Prior presentation
-
•
Poster presentation at the USCAP 108th Annual Meeting, March 16–21, 2019.
Keywords: Early-stage non-small cell lung cancer, Computational pathology, Prognostic and predictive
Abstract
Background
We developed and validated a prognostic and predictive computational pathology risk score (CoRiS) using H&E stained tissue images from patients with early-stage non-small cell lung cancer (ES-NSCLC).
Methods
1330 patients with ES-NSCLC were acquired from 3 independent sources and divided into four cohorts D1-4. D1 comprised 100 surgery treated patients and was used to identify prognostic features via an elastic-net Cox model to predict overall and disease-free survival. CoRiS was constructed using the Cox model coefficients for the top features. The prognostic performance of CoRiS was evaluated on D2 (N=331), D3 (N=657) and D4 (N=242). Patients from D2 and D3 which comprised surgery + chemotherapy were used to validate CoRiS as predictive of added benefit to adjuvant chemotherapy (ACT) by comparing survival between different CoRiS defined risk groups.
Findings
CoRiS was found to be prognostic on univariable analysis, D2 (hazard ratio (HR) = 1.41, adjusted (adj.) P = .01) and D3 (HR = 1.35, adj. P < .001). Multivariable analysis showed CoRiS was independently prognostic, D2 (HR = 1.41, adj. P < .001) and D3 (HR = 1.35, adj. P < .001), after adjusting for clinico-pathologic factors. CoRiS was also able to identify high-risk patients who derived survival benefit from ACT D2 (HR = 0.42, adj. P = .006) and D3 (HR = 0.46, adj. P = .08).
Interpretation
CoRiS is a tissue non-destructive, quantitative and low-cost tool that could potentially help guide management of ES-NSCLC patients.
Research in context.
Evidence before this study
Complete surgical excision is the standard of care treatment for early-stage (stage I and II) non-small cell lung cancer (ES-NSCLC). While current guidelines recommend adjuvant cisplatin based chemotherapy (ACT) for stage II patients, stage I patients continue to be treated with surgery alone. Benefit of ACT following surgical resection has been shown in multiple large clinical trials, with significant improvement in overall survival (OS) and Disease-free survival (DFS). Clinical parameters such as tumor stage, nodal status, age, and performance score have been traditionally shown to be prognostic, but presently there is a paucity of accurate and validated biomarkers based off clinicopathologic factors which can identify patients who would benefit from ACT. With the advent of digital pathology and the corresponding increase in machine learning and computerized pathology image analysis, there is the opportunity to mine and associate quantitative features relating to tumor morphology to cancer prognosis and outcome. A few groups have recently shown that machine learning based prognostic classifiers can predict disease recurrence and survival in the context of NSCLC.
Added value of this study
In this manuscript, we present a computational derived image risk score (CoRiS) from hematoxylin and eosin (H&E) stained whole slide images derived from surgical specimens, that is prognostic of disease-free and overall survival, and also predictive of added benefit of adjuvant chemotherapy (ACT) in early stage (stage I and II) non-small cell lung cancer (ES-NSCLC). To the best of our knowledge, this is the first computational pathology based work that was validated to be not only prognostic but also predictive of added benefit of ACT on multi-site ES-NSCLC data set with over 1000 cases.
Implications of all the available evidence
The CoRiS presented in this study could be potentially used as an inexpensive, tissue non-destructive, prognostic and predictive companion diagnostic for ES-NSCLC to identify patients with high risk for ACT.
Alt-text: Unlabelled box
1. Introduction
Early-stage non-small cell lung cancer (ES-NSCLC) usually comprises of stage I and II cancers and complete surgical excision is the standard of care treatment for these patients [1], [2], [3]. While current guidelines recommend adjuvant cisplatin based chemotherapy (ACT) for stage II patients, stage I patients continue to be treated with surgery alone. Benefit of ACT following surgical resection has been shown in multiple large clinical trials [1], [2], [3], with significant improvement in overall survival (OS) and Disease-free survival (DFS) for the ACT group in ES-NSCLC. A large pooled meta-analysis of these trials – the Lung Adjuvant Cisplatin Evaluation (LACE) [8] including 4584 patients across five trials revealed a 5-year benefit of 5.4% from ACT with the hazard ratio (HR) for OS of 0.89 (95% CI, 0.82–0.96; P = .005), with a median follow-up time of 5.2 years. Interestingly, some trials including the Adjuvant Lung Project Italy (N=1209) and European Big Lung (N=381) trials failed to find statistically significant differences in survival between the surgery only and the ACT group in ES-NSCLC [9,10]. One possible reason is the lack of predictive biomarkers to identify patients who would derive benefit from ACT.
Stratified subgroup analysis of these trials based on the American Joint Committee on Cancer (AJCC) 6th edition tumor stage meanwhile has shown that ACT does not lead to a significantly improved OS in stage IB (T2aN0M0) patients (HR =0.92; 95% CI 0.78–1.10) [8]. Based on the lack of significant survival benefit demonstrated in stage I (and sometimes detriment in stage IA – HR>1) with ACT, ACT is currently not recommended following surgery in stage I patients [11]. However, even after curative resection about 40% of stage I patients tend to recur [12,13], possibly indicating these are patients at increased risk of disease recurrence and therefore might benefit from ACT.
While clinical parameters such as tumor stage, nodal status, age, and performance score have been traditionally shown to be prognostic [14], presently there is a paucity of accurate and validated biomarkers based off clinicopathologic factors which can identify patients who would benefit from ACT. Companion diagnostic assays like those from Myriad [15] are tissue destructive and expensive and not routinely ordered for every lung cancer patient. While there are a number of multi-gene based prognostic biomarkers, the few existing biomarkers for predicting survival benefit of ACT are molecular or multi-gene based assays [16], [17], [18], [19].
With the advent of digital pathology and the corresponding increase in machine learning and computerized pathology image analysis, there is the opportunity to mine and associate quantitative features relating to tumor morphology to cancer prognosis and outcome. A few groups have recently shown that machine learning based prognostic classifiers can predict disease recurrence and survival [4], [5], [6], [7] in the context of NSCLC. However, none of these approaches has been evaluated in their ability to predict added benefit of ACT in ES-NSCLC.
In this work, we present a computational pathology risk score (CoRiS) that employs quantitative image features relating to shape, size, and morphology of cancer nuclei derived from digitalized hematoxylin and eosin (H&E) stained images of resected ES-NSCLC tissue specimens to predict OS and DFS. Using a total of 1330 ES-NSCLC patients from 3 sites, treated either with surgery+ACT or surgery alone, we demonstrate that CoRiS is both (a) prognostic of OS and DFS and (b) associated with added benefit of ACT in ES-NSCLC patients.
2. Patients and methods
2.1. Ethics statement
An Institutional review board (IRB) approved protocol was used for the retrospective analysis, and the informed consent requirement was waived by the IRB. The study was compliant with the Health Insurance Portability and Accountability Act (HIPPA). All data used in this study were de-identified and no protected health data was needed.
2.2. Patients
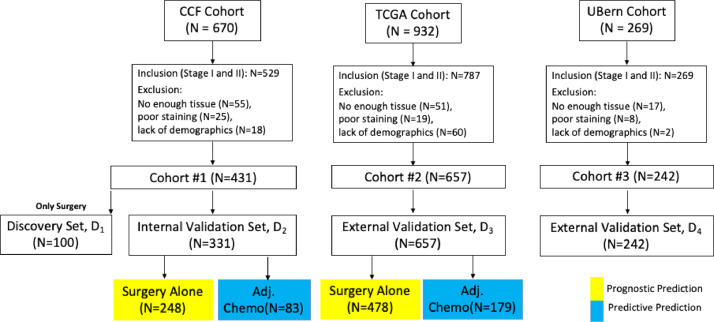
Retrospective chart review of patients continuously admitted in the Cleveland Clinic Foundation (CCF) with NSCLC between 2005–15 yielded 670 patients. All resected stage I and II NSCLC were included in the study, however those tissue slides which following pathological evaluation did not meet quality requirements such as poor staining and lack of sufficient tissue were excluded (flow diagram, Fig. 1). This process resulted in rendering 431 ES-NSCLC patients suitable for the analysis. Out of these, 83 patients received ACT. 100 patients with surgery alone formed the discovery cohort (D1) while the remaining N=331 formed the validation cohort (D2). The Cancer Genome Archive (TCGA)-lung adenocarcinomas (ADC; N=523) and TCGA-lung squamous cell carcinomas (SCC; N=409) cohorts are publicly available datasets, assembled from different institutions [20]. After applying the inclusion and exclusion criteria, a TCGA derived independent validation cohort D3 (N=657; ADC=378, SCC=279) was identified. In D3, 179 patients received ACT. Additionally, a cohort of N=269 consecutive, primary resected ES-NSCLC patients from the University of Bern was used for prognostic validation and formed D4 (N=242 after applying the inclusion and exclusion criteria). D4 comprised only SCC cases and all of the patients underwent surgery with 62 patients received ACT. Patient demographics and characteristics are summarized in Table 1. (Fig. 2).
Fig. 1.
Patient Selection Diagram from 3 different cohorts: D1 with 100 surgery alone patients were used as discovery set; and D2-4 were used as validation set.
Table 1.
Patients’ demographics and characteristics.
| Variable | Sub variables | D1N(%) | D2N(%) | D3N(%) | D4N(%) | Total N | Adj. P |
|---|---|---|---|---|---|---|---|
| Number of patients | 100 | 331 | 657 | 242 | 1330 | ||
| Age (mean+/-std year) | unknown | unknown | 64 +/-15 | 67+/-8 | 65+/-13 | <0.001 | |
| Gender | Male Female |
unknown | unknown | 438(66.7) 219(33.3) |
207(85.5) 35(14.5) |
645 254 |
<0.001 |
| Tumor Size (mean+/-std mm) | 39.6+/- 98.6 | 33.3+/-34.0 | unknown | 46.61+/-23.38 | 39.36+/-46.66 | 0.002 | |
| Smoking Status | Previous/Current Never |
88(88) 12(12) |
267(80.7) 64(19.3) |
580(88.3) 77(11.7) |
unknown | 935 153 |
0.199 |
| pN | 0 1 Unknown |
82(82) 8(8) 10(10) |
230(69.5) 61(18.4) 40(12.1) |
491(74.7) 155(23.6) 11(1.7) |
152(62.8) 88(36.4) 2(0.8) |
955 312 63 |
0.294 |
| pT | 1 2 3 Unknown |
47(47) 44(44) 8(8) 1(1) |
163(49.2) 134(40.5) 33(10.0) 1(0.3) |
295(44.9) 310(47.2) 52(7.9) |
60(24.8) 162(66.9) 20(8.3) |
565 650 113 2 |
0.332 |
| Overall Stage | IA IB I IIA IIB II |
45(45) 32(32) 0(0) 14(14) 8(8) 1(1) |
108(32.6) 110(33.2) 3(0.9) 66(19.9) 41(12.4) 3(0.9) |
223(33.9) 191(29.1) 5(0.8) 141(21.5) 90(13.7) 7(1.1) |
65(26.9) 31(12.8) 1(0.4) 75(31.0) 64(26.4) 6(2.5) |
441 364 9 296 203 17 |
<0.001 |
| Treatment |
Surgery Only Surg. + Chemo |
100(100) 0(0) |
248(74.9) 83(25.1) |
478(72.8) 179(27.2) |
180(74.4) 62(25.6) |
1006 324 |
<0.001 |
| Recurrence | Non-recurrence Recurrence |
78(78) 22(22) |
249(75.2) 82(24.8) |
519(79.0) 138(21.0) |
145(59.9) 97(40.1) |
991 339 |
0.233 |
| Tumor types | Adenocarcinoma SCC Others |
10(10) 88(88) 2(2) |
130(39.3) 20(6.0) 181(54.7) |
378(57.5) 279(42.5) |
0(0) 242(100) 0(0) |
518 629 183 |
0.284 |
Abbreviations: SCC, squamous cell carcinoma; Surg. + Chemo: surgery and chemotherapy; Adj. p: adjusted p; pT: pathological tumor stage; pN: pathological nodal stage.
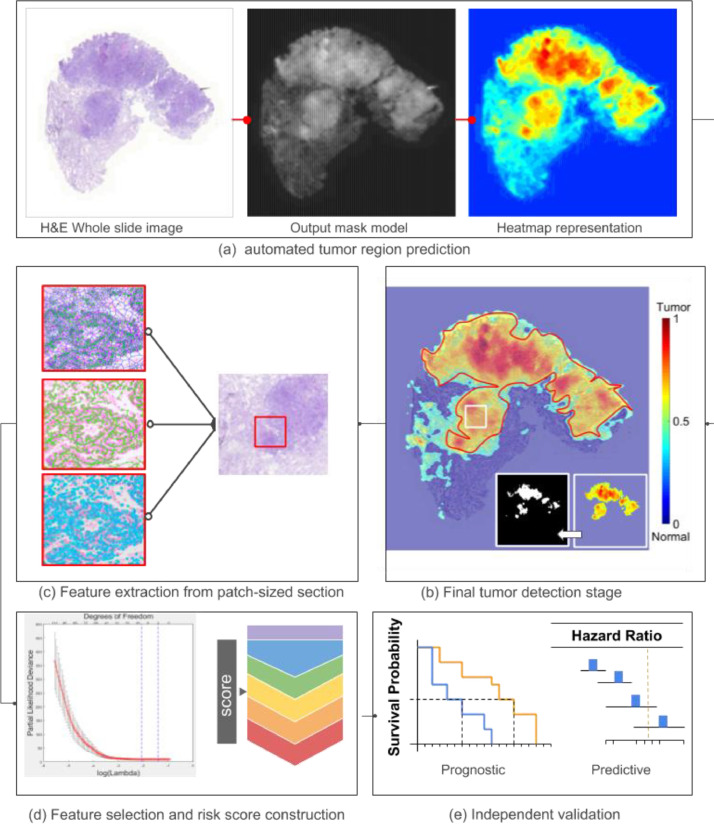
Fig. 2.
Flowchart of overall workflow: (a) automated tumor region prediction; (b) image tile extraction from tumor and (c) nuclei and peri-nuclei features extraction; (d) feature selection and CoRiS calculation; (e) prognostic and predictive validation of CoRiS.
2.3. Image acquisition
Whole slide images (WSI) obtained from routine H&E diagnostic tissue slides of the primary tumor were collected for D1, D2 and D3. D4 was collected in the form of tissue microarrays (TMA) to represent the core of the tumor. The H&E slides in D1 and D2 were scanned using a Roche-Ventana iScan HT scanner (serial #: BI15N7205) at a magnification of 20x. D3 was a publicly acquired dataset from multiple institutions and heterogeneous scanners. The different pathology labs that contributed studies to the TCGA likely used different vendors for the whole slide scanning, unfortunately the specific scanner make and model for the individual TCGA sites were not available. For our analysis, the image and feature analysis were consistently performed at 20x magnification for all datasets. D4 was in the form of TMAs scanned at 40x (down-sampled to 20x) by Panoramic Digital Slide Scanner 250 (version: 1.23.1.71684).
2.4. Automatic tumor detection and segmentation of cancer nuclei and perinuclear region
A U-Net based convolutional neural network was employed for segmentation with adversarial training (training rates are 0.001 and 0.01 for regular and adversarial training, respectively) [21]. Two different U-net based models were trained for tumor detection and nuclei segmentation respectively. While the tumor detector was used to generate the heat map for WSI to indicate the probability of tumor, the nuclei segmentation model was used to delineate the boundary pixels of each nuclei. The perinuclear region was segmented by taking 15 pixels at 20x magnification outward from the boundaries of nuclei. Each of WSI was sliced into 2000 by 2000 pixels consecutive tiles and only the tiles from the detected tumor regions were used to represent each patient. The ground truth set of nuclei and tumor was generated by two pathologists from University Hospital. For nuclei segmentation, 8000 nuclei were annotated from 100 digitized H&E images of breast (40) and lung cancer (60). For tumor annotation, 125 whole slide sections of lung cancer were manually annotated, 80 of them were used for training and rest 45 were used for validation. The tumor detector achieved 90.6% patch-level accuracy on a ground truth set curated by pulmonary pathologists. Meanwhile, nuclei segmentation model yielded an F-score 0.88, comparable to current state-of-the-art nuclear segmentation algorithm [22]. All U-Net based segmentations were implemented in Tensorflow 1.6 on Nvidia Titan XP GPU clusters (network details are specified in supplementary Table S3).
2.5. Quantitative feature extraction
We extracted 242 nuclear descriptors from previously segmented nuclei. These features corresponded to five categories: nuclei shape [23], orientation entropy [24], texture, local and global graph [25]. On the one hand, shape features included basic measurements of nuclear area, perimeters and mathematical descriptors of contour. Orientation entropy and texture on the other hand characterized directionality coherence and pixel intensity distribution of the nuclei [24]. While local graph measured the architecture in relation to neighborhood cells, global graph captured the arrangement in relation to the entire WSI [6]. A graph was a mathematical operation which included a set of nodes (nucleus) to capture relationships through pairwise edges formed between the nodes (Details in supplementary Table S1).
Another 35 peri-nuclear features from the adjoining cytoplasmic area were also extracted. These included quantitative measurements including area, area ratio of the peri-nuclear region to the nuclear region, pixel intensities as well as texture [26]. These features would not only characterize the space adjoining the nuclei in the cancer cell but would also highlight relationships between the nucleus and cytoplasm in the cancer cell (Details in supplementary Table S2). Finally mean, standard deviation, min and max value of patch-level features were calculated and concatenated to generate a patient level image signature. The feature extraction was implemented in MatLab 2020a.
2.6. Constructing the CoRiS risk score
A total of 277 nuclear and peri-nuclear features were extracted for each patient in D1. In order to regularize the number of features proportionate to samples size [27], the top discriminative features were selected by Elastic-Net regularization with non-zero coefficients and these features were fit into Cox Proportional Hazard model with OS and DFS as the outcomes of interest, respectively. The CoRiS was computed by a weighted linear combination of selected features and their corresponding coefficients. The tradeoff value alpha (mixing parameter) between L1 and L2 for elastic net was evaluated from 0 to 1 with step size of 0.1. It was determined that 0.8 would be the optimal value. The optimal value of the tuning parameter in the Elastic-Net Cox (lambda) was determined by 10-fold cross validation in D1.
2.7. Statistical analysis
OS was measured from the date of diagnosis to the date of death and censored at the date of last follow-up for survivors. DFS was calculated from the date of surgery to the date of recurrence or death whichever occurred earlier and censored at the date of last follow-up for those still alive without recurrence. The CoRiS was divided into low and high risk based on the median value of CoRiS classifying OS/DFS obtained on D1. Further stratification of CoRiS was done by dividing it into four groups (H, IH, IL, L) based on quartile values from training CoRiS. High-risk group comprised the upper two quartiles, high (H) and intermediate high (IH), while low risk comprised lower two quartiles, intermediate low (IL) and low (L). Univariable analysis of CoRiS and the clinicopathologic variables (i.e. smoking history, tumor subtypes, pathological stage) were conducted. Multivariable Cox-regression models were built to assess the relationships between the various covariates and OS/DFS while adjusting for baseline factors [28]. Forest plots were constructed to show the HRs comparing OS between ACT and the surgery alone group in all the cohorts with patients being stratified based on the quartiles of CoRiS. Further subset analysis involved looking at survival differences in the different AJCC stages and subtypes of tumor, i.e. SCC and ADC. Kaplan-Meier survival curves were obtained to visualize the differences based on CoRiS and Hazard Ratios were computed. All p-values were adjusted based on Benjamin and Hochberg's procedure[29,30] and a significance level of 0.05 was set to be statistically significant. Statistical analysis was implemented in R 3.6.1.
3. Experiments and results
3.1. CoRiS predicts OS and DFS on validation sets, independent of clinicopathologic factors
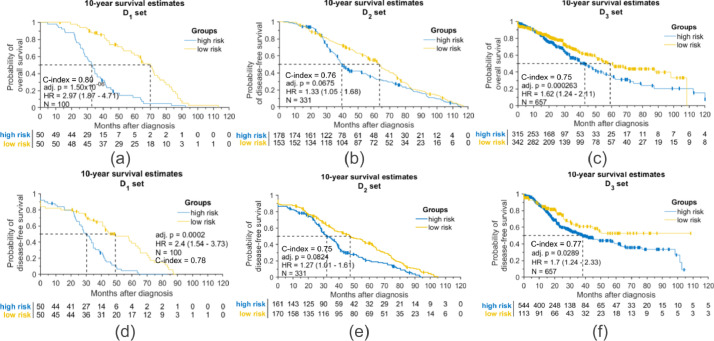
Eleven most discriminative features were selected to construct CoRiS (Figs. S1–S2). CoRiS, (Fig. 3) was found to be prognostic of both OS (D1, HR = 2.97, 95% CI: 1.87–4.71, adjusted (adj.) P < .001; D2, HR = 1.33, 95% CI: 1.05–1.68, adj. P = .067; D3, HR = 1.62, 95% CI: 1.24–2.11, adj. P < .001; D4, HR = 1.54, 95% CI: 1.03-2.3, adj. P = .082, Figs. 3 and S5) and DFS (D1, HR = 2.4, 95% CI: 1.54–3.73, adj. P < .001; D2, HR = 1.27, 95% CI: 1.01–1.61, adj. P = .082; D3, HR = 1.7, 95% CI: 1.24-2.33, adj. P = .028; D4, HR = 1.54, 95% CI: 1.05–2.25, adj. P = .082, Figs. 3 and S6). On univariable analysis, only CoRiS was significantly prognostic among all test sets (Table 2). Kaplan-Meier (KM) curves for predicting OS/DFS by CoRiS are shown in Fig. 3 for D1–D3. On multivariable analysis with controlling covariates, CoRiS was found to be independently prognostic (D2, HR = 1.24, 95% CI: 1.06-1.39, adj. P = 0.01 and D3, HR = 1.14, 95% CI: 1.02–1.28, adj. P = 0.01 Table 2). In addition, CoRiS could separately predict the OS/DFS in two major NSCLC subtypes, ADC and SCC. Details of the subtype analysis are included in Figs. S5–S8.
Fig. 3.
Kaplan-Meier plots showing prognostic effect of CoRiS with (a–c) patients’ overall survival in D1-3 and (d–f) disease free survival in D1-3. (Interaction p-value between treatment and risk score in overall survival for D1, D2, D3 was 0.17, 0.37, and 0.51, respectively; in disease free survival, interaction p-values were 0.2 for D1, 0.35 for D2 and 0.46 for D3.)
Table 2.
Univariable and multivariable with overall survival analysis on validation set (D2 and D3).
| Dataset | Variable | Univariable Analysis |
Multivariable Analysis |
||
|---|---|---|---|---|---|
| HR (95% CI) | Adj. P | HR (95% CI) | Adj. P | ||
| D2 | Nonsmoker vs Previous/Current Smoker | 1.34(1.00-1.80) | 0.05 | 1.28(0.94-1.76) | 0.14 |
| Subtypes ADC vs SCC |
0.97(0.84-1.12) | 0.64 | 0.93(0.80-1.09) | 0.40 | |
| Overall Stage IA IB IIA IIB |
Reference 1.10(0.75-1.63) 1.27(0.94-1.71) 1.45(1.06-1.98) |
0.68 0.20 0.10 |
Reference 1.05(0.67-1.67) 1.23(0.90-1.68) 1.41(0.98-1.98) |
0.81 0.24 0.08 |
|
| Tumor Size (mm) | 1.00(1.00-1.01) | 0.21 | 1.00(0.99-1.01) | 0.95 | |
| Treatment Surg vs Surg + ACT |
1.33(0.99-1.79) | 0.06 | 1.38(0.97-1.83) | 0.10 | |
| Risk score (CoRiS) | 1.41(1.08-1.84) | 0.01 | 1.24(1.06-1.39) | 0.01 | |
| D3 | Nonsmoker vs Previous/Current Smoker | 1.04(0.56-1.41) | 0.80 | 1.07(0.56-2.05) | 0.85 |
| Gender Male vs Female |
1.09(0.76-1.57) | 0.64 | 1.24(0.56-2.01) | 0.30 | |
| Age (years) | 1.00(0.99-1.02) | 0.55 | 1.01(0.99-1.02) | 0.68 | |
| Subtypes ADC vs SCC |
0.89(0.68-1.16) | 0.38 | 0.80(0.53-1.20) | 0.30 | |
| Overall Stage IA IB IIA IIB |
Reference 1.11(0.57-2.17) 1.35(0.83-2.19) 1.57(0.97-2.55) |
0.81 0.30 0.16 |
Reference 1.13(0.58-2.21) 1.29(0.79-2.11) 1.64(0.97-2.67) |
0.80 0.51 0.11 |
|
| Treatment Surg vs Surg + ACT |
1.02(0.76-1.37) | 0.89 | 1.21(0.71-1.98) | 0.51 | |
| Risk score (CoRiS) | 1.35(1.15-1.59) | 2.52e-4 | 1.14(1.02-1.28) | 0.01 | |
Abbreviations: ADC, adenocarcinoma; SCC, squamous cell carcinoma; Surg, surgery; ACT: adjuvant chemotherapy; HR, hazards ratio; CI, confidence interval; adj. p: adjusted p.
Note: HR standards for hazard ratio; values in bold are statistically significant by two-tailed test, p < 0.05
3.2. CoRiS predicts ACT benefit in two independent validation sets
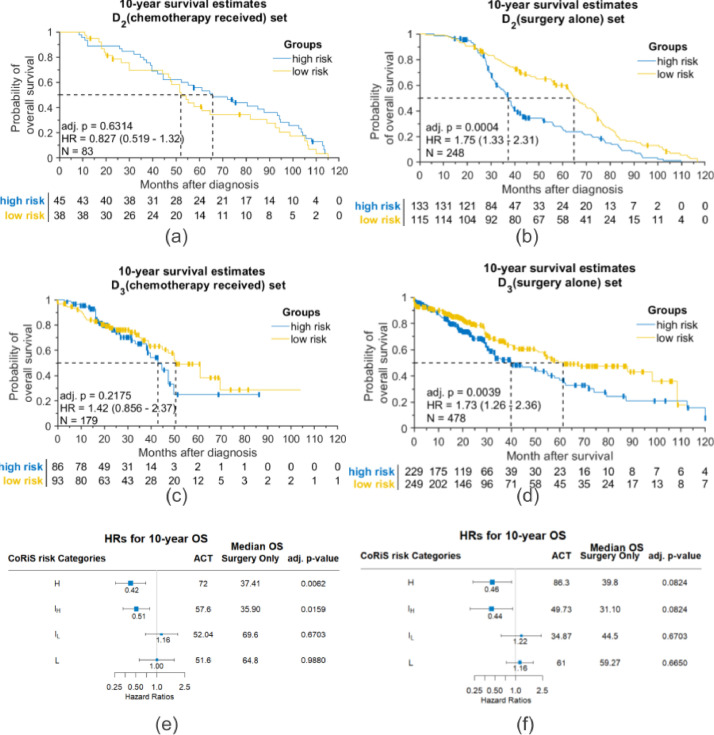
CoRiS classified 38 and 45 patients who received ACT into low-risk and high-risk groups based on median CoRiS in D2 (Fig. 4a). Similarly, in D3, CoRiS classified 93 and 86 of those patients who received ACT into low-risk and high-risk groups respectively (Fig. 4c). Survival comparisons between the groups (low-and high risk) in patients who received ACT showed no statistically significant difference in OS for both D2 (HR = 0.83 95% CI: 0.52–1.32, adj. P = .631, Fig. 4a) and D3 (HR = 1.42, 95% CI: 0.86–2.37, adj. P = .218, Fig. 4c). In contrast, for patients who underwent surgery alone without ACT, there was a statistically significant difference in OS between the low and high risk groups in D2 (HR = 1.75, 95% CI: 1.33–2.31, adj. P < .001, Fig. 4b) and D3 (HR = 1.73, 95% CI: 1.26–2.36, adj. P = .004, Fig. 4d). Results for DFS were similarly significant (see Supplementary Fig. S9). Granular analysis of CoRiS showed the patients with increased risk (H and IH) tended to have longer survival when ACT was administered. The H group showed improved median OS by about 35 months (95% longer) in D2 (HR=0.42, 95% CI: 0.26–0.69, adj. P = .006, Fig. 4e) and 46 months (115% longer) in D3 (HR=0.46, 95% CI: 0.24–0.87, adj. P = .082, Fig. 4f) between ACT and surgery alone patients. In IH group, median OS was found to be higher by 21 months (58% longer) in D2 (HR=0.51, 95% CI: 0.33–0.78, adj. P = .016, Fig. 4e) and 19 months (61% longer) in D3 (HR=0.44, 95% CI: 0.22–0.91, adj. P = .082, Fig. 4f) when ACT was given. In the IL and L groups, the ACT population showed worse survival as compared to the surgery alone group but was not statistically significant (adj. P > 0.05) in both D2 and D3 (Fig. 4 e, f). Estimated survival benefit according to DFS was similar to OS, higher CoRiS showing an estimated DFS benefit in D2 (HR=0.36, 95% CI:0.20–0.66, adj. P = .015) and D3 (HR=0.45, 95% CI: 0.24–0.86, adj. P = 0.082), respectively (Fig. S9 e, f).
Fig. 4.
Kaplan-Meier plots showing predictive effect in CoRiS defined different risk of overall survival groups: (a) patients received adjuvant chemotherapy in D2; (b) patients only received surgery in D2; (c) patients received adjuvant chemotherapy in D3; (d) patients only received surgery in D3. Forest plots of different CoRiS defined risk of overall survival groups in (e) D2 and (f) D3.
On subset analysis by stage (stage IA, IB and II), CoRiS was predictive of survival benefit to ACT, suggesting that only high-risk patients received benefit to ACT, with either no advantage or potential negative impact of ACT in the low-risk group (Figs. S3 and S4, complete results by stage).
4. Discussion
Due to contradictory results from multiple clinical trials, ACT is currently not recommended in stage IA while there is controversy regarding its use in stage IB patients [1,2,11,[16], [17], [18], [19]]. While the American Society of Clinical Oncology guidelines do not recommend ACT in stage IB patients, the NCCN guidelines currently only recommend ACT in stage IB patients with a high risk of recurrence[11,31]. There is thus a need for predictive biomarkers to identify tumors at higher risk of recurrence and would be potential candidates for ACT. Identifying the low risk patients who will do well with surgery alone would spare them from toxicity of ACT.
Amongst the existing biomarkers in NSCLC, most are prognostic and reliant on molecular or multi-gene assays. For instance, many studies showed that class III β-tubulin expression, abnormalities in the k-ras oncogene and p53 tumor suppressor gene, and DNA methylation markers could potentially identify the high-risk patients who would benefit from ACT [32,33]. The only known molecular assay predictive of benefit to ACT was published by Zhu et al. [19] who showed that a 15-gene signature was not only prognostic but predicted improved survival after ACT in signature defined high-risk patients (HR = 0.33; 95% CI, 0.17 to 0.63; P = .0005), but not in low-risk patients (HR = 3.67; 95% CI, 1.22 to 11.06; P = .0133; interaction P < .001) [19]. However, all the mentioned biomarkers are tissue destructive, expensive and time-consuming involving RNA expression and microarray profiling analysis.
In this work we presented CoRiS, the first of its kind digital pathology based companion diagnostic test, which is not only prognostic but also predictive of added benefit of ACT in ES-NSCLC. The CoRiS comprises 11 features relating to nuclei and peri-nuclear histomorphometric attributes obtained from digitized H&E tissue images. We used a group of resected ES-NSCLC without ACT to train CoRiS as prognostic model. CoRiS was further independently validated on multiple sets (independent of clinical factors such as tumor stage and smoking history; see Table 2).
For predicting benefit to ACT, the two top CoRiS groups (H, IH) showed statistically significant survival benefit for validation set D2. While CoRiS did not yield the same significant survival difference after p-value adjustment (adj. P<0.1) on D3, there was a clear trend that patients who received ACT had a longer median survival time. In fact, the highest risk CoRiS group (H) showed >90% median OS improvement (Fig. 4e, f) for the ACT as compared to the surgery alone patients. Interestingly, the low CoRiS groups (IL, L) across the validation cohorts showed no statistically significant differences in HR between the surgery alone and ACT groups, and in some cases showed detrimental effects of ACT (HR>1; Fig. 4e, f). This seems to suggest that patients in the CoRiS low group (L) would do equally well with surgery alone and can be spared the deleterious effects of ACT.
A subgroup analysis on stage IB patients showed that CoRiS divided D2-4 into low risk (Fig. S3d) and high risk (Fig. S3c) groups. This illustrates that the patients who received ACT had reduced hazard of dying in high risk group but in the low risk group, no survival difference between two cohorts of patients (with or without ACT) was identified with HR = 0.96 and adj. P = 0.899. In addition, the CoRiS defined high-risk group had significantly improved OS with ACT versus surgery alone (Fig. 4e). However, the low risk CoRiS group had no additional benefit with ACT (Fig. 4e). While the CALGB9663 and the LACE meta-analysis showed a small but non-significant statistical benefit to HR in stage IB with ACT, the IALT and the JBR10 trials did not show OS differences in stage IB [2,4,5]. These results thus seem to suggest a basis for the non-significant benefit to ACT in the completed clinical trials in stage IB patients. The combination of two distinct risk groups within a homogenous clinically defined stage could be a possible reason for the low benefit to ACT seen in published studies.
Meanwhile, in stage II patients where the present recommendation is ACT following resection, the CoRiS signature identified a low-risk group that did not have a significantly different HR when compared to the surgery alone group (Fig. S3f), thus potentially identifying and unveiling a group with relatively good survival that might be spared the toxicity of ACT.
Machine learning approaches have been applied to digital pathology images for different cancer types to prognosticate patients’ outcome[21,22]. To the best of our knowledge, CoRiS, is different from previous works[19,34] in that it is not just prognostic of risk of recurrence but also predictive of added benefit of ACT for early stage NSCLC. Wang et al. demonstrated that nuclear shape and texture features based off H&E biopsy TMAs could identify patients who would recur following surgery in early-stage NSCLC [6]. Meanwhile, Corredor et al. showed that the spatial architecture of tumor infiltrating lymphocytes (TILs) was prognostic of recurrence free survival in several independent validation datasets, pathologist based TIL estimation by comparison was not prognostic for those datasets [4]. The selected features for CoRiS includes descriptors characterizing nuclei arrangement and abundance. For example, the average area of the Voronoi diagram obtained by connecting nuclei could allow for capture of the number and proximity of nuclei within the tumor. In addition, the texture feature (Haralick) characterizing the peri-nuclei region might be reflective of the coherence of extracellular staining, in other word, less aggressive tumor might present with more homogenous cytoplasm formation. This work was significantly different from previous related publications [4,6,23] by (a) the features comprising CoRiS included not just the morphology and spatial arrangement (i.e. Voronoi and cell cluster graph) of nuclei but also a set of innovative peri-nuclear features (texture features from peri-nuclei region); (2) CoRiS has shown to be not only prognostic to patients’ outcome but also predictive to benefits of ACT; and (3) CoRiS was validated on over 1000 patients from multiple different institutions.
The study did have its limitations. Firstly, CoRiS was developed and validated using retrospective data from different institutions, which means the pathological staging criteria applied might have varied at the time of tissue examination [35], additionally at least a few demographic related parameters were not available for some of the datasets. Secondly, for predicting benefit to ACT, the surgery only and the surgery+ACT groups used in the analysis were not strictly and homogeneously controlled (including ACT protocol), it is likely that the assignment and protocol of ACT might have differed across the institutions considered in this study. Recently, transfer learning based approaches have been applied in tumor detection and classification [36,37]. An avenue for future investigation might involve the use of transfer learning, potentially leveraging other data streams like quantitative immunofluorescence, for the problems of cancer prognosis and response prediction. While the difference of median OS between CoRiS defined low and high risk group is over 90% in D3, the survival benefit between two groups is not significant after p-value adjustment. However, the effect sizes (HRs) of CoRiS across the validation datasets (D2 and D3) are similar (see Table 2). Multiple test correction using FDR approach was done only for each task separately rather than done considering all related survival prediction tasks together. As a result, the family-wise error rate was not controlled at 0.05 level for all classification tasks. In addition, we focused on features relating to nuclei from within cancer-identified regions on H&E images without differentiating cell types (i.e. tumor and lymphocyte cells). Computerized discrimination of the different cell type categories (i.e. lymphocytes, cancer nuclei, fibroblasts, macrophages) can be challenging on H&E images alone, and we were cautious to include another possible confounding variable to our predictor. Manually checking the fidelity of the detected lymphocytes on over 1,000 H&E WSIs from multiple institutions was clearly not feasible. Additionally, we did not have access to immunohistochemistry (IHC) or quantitative multiplex immunofluorescence (qmIF) images that would have allowed us to better define and employ features from different immune cell subtypes (e.g. CD4, CD3, and CD20). Clearly an avenue for future investigation will be the possible combination of features from H&E images along with corresponding features from IHC and qmIF images. Deep learning has shown better performance in different tumor segmentation tasks compared to hand-crafted based approaches [38,39]. However, in detecting tumor regions directly from whole slide images (WSI), our approach based on U-Net and adversarial training achieves comparable results (Table S4) to recently published deep learning methods in terms of both accuracy and computational efficiency [40]. For clinical utility and deployment, CoRiS needs to be prospectively validated, and needs to be applied on clinical trials with randomly assigned patients to surgery and surgery+ACT to truly validate its utility in predicting benefit to ACT.
In summary, we developed and validated an 11-feature prognostic and predictive signature for ACT benefit in patients with ES-NSCLC. With additional validation, possibly in the context of clinical trials like JBR10 and IALT[4,7], CoRiS could be validated as an inexpensive, tissue non-destructive, prognostic and predictive companion diagnostic for ES-NSCLC that could possibly have global impact.
Data sharing statement
The CoRiS codes and related data during the current study were available at:
Nuclei segmentation codes: https://github.com/maberyick/nucleiSegmentationHEDL
Nuclei data: https://cwru.app.box.com/s/3pf10foxvpngzgznfwztviif9b5uaxr4
Tumor detection codes: https://github.com/maberyick/TumorSegmentationHE_UNET
Tumor data: https://cwru.app.box.com/s/vq1q01xd6cifjlb8vv56tqbn1nb340dp
Quantitative feature codes: https://github.com/maberyick/periNuclearHE
Declaration of Competing Interest
X.W., K.B., C.B., Y. Z., C.L., P.V., P. F., M.Y., H.C. disclosed no conflicts of interest. S.B. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: institution has grants or grants pending with Roche and Basilea; institution has received payment for conference organization from Roche, MSD, AstraZeneca, Agilent and Biosystems; is on the speakers bureaus of MSD, AZ and Roche; has received payment for the development of educational presentations from Roche. Other relationships: disclosed no relevant relationships. V.V. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a consultant for BMS, Genentech, Astrazeneca, Celgene, Foundation Medicine, Taekeda, Merck, Alkermes, and Nektar Therapeutics; institution has grants or grants pending with Astrazeneca, Merck, BMS, Genentech, and Alkermes; is on the speakers bureaus of Novartis, BMS, Celgene, and Foundation Medicine; has received payment for the development of educational presentations from BMS and Foundation Medicine. Other relationships: disclosed no relevant relationships. A.M. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is on the board of and is a consultant to Inspirata; institution has three NCI RO1 grants with Inspirata and one ongoing U24 with PathCore; Elucid Bioimaging and Inspirata has licensed some of institutions’ patents (both Case Western Reserve University and Rutgers University); receives royalties from Elucid Bioimaging and Inspirata; holds stock equity in Elucid Bioimaging and Inspirata. Other relationships: disclosed no relevant relationships.
Acknowledgments
Contributors
X.W., C.B., Y. Z., C.L., P.V., P. F., A.M. designed and conducted the experiments. X.W., K.B., C.B., A.M. wrote the manuscripts. M.Y., R.S., S.B., H.C., V.V. acquired and provided the image and clinical data and also the medical writing guidance. All authors read and approved the final version of the manuscript and have verified the underlying data.
The funders of this study had no role in the research design, data collection, data analysis, data interpretation, and paper writing. The corresponding authors had full access to all data obtained from this study and had final responsibility for the decision to submit for publication.
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute under award numbers 1U24CA199374-01, R01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01, 1U54CA254566-01, National Heart, Lung and Blood Institute 1R01HL15127701A1, National Institute of Biomedical Imaging and Bioengineering 1R43EB028736-01, National Center for Research Resources under award number 1 C06 RR12463-01, VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404), the Kidney Precision Medicine Project (KPMP) Glue Grant, the Ohio Third Frontier Technology Validation Fund, the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, The Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. Sponsored research agreements from Bristol Myers-Squibb, Boehringer-Ingelheim, and Astrazeneca.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103481.
Appendix. Supplementary materials
References
- 1.Arriagada R., Dunant A., Pignon J.P. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J Clin Oncol. 2010;28:35–42. doi: 10.1200/JCO.2009.23.2272. [DOI] [PubMed] [Google Scholar]
- 2.Arriagada R., Bergman B., Dunant A. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 3.Winton T., Livingston R., Johnson D. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 4.Corredor G., Wang X., Zhou Y. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2019;25:1526–1534. doi: 10.1158/1078-0432.CCR-18-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu C., Wang X., Prasanna P. Proceedings of the 21st international conference medical image computing and computer assisted intervention. Vol. 2018. Springer Verlag; 2018. Feature driven local cell graph (FeDeG): predicting overall survival in early stage lung cancer. pp. 407–416. [Google Scholar]
- 6.Wang X., Janowczyk A., Zhou Y. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci Rep. 2017;7 doi: 10.1038/s41598-017-13773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu K.H., Zhang C., Berry G.J. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pignon J.P., Tribodet H., Scagliotti G.V. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE collaborative group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 9.Scagliotti G.V., Fossati R., Torri V. Randomized study of adjuvant chemotherapy for completely resected stage I, II, or IIIA non-small-cell Lung cancer. J Natl Cancer Inst. 2003;95:1453–1461. doi: 10.1093/jnci/djg059. [DOI] [PubMed] [Google Scholar]
- 10.Waller D., Peake M.D., Stephens R.J. Chemotherapy for patients with non-small cell lung cancer: the surgical setting of the big lung trial. Eur J Cardiothorac Surg. 2004;26:173–182. doi: 10.1016/j.ejcts.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 11.Ettinger D.S., Aisner D.L., Wood D.E. NCCN guidelines insights: non-small cell lung cancer, Version 5.2018. J Natl Compr Cancer Netw. 2018;16:807–821. doi: 10.6004/jnccn.2018.0062. [DOI] [PubMed] [Google Scholar]
- 12.Kelsey C.R., Marks L.B., Hollis D. Local recurrence after surgery for early stage lung cancer: an 11-year experience with 975 patients. Cancer. 2009;115:5218–5227. doi: 10.1002/cncr.24625. [DOI] [PubMed] [Google Scholar]
- 13.Nesbitt J.C., Putnam J.B., Walsh G.L., Roth J.A., Mountain C.F. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg. 1995;60:466–472. doi: 10.1016/0003-4975(95)00169-l. [DOI] [PubMed] [Google Scholar]
- 14.Wisnivesky J.P., Henschke C., McGinn T., Iannuzzi M.C. Prognosis of stage II non-small cell lung cancer according to tumor and nodal status at diagnosis. Lung Cancer. 2005;49:181–186. doi: 10.1016/j.lungcan.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Bueno R., Hughes E., Wagner S. Validation of a molecular and pathological model for five-year mortality risk in patients with early stage lung adenocarcinoma. J Thorac Oncol. 2015;10:67–73. doi: 10.1097/JTO.0000000000000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Custodio A.B., González-Larriba J.L., Bobokova J. Prognostic and predictive markers of benefit from adjuvant chemotherapy in early-stage non-small cell lung cancer. J Thorac Oncol. 2009;4:891–910. doi: 10.1097/JTO.0b013e3181a4b8fb. [DOI] [PubMed] [Google Scholar]
- 17.Kim M.H., Kim Y.K., Shin D.H. Yes associated protein is a poor prognostic factor in well-differentiated lung adenocarcinoma. Int J Clin Exp Pathol. 2015;8:15933–15939. [PMC free article] [PubMed] [Google Scholar]
- 18.Roepman P., Jassem J., Smit E.F. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res. 2009;15:284–290. doi: 10.1158/1078-0432.CCR-08-1258. [DOI] [PubMed] [Google Scholar]
- 19.Zhu C.Q., Ding K., Strumpf D. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang J.T.H., Lee Y.M., Huang R.S. The impact of the cancer genome atlas on lung cancer. Transl Res. 2015;166:568–585. doi: 10.1016/j.trsl.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganin Y., Ustinova E., Ajakan H. Domain-adversarial training of neural networks. In: Csurka G., editor. Domain adaptation in computer vision applications. Springer International Publishing; Cham: 2017. pp. 189–209. [Google Scholar]
- 22.Janowczyk A., Doyle S., Gilmore H., Madabhushi A. A resolution adaptive deep hierarchical (RADHicaL) learning scheme applied to nuclear segmentation of digital pathology images. Comput Methods Biomech Biomed Eng Imaging Vis. 2016:1–7. doi: 10.1080/21681163.2016.1141063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C., Romo-Bucheli D., Wang X. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab Investig. 2018 doi: 10.1038/s41374-018-0095-7. published online June 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee G., Ali S., Veltri R., Epstein J.I., Christudass C., Madabhushi A. Cell orientation entropy (COrE): predicting biochemical recurrence from prostate cancer tissue microarrays. Med Image Comput Comput Assist Interv. 2013;16:396–403. doi: 10.1007/978-3-642-40760-4_50. [DOI] [PubMed] [Google Scholar]
- 25.Leo P., Lee G., Shih N.N.C., Elliott R., Feldman M.D., Madabhushi A. Evaluating stability of histomorphometric features across scanner and staining variations: prostate cancer diagnosis from whole slide images. J Med Imaging (Bellingham) 2016;3 doi: 10.1117/1.JMI.3.4.047502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haralick R.M. Statistical and structural approaches to texture. Proc IEEE. 1979;67:786–804. [Google Scholar]
- 27.Wu Y. Elastic net for Cox's proportional hazards model with a solution path algorithm. Stat Sin. 2012;22:27–294. doi: 10.5705/ss.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C.H., George SL. The bootstrap and identification of prognostic factors via Cox's proportional hazards regression model. Stat Med. 1985;4:39–46. doi: 10.1002/sim.4780040107. [DOI] [PubMed] [Google Scholar]
- 29.Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y., Hochberg Y. Controlling the false discovery rate-a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 31.Pisters K.M.W., Evans W.K., Azzoli C.G. Cancer care ontario and american society of clinical oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I-IIIA resectable non small-cell lung cancer guideline. J Clin Oncol. 2007;25:5506–5518. doi: 10.1200/JCO.2007.14.1226. [DOI] [PubMed] [Google Scholar]
- 32.Schneider P.M., Praeuer H.W., Stoeltzing O. Multiple molecular marker testing (p53, C-Ki-ras, c-erbB-2) improves estimation of prognosis in potentially curative resected non-small cell lung cancer. Br J Cancer. 2000;83:473–479. doi: 10.1054/bjoc.2000.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endoh H., Tomida S., Yatabe Y. Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol. 2004;22:811–819. doi: 10.1200/JCO.2004.04.109. [DOI] [PubMed] [Google Scholar]
- 34.Lu C., Koyuncu C., Corredor G. Feature-driven local cell graph (FLocK): new computational pathology-based descriptors for prognosis of lung cancer and HPV status of oropharyngeal cancers. Med Image Anal. 2021;68 doi: 10.1016/j.media.2020.101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Detterbeck F.C. The eighth edition TNM stage classification for lung cancer: What does it mean on main street? J Thorac Cardiovasc Surg. 2018;155:356–359. doi: 10.1016/j.jtcvs.2017.08.138. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y.-.G., Kim S., Cho C.E. Effectiveness of transfer learning for enhancing tumor classification with a convolutional neural network on frozen sections. Sci Rep. 2020;10:21899. doi: 10.1038/s41598-020-78129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehrotra R., Ansari M.A., Agrawal R., Anand R.S. A transfer learning approach for AI-based classification of brain tumors. Mach Learn Appl. 2020;2 [Google Scholar]
- 38.Qaiser T., Tsang Y.-.W., Taniyama D. Fast and accurate tumor segmentation of histology images using persistent homology and deep convolutional features. Med Image Anal. 2019;55:1–14. doi: 10.1016/j.media.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Roy M., Kong J., Kashyap S. Convolutional autoencoder based model HistoCAE for segmentation of viable tumor regions in liver whole-slide images. Sci Rep. 2021;11:139. doi: 10.1038/s41598-020-80610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Chen H., Gan C. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans Cybern. 2020;50:3950–3962. doi: 10.1109/TCYB.2019.2935141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.