Abstract
The inter-fragment interactions at various binding sites and the overall cluster stability of quinolone (QNOL), cinnoline (CNOL), quinazoline (QNAZ), and quinoxaline (QNOX) complexes with H2O were studied using the density functional theory (DFT) approach. The adsorption and H-bond binding energies, and the energy decomposition mechanism was considered to determine the relative stabilization status of the studied clusters. Scanning tunneling microscopy (STM), natural bonding orbitals (NBO) and charge decomposition were studied to expose the electronic distribution and interaction between fragments. The feasibility of formations of the various complexes were also studied by considering their thermodynamic properties. Results from adsorption studies confirmed the actual adsorption of H2O molecules on the various binding sites studied, with QNOX clusters exhibiting the best adsorptions. Charge decomposition analysis (CDA) revealed significant charge transfer from substrate to H2O fragment in most complexes, except in QNOL, CNOL and QNAZ clusters with H2O at binding position 4, where much charges are back-donated to substrate. The O---H inter-fragment bonds was discovered to be stronger than counterpart N---H bonds in the complexes, whilst polarity indices confirmed N---H as more polar covalent than O---H bonds. Thermodynamic considerations revealed that the formation process of all studied complexes are endothermic (+ve ΔHf) and non-spontaneous (+ve ΔGf).
Keywords: Diazanaphthalenes, Quinolone, Aqueous, DFT, Adsorption
Diazanaphthalenes; Quinolone; Aqueous; DFT; Adsorption.
1. Introduction
The agrochemical, pharmaceutical, corrosion and vertinary industries are few of the vast application fields for heterocyclic compounds (HCs) [1, 2, 3, 4]. In other to expand the domain of consumption of heterocyclic compounds and its derivatives, much energy have been channeled into the development [2], synthesis [2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12] and interaction studies of these compounds [13, 14, 15] in various solvent systems. Many important organic-based chemical industries utilizes HCs as their feedstock, for special and multipurpose chemicals manufacture. They have been used in corrosion inhibitor products [10], developers, sanitizers [3], etc. It is also noteworthy that many enzymes and living tissues contain HCs in their backbone [1]. Heterocyclic compounds are cyclic compounds having at least two kinds of atoms in its ring skeleton (mostly carbon and non-metals like Nitrogen, Sulphur, Oxygen, etc.). HCs are classified as aromatic or non-aromatic, depending on the electronic composition of the rings [1]. Aromatic HCs must obey the general Huckels rule, which states that any aromatic compound must have 4n + 2 amount of π electrons in its ring system, examples are furan, pyridine, thiophene, quinoline, etc. The non-aromatic heterocycles are those that don't exhibit the aromatic ring structure, e.g., tetrahydrofuran, thiirane, thietane, etc. Quinoline, cinnoline, quinazoline and quinoxaline are aromatic HCs of nitrogen [1, 16, 17, 18]. They are generally obtained structurally by doping naphthalene with nitrogen atom(s). A mono doping of naphthalene with N at position 1 yields a quinolone molecule [19, 20]. While cinnoline, quinazoline and quinoxaline are derived via di-doping of naphthalene with N atoms at positions 1 and 2, 1 and 3, and 1 and 4, respectively [21, 22, 23, 24]. The inclusion of N atoms in the backbone of naphthalene chemically alters the electronic make-up, hence the emergence of new properties of interest.
Diazanaphthalenes are building blocks for many medical and structural valuable chemicals. Some quinoline and cinnoline derivatives for instance are recently explored in the medical industry, many are under clinical test for their antifungal, antimalarial, anti-inflammatory, analgesic, antitumor, etc., activities [25, 26, 27, 28].
The density functional theory (DFT) method of analysis, which exposes the electronic compositions, hence properties, is an important approach to the study of interacting molecules in conjugate systems [29], unlike when molecules interact, adhesive bonds are formed. So, for an appreciable understanding of the system, the nature and energies of the interaction bonds, and the charge transfers are required [30]. Also the thermodynamic properties (ΔH, ΔS and ΔG) of the process is demanded [31].
Relevant DFT works have been conducted on the activities of quinoline and other heterocycle derivatives [32, 33]; derivatives of quinoline tested as anti-tuberculosis agents showed positive results, pending in-vitro and in-vivo confirmation test [32]. Also, the anti-corrosion properties of some heterocycles are reported [34, 35]. In decision making of the applicability of molecules in a specific field, the interaction properties of the molecule and intended system composition is crucial. Hence, the aqueous chemistry of valuable active materials is imperative. Several DFT researches have been reported on the interactions of H2O with cluster systems [36, 37, 38, 39, 40, 41].
Understanding the aqueous chemistry of quinoline and the diazanaphthalenes will further explain their pharmacodynamics and pharmacokinetics in biological systems [42, 43]. Those of their derivatives can also be predicted considering the groups accommodated. This current study will expose in details, the adsorption properties of quinolone and some selected diazanaphthalene compounds (cinnoline, quinazoline and quinoxaline), which will be achieved through a systematic merge of H2O molecule on different binding sites on the activated ring of the pseudo-naphthalene compounds, to form a complex. The hydrogen bond characteristics are discussed in detail, the adsorption energies, significance and applicability are also exhaustively treated. Furthermore, the thermodynamics of formation of the substrate + H2O complexes are studied. The various studied complexes are titled in Table 1., for easy discussion and reference.
Table 1.
Structures, compositions, position of attachment and titles of various complexes studied with DFT/3BLYP method and 631 G (d) subset.
| S/N | Optimized structures | Composition/interaction position | Title |
|---|---|---|---|
| 1 |  |
Quinoline + H2O (1) | QNOL1 |
| 2 |  |
Quinoline + H2O (2) | QNOL2 |
| 3 |  |
Quinoline + H2O (3) | QNOL3 |
| 4 |  |
Quinoline + H2O (4) | QNOL4 |
| 5 |  |
Cinnoline + H2O (2) | CNOL2 |
| 6 |  |
Cinnoline + H2O (3) | CNOL3 |
| 7 |  |
Cinnoline + H2O (4) | CNOL4 |
| 8 |  |
Quinazoline + H2O (2) | QNAZ2 |
| 9 |  |
Quinazoline + H2O (3) | QNAZ3 |
| 10 |  |
Quinazoline + H2O (4) | QNAZ4 |
| 11 |  |
Quinoxaline + H2O (2) | QNOX2 |
| 12 |  |
Quinoxaline + H2O (3) | QNOX3 |
| 13 |  |
Quinoxaline + H2O (4) | QNOX4 |
N/B→ Binding site position numbers starts with the first N atom on ring 1, and proceeds in a clockwise direction.
2. Computational methods
For the fragment-fragment interaction studies of various titled complexes of water with quinoline and some selected di-azanaphthalene moieties, the Gaussian 09 software [44] coupled with Gauss-view interface was employed for the ground state optimizations, frequency calculations, natural bonding orbitals (NBO) analysis and surfaces and contour plotting, employing the DFT/B3LYP method and 631-G (d) basis set [45]. The basis set; 631-G(d) is relatively good for energy determinations, a heavy atom polarization term, d, was included to improve the energy output for large atoms [45]. H2O molecule and the various adsorption substrates were optimized individually, after which H2O was coupled on different positions of each substrate for optimization, observing the basic chemistry demand for the interactions of electronegative and electropositive terminals of molecules. Gaussian output files were booted on Multiwfn program [46] for topology analysis, Charge decomposition analysis (CDA), Density of State (DOS) plots, Simulating Scanning Tunneling Microscopy (STM) imaging, energy index (EI) and bond polarity index (BPI) analyses. While the intermolecular binding or adsorption energy (BE or AE) calculations, energy decomposition analysis (EDA), natural bonding orbital (NBO) analysis and thermodynamic studies were carried out on the Gaussian 09 software. The amount of energy required to disengage water molecules from the respective complexes is known as the adsorption energy [47]. The adsorption energies of water molecule on the various studied substrates were obtained through ground state energy determination of the various optimized complexes, and their corresponding fragments (H2O and quinolone or di-azanaphthalenes) on Gaussian. Calculated energies was transformed to BE using the traditional expression for adsorption energy, as indicated in Eq. (1).
| B.E = E(substrate) + E(H2O) – E(complex) | (1) |
Where E(substrate), E(H2O) and E(complex) are the energies of the substrate, water molecule and complex respectively. For a fair understanding of the energy decomposition between specific fragments of the various molecular clusters, we employed the EDA method based on classical forcefield (EDA-FF). The B3LYP/631-G (d) optimized clusters were disintegrated into its corresponding fragments, which were loaded into Multiwfn program for EDA-FF analysis. The total interaction energy (Etot), electrostatic interaction (Eels), exchange repulsion (Eex) and orbital interaction (Eorb) terms obtained for the substrates, H2O and corresponding complexes as defined in Eqs. (2), (3), (4), and (5), were used to explain the interactions.
| ΔEtot= Ecomplex- ΣEfragment | (2) |
| ΔEtot= ΔEels+ ΔEex+ ΔEorb | (3) |
| ΔEorb= ΔESCF_last– ΔESCF_1st | (4) |
| ΔEels+ ΔEex= ΔEtot– ΔEorb | (5) |
Where ΔESCF_last and ΔESCF_1st are the self-consistent field (SCF) energies for the last and first cycles respectively, of the SCF procedure as obtained [48]. In order to understand the donor-acceptor interactions of the fragments in the complexes, CDA was carried out. Intermolecular interactions in our current study are all N---H and O---H in nature, and for each case N and O atoms employ their lone pair of electrons in interacting with the unoccupied orbitals of H atom. The next transfer of electron density from fragment 1 (substrate) to 2 (H2O) is obtainable from extended CDA [49]. Topology analysis (through Atom in molecules (AIM) analysis) were carried out on Multiwfn program, in other to understand the hydrogen bonds formed between fragments in the complexes. Hydrogen bond B.E were calculated from the electron densities (ρ) of critical points (CPs) of the various intermolecular H-bonds, using Eq. (6), peculiar for neutral H-bond. CP properties were used to calculate Shannon aromacity of the rings.
| H-Bond B.E = -223.08 × ρ(rBCP) + 0.7423 | (6) |
Where ρ and B.E are obtained in a.u. and kcal/mol respectively [50].
The polarity on the intermolecular bonds between the two fragments was studied by determining the bond polarity index (BPI) and energy index (EI) of the bonds. Energy index of reference and fragment atoms of the binding sites of fragments groups were obtained from Multiwfn program. While the BPI were obtained using Eq. (7).
| BPI = (EIfrag1– EIfrag1ref) – (EIfrag2– EIfrag2ref) | (7) |
Where EIfrag1, EIfrag1ref, EIfrag2 and EIfrag2ref are the energy indices for fragment 1, fragment 1 reference, fragment 2 and fragment 2 reference, respectively [51]. The thermodynamic properties; Enthalpy and Gibbs free energy of formation of the various complexes were considered in this study. This was carried out through frequency calculations on the complexes, substrate and H2O, using B3LYP/631-G(d) method. ϵ0 (total electron energy), ϵZPE (zero-point energy), Etot (thermal energy), Hcorr (thermal enthalpy) and Gcorr (thermal free energy) terms are obtained from the Gaussian outputs for H2O, complexes and corresponding substrates.
| Hcorr= Etot+ KBT | (8) |
Where KB and T are Boltzman constant and temperature respectively,
| Gcorr= Hcorr– TStot | (9) |
Where Stot is the total entropy of the system, which is defined in Eq. (10) below.
| Stot= St+ Sr+ Sv+ Se | (10) |
Where St, Sr, Sv and Se are the translational, rotational, vibrational and electronic entropy contributions.
The enthalpies and Gibbs free energies of formation of the various titled clusters were obtained using Eqs. (11), (12), and (13). Where calculations were performed at STP (T = 298K; P = 1 atm)
| ΔfH0(298K) = [ΣΔfH0products(298K) - ΣΔfH0reactants(298K)]× 627.5095 | (11) |
The constant 627.5095 is a factor to covert energy from Hartree to kcal/mol. But from the electronic and thermal energy, which are obtainable from the Gaussian 09 calculations, one can easily determine the Δf H0 for the clusters.
| Δf H0 (298K) = [Σ(ϵ0 + Hcorr)products – Σ(ϵ0 + Hcorr)reactants]× 627.5095 | (12) |
| ΔfG0(298K) = [Σ(ϵ0+ Gcorr)products– Σ(ϵ0+ Gcorr)reactants]× 627.5095 | (13) |
Eqs. (8), (9), (10), (11), (12), and (13) was adopted from the reports from Ochterski in 2000 [52,53].
Natural bonding orbital (NBO) analysis was carried out on Gaussian 09, in order to study the orbital interactions leading to complex stabilization. Interaction with significant energy contributions are considered and interpreted. The electrostatic potential (ESP) and STM plots was done on Gaussview software and Multiwfn program respectively, using total electron distribution.
3. Results and discussion
3.1. Adsorption energy studies
Adsorption energy or binding energy of a complex is defined as the amount of energy required to dissociate the complex into its basic components, this is equal the magnitude of energy required to bind the components [54]. The intermolecular binding energy was analyzed to expose details of grip strength, bond strength and relative stability of the complexes. Positive values of adsorption energy imply that there is desorption, while negative values are indicative of adsorption. The adsorption energies as reported in Table 2., were calculated using Eq. (1), from the individual energy values of the clusters and components, obtained from single point energy calculations of the DFT/B3LYP optimized structures.
Table 2.
Adsorption energies, energy decomposition terms from energies of clusters, substrates and H2O of various complexes studied with DFT/3BLYP method and 631 G (d) basis.
| Clusters | Adsorption energy (EV) | Total energy term ΔE (TOT) KCAL/MOL |
Polarization term ΔE(ORB) KCAL/MOL |
Steric term ΔE(ELS)+ΔE(EX) KCAL/MOL |
|---|---|---|---|---|
| QNOL1 | -3.66 | -9.73 | -159.89 | 150.16 |
| QNOL2 | -3.71 | -8.53 | -160.39 | 151.86 |
| QNOL3 | -3.95 | -2.95 | -162.02 | 159.07 |
| QNOL4 | -3.91 | -3.77 | -161.14 | 157.38 |
| CNOL2 | -3.81 | -8.66 | -148.47 | 139.81 |
| CNOL3 | -3.81 | -8.72 | -148.47 | 139.74 |
| CNOL4 | -3.99 | -4.45 | -145.96 | 141.50 |
| QNAZ2 | -3.85 | -7.66 | -145.08 | 137.42 |
| QNAZ3 | -3.83 | -8.09 | -145.52 | 137.42 |
| QNAZ4 | -3.98 | -4.52 | -144.76 | 140.25 |
| QNOX2 | -3.98 | -7.66 | -143.20 | 135.54 |
| QNOX3 | -3.98 | -7.66 | -143.26 | 135.60 |
| QNOX4 | -3.93 | -9.04 | -143.20 | 134.16 |
The figures in bold are significant in the discussion, so they were segregated for easy visualizing.
From the adsorption energies in Table 2., where negative BE values was obtained for all studied complexes, one can infer that there was an overall adsorption of H2O molecules on the various substrates at all binding sites. Relatively, the diazanaphthalene-H2O complexes are more chemically held together than the various quinoline-H2O complexes, owing to the less negative values of BE for quinoline based complexes. Highest BE value of -3.99 eV is recorded for CNOL4, which implies that water molecule is strongly adsorbed at position 4 of cinnoline. This is predicted to be due to the formation of a di-hydrogen bonding between O atom of water molecule and two H atoms of cinnoline, as can be visualized on the CNOL4 structure in Table 1. Best overall binding is noticeable for quinoxaline + H2O (QNOX) clusters, with B.E < -3.90 eV for all studied binding sites, as can be visualized in Figure 1.
Figure 1.
Plots of Adsorption energies for the various interactions positions in QNOL, CNOL, QNAZ and QNOX studied with DFT/B3LYP method and 631-G (d) basis set.
3.2. Energy decomposition studies
EDA was carried out in order to understand the extent of interactions between adsorbed H2O molecules and the various substrates. The total interaction energy term (ΔE(tot)) explains the amount of energy existing between two fragments, with which one can understand the linkage chemistry [55, 56]. Orbital interaction energy term (ΔE(orb)), also known as the polarization term is used to determine the level of orbital interactions between the molecular orbitals (MOs) of the fragment, which stabilizes the adduct. Also, the sum of the electrostatic and exchange-repulsion energy terms (ΔE(els)+ ΔE(ex)) defines the level of steric hindrance around the interaction sites of clusters. This hindrance is as a result of the arrangement of the vicinal groups ([N or C] and their substituents) to the binding sites. The ΔE(tot), ΔE(orb), and ΔE(els)+ ΔE(ex) for the various complexes at B3LYP/631-G (d) are reported in Table 2. From the ΔE(tot) values in Kcal/mol as reported in Table 2., it is evident that QNOL1 (-9.73), CNOL2 (-8.66), CNOL3 (-8.72), QNAZ3 (-8.09), QNOX2 (-7.66), QNOX3 (-7.66) and QNOX4 (-9.04), where dual interaction bonds of N---H and O---H are confirmed to exist between H2O and substrate, exhibited good interactions, hence are classified to be relatively stable clusters. A relatively weak dual interaction where the oxygen atom of H2O form intermolecular bonds with two hydrogens of the substrate is evident in QNAZ4 (-4.52). This weakness in dual bond is expected because it is a projection from the oxygen of H2O to two hydrogen atoms of the substrate, exhibiting a lower electron density. Very weak interaction energies were obtained for QNOL3, QNOL4, CNOL4 and QNAZ4. Overall, the high ΔE(tot) values of quinoxaline-H2O clusters indicates that the complexes are more stable. Better stabilization is predicted CNOL2, CNOL3 and QNAZ3 in their various groups, owing to their higher ΔE(orb) values, although the differences recorded for the different clusters aren't obvious. High ΔE(els)+ ΔE(ex) are observed for CNOL4, QNAZ4 and QNOX3 clusters, this is interpreted to represent a high steric hindrance around their binding sites.
3.3. Density of state (DOS) analysis
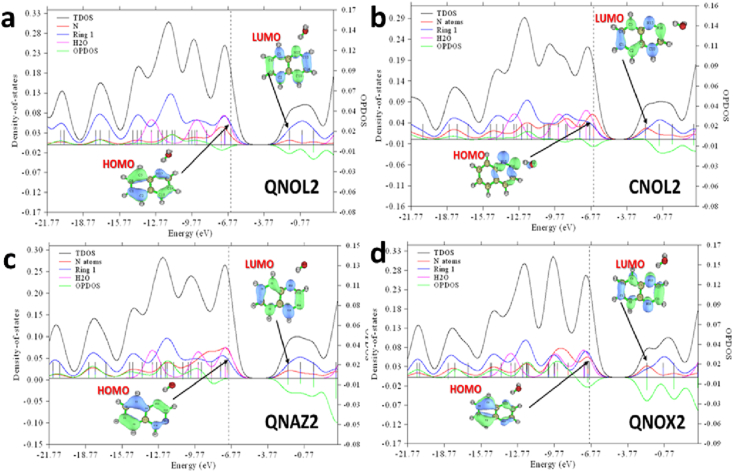
The partial, total and overlap population density of states (PDOS, TDOS and OPDOS) were plotted for all cluster interactions to understand the orbital compositions of the different MOs in the complexes [57]. The PDOS, TDOS and OPDOS plots for QNOL2, CNOL2, QNAZ2 and QNOX2 (representative of their various groups) are presented in Figure 2 for a comparative study of the various substrates. The complexes are split into fragments of interest, so as to visualize the contributions of the various groups in the studied complex. The Nitrogen atoms contribution is represented with orange curve, H2O with pink, and the heterocyclic ring with blue curve.
Figure 2.
PDOS, TDOS and OPDOS Plots for (a) QNOL2, (b) CNOL2, (c) QNAZ2 and (d) QNOX2 studied with DFT/B3LYP method and 631-G (d) basis set.
One can visualize from Figure 2., that for QNOL2, CNOL2, QNAZ2 and QNOX3, ring 1 (blue) and N atoms (orange) correspond to the PDOS curve (green), hence the heterocyclic ring and nitrogen groups contribute largely to the stability of the clusters. Negative value for PDOS is indicative of antibonding interaction at the corresponding MO. This implies that CNOL2 and QNOX2 exhibits antibonding characters at their HOMO levels, which arises from the inadequate overlap of MOs. All clusters discussed experienced antibonding properties at LUMO, as visualized from the polarity of PDOS curve at this point. The effect of the heterocyclic ring is maximum between -12.77— -9.77 eV for all studied complexes, this implies that the aromatic ring has its highest contribution to the MOs within this range. It is evident that H2O molecular orbital contribution is highest at HOMO for all complexes, as can be visualized from its high curve at HOMO levels.
3.4. Scanning tunneling microscopy (STM)
STM technique is employed in imaging chemical systems at their atomic level, it is directly related with the electronic composition of the chemical system. STM plot is a special tool in understanding the tunneling current (I) distribution around molecules. Tunneling current is directly proportional to local density of state (LDOS) [58]. In visualizing STM diagrams, the bright white coloration indicates regions with high LDOS, hence strong tunneling current. In this study, the STM plots for QNOL2, CNOL2, QNAZ2 and QNOX2 (representative of their various groups) are presented in Figure 3.
Figure 3.
STM images of (a) QNOL2, (b) CNOL2, (c) QNAZ2 and (d) QNOX2 studied with DFT/B3LYP method and 631-G (d) basis set.
A high tunneling current is observable around the C atoms of Ring 2 of QNOL2, QNAZ2 and QNOX2 (Figure 3), as can be inferred from the bright white coloration around these regions. Mild distribution of I around C–N bonds in the various complexes is visible, except for CNOL2, where a bright white surface is concentrated on the N atoms. It is evident that tunneling current (I), hence electron density and LDOS is highly concentrated on the N heteroatoms of CNOL2 complex. Much similarity is confirmed for the electronic distribution of I around complex molecules in QNOL, QNAZ and QNOX, but CNOL.
3.5. Charge decomposition studies
Charge decomposition analysis (CDA) and extended CDA are carried out to understand the mechanism of charge transfer between H2O fragment and substrates in the various studied complexes to obtain charge equilibrium [59]. The studied interactions in the project are basically N---H and O---H in nature, where N and O uses their lone pair of electrons to interact with the unoccupied orbital of H. Extended CDA is used to determine the net transfer of electrons from substrate to H2O in the complex [49]. ECDA excludes the electronic contributions from polarization effect (PL) making it a better approach than the general ECDA. The ECDA results for all studied clusters are presented in Table 3. The substrates are defined as fragment 1, and H2O moiety as fragment 2 for the various ECDA determinations (Figure 4).
Table 3.
Net electron transfer between fragments from ECDA for clusters studied with DFT/3BLYP method and 631 G (d) basis.
| Cluster | Net Electron Transfer Frag 1→2 | Net Electron Transfer Frag 2→1 | Net Electrons Obtained by Frag 2 |
|---|---|---|---|
| QNOL1 | 0.0762 | 0.0492 | 0.0270 |
| QNOL2 | 0.0677 | 0.0308 | 0.0369 |
| QNOL3 | 0.0023 | 0.0384 | -0.0361 |
| QNOL4 | 0.0030 | 0.0414 | -0.0385 |
| CNOL2 | 0.0607 | 0.0323 | 0.0284 |
| CNOL3 | 0.0608 | 0.0323 | 0.0285 |
| CNOL4 | 0.0031 | 0.0437 | -0.0405 |
| QNAZ2 | 0.0626 | 0.0305 | 0.0321 |
| QNAZ3 | 0.0562 | 0.0292 | 0.0270 |
| QNAZ4 | 0.0033 | 0.0470 | -0.0437 |
| QNOX2 | 0.0612 | 0.0328 | 0.0284 |
| QNOX3 | 0.0635 | 0.0351 | 0.0284 |
| QNOX4 | 0.0601 | 0.0317 | 0.0284 |
The figures in bold are significant in the discussion, so they were segregated for easy visualizing.
Figure 4.
Plots of Electron Obtained by Fragment 2 (H2O) from CDA for the various interactions positions in QNOL, CNOL, QNAZ and QNOX studied with DFT/B3LYP method and 631-G (d) basis set.
From Table 3., the net amount of electrons transferred to fragment 2 is deduced by subtracting the electron density back-donated from fragment 2 to 1 (CT 2→1) from the amount donated from fragment 1 to 2 (CT 1→2). Overall, there is net electron transfer from fragment 2 to 1 in QNOL3, QNOL4, CNOL4 and QNAZ4, as depicted in the negative values of net electron obtained. This is because the O atom in H2O was used in binding for these complexes. The other complexes exhibited a net electron transfer from fragment 1 to 2, which is traced to the N→H binding that took place in many clusters studied. It is worthy to note that the QNOL2 (0.0369) exhibit the highest net electron transfer from substrate to H2O. Also, appreciable π-back donation properties are predicted for QNOL3, QNOL4, CNOL4 and QNAZ4, owing to their high net electron transfer from fragment 2→1, which is as a result of depleted overlap region between the two fragments in such systems [59]. This observation can be visualized clearly in Figure 5. Orbital interaction diagrams (Figure 5.) were plotted for QNOL2, CNOL2, QNAZ2 and QNOX2, in other to visualize the mixing of fragments orbitals as the complexes are obtained.
Figure 5.
Orbital interaction diagrams from CDA for the various interactions positions in QNOL, CNOL, QNAZ and QNOX studied with DFT/B3LYP method and 631-G (d) basis set.
The fragment orbitals of fragment 1 (substrate) are aligned to the left, fragment 2 (H2O) to the right and cluster orbitals at the middle. Linking (red lines) connects fragment orbitals (FOS) with electron composition higher than 10% [59, 60]. Substrate FOs 16, 17 and 18 are linked with complex orbitals (COs) 18, 19 and 20 respectively in QNOL2, CNOL2, QNAZ2 and QNOX2, which implies that FOs 16, 17 and 18 are unperturbed during the formation of the respective complexes. They are π in nature, hence cannot partake in σ-type donor-acceptor interactions between complex fragments. The interactions of FOs of H2O with the COs as shown in Figure 5., are equivalent in QNAZ2 and QNOX2, were FOs 3 and 4 of H2O are linked with COs 33 and 35 respectively.
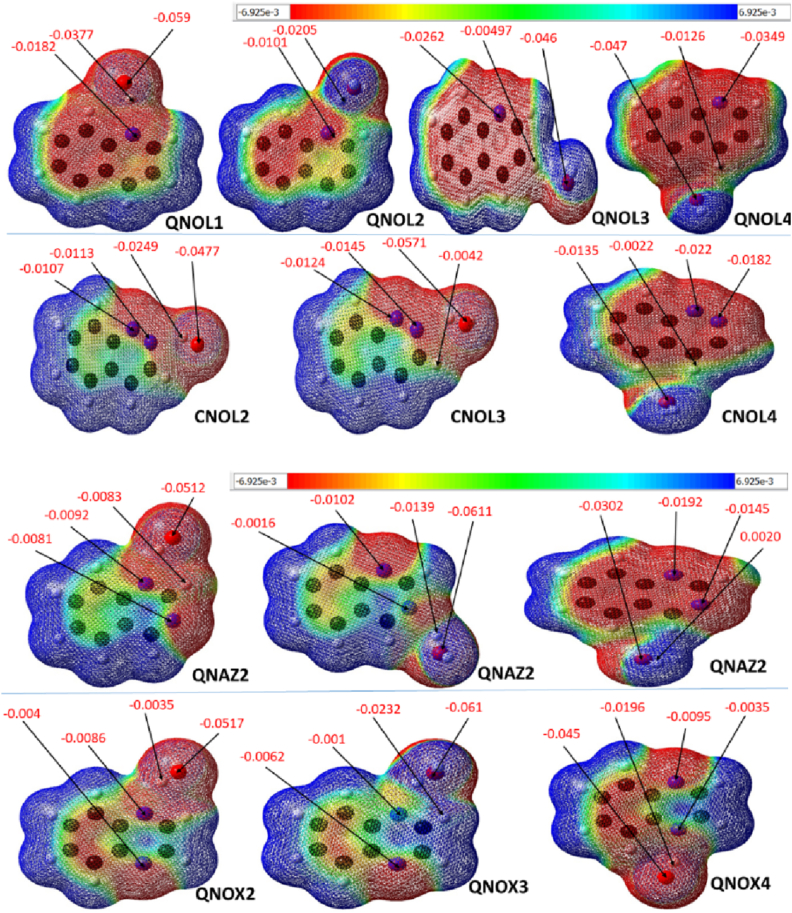
3.6. Electrostatic potential (ESP) studies
The electrostatic potential (ESP) images were plotted in order to visualize the electron density distribution around the atoms in the complexes. The ESP surface plots are colored from red through green to blue. Red regions represent areas with high presence of electrons, which are characterized with negative values for isosurfaces, while blue depicts deficiency in electron density (positive isosurface values are recorded for such surfaces) [61]. Isosurface plots for all studied clusters are presented in Figure 6. From the isosurfaces in Figure 6 it is visible that high electron density is located around the N and O atoms, this corresponds to their electron pulling capacity as highly electronegative atoms. An appreciable amount of electrons is visible around the aromatic ring core for all studied clusters, this is inferred from the light green to dark red colorations around the rings. Hydrogen atoms are surrounded by deep blue colorations, which is indicative of deficiency in electron density, which results from the highly distorted bonding electron distribution in hydrogen to electronegative groups. This is evident for H on aromatic rings and on oxygen, the blue surfaces represent regions where electrons are depleted. Areas around the oxygen of H2O fragment exhibited the highest isosurface values in all studied clusters.
Figure 6.
ESP Isosurface plots for all Clusters studied with DFT/B3LYP method and 631-G (d) basis set.
3.7. Topology studies
The individual hydrogen bonding systems at the various binding sites are studies using atoms in molecule (AIM) basins approach [62]. All properties measured are dependent on the critical point (CP), which is considered as a point of reference in topology studies. The CP is a point between two neighboring atoms, which defines a bond or interaction found along the connection axis, it is located along the path of an existing or none-existing bond [62]. It was observed that during adsorption of H2O on the various studied substrates, one or two Hydrogen bonds are formed between the fragments. In each case, distinct H-bonds are treated in other to fully understand their individual contributions to the stability of the clusters obtained. Hydrogen bond length, electron densities and binding energies are calculated for every hydrogen bond CP identified, these are reported in Table 4. In extension, the shannon aromaticity (SA) of the rings of studied complexes were measured, adopting the reports of Noorizadeh and Shakerzadeh in 2010 [63,64], the results are reported in Table 5., in the supporting information. Shannon aromaticity is relative; a reason it was employed in our study. From the figures reported on Table 5., QNAZ2 exhibited the highest aromaticity (SA = 0.0745), representing a good stabilization around the ring structure of the substrate fragment. It is observed that QNOX4 (with SA = 0.00192) is the most aromatic in its group, this supports the claim that position 4 of QNOX is the most suitable for H2O adsorption. Relatively low aromaticity is shown by all QNOL complexes, but QNOL (with SA = 0.00198), hence H2O adsorption on position 1 stabilized the aromatic rings of QNOL.
Table 4.
Measured H-bond lengths, H-bond electron densities, and H-bond binding energy from AIMs analysis of all clusters studied with DFT/B3LYP method and 631-G (d) basis set.
| Clusters | Identified H-bonds |
CPS number | Bond length (Å) | Density (A.U) | H-Bond B.E (KCAL/MOL) |
|---|---|---|---|---|---|
| QNOL1 | O18 – H13 | 26 | 2.42 | 0.12 | -26.23 |
| N16 – H19 | 34 | 1.98 | 0.30 | -65.36 | |
| QNOL2 | N17 – H20 | 26 | 1.98 | 0.29 | -64.02 |
| QNOL3 | O18 – H7 | 32 | 2.39 | 0.13 | -28.15 |
| QNOL4 | O18 – H8 | 27 | 2.49 | 0.82 | -181.13 |
| O18 – H6 | 36 | 2.55 | 0.10 | -21.70 | |
| CNOL2 | N16 – H19 | 28 | 2.01 | 0.27 | -60.43 |
| O17 – H14 | 38 | 2.58 | 0.90 | -200.12 | |
| CNOL3 | O17 – H14 | 25 | 2.58 | 0.90 | -199.76 |
| N16 – H19 | 35 | 2.00 | 0.27 | -60.58 | |
| CNOL4 | O17 – H7 | 26 | 2.44 | 0.85 | -189.46 |
| O17 – H9 | 35 | 2.54 | 0.11 | -23.48 | |
| QNAZ2 | N15 – H19 | 29 | 2.01 | 0.27 | -60.02 |
| QNAZ3 | O17 – H16 | 26 | 2.64 | 0.80 | -178.61 |
| N14 – H19 | 36 | 2.06 | 0.24 | -53.51 | |
| QNAZ4 | O17 – H7 | 25 | 2.50 | 0.80 | -176.65 |
| O17 – H9 | 34 | 2.57 | 0.11 | -22.79 | |
| QNOX2 | O14 – H18 | 28 | 2.61 | 0.27 | -58.82 |
| N13 – H16 | 34 | 2.01 | 0.87 | -193.09 | |
| QNOX3 | N16 – H18 | 25 | 2.02 | 0.27 | -58.71 |
| O17 – H14 | 29 | 2.61 | 0.87 | -192.76 | |
| QNOX4 | O14 – H8 | 22 | 2.41 | 0.12 | -26.63 |
| N19 – HI6 | 32 | 2.01 | 0.28 | -60.74 |
The figures in bold are significant in the discussion, so they were segregated for easy visualizing.
Table 5.
Measured Shannon aromaticity indices from AIMs analysis of all clusters studied with DFT/B3LYP method and 631-G (d) basis set.
| Clusters | Shannon aromaticity |
|---|---|
| QNOL1 | 0.00198 |
| QNOL2 | 0.00138 |
| QNOL3 | 0.00153 |
| QNOL4 | 0.00154 |
| CNOL2 | 0.00422 |
| CNOL3 | 0.00421 |
| CNOL4 | 0.00411 |
| QNAZ2 | 0.07454 |
| QNAZ3 | 0.00216 |
| QNAZ4 | 0.00221 |
| QNOX2 | 0.00188 |
| QNOX3 | 0.00188 |
| QNOX4 | 0.00192 |
N/B; Shannon aromaticity is relative.
3.7.1. Hydrogen bond length
From the measured bond length in Angstrom (Å) reported in Table 4., it is evident that N---H bonds are shorter than O---H bonds in the various studied clusters. CNOL1 and CNOL2 exhibited the shortest N---H bond in this study, this implies that if we consider all H2O fragments binding on the substrates, CNOL1 and CNOL2 are more compacted at N sites. O---H bonds are perceived to naturally be longer than other counterparts [65].
3.7.2. Hydrogen bond binding energy
The H-bond B.E were calculated from the electron density values determined for various CPs, employing Eq. (6). They are presented in Table 4., as obtained in Kcal/mol. It is conspicuous that O---H interactions are stronger than N---H, inferable from their relative binding energies, despite the supposed contradictions from the bond length values. Hence, bond strength is not only dependent on the length but also on the electronic composition of the bonding atoms. With a closer at Table 4., one can observe a high deal of stabilizing binding energies for the O---H interactions in QNOL4 (-181.13 kcal/mol), CNOL2 (-200.12 kcal/mol), CNOL3 (-199.76 kcal/mol), CNOL4 (-189.46 kcal/mol), QNAZ4 (-176.65 kcal/mol), QNOX2 (-193.09 kcal/mol) and QNOX3 (-192.72 kcal/mol). These binding energies are crucial components of the overall cluster stabilization energies (a reason for the high cluster B.E for CNOL4 and QNAZ4 as observed from the adsorption energy study).
3.8. Energy index (EI) and bond polarity index (BPI)
Energy index is a useful tool in the description of the nature of electron sharing between atoms in a covalent system. Traditionally, electrons are shared equally between bonding atoms in nonpolar covalent bond, while polar covalent bond involves a distorted electron distribution density. On the other hand, bond polarity index (BPI) explains the polarity of a bond [66]. BPI was analyzed to ascertain hydrogen bond polarity between fragments in the various studied clusters. To calculate the BPI for N---H and O---H bonds in substrate + H2O complexes, the reference EI values of N and H atoms for each substrate was initially calculated, and O and H atoms in H2O molecule. The BPI were determine by invoking Eq. (7)., and the results are presented in Table 6. The EI of the various references and fragments in clusters are reported in Table 7.
Table 6.
Calculated BPI values for various intermolecular bonds of all clusters studied with DFT/B3LYP method and 631-G (d) basis set.
| Clusters | Bond type rEF1---rEF2 | BPI |
|---|---|---|
| QNOL1 | N---H | -0.110 |
| QNOL2 | H---O | -0.014 |
| QNOL3 | H---O | 0.055 |
| QNOL4 | H---O | 0.065 |
| CNOL2 | N---H | -0.069 |
| CNOL3 | H---O | -0.011 |
| CNOL4 | H---O | 0.072 |
| QNAZ2 | H---O | 0.076 |
| QNAZ3 | N---H | -0.074 |
| QNAZ4 | H---O | 0.067 |
| QNOX2 | H---O | -0.0003 |
| QNOX3 | H---O | -0.0003 |
| QNOX4 | N---H | -0.090 |
The figures in bold are significant in the discussion, so they were segregated for easy visualizing.
Table 7.
Calculated fragment and reference EI and BPI values for various intermolecular bonds of all clusters studied with DFT/B3LYP method and 631-G (d) basis set.
| Clusters | Bond type REF1-REF2 | EI of binding site on REF1 | EI of binding site on REF2 | EI of binding sites on FRAG1 | EI of binding site on FRAG2 | BPI |
|---|---|---|---|---|---|---|
| QNOL1 | N–H | -0.5 | -0.605 | -0.542 | -0.537043 | -0.11012 |
| QNOL2 | H–O | -0.48 | -0.534 | -0.465 | -0.500291 | -0.0135 |
| QNOL3 | H–O | -0.49 | -0.534 | -0.461 | -0.554698 | 0.054805 |
| QNOL4 | H–O | -0.5 | -0.534 | -0.458 | -0.560775 | 0.065366 |
| CNOL2 | N–H | -0.53 | -0.605 | -0.54 | -0.542198 | -0.06919 |
| CNOL3 | H–O | -0.5 | -0.534 | -0.477 | -0.50337 | -0.01098 |
| CNOL4 | H–O | -0.51 | -0.534 | -0.471 | -0.567065 | 0.072141 |
| QNAZ2 | H–O | -0.57 | -0.534 | -0.469 | -0.505136 | 0.076012 |
| QNAZ3 | N–H | -0.52 | -0.605 | -0.545 | -0.55447 | -0.07363 |
| QNAZ4 | H–O | -0.5 | -0.534 | -0.466 | -0.562777 | 0.067252 |
| QNOX2 | H–O | -0.5 | -0.534 | -0.478 | -0.51091 | -0.00026 |
| QNOX3 | H–O | -0.5 | -0.534 | -0.478 | -0.510929 | -0.00026 |
| QNOX4 | N–H | -0.53 | -0.605 | -0.56 | -0.54952 | -0.09003 |
N/B; Fragment 1 = Substrate (QNOL, CNOL, QNAZ and QNOX).
Fragment 2 = H2O.
EI of Ref1 = EI of N or H atoms in substrate as applicable, depended on binding site.
EI of Ref2 = EI of O or H atoms in H2O as applicable.
As can be seen from the BPI values for the various inter-fragment bond in Table 6., highly negative BPI are derived for N---H intermolecular linkages, which implies that they are more polar, so can be relatively classified as polar covalent bonds. This is due to distortion in the electron distribution along the binding part in such clusters. QNOL1, QNOL2, CNOL2, CNOL3, QNAZ3 and QNOX4 with bond polarity indices; -0.11, -0.014, -0.069, -0.074 and -0.09 respectively are appreciably polar at binding sites. Hence, they are predicted to be more soluble in aqueous systems. O---H linkages are less polar to N---H bonds in our studied clusters. This is due to the extended system (C–H---O–H), where electrons are more evenly distributed than C=N---H–O for N adsorption sites.
3.9. Thermodynamics considerations
Thermodynamics studies the relationship between work and heat energy, as both changes on the course of a chemical reaction [67]. In other to understand the feasibility of the formations and stabilities of the studied clusters, we considered the enthalpies (ΔH) and Gibbs free energies (ΔG) of formation. The Gibbs free energy; a function of enthalpy change (ΔH), entropy change (ΔS), and temperature, explains the spontaneity of a chemical reactions [52]. The total electron energy (ϵ0), zero-point energy (ϵzpe), thermal energy (Etot), total entropy (Stot) thermal enthalpy (Hcorr), and thermal free energy (Gcorr) terms required from Gaussian 09 output, in the calculations of ΔHf and ΔGf of the various complexes were calculated. All thermodynamic calculations are done at 298.15K and 1atm, considering one mole of complex, by applying Eqs. (8), (9), (10), (11), (12), and (13). The thermodynamics parameters of all studied fragments were calculated with DFT/B3LYP method at 631-G (d) basis, the ΔHf of water at STP is calculated to be -76.384 eV at this level.
Positive ΔHf values are obtained for all studied clusters (Table 8), which is interpreted as endothermic, this implies that heat is absorbed from the surrounding during their formation process. It is notable that QNOL, CNOL, QNAZ and QNOX interacts with H2O molecules by absorbing a deal of energy from the surroundings, hence a cooling effect is felt when they are solvated. The change in enthalpy for the formation of QNOX4 (+59.17 kcal/mol) is the least for the studied binding sites in QNOX, hence binding at position 4 of QNOX absorbs the least amount of energy. CNOL4 formation is accompanied with the highest energy absorption, this is explained by its high ΔHf value (62.85 kcal/mol).
Table 8.
Change in enthalpies (ΔHf) and Gibbs free energies (ΔGf) of formation from Gaussian 09 calculations at 298.15K and 1atm for all clusters studied with DFT/B3LYP method and 631-G (d) basis set.
| Clusters | ΔHF (KCAL/MOL) | ΔGF (KCAL/MOL) |
|---|---|---|
| QNOL1 | 55.81 | 64.60 |
| QNOL2 | 56.99 | 65.38 |
| QNOL3 | 61.90 | 66.75 |
| QNOL4 | 60.61 | 66.99 |
| CNOL2 | 59.09 | 67.55 |
| CNOL3 | 59.09 | 67.55 |
| CNOL4 | 62.85 | 68.33 |
| QNAZ2 | 60.31 | 68.61 |
| QNAZ3 | 59.84 | 68.21 |
| QNAZ4 | 63.01 | 69.35 |
| QNOX2 | 60.31 | 68.65 |
| QNOX3 | 60.31 | 68.64 |
| QNOX4 | 59.17 | 67.89 |
The figures in bold are significant in the discussion, so they were segregated for easy visualizing.
The calculated ΔGf values for all complexes at 298.15K and 1atm, as can be seen in Table 8., are positive, this is an implication that the formation process of all the studied complexes are nonspontaneous in nature. Hence a given amount of energy is required to propel the various reactions leading to our studied clusters. It is evident that ΔS is negative, due to the positive values obtained for ΔGf and ΔHf (eqn 9) respectively. In comparison, the formation of QNOX4 is an easier process than of other QNOX complexes, because of its lower ΔGf value (+67.89 kcal/mol) in the group. CNOL4 is more difficult to obtained, comparing the ΔGf value (+68.33), which is the highest for CNOL complexes. Hence, binding of H2O molecule are position 4 of CNOL is more difficult, although, it gives the most stable complex (as discovered from B.E studies). It is visible from ΔGf values in Table 8., that QNOL + H2O are the easiest to obtain, they exhibited a comparably low ΔGf values.
3.10. Natural bonding orbitals (NBO) analysis
The stabilization energies (second perturbation energies) of the interacting NBOs of the studied complexes with significant energies are considered in this research [68, 69]. It was inferred from the Shannon aromaticity studies that the presence on N atoms in the aromatic ring backbone improved the overall aromaticity of the studied complexes, where QNOL + H2O clusters exhibited lower aromaticity, hence less stable rings. Very important interactions are expected from the bonding orbitals of N atoms with other orbitals to attain more stability for complexes. The second perturbation energies of the natural bonding orbitals of the studied complexes are presented in Table 9 n→σ∗ and n→π∗ donor acceptor resonance interactions of heteroatoms leads to stability. This type of interaction is evident in for all studied complexes; e.g. nC3→π∗C1–C2 (49.96 kcal/mol)/π∗C10–C13 (54.43 kcal/mol) nN15→σ∗C3–C4 (10.34 kcal/mol) in CNOL2 and nC4→π∗C5–C6 (48.69 kcal/mol)/π∗N13–C17 (60.81 kcal/mol) in QNOX2, nC4→π∗C5–C6 (49.44 kcal/mol)/π∗C11–N15 (59.18 kcal/mol) in QNOX3 and nC3→π∗C1–C2 (Kcal/mol)/π∗C11–N19 (Kcal/mol) in QNOX4 are good stabilizing interactions with significant energies of stabilization. Other very important second order perturbations are the π→n∗ and π→π∗ types, they correspond to the hyper-conjugative interactions of the aromatic rings. Our utmost interest is in those involving N atoms of the ring; πN15-N16→n∗C4 (28.99 kcal/mol)/π∗C10–C13 (16.25 kcal/mol) in CNOL2, πN15-N16→n∗ C4 (29.00 kcal/mol)/π∗C10–C13 (16.26 kcal/mol) in CNOL3, πN15-N16→π∗C3–C4 (15.18 kcal/mol)/π∗C10–C13 (16.21 kcal/mol) in CNOL4, πC13-N15→n∗C4 (43.16 kcal/mol)/π∗C10–N16 (8.97 kcal/mol) in QNAZ2, πN11-C12→n∗C4 (43.53 kcal/mol)/π∗N14–C15 (Kcal/mol) in QNAZ3, πN13-C17→n∗C4 (39.07 kcal/mol)/π∗C11–N19 (14.82 kcal/mol) in QNOX2 and πN13-C17→n C4 (39.43 kcal/mol)/π∗C11–N19 (16.73 kcal/mol) in QNOX4 are necessary interactions which contribute to the stabilization of the studied complexes. No orbital interaction with significant stabilization energy was observed involving H2O fragment orbitals nor fragment bonding orbitals, hence the inter-fragment interactions are generally weak. This claim agrees with the thermodynamic results of the studied complexes; All formation processes are endothermic (requires high deal of energy to proceed) and nonspontaneous (due to large ΔG values).
Table 9.
Results from the second order perturbation theory analysis performed with Gaussian 09 for all clusters studied with DFT/B3LYP method and 631-G (d) basis set.
| Donor | Occupancy | Acceptor | Occupancy | E (2) Kcal/mol | E(j)-E(i) a.u. | F (j.i) a.u. |
|---|---|---|---|---|---|---|
|
QNOL1 | ||||||
| πC1 - C2 | 1.72237 | π∗C3 – C4 | 0.45392 | 14.45 | 0.29 | 0.061 |
| π∗C5 – N16 | 0.31039 | 25.11 | 0.27 | 0.075 | ||
| πC3 - C4 | 1.52055 | π∗C1 – C2 | 0.23661 | 18.47 | 0.27 | 0.067 |
| π∗C5 – N16 | 0.31039 | 14.61 | 0.25 | 0.056 | ||
| π∗C9 – C12 | 0.24845 | 16.13 | 0.28 | 0.064 | ||
| π∗C10 – C11 | 0.23232 | 14.53 | 0.29 | 0.062 | ||
| πC5 - N16 | 1.80242 | π∗C1 – C2 | 0.23661 | 10.84 | 0.34 | 0.055 |
| π∗ C3 – C4 | 0.45392 | 18.70 | 0.34 | 0.076 | ||
| πC9 - C12 | 1.74010 | π∗C3 – C4 | 0.45392 | 16.55 | 0.28 | 0.064 |
| π∗C10 – C11 | 0.23232 | 16.58 | 0.30 | 0.063 | ||
| πC10 - C11 | 1.72058 | π∗ C3 – C4 | 0.45392 | 17.85 | 0.28 | 0.066 |
| π∗C9 – C12 | 0.24845 | 17.90 | 0.29 | 0.065 | ||
| π∗C3 – C4 | 0.45392 | π∗C9 – C12 | 0.24845 | 238.20 | 0.01 | 0.077 |
| π∗C10 – C11 | 0.23232 | 151.54 | 0.02 | 0.079 | ||
| π∗C5 – N16 | 0.31039 | π∗C1 – C2 | 0.23661 | 114.62 | 0.02 | 0.082 |
| π∗C3 – C4 |
0.45392 |
135.33 |
0.02 |
0.074 |
||
|
QNOL2 | ||||||
| πC1 – C2 | 1.73957 | LP (1) C3 | 1.00879 | 40.26 | 0.15 | 0.087 |
| π∗C5 – C6 | 0.24062 | 17.12 | 0.30 | 0.064 | ||
| πC5 – C6 | 1.72983 | LP∗ (1) C4 | 0.96601 | 43.15 | 0.15 | 0.088 |
| π∗C1 – C2 | 0.24823 | 17.42 | 0.29 | 0.064 | ||
| πC10 – C14 | 1.72280 | LP (1) C3 | 1.00879 | 37.16 | 0.16 | 0.086 |
| π∗C13 – N17 | 0.30197 | 24.96 | 0.28 | 0.075 | ||
| πC13 – N17 | 1.79733 | LP∗ (1) C4 | 0.96601 | 40.26 | 0.20 | 0.101 |
| π∗C10 – C14 | 0.23279 | 10.87 | 0.34 | 0.054 | ||
| LP (1) C3 | 1.00879 | π∗C1 – C2 | 0.24823 | 54.35 | 0.15 | 0.100 |
| π∗C10 – C14 | 0.23279 | 59.05 | 0.14 | 0.102 | ||
| LP∗ (1) C4 | 0.96601 | π∗C5 – C6 | 0.24062 | 50.77 | 0.15 | 0.011 |
| π∗C13 – N17 | 0.30197 | 56.74 | 0.12 | 0.093 | ||
| π∗C13 – N17 |
0.30197 |
π∗C10 – C14 |
0.23279 |
125.39 |
0.02 |
0.082 |
|
QNOL3 | ||||||
| πC1 – C2 | 1.72094 | π∗C3 – C4 | 0.45327 | 15.05 | 0.29 | 0.062 |
| π∗C5 – N16 | 0.29651 | 24.00 | 0.28 | 0.073 | ||
| πC3 – C4 | 1.52242 | π∗C1 – C2 | 0.23081 | 17.70 | 0.27 | 0.066 |
| π∗C5 – N16 | 0.29651 | 14.45 | 0.26 | 0.057 | ||
| π∗C9 – C12 | 0.25240 | 16.63 | 0.28 | 0.064 | ||
| π∗C10 – C11 | 0.24246 | 15.26 | 0.28 | 0.062 | ||
| πC5 – N16 | 1.79061 | π∗C1 – C2 | 0.23081 | 11.58 | 0.33 | 0.056 |
| π∗C3 – C4 | 0.45327 | 19.48 | 0.33 | 0.076 | ||
| πC9 – C12 | 1.74418 | π∗C3 – C4 | 0.45327 | 16.14 | 0.29 | 0.064 |
| π∗C10 – C11 | 0.24246 | 16.96 | 0.30 | 0.064 | ||
| πC10 – C11 | 1.73408 | π∗C3 – C4 | 0.45327 | 16.94 | 0.28 | 0.065 |
| π∗C9 – C12 | 0.25240 | 17.66 | 0.29 | 0.064 | ||
| σC12 – H15 | 1.98272 | σ∗C3 – C9 | 0.02223 | 4.11 | 1.07 | 0.059 |
| LP (1) N16 | 1.92371 | σ∗C1 – C5 | 0.02872 | 10.52 | 0.87 | 0.087 |
| σ∗C3 – C4 | 0.04271 | 10.58 | 0.87 | 0.086 | ||
| π∗C3 – C4 | 0.45327 | π∗C10 – C11 | 0.24246 | 248.07 | 0.01 | 0.079 |
| π∗C5 – N16 | 0.29651 | π∗C1 – C2 | 0.23081 | 131.35 | 0.02 | 0.081 |
| π∗C3 – C4 |
0.45327 |
219.29 |
0.01 |
0.079 |
||
|
QNOL4 | ||||||
| πC1 – C2 | 1.72537 | π∗C3 – C4 | 0.45197 | 14.38 | 0.29 | 0.061 |
| π∗C5 – N16 | 0.29624 | 24.27 | 0.28 | 0.074 | ||
| σC1 – H7 | 1.98253 | σ∗C5 – N16 | 0.01143 | 4.06 | 1.11 | 0.060 |
| πC3 – C4 | 1.52284 | π∗C1 – C2 | 0.23405 | 18.14 | 0.27 | 0.067 |
| π∗C5 – N16 | 0.29624 | 14.42 | 0.26 | 0.057 | ||
| π∗C9 – C12 | 0.24665 | 16.47 | 0.28 | 0.064 | ||
| π∗C10 – C11 | 0.24492 | 15.21 | 0.28 | 0.062 | ||
| πC5 – N16 | 1.79140 | π∗C1 – C2 | 0.23405 | 11.39 | 0.33 | 0.055 |
| π∗C3 – C4 | 0.45197 | 19.41 | 0.33 | 0.076 | ||
| πC9 – C12 | 1.73835 | π∗C3 – C4 | 0.45197 | 16.11 | 0.28 | 0.064 |
| π∗C10 – C11 | 0.24492 | 17.49 | 0.29 | 0.064 | ||
| πC10 – C11 | 1.73596 | π∗C3 – C4 | 0.45197 | 16.99 | 0.23 | 0.065 |
| π∗C9 – C12 | 0.24665 | 17.13 | 0.29 | 0.064 | ||
| LP (1) N16 | 1.92350 | σ∗C1 – C5 | 0.02899 | 10.54 | 0.87 | 0.086 |
| σ∗C3 – C4 | 0.04244 | 10.50 | 0.87 | 0.086 | ||
| π∗C5 – N16 | 0.29624 | π∗C1 – C2 | 0.23405 | 142.15 | 0.02 | 0.080 |
| π∗C3 – C4 |
0.45197 |
197.30 |
0.01 |
0.075 |
||
|
CNOL2 | ||||||
| πC1 – C2 | 1.73877 | LP (1) C3 | 0.97922 | 42.49 | 0.15 | 0.089 |
| πC1 – C2 | 1.73877 | π∗C5 – C6 | 0.21735 | 15.68 | 0.30 | 0.062 |
| σC2 – H9 | 1.98148 | σ∗C3 – C4 | 0.04221 | 4.22 | 1.06 | 0.060 |
| πC5 – C6 | 1.73199 | LP∗ (1) C4 | 0.97587 | 41.56 | 0.15 | 0.086 |
| π∗C1 – C2 | 0.22920 | 17.76 | 0.29 | 0.065 | ||
| σH7 – C10 | 1.98116 | σ∗C13 – N16 | 0.03000 | 4.53 | 1.03 | 0.061 |
| πC10 – C13 | 1.67309 | LP (1) C3 | 0.97922 | 45.81 | 0.15 | 0.091 |
| π∗N15 – N16 | 0.39966 | 23.30 | 0.24 | 0.068 | ||
| σC13 – H14 | 1.97964 | π∗N15 – N16 | 0.39966 | 5.48 | 1.03 | 0.067 |
| πN15 – N16 | 1.80908 | LP∗ (1) C4 | 0.97587 | 28.99 | 0.22 | 0.092 |
| π∗C10 – C13 | 0.23329 | 16.25 | 0.37 | 0.070 | ||
| LP (1) C3 | 0.97922 | π∗C1 – C2 | 0.22920 | 49.96 | 0.15 | 0.098 |
| π∗C10 – C13 | 0.23329 | 54.43 | 0.14 | 0.101 | ||
| LP∗ (1) C4 | 0.97587 | π∗C5 – C6 | 0.21735 | 48.61 | 0.15 | 0.099 |
| π∗N15 – N16 | 0.39966 | 112.98 | 0.09 | 0.107 | ||
| LP (1) N15 | 1.93896 | σ∗C3 – C4 | 0.04221 | 10.34 | 0.89 | 0.086 |
| π∗C13 – N16 | 0.03000 | 10.33 | 0.86 | 0.085 | ||
| LP (1) N16 | 1.91904 | σ∗C4 – N15 | 0.03336 | 10.17 | 0.86 | 0.084 |
| π∗N15 – N16 |
0.39966 |
π∗C10 – C13 |
0.23329 |
45.45 |
0.06 |
0.080 |
|
CNOL3 | ||||||
| πC1 – C2 | 1.73879 | LP (1) C3 | 0.97921 | 42.49 | 0.15 | 0.089 |
| π∗C5 – C6 | 0.21731 | 15.68 | 0.30 | 0.062 | ||
| σC1 – H8 | 1.98247 | σ∗C2 – C3 | 0.02297 | 4.19 | 1.07 | 0.060 |
| πC5 – C6 | 1.73202 | LP∗ (1) C4 | 0.97585 | 41.56 | 0.15 | 0.086 |
| π∗C1 – C2 | 0.22915 | 17.76 | 0.29 | 0.065 | ||
| πC10 – C13 | 1.73202 | LP (1) C3 | 0.97921 | 45.83 | 0.15 | 0.091 |
| π∗N15 – N16 | 0.39981 | 23.31 | 0.24 | 0.068 | ||
| πN15 – N16 | 1.80902 | LP∗ (1) C4 | 0.97585 | 29.00 | 0.22 | 0.092 |
| π∗C10 – C13 | 0.23338 | 16.26 | 0.37 | 0.070 | ||
| LP (1) C3 | 0.97921 | π∗C1 – C2 | 0.22915 | 49.95 | 0.15 | 0.098 |
| π∗C10 – C13 | 0.23338 | 54.45 | 0.14 | 0.101 | ||
| LP∗ (1) C4 | 0.97585 | π∗C5 – C6 | 0.21731 | 48.60 | 0.15 | 0.099 |
| π∗N15 – N16 | 0.39981 | 113.06 | 0.09 | 0.107 | ||
| LP (1) N15 | 1.93896 | σ∗C3 – C4 | 0.04221 | 10.34 | 0.89 | 0.086 |
| σ∗C13 – N16 | 0.03000 | 10.32 | 0.86 | 0.085 | ||
| LP (1) N16 | 1.91896 | σ∗C4 – N15 | 0.03335 | 10.17 | 0.86 | 0.084 |
| π∗N15 – N16 |
0.39981 |
π∗C10 – C13 |
0.23338 |
45.47 |
0.06 |
0.080 |
|
CNOL4 | ||||||
| πC1 – C2 | 1.73816 | π∗C3 – C4 | 0.45387 | 16.88 | 0.29 | 0.065 |
| π∗C5 – C6 | 0.22457 | 16.30 | 0.30 | 0.063 | ||
| σC1 – H8 | 1.98280 | σ∗C2 – C3 | 0.02301 | 4.12 | 1.07 | 0.059 |
| σC2 – H9 | 1.98057 | σ∗C3 – C4 | 0.04209 | 4.29 | 1.06 | 0.060 |
| πC3 – C4 | 1.50306 | π∗C1 – C2 | 0.22824 | 14.76 | 0.28 | 0.062 |
| π∗C5 – C6 | 0.22457 | 14.88 | 0.28 | 0.062 | ||
| π∗C10 – C13 | 0.23666 | 16.62 | 0.28 | 0.065 | ||
| π∗N15 – N16 | 0.38100 | 23.33 | 0.23 | 0.067 | ||
| πC5 – C6 | 1.73793 | π∗C1 – C2 | 0.22824 | 17.28 | 0.29 | 0.064 |
| π∗C3 – C4 | 0.45387 | 16.33 | 0.28 | 0.064 | ||
| πC10 – C13 | 1.68207 | π∗C3 – C4 | 0.45387 | 16.38 | 0.29 | 0.064 |
| π∗N15 – N16 | 0.38100 | 23.54 | 0.25 | 0.069 | ||
| πN15 – N16 | 1.80166 | π∗C3 – C4 | 0.45387 | 15.18 | 0.35 | 0.070 |
| π∗C10 – C13 | 0.23666 | 16.21 | 0.36 | 0.069 | ||
| LP (1) N15 | 1.94001 | σ∗C3 – C4 | 0.04209 | 10.40 | 0.89 | 0.086 |
| σ∗C13 – N16 | 0.02945 | 10.07 | 0.86 | 0.084 | ||
| LP (1) N16 | 1.94235 | σ∗C4 – N15 | 0.03511 | 10.54 | 0.85 | 0.085 |
| π∗C3 – C4 | 0.45387 | π∗C5 – C6 | 0.22457 | 224.61 | 0.01 | 0.078 |
| π∗N15 – N16 | 0.38100 | π∗C3 – C4 | 0.45387 | 76.02 | 0.04 | 0.081 |
| π∗C10 – C13 |
0.23666 |
50.37 |
0.05 |
0.080 |
||
|
QNAZ2 | ||||||
| πC1 – C2 | 1.73107 | LP (1) C3 | 1.02311 | 40.84 | 0.15 | 0.087 |
| π∗C5 – C6 | 0.23261 | 17.59 | 0.30 | 0.065 | ||
| σC1 – H8 | 1.98256 | σ∗C2 – C3 | 0.02248 | 4.05 | 1.07 | 0.059 |
| σC1 – H9 | 1.97508 | σ∗C3 – C4 | 0.04072 | 4.31 | 1.06 | 0.061 |
| πC5 – C6 | LP∗ (1) C4 | 0.94559 | 46.20 | 0.14 | 0.089 | |
| π∗C1 – C2 | 0.23865 | 16.64 | 0.29 | 0.063 | ||
| πC10 – N16 | 1.72004 | LP (1) C3 | 1.02311 | 23.05 | 0.19 | 0.076 |
| π∗C13 – N15 | 0.27606 | 29.38 | 0.32 | 0.087 | ||
| πC13 – N15 | 1.79101 | LP∗ (1) C4 | 0.94559 | 43.16 | 0.20 | 0.104 |
| π∗C10 – N16 | 0.26562 | 8.97 | 0.32 | 0.049 | ||
| LP (1) C3 | 1.02311 | π∗C1 – C2 | 0.23865 | 54.20 | 0.15 | 0.101 |
| π∗C10 – N16 | 0.26562 | 79.51 | 0.13 | 0.111 | ||
| LP∗ (1) 4 | 0.94559 | π∗C5 – C6 | 0.23261 | 48.50 | 0.15 | 0.099 |
| π∗C13 – N15 | 0.27606 | 49.94 | 0.12 | 0.090 | ||
| LP (1) N15 | 1.90080 | σ∗C13 – N16 | 0.03394 | 12.30 | 0.85 | 0.093 |
| LP (1) N16 | 1.92323 | σ∗C3 – C10 | 0.03426 | 10.60 | 0.88 | 0.087 |
| σ∗C13 – N15 |
0.01123 |
11.62 |
0.91 |
0.093 |
||
|
QNAZ3 | ||||||
| πC1 – C2 | 1.73081 | LP (1) C3 | 1.02277 | 40.79 | 0.15 | 0.087 |
| π∗C5 – C6 | 0.23039 | 17.49 | 0.30 | 0.065 | ||
| σC2 – H8 | 1.98148 | σ∗C3 – C4 | 0.04245 | 4.33 | 1.06 | 0.061 |
| πC5 – C6 | 1.71924 | LP∗ (1) C4 | 0.94422 | 45.72 | 0.15 | 0.089 |
| π∗C1 – C2 | 0.23718 | 16.68 | 0.29 | 0.063 | ||
| πN11 – C12 | 1.98980 | LP∗ (1) C4 | 0.94422 | 43.53 | 0.20 | 0.103 |
| π∗N14 – C15 | 0.27897 | 9.46 | 0.31 | 0.049 | ||
| πN14 – C15 | 1.77511 | LP (1) C3 | 1.02277 | 21.59 | 0.20 | 0.075 |
| π∗N11 – C12 | 0.26636 | 27.91 | 0.33 | 0.086 | ||
| LP (1) C3 | 1.02277 | π∗C1 – C2 | 0.23718 | 53.99 | 0.15 | 0.101 |
| π∗N14 – C15 | 0.27897 | 86.29 | 0.12 | 0.113 | ||
| LP∗ (1) C4 | 0.94422 | π∗C5 – C6 | 0.23039 | 48.41 | 0.15 | 0.099 |
| π∗N11 – C12 | 0.26636 | 50.44 | 0.13 | 0.091 | ||
| LP (1) N11 | 1.91819 | σ∗C3 – C4 | 0.02258 | 10.23 | 0.87 | 0.085 |
| σ∗C12 – N14 | 0.03826 | 13.51 | 0.83 | 0.096 | ||
| LP (1) N14 | 1.90903 | σ∗C3 – C15 | 0.03272 | 9.62 | 0.89 | 0.084 |
| σ∗N11 – C12 | 0.02411 | 10.84 | 0.92 | 0.091 | ||
| σ∗O17 – H19 |
1.99824 |
9.20 |
0.85 |
0.080 |
||
|
QNAZ4 | ||||||
| πC1 – C2 | 1.72813 | π∗C3 – C4 | 0.44611 | 16.16 | 0.28 | 0.063 |
| π∗C5 – C6 | 0.23643 | 17.99 | 0.29 | 0.065 | ||
| σC2 – H9 | 1.98056 | σ∗C3 – C4 | 0.04195 | 4.41 | 1.06 | 0.061 |
| πC3 – C4 | 1.52256 | π∗C1 – C2 | 0.23591 | 16.69 | 0.28 | 0.065 |
| π∗C5 – C6 | 0.23643 | 13.99 | 0.28 | 0.060 | ||
| π∗C10 – N16 | 0.26875 | 25.36 | 0.26 | 0.076 | ||
| π∗C13 – N15 | 0.27018 | 12.49 | 0.26 | 0.054 | ||
| πC5 – C6 | 1.72548 | π∗C1 – C2 | 0.23591 | 16.33 | 0.29 | 0.062 |
| π∗C3 – C4 | 0.44611 | 18.42 | 0.28 | 0.067 | ||
| πC10 – N16 | 1.76831 | π∗C3 – C4 | 0.44611 | 9.67 | 0.33 | 0.054 |
| π∗C13 – N15 | 0.27018 | 28.27 | 0.32 | 0.086 | ||
| πC13 – N15 | 1.78563 | π∗C3 – C4 | 0.44611 | 20.70 | 0.33 | 0.079 |
| π∗C10 – N16 | 0.01144 | 9.43 | 0.32 | 0.050 | ||
| LP (1) N15 | 1.91942 | σ∗C3 – C4 | 0.04195 | 10.11 | 0.88 | 0.085 |
| σ∗C13 – N16 | 0.03669 | 13.43 | 0.83 | 0.095 | ||
| LP (1) N16 | 1.92572 | σ∗C3 – C10 | 0.03407 | 10.42 | 0.88 | 0.086 |
| σ∗C13 – N15 | 0.02601 | 11.45 | 0.92 | 0.093 | ||
| π∗C3 – C4 | 0.04195 | π∗C1 – C2 | 0.23591 | 234.45 | 0.01 | 0.077 |
| π∗C5 – C6 | 0.23643 | 247.84 | 0.01 | 0.079 | ||
| π∗C10 – N16 | 0.26875 | π∗C3 – C4 | 0.44611 | 213.43 | 0.01 | 0.081 |
| π∗C13 – N15 |
0.27018 |
π∗C3 – C4 |
0.44611 |
225.26 |
0.01 |
0.074 |
|
QNOX2 | ||||||
| πC1 – C2 | 1.72706 | LP (1) C3 | 0.97636 | 44.14 | 0.14 | 0.088 |
| π∗C5 – C6 | 0.23909 | 17.35 | 0.29 | 0.064 | ||
| σC2 – H8 | 1.98041 | σ∗C3 – C4 | 0.04996 | 4.34 | 1.04 | 0.060 |
| πC5 – C6 | 1.72873 | LP∗ (1) C4 | 0.97411 | 44.91 | 0.14 | 0.088 |
| π∗C1 – C2 | 0.23546 | 16.85 | 0.30 | 0.064 | ||
| σC11 – H12 | 1.98225 | σ∗C3 – N19 | 0.02212 | 5.08 | 1.03 | 0.065 |
| πC11 – N19 | 1.77335 | LP (1) C3 | 0.97636 | 39.76 | 0.20 | 0.100 |
| π∗N13 – C17 | 0.27948 | 16.64 | 0.31 | 0.065 | ||
| πN13 – C17 | 1.78621 | LP∗ (1) C4 | 0.97411 | 39.07 | 0.20 | 0.100 |
| π∗C11 – N19 | 0.26772 | 14.82 | 0.32 | 0.063 | ||
| LP (1) C3 | 0.97636 | π∗C1 – C2 | 0.23546 | 49.46 | 0.15 | 0.100 |
| π∗C11 – N19 | 0.26772 | 59.17 | 0.12 | 0.096 | ||
| LP∗ (1) C4 | 0.97411 | π∗C5 – C6 | 0.23909 | 49.47 | 0.15 | 0.100 |
| π∗N13 – C17 | 0.27948 | 59.19 | 0.12 | 0.094 | ||
| LP (1) N13 | 1.90854 | σ∗C3 – C4 | 0.04996 | 9.48 | 0.88 | 0.082 |
| LP (1) N19 | 1.92659 | σ∗C3 – C4 | 0.04996 | 10.12 | 0.87 | 0.084 |
| σ∗C11 – C17 |
0.03855 |
10.14 |
0.87 |
0.085 |
||
|
QNOX3 | ||||||
| πC1 – C2 | 1.72877 | LP∗ (1) C3 | 0.97413 | 44.91 | 0.14 | 0.088 |
| π∗C5 – C6 | 0.23539 | 16.84 | 0.30 | 0.064 | ||
| σC2 – H8 | 1.98054 | σ∗C3 – C4 | 0.04996 | 4.34 | 1.04 | 0.060 |
| πC5 – C6 | 1.72711 | LP (1) C4 | 0.97635 | 44.13 | 0.14 | 0.088 |
| π∗C1 – C2 | 0.23901 | 17.35 | 0.29 | 0.064 | ||
| πC11 – N15 | 1.77333 | LP (1) C4 | 0.97635 | 39.77 | 0.20 | 0.100 |
| π∗C12 – N16 | 0.27955 | 16.64 | 0.31 | 0.065 | ||
| πC12 – N16 | 1.78619 | LP∗ (1) C3 | 0.97413 | 39.06 | 0.20 | 0.100 |
| π∗C11 – N15 | 0.26778 | 14.83 | 0.32 | 0.063 | ||
| LP∗ (1) C3 | 0.97413 | π∗C1 – C2 | 0.23901 | 49.44 | 0.15 | 0.100 |
| π∗C12 – N16 | 0.27955 | 59.21 | 0.12 | 0.094 | ||
| LP (1) 4 | 0.97635 | π∗C5 – C6 | 0.23539 | 49.44 | 0.15 | 0.100 |
| π∗C11 – N15 | 0.26778 | 59.18 | 0.12 | 0.096 | ||
| LP (1) N15 | 1.92660 | σ∗C3 – C4 | 0.04996 | 10.12 | 0.87 | 0.084 |
| σ∗C11 – H12 |
0.03854 |
10.14 |
0.87 |
0.085 |
||
|
QNOX4 | ||||||
| πC1 – C2 | 1.71899 | LP∗ (1) C3 | 0.97467 | 46.54 | 0.14 | 0.088 |
| π∗C5 – C6 | 0.01314 | 17.34 | 0.29 | 0.064 | ||
| σC1 – H7 | 1.98254 | σ∗C2 – C3 | 0.02416 | 4.06 | 1.07 | 0.059 |
| πC5 – C6 | 1.72734 | LP (1) C4 | 0.97492 | 44.90 | 0.14 | 0.088 |
| π∗C1 – C2 | 0.22984 | 16.73 | 0.30 | 0.064 | ||
| πC11 – N19 | 1.79101 | LP∗ (1) C3 | 0.97467 | 37.45 | 0.20 | 0.100 |
| π∗N13 – C17 | 0.27222 | 14.81 | 0.33 | 0.063 | ||
| πN13 – C17 | 1.77284 | LP (1) C4 | 0.97492 | 39.43 | 0.20 | 0.100 |
| π∗C11 – N19 | 0.29012 | 16.73 | 0.31 | 0.065 | ||
| LP∗ (1) C3 | 0.97467 | π∗C1 – C2 | 0.22984 | 47.54 | 0.16 | 0.100 |
| π∗C11 – N19 | 0.29012 | 62.59 | 0.11 | 0.095 | ||
| LP (1) C4 | 0.97492 | π∗C5 – C6 | 0.01314 | 48.69 | 0.15 | 0.099 |
| π∗N13 – C17 | 0.27222 | 60.81 | 0.12 | 0.097 | ||
| LP (1) N13 | 1.92617 | σ∗C3 – C4 | 0.04970 | 10.11 | 0.87 | 0.084 |
| σ∗C11 – C17 | 0.03812 | 10.19 | 0.87 | 0.085 | ||
4. Conclusion
The inter-fragment chemistry and stability studies of complexes of quinoline, cinnoline, quinazoline and quinoxaline with H2O was achieved in this work. We discovered that H2O molecules are adsorbed on the various studied binding sites of substrates, although no much interactions are recorded between the fragments. From thermodynamic studies, we observed that the formation of the studied complexes at STP are significantly endothermic and nonspontaneous, with the complex of cinnoline with H2O at position 4 being more stable. Generally, the formation of quinoxaline + H2O complexes are more easily obtainable and are also fairly stable. Appreciable tunneling current (I) is concentrated on the N atoms of CNOL, as revealed by visualized STM images, while much similarities exist for QNOL, QNAZ and QNOX, where I are distributed about the aromatic C atoms. We also observed from the visualized ESP diagrams of the studied complexes that electron densities are concentrated around N and O atoms of the complexes, implying that they are relatively more electronegative than other groups in the clusters. The N---H inter-fragment linkages were found to be highly polar covalent in nature from bond polarity index studies, while electronic distribution around O---H inter-fragment bonds are very even, making them less polar. From CDA results, a net charge transfers from fragment 1 (substrate) to fragment 2 (H2O) (CT 1→2) was revealed for most complexes, except QNAZ4, CNOL4, QNOL4 and QNOL2, where CT 2→1 is exhibited. The second perturbation energies result of the various interacting NBOs exposed very important n→σ∗, n→π∗, π→n∗ and π→π∗ types of interaction between natural orbitals around the cluster structures, contributing to total stabilization energies. Though, no significant inter-fragment stabilization interactions occurred between NBOs of fragments. We conclude that the formation of clusters of H2O on quinoline, cinnoline, quinazoline and quinoxaline is possible but non-spontaneous, but little aqueous chemistry is predictable.
Declarations
Author contribution statement
Obieze C. Enudi: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Hitler Louis: Conceived and designed the experiments.
Moses M. Edim, John A. Agwupuye, Francis O. Ekpen: Contributed reagents, materials, analysis tools or data.
Emmanuel A. Bisong, Patrick M. Utsu: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors wish to acknowledge the Computational Chemistry and Bio-simulation Research group leader, Hitler Louis for designing and supervising this research. Obieze C. Enudi is also grateful to his PI, Moses M. Edim, for his undisputed input to the success of this project.
Contributor Information
Obieze C. Enudi, Email: enudij@gmail.com.
Hitler Louis, Email: louismuzong@gmail.com.
References
- 1.Kumari J. Application of heterocyclic compounds in everyday life. J. Mod. Chem. Chem. Technol. 2018;9(1):1–7. [Google Scholar]
- 2.Fei T., Lv P., Liu Y., He C., Sun C., Pang S. Design and synthesis of a series of CL-20 cocrystals: six-membered symmetrical N-heterocyclic compounds as effective coformers. Cryst. Growth Des. 2019;19(5):2779–2784. [Google Scholar]
- 3.Shaabani A., Nazeri M.T., Afshari R. 5-Amino-pyrazoles: potent reagents in organic and medicinal synthesis. Mol. Divers. 2019;23(3):751–807. doi: 10.1007/s11030-018-9902-8. [DOI] [PubMed] [Google Scholar]
- 4.De Oliveira Silva A., McQuade J., Szostak M. Recent advances in the synthesis and reactivity of isothiazoles. Adv. Synth. Catal. 2019;361(13):3050–3067. [Google Scholar]
- 5.De Andrade V.S., de Mattos M.C. N-halo reagents: modern synthetic approaches for heterocyclic synthesis. Synthesis. 2019;51(9):1841–1870. [Google Scholar]
- 6.Atanasova-Stamova S.Y., Georgieva S.F., Georgieva M.B. Reaction strategies for synthesis of imidazole derivatives: a review. Scripta Scientifica Pharmaceutica. 2018;5(2):7–13. [Google Scholar]
- 7.Kapri K.P., Singar S.B., Khanal S., Shakya B. Synthesis of schiff bases of 4-amino-5-(2-hydroxyphenyl)-4H-1, 2, 4-triazole-3-thiol as potent antimicrobial agents. Amrit Res. J. 2020;1(1):29–36. [Google Scholar]
- 8.Budhwan R., Garg G., Namboothiri I.N., Murarka S. Hypervalent iodine (III) reagents in the synthesis of heterocyclic compounds. Targets Heterocycl. Syst. 2020;20:27–52. [Google Scholar]
- 9.Ferlin F., Luciani L., Viteritti O., Brunori F., Piermatti O., Santoro S., Vaccaro L. Polarclean as a sustainable reaction medium for the waste minimized synthesis of heterocyclic compounds. Front. Chem. 2019;6:659. doi: 10.3389/fchem.2018.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed S.K., Ali W.B., Khadom A.A. Synthesis and investigations of heterocyclic compounds as corrosion inhibitors for mild steel in hydrochloric acid. Int. J. Ind. Chem. 2019;10(2):159–173. [Google Scholar]
- 11.Cabrele C., Reiser O. The modern face of synthetic heterocyclic chemistry. J. Org. Chem. 2016;81(21):10109–10125. doi: 10.1021/acs.joc.6b02034. [DOI] [PubMed] [Google Scholar]
- 12.Edim M.M., Enudi O.C., Asuquo B.B., Louis H., Bisong E.A., Agwupuye J.A., Bassey F.I. Aromaticity indices, electronic structural properties, and fuzzy atomic space investigations of naphthalene and its aza-derivatives. Heliyon. 2021;7(2) doi: 10.1016/j.heliyon.2021.e06138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidal-Vidal A., Faza O.N., Silva Lopez C. CO2 complexes with five-membered heterocycles: structure, topology, and spectroscopic characterization. J. Phys. Chem. 2017;121(47):9118–9130. doi: 10.1021/acs.jpca.7b09394. [DOI] [PubMed] [Google Scholar]
- 14.Ren J., Li J., Li J., Chen Z., Cheng F. Tracking multiple aromatic compounds in a full-scale coking wastewater reclamation plant: interaction with biological and advanced treatments. Chemosphere. 2019;222:431–439. doi: 10.1016/j.chemosphere.2019.01.179. [DOI] [PubMed] [Google Scholar]
- 15.Tyunina E.Y., Badelin V.G., Mezhevoi I.N. Observation of complex formation between L-histidine and heterocyclic compounds in water and aqueous buffer solution using calorimetric and spectroscopic methods. J. Mol. Liq. 2019;278:505–511. [Google Scholar]
- 16.Bamdad, H., Papari, S., MacQuarrie, S., & Hawboldt, K. Study of surface heterogeneity and nitrogen functionalizing of biochars: molecular modeling approach. Carbon, 171, 161-170.
- 17.Vidal Á.V., López C.S., Faza O.N. Nitrogen doped nanohoops as promising CO 2 capturing devices. Phys. Chem. Chem. Phys. 2018;20(13):8607–8615. doi: 10.1039/c7cp08498f. [DOI] [PubMed] [Google Scholar]
- 18.Prieschl D., Belanger-Chabot G., Guo X., Dietz M., Muller M., Krummenacher I., Braunschweig H. Synthesis of complex boron–nitrogen heterocycles comprising borylated triazenes and tetrazenes under mild conditions. J. Am. Chem. Soc. 2019;142(2):1065–1076. doi: 10.1021/jacs.9b12336. [DOI] [PubMed] [Google Scholar]
- 19.Xie L.Y., Fang T.G., Tan J.X., Zhang B., Cao Z., Yang L.H., He W.M. Visible-light-induced deoxygenative C2-sulfonylation of quinoline N-oxides with sulfinic acids. Green Chem. 2019;21(14):3858–3863. [Google Scholar]
- 20.Zhang J., Zheng C., Zhang M., Qiu Y., Xu Q., Cheong W.C., Wang D. Controlling N-doping type in carbon to boost single-atom site Cu catalyzed transfer hydrogenation of quinoline. Nano Res. 2020;13(11):3082–3087. [Google Scholar]
- 21.Szumilak M., Stanczak A. Cinnoline scaffold—a molecular heart of medicinal chemistry? Molecules. 2019;24(12):2271. doi: 10.3390/molecules24122271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Azab A.S., Alaa A.M., Bua S., Nocentini A., El-Gendy M.A., Mohamed M.A., Supuran C.T. Synthesis of benzensulfonamides linked to quinazoline scaffolds as novel carbonic anhydrase inhibitors. Bioorg. Chem. 2019;87:78–90. doi: 10.1016/j.bioorg.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z., Li H., Peng Z., Wang Z., Wang Y., Lu P. Preparation and photophysical properties of quinazoline-based fluorophores. RSC Adv. 2020;10(51):30297–30303. doi: 10.1039/d0ra05701k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montana M., Mathias F., Terme T., Vanelle P. Antitumoral activity of quinoxaline derivatives: a systematic review. Eur. J. Med. Chem. 2019;163:136–147. doi: 10.1016/j.ejmech.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 25.Evangelin M.P., Gold T.S., Elisha Y., Radhika G., Vamsi G.K., Arathi K. A concise review on cinnolines. Innovat. J. Med. Health Sci. 2020;10(4):897–901. [Google Scholar]
- 26.Varshney Shruti, Saxena V. Design, synthesis, characterization and biological evaluation of some novel cinnolo piperazine derivatives. Int. J. Pharm. Pharmaceut. Sci. 2014;6:245–248. [Google Scholar]
- 27.Zoidis G., Sosic A., Da Ros S., Gatto B., Sissi C., Palluotto F.,., Catto M. Indenocinnoline derivatives as G-quadruplex binders, topoisomerase IIα inhibitors and antiproliferative agents. Bioorg. Med. Chem. 2017;25(9):2625–2634. doi: 10.1016/j.bmc.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Awad E.D., El-Abadelah M.M., Matar S., Zihlif M.A., Naffa R.G., Al-Momani E.Q., Mubarak M.S. Synthesis and biological activity of some 3-(4-(Substituted)-piperazin-1-yl) cinnolines. Molecules. 2012;17(1):227–239. doi: 10.3390/molecules17010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan I.M., Alam K., Alam M.J., Ahmad M. Spectrophotometric and photocatalytic studies of H-bonded charge transfer complex of oxalic acid with imidazole: single crystal XRD, experimental and DFT/TD-DFT studies. New J. Chem. 2019;43(23):9039–9051. [Google Scholar]
- 30.Kavitha R., Nirmala S., Nithyabalaji R., Sribalan R. Biological evaluation, molecular docking and DFT studies of charge transfer complexes of quinaldic acid with heterocyclic carboxylic acid. J. Mol. Struct. 2020;1204:127508. [Google Scholar]
- 31.Zhang X., Chen J., Lou G., Li J., Wang F. Theoretical prediction of new structure, mechanical properties, anisotropy in elasticity and thermodynamic properties of Mo3Ge material. Vacuum. 2019;170:108978. [Google Scholar]
- 32.Adeniji S.E., Shallangwa G.A., Arthur D.E., Abdullahi M., Mahmoud A.Y., Haruna A. Quantum modelling and molecular docking evaluation of some selected quinoline derivatives as anti-tubercular agents. Heliyon. 2020;6(3) doi: 10.1016/j.heliyon.2020.e03639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flores-Holguín N., Frau J., Glossman-Mitnik D. Chemical reactivity and bioactivity properties of the Phallotoxin family of fungal peptides based on Conceptual Peptidology and DFT study. Heliyon. 2019;5(8) doi: 10.1016/j.heliyon.2019.e02335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma C., Quraishi M.A., Ebenso E.E. Quinoline and its derivatives as corrosion inhibitors: a review. Surface. Interfac. 2020:100634. [Google Scholar]
- 35.Alamshany Z.M., Ganash A.A. Synthesis, characterization, and anti-corrosion properties of an 8-hydroxyquinoline derivative. Heliyon. 2019;5(11) doi: 10.1016/j.heliyon.2019.e02895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uludağ N., Serdaroğlu G. New route for synthesis of 2-(2, 2-dimethoxyethyl)-1, 2, 3, 4, 5, 6-hexahydro-1, 5-methanoazocino [4, 3-b] indole and DFT investigation. Heliyon. 2020;6(6) doi: 10.1016/j.heliyon.2020.e04105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasitha T., John W.J. Design, docking, and DFT investigations of 2, 6-bis (3, 4-dihydroxyphenyl)-3-phenethylpiperidin-4-one. Heliyon. 2021;7(2) doi: 10.1016/j.heliyon.2021.e06127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurtado-Aular O., Vidal A.B., Peña-Mena J., Sierraalta A., Añez R. DFT thermodynamic study of the adsorption of CO2 and H2O on W3Ox/M (1 1 1)(x= 6 or 9 and M= Cu, Ag or Au). Insight for the water-gas shift reaction. Appl. Surf. Sci. 2020;531:147337. [Google Scholar]
- 39.Caglioti C., Palazzetti F. Potential energy surfaces for water interacting with diatomic heteronuclear molecules: H2O–HF as a case study. Chem. Phys. Lett. 2021:138692. [Google Scholar]
- 40.Warad I., Musameh S., Badran I., Nassar N.N., Brandao P., Tavares C.J., Barakat A. Synthesis, solvatochromism and crystal structure of trans-[Cu (Et2NCH2CH2NH2) 2. H2O](NO3) 2 complex: experimental with DFT combination. J. Mol. Struct. 2017;1148:328–338. [Google Scholar]
- 41.Carlotto S., Pandolfo L., Casarin M. Trinuclear Cu (II) complexes from the classic [Cu2 (RCOO) 4 (H2O) 2] lantern complex and pyrazole: a DFT modelling of the reaction path. Inorg. Chim. Acta. 2018;470:93–99. [Google Scholar]
- 42.Centanni M., Moes D.J.A., Trocóniz I.F., Ciccolini J., van Hasselt J.C. Clinical pharmacokinetics and pharmacodynamics of immune checkpoint inhibitors. Clin. Pharmacokinet. 2019:1–23. doi: 10.1007/s40262-019-00748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alffenaar J.W.C., Gumbo T., Dooley K.E., Peloquin C.A., McIlleron H., Zagorski A.,., Migliori G.B. Integrating pharmacokinetics and pharmacodynamics in operational research to end tuberculosis. Clin. Infect. Dis. 2020;70(8):1774–1780. doi: 10.1093/cid/ciz942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trucks G.W., Frisch M.J., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., Peralta J.E., Jr., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas O., Foresman J.B., Ortiz J.V., Fox J. Cioslowskiand D.J. Gaussian, Inc.; Wallingford CT: 2013. Gaussian09, Revision D. 01. [Google Scholar]
- 45.Scior T., Abdallah H.H., Salvador-Atonal K., Laufer S. Dapsone is not a pharmacodynamic lead compound for its aryl derivatives. Curr. Comput. Aided Drug Des. 2020;16(3):327–339. doi: 10.2174/1573409915666191010104527. [DOI] [PubMed] [Google Scholar]
- 46.Tian Lu., Feiwu Chen. Multiwfn: a multifunctional analyzer. J. Comput. Chem. 2012;33(5):580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- 47.Emamian S., Lu T., Kruse H., Emamian H. Exploring nature and predicting strength of hydrogen bonds: a correlation analysis between atoms-in-molecules descriptors, binding energies, and energy components of symmetry-adapted perturbation theory. J. Comput. Chem. 2019;40(32):2868–2881. doi: 10.1002/jcc.26068. [DOI] [PubMed] [Google Scholar]
- 48.Brandenburg J.G., Grimme S. Accurate modeling of organic molecular crystals by dispersion-corrected density functional tight binding (DFTB) J. Phys. Chem. Lett. 2014;5(11):1785–1789. doi: 10.1021/jz500755u. [DOI] [PubMed] [Google Scholar]
- 49.Xiao Meng, Lu Tian. Generalized charge decomposition analysis (GCDA) method. J. Adv. Phys. Chem. 2015;4:111–124. [Google Scholar]
- 50.Emamian S., Lu T., Kruse H., Emamian H. Exploring nature and predicting strength of hydrogen bonds: a correlation analysis between atoms-in-molecules descriptors, binding energies, and energy components of symmetry-adapted perturbation theory. J. Comput. Chem. 2019;40(32):2868–2881. doi: 10.1002/jcc.26068. [DOI] [PubMed] [Google Scholar]
- 51.Mierzwa G., Gordon A.J., Berski S. Topological analysis of electron localisation function: unlocking the nature of BC chemical bond. Possible existence of multiple bonds BC and BC. Polyhedron. 2019;170:180–187. [Google Scholar]
- 52.Ochterski J.W. Vol. 1. Gaussian Inc; 2000. Thermochemistry in Gaussian; p. 19. [Google Scholar]
- 53.Zhao J., Khalizov A., Zhang R., McGraw R. Hydrogen-bonding interaction in molecular complexes and clusters of aerosol nucleation precursors. J. Phys. Chem. 2009;113(4):680–689. doi: 10.1021/jp806693r. [DOI] [PubMed] [Google Scholar]
- 54.Xie C., Yan D., Chen W., Zou Y., Chen R., Zang S.,., Wang S. Insight into the design of defect electrocatalysts: from electronic structure to adsorption energy. Mater. Today. 2019;31:47–68. [Google Scholar]
- 55.Mao Y., Head-Gordon M. Probing blue-shifting hydrogen bonds with adiabatic energy decomposition analysis. J. Phys. Chem. Lett. 2019;10(14):3899–3905. doi: 10.1021/acs.jpclett.9b01203. [DOI] [PubMed] [Google Scholar]
- 56.Stasyuk O.A., Sedlak R., Guerra C.F., Hobza P. Comparison of the DFT-SAPT and canonical EDA schemes for the energy decomposition of various types of noncovalent interactions. J. Chem. Theor. Comput. 2018;14(7):3440–3450. doi: 10.1021/acs.jctc.8b00034. [DOI] [PubMed] [Google Scholar]
- 57.Sadeghi S., Shiri H.M., Ehsani A., Oftadeh M. Solid State Sciences; 2020. Electrosynthesis of high-purity TbMn2O5 nanoparticles and its nanocomposite with conjugated polymer: surface, density of state and electrochemical investigation; p. 106227. [Google Scholar]
- 58.Barborini M., Sorella S., Rontani M., Corni S. Correlation effects in scanning tunneling microscopy images of molecules revealed by quantum Monte Carlo. J. Chem. Theor. Comput. 2016;12(11):5339–5349. doi: 10.1021/acs.jctc.6b00710. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Ying, Chen Hongbin. Phenylalanine system fragment CDA and orbital interaction. J. Jilin Univ. (Sci. Ed.) 2019;57(1):145–150. [Google Scholar]
- 60.Lu T., Manzetti S. Wavefunction and reactivity study of benzo [a] pyrene diol epoxide and its enantiomeric forms. Struct. Chem. 2014;25(5):1521–1533. [Google Scholar]
- 61.Emamian S., Lu T., Kruse H., Emamian H. Exploring nature and predicting strength of hydrogen bonds: a correlation analysis between atoms-in-molecules descriptors, binding energies, and energy components of symmetry-adapted perturbation theory. J. Comput. Chem. 2019;40(32):2868–2881. doi: 10.1002/jcc.26068. [DOI] [PubMed] [Google Scholar]
- 62.Shahbazian S. Why bond critical points are not “bond” critical points. Chem. A Eur. J. 2018;24(21):5401–5405. doi: 10.1002/chem.201705163. [DOI] [PubMed] [Google Scholar]
- 63.Noorizadeh S., Shakerzadeh E. Shannon entropy as a new measure of aromaticity, Shannon aromaticity. Phys. Chem. Chem. Phys. 2010;12(18):4742–4749. doi: 10.1039/b916509f. [DOI] [PubMed] [Google Scholar]
- 64.Matrodi A., Noorizadeh S. N-Derivatives of Shannon entropy density as response functions. Phys. Chem. Chem. Phys. 2020;22(37):21535–21542. doi: 10.1039/d0cp03808c. [DOI] [PubMed] [Google Scholar]
- 65.Gould I.R., Hillier I.H. Solvation of alanine dipeptide: a quantum mechanical treatment. J. Chem. Soc., Chem. Commun. 1993;(11):951–952. [Google Scholar]
- 66.Allen L.C., Egolf D.A., Knight E.T., Liang C. Bond polarity index. J. Phys. Chem. 1990;94(14):5602–5607. [Google Scholar]
- 67.Ghasemi A.S., Taghartapeh M.R., Soltani A., Mahon P.J. Adsorption behavior of metformin drug on boron nitride fullerenes: thermodynamics and DFT studies. J. Mol. Liq. 2019;275:955–967. [Google Scholar]
- 68.Bamba K., Patrice O.W., Ziao N. NBO population analysis and electronic calculation of four azopyridine ruthenium complexes by DFT method. Comput. Chem. 2017;5(1):51. [Google Scholar]
- 69.Günay N., Tamer Ö., Kuzalic D., Avci D., Atalay Y. Theoretical investigation of N-methyl-N'-(4-nitrobenzylidene) pyrazine-2-carbohydrazide: conformational study, NBO analysis, molecular structure and NMR spectra. Acta Phys. Pol., A. 2015;127(3) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.