Abstract
Background
Inevitable resistance to chemotherapeutic drugs has become a major obstacle for the clinical treatment of multiple myeloma (MM). Circular RNAs (circRNAs) can regulate the chemoresistance in different tumors. Our study was to explore the regulation of circRNA arginine-glutamic acid dipeptide repeats (circRERE) in bortezomib (BTZ) resistance of MM.
Methods
CircRERE, microRNA-152-3p (miR-152-3p) and cluster of differentiation 47 (CD47) levels were assayed through the quantitative real-time polymerase chain reaction (qRT-PCR). Cell sensitivity to BTZ was analyzed using Cell Counting Kit-8 (CCK-8) assay. Cell proliferation and apoptosis were determined via colony formation assay and flow cytometry, respectively. The detection of all proteins was conducted by western blot. The target binding was analyzed via the dual-luciferase reporter assay and RIP assay.
Results
We found the upregulation of circRERE in BTZ-resistant MM samples and cells. BTZ resistance was inhibited after circRERE expression was downregulated in MM cells. CircRERE was identified to act as a miR-152-3p sponge. The effect of circRERE on the BTZ resistance was associated with the sponge function for miR-152-3p. CD47 was a target for miR-152-3p and circRERE could sponge miR-152-3p to generate the expression regulation of CD47. MiR-152-3p facilitated the susceptibility of MM cells to BTZ by targeting CD47.
Conclusion
These results suggested that circRERE could suppress the BTZ resistance in MM cells by mediating the miR-152-3p/CD47 axis.
Keywords: CircRERE, Bortezomib, Multiple myeloma, miR-152-3p, CD47
1. Introduction
Multiple myeloma (MM) ranks the second place in hematologic malignancies, and it is mainly diagnosed in elderly people with a median age of 70 years old [1]. The median survival for MM has been increased to almost 50% following the advancement of modern and diversified therapies [2]. Chemotherapy plays a vital role in the clinal MM treatment, and bortezomib (BTZ) is one of the most commonly used chemotherapeutic drugs [3]. BTZ is a proteasome inhibitor that can improve the prognosis and treatment of MM, but the therapeutic effect has been largely affected due to the intrinsic and acquired chemoresistance [4]. To discover the regulatory targets for BTZ resistance may be beneficial for resistant MM patients.
Noncoding RNAs (ncRNAs) participate in all aspects of cancer biology, such as cell apoptosis, proliferation and drug resistance [5]. Circular RNAs (circRNAs) are covalently closed-loop ncRNAs without 5′ and 3′ ends, and they are produced by back-splicing of premessenger RNAs. The circular structures confer circRNAs high stability relative to linear RNAs. CircRNAs can modulate the gene expression to affect the biological processes of cancers by functioning as “microRNA (miRNA) sponges” [6]. CircITCH could promote the sensitivity of BTZ to MM cells by the mediation of miR-615-3p/PRKCD axis [7], and circ_0007841 increased the resistance of BTZ via relying on the miR-129-5p/JAG1 pathway in MM [8]. The research of Zhou et al. has indicated that circRNA arginine-glutamic acid dipeptide repeats (circRERE) was upregulated in MM patients [9]. It is unknown whether circRERE is implicated in the chemoresistance regulation in MM.
MicroRNA-152-3p (miR-152-3p) expression was downregulated in MM patient samples, and it acted as a tumor repressor in the malignant progression of MM by targeting BRD4 [10]. Cluster of differentiation 47 (CD47) is an immune checkpoint and anti-CD47 therapeutic strategy has been used for killing MM cells [11], [12]. MiRNAs evoke the expression alteration of downstream targets by combining with the 3′untranslated regions (3′UTRs) of mRNAs in cancer regulation [13]. Drug resistance in MM has been overcome by miR-155/CD47 axis [14]. It is potential that miR-152-3p participates in the modulation of the BTZ resistance in MM by targeting CD47.
The functional role of circRERE and the regulation of circRERE/miR-152-3p/CD47 network in the chemoresistance of BTZ are two key points of this study. Our findings were devoted to explore the molecular understanding of the resistance mechanism in MM and develop the probable therapeutic targets for drug-resistant MM patients.
2. Materials and methods
2.1. Serum samples
Normal serum samples (NC, n = 30) were donated by healthy populations in the physical examination center of the First Affiliated Hospital of Chongqing Medical University. 125 MM patients at the First Affiliated Hospital of Chongqing Medical University have enrolled in this study and provided the written informed consent forms. These patients were diagnosed at I stage (n = 20), II stage (n = 45) and III stage (n = 60) according to the International Staging System (ISS). 65 patients have not received any treatment, and the serum samples were considered as BTZ-sensitive group (normal MM, N-MM; n = 65). 60 patients with BTZ treatment has generated chemoresistance through the clinical observation, and the serum samples were considered as BTZ-resistant group (resistant MM, R-MM; n = 60). The patients with other types of treatments have been excluded from this study. Then, all specimens were stored in liquid nitrogen for later use. The Declaration of Helsinki was followed in overall operations, and this study was approved by the Institutional Review Board of the First Affiliated Hospital of Chongqing Medical University.
2.2. Cell lines
The parental MM cell lines 8226 and MM1.S were purchased from BioVector NTCC Inc. (Beijing, China). 8226R5 that is a multidrug-resistant MM cell line with cross-resistance to BTZ has been kindly donated by Dr. R Buzzeo [15]. MM1.R (BioVector NTCC Inc.) with resistance to dexamethasone has also been shown to be resistant to BTZ [16]. These cells were cultivated in Roswell Park Memorial Institute-1640 (RPMI-1640; Gibco, Carlsbad, CA, USA) complemented with 10% fetal bovine serum (FBS; Gibco) and 1% antibiotics (Gibco) in a 37 °C, 5% CO2 incubator. 8226R5 and MM1.R cells were subjected to the treatment of 10 nM BTZ (PS-341; Selleck, Houston, TX, USA) for 24 h.
2.3. Cell transfection
To construct the stable cell lines, lentiviral vector containing short hairpin RNA (shRNA) of circRERE (sh-circRERE) and that containing shRNA of negative control (sh-NC) were provided by GenePharma (Shanghai, China). MiR-152-3p mimic, mimic NC, miR-152-3p inhibitor and inhibitor NC were also from GenePharma. The pcDNA expression vector and the pcDNA-CD47 recombinant vector were obtained from Invitrogen (Carlsbad, CA, USA). Lipofectamine™ 3000 (Invitrogen) was used for the transfection of RNAs or vectors in 8226R5 and MM1.R cells, following the user guide supplied by the manufacturer.
2.4. The quantitative real-time polymerase chain reaction (qRT-PCR) assay
As per the instruction book, Beyozol reagent (Beyotime, Shanghai, China) was used for the RNA purification. RNA was then reversely transcribed into the complementary RNA (cDNA) by the miScript II RT Kit and QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). The quantitative expression was detected using miScript SYBR Green PCR Kit and QuantiTect SYBR® Green PCR Kit (Qiagen), followed by the data analysis by the 2−ΔΔCt method [17]. The sequences for all primers were presented as below: circRERE, 5′-TTTGCAGGAATGTGTGATGG-3′ (forward, F) and 5′-ATGAATCCACTCGGGCTTTA-3′ (reverse, R); RERE, 5′-GCAGAGACAGTGAAGAAGTCGG-3′ (F) and 5′-CTTCTTGGAGCTGGTCCTGTCA-3′ (R); miR-152-3p, 5′-GCGCTCAGTGCATGACAGA-3′ (F) and 5′-GTCGTATCCAGTGCAGGGTC-3′ (R); CD47, 5′-TATCCTCGCTGTGGTTGGACTG-3′ (F) and 5′-TAGTCCAAGTAATTGTGCTAGAGC-3′ (R); glyceraldehyde-phosphate dehydrogenase (GAPDH), 5′-GTCTCCTCTGACTTCAACAGCG-3′ (F) and 5′-ACCACCCTGTTGCTGTAGCCAA-3′ (R); U6, 5′-CTCGCTTCGGCAGCACA-3′ (F) and 5′-AACGCTTCACGAATTTGCGT-3′ (R). The normalization of expression level was performed by using GAPDH and U6 as the internal controls.
2.5. RNase R and Actinomycin D treatment
To analyze the stability of circRERE and linear RERE, total RNA was digested with RNase R (Epicentre Technologies, Madison, WI, USA) for 1 h and cells were incubated with Actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) for different times (0 h, 4 h, 8 h, 12 h). Then, the circRERE and RERE level examination was administrated via the qRT-PCR.
2.6. Cell sensitivity assay
The different concentrations (0 nM, 1 nM, 5 nM, 10 nM, 20 nM, 40 nM, 80 nM, 160 nM) of BTZ treatment were performed for 8225R5 and MM1.R cells, followed by the incubation of CCK-8 solution (Beyotime) with 10 μL/well for 120 min. The absorbance at 450 nm was determined by the microplate reader (Bio-Rad, Hercules, CA, USA). When cell viability was reduced to 50%, the BTZ concentration represented the maximum half inhibitory concentration (IC50) for BTZ.
2.7. Colony formation assay
8226R5 and MM1.R cells with different treatment were seeded onto the 12-well plates at the cell density of 500 cells/well, then these cells were incubated in the 37 °C incubator for two weeks. The colonies were counted by a microplate reader (Bio-Rad) after cell staining using crystal violet (Sigma-Aldrich).
2.8. Flow cytometry
Cell apoptosis was evaluated via the Apoptosis Detection Kit (BD Biosciences, San Diego, CA, USA). Briefly, the harvested cells were stained with Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and Propidium Iodide (PI) for 30 min at room temperature in the dark environment. Ultimately, cell status was differentiated by a flow cytometer (BD Biosciences). The cell percentages at the first quadrant and the fourth quadrant were used to exhibit the apoptotic rate.
2.9. Western blot
Western blot assay was applied to assay the expression levels of different proteins in accordance with the previous descriptions [18], [19]. The primary antibodies were purchased from Cell Signaling Technology (CST, Boston, MA, USA), including anti-pro-caspase3 (#14220), anti-cleaved-caspase3 (#9664), anti-proliferating cell nuclear antigen (anti-PCNA; #13110), anti-ki-67 (#9129), anti-CD47 (#63000) and anti-GAPDH (#2118). All primary antibodies were incubated at the ratio of 1:1000, and Anti-rabbit IgG, HRP-linked Secondary Antibody (CST, #7074) was incubated with 1:3000. Finally, the protein level was quantified using the ImageJ software (NIH, Bethesda, MD, USA).
2.10. Dual-luciferase reporter assay
Starbase3.0 (http://starbase.sysu.edu.cn) was used to predict the binding between miR-152-3p and circRERE or CD47. To analyze the interaction between miR-152-3p and circRERE, the circRERE luciferase plasmids of wild-type (circRERE wt) and mutant-type (circRERE mut) were generated by the molecular cloning into the pmirGLO plasmid (Promega, Madison, WI, USA). For miR-152-3p and CD47, the luciferase plasmids CD47 3′UTR wt and CD47 3′UTR mut were constructed. These plasmids were respectively co-transfected with mimic NC or miR-152-3p mimic into 293 T cells (BioVector NTCC Inc.) for 48 h, then the luciferase activity examination was carried out through the Luc-Pair™ Duo-Luciferase HS Assay Kit (GeneCopoeia, Rockville, MA, USA).
2.11. RNA immunoprecipitation (RIP) assay
Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was applied to investigate whether circRERE could bind to miR-152-3p. The magnetic beads were pre-coated with anti-Argonaute-2 (anti-Ago2) and anti-immunoglobulin G (anti-IgG). 8226R5 and MM1.R cells were collected and incubated with the antibody-coated magnetic beads, and the unincubated cells acted as the positive Input group. The magnetic beads were washed using phosphate buffer solution (PBS; Gibco) and the immunoprecipitated RNAs were extracted, then the levels of circRERE and miR-152-3p were assayed via qRT-PCR.
2.12. Statistical analysis
Data of three repetitions were indicated as the mean ± standard deviation (SD). The relationship of target was analyzed by Pearson’s correlation coefficient. The survival analysis was performed by log-rank test. SPSS 22.0 (SPSS Inc., Chicago, IL, USA) was exploited for the statistical detection, and the difference was analyzed using Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test. If P value was less than 0.05, the difference was regarded as significant.
3. Results
3.1. CircRERE was highly expressed in BTZ-resistant MM samples and cells
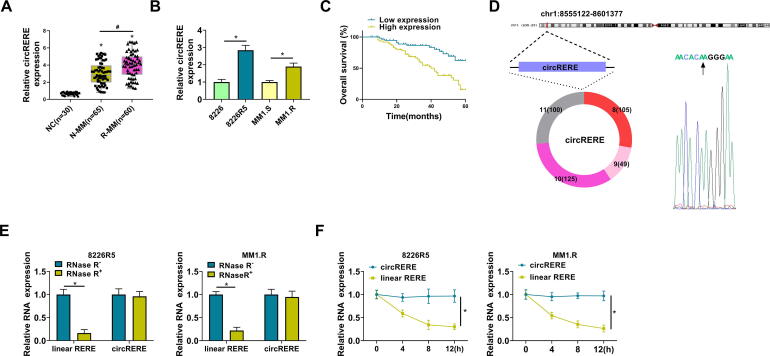
The RNA was extracted from serum samples, followed by the qRT-PCR analysis for circRERE. As shown in Fig. 1A, the circRERE level was upregulated in BTZ-resistant MM serum samples (n = 60) contrasted with N-MM (n = 65) and NC (n = 30) groups. The expression detection in cells also indicated that circRERE expression was higher in 8226R5 and MM1.R cells than that in 8226 and MM1.S cells (Fig. 1B). The clinical features indicated that high expression of circRERE was associated with disease stage and chemoresistance, and the detailed information was shown in Table 1. In addition, the survival analysis showed that circRERE upregulation could predict poor prognosis of MM patients (Fig. 1C). CircRERE (has_circ_0009581) is derived from the back-splicing of exon 8–11 of RERE gene with the location of chr1: 8555122–8601377 and splicing junction of AACACAAGGGAA (Fig. 1D). CircRNAs exhibit high stability in comparison to linear RNAs [20]. Our data demonstrated that circRERE was not affected by the treatment of RNase R (Fig. 1E) and Actinomycin D (Fig. 1F), while the RERE level was significantly reduced. Thus, circRERE was more stable than the linear RERE. CircRERE was aberrantly expressed in BTZ-resistant MM samples and cells.
Fig. 1.
CircRERE was highly expressed in BTZ-resistant MM samples and cells. (A) The expression of circRERE was determined by qRT-PCR in NC, N-MM and R-MM serum samples. (B) The detection of circRERE was performed by qRT-PCR in the parental and resistant MM cells. (C) The survival analysis was performed by log-rank test in MM patients. (D) The genic information of circRERE. (E-F) The circRERE and linear RERE levels were assayed using qRT-PCR after the treatment of RNase R (E) or Actinomycin D (F). *P < 0.05.
Table 1.
Clinical and laboratory features of MM patients with circRERE expression.
| Characteristics | Total | circRERE (hsa_circ_0009581) |
P value | |
|---|---|---|---|---|
| Low | High | |||
| Age | ||||
| <60 | 66 | 36 | 30 | 0.3725 |
| ≥60 | 59 | 27 | 32 | |
| Gender | ||||
| Female | 65 | 35 | 30 | 0.476 |
| Male | 60 | 28 | 32 | |
| M protein | ||||
| IgG | 34 | 20 | 14 | 0.0153* |
| IgA | 26 | 18 | 8 | |
| Light chain | 65 | 25 | 40 | |
| ISS stage | ||||
| I | 20 | 14 | 6 | 0.0003* |
| II | 45 | 30 | 15 | |
| III | 60 | 19 | 41 | |
| Durie-Salmon stage | ||||
| I | 18 | 11 | 7 | 0.0027* |
| II | 35 | 25 | 10 | |
| III | 72 | 27 | 45 | |
| Hypercalcemia | ||||
| Yes | 70 | 40 | 30 | 0.0889 |
| No | 55 | 23 | 32 | |
| Renal insufficiency | ||||
| Yes | 51 | 22 | 29 | 0.1776 |
| No | 74 | 41 | 33 | |
| Bone lesion | ||||
| Yes | 69 | 37 | 32 | 0.4237 |
| No | 56 | 26 | 30 | |
| Cytogenetic abnormality | ||||
| Yes | 44 | 18 | 26 | 0.1178 |
| No | 81 | 45 | 36 | |
| Chemotherapy | ||||
| Chemoresistance | 60 | 23 | 37 | 0.0095* |
| Chemosensitivity | 65 | 40 | 25 | |
3.2. Knockdown of circRERE reduced the resistance of BTZ to MM cells
The knockdown efficiency was detected by qRT-PCR in sh-NC and sh-circRERE-transfected cells. The results manifested that circRERE was downregulated in sh-circRERE-transfected 8226R5 and MM1.R cells compared with sh-NC-transfected cells (Fig. 2A). CCK-8 assay revealed that the IC50 of BTZ was lower in sh-circRERE group than that in sh-NC group of 8226R5 and MM1.R cells (Fig. 2B). The stably transfected cells were treated with PBS or BTZ for 24 h. Colony formation assay and flow cytometry showed that silencing circRERE inhibited cell proliferation (Fig. 2C-D) and promoted cell apoptosis (Fig. 2E-F) after the treatment of PBS or BTZ. The proliferative and apoptotic markers were examined using western blot. Downregulation of circRERE was observed to upregulate the protein expression of cleaved-caspase3/pro-caspase3 but decrease the protein levels of PCNA and ki-67 in PBS and BTZ groups (Fig. 2G-H). Taken together, circRERE knockdown repressed the BTZ resistance to enhance the anti-tumor effect of BTZ on MM cells.
Fig. 2.
Knockdown of circRERE reduced the resistance of BTZ to MM cells. (A) The qRT-PCR was applied to evaluate the knockdown efficiency of circRERE in sh-NC/sh-circRERE-transfected 8226R5 and MM1.R cells. (B) CCK-8 assay was applied to examine the IC50 of BTZ. (C-D) Colony formation assay was applied to assess cell proliferation after the transfected cells were treated with PBS or BTZ. (E-F) Flow cytometry was used for the analysis of cell apoptosis. (G-H) Western blot was used for the quantification of pro-caspase3/cleaved-caspase3, PCNA and ki-67. *P < 0.05.
3.3. CircRERE was a natural miR-152-3p sponge
By performing the prediction of starbase3.0, the complementary sites for miR-152-3p were noticed in the sequence of circRERE (Fig. 3A). Dual-luciferase reporter assay in 293 T cells indicated that miR-152-3p mimic transfection suppressed the luciferase activity in circRERE wt plasmid relative to mimic NC transfection, but no change of luciferase activity was exhibited in circRERE mut plasmid after transfection of mimic NC and miR-152-3p mimic (Fig. 3B). As the results of RIP assay in Fig. 3C, circRERE and miR-152-3p levels were simultaneously increased in anti-Ago group but not anti-IgG group. The expression of miR-152-3p was shown to be downregulated in BTZ-resistant MM serum samples (Fig. 3D) and MM cells (Fig. 3E), in contrast with the sensitive MM serum samples and the parental MM cells. Through the analysis of Pearson’s correlation coefficient, we found there was a negative relation (r = −0.5198, P < 0.001) between the expression levels of circRERE and miR-152-3p in MM samples (Fig. 3F). These findings could prove the sponge effect of circRERE on miR-152-3p.
Fig. 3.
CircRERE was a natural miR-152-3p sponge. (A) CircRERE was predicted to be combined with miR-152-3p by starbase3.0. (B-C) The binding analysis between miR-152-3p and circRERE was conducted by dual-luciferase reporter assay (B) and RIP assay (C). (D-E) The miR-152-3p expression detection was performed using qRT-PCR in NC/N-MM/R-MM serum samples (D) and the parental or resistant cell lines (E). (F) The correlation between circRERE and miR-152-3p was analyzed via the Pearson’s correlation coefficient. *P < 0.05.
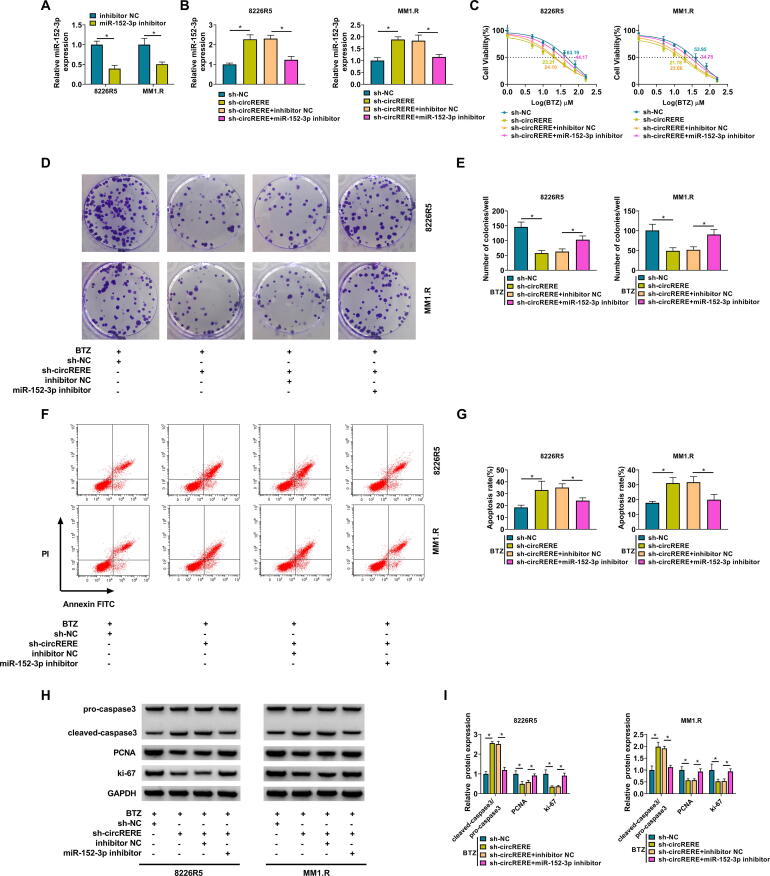
3.4. The inhibitory effect of circRERE knockdown on the BTZ resistance was attenuated by miR-152-3p inhibitor
To confirm that the function of circRERE was associated with miR-152-3p sponging, the reverted experiments were performed. Firstly, the qRT-PCR has exhibited the great repression of miR-152-3p expression by miR-152-3p inhibitor compared to inhibitor NC group (Fig. 4A). In addition, the upregulation of miR-152-3p caused by sh-circRERE was weakened after the transfection of miR-152-3p inhibitor (Fig. 4B). The sh-circRERE-mediated suppressive influences on IC50 of BTZ (Fig. 4C) and cell proliferation (Fig. 4D-E), as well as the stimulative influence on cell apoptosis (Fig. 4F-G), were alleviated following the expression inhibition of miR-152-3p in 8226R5 and MM1.R cells. The protein regulation of pro-caspase3/pro-caspase3, PCNA and ki-67 by the knockdown of circRERE was also blocked with the downregulation of miR-152-3p (Fig. 4H-I). The above evidence affirmed that the regulatory effect of circRERE on the BTZ resistance in MM cells was concerned with acting as a miR-152-3p sponge.
Fig. 4.
The inhibitory effect of circRERE knockdown on the BTZ resistance was attenuated by miR-152-3p inhibitor. (A) The transfection efficiency of miR-152-3p inhibitor was tested via qRT-PCR in 8226R5 and MM1.R cells. (B) The expression of miR-152-3p was assayed via qRT-PCR in the sh-NC, sh-circRERE, sh-circRERE + inhibitor NC, and sh-circRERE + miR-152-3p inhibitor groups. (C) The IC50 for BTZ was determined by CCK-8 assay. (D-G) Cell proliferation (D-E) and apoptosis (F-G) were respectively measured by CCK-8 assay and flow cytometry after the above transfection and 10 nM BTZ treatment. (H-I) The protein levels of pro-caspase3/cleaved-caspase3, PCNA and ki-67 were detected using western blot. *P < 0.05.
3.5. CircRERE induced the positive regulation of CD47 by targeting miR-152-3p
CD47 3′UTR sequence contained the binding sites for miR-152-3p (Fig. 5A), implying that CD47 might be a downstream target of miR-152-3p. In 293 T cells, the luciferase activity inhibition was only found in CD47 3′UTR wt group rather than CD47 3′UTR mut group after the miR-152-3p was overexpressed (Fig. 5B). Western blot has revealed that CD47 protein expression was elevated in 8226R5 and MM1.R cells by comparison to the parental cells (Fig. 5C). The high expression of CD47 was also confirmed by qRT-PCR and western blot in BTZ-resistant MM serum samples, contrasted to N-MM and NC serum samples (Fig. 5D-E). Pearson’s correlation coefficient demonstrated that CD47 expression was negatively related to miR-152-3p expression (r = −0.5350, P < 0.0001) (Fig. 5F). More interestingly, there was a positive relationship (r = −0.5090, P < 0.001) between the expression of circRERE and CD47 (Fig. 5G). The protein level of CD47 was reduced by the downregulation of circRERE but this effect was restored by miR-152-3p inhibitor (Fig. 5H). Hence, miR-152-3p targeted CD47 and circRERE regulated CD47 in a positive way by targeting miR-152-3p.
Fig. 5.
CircRERE induced the positive regulation of CD47 by targeting miR-152-3p. (A) Starbase3.0 has exhibited the binding sites between CD47 3′UTR and miR-152-3p. (B) Dual-luciferase reporter assay was performed to validate the interaction between CD47 3′UTR and miR-152-3p. (C) CD47 protein analysis was conducted by western blot in the parental and resistant MM cells. (D-E) The qRT-PCR and western blot were used to examine the mRNA and protein levels of CD47 in NC, N-MM and R-MM serum samples. (F-G) The analysis of linear relation between CD47 and miR-152-3p (F) or circRERE (G) was carried out using Pearson’s correlation coefficient. (H) The protein level of CD47 was assayed via western blot after transfection of sh-NC, sh-circRERE, sh-circRERE + inhibitor NC or sh-circRERE + miR-152-3p inhibitor. *P < 0.05.
3.6. MiR-152-3p acted as a sensitizer of MM cells to BTZ by decreasing the CD47 level
The involvement of miR-152-3p and CD47 in BTZ resistance was further researched. The qRT-PCR and western blot respectively indicated that the overexpression effects of miR-152-3p mimic on the miR-152-3p expression (Fig. 6A) and pcDNA-CD47 on the CD47 protein expression (Fig. 6B) were conspicuous. Additionally, the CD47 protein downregulation mediated by miR-152-3p mimic was reversed by the introduction of pcDNA-CD47 (Fig. 6C). The overexpression of miR-152-3p led to the repression of the IC50 for BTZ (Fig. 6D) and cell proliferative ability (Fig. 6E-F) but the acceleration of cell apoptosis (Fig. 6G-H), which were then abolished by the promotion of CD47 expression. Meanwhile, the protein detection by western blot manifested also that CD47 upregulation countervailed the miR-152-3p mimic-induced pro-apoptotic and anti-proliferative effects in 8226R5 and MM1.R cells (Fig. 6I-J). Overall, miR-152-3p could sensitize MM cells to BTZ by targeting CD47.
Fig. 6.
MiR-152-3p acted as a sensitizer of MM cells to BTZ by decreasing the CD47 level. (A) The promoting effects of miR-152-3p mimic and pcDNA-CD47 on miR-152-3p (A) and CD47 (B) were respectively assessed by qRT-PCR and western blot. (C) CD47 protein expression was measured using western blot after 8226R5 and MM1.R cells were transfected with mimic NC, miR-152-3p mimic, miR-152-3p mimic + pcDNA or miR-152-3p mimic + pcDNA-CD47. (D) The calculation of IC50 for BTZ was conducted via CCK-8 assay. (E-H) CCK-8 assay and flow cytometry were used for the respective detection of cell proliferation (E-F) and apoptosis (G-H) after treatment of 10 nM BTZ in the above transected cells. (I-J) The protein analysis for pro-caspase3/cleaved-caspase3, PCNA and ki-67 was performed by western blot. *P < 0.05.
4. Discussion
MM is responsible for more than 20% of human death from hematological cancers worldwide [10]. BTZ is the first-line chemotherapeutic agent for MM, but the failure of treatment for MM is frequently caused by the BTZ resistance [7]. This study found that circRERE facilitated the resistance of MM to BTZ by sponging miR-152-3p and upregulating CD47, indicating that circRERE/miR-152-3p/CD47 could be used to enhance the BTZ sensitivity in MM.
CircRERE was confirmed as a stable circular RNA in MM cells. Its expression pattern was significantly increased in BTZ-resistant MM samples and cells. Functionally, the expression knockdown of circRERE impeded cell proliferation and BTZ resistance but facilitated apoptosis of MM cells. In the issued studies, circRNAs have been revealed to serve as the regulators for drug resistance in MM. For example, circMYC was associated with the BTZ resistance and the recurrence of MM [21]. Circ_0007841 contributed to the resistance of doxorubicin in MM cells [22]. The regulation of circRNAs for chemoresistance was also manifested in other cancers. Zhang et al. have reported that circ_0003998 repressed the chemotherapeutic sensitivity of non-small cell lung cancer (NSCLC) cells to docetaxel via acting on the miR-136-5p/CORO1C axis [23]. Zheng et al. asserted that circGFRA1 reduced the paclitaxel resistance of triple-negative breast cancer cells by affecting the miR-361-5p/TLR4 regulatory axis [24]. Li et al. reported that cisplatin susceptibility was restrained by circARNT2 in hepatocellular carcinoma cells via targeting the miR-155-5p/PDK1 axis [25]. These studies supported that the dysregulated circRNAs exerted the regulatory effects on drug resistance in cancers. Our evidence demonstrated that circRERE facilitated the resistance of MM cells to BTZ in vitro. The miRNA/mRNA axis for circRERE in the resistance regulation of MM was to be explored by further experiments.
The prediction software has shown the binding sites of miR-152-3p in circRERE sequence. Subsequently, the molecular binding between circRERE and miR-152-3p was validated. Moreover, the sh-circRERE-mediated inhibitory effect on BTZ resistance in MM cells was counterbalanced by the inhibition of miR-152-3p. Increasing circRNAs have been identified as the sponges for miRNAs in regulating various biological behaviors. For instance, circ_0071036 promoted cell invasion and proliferation by sponging miR-489 in pancreatic cancer [26]. CircSMYD4 was affirmed as a miRNA sponge to target miR-584-5p and regulate cell metastasis in hepatocellular carcinoma [27]. Hsa_circ_0002483 retarded the cancer progression and elevated the taxol sensitivity by targeting miR-182-5p in NSCLC [28]. In accordance with the sponge effects of these circRNAs on miRNAs, circRERE regulated the BTZ resistance in MM cells by suppressing miR-152-3p as a natural sponge.
Furthermore, CD47 was proved as a downstream target for miR-152-3p and BTZ resistance was inhibited by miR-152-2p through reducing the CD47 level in MM cells. A variety of miRNA/mRNA axes have been involved in the BTZ resistance regulation in MM, such as miR-631/UbcH10, miR-218/LRRC28 and miR-1252-5p/HPSE [29], [30], [31]. In addition, Sun et al. have shown that hsa_circ_0020095 improved oncogenesis and cisplatin resistance by sponging miR-487a-3p to modulate the SOX9 level in colon cancer [32]. Hsa_circ_0000714 was displayed to trigger the expression promotion of RAB17 by absorbing miR-370-3p to inhibit the paclitaxel resistance in ovarian cancer [33]. Our analysis also exhibited that circRERE could sponge the miR-152-3p to regulate the expression pattern of CD47. The miR-152-3p-dependent CD47 expression change was considered to be accountable for the regulatory function of circRERE in carcinogenesis and BTZ resistance of MM.
This study still has some limitations. For instance, all experiments were performed in vitro. Animal model should be constructed to explore the function of circRERE in vivo. In addition, the expression detection in bone marrow samples might provide more information in clinic. All in all, our results validated that the function and mechanism of circRERE in chemoresistance. The further study will be conducted in future.
5. Conclusion
In conclusion, circRERE was associated with the emergence of BTZ resistance in MM by targeting the miR-152-3p/CD47 axis. More importantly, the downregulation of circRERE might be a therapeutic strategy to overcome the BTZ resistance in MM patients.
6. Data availability statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Schoenbeck K.L., Wildes T.M. Updated perspectives on the management of multiple myeloma in older patients: focus on lenalidomide. Clin. Interv. Aging. 2020;15:619–633. doi: 10.2147/CIA.S196087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond B., Maclachlan K., Chung D.J., Lesokhin A.M., Ola Landgren C. Maintenance therapy and need for cessation studies in multiple myeloma: focus on the future. Best Pract. Res. Clin. Haematol. 2020;33(1) doi: 10.1016/j.beha.2020.101140. [DOI] [PubMed] [Google Scholar]
- 3.Tabchi S., Nair R., Kunacheewa C., Patel K.K., Lee H.C., Thomas S.K., Amini B., Ahmed S., Mehta R.S., Bashir Q., Qazilbash M.H., Weber D.M., Orlowski R.Z., Alexanian R., Feng L., Manasanch E.E. Retrospective review of the use of high-dose cyclophosphamide, bortezomib, doxorubicin, and dexamethasone for the treatment of multiple myeloma and plasma cell leukemia. Clin. Lymphoma Myeloma Leuk. 2019;19(9):560–569. doi: 10.1016/j.clml.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Santamarta M., Quinet G., Reyes-Garau D., Sola B., Roue G., Rodriguez M.S. Resistance to the proteasome inhibitors: lessons from multiple myeloma and mantle cell lymphoma. Adv. Exp. Med. Biol. 2020;1233:153–174. doi: 10.1007/978-3-030-38266-7_6. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Liu X., Lin C., Jia X., Zhu H., Song J., Zhang Y. Noncoding RNAs regulate alternative splicing in Cancer. J. Exp. Clin. Cancer Res. 2021;40(1):11. doi: 10.1186/s13046-020-01798-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eger N., Schoppe L., Schuster S., Laufs U., Boeckel J.N. Circular RNA splicing. Adv. Exp. Med. Biol. 2018;1087:41–52. doi: 10.1007/978-981-13-1426-1_4. [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Du F., Chen C., Li D., Chen Y., Xiao X., Hou X. CircRNA ITCH increases bortezomib sensitivity through regulating the miR-615-3p/PRKCD axis in multiple myeloma. Life Sci. 2020;262 doi: 10.1016/j.lfs.2020.118506. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Lin Q., Song C., Ma R., Li X. Depletion of circ_0007841 inhibits multiple myeloma development and BTZ resistance via miR-129-5p/JAG1 axis. Cell Cycle. 2020;19(23):3289–3302. doi: 10.1080/15384101.2020.1839701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan C., Liu X., Li W., Wang H., Teng Y., Ren J., Huang Y. Circular RNA circ KMT2E is up-regulated in diabetic cataract lenses and is associated with miR-204-5p sponge function. Gene. 2019;710:170–177. doi: 10.1016/j.gene.2019.05.054. [DOI] [PubMed] [Google Scholar]
- 10.Zheng J.F., Guo N.H., Zi F.M., Cheng J. Long Noncoding RNA H19 promotes tumorigenesis of multiple myeloma by activating BRD4 signaling by targeting MicroRNA 152–3p. Mol. Cell. Biol. 2020;40(3) doi: 10.1128/MCB.00382-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. Sun, B. Muz, K. Alhallak, M. Markovic, S. Gurley, Z. Wang, N. Guenthner, K. Wasden, M. Fiala, J. King, D. Kohnen, N.N. Salama, R. Vij, A.K. Azab, Targeting CD47 as a Novel Immunotherapy for Multiple Myeloma, Cancers (Basel) 12(2) (2020). [DOI] [PMC free article] [PubMed]
- 12.P. Storti, R. Vescovini, F. Costa, V. Marchica, D. Toscani, B. Dalla Palma, L. Craviotto, F. Malavasi, N. Giuliani, CD14(+) CD16(+) monocytes are involved in daratumumab-mediated myeloma cells killing and in anti-CD47 therapeutic strategy, Br J Haematol 190(3) (2020) 430-436. [DOI] [PubMed]
- 13.Schuster S.L., Hsieh A.C. The untranslated regions of mRNAs in cancer. Trends Cancer. 2019;5(4):245–262. doi: 10.1016/j.trecan.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rastgoo N., Wu J., Liu A., Pourabdollah M., Atenafu E.G., Reece D., Chen W., Chang H. Targeting CD47/TNFAIP8 by miR-155 overcomes drug resistance and inhibits tumor growth through induction of phagocytosis and apoptosis in multiple myeloma. Haematologica. 2020;105(12):2813–2823. doi: 10.3324/haematol.2019.227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buzzeo R., Enkemann S., Nimmanapalli R., Alsina M., Lichtenheld M.G., Dalton W.S., Beaupre D.M. Characterization of a R115777-resistant human multiple myeloma cell line with cross-resistance to PS-341. Clin. Cancer Res. 2005;11(16):6057–6064. doi: 10.1158/1078-0432.CCR-04-2685. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L., Rastgoo N., Wu J., Zhang M., Pourabdollah M., Zacksenhaus E., Chen Y., Chang H. MARCKS inhibition cooperates with autophagy antagonists to potentiate the effect of standard therapy against drug-resistant multiple myeloma. Cancer Lett. 2020;480:29–38. doi: 10.1016/j.canlet.2020.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Ogoyama M., Ohkuchi A., Takahashi H., Zhao D., Matsubara S., Takizawa T. LncRNA H19-Derived miR-675-5p accelerates the invasion of extravillous trophoblast cells by inhibiting GATA2 and subsequently activating matrix metalloproteinases. Int. J. Mol. Sci. 2021;22(3) doi: 10.3390/ijms22031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng F., Zhang J.X., Chen Y., Shen X.D., Su C., Guo Y.J., Wang P.H., Shi C., Lei M., Cao Y.O., Liu S.Q. LncRNA NKX2-1-AS1 promotes tumor progression and angiogenesis via upregulation of SERPINE1 expression and activation of the VEGFR-2 signaling pathway in gastric cancer. Mol. Oncol. 2021 doi: 10.1002/1878-0261.12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Zhang Y., Wang Y., Zhao Y., Ding H., Li P. Circular RNAs: functions and clinical significance in cardiovascular disease. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.584051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y., Gui R. Circulating Exosomal CircMYC is associated with recurrence and bortezomib resistance in patients with multiple myeloma. Turk. J. Haematol. 2020;37(4):248–262. doi: 10.4274/tjh.galenos.2020.2020.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Y. Song, N. Hu, X. Song, J. Yang, Hsa_Circ_0007841 Enhances Multiple Myeloma Chemotherapy Resistance Through Upregulating ABCG2, Technol Cancer Res Treat 19 (2020) 1533033820928371. [DOI] [PMC free article] [PubMed]
- 23.W. Zhang, C. Song, X. Ren, Circ_0003998 Regulates the Progression and Docetaxel Sensitivity of DTX-Resistant Non-Small Cell Lung Cancer Cells by the miR-136-5p/CORO1C Axis, Technol Cancer Res Treat 20 (2021) 1533033821990040. [DOI] [PMC free article] [PubMed]
- 24.Zheng S.R., Huang Q.D., Zheng Z.H., Zhang Z.T., Guo G.L. circGFRA1 affects the sensitivity of triple negative breast cancer cells to paclitaxel via the miR-361-5p/TLR4 pathway. J. Biochem. 2021 doi: 10.1093/jb/mvaa148. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Zhang Y., Zhang S., Huang D., Li B., Liang G., Wu Y., Jiang Q., Li L., Lin C., Wei Z., Meng L. circRNA circARNT2 suppressed the sensitivity of hepatocellular carcinoma cells to cisplatin by targeting the miR-155-5p/PDK1 Axis. Mol. Ther. Nucl. Acids. 2021;23:244–254. doi: 10.1016/j.omtn.2020.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han X., Fang Y., Chen P., Xu Y., Zhou W., Rong Y., Li J.A., Chen W., Lou W. Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis. Cell Cycle. 2021:1–14. doi: 10.1080/15384101.2021.1874684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Wang H., Li C., Gao L., Zheng Y., Chang W., Lu C., Zhao X. CircSMYD4 regulates proliferation, migration and apoptosis of hepatocellular carcinoma cells by sponging miR-584-5p. Cancer Cell Int. 2020;20(1):556. doi: 10.1186/s12935-020-01648-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Yang B., Ren H., Xiao T., Zhang L., Li L., Li M., Wang X., Zhou H., Zhang W. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10(12):953. doi: 10.1038/s41419-019-2180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xi H., Li L., Du J., An R., Fan R., Lu J., Wu Y.X., Wu S.X., Hou J., Zhao L.M. hsa-miR-631 resensitizes bortezomib-resistant multiple myeloma cell lines by inhibiting UbcH10. Oncol. Rep. 2017;37(2):961–968. doi: 10.3892/or.2016.5318. [DOI] [PubMed] [Google Scholar]
- 30.Chen H., Cao W., Chen J., Liu D., Zhou L., Du F., Zhu F. miR-218 contributes to drug resistance in multiple myeloma via targeting LRRC28. J. Cell. Biochem. 2021 doi: 10.1002/jcb.29684. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues-Junior D.M., Pelarin M.F.A., Nader H.B., Vettore A.L., Pinhal M.A.S. MicroRNA-1252-5p associated with extracellular vesicles enhances bortezomib sensitivity in multiple myeloma cells by targeting heparanase. Onco. Targets Ther. 2021;14:455–467. doi: 10.2147/OTT.S286751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y., Cao Z., Shan J., Gao Y., Liu X., Ma D., Li Z. Hsa_circ_0020095 promotes oncogenesis and cisplatin resistance in colon cancer by sponging miR-487a-3p and modulating SOX9. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.604869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo M., Li S., Zhao X., Yuan Y., Zhang B., Guan Y. Knockdown of Circular RNA Hsa_circ_0000714 can regulate RAB17 by sponging miR-370-3p to reduce paclitaxel resistance of ovarian cancer through CDK6/RB pathway. Onco. Targets Ther. 2020;13:13211–13224. doi: 10.2147/OTT.S285153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.