Graphical abstract

Keywords: Gene expression signature, Immune subtypes, Tumor heterogeneity, Immune invasion, Hybrid epithelial/mesenchymal cells
Abstract
Epithelial-mesenchymal plasticity plays a critical role in many solid tumor types as a mediator of metastatic dissemination and treatment resistance. In addition, there is also a growing appreciation that the epithelial/mesenchymal status of a tumor plays a role in immune evasion and immune suppression. A deeper understanding of the immunological features of different tumor types has been facilitated by the availability of large gene expression datasets and the development of methods to deconvolute bulk RNA-Seq data. These resources have generated powerful new ways of characterizing tumors, including classification of immune subtypes based on differential expression of immunological genes. In the present work, we combine scoring algorithms to quantify epithelial-mesenchymal plasticity with immune subtype analysis to understand the relationship between epithelial plasticity and immune subtype across cancers. We find heterogeneity of epithelial-mesenchymal transition (EMT) status both within and between cancer types, with greater heterogeneity in the expression of EMT-related factors than of MET-related factors. We also find that specific immune subtypes have associated EMT scores and differential expression of immune checkpoint markers.
1. Introduction
Epithelial-to-mesenchymal transition (EMT) is a cellular process in which epithelial cells lose epithelial characteristics, such as apical-basal polarity and tight cell–cell adhesions, and gain mesenchymal features, such as anterior-posterior polarization, focal cell contacts, and enhanced motility and invasiveness [1], [2], [3]. Initially characterized in the field of embryology as a feature of normal development, EMT is now also known to play fundamental roles in multiple cellular processes, such as wound healing, fibrosis, and cancer [4]. The regulation of EMT is complex and controlled by the combined action of core EMT transcription factors (EMT-TFs, such as SNAI1/SNAI2, Twist1, and ZEB1/2) [2], [3], epithelial factors such as GRHL2 [5]; Jason A. [6] and OVOL1/2 [7], post-transcriptional regulation, including microRNAs [8], [9] and alternative splicing [10], [11], [12], epigenetic modifications [13], and post-translational regulation [14]. In addition to these cell-intrinsic mechanisms, features of the tumor microenvironment, such as hypoxia [15], [16], [17], [18] and interactions between cancer cells and stromal [19], [20], [21], [22] and immune cells [23], [24], [25] also promote EMT.
Early work on EMT in cancer focused on EMT as a driver of metastatic dissemination. The reversion of cancer cells from a post-EMT-like state back to an epithelial-like phenotype by mesenchymal-to-epithelial transition (MET) was shown to be important for metastatic colonization subsequent to dissemination and seeding [6], [26], [27], [28], [29]. In addition to metastatic potential, however, EMT has also been shown to function in several other key cancer processes, including tumor-propagating/stemness-like phenotypes [30], [31], [32], [33], [34] and treatment resistance [35], [36]. Consistent with the role of EMT in metastatic dissemination and therapy resistance the number and EMT phenotype of circulating tumor cells (CTCs) varies depending on a patient’s treatment response status, with treatment-refractory patients having more overall CTCs with a higher proportion of mesenchymal-type CTCs, and treatment responders having fewer overall CTCs with a higher proportion of epithelial-type CTCs [37].
Although early studies on EMT and MET viewed these phenotypic transitions as binary states, as this field of study has matured the simplified view of EMT/MET dynamics and roles in cancer progression has been shown to be more complex than originally described [38], [2]. EMT, in some contexts, has now been shown to be dispensable for metastatic dissemination [35], [2], [36]. Similarly, MET has also been demonstrated to be dispensable for metastatic colonization, with two distinct paths to metastatic colonization [1], [6] and predicted by [39]. Likewise, while the importance of these processes is more complex, the dynamics of this gene regulatory program are also more nuanced than originally appreciated. Rather than a binary switch between states, EMT/MET is now viewed as a spectrum along which cells can have various hybrid E/M phenotypes [38], [1], [2]. In fact, hybrid E/M phenotypes may be marks of increased cancer aggressiveness and metastatic ability [40]. This has been observed not only in cancer cell lines, but also in clinical CTC samples [41], [40], [37].
Just as EMT promotes chemoresistance in multiple cancer types, the EMT/MET status of cancers has also been linked to resistance to novel immunotherapies. Immune checkpoint inhibitors have revolutionized the treatment landscape and vastly improved patient outcomes of several cancer subtypes, including non-small cell lung cancer (NSCLC) [42], melanoma [43], and renal cell carcinoma (RCC) [44], [43]. However, resistance to these agents is common, and there are several cancer subtypes that do not respond well to immune checkpoint blockade.
One possible explanation for poor immunotherapy response may be the relationship between the EMT status of a tumor and the expression of immune checkpoint molecules [45]. For example, mesenchymal-like cancer cells have been shown to be capable of immunosuppression via interactions with stromal immune cells in the tumor microenvironment. In particular, SNAI1, a core EMT-TF, upregulates expression of CXCL2, a major mediator of myeloid-derived suppressor cell (MDSC) infiltration [46]. MDSCs are major drivers of immunosuppression in the tumor microenvironment. MDSC infiltration renders cancer cells less susceptible to attack by CD8 + T cells and NK cells and leads to an increased ratio of immunosuppressive CD4 + Foxp3 + Treg-like cells, which facilitates tumor growth [47]. SNAI1 and vimentin, a mesenchymal cytoskeletal marker, are positively correlated with programmed death-ligand 1 (PD-L1) scores [48]. Furthermore, PD-L1 transcription is regulated by SNAI1 and the miR200/ZEB1 axis [49]. In contrast, E-cadherin (an epithelial marker) is negatively correlated with PD-L1 scores and epithelial-like cancer cells have higher numbers of infiltrating M1 macrophages and CD8 + T cells in the tumor microenvironment, which allows for greater susceptibility to immune checkpoint inhibitors [50]. It has also been proposed that the regulation between EMT and PD-L1 is actually bidirectional [50], [51]. This is supported by the observation that PD-L1 upregulates the EMT-TF and Twist [52]. In addition to this bidirectional regulation, there may be common inducers of EMT and immune evasion, including chronic inflammation, hypoxia, and metabolic reprogramming. There may also be non-cell-autonomous factors contributing to immune responses; for instance, EMT factors, such as ZEB1, driving an M2 macrophage-enriched environment by inducing both PD-L1 and CD47, the latter of which drives M2 polarization [53]. OVOL1, on the other hand, can enable M1 macrophage enrichment by controlling the transcription of IL-10 [54].
Our understanding of EMT, immune evasion, and other facets of cancer biology has been greatly aided by large, publicly-available gene expression datasets [55], [56]. Analyses of these datasets have allowed researchers to characterize cancer types to an unprecedented level of detail. The coupling of these large genomics data sets with innovative algorithms that can uncouple bulk RNA-Seq data has also enabled inference of immune subtypes based on differential expression of immune-related genes within tumors [57], [58], [59]. These immune subtypes have prognostic value and can be used for survival stratification. Differences in outcome exist within and between cancers when stratified by immune subtypes.
In this study, we combined three novel EMT scoring metrics [60] with immune subtype analysis from the iATLAS algorithm [60], [59], [61] to understand the relationships between EMT and immune signature within and between cancer types. Our analyses demonstrate heterogeneity in both E/M phenotype and immune signature within a single cancer type. By comparing known drivers of epithelial and mesenchymal lineages across cancers, we reveal consistency in gene expression of epithelial factors across epithelial tumors, but cancer type-specific expression of a subset of mesenchymal drivers, suggesting that epithelial-derived cancers have convergence of key epithelial driver genes while mesenchymal-derived cancers may be driven by heterogeneous expression of one or more EMT-TFs. These drivers of E/M phenotype are also associated with distinct immune subtypes, with enrichment for specific immune subtypes across the EMT spectrum, illustrating the relationships between E/M status, immune subtype, with potential implications for patient response to immunotherapy.
2. Results
2.1. The EMT status of cancers is heterogeneous within and between cancer types
The EMT status of specific tumor types has been quantified using a variety of previously-established signatures, most of which use a limited set of molecules or functional traits and/or individual algorithms to measure the extent of EMT on a continuum [62], [63], [64], [65], [66], [67], [68]. Here, to provide a robust comparison of EMT-like status across cancer types, we calculated the EMT scores with available RNA-Seq data from the Cancer Cell Encyclopedia (CCLE) using three distinct EMT scoring algorithms [69], [40], [70]. Each of these three metrics – KS, MLR and GS76 - score the extent of EMT on a continuum, based on the expression of EMT-specific genes identified by various groups. These three methods use different gene lists and scoring methods: the GS76 method uses a weighted sum of expression levels of 76 genes, the KS method compares the cumulative distribution functions of epithelial and mesenchymal signatures, and the MLR method uses a multinomial logistic regression to calculate the probability of a sample to belong to varying EMT categories. KS and MLR score samples on a scale of [-1, 1] and [0,2], respectively, while the GS76 metric has no pre-defined scale. Higher MLR or KS scores represent more mesenchymal samples while this is the inverse for GS76 scores (lower = more mesenchymal). Thus, KS and MLR scores of samples in a given dataset correlate positively with one another, and both KS and MLR correlate negatively with GS76 scores, as observed across multiple datasets [60].
Consistent with their lineages of origin, carcinoma cell lines, such as colorectal, breast, stomach and prostate lines have lower median KS and MLR scores and higher GS76 scores as compared to mesenchymally-derived cancer cell lines, such as sarcoma, melanomas and glioma (Fig. 1A, S1A, S2A). It is also noteworthy that the variance in EMT scores is higher in cell lines of epithelial lineages as compared to mesenchymally-derived cell lines (Fig. 1A, S1A, S2A).
Fig. 1.
Carcinomas have more epithelial EMT scores. The KS algorithm scores samples on a scale of [-1, 1], with more positive scores representing more mesenchymal samples. (A) KS algorithm applied to RNA-Seq data from the Cancer Cell Encyclopedia (CCLE). (B) KS algorithm applied to RNA-Seq data from the Cancer Genome Atlas (TCGA); COAD = Colon adenocarcinoma, READ = Rectum adenocarcinoma, CESC = Cervical squamous cell carcinoma and endocervical adenocarcinoma, BLCA = Bladder urothelial carcinoma, STAD = stomach adenocarcinoma, ESCA = esophageal carcinoma, UCEC = uterine corpus endometrial carcinoma, LUAD = lung adenocarcinoma, HNSC = head and neck squamous cell carcinoma, PRAD = prostate adenocarcinoma, CHOL = cholangiocarcinoma, PAAD = pancreatic adenocarcinoma, KICH = kidney chromophobe, OV = ovarian serous cystadenocarcinoma, LUSC = lung squamous cell carcinoma, THCA = thyroid carcinoma, BRCA = breast invasive carcinoma, KIRP = kidney renal papillary cell carcinoma, LIHC = liver hepatocellular carcinoma, THYM = thymoma, LAML = acute myeloid leukemia, KIRC = kidney renal clear cell carcinoma, TGCT = testicular germ cell tumors, MESO = mesothelioma, UCS = uterine carcinosarcoma, DLBC = lymphoid neoplasm diffuse large B-cell lymphoma, UVM = uveal melanoma, SKCM = skin cutaneous melanoma, PCPG = pheochromocytoma and paraganglioma, LGG = brain lower grade glioma, SARC = sarcoma, GBM = glioblastoma multiforme. Violin plots show the median and interquartile range.
We next investigated the distribution of EMT scores across tumor types from TCGA. Similar to the CCLE data, epithelial tumors (adenocarcinomas of the colon, rectum, stomach, prostate, etc.) have lower mean value of KS and MLR scores and a higher mean value GS76 score as compared to tumors derived from mesenchymal lineages (glioblastoma, glioma, sarcoma) (Fig. 1B, S1B, S2B). Also consistent with CCLE data, mesenchymally-derived tumors have a lower variance in scores whereas carcinomas display higher variability in EMT score (Fig. 1B, S1B, S2B), suggesting less heterogeneity in the EMT status of mesenchymally-derived cancers. To account for any potential bias in EMT score calculation that may be due to excess stromal contamination we applied a previously-reported combination of five tumor purity prediction methods [71]. EMT score was weakly correlated with tumor purity (Figure S3A). The ESTIMATE algorithm was the most highly correlated to EMT score across cancers, with correlations of − 0.47, −0.23, and 0.17 to KS, MLR, and GS76, respectively (Figure S3A). Other metrics of tumor purity, such as immunohistochemistry and the LUMP, ABSOLUTE, and CPE algorithms, had a maximum correlation of − 0.23 (CPE vs. KS; Figure S3A). We also re-analyzed TCGA samples in the 50th and 75th percentiles of tumor purity. While samples with higher tumor purity tended to have lower variance, samples with higher purity were not markedly different in EMT score compared to all samples in a given cancer type (Figure S3B). Together, these analyses suggest that, while single samples may be influenced to a moderate extent by tumor purity, EMT score for a given cancer type is driven predominantly by tumor cell lineage and not a consequence of high stromal contamination.
2.2. EMT-TFs display heterogeneous patterns across cancers while MET factors are more highly correlated
Given the heterogeneity in EMT scores within and between carcinomas, we asked if this heterogeneity was correlated with differences in EMT-TFs and MET-associated factors. To address this question we assessed the levels of a panel of five well-studied EMT-inducing or EMT-associated factors – SNAI1, SNAI2, ZEB1, ZEB2, and TWIST1 [72], [73], [74], [75] – and five MET-inducing or MET-associated factors – GRHL2, OVOL1, OVOL2, ESRP1, and ESRP2 [76], [77], [78], [11], [79] – across tumor types. These factors are well-studied regulators of EMT and MET in vitro and in vivo, and many of them are involved through double negative feedback loops enabling multiple phenotypes; for instance ZEB1 forms such loops with OVOL1/2, GRHL2 and ESRP1 [11], [7], [80]. Such feedback loops drive decision-making during embryonic development and cancer progression [81]. The breakdown of these feedback loops can restrict cellular plasticity, with consequent impact on metastatic potential [82], [83]. While GRHL2 and OVOL1/2 are transcription factors, ESRP1 and ESRP2 are splicing regulatory players associated with epithelial splicing programs in development and in cancer [10], [84], [85].
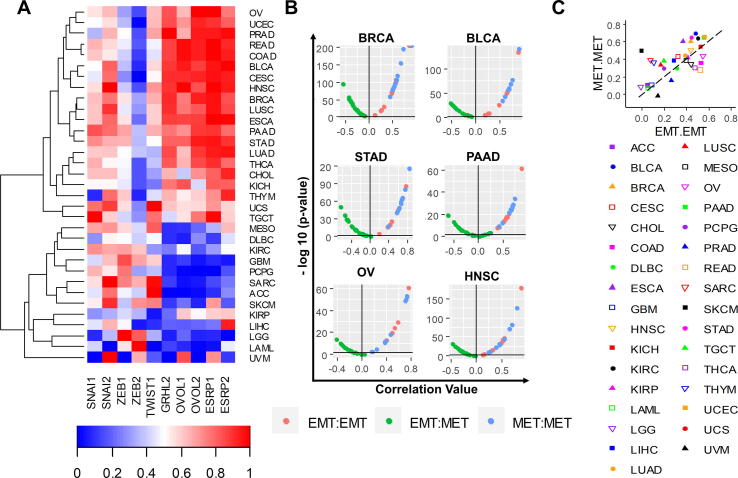
From this analysis of EMT and MET factors, we noted the emergence of two large clusters: one set of tumors comprised predominantly of carcinomas, with higher levels of MET factors and low levels of EMT factors (top cluster, Fig. 2A), and another set of tumors with the opposite trend, comprised mostly of mesenchymally-derived cancers (glioblastoma, glioma, and sarcoma). We also noted a distinct difference in the relationship between MET factors and EMT factors in these groups: In the carcinoma subset MET factors are all highly expressed, while this consistency across EMT factors is not observed in the predominantly-mesenchymal cancers. Instead, mesenchymal tumors are characterized by upregulation of one or two predominant EMT-factors, such as SNAI2 for uveal melanoma and TWIST1 and SNAI2 for sarcoma (Fig. 2A). Conversely, while the mesenchymal-like cluster displays consistently low levels of MET factors, the epithelial-like cluster displays high expression of EMT factors in several cases. Principal component analysis indicated that expression of MET factors is the predominant contributor to the formation of the epithelial-like and mesenchymal-like clusters, while the EMT factors alone are unable to segregate clusters by epithelial/mesenchymal lineage (Figure S4).
Fig. 2.
EMT factors are more heterogeneous across cancers than MET factors. EMT and MET marker expression across TCGA tumor types; COAD = Colon adenocarcinoma, READ = Rectum adenocarcinoma, CESC = Cervical squamous cell carcinoma and endocervical adenocarcinoma, BLCA = Bladder urothelial carcinoma, STAD = stomach adenocarcinoma, ESCA = esophageal carcinoma, UCEC = uterine corpus endometrial carcinoma, LUAD = lung adenocarcinoma, HNSC = head and neck squamous cell carcinoma, PRAD = prostate adenocarcinoma, CHOL = cholangiocarcinoma, PAAD = pancreatic adenocarcinoma, KICH = kidney chromophobe, OV = ovarian serous cystadenocarcinoma, LUSC = lung squamous cell carcinoma, THCA = thyroid carcinoma, BRCA = breast invasive carcinoma, KIRP = kidney renal papillary cell carcinoma, LIHC = liver hepatocellular carcinoma, THYM = thymoma, LAML = acute myeloid leukemia, KIRC = kidney renal clear cell carcinoma, TGCT = testicular germ cell tumors, MESO = mesothelioma, UCS = uterine carcinosarcoma, DLBC = lymphoid neoplasm diffuse large B-cell lymphoma, UVM = uveal melanoma, SKCM = skin cutaneous melanoma, PCPG = pheochromocytoma and paraganglioma, LGG = brain lower grade glioma, SARC = sarcoma, GBM = glioblastoma multiforme (A) Normalized EMT and MET marker expression across all TCGA tumor types. (B) Pairwise correlations between expression values of all EMT-EMT, MET-MET, and EMT-MET factor pairs across a subset of TCGA tumor types. (C) Plot of mean pairwise correlation coefficients for all EMT-EMT factor pairs (x-axis) versus mean pairwise correlation coefficients for all MET-MET factor pairs (y-axis) across all TCGA tumor types.
To further analyse this trend quantitatively, we calculated the pairwise correlations between expression values of all 10 EMT and MET factors across a subset of tumor types. Consistent with our qualitative observations above, the five MET factors are significantly positively correlated with each other, while the EMT and MET factors are significantly negatively correlated. However, unlike the MET factors, the correlation among different EMT factors is less consistent and often not statistically significant (Fig. 2B). We also quantified the mean of the correlation coefficients for all pairwise correlations between any two EMT factors or for any two MET factors. These values are shown as x- and y-axes on a scatter plot, where each dot represents a tumor type. The majority of the tumors are above the × = y line, signifying that the average correlation between any two MET factors is greater than that between two EMT factors (Fig. 2C). Such correlation is reminiscent of recent observations of groups of factors engaged in various feedback loops, driving phenotypic plasticity and heterogeneity in many cancers [86], [87]. Similarly, visualization of the variance of the pairwise correlation coefficients for EMT and MET factors revealed a lower variability in MET factors as compared to EMT factors (Figure S5). These trends are also observed in CCLE cancer types (Figures S6, S7). Together, these analyses suggest that more heterogeneity in gene expression exists for EMT factors as compared to MET factors.
2.3. EMT score is associated with specific immune subtypes in cancer
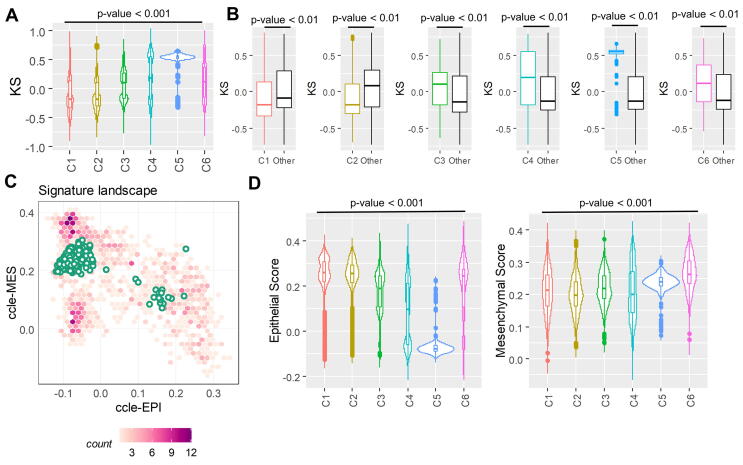
Activation of a partial or complete EMT has been associated with immune evasion across multiple cancers [88], [89]. A recent approach characterized immune tumor microenvironment across 33 cancer types in TCGA and defined six major immune subtypes spanning cancer types and molecular subtypes – C1 (wound healing), C2 (IFN-γ dominant), C3 (inflammatory), C4 (lymphocyte depleted), C5 (immunologically quiet), and C6 (TGF-β dominant) [61]. Given the relationship between EMT and immune checkpoint molecules [49], [48], [88] as well as immunomodulatory cytokines, we sought to understand the relationship between EMT status and immune subtype within and across cancers. To do this, we calculated EMT scores for all TCGA samples for which immune subtypes have been assigned. Overall, cancers with the C1 wound healing subtype and C2 IFN-γ dominant subtype tended to have more epithelial-like scores, while all other subtypes were more mesenchymal (Fig. 3A, S8A, C). In particular, the C5 quiescence subtype is the most mesenchymal subtype (Fig. 3A-B, S8). This observation is consistent with its sample composition, as C5 contained almost exclusively low-grade glioma samples [61]. A leave-one-out analysis further demonstrated these trends, with the wound healing (C1) and IFN-γ (C2) signatures more epithelial than all other cancer samples, and the other immune signatures more mesenchymal than other signatures/samples (Fig. 3B, S8B, D). We applied Singscore, a recently-developed rank-based single sample method to quantify the epithelial and mesenchymal scores separately [90](Fig. 3C). Singscore is a non-parametric method that calculates scores for ensembles of gene sets corresponding to epithelial and mesenchymal phenotypes. Using this method, the C5 samples showed high mesenchymal scores and very low epithelial scores. Interestingly, the mesenchymal scores for all six immune subtypes do not vary much in terms of fold-change (despite being statistically significantly different) while the epithelial score is the lowest for C5 (Fig. 3D). The C6 TGF-β dominant immune subtype shows a high score for both the axes, which may suggest that cancers with this immune signature would have a more hybrid epithelial/mesenchymal (E/M) phenotype (Fig. 3D).
Fig. 3.
Immune subtypes are associated with EMT scores. EMT scores across TCGA immune subtypes; C1 = wound healing, C2 = IFN-γ dominant, C3 = inflammatory, C4 = lymphocyte depleted, C5 = immunologically quiet, C6 = TGF-β dominant. (A) Plot of calculated KS score across all immune subtypes. (B) Leave-one-out-analysis: pairwise comparison of each cancer immune subtype’s KS score to the KS scores of all other immune subtypes. (C) Application of Singscore to C5 immune subtype samples to quantify epithelial and mesenchymal scores separately. (D) Singscore epithelial and mesenchymal scores across immune subtypes.
Given the marked heterogeneity in EMT scores and immune subtypes, we sought to understand whether particular cancer types are enriched in specific EMT scores and immune subtypes. Analysis of the upper and lower quartiles of EMT score across immune subtypes revealed distinct cancer types within each of the immune subtypes. For example, tumors in the lower quartile of the C1 wound healing subtype are enriched in colorectal cancer specimens as compared to the lower quartile of C1 (Figure S9A). Conversely, the composition of cancer types between upper quartile of EMT scores within the IFNγ-dominant C2 subtype are spread across multiple cancer types of diverse lineages, including, but not limited to, breast cancer, head and neck cancer, sarcomas, melanomas, and testicular germ cell tumors (Figure S9A, B). Likewise, the cancer types in upper and lower quartiles of EMT scores within the inflammatory (C3) subtype also differ substantially. More mesenchymal-like tumors within the C3 subtype (inflammatory) are enriched in renal clear cell carcinomas as compared to the lower quartile of C3 tumors, which is comprised of lung, prostate, breast, and thyroid cancers (Figure S9A, B). Analysis of tumor microenvironment cell populations within each immune subtype using the microenvironment cell populations (MCP)-counter method revealed enrichment of gene expression signatures consistent with fibroblasts, endothelial cells, and cells of a monocyte lineage (Figure S10). The lymphocyte-depleted (C4) and immunologically quiescent (C5) subtypes are the most depleted of immune subtypes, while the IFN-γ dominant subtype (C2) has the highest enrichment in T cell signatures and the TGF-β-dominant subtype (C6) has the greatest enrichment in signatures of myeloid-derived cells (Figure S10).
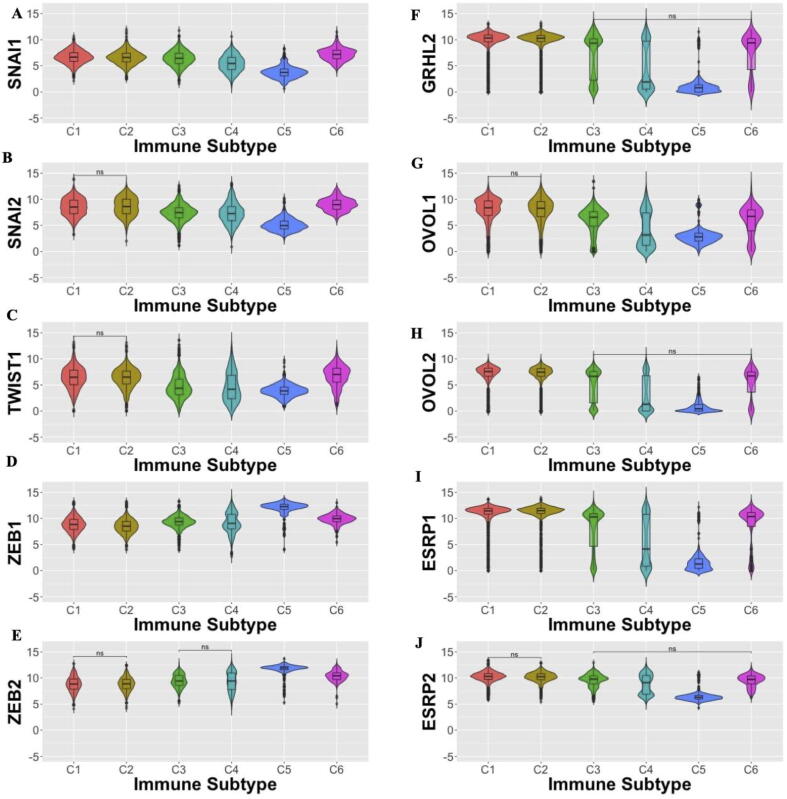
To further understand how EMT and MET factors may be associated with specific immune signatures we performed separate analyses of EMT factors for each immune subtype. EMT factors SNAI1, SNAI2, and Twist are most highly expressed in the C6 TGF-β dominant immune signature, with ZEB1 and ZEB2 most upregulated in the C5 immunologically quiescent signature (Fig. 4A-E). Other immune subtypes have low to moderate levels of EMT factors, with wide distributions in expression across these subtypes. MET factors are predominantly expressed in C1-C3 signatures, with a bimodal distribution of expression in C4, low expression in C5, and relatively high expression in C6 (Fig. 4F-J). Such co-expression of EMT and MET factors in C6 samples suggests that samples in the TGF-β dominant C6 immune subtype correspond to a more hybrid epithelial/ mesenchymal phenotype(s).
Fig. 4.
EMT and MET factor gene expression varies across immune subtypes. (A-E) Expression of EMT factors across immune subtypes; (F-J) Expression of MET factors across immune subtypes; (A) SNAI1, (B) SNAI2, (C) TWIST1, (D) ZEB1, (E) ZEB2, (F) GRHL2, (G) OVOL1, (H) OVOL2, (I) ESRP1, and (J) ESRP2 expression. Unless specified by n.s. (not significant), all comparisons are statistically reliable (p < 0.05; Wilcoxon rank means test with Benjamini-Hochberg adjustment for multiple comparisons).
We also calculated the pairwise correlations between the EMT scores from the different scoring metrics (GS76, KS, MLR) and expression of EMT and MET factors from samples in each immune subtype. Across all immune signatures, these pairwise correlations showed consistent trends: GS76 scores correlated positively with levels of MET-factors and negatively with those of EMT-factors; KS and MLR scores followed the inverse trend (Figure S11). Among the EMT factors, ZEB1 and ZEB2 had the highest correlations with all EMT scoring metrics (Fig S11). The upregulation of ZEB1 and ZEB2 in C5, and the strong correlations of ZEB1 and ZEB2 with EMT scoring metrics further supports observations about the key roles of these two EMT-TFs in maintenance of a mesenchymal phenotype [91].
2.4. Immune checkpoint markers across immune subtypes
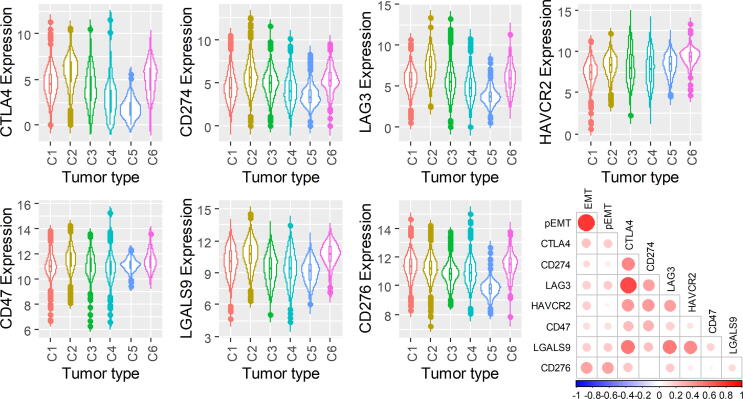
We next investigated the levels of different immune checkpoint markers across the immune subtypes: CTLA-4 (cytotoxic T lymphocyte antigen-4), CD274 (PD-L1; programmed death-ligand 1) [92], LAG3 (lymphocyte activation gene 3) [93], CD276 (B7-H3) ([94], CD47 (cluster of differentiation 47) [95], and HAVCR2 (TIM-3) and its ligand LGASL9 (galectin 9) [96] (Fig. 5A-G). While expression of most immune checkpoint molecules varies across all immune subtypes, the immunologically quiescent C5 immune subtype has the lowest levels of CTLA-4, CD274, CD276, and LAG3, but with HAVCR2 and CD47 expression similar to other immune subtypes (Fig. 5). The TGF-β-dominant subtype (C6) displays elevated HAVCR2, LGALS9, and CD276 (Fig. 5). The expression levels of these immune checkpoint markers are positively correlated with one another as well as with the single-sample GSEA scores for EMT and partial EMT [65] signatures (Fig. 5H). These results suggest that common signalling pathways implicated in EMT may be associated with changes in levels of various immune checkpoint markers.
Fig. 5.
Immune checkpoint expression varies across immune subtypes. (A) CTLA4, (B) CD274, (C) LAG3, (D) HAVCR2, (E) CD47, (F) LGALS9, and (G) CD276 expression across immune subtypes. (H) Correlation matrix of EMT and pseudo-EMT scores with immune checkpoint markers.
3. Discussion
Applying three distinct RNA-based EMT scoring algorithms – KS, MLR, and GS76 – we characterized the diversity and heterogeneity of EMT status both within and across cancer types. These analyses revealed a higher variance in EMT score among cancers of an epithelial lineage as compared to mesenchymally-derived tumors. Similarly, analysis of EMT- and MET-associated factors revealed more heterogeneity in gene expression for EMT factors as compared to MET factors. While these data suggest that the EMT program is more heterogeneous than the MET program across cancers, the reasons for this are not clear. One possible explanation for this may be that the selected MET factors, such as GRHL2, OVOL1, and OVOL2 are indicative of epithelial lineages [97], [98], [99], while the selected EMT factors may be more unique to a specific cellular lineage. It is possible that greater heterogeneity in the MET program would perhaps be more accurately reflected by analysing tissue-specific cadherins and keratins that mark specific lineages [100]. It is also possible that consideration of other aspects of the cellular response (post-transcriptional, translational/ post-translational, epigenetic [6], metabolic [101], [102], etc. could reveal additional layers of regulation and perhaps more conserved circuitry across the MET program.
Another possible explanation may be that the MET program represents a more fixed, derived phenotype while the EMT program is more plastic in nature. This notion is supported by the observations that the EMT program is coupled to cancer stemness-like pathways [103], [104], [34]. Although it is possible that EMT scores may be skewed by differences in tumor:stromal ratios across samples and cancer types [21], [71], our analyses revealed low correlations between EMT score and tumor purity scores [71], suggesting that the EMT scores were not substantially skewed by high stromal content. In addition, the scoring metrics were also consistent when applied to both TCGA and CCLE data, suggesting that these scoring metrics can be applied to both potentially-heterogeneous cancer samples and more homogeneous cancer cell lines. The remarkable heterogeneity in EMT and MET scores and EMT/MET factors across cancers underscores that EMT/MET dynamics are context-specific and more of a spectrum of states instead of a binary classification [2], [3].
The present work sheds further light on our collective understanding of the potential cross-talk between cancer cells and immune subsets across the EMT spectrum. Both EMT scores and known EMT/MET factors were associated with certain immune subtypes. For example, cancers with C1 (wound healing) and C2 (IFN-γ dominant) subtypes tended to have more epithelial scores with high expression of MET factors, C5 (quiescent) was dominated by a high mesenchymal score with upregulation of ZEB1 and ZEB2, and C6 (TGF-β dominant) was enriched for a more hybrid epithelial/mesenchymal score. Consistent with this, the TGF-β dominant subtype shows upregulation of ZEB1 and ZEB2 as well as GRHL2, ESRP1, and ESRP2 (Fig. 4). The hybrid E/M state plays an important role in therapeutic resistance and formation of highly tumorigenic CTC clusters [105], [98]. TGF-β has been shown to activate ZEB1 through DNA methylation of the ZEB1 repressive microRNA, miR-200 [106]. Our prior work indicated that both GRHL2 and miR200s are necessary to induce MET in sarcomas [6], and it is possible that the TGF-β-mediated subtype additional signals to drive the phenotype toward a more complete EMT. One limitation of this study is that the analysis of TCGA data was from almost exclusively primary tumor sites rather than metastases. Metastatic samples comprise just 3.4% of all TCGA solid tumor samples (https://portal.gdc.cancer.gov/). Given the considerable differences in EMT biology between primary tumors and metastases [27], [29], [107], [108], it is likely that the biology of the immune subsets within metastatic microenvironments also differs.
Analysis of seven immune checkpoint molecules – CTLA-4, CD274/PD-L1, LAG3, CD276/B7-H3, CD47, HAVCR2/TIM-3, and LGAL29/galectin-9 – across the immune subtypes revealed the highest levels among C2 (IFN-γ dominant) and C6 (TGF-β dominant) subtypes and lowest in the C5 (quiescent) subtype. IFN-γ is a potent inducer of PD-L1 expression [109], which may explain the high levels of immune checkpoint molecules in the C2 subtype despite its more epithelial-like score. In addition, epithelial-like cancer cells are known to have higher numbers of infiltrating M1 macrophages and CD8 + T cells in the tumor microenvironment [50], which is consistent with the observation that the C2 (IFN-γ dominant) subtype had the highest M1/M2 macrophage polarization and strong CD8 signal among all six subtypes [61]. The TGF-β dominant subtype (C6) displayed increased levels of HAVCR2 (also known as Tim-3), and LGALS2 (galectin-9). Interestingly, HAVCR2 expression is upregulated by TGF-β [110], [111], and galectin-9 promotes TGF-β-induced signalling and subsequent conversion of CD4 + CD25- T cells into regulatory T-cells [112], [113]. These observations suggest that patients enriched for the C6 subtype may benefit from TGF-β inhibitors in combination with HAVCR2 or galectin-9 suppression.
4. Methods
All analyses were performed in R (version 3.4.4), and data was plotted using the ggplot2 package. Heatmaps were plotted using the gplots R package.
TCGA datasets: TCGA gene expression datasets were obtained from https://xenabrowser.net/datapages/.
RNA Seq data pre-processing: Normalized gene expression counts of all the tumor datasets from TCGA website were further pre-processed to ensure a robust approximation of EMT scores. To do this, a regression analysis was used to convert all RNA-Seq count data to a standard EMT metric score based on previous microarray data. The regression parameters to transform RNASeq to microarray data were estimated as described previously [114].
CCLE dataset: CCLE gene expression data was downloaded from https://portals.broadinstitute.org/ccle/data
EMT scoring: Three different EMT scoring methods – KS, MLR, GS76 – were used to score samples separately in all the datasets as previously described [60]. Unless noted otherwise, the KS score was used for analyses of immune subtypes and EMT/MET factors.
ssGSEA analysis: ssGSEA analysis for various different gene sets were performed using GSVA R Bioconductor package with “ssgsea” option for method argument [115].
Statistical analysis: All the pairwise comparison significance was tested using student’s t-test and the multiple group comparisons significance was tested using ANOVA. A Wilcoxon rank means test with Benjamini-Hochberg adjustment for multiple comparisons was used for the analysis of immune subtypes in Fig. 4.
Min-max standardization: The gene expression values and EMT scores were standardized in the range of 0 to 1 as following:
Where, Xscaled is the min–max standardized value of a gene X.
Principal component analysis: Principal component analysis was used to visualize the gene expression data of multiple variables (5 EMT and/or 5 MET factors). multidimensional gene expression and simulation data. To determine the correlation between variables and the representation of variables by the principal components, a correlation circle with squared cosines was plotted.
CRediT authorship contribution statement
Priyanka Chakraborty: Conceptualization, Data curation, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Emily L. Chen: Writing - original draft, Writing - review & editing. Isabelle McMullen: Conceptualization, Data curation, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Andrew J. Armstrong: Conceptualization, Writing - original draft, Writing - review & editing. Mohit Kumar Jolly: Conceptualization, Data curation, Formal analysis, Visualization, Supervision, Writing - original draft, Writing - review & editing. Jason A. Somarelli: Conceptualization, Data curation, Formal analysis, Visualization, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
JAS is supported by the Department of Defense (W81XWH-18-1-0189). AJA and JAS acknowledge funding support from NCI 1R01CA233585-02. MKJ was supported by Ramanujan Fellowship awarded by Science and Engineering Research Board, Department of Science and Technology, Government of India (SB/S2/RJN-049/2018). The results published here are based, in part, upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.023.
Contributor Information
Mohit Kumar Jolly, Email: mkjolly@iisc.ac.in.
Jason A. Somarelli, Email: jason.somarelli@duke.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jolly M.K., Ware K.E., Gilja S., Somarelli J.A., Levine H. EMT and MET: necessary or permissive for metastasis? Mol Oncol. 2017;11(7):755–769. doi: 10.1002/1878-0261.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nieto M.A., Huang R.-Y.-J., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M., Campbell, K., Cano, A., Casanova, J., Christofori, G., Dedhar, S., Derynck, R., Ford, H. L., Fuxe, J., García de Herreros, A., Goodall, G. J., Hadjantonakis, A.-K., Huang, R. J. Y., Kalcheim, C., … EMT International Association (TEMTIA). (2020). Guidelines and definitions for research on epithelial-mesenchymal transition. Nature Reviews. Molecular Cell Biology, 21(6), 341–352. [DOI] [PMC free article] [PubMed]
- 4.Stone R.C., Pastar I., Ojeh N., Chen V., Liu S., Garzon K.I. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 2016;365(3):495–506. doi: 10.1007/s00441-016-2464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao X., Bali A.S., Randell S.H., Hogan B.L.M. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. The Journal of Cell Biology. 2015;211(3):669–682. doi: 10.1083/jcb.201506014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somarelli J.A., Shetler S., Jolly M.K., Wang X., Bartholf Dewitt S., Hish A.J. Mesenchymal-Epithelial Transition in Sarcomas Is Controlled by the Combinatorial Expression of MicroRNA 200s and GRHL2. Mol Cell Biol. 2016;36(19):2503–2513. doi: 10.1128/MCB.00373-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roca H., Hernandez J., Weidner S., McEachin R.C., Fuller D., Sud S. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0076773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory P.A., Bracken C.P., Bert A.G., Goodall G.J. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7(20):3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 9.Paterson E.L., Kolesnikoff N., Gregory P.A., Bert A.G., Khew-Goodall Y., Goodall G.J. The microRNA-200 family regulates epithelial to mesenchymal transition. TheScientificWorldJournal. 2008;8:901–904. doi: 10.1100/tsw.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bebee T.W., Park J.W., Sheridan K.I., Warzecha C.C., Cieply B.W., Rohacek A.M. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. eLife. 2015:4. doi: 10.7554/eLife.08954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolly M.K., Preca B.-T., Tripathi S.C., Jia D., George J.T., Hanash S.M. Interconnected feedback loops among ESRP1, HAS2, and CD44 regulate epithelial-mesenchymal plasticity in cancer. APL Bioengineering. 2018;2(3) doi: 10.1063/1.5024874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warzecha C.C., Sato T.K., Nabet B., Hogenesch J.B., Carstens R.P. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33(5):591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skrypek N., Goossens S., De Smedt E., Vandamme N., Berx G. Epithelial-to-Mesenchymal Transition: Epigenetic Reprogramming Driving Cellular Plasticity. Trends in Genetics: TIG. 2017;33(12):943–959. doi: 10.1016/j.tig.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Serrano-Gomez S.J., Maziveyi M., Alahari S.K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Molecular Cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannito S., Novo E., Compagnone A., Valfrè di Bonzo L., Busletta C., Zamara E. Redox mechanisms switch on hypoxia-dependent epithelial-mesenchymal transition in cancer cells. Carcinogenesis. 2008;29(12):2267–2278. doi: 10.1093/carcin/bgn216. [DOI] [PubMed] [Google Scholar]
- 16.Choi B.-J., Park S.-A., Lee S.-Y., Cha Y.N., Surh Y.-J. Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9. Sci Rep. 2017;7(1):15918. doi: 10.1038/s41598-017-15139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxena K., Jolly M.K. Acute vs. Chronic vs. Cyclic Hypoxia: Their Differential Dynamics, Molecular Mechanisms, and Effects on Tumor Progression. Biomolecules. 2019;9(8) doi: 10.3390/biom9080339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M.-H., Wu M.-Z., Chiou S.-H., Chen P.-M., Chang S.-Y., Liu C.-J. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10(3):295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 19.Gao M.-Q., Kim B.G., Kang S., Choi Y.P., Park H., Kang K.S. Stromal fibroblasts from the interface zone of human breast carcinomas induce an epithelial-mesenchymal transition-like state in breast cancer cells in vitro. J Cell Sci. 2010;123(Pt 20):3507–3514. doi: 10.1242/jcs.072900. [DOI] [PubMed] [Google Scholar]
- 20.Giannoni E., Bianchini F., Masieri L., Serni S., Torre E., Calorini L. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial-mesenchymal transition and cancer stemness. Cancer Res. 2010;70(17):6945–6956. doi: 10.1158/0008-5472.CAN-10-0785. [DOI] [PubMed] [Google Scholar]
- 21.Wang L., Saci A., Szabo P.M., Chasalow S.D., Castillo-Martin M., Domingo-Domenech J. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun. 2018;9(1):3503. doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Lan W., Xu M., Song J., Mao J., Li C. Cancer-associated fibroblast-derived SDF-1 induces epithelial-mesenchymal transition of lung adenocarcinoma via CXCR4/β-catenin/PPARδ signalling. Cell Death Dis. 2021;12(2):214. doi: 10.1038/s41419-021-03509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedele V., Melisi D. Permissive State of EMT: The Role of Immune Cell Compartment. Front Oncol. 2020;10:587. doi: 10.3389/fonc.2020.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romeo E., Caserta C.A., Rumio C., Marcucci F. The Vicious Cross-Talk between Tumor Cells with an EMT Phenotype and Cells of the Immune System. Cells. 2019;8(5) doi: 10.3390/cells8050460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toh B., Wang X., Keeble J., Sim W.J., Khoo K., Wong W.-C. Mesenchymal transition and dissemination of cancer cells is driven by myeloid-derived suppressor cells infiltrating the primary tumor. PLoS Biol. 2011;9(9) doi: 10.1371/journal.pbio.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esposito M., Mondal N., Greco T.M., Wei Y., Spadazzi C., Lin S.-C. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat Cell Biol. 2019;21(5):627–639. doi: 10.1038/s41556-019-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ocaña O.H., Córcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22(6):709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Stankic M., Pavlovic S., Chin Y., Brogi E., Padua D., Norton L. TGF-β-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell Reports. 2013;5(5):1228–1242. doi: 10.1016/j.celrep.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai J.H., Donaher J.L., Murphy D.A., Chau S., Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(6):725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guen V.J., Chavarria T.E., Kröger C., Ye X., Weinberg R.A., Lees J.A. EMT programs promote basal mammary stem cell and tumor-initiating cell stemness by inducing primary ciliogenesis and Hedgehog signaling. PNAS. 2017;114(49):E10532–E10539. doi: 10.1073/pnas.1711534114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo D., Xu B.-L., Zhang X.-H., Dong M.-M. Cancer stem-like side population cells in the human nasopharyngeal carcinoma cell line cne-2 possess epithelial mesenchymal transition properties in association with metastasis. Oncol Rep. 2012;28(1):241–247. doi: 10.3892/or.2012.1781. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Wang L., Pappan L., Galliher-Beckley A., Shi J. IL-1β promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Molecular Cancer. 2012;11:87. doi: 10.1186/1476-4598-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasani S., Sahoo S., Jolly M.K. Hybrid E/M Phenotype(s) and Stemness: A Mechanistic Connection Embedded in Network Topology. J Clin Med Res. 2020;10(1) doi: 10.3390/jcm10010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 35.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T.C. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu M., Bardia A., Wittner B.S., Stott S.L., Smas M.E., Ting D.T. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biswas K., Jolly M.K., Ghosh A. Stability and mean residence times for hybrid epithelial/mesenchymal phenotype. Phys Biol. 2019;16(2) doi: 10.1088/1478-3975/aaf7b7. [DOI] [PubMed] [Google Scholar]
- 39.Brabletz T. To differentiate or not–routes towards metastasis. Nat Rev Cancer. 2012;12(6):425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 40.George J.T., Jolly M.K., Xu S., Somarelli J.A., Levine H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res. 2017;77(22):6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armstrong A.J., Marengo M.S., Oltean S., Kemeny G., Bitting R.L., Turnbull J.D. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Molecular Cancer Research: MCR. 2011;9(8):997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brahmer J.R., Govindan R., Anders R.A., Antonia S.J., Sagorsky S., Davies M.J. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (NSCLC) J ImmunoTher Cancer. 2018;6(1):75. doi: 10.1186/s40425-018-0382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss S.A., Wolchok J.D., Sznol M. Immunotherapy of Melanoma: Facts and Hopes. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2019;25(17):5191–5201. doi: 10.1158/1078-0432.CCR-18-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rini B.I., Battle D., Figlin R.A., George D.J., Hammers H., Hutson T. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of advanced renal cell carcinoma (RCC) J ImmunoTher Cancer. 2019;7(1):354. doi: 10.1186/s40425-019-0813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y., Zhang L., Kamimura Y., Ritprajak P., Hashiguchi M., Hirose S. B7–H1 overexpression regulates epithelial-mesenchymal transition and accelerates carcinogenesis in skin. Cancer Res. 2011;71(4):1235–1243. doi: 10.1158/0008-5472.CAN-10-2217. [DOI] [PubMed] [Google Scholar]
- 46.Taki M., Abiko K., Baba T., Hamanishi J., Yamaguchi K., Murakami R. Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat Commun. 2018;9(1):1685. doi: 10.1038/s41467-018-03966-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian L., Liu Y., Wang S., Gong W., Jia X., Liu L. NKG2D ligand RAE1ε induces generation and enhances the inhibitor function of myeloid-derived suppressor cells in mice. J Cell Mol Med. 2017;21(9):2046–2054. doi: 10.1111/jcmm.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S., Koh J., Kim M.-Y., Kwon D., Go H., Kim Y.A. PD-L1 expression is associated with epithelial-to-mesenchymal transition in adenocarcinoma of the lung. Hum Pathol. 2016;58:7–14. doi: 10.1016/j.humpath.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.-H., Byers L.A. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dongre A., Rashidian M., Reinhardt F., Bagnato A., Keckesova Z., Ploegh H.L. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res. 2017;77(15):3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Y., Zhan H. Communication between EMT and PD-L1 signaling: New insights into tumor immune evasion. Cancer Lett. 2020;468:72–81. doi: 10.1016/j.canlet.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Cao Y., Zhang L., Ritprajak P., Tsushima F., Youngnak-Piboonratanakit P., Kamimura Y. Immunoregulatory molecule B7–H1 (CD274) contributes to skin carcinogenesis. Cancer Res. 2011;71(14):4737–4741. doi: 10.1158/0008-5472.CAN-11-0527. [DOI] [PubMed] [Google Scholar]
- 53.Guo Y., Lu X., Chen Y., Rendon B., Mitchell R.A., Cuatrecasas M. Zeb1 induces immune checkpoints to form an immunosuppressive envelope around invading cancer cells. Science. Advances. 2021;7(21) doi: 10.1126/sciadv.abd7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.OVOL2 inhibits macrophage M2 polarization by regulating IL-10 transcription, and thus inhibits the tumor metastasis by modulating the tumor microenvironment. (2021). Immunology Letters. https://doi.org/10.1016/j.imlet.2021.05.004 [DOI] [PubMed]
- 55.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding, L., Bailey, M. H., Porta-Pardo, E., Thorsson, V., Colaprico, A., Bertrand, D., Gibbs, D. L., Weerasinghe, A., Huang, K.-L., Tokheim, C., Cortés-Ciriano, I., Jayasinghe, R., Chen, F., Yu, L., Sun, S., Olsen, C., Kim, J., Taylor, A. M., Cherniack, A. D., … Cancer Genome Atlas Research Network. (2018). Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell, 173(2), 305–320.e10. [DOI] [PMC free article] [PubMed]
- 57.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eddy J.A., Thorsson V., Lamb A.E., Gibbs D.L., Heimann C., Yu J.X. CRI iAtlas: an interactive portal for immuno-oncology research. F1000Research. 2020;9:1028. doi: 10.12688/f1000research.25141.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chakraborty P., George J.T., Tripathi S., Levine H., Jolly M.K. Comparative Study of Transcriptomics-Based Scoring Metrics for the Epithelial-Hybrid-Mesenchymal Spectrum. Front Bioeng Biotechnol. 2020;8:220. doi: 10.3389/fbioe.2020.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.-H. The Immune Landscape of Cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhatia S., Monkman J., Blick T., Pinto C., Waltham M., Nagaraj S.H. Interrogation of Phenotypic Plasticity between Epithelial and Mesenchymal States in Breast Cancer. J Clin Med Res. 2019;8(6) doi: 10.3390/jcm8060893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Devaraj V., Bose B. Morphological State Transition Dynamics in EGF-Induced Epithelial to Mesenchymal Transition. J Clin Med Res. 2019;8(7) doi: 10.3390/jcm8070911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandal M., Ghosh B., Anura A., Mitra P., Pathak T., Chatterjee J. Modeling continuum of epithelial mesenchymal transition plasticity. Integrative Biology: Quantitative Biosciences from Nano to Macro. 2016;8(2):167–176. doi: 10.1039/c5ib00219b. [DOI] [PubMed] [Google Scholar]
- 65.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171(7):1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruscetti M., Dadashian E.L., Guo W., Quach B., Mulholland D.J., Park J.W. HDAC inhibition impedes epithelial-mesenchymal plasticity and suppresses metastatic, castration-resistant prostate cancer. Oncogene. 2016;35(29):3781–3795. doi: 10.1038/onc.2015.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schliekelman M.J., Taguchi A., Zhu J., Dai X., Rodriguez J., Celiktas M. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015;75(9):1789–1800. doi: 10.1158/0008-5472.CAN-14-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tripathi S., Chakraborty P., Levine H., Jolly M.K. A mechanism for epithelial-mesenchymal heterogeneity in a population of cancer cells. PLoS Comput Biol. 2020;16(2) doi: 10.1371/journal.pcbi.1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Byers L.A., Diao L., Wang J., Saintigny P., Girard L., Peyton M. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2013;19(1):279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan T.Z., Miow Q.H., Miki Y., Noda T., Mori S., Huang R.-Y.-J. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6(10):1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drápela S., Bouchal J., Jolly M.K., Culig Z., Souček K. ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance. Frontiers in Molecular Biosciences. 2020;7:36. doi: 10.3389/fmolb.2020.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Subbalakshmi A.R., Sahoo S., Biswas K., Jolly M.K. Cells; Tissues, Organs: 2021. A Computational Systems Biology Approach Identifies SLUG as a Mediator of Partial Epithelial-Mesenchymal Transition (EMT) pp. 1–14. [DOI] [PubMed] [Google Scholar]
- 74.Vandewalle C. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res (Vol. 2005;33(20):6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 76.Cieply B., Riley P., Pifer P.M., Widmeyer J., Addison J.B. Suppression of the epithelial–mesenchymal transition by Grainyhead-like-2. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-4038. https://cancerres.aacrjournals.org/content/72/9/2440.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fici P., Gallerani G., Morel A.-P., Mercatali L., Ibrahim T., Scarpi E. Splicing factor ratio as an index of epithelial-mesenchymal transition and tumor aggressiveness in breast cancer. Oncotarget. 2017;8(2):2423–2436. doi: 10.18632/oncotarget.13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeong H.M., Han J., Lee S.H., Park H.J., Lee H.J., Choi J.S. ESRP1 is overexpressed in ovarian cancer and promotes switching from mesenchymal to epithelial phenotype in ovarian cancer cellsThis article has been corrected since Advance Online Publication and an erratum is also printed in this issue. Oncogenesis. 2017;6(10):e389. doi: 10.1038/oncsis.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saxena K., Srikrishnan S., Celia-Terrassa T., Jolly M.K. Cells; Tissues, Organs: 2020. OVOL1/2: Drivers of Epithelial Differentiation in Development, Disease, and Reprogramming; pp. 1–10. [DOI] [PubMed] [Google Scholar]
- 80.Mooney S.M., Talebian V., Jolly M.K., Jia D., Gromala M., Levine H. The GRHL2/ZEB Feedback Loop-A Key Axis in the Regulation of EMT in Breast Cancer. J Cell Biochem. 2017;118(9):2559–2570. doi: 10.1002/jcb.25974. [DOI] [PubMed] [Google Scholar]
- 81.Zhou J.X., Huang S. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends in Genetics: TIG. 2011;27(2):55–62. doi: 10.1016/j.tig.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Hari K., Sabuwala B., Subramani B.V., La Porta C.A.M., Zapperi S., Font-Clos F. Identifying inhibitors of epithelial-mesenchymal plasticity using a network topology-based approach. npj Syst Biol Appl. 2020;6(1):15. doi: 10.1038/s41540-020-0132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Celià-Terrassa T., Bastian C., Liu D.D., Ell B., Aiello N.M., Wei Y. Hysteresis control of epithelial-mesenchymal transition dynamics conveys a distinct program with enhanced metastatic ability. Nat Commun. 2018;9(1):5005. doi: 10.1038/s41467-018-07538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Warzecha C.C., Shen S., Xing Y., Carstens R.P. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6(5):546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ishii H., Saitoh M., Sakamoto K., Kondo T., Katoh R., Tanaka S. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. The Journal of Biological Chemistry. 2014;289(40):27386–27399. doi: 10.1074/jbc.M114.589432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pillai M., Jolly M.K. Systems-level network modeling deciphers the master regulators of phenotypic plasticity and heterogeneity in melanoma. bioRxiv. 2021 doi: 10.1101/2021.03.11.434533. (p. 2021.03.11.434533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chauhan, L., Ram, U., Hari, K., & Jolly, M. K. (n.d.). 2021. Topological signatures in regulatory network enable phenotypic heterogeneity in small cell lung cancer. eLife. https://doi.org/10.1101/2020.10.30.362228 [DOI] [PMC free article] [PubMed]
- 88.Terry S., Savagner P., Ortiz-Cuaran S., Mahjoubi L., Saintigny P., Thiery J.-P. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11(7):824–846. doi: 10.1002/1878-0261.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tripathi S.C., Peters H.L., Taguchi A., Katayama H., Wang H., Momin A. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. PNAS. 2016;113(11):E1555–E1564. doi: 10.1073/pnas.1521812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Foroutan M., Bhuva D.D., Lyu R., Horan K., Cursons J., Davis M.J. Single sample scoring of molecular phenotypes. BMC Bioinf. 2018;19(1):404. doi: 10.1186/s12859-018-2435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Addison J.B., Voronkova M.A., Fugett J.H., Lin C.-C., Linville N.C., Trinh B. Functional Hierarchy and Cooperation of EMT Master Transcription Factors in Breast Cancer Metastasis. MCR Molecular Cancer Research. 2021 doi: 10.1158/1541-7786.MCR-20-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei S.C., Anang N., Sharma R. Proceedings of the. 2019. Combination anti–CTLA-4 plus anti–PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Solinas, C., Migliori, E., De Silva, P., & Willard-Gallo, K. (2019). LAG3: The Biological Processes That Motivate Targeting This Immune Checkpoint Molecule in Human Cancer. Cancers, 11(8). https://doi.org/10.3390/cancers11081213 [DOI] [PMC free article] [PubMed]
- 94.Yang S., Wei W., Zhao Q. B7–H3, a checkpoint molecule, as a target for cancer immunotherapy. International Journal of Biological Sciences (Vol. 2020;16(11):1767–1773. doi: 10.7150/ijbs.41105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu X., Pu Y., Cron K., Deng L., Kline J., Frazier W.A. CD47 blockade triggers T cell–mediated destruction of immunogenic tumors. Nat Med (Vol. 2015;21(10):1209–1215. doi: 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holderried T.A.W., de Vos L., Bawden E.G., Vogt T.J., Dietrich J., Zarbl R. Molecular and immune correlates of TIM-3 (HAVCR2) and galectin 9 (LGALS9) mRNA expression and DNA methylation in melanoma. In Clinical. Epigenetics. 2019;Vol. 11, Issue 1 doi: 10.1186/s13148-019-0752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hong T., Watanabe K., Ta C.H., Villarreal-Ponce A., Nie Q., Dai X. An Ovol2-Zeb1 Mutual Inhibitory Circuit Governs Bidirectional and Multi-step Transition between Epithelial and Mesenchymal States. PLoS Comput Biol. 2015;11(11) doi: 10.1371/journal.pcbi.1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jolly M.K., Tripathi S.C., Jia D., Mooney S.M., Celiktas M., Hanash S.M. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7(19):27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee B., Villarreal-Ponce A., Fallahi M., Ovadia J., Sun P., Yu Q.-C. Transcriptional mechanisms link epithelial plasticity to adhesion and differentiation of epidermal progenitor cells. Dev Cell. 2014;29(1):47–58. doi: 10.1016/j.devcel.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wahl G.M., Spike B.T. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. npj Breast Cancer. 2017;3:14. doi: 10.1038/s41523-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Giudetti A.M., De Domenico S., Ragusa A., Lunetti P., Gaballo A., Franck J. A specific lipid metabolic profile is associated with the epithelial mesenchymal transition program. Biochimica et Biophysica Acta, Molecular and Cell Biology of Lipids. 2019;1864(3):344–357. doi: 10.1016/j.bbalip.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 102.Ochoa S., de Anda-Jáuregui G., Hernández-Lemus E. Multi-Omic Regulation of the PAM50 Gene Signature in Breast Cancer Molecular Subtypes. Front Oncol. 2020;10:845. doi: 10.3389/fonc.2020.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo W., Keckesova Z., Donaher J.L., Shibue T., Tischler V., Reinhardt F. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mani S.A., Guo W., Liao M.-J., Eaton E.N., Ayyanan A., Zhou A.Y. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jolly M.K. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. In Frontiers in Oncology. 2015;Vol. 5 doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gregory P.A., Bracken C.P., Smith E., Bert A.G., Wright J.A., Roslan S. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22(10):1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bonnomet A., Syne L., Brysse A., Feyereisen E., Thompson E.W., Noël A. A dynamic in vivo model of epithelial-to-mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene. 2012;31(33):3741–3753. doi: 10.1038/onc.2011.540. [DOI] [PubMed] [Google Scholar]
- 108.Somarelli J.A., Schaeffer D., Marengo M.S., Bepler T., Rouse D., Ware K.E. Distinct routes to metastasis: plasticity-dependent and plasticity-independent pathways. Oncogene. 2016;35(33):4302–4311. doi: 10.1038/onc.2015.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zou W., Wolchok J.D., Chen L. PD-L1 (B7–H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wiener Z., Kohalmi B., Pocza P., Jeager J., Tolgyesi G., Toth S. TIM-3 is expressed in melanoma cells and is upregulated in TGF-beta stimulated mast cells. J Invest Dermatol. 2007;127(4):906–914. doi: 10.1038/sj.jid.5700616. [DOI] [PubMed] [Google Scholar]
- 111.Yan W., Liu X., Ma H., Zhang H., Song X., Gao L. Tim-3 fosters HCC development by enhancing TGF-β-mediated alternative activation of macrophages. Gut. 2015;64(10):1593–1604. doi: 10.1136/gutjnl-2014-307671. [DOI] [PubMed] [Google Scholar]
- 112.Lv K., Zhang Y., Zhang M., Zhong M., Suo Q. Galectin-9 promotes TGF-β1-dependent induction of regulatory T cells via the TGF-β/Smad signaling pathway. Mol Med Rep. 2013;7(1):205–210. doi: 10.3892/mmr.2012.1125. [DOI] [PubMed] [Google Scholar]
- 113.Wu C., Thalhamer T., Franca R.F., Xiao S., Wang C., Hotta C. Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity. 2014;41(2):270–282. doi: 10.1016/j.immuni.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao S., Fung-Leung W.-P., Bittner A., Ngo K., Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature (Vol. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.