model of EAE
Specific components in whole milk, which is the base of the yogurt-based supplement (YBS), may have interacted with the immune system and the gut microbiome to decrease disease incidence and severity in the YBS-fed marmosets. (Left) Butyrophilin, the most abundant protein in the fat globule membrane in milk, has been shown to be immunomodulatory; it increases levels of anti-inflammatory IL-10 and decreases levels of pro-inflammatory IFNΥ in EAE44. (Center left) Members of the gut microbiome, such as Lactobacilli and Bifidobacterium spp., are capable of metabolizing dietary tryptophan into ligands of the aryl hydrocarbon receptor, such as indole-3-aldehyde and indole-3-lactic acid. These ligands influence glial cells, including astrocytes, and thereby reduce CNS inflammation and EAE disease scores52,53. (Center right) N-glycans (carbohydrates that consist of several sugars linked to the nitrogen atom in the side chain of asparagine) present in milk serve as substrates for Bifidobacterium spp., which in turn produce SCFA as end products of carbohydrate fermentation. SCFAs have a direct impact on EAE by reducing Th1 cells and stimulating the proliferation of lamina propria-derived Treg56,58. (Right) Proteolytic bacteria, such as Prevotella and Megasphaera spp., produce BCFA (iso-butyrate, valerate, and iso-valerate) as metabolic end products. Valproic acid is a valerate derivative that has anti-epileptic, neuro-protective, and anti-inflammatory effects and is capable of ameliorating symptoms in EAE and optic neuritis64–66. EAE, experimental autoimmune encephalomyelitis; CNS, central nervous system; SCFAs, short-chain fatty acids; BCFAs, branched-chain fatty acids.
Table 1.
Macronutrients and energy provided per week by the WBS and the YBS to the marmosets.
| Macronutrients | WBS |
YBS |
||
|---|---|---|---|---|
| Weight (g) | Percentage (%) | Weight (g) | Percentage (%) | |
| Carbohydrates | 11.4 | 22.7 | 11.9 | 22.0 |
| Protein | 7.6 | 15.5 | 9.8 | 18.1 |
| Fat | 13.8 | 61.9 | 14.4 | 60.0 |
| Energy (kcal(Kj)) | 200.4 (838.5) | 66.9∗ | 216.2 (904.6) | 72.2∗ |
Table shows the amount in grams of carbohydrates, protein, and fat provided by the dietary supplements to the marmosets, the percentage of the total energy the macronutrients represent in the supplement, and the percentage of the field metabolic rate that the supplements meet. The asterisk (∗) denotes percentage of field metabolic rate. g, grams; kcal, kilocalories; Kj, kilojoules.
Microbiome-dependent diet effects
Putative microbiome-dependent dietary effects are effects either caused by diet-induced changes in gut microbial composition or that correlate with changes in the production of bacterial-derived metabolites capable of impacting the host. Here, we discuss both diet-induced microbial changes and bacterially derived metabolites that could have affected the development of pathology in YBS-fed marmosets.
N-glycans
One microbiome-dependent effect implicated in the data from Kap et al. relates to N-glycans in YBS and the abundance of Bifidobacterium spp. The main energy sources for gut bacteria are carbohydrates, which can be diet or host derived. Bovine milk, the basic ingredient of the YBS, contains lactose and 51 different types of N-glycans (carbohydrates that consist of several sugars linked to the nitrogen atom in the side chain of asparagine). In comparison, human milk only contains 38 N-glycans (Nwosu et al., 2012). Both lactose and N-glycans have been associated with higher abundances of Bifidobacteria (Schmidt et al., 2020; Karav et al., 2016). Karav et al. (2016) reported that N-linked glycoproteins from milk serve as selective substrates for the growth of Bifidobacterium species in the host gut. They concluded that the variety of N-glycans released from bovine milk glycoproteins “may serve as prebiotic substrates with selective properties similar to those of human milk oligosaccharides” (Karav et al., 2016).
In line with this, our analysis of the marmoset gut microbiome revealed that Bifidobacterium spp. were more abundant in the gut communities of YBS-fed marmosets and decreased over the course of the experiment in the WBS-fed group. The shift from YBS to WBS caused the relative abundance of Bifidobacterium to drop from 57.9% ± 13.2% at baseline to 49.2% ± 20.5% by the end of experiment, while it remained at a relatively stable average of 63.0% ± 12.1% in the YBS group (Table 1). The decrease in Bifidobacterium could be attributed to the decrease in the availability of dietary N-glycans and carbohydrate sources when the YBS was removed from the diet.
The YBS-induced enhancement of Bifidobacteria might contribute to the beneficial health effects of the diet. Bifidobacteria, when consumed as probiotics, are capable of delaying the onset and ameliorating the severity of MS by enhancing numbers of CD4+ CD25 + FoxP3+ expressing T cells and modulating immune cell subsets such as monocytes and dendritic cells (Salehipour et al., 2017; Kobayashi et al., 2012). Intriguingly, even though some research groups have reported increases in Bifidobacterium species as part of the gut dysbiosis seen in patients with MS (Miyake et al., 2015; Chu et al., 2018), probiotic supplementation with the species has been recommended as a potential therapeutic for MS because Bifidobacterium produces γ-aminobutiric acid, folate, conjugated linoleic acids (Toghi et al., 2019), and other metabolites capable of impacting the immune system and the CNS.
Short-chain fatty acids
Short-chain fatty acids (SCFAs), specifically propionate, butyrate, and acetate, are produced by the gut microbiome as a product of the fermentation of carbohydrate sources such as dietary fiber and could also represent a microbiome-dependent health benefit in this marmoset model. SCFAs are associated with health benefits in a myriad of disorders, including the amelioration of disease in other EAE models (Mizuno et al., 2017; Chen et al., 2019b; Haghikia et al., 2015). Treatment with SCFAs has a direct impact in EAE by reducing Th1 cells and stimulating proliferation of lamina propria-derived Treg cells (Mizuno et al., 2017; Haghikia et al., 2015). Patients with MS correspondingly have lower levels of fecal SCFAs, as well as reduced numbers of SCFA-producing bacteria in their gut microbiome (Zeng et al., 2019; Miyake et al., 2015).
The role of SCFAs in marmoset EAE remains completely unexplored and was not assessed by Kap et al. Clostridium spp., which are commonly found in the human gut and are some of the main producers of SCFAs, could not be detected in the gut of the marmosets included in this study. Several studies have found Clostridium spp. to be depleted in patients with MS (Tremlett et al., 2016; Miyake et al., 2015), but the role of this genus in MS/EAE seems to be controversial. For example, while the epsilon toxin produced by Clostridium perfringens was shown to cause death of oligodendrocytes and induce demyelination (Rumah et al., 2013; Linden et al., 2015), Clostridium butirycum was shown to ameliorate EAE by decreasing Th17 responses and increasing regulatory T-cell responses (Chen et al., 2019a, 2019b). Since some Clostridium species are capable of immunomodulation, it would be interesting to test whether introducing this genus to the marmoset gut influences the progression of EAE.
Tryptophan
In addition to SCFAs, ω-3 fatty acids and metabolites derived from tryptophan (Trp) have been implicated as drivers or modulators of diet-microbiome-host interactions in EAE/MS (Thorburn et al., 2014). Whether they derive from food or from bacteria, it seems that the effects of these metabolites are exerted through shared mechanisms: G-protein-coupled receptor signaling, epigenetic regulation of gene expression through inhibition of histone deacetylases, or the effects of transcription factors such as the aryl hydrocarbon receptor (AHR) (reviewed by Katz Sand, 2018). Milk proteins are generally a source of Trp-containing peptides (Nongonierma and FitzGerald, 2015). Trp, an essential amino acid primarily used in protein synthesis, has numerous physiological functions, as it is a precursor of bioactive molecules produced by gut bacteria that are fundamental to human health (Nongonierma and FitzGerald, 2015). Rothhammer et al. (Rothhammer et al., 2016, 2018) reported that Trp metabolites, such as kynurenine, are able to downregulate CNS immune responses through activation of the AHR on glial cells. Dietary Trp is metabolized by members of the gut microbiota (e.g., Lactobacillaceae and Bifidobacterium species) into AHR ligands, such as indole-3-aldehyde and indole-3-lactic acid (Zelante et al., 2013), which have an effect on astrocytes, reducing CNS inflammation and disease scores in EAE. Given that Bifidobacterium species were the predominant taxa in the marmoset fecal microbiota, there is the potential that Bifidobacterium mediated health benefits in the YBS-fed marmosets via Trp-derived metabolites. In line with this, higher abundances of gut microbiota-derived Trp metabolites have been associated with lower disability scores in pediatric MS (Nourbakhsh et al., 2018).
Branched chain fatty acids
Another dietary metabolite related to amino acids and proteins is branched chain fatty acids (BCFAs), which include iso-butyrate, valerate, and iso-valerate. Small amounts of BCFAs are also formed as metabolites from protein and amino acid degradation by the gut microbiome (Neis et al., 2015). To our knowledge, there are no studies assessing the direct effect of raw BCFA supplementation in MS or EAE. However, valproate or valproic acid is a valerate derivate with anti-epileptic, neuro-protective, and anti-inflammatory effects and has been shown to ameliorate symptoms in EAE and optic neuritis (Liu et al., 2017; Castelo-Branco et al., 2014; Zhang et al., 2012). BCFAs are formed by the bacterial fermentation of proteins, and the YBS provided higher amounts of protein than the WBS. In our re-analysis, we found that OTUs of protein degraders belonging to Prevotellaceae and Megasphaera exhibited a 59% increase in abundance in the YBS cohort compared to only a 38% increase in the WBS cohort. The increase in both experimental groups was likely attributable to the availability of carbohydrates and fiber sources, but the greater increase in YBS-fed monkeys presumably reflects the increased amount of protein provided by the YBS.
Microbiome-independent diet effects
Given that both Kap et al. and our more detailed re-analysis suggested that many of the microbial signatures of pathology are independent of diet and the study design itself does not allow for causal relationships to be reliably established, it is also important to consider how diet alone may have influenced the development of pathology. Diet-related benefits in mammalian hosts can be microbiome independent even if strong correlations are detected between the microbiome and host phenotypes (Bindels et al., 2017). These direct diet-related benefits include the general fulfillment of the host's nutritional needs for proper development and maintenance of the immune system, as well as the more focused impact of specific vitamins and minerals on the immune system. Both the water- and yogurt-based dietary supplements, combined with the other food sources Kap et al. provided as part of the marmoset's daily diets (e.g., nuts, fruits, and gums), broadly met the nutritional demands of moderately active marmosets (Table 1; see also supplemental information). We therefore examined how specific components of the supplements, such as vitamins, minerals, and trace elements, among others, may have influenced the onset of the disease and its progression.
Vitamins
The supplements used by Kap et al. differed in their vitamin content. Compared to the WBS, the YBS had almost double the amount of the water-soluble complex B vitamins and the fat-soluble vitamins D and E. Here, we discuss how the increased amounts of vitamins B and E may have ameliorated disease progression in YBS-fed marmosets, as the controversial role of vitamin D supplementation in EAE and MS has already been extensively reviewed elsewhere (Bivona et al., 2019; Caballero-Villarraso et al., 2019; McLaughlin et al., 2018; Jagannath et al., 2018; Ismailova et al., 2019).
The complex B vitamins added to the nutritional supplements described by Kap et al. include thiamine (B1), riboflavin (B2), niacin (B3), pantothenic acid (B5), pyridoxine (B6), folic acid (B11), and cobalamine (B12). Nemazannikova et al. (2018) recently published an excellent review on the role of the complex B vitamins in MS. They concluded that all the complex B vitamins play crucial roles in cell metabolism and are involved, either directly or indirectly, in neurological processes and myelin formation (Nemazannikova et al., 2018). In the case of riboflavin, riboflavin supplementation by itself or in combination with IFNβ-1a was found to decrease disease scores in MOG-induced EAE in C57BL/6 mice by modulating gene expression and protein levels of both brain-derived neurotrophic factor and IL-6.
Vitamin E is a potent antioxidant found in vegetable oils, nuts, seeds, and non-citrus fruits, which the marmosets received as part of their diet and/or as ingredients of the YBS (Kap et al., 2018). It occurs either as tocopherol or tocotrienol isoforms, with tocopherol having higher bioavailability after oral ingestion (van Meeteren et al., 2005). Supplementation with vitamin E decreases demyelination and potentially promotes myelination in animal models of ethidium bromide-induced demyelination (Goudarzvand et al., 2010). Additionally, intraperitoneal administration of α-tocopherol or a synthetic vitamin E derivative reduces disease incidence and delays disease onset and progression in MOG-induced EAE (Xue et al., 2016; Blanchard et al., 2013).
Minerals and trace elements
Kap et al. did not report the mineral or trace element content of their two dietary supplements, but both are potentially relevant to disease outcomes. Several studies have found magnesium intake to be important for physical performance and fatigue management in patients with MS (Bitarafan et al., 2014; Bromley et al., 2019; Pommerich et al., 2018). Indeed, oral magnesium supplements have been reported to decrease the relapse rate in patients with MS (Goldberg et al., 1986). Additionally, a growing body of evidence suggests that ionized magnesium has a crucial role in ameliorating disruptions to the BBB during neurological diseases (Kaya and Ahishali, 2011), and the BBB is known to be affected in MS/EAE. Interestingly, vitamin D and magnesium have a mutualistic relationship: magnesium absorption is increased by vitamin D (Sizar and Givler, 2019), which in turn requires magnesium to be active and functional (Uwitonze and Razzaque, 2018). Nuts and broad beans, which are considered high sources of magnesium, were among the additional foods received by the YBS-supplemented marmosets but not by the WBS-supplemented marmosets. This discrepancy could have contributed to the decreased disease incidence in YBS-fed marmosets.
Vitamin D also has a close relationship with calcium, as vitamin D is needed for calcium absorption. In fact, vitamin D supplementation was found to prevent EAE development in SJL/J and B10.PL mice while on a high calcium diet, but the protective effect was lost when low calcium diets were used (Cantorna et al., 1999; Lemire and Clay Archer, 1991; Ramsaransing et al., 2009). Moreover, supplementation with magnesium, calcium, and vitamin D resulted in decreased relapse rate in a group of patients with MS (Goldberg et al., 1986). In line with these results, the higher levels of dietary calcium provided by the YBS (a dairy product) may have contributed to a decreased rate of disease.
Milk proteins
The recommendation of dairy products to patients with MS has been controversial. Some studies suggest that certain proteins in milk, such as BSAbovine serum albumin (BSA) and butyrophilin (BTN), a protein that constitutes 40% of the milk's fat globule membrane (MFGM), increase the risk of developing MS (Winer et al., 2001; Guggenmos et al., 2004; Mana et al., 2004). These studies have therefore suggested that decreasing milk consumption could be a non-invasive intervention strategy in MS (Winer et al., 2001). However, this recommendation is not universally accepted. Other studies have suggested that milk, and specifically the MFGM, should be considered a potential nutraceutical, as it has beneficial health effects in connection to several diseases, including MS (Spitsberg, 2005).
In the case of BSA, the peptide BSA193 was found to induce the development of EAE in SJL/J mice (Winer et al., 2001). The authors of this study reported that, despite the structural homology between BSA193 and exon 2 of the myelin basic protein (MBP, a known antigen commonly used to induce EAE), they were unable to establish T-cell cross-reactivity or mimicry when stimulating PBMCs in patients with MS using BSA193 as the antigen. Instead, they found cross-reactivity between BSA and MBP antibodies. Cross-reactivity of B- and T-lymphocyte receptors has important consequences for the host's health. However, the causes of cross-reactivity among immune responses are not always known, and there might be “potential proximate and evolutionary explanations” (Fairlie-Clarke et al., 2009) for this event. Cross-reactivity could be a consequence of the immune system's inability to process information about the antigens or a vital feature of the immune system that allows it to acquire biological robustness. Development of cross-reactivity could represent an advantageous path in the evolution of the host and a mechanism by which the host improves its fitness. In the case of MS and EAE, unknown or unexplored factors could be involved in determining whether subsequent immune responses to cross-reactivity are pathogenic or preventive, but diet components could not be discarded as either potential offenders or as redeemers that decide the fate of those responses.
There is a renewed interest in BTN, as this protein also seems to regulate immune responses in several diseases, including MS and EAE (Afrache et al., 2012; Rhodes et al., 2016). As with BSA, cross-reactivity of human T-cell responses has also been reported for BTN, as many amino acid residues are conserved between the MOG and the BTN protein (Guggenmos et al., 2004). BTN domains are also structurally similar to the B7 proteins, a family of costimulatory molecules that are essential for the regulation of T-cell activation and tolerance (Afrache et al., 2012). Indeed, BTN has been shown to modulate encephalitogenic T-cell responses to MOG in rodents with EAE (Stefferl et al., 2000; Guggenmos et al., 2004). This property seems to be rodent strain specific, suggesting a role of genetics in how BTN affects the development of disease. For example, immunization of Dark Agouti, Brown Norway, or Lewis rats with BTNIgV did not induce overt clinical EAE, but it did induce subclinical CNS inflammation in Dark Agouti rats (Stefferl et al., 2000). BTN's pro-inflammatory activity was attributed to the extracellular IgV-like domain, which is conserved in MOG, and to antigen mimicry with the MOG76-87 peptide sequence. However, it was found that BTN could either trigger or suppress the development of EAE (Stefferl et al., 2000). Studies in C57BL/6 mice have also demonstrated that BTN, administered either before or after immunization with MOG, prevented and/or ameliorated the clinical manifestations of EAE (Mana et al., 2004) by inhibiting production of pro-inflammatory IFN-γ, IL-2, and IL-12, while increasing anti-inflammatory IL-10 (Mana et al., 2004). This raises the question of which specific features of the host, the diet, or the host's gut microbiome are able to modulate the effect of BTN and/or other milk components in disease development.
Due to the duality (induce vs. prevent EAE) shown by these proteins (BSA and BTN), it has been suggested that the cross-reactivity mentioned above could reduce the risk of MS in humans (Stefferl et al., 2000). Within hours of ingestion, immunogenic peptides are capable of crossing gut mucosal surfaces and entering the gut-associated lymphoid tissue, where they are presented to T cells and can influence the selection, survival, and function of the T-cell repertoire. This process contributes to the development of “mucosal tolerance” or the systemic suppression of potentially inflammatory responses to dietary antigens. Indeed, individuals who consumed a large quantity of milk and dairy products during infancy and childhood but then dramatically and suddenly reduced dairy consumption during adolescence have been found to have a higher risk of developing MS in young adulthood than individuals whose consumption of milk and dairy products remained steady throughout life (Butcher, 1986).
Overall, our review of the potential health benefits of milk proteins in attenuating MS/EAE indicates that the consumption of dairy products, including fermented alternatives, may be able to positively modulate immune responses. Remarkably, an analysis of transcriptional changes in the lymph nodes of mice with EAE revealed increased expression of genes coding for milk components, including casein (Csn1s1, Csn2, Csn1s2a, and Csn3) and lactalbumin α, at the time of recovery from an attack (Otaegui et al., 2007). Moreover, expression of Csn3 was found to be elevated in the blood of patients with MS 72 hr after a relapse (Otaegui et al., 2007). Casein has immunosuppressive effects both in vivo and in vitro (Daddaoua et al., 2005; Otani et al., 2001) and could therefore play a protective role in MS. Although there is the potential that some of these immunomodulatory effects of milk proteins are related to the microbiome in as yet unknown ways, it appears likely that, with respect to the YBS diet in the marmoset model of MS, milk proteins alone have important and microbiome-independent physiological effects that attenuated disease onset and progression.
Challenges in identifying cause-and-effect relationships and physiological mechanisms in animal models
In the previous two sections, we showed that (1) there were significant associations among diet, the gut microbiota, and the development of pathology in a marmoset model of EAE and (2) the few taxa that were affected by diet are known to participate in host-microbe interactions that contribute to immunopathologies. However, because the data obtained from studies like that of Kap et al. (Kap et al., 2018) are correlational, it is impossible to assign causality to these diet- and disease-associated changes in the gut microbiome. For example, although altered microbiomes might contribute to the development of pathology, they might instead be the consequence of it. In this respect, it is noteworthy that the abundance of some MS-associated bacterial species is restored to levels seen in healthy controls when patients are treated with disease-modifying drugs (Jangi et al., 2016; Chen et al., 2016). Although these observations are also correlative, the fact that these drugs affect the microbiome, despite being targeted against the host immune system, suggests that some microbiome changes associated with MS might be secondary to the disease.
Our re-analysis of the Kap et al. (Kap et al., 2018) data generally supports the hypothesis that the majority of the microbiome changes associated with EAE in the marmoset model were caused by the disease, not the diet, as associations between taxon abundances and pathology were stronger and more common than associations between taxon abundances and diet (Figures 2 and 3). Very few taxa were associated with both diet and pathology, which would be expected if the diet ameliorated pathology via the microbiome. Broadly, our findings suggest that the diet influenced the development of pathology, which, in turn, was the main driver of microbiome changes in this marmoset colony. We can only speculate about the reason for these associations. Inflammation (Bander et al., 2020) and metabolic aberrancies (Martínez et al., 2013) have both been shown to alter gut microbiomes, and the enteric nervous system could influence peristaltic movement having an effect on transit time and stool consistency (Vandeputte et al., 2016). However, although our findings do not support a major role of diet-induced shifts in microbiota composition in pathology, it remains possible that some of the health benefits of the YBS diet were mediated via bacterially derived metabolites, which may not be detected in taxonomy-based assessments of microbiome composition.
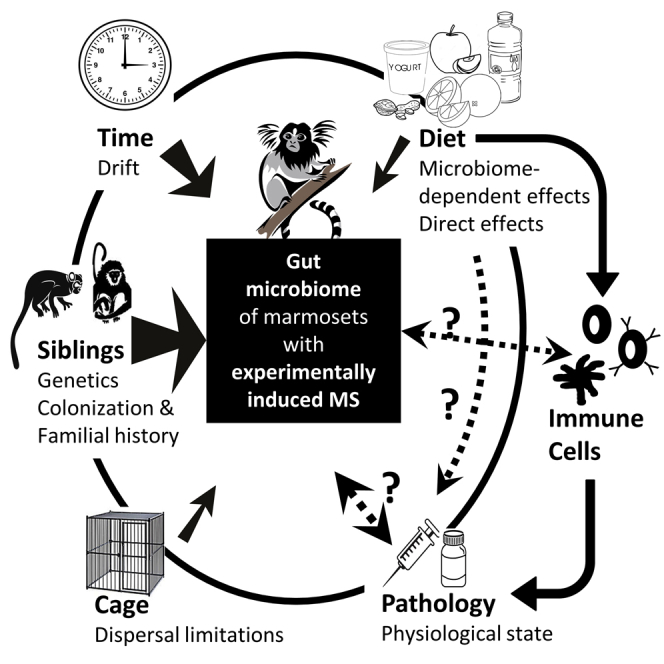
An ecological perspective on the factors that shape the marmoset gut microbiome
In addition to the limitations of correlative and taxonomy-focused data, another challenge to elucidating mechanistic interactions and cause-and-effect relationships in gut microbiome studies is that it is difficult to experimentally control for all the factors that shape the gut microbiome and its interactions with the host (Vujkovic-Cvijin et al., 2020). Animal experiments have the advantage of controlling for environmental (e.g., diet, cage environment, bedding, and handling) and host (e.g., age, genotype, etc.) factors that potentially shape the gut microbiome, but even so, microbiome research in animal models of disease has often neglected or underestimated the importance of ecological processes that shape gut microbial communities (Martínez et al., 2018; Ubeda et al., 2012). For example, dispersal limitation, familial transmission, and colonization history can all lead to facility, cage, and litter effects that cause stochastic differences in gut microbiota composition that are completely independent of the treatments tested (Martínez et al., 2018; McCoy et al., 2017; Ubeda et al., 2012; Walter and Ley, 2011; Walter, 2015). The microbiome field is full of studies (Ubeda et al., 2012) where microbiome changes presumably associated with pathology could instead be independently caused by a confounding factor. Sophisticated experimental designs are required to avoid these effects (McCoy et al., 2017). It is therefore essential that animal experiments designed to test hypotheses about how specific environmental factors may shape the gut microbiota's role in pathology are conducted and evaluated within a broader ecological framework. Below we use the Kap et al. data as an example of how much some of these ecological variables can influence experimental results.
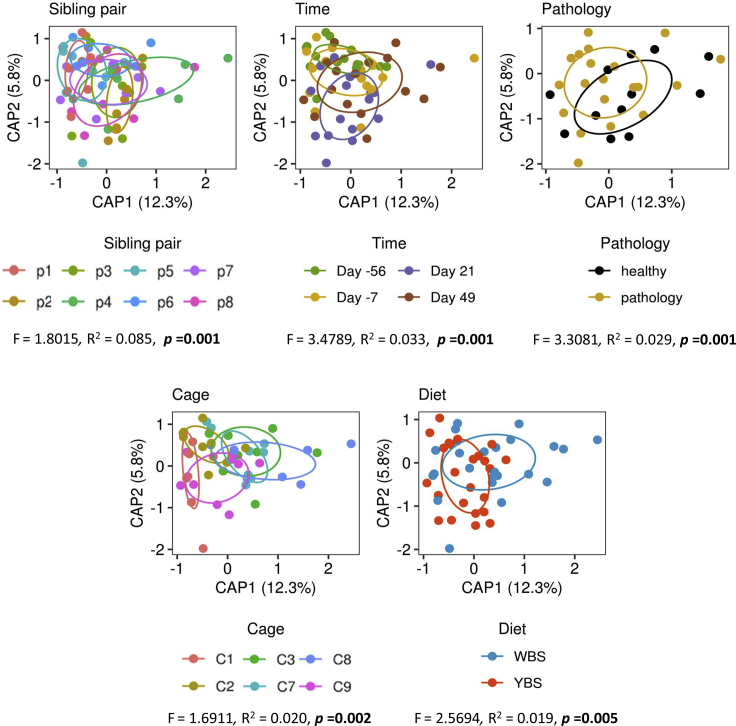
To first examine how additional experimental and ecological factors, such as marmoset sibling pair and cage, could also have broadly affected variation in microbiome composition, we used a Bray-Curtis distance-based redundancy analysis (dbRDA) based not only on the experimentally manipulated variables (diet, pathology, and experimental time point) but also on environmental factors (sibling pair and cage). We identified the best predictors of microbiome composition based on the adjusted R2 value for each predictor in a forward selection procedure (see Data S1). Our results suggested that the ecological variables that affected microbial structures were, in order of decreasing explanatory power: sibling pair, time, pathology, and lastly diet and cage, with the latter two variables, which are nested, having a similar effect (R2 values of 0.085, 0.033, 0.029, 0.020, and 0.019, respectively; Figure 5). These findings emphasize that while diet indeed influenced the gut microbiota in this marmoset model of MS, almost every other variable had a stronger effect than diet. Sibling pair was the most important factor driving microbiota composition despite the fact that siblings were separated and re-paired with another marmoset. Its overwhelming importance suggests the significance of host genetic factors (Goodrich et al., 2014, 2016; Turnbaugh et al., 2009), shared colonization history (Martínez et al., 2018), or familial history (Ubeda et al., 2012) in shaping the gut microbiome. Its importance likely goes beyond relatedness and might involve priority effects and joint vertical transmission and might also be impacted by ecological processes such as dispersal. Time also had a large effect, emphasizing the importance of stochastic ecological processes, such as drift (the time effect) and, to some extent, dispersal limitations (the cage effect). Most importantly, disease induction and the development of pathology shaped the microbiome more than diet, supporting our conclusions above that associations between microbiome and pathology might be primarily driven by the host's physiological state and not by diet altering pathology via the microbiome.
Figure 5.
Ecological factors that potentially affected β-diversity in the study
Bray-Curtis distance-based redundancy analyses (dbRDA) of all ecological factors that could have affected outcomes in the study. Significance was established by ANOVA at p < 0.05.
We additionally examined the effects of sibling pair and individual identity in predictions of taxon abundances. All our mixed-effect modeling procedures included individual identity nested within sibling pair as a random effect term to account for this variation. When we compared the marginal and conditional R2 values for our models, which can be used to calculate the amount of variance explained by the fixed effects (diet, pathology, time) and the random term (individual nested in sibling pair) (Nakagawa and Schielzeth, 2013), we found that the random term explained more variance than all the fixed terms in the majority of our models (56% of abundance-based models [Figure 2 and Table S4] and 61% of delta abundance-based models [Figure 3 and Table S5]). On average, fixed and random effects explained 10.2% and 15.8% of variance in our abundance-based models and 11.1% and 25.1% of variance in our delta abundance-based models, respectively, indicating the importance of these additional ecological factors. The fact that many of these models explain less than 40% of overall variation in each taxon further emphasizes how other ecological factors, along with stochastic variation, can play a strong role in microbiome research.
The dbRDA and mixed model results underscore how, in experimental studies that collect correlational data, appropriate consideration must be given to controlling for as much natural ecological variation as possible in the experimental design and adequately accounting for the remaining ecological factors that cannot be experimentally controlled. Poorly designed or inappropriate analyzed studies that do not address the complex factors that shape the marmoset microbiome can impede the identification of true associations among diet, microbiome composition, and the development of MS. In our re-analysis of Kap et al., we accounted for sibling pair and individual identity as a random effect in all mixed-effect models, but our statistical power to control for any other effects was limited by the modest sample size. We therefore recommend that future studies also consider the sample size that is needed to obtain enough statistical power to reliably identify true biological associations amid the unavoidable noise variables.
Conclusions and future directions
Diet has the exciting potential to be a supplementary therapy in human MS (Janakiraman and Krishnamoorthy, 2018). However, there is currently not enough information available to make reliable dietary recommendations for patients with MS. Obtaining this information will require a combination of nutrition-related studies in patients with MS and mechanistic studies in animal models. As we show in this review, these studies should consider the three-way relationships among diet, gut microbiome composition, and the immune system: diet impacts the structure and composition of the microbiota, which can indirectly mediate the health effects of diet, and both diet and the microbiota can act independently or in a synergistic way to directly affect the immune system.
Marmosets are an attractive model for this research due to their physiological similarities with humans. An important advantage of the marmoset dizygotic bone marrow chimera twins is that they allow us to partially control for genetic diversity in outbred animals evolutionarily close to humans, by dividing twin members over control versus treatment groups. This allows for experiments that are similar to controlled trials in human twin studies (Beltrán et al., 2019; Gerdes et al., 2020). In addition, the marmoset EAE model also allows us to faithfully mimic one of the best established environmental risk factors for MS, namely Epstein-Barr virus (EBV). Kap et al. demonstrated that the levels of the EBV counterpart in marmosets, CalHV3, in different anatomical compartments were reduced upon changing the dietary supplement (Kap et al., 2018). Although marmoset research has already provided novel insights, the experiments done to date have been small and were not designed to unravel mechanisms or cause-and-effect relationships. To robustly explore these more nuanced questions in the marmoset model of MS, we recommend three key considerations for future research in this model.
First, it is important to design studies that explicitly test for causal relationships among diet, the gut microbiome, and MS. For example, future studies could use gnotobiotic approaches (experiments with defined microbiomes) or specifically target Actinobacteria, which were implicated in the Kap et al. data, using an interventionist approach (Walter et al., 2020). The marmoset model provides exciting prospects for conducting such definitive studies. Ideally, these studies will incorporate techniques that determine not only fecal microbiota composition but also metabolic function, such as metagenomics and metabolomics. Even in cases where the taxonomic composition of the gut microbiome is not affected by a particular disease or treatment, diet might impact disease by changing the metabolic activity of the gut microbiota, which could be detected using these more sensitive techniques. Longitudinal studies with frequent sampling of the gut microbiota —even more frequently than in Kap et al.—could also allow for statistical approaches that can assign causality, such as mediation (Leong et al., 2018) or Bayesian inference analysis (Schluter et al., 2020). Only a mechanistic understanding of the diet-microbiome-immune system triad will allow for reliable diet recommendations for patients with MS.
Second, the findings from our re-analysis warrant the use of a broader ecological framework when evaluating these mechanistic interactions among diet, the microbiome, and the immune system in MS. Any animal study must consider the ecological factors that are not directly linked to experimental variables being tested but have the potential to confound the results, specifically by introducing stochastic differences in the gut microbiota that are not linked to the treatment. Studies that use larger numbers of animals under more standardized conditions will be better able to account for these ecological factors.
Third, the consideration of ecological processes is also important for studies of MS in humans. Discrepancies have been reported in the differences in microbiome composition between patients with MS and healthy controls, and although consistent patterns were observed, a microbiome signature that could serve as a biomarker for MS has not been confirmed to date (Mirza et al., 2020). Studies on the drivers of disease-associated microbiome alterations, their cause-and-effect relationships with host pathology, and the ecological mechanisms involved in diet-microbiome-immune system interaction require knowledge of which previous environmental exposures and lifestyle factors could be relevant to the results in the affected population. In this regard, marmosets are an attractive model for studying the ecological processes that shape the gut microbiome in relation to pathology due to their evolutionarily close similarities with humans. We hope that our re-analysis of the Kap et al. marmoset data, though limited by the study's modest sample size, provides valuable future hypotheses to test in the marmoset model of MS and models how larger future studies should be designed and analyzed.
Acknowledgments
The marmoset EAE experiment was exclusively funded by an internal subsidy of the Biomedical Primate Research Center. The APC Microbiome Ireland is a research institute funded by Science Foundation Ireland (SFI) through the Irish Government's National Development Plan. J.W. is supported by SFI through grants SFI/12/RC/2273 and 19/RP/6853. M.E.P.-M. and J.W. acknowledge support from the Weston Family Microbiome Initiative POP 2018 from the Weston Family Foundation.
Author contributions
Conceptualization, J.D.L., J.W., and M.E.P.-M.; data curation, M.E.P.-M. and S.S.; formal analysis, M.E.P.-M. and S.S.; writing – original draft, M.E.P.-M., S.S., and J.W.; writing – review and editing, M.E.P.-M., S.S., J.W., J.D.L., H.J.M.H., and B.A.t’H.; visualizations, M.E.P.-M. and S.S.
Declaration of interests
The authors declare no conflicts of interest.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102709.
Contributor Information
Jon D. Laman, Email: j.d.laman@umcg.nl.
Jens Walter, Email: jenswalter@ucc.ie.
Supporting citations
Altschul et al., 1990, Bates et al., 2015, Deehan et al., 2020, Edgar, 2013, Eren et al., 2013, McMurdie and Holmes, 2013, , Nguyen et al., 2020, Oksanen et al., 2019, Patry et al., 2019,Quast et al., 2013, R Core Team, 2018, Wang et al., 2007, Yoon et al., 2017.
Supplemental information
Coefficients were considered significant (∗∗) if the 95% confidence interval did not overlap zero. Variables with no coefficient shown for a given taxon were not present in top-ranked models (ΔAICc <2) predicting that taxon's abundance. Positive and negative coefficients indicate YBS and WBS, respectively, for diet; symptomatic and healthy marmosets, respectively, for pathology; and increases and decreases over the course of the experiment, respectively, for time.
Coefficients were considered significant (∗∗) if the 95% confidence interval did not overlap zero. Variables with no coefficient shown for a given taxon were not present in top-ranked models (ΔAICc <2) predicting that taxon's change in abundance. Positive and negative coefficients indicate YBS and WBS, respectively, for diet; symptomatic and healthy marmosets, respectively, for pathology; and increases and decreases in the magnitude of shifts in abundance, respectively, for Δ time.
References
- Afrache H., Gouret P., Ainouche S., Pontarotti P., Olive D. The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics. 2012;64:781–794. doi: 10.1007/s00251-012-0619-z. [DOI] [PubMed] [Google Scholar]
- Al-Ghezi Z.Z., Busbee P.B., Alghetaa H., Nagarkatti P.S., Nagarkatti M. Combination of cannabinoids, delta-9-tetrahydrocannabinol (THC) and cannabidiol (CBD), mitigates experimental autoimmune encephalomyelitis (EAE) by altering the gut microbiome. Brain Behav. Immun. 2019;82:25–35. doi: 10.1016/j.bbi.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlAmmar W.A., Albeesh F.H., Ibrahim L.M., Algindan Y.Y., Yamani L.Z., Khattab R.Y. Effect of omega-3 fatty acids and fish oil supplementation on multiple sclerosis: a systematic review. Nutr. Neurosci. 2019 doi: 10.1080/1028415X.2019.1659560. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez J.I., Cayrol R., Prat A. Disruption of central nervous system barriers in multiple sclerosis. Biochim. Biophys. Acta. 2011;1812:252–264. doi: 10.1016/j.bbadis.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Arboleya S., Watkins C., Stanton C., Ross R.P. Gut Bifidobacteria populations in human health and aging. Front. Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bander Z. Al, Nitert M.D., Mousa A., Naderpoor N. The gut microbiota and inflammation: an overview. Int. J. Environ. Res. Public Health. 2020;17:7618. doi: 10.3390/ijerph17207618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D., Knapka J., Renquist D. The apparent reversal of a wasting syndrome by nutritional intervention in Saguinus mystax. Lab. Anim. Sci. 1988;38:282–288. [PubMed] [Google Scholar]
- Barnes C.J., Rasmussen L., Asplund M., Knudsen S.W., Clausen M.-L., Agner T., Hansen A.J. Comparing DADA2 and OTU clustering approaches in studying the bacterial communities of atopic dermatitis. J. Med. Microbiol. 2020;69:1293–1302. doi: 10.1099/jmm.0.001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B.M., Walker S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- Beltrán E., Gerdes L.A., Hansen J., Flierl-Hecht A., Krebs S., Blum H., Ertl-Wagner B., Barkhof F., Kümpfel T., Hohlfeld R., Dornmair K. Early adaptive immune activation detected in monozygotic twins with prodromal multiple sclerosis. J. Clin. Invest. 2019;129:4758–4768. doi: 10.1172/JCI128475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K., Mues M., Koutrolos M., Rasbi Z. Al, Boziki M., Johner C., Wekerle H., Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- Berer K., Gerdes L.A., Cekanaviciute E., Jia X., Xiao L., Xia Z., Liu C., Klotz L., Stauffer U., Baranzini S.E. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10719–10724. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K., Martínez I., Walker A., Kunkel B., Schmitt-Kopplin P., Walter J., Krishnamoorthy G. Dietary non-fermentable fiber prevents autoimmune neurological disease by changing gut metabolic and immune status. Sci. Rep. 2018;8:10431. doi: 10.1038/s41598-018-28839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff S.A., Gerndt N., Winchenbach J., Stumpf S.K., Hosang L., Odoardi F., Ruhwedel T., Böhler C., Barrette B., Stassart R. Dietary cholesterol promotes repair of demyelinated lesions in the adult brain. Nat. Commun. 2017;8:14241. doi: 10.1038/ncomms14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels L.B., Segura Munoz R.R., Gomes-Neto J.C., Mutemberezi V., Martínez I., Salazar N., Cody E.A., Quintero-Villegas M.I., Kittana H., de los Reyes-Gavilán C.G. Resistant starch can improve insulin sensitivity independently of the gut microbiota. Microbiome. 2017;5:12. doi: 10.1186/s40168-017-0230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitarafan S., Harirchian M.-H., Nafissi S., Sahraian M.-A., Togha M., Siassi F., Saedisomeolia A., Alipour E., Mohammadpour N., Chamary M. Dietary intake of nutrients and its correlation with fatigue in multiple sclerosis patients. Iran. J. Neurol. 2014;13:28–32. [PMC free article] [PubMed] [Google Scholar]
- Bivona G., Gambino C.M., Iacolino G., Ciaccio M. Vitamin D and the nervous system. Neurol. Res. 2019;41:827–835. doi: 10.1080/01616412.2019.1622872. [DOI] [PubMed] [Google Scholar]
- Blanchard B., Heurtaux T., Garcia C., Moll N.M., Caillava C., Grandbarbe L., Klosptein A., Kerninon C., Frah M., Coowar D. Tocopherol derivative tfa-12 promotes myelin repair in experimental models of multiple sclerosis. J. Neurosci. 2013;33:11633–11642. doi: 10.1523/JNEUROSCI.0774-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Caporaso J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley L., Horvath P.J., Bennett S.E., Weinstock-Guttman B., Ray A.D. Impact of nutritional intake on function in people with mild-to-moderate multiple sclerosis. Int. J. MS Care. 2019;21:1–9. doi: 10.7224/1537-2073.2017-039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher P.J. Milk consumption and multiple sclerosis--an etiological hypothesis. Med. Hypotheses. 1986;19:169–178. doi: 10.1016/0306-9877(86)90057-5. [DOI] [PubMed] [Google Scholar]
- Caballero-Villarraso J., Jiménez-Jiménez M.J., Escribano B.M., Agüera E., Santamaría A., Túnez I. Role of vitamin D in multiple sclerosis and other neurodegenerative processes: bibliometric analysis and systematic review. CNS Neurol. Disord. Drug Targets. 2019;18 doi: 10.2174/1871527318666190703102330. [DOI] [PubMed] [Google Scholar]
- Cantorna M.T., Humpal-Winter J., DeLuca H.F. Dietary calcium is a major factor in 1,25-dihydroxycholecalciferol suppression of experimental autoimmune encephalomyelitis in mice. J. Nutr. 1999;129:1966–1971. doi: 10.1093/jn/129.11.1966. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G., Stridh P., Guerreiro-Cacais A.O., Adzemovic M.Z., Falcão A.M., Marta M., Berglund R., Gillett A., Hamza K.H., Lassmann H. Acute treatment with valproic acid and l-thyroxine ameliorates clinical signs of experimental autoimmune encephalomyelitis and prevents brain pathology in DA rats. Neurobiol. Dis. 2014;71:220–233. doi: 10.1016/j.nbd.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Cekanaviciute E., Yoo B.B., Runia T.F., Debelius J.W., Singh S., Nelson C.A., Kanner R., Bencosme Y., Lee Y.K., Hauser S.L. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U S A. 2017;114:10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chia N., Kalari K.R., Yao J.Z., Novotna M., Soldan M.M.P., Luckey D.H., Marietta E.V., Jeraldo P.R., Chen X. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Ma X.X., Liu Y., Ma L., Chen Z., Lin X., Si L., Ma X.X., Chen X. Gut microbiota interventions with Clostridium butyricum and norfloxacin modulate immune response in experimental autoimmune encephalomyelitis mice. Front. Immunol. 2019;10:1662. doi: 10.3389/fimmu.2019.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Noto D., Hoshino Y., Mizuno M., Miyake S. Butyrate suppresses demyelination and enhances remyelination. J. Neuroinflammation. 2019;16:165. doi: 10.1186/s12974-019-1552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F., Shi M., Lang Y., Shen D., Jin T., Zhu J., Cui L. Gut microbiota in multiple sclerosis and experimental autoimmune encephalomyelitis: current applications and future perspectives. Mediat. Inflamm. 2018;2018:8168717. doi: 10.1155/2018/8168717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M.J., Cusack S., O’Sullivan O., Greene-Diniz R., De Weerd H., Flannery E., Marchesi J.R., Falush D., Dinan T., Fitzgerald G. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U S A. 2011;108:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Costa G., Romeo M., Esposito F., Sangalli F., Colombo B., Radaelli M., Moiola L., Comi G., Martinelli V. Caesarean section and infant formula feeding are associated with an earlier age of onset of multiple sclerosis. Mult. Scler. Relat. Disord. 2019;33:75–77. doi: 10.1016/j.msard.2019.05.010. [DOI] [PubMed] [Google Scholar]
- Cree B.A.C., Spencer C.M., Varrin-Doyer M., Baranzini S.E., Zamvil S.S. Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann. Neurol. 2016;80:443–447. doi: 10.1002/ana.24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J.F., O’riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V. The microbiota-gut-brain axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- Daddaoua A., Puerta V., Zarzuelo A., Suárez M.D., Sánchez de Medina F., Martínez-Augustin O. Bovine glycomacropeptide is anti-inflammatory in rats with hapten-induced colitis. J. Nutr. 2005;135:1164–1170. doi: 10.1093/jn/135.5.1164. [DOI] [PubMed] [Google Scholar]
- Dargahi N., Matsoukas J., Apostolopoulos V. Streptococcus thermophilus ST285 alters pro-inflammatory to anti-inflammatory cytokine secretion against multiple sclerosis peptide in mice. Brain Sci. 2020;10:126. doi: 10.3390/brainsci10020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deehan E.C., Yang C., Perez-Muñoz M.E., Nguyen N.K., Cheng C.C., Triador L., Zhang Z., Bakal J.A., Walter J. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe. 2020;27:389–404.e6. doi: 10.1016/j.chom.2020.01.006. [DOI] [PubMed] [Google Scholar]
- Dobbing J. The influence of early nutrition on the development and myelination of the brain. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 1964;159:503–509. doi: 10.1098/rspb.1964.0016. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Eren A.M., Vineis J.H., Morrison H.G., Sogin M.L. A filtering method to generate high quality short reads using illumina paired-end technology. PLoS One. 2013;8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., Sparaco M., Maniscalco G.T., Signoriello E., Lanzillo R., Russo C., Carmisciano L., Cepparulo S., Lavorgna L., Gallo A. Lifestyle and Mediterranean diet adherence in a cohort of Southern Italian patients with multiple sclerosis. Mult. Scler. Relat. Disord. 2021;47:102636. doi: 10.1016/j.msard.2020.102636. [DOI] [PubMed] [Google Scholar]
- Fairlie-Clarke K.J., Shuker D.M., Graham A.L. Why do adaptive immune responses cross-react? Evol. Appl. 2009;2:122. doi: 10.1111/J.1752-4571.2008.00052.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Zhang J. Dietary modulation of intestinal microbiota: future opportunities in experimental autoimmune encephalomyelitis and multiple sclerosis. Front. Microbiol. 2019;10:740. doi: 10.3389/fmicb.2019.00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M.A. Microbiome: focus on causation and mechanism. Cell. 2018;174:785–790. doi: 10.1016/j.cell.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe P., Kunze W.A. Voices from within: gut microbes and the CNS. Cell. Mol. Life Sci. 2013;70:55–69. doi: 10.1007/s00018-012-1028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman S.N., Shahi S.K., Mangalam A.K. The “gut feeling”: breaking down the role of gut microbiome in multiple sclerosis. Neurotherapeutics. 2018;15:109–125. doi: 10.1007/s13311-017-0588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandy K.A.O., Zhang J., Nagarkatti P., Nagarkatti M. The role of gut microbiota in shaping the relapse-remitting and chronic-progressive forms of multiple sclerosis in mouse models. Sci. Rep. 2019;9:6923. doi: 10.1038/s41598-019-43356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes L.A., Janoschka C., Eveslage M., Mannig B., Wirth T., Schulte-Mecklenbeck A., Lauks S., Glau L., Gross C.C., Tolosa E. Immune signatures of prodromal multiple sclerosis in monozygotic twins. Proc. Natl. Acad. Sci. U S A. 2020;117:21546–21556. doi: 10.1073/pnas.2003339117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg P., Fleming M.C., Picard E.H. Multiple sclerosis: decreased relapse rate through dietary supplementation with calcium, magnesium and vitamin D. Med. Hypotheses. 1986;21:193–200. doi: 10.1016/0306-9877(86)90010-1. [DOI] [PubMed] [Google Scholar]
- Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Davenport E.R., Beaumont M., Jackson M.A., Knight R., Ober C., Spector T.D., Bell J.T., Clark A.G., Ley R.E. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzvand M., Javan M., Mirnajafi-Zadeh J., Mozafari S., Tiraihi T. Vitamins E and D3 attenuate demyelination and potentiate remyelination processes of hippocampal formation of rats following local injection of ethidium bromide. Cell. Mol. Neurobiol. 2010;30:289–299. doi: 10.1007/s10571-009-9451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E.A., Segre J.A. The human microbiome: our second genome. Annu. Rev. Genomics Hum. Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggenmos J., Schubart A.S., Ogg S., Andersson M., Olsson T., Mather I.H., Linington C. Antibody cross-reactivity between myelin oligodendrocyte glycoprotein and the milk protein butyrophilin in multiple sclerosis. J. Immunol. 2004;172:661–668. doi: 10.4049/jimmunol.172.1.661. [DOI] [PubMed] [Google Scholar]
- Hadley K., Ryan A., Forsyth S., Gautier S., Salem N. The essentiality of arachidonic acid in infant development. Nutrients. 2016;8:216. doi: 10.3390/nu8040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghikia A., Jörg S., Duscha A., Berg J., Manzel A., Waschbisch A., Hammer A., Lee D.-H., May C., Wilck N. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity. 2015;43:817–829. doi: 10.1016/j.immuni.2015.09.007. [DOI] [PubMed] [Google Scholar]
- He B., Hoang T.K., Tian X., Taylor C.M., Blanchard E., Luo M., Bhattacharjee M.B., Freeborn J., Park S., Couturier J. Lactobacillus reuteri reduces the severity of experimental autoimmune encephalomyelitis in mice by modulating gut microbiota. Front. Immunol. 2019;10:385. doi: 10.3389/fimmu.2019.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M.O. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–432. doi: 10.2307/1934352. [DOI] [Google Scholar]
- Hirschberg S., Gisevius B., Duscha A., Haghikia A. Implications of diet and the gut microbiome in neuroinflammatory and neurodegenerative diseases. Int. J. Mol. Sci. 2019;20:3109. doi: 10.3390/ijms20123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismailova K., Poudel P., Parlesak A., Frederiksen P., Heitmann B.L. Vitamin D in early life and later risk of multiple sclerosis—a systematic review, meta-analysis. PLoS One. 2019;14:e0221645. doi: 10.1371/journal.pone.0221645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath V.A., Filippini G., Di Pietrantonj C., Asokan G.V., Robak E.W., Whamond L., Robinson S.A. Vitamin D for the management of multiple sclerosis. Cochrane Database Syst. Rev. 2018;9:CD008422. doi: 10.1002/14651858.CD008422.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman M., Krishnamoorthy G. Emerging role of diet and microbiota interactions in neuroinflammation. Front. Immunol. 2018;9:2067. doi: 10.3389/fimmu.2018.02067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi S., Gandhi R., Cox L.M., Li N., Von Glehn F., Yan R., Patel B., Mazzola M.A., Liu S., Glanz B.L. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S.M., Ochoa-Repáraz J., Zlotkowska D., Hoyt T., Pascual D.W. Bystander-mediated stimulation of proteolipid protein-specific regulatory T (Treg) cells confers protection against experimental autoimmune encephalomyelitis (EAE) via TGF-β. J. Neuroimmunol. 2012;245:39–47. doi: 10.1016/j.jneuroim.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kap Y.S., Bus-Spoor C., van Driel N., Dubbelaar M.L., Grit C., Kooistra S.M., Fagrouch Z.C., Verschoor E.J., Bauer J., Eggen B.J.L. Targeted diet modification reduces multiple sclerosis–like disease in adult marmoset monkeys from an outbred colony. J. Immunol. 2018;201:3229–3243. doi: 10.4049/jimmunol.1800822. [DOI] [PubMed] [Google Scholar]
- Karav S., Le Parc A., Leite Nobrega de Moura Bell J.M., Frese S.A., Kirmiz N., Block D.E., Barile D., Mills D.A. Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated Bifidobacteria. Appl. Environ. Microbiol. 2016;82:3622–3630. doi: 10.1128/AEM.00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Sand I. The role of diet in multiple sclerosis: mechanistic connections and current evidence. Curr. Nutr. Rep. 2018;7:150–160. doi: 10.1007/s13668-018-0236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya M., Ahishali B. The role of magnesium in edema and blood brain barrier disruption. In: Vink R., Nechifor M., editors. Magnesium in the Central Nervous System. University of Adelaide Press; 2011. pp. 135–144. [PubMed] [Google Scholar]
- Kim M., Kim C.H. Regulation of humoral immunity by gut microbial products. Gut Microbes. 2017;8:392–399. doi: 10.1080/19490976.2017.1299311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross J.M., Darzi A.W., Nicholson J.K. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Suzuki T., Kaji R., Serata M., Nagata T., Ando M., Iizuka R., Tsujibe S., Murakami J., Kiyoshima-Shibata J. Probiotic upregulation of peripheral IL-17 responses does not exacerbate neurological symptoms in experimental autoimmune encephalomyelitis mouse models. Immunopharmacol. Immunotoxicol. 2012;34:423–433. doi: 10.3109/08923973.2010.617755. [DOI] [PubMed] [Google Scholar]
- Lee Y.K., Menezes J.S., Umesaki Y., Mazmanian S.K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U S A. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y.M.K., Nair L., Alegre M.-L. The interplay between the intestinal microbiota and the immune system. Clin. Res. Hepatol. Gastroenterol. 2015;39:9–19. doi: 10.1016/j.clinre.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire J.M., Clay Archer D. 1,25-dihydroxyvitamin D3 prevents the in vivo induction of murine experimental autoimmune encephalomyelitis. J. Clin. Invest. 1991;87:1103–1107. doi: 10.1172/JCI115072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong C., Haszard J.J., Lawley B., Otal A., Taylor R.W., Szymlek-Gay E.A., Fleming E.A., Daniels L., Fangupo L.J., Tannock G.W., Heath A.L.M. Mediation analysis as a means of identifying dietary components that differentially affect the fecal microbiota of infants weaned by modified baby-led and traditional approaches. Appl. Environ. Microbiol. 2018;84:e00914–e00918. doi: 10.1128/AEM.00914-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden J.R., Ma Y., Zhao B., Harris J.M., Rumah K.R., Schaeren-Wiemers N., Vartanian T. Clostridium perfringens epsilon toxin causes selective death of mature oligodendrocytes and central nervous system demyelination. mBio. 2015;6:e02513. doi: 10.1128/mBio.02513-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Li H., Yang J., Niu X., Zhao C., Zhao L., Wang Z. Valproic acid attenuates inflammation of optic nerve and apoptosis of retinal ganglion cells in a rat model of optic neuritis. Biomed. Pharmacother. 2017;96:1363–1370. doi: 10.1016/j.biopha.2017.11.066. [DOI] [PubMed] [Google Scholar]
- Liu S., Rezende R.M., Moreira T.G., Tankou S.K., Cox L.M., Wu M., Song A., Dhang F.H., Wei Z., Costamagna G., Weiner H.L. Oral administration of miR-30d from feces of MS patients suppresses MS-like symptoms in mice by expanding Akkermansia muciniphila. Cell Host Microbe. 2019;26:779–794.e8. doi: 10.1016/j.chom.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K.E., Parke E.C., O’Malley M.A. How causal are microbiomes? A comparison with the Helicobacter pylori explanation of ulcers. Biol. Philos. 2019;34:62. doi: 10.1007/s10539-019-9702-2. [DOI] [Google Scholar]
- Maghzi A.-H., Etemadifar M., Heshmat-Ghahdarijani K., Nonahal S., Minagar A., Moradi V. Cesarean delivery may increase the risk of multiple sclerosis. Mult. Scler. 2012;18:468–471. doi: 10.1177/1352458511424904. [DOI] [PubMed] [Google Scholar]
- Mana P., Goodyear M., Bernard C., Tomioka R., Freire-Garabal M., Liñares D. Tolerance induction by molecular mimicry: prevention and suppression of experimental autoimmune encephalomyelitis with the milk protein butyrophilin. Int. Immunol. 2004;16:489–499. doi: 10.1093/intimm/dxh049. [DOI] [PubMed] [Google Scholar]
- Mangalam A., Shahi S.K., Luckey D., Karau M., Marietta E., Luo N., Choung R.S., Ju J., Sompallae R., Gibson-Corley K. Human gut-derived commensal bacteria suppress CNS inflammatory and demyelinating disease. Cell Rep. 2017;20:1269–1277. doi: 10.1016/j.celrep.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Perdicaro D.J., Brown A.W., Hammons S., Carden T.J., Carr T.P., Eskridge K.M., Walter J. Diet-induced alterations of host cholesterol metabolism are likely to affect the gut microbiota composition in hamsters. Appl. Environ. Microbiol. 2013;79:516–524. doi: 10.1128/AEM.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Maldonado-Gomez M.X., Gomes-Neto J.C., Kittana H., Ding H., Schmaltz R., Joglekar P., Cardona R.J., Marsteller N.L., Kembel S.W. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife. 2018;7:e36521. doi: 10.7554/eLife.36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy K.D., Geuking M.B., Ronchi F. Gut microbiome standardization in control and experimental mice. Curr. Protoc. Immunol. 2017;117 doi: 10.1002/cpim.25. 23.1.1–23.1.13. [DOI] [PubMed] [Google Scholar]
- McLaughlin L., Clarke L., Khalilidehkordi E., Butzkueven H., Taylor B., Broadley S.A. Vitamin D for the treatment of multiple sclerosis: a meta-analysis. J. Neurol. 2018;265:2893–2905. doi: 10.1007/s00415-018-9074-6. [DOI] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meeteren M.E., Teunissen C.E., Dijkstra C.D., van Tol E.A.F. Antioxidants and polyunsaturated fatty acids in multiple sclerosis. Eur. J. Clin. Nutr. 2005;59:1347–1361. doi: 10.1038/sj.ejcn.1602255. [DOI] [PubMed] [Google Scholar]
- Mirza A., Forbes J.D., Zhu F., Bernstein C.N., Van Domselaar G., Graham M., Waubant E., Tremlett H. The multiple sclerosis gut microbiota: a systematic review. Mult. Scler. Relat. Disord. 2020;37:101427. doi: 10.1016/j.msard.2019.101427. [DOI] [PubMed] [Google Scholar]
- Miyake S., Kim S., Suda W., Oshima K., Nakamura M., Matsuoka T., Chihara N., Tomita A., Sato W., Kim S.-W. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno M., Noto D., Kaga N., Chiba A., Miyake S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS One. 2017;12:e0173032. doi: 10.1371/journal.pone.0173032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Schielzeth H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013;4:133–142. doi: 10.1111/j.2041-210x.2012.00261.x. [DOI] [Google Scholar]
- Neis E.P.J.G., Dejong C.H.C., Rensen S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazannikova N., Mikkelsen K., Stojanovska L., Blatch G.L., Apostolopoulos V. Is there a link between vitamin B and multiple sclerosis? Med. Chem. 2018;14:170–180. doi: 10.2174/1573406413666170906123857. [DOI] [PubMed] [Google Scholar]
- Nguyen N.K., Deehan E.C., Zhang Z., Jin M., Baskota N., Perez-Muñoz M.E., Cole J., Tuncil Y.E., Seethaler B., Wang T. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome. 2020;8:118. doi: 10.1186/s40168-020-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongonierma A.B., FitzGerald R.J. Milk proteins as a source of tryptophan-containing bioactive peptides. Food Funct. 2015;6:2115–2127. doi: 10.1039/C5FO00407A. [DOI] [PubMed] [Google Scholar]
- Norgaard M., Nielsen R.B., Jacobsen J.B., Gradus J.L., Stenager E., Koch-Henriksen N., Lash T.L., Sorensen H.T. Use of penicillin and other antibiotics and risk of multiple sclerosis: a population-based case-control study. Am. J. Epidemiol. 2011;174:945–948. doi: 10.1093/aje/kwr201. [DOI] [PubMed] [Google Scholar]
- Nourbakhsh B., Bhargava P., Tremlett H., Hart J., Graves J., Waubant E. Altered tryptophan metabolism is associated with pediatric multiple sclerosis risk and course. Ann. Clin. Transl. Neurol. 2018;5:1211–1221. doi: 10.1002/acn3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri M., Bredberg A., Weström B., Lavasani S. Intestinal barrier dysfunction develops at the onset of experimental autoimmune encephalomyelitis, and can be induced by adoptive transfer of auto-reactive T cells. PLoS One. 2014;9:e106335. doi: 10.1371/journal.pone.0106335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu C.C., Aldredge D.L., Lee H., Lerno L.A., Zivkovic A.M., German J.B., Lebrilla C.B. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2012;11:2912–2924. doi: 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Repáraz J., Riccardi C., Rynda A., Jun S., Callis G., Pascual D.W. Regulatory T cell vaccination without autoantigen protects against experimental autoimmune encephalomyelitis. J. Immunol. 2007;178:1791–1799. doi: 10.4049/jimmunol.178.3.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J., Mielcarz D.W., Ditrio L.E., Burroughs A.R., Foureau D.M., Haque-Begum S., Kasper L.H. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- Ochoa-Repáraz J., Mielcarz D.W., Ditrio L.E., Burroughs A.R., Begum-Haque S., Dasgupta S., Kasper D.L., Kasper L.H. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- Ochoa-Repáraz J., Mielcarz D.W., Wang Y., Begum-Haque S., Dasgupta S., Kasper D.L., Kasper L.H. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–495. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- Ochoa-Reparaz J., Magori K., Kasper L.H. The chicken or the egg dilemma: intestinal dysbiosis in multiple sclerosis. Ann. Transl. Med. 2017;5:145. doi: 10.21037/atm.2017.01.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunrinola G.A., Oyewale J.O., Oshamika O.O., Olasehinde G.I. The human microbiome and its impacts on health. Int. J. Microbiol. 2020;2020:8045646. doi: 10.1155/2020/8045646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., Mcglinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P. 2019. “vegan”: Community Ecology Package Version 2.5-6. [Google Scholar]
- Oshida K., Shimizu T., Takase M., Tamura Y., Shimizu T., Yamashiro Y. Effects of dietary sphingomyelin on central nervous system myelination in developing rats. Pediatr. Res. 2003;53:589–593. doi: 10.1203/01.PDR.0000054654.73826.AC. [DOI] [PubMed] [Google Scholar]
- Otaegui D., Mostafavi S., Bernard C.C.A., Lopez de Munain A., Mousavi P., Oksenberg J.R., Baranzini S.E. Increased transcriptional activity of milk-related genes following the active phase of experimental autoimmune encephalomyelitis and multiple sclerosis. J. Immunol. 2007;179:4074–4082. doi: 10.4049/jimmunol.179.6.4074. [DOI] [PubMed] [Google Scholar]
- Otani H., Watanabe T., Tashiro Y. Effects of bovine beta-casein (1-28) and its chemically synthesized partial fragments on proliferative responses and immunoglobulin production in mouse spleen cell cultures. Biosci. Biotechnol. Biochem. 2001;65:2489–2495. doi: 10.1271/bbb.65.2489. [DOI] [PubMed] [Google Scholar]
- O’Rourke D.R., Bokulich N.A., Jusino M.A., MacManes M.D., Foster J.T. A total crapshoot? Evaluating bioinformatic decisions in animal diet metabarcoding analyses. Ecol. Evol. 2020;10:9721–9739. doi: 10.1002/ece3.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry R.T., Stahl M., Perez-Munoz M.E., Nothaft H., Wenzel C.Q., Sacher J.C., Coros C., Walter J., Vallance B.A., Szymanski C.M. Bacterial AB 5 toxins inhibit the growth of gut bacteria by targeting ganglioside-like glycoconjugates. Nat. Commun. 2019;10:1390. doi: 10.1038/s41467-019-09362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielou E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966;13:131–144. doi: 10.1016/0022-5193(66)90013-0. [DOI] [Google Scholar]
- Pommerich U.M., Brincks J., Christensen M.E. Is there an effect of dietary intake on MS-related fatigue? – a systematic literature review. Mult. Scler. Relat. Disord. 2018;25:282–291. doi: 10.1016/j.msard.2018.08.017. [DOI] [PubMed] [Google Scholar]
- Pröbstel A.K., Baranzini S.E. The role of the gut microbiome in multiple sclerosis risk and progression: towards characterization of the “MS microbiome. Neurotherapeutics. 2018;15:126–134. doi: 10.1007/s13311-017-0587-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D59–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rajan S.K., Lindqvist M., Brummer R.J., Schoultz I., Repsilber D. Phylogenetic microbiota profiling in fecal samples depends on combination of sequencing depth and choice of NGS analysis method. PLoS One. 2019;14:e0222171. doi: 10.1371/journal.pone.0222171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsaransing G.S.M., Mellema S.A., De Keyser J. Dietary patterns in clinical subtypes of multiple sclerosis: an exploratory study. Nutr. J. 2009;8:36. doi: 10.1186/1475-2891-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rensburg S.J., Kotze M.J., Hon D., Haug P., Kuyler J., Hendricks M., Botha J., Potocnik F.C.V., Matsha T., Erasmus R.T. Iron and the folate-vitamin B12-methylation pathway in multiple sclerosis. Metab. Brain Dis. 2006;21:117–133. doi: 10.1007/s11011-006-9019-0. [DOI] [PubMed] [Google Scholar]
- Rhodes D.A., Reith W., Trowsdale J. Regulation of immunity by butyrophilins. Annu. Rev. Immunol. 2016;34:151–172. doi: 10.1146/annurev-immunol-041015-055435. [DOI] [PubMed] [Google Scholar]
- Rothhammer V., Mascanfroni I.D., Bunse L., Takenaka M.C., Kenison J.E., Mayo L., Chao C.-C., Patel B., Yan R., Blain M. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V., Borucki D.M., Tjon E.C., Takenaka M.C., Chao C.-C., Ardura-Fabregat A., de Lima K.A., Gutiérrez-Vázquez C., Hewson P., Staszewski O. Microglial control of astrocytes in response to microbial metabolites. Nature. 2018;557:724–728. doi: 10.1038/s41586-018-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- Rumah K.R., Linden J., Fischetti V.A., Vartanian T. Isolation of Clostridium perfringens type B in an individual at first clinical presentation of multiple sclerosis provides clues for environmental triggers of the disease. PLoS One. 2013;8:e76359. doi: 10.1371/journal.pone.0076359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saher G., Brügger B., Lappe-Siefke C., Möbius W., Tozawa R., Wehr M.C., Wieland F., Ishibashi S., Nave K.-A. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Iino T., Hamada M., Ohkuma M. Parolsenella catena gen. nov., sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2018;68:1165–1172. doi: 10.1099/ijsem.0.002645. [DOI] [PubMed] [Google Scholar]
- Salehipour Z., Haghmorad D., Sankian M., Rastin M., Nosratabadi R., Soltan Dallal M.M., Tabasi N., Khazaee M., Nasiraii L.R., Mahmoudi M. Bifidobacterium animalis in combination with human origin of Lactobacillus plantarum ameliorate neuroinflammation in experimental model of multiple sclerosis by altering CD4+ T cell subset balance. Biomed. Pharmacother. 2017;95:1535–1548. doi: 10.1016/j.biopha.2017.08.117. [DOI] [PubMed] [Google Scholar]
- Saresella M., Mendozzi L., Rossi V., Mazzali F., Piancone F., LaRosa F., Marventano I., Caputo D., Felis G.E., Clerici M. Immunological and clinical effect of diet modulation of the gut microbiome in multiple sclerosis patients: a pilot study. Front. Immunol. 2017;8:1391. doi: 10.3389/fimmu.2017.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Jansen T., Jacobs L., Bonder M.J., Kurilshikov A. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136.e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter J., Peled J.U., Taylor B.P., Markey K.A., Smith M., Taur Y., Niehus R., Staffas A., Dai A., Fontana E. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588:303–307. doi: 10.1038/s41586-020-2971-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt V., Enav H., Spector T.D., Youngblut N.D., Ley R.E. Strain-level analysis of Bifidobacterium spp. from gut microbiomes of adults with differing lactase persistence genotypes. mSystems. 2020;5 doi: 10.1128/msystems.00911-20. e00911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi S.K., Freedman S.N., Mangalam A.K. Gut microbiome in multiple sclerosis: the players involved and the roles they play. Gut Microbes. 2017;8:607–615. doi: 10.1080/19490976.2017.1349041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi S.K., Freedman S.N., Murra A.C., Zarei K., Sompallae R., Gibson-Corley K.N., Karandikar N.J., Murray J.A., Mangalam A.K. Prevotella histicola, a human gut commensal, is as potent as COPAXONE® in an animal model of multiple sclerosis. Front. Immunol. 2019;10:462. doi: 10.3389/fimmu.2019.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizar O., Givler A. StatPearls; 2019. Vitamin D Deficiency. [PubMed] [Google Scholar]
- Skripuletz T., Manzel A., Gropengießer K., Schäfer N., Gudi V., Singh V., Salinas Tejedor L., Jörg S., Hammer A., Voss E. Pivotal role of choline metabolites in remyelination. Brain. 2015;138:398–413. doi: 10.1093/brain/awu358. [DOI] [PubMed] [Google Scholar]
- Sonner J.K., Keil M., Falk-Paulsen M., Mishra N., Rehman A., Kramer M., Deumelandt K., Röwe J., Sanghvi K., Wolf L. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat. Commun. 2019;10:4877. doi: 10.1038/s41467-019-12776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitsberg V.L. Invited review: bovine milk fat globule membrane as a potential nutraceutical. J. Dairy Sci. 2005;88:2289–2294. doi: 10.3168/jds.S0022-0302(05)72906-4. [DOI] [PubMed] [Google Scholar]
- Stanisavljević S., Dinić M., Jevtić B., Đedović N., Momčilović M., Đokić J., Golić N., Mostarica Stojković M., Miljković Đ. Gut microbiota confers resistance of albino oxford rats to the induction of experimental autoimmune encephalomyelitis. Front. Immunol. 2018;9:942. doi: 10.3389/fimmu.2018.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefferl A., Schubart A., Storch M., Amini A., Mather I., Lassmann H., Linington C. Butyrophilin, a milk protein, modulates the encephalitogenic T cell response to myelin oligodendrocyte glycoprotein in experimental autoimmune encephalomyelitis. J. Immunol. 2000;165:2859–2865. doi: 10.4049/jimmunol.165.5.2859. [DOI] [PubMed] [Google Scholar]
- Takiishi T., Fenero C.I.M., Câmara N.O.S. Intestinal barrier and gut microbiota: shaping our immune responses throughout life. Tissue Barriers. 2017;5:e1373208. doi: 10.1080/21688370.2017.1373208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tankou S.K., Regev K., Healy B.C., Cox L.M., Tjon E., Kivisakk P., Vanande I.P., Cook S., Gandhi R., Glanz B. Investigation of probiotics in multiple sclerosis. Mult. Scler. Jour. 2018;24:58–63. doi: 10.1177/1352458517737390. [DOI] [PubMed] [Google Scholar]
- Thorburn A.N., Macia L., Mackay C.R. Diet, metabolites, and “Western-lifestyle” inflammatory diseases. Immunity. 2014;40:P833–P842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- Toghi M., Bitarafan S., Kasmaei H.D., Ghafouri-Fard S. Bifidobacteria: a probable missing puzzle piece in the pathogenesis of multiple sclerosis. Mult. Scler. Relat. Disord. 2019;36:101378. doi: 10.1016/j.msard.2019.101378. [DOI] [PubMed] [Google Scholar]
- Tremlett H., Fadrosh D.W., Faruqi A.A., Hart J., Roalstad S., Graves J., Lynch S., Waubant E., US Network of Pediatric MS Centers Gut microbiota composition and relapse risk in pediatric MS: a pilot study. J. Neurol. Sci. 2016;363:153–157. doi: 10.1016/j.jns.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity. Ecography (Cop.). 2010;33:2–22. doi: 10.1111/j.1600-0587.2009.05880.x. [DOI] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Jason P., Egholm M. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F., Peano C., Pass D.A., Foroni E., Severgnini M., Claesson M.J., Kerr C., Hourihane J., Murray D., Fuligni F. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy R. Academic-industry partnerships in addressing nutrition--[infection-immunity-inflammation] interactions. Br. J. Nutr. 2007;98(Suppl 1):S17–S23. doi: 10.1017/S0007114507832892. [DOI] [PubMed] [Google Scholar]
- Ubeda C., Lipuma L., Gobourne A., Viale A., Leiner I., Equinda M., Khanin R., Pamer E.G. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwitonze A.M., Razzaque M.S. Role of magnesium in Vitamin D activation and function. J. Am. Osteopath. Assoc. 2018;118:181. doi: 10.7556/jaoa.2018.037. [DOI] [PubMed] [Google Scholar]
- Vallet J.L., Rempel L.A., Miles J.R., Webel S.K. Effect of essential fatty acid and zinc supplementation during pregnancy on birth intervals, neonatal piglet brain myelination, stillbirth, and preweaning mortality. J. Anim. Sci. 2014;92:2422–2432. doi: 10.2527/jas.2013-7130. [DOI] [PubMed] [Google Scholar]
- Vandeputte D., Falony G., Vieira-Silva S., Tito R.Y., Joossens M., Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voreades N., Kozil A., Weir T.L. Diet and the development of the human intestinal microbiome. Front. Microbiol. 2014;5:494. doi: 10.3389/fmicb.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I., Sklar J., Jiang L., Natarajan L., Knight R., Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature. 2020;587:448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J. Murine gut microbiota-diet trumps genes. Cell Host Microbe. 2015;17:3–5. doi: 10.1016/j.chom.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Walter J., Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Walter J., Armet A.M., Finlay B.B., Shanahan F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020 doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wekerle H. Nature plus nurture: the triggering of multiple sclerosis. Swiss Med. Wkly. 2015;145:w14189. doi: 10.4414/smw.2015.14189. [DOI] [PubMed] [Google Scholar]
- Wekerle H. Brain autoimmunity and intestinal microbiota: 100 trillion game changers. Trends Immunol. 2017;38:483–497. doi: 10.1016/j.it.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Wesemann D.R., Portuguese A.J., Meyers R.M., Gallagher M.P., Cluff-Jones K., Magee J.M., Panchakshari R.A., Rodig S.J., Kepler T.B., Alt F.W. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer S., Astsaturov I., Cheung R.K., Schrade K., Gunaratnam L., Wood D.D., Moscarello M.A., O’Connor P., McKerlie C., Becker D.J., Dosch H.M. T cells of multiple sclerosis patients target a common environmental peptide that causes encephalitis in mice. J. Immunol. 2001;166:4751–4756. doi: 10.4049/jimmunol.166.7.4751. [DOI] [PubMed] [Google Scholar]
- Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H., Ren H., Zhang L., Sun X., Wang W., Zhang S., Zhao J., Ming L. Alpha-tocopherol ameliorates experimental autoimmune encephalomyelitis through the regulation of Th1 cells. Iran. J. Basic Med. Sci. 2016;19:561–566. [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Kable M.E., Marco M.L. Impact of DNA sequencing and analysis methods on 16s rRNA gene bacterial community analysis of dairy products. mSphere. 2018;3 doi: 10.1128/msphere.00410-18. 00410–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokote H., Miyake S., Croxford J.L., Oki S., Mizusawa H., Yamamura T. NKT cell-dependent amelioration of a mouse model of multiple sclerosis by altering gut flora. Am. J. Pathol. 2008;173:1714–1723. doi: 10.2353/ajpath.2008.080622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapatera B., Prados A., Gómez-Martínez S., Marcos A. Immunonutrition: methodology and applications. Nutr. Hosp. 2015;31(Suppl 3):145–154. doi: 10.3305/nh.2015.31.sup3.8762. [DOI] [PubMed] [Google Scholar]
- Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D’Angelo C., Massi-Benedetti C., Fallarino F. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zeng Q., Gong J., Liu X., Chen C., Sun X., Li H., Zhou Y., Cui C., Wang Y., Yang Y. Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem. Int. 2019;129:104468. doi: 10.1016/j.neuint.2019.104468. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang Z.-Y., Wu Y., Schluesener H.J. Valproic acid ameliorates inflammation in experimental autoimmune encephalomyelitis rats. Neuroscience. 2012;221:140–150. doi: 10.1016/j.neuroscience.2012.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coefficients were considered significant (∗∗) if the 95% confidence interval did not overlap zero. Variables with no coefficient shown for a given taxon were not present in top-ranked models (ΔAICc <2) predicting that taxon's abundance. Positive and negative coefficients indicate YBS and WBS, respectively, for diet; symptomatic and healthy marmosets, respectively, for pathology; and increases and decreases over the course of the experiment, respectively, for time.
Coefficients were considered significant (∗∗) if the 95% confidence interval did not overlap zero. Variables with no coefficient shown for a given taxon were not present in top-ranked models (ΔAICc <2) predicting that taxon's change in abundance. Positive and negative coefficients indicate YBS and WBS, respectively, for diet; symptomatic and healthy marmosets, respectively, for pathology; and increases and decreases in the magnitude of shifts in abundance, respectively, for Δ time.