Summary
The organization of chromatin structure plays a crucial role in gene expression, DNA replication, and repair. Chromatin alterations influence gene expression, and modifications could be associated with genomic instability in the cells during aging or diseases. Here, we provide a modified protocol to isolate fixed neuronal nuclei from a single mouse cortex to investigate the spatial organization of chromatin structure on a genome-wide scale by ATAC-seq (the assay for transposase-accessible chromatin with high-throughput sequencing) and chromatin conformation by Hi-C (high-throughput chromosome conformation capture).
Subject areas: Cell Biology, Flow Cytometry/Mass Cytometry, Cell separation/fractionation, Genomics, Neuroscience
Graphical abstract

Highlights
-
•
Isolation of nuclei from mouse cortical tissue
-
•
Immunolabeling of nuclei and sorting by FACS
-
•
Fixed nuclei protocol
-
•
Preparation of ATAC-seq and Hi-C libraries
The organization of chromatin structure plays a crucial role in gene expression, DNA replication, and repair. Chromatin alterations influence gene expression, and modifications could be associated with genomic instability in the cells during aging or diseases. Here, we provide a modified protocol to isolate fixed neuronal nuclei from a single mouse cortex to investigate the spatial organization of chromatin structure on a genome-wide scale by ATAC-seq and chromatin conformation by Hi-C.
Before you begin
The Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) method is based on the construction of a next-generation sequencing library using the transposase Tn5 (Buenrostro et al., 2013; Chen et al., 2016). The transposase enables the generation of the ATAC-seq library by simultaneous fragmentation of chromatin and integrating specific adapters into open chromatin regions. Genomic regions with accessible or open chromatin can be sequenced by next-generation sequencing and analyzed using bioinformatics tools. Most of the current epigenetic approaches allow profiling of chromatin landscapes from naive nuclei (Buenrostro et al., 2013, Buenrostro et al., 2015). Fixation and mechanical homogenization of the tissue tend to reduce the quality of the data. Here, we present an ATAC-seq method capable of generating high-quality chromatin organization and accessibility data from formaldehyde-fixed nuclei. The protocol includes several steps, such as extraction of the mouse cortical nuclei in low sucrose buffer and centrifugation in gradient iodixanol solutions, and immunolabeling and sorting of NeuN-positive nuclei for further analysis of chromatin structure (Hempel et al., 2007; Marion-Poll et al., 2014; Javier Rubio et al., 2014; Policicchio et al., 2020). Hi-C (high-throughput chromosome conformation capture) is a method to study 3-D genome structure and gene regulation, which facilitates the understanding of the correlation between chromatin organization and gene expression (Rao et al., 2014). Fixation is a critical step to preserve nuclear and cellular architecture. In Hi-C, DNA-protein complexes are fixed with formaldehyde, causing interacting loci to be bound to one another with covalent DNA-protein cross-links. Our modifications allow us to use a reduced amount of starting material, compared to the original paper (Belaghzal et al., 2017). Moreover, we optimized the current protocol for ATAC-seq and Hi-C for a single mouse cortex.
We recommend that you conduct nuclei isolation, sorting and several first steps of ATAC-seq and Hi-C in one day in order not to freeze nuclei (steps 28–31 for ATAC-seq and 52–69 for Hi-C). Then, continue experiments in parallel. The minimum is 4 days to finish library preparation. Of course, you can distribute all steps as it is convenient for you.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Isofluorane | Piramal Critical Care | NDC66794-017 |
| PBS | Biological Industries | 02-023-1A |
| Sucrose | Sigma-Aldrich | S0389 |
| HEPES sodium salt | Sigma-Aldrich | H7006 |
| CaCl2 | Sigma-Aldrich | C5080 |
| Mg(CH3COO)2 | Sigma-Aldrich | M5661 |
| EDTA | Merck | 8414 |
| Triton X-100 | Sigma-Aldrich | T8787 |
| DTT | Sigma-Aldrich | D0632 |
| KCL | Sigma-Aldrich | P9541 |
| MgCl2 | Sigma-Aldrich | M8266 |
| Tris (hydroxymethyl)aminomethane | Bio-Lab ltd | 002009239100 |
| NaCl | DAEJUNG | 7646-15-5 |
| SDS | TCI | D0996 |
| Protease Inhibitor Cocktail | Sigma-Aldrich | P5726 |
| Formaldehyde | Sigma-Aldrich | F-1635 |
| Glycine | Bio-Lab ltd | 000713239100 |
| OptiPrep™ Density Gradient Medium | Sigma-Aldrich | D1556 |
| BSA | BioWorld | 22070008-2 |
| Goat serum | Sigma-Aldrich | G9023 |
| Mouse Anti-NeuN Antibody (clone A60) | EMD Millipore | FCMAB317PE |
| Illumina Tagment DNA TDE1 Enzyme and Buffer Small Kit | Illumina | 20034197 |
| Agarose | Lonza, SeaKem | 50004 |
| Proteinase K | NEB | P8107S |
| BSA, Molecular Biology Grade | NEB | B9000S |
| Ultrapure water | Biological Industries | 01-866-1B |
| iTaq™ Universal SYBR® Green One-Step Kit | Bio-Rad | 1725150 |
| NucleoSpin Gel and PCR Clean-up Kit | MACHEREY-NAGEL | 740609.50 |
| NEBNext® High-Fidelity 2X PCR Master Mix | NEB | #M0541 |
| AMPure XP beads | Beckman Coulter | A63881 |
| DpnII buffer | NEB | B0543S |
| DpnII | NEB | R0543S |
| dCTP | Invitrogen | 10217016 |
| dGTP | Invitrogen | 10218014 |
| dTTP | Invitrogen | 10219012 |
| Biotin-14-dATP | Invitrogen | 2067549 |
| DNA Polymerase I, Large (Klenow) Fragment | NEB | M0210S |
| NEB 2.1 buffer | NEB | B7202S |
| Chloroform | Frutarom | 5551030 |
| Phenol | Bio-Lab ltd | 0016912344006 |
| Glycogen | Sigma-Aldrich | R0561 |
| T4 Polynucleotide Kinase | NEB | M0201S |
| T4 DNA Polymerase | NEB | M0203S |
| MyOne Streptavidin C1 beads | Invitrogen | 65001 |
| Klenow Fragment (3'→5' exo-) | NEB | M0212S |
| T4 DNA Ligase | NEB | M0202S |
| T4 DNA Ligase Reaction Buffer | NEB | B0202S |
| Quick Ligation™ Kit | NEB | M2200S |
| dNTP Mix | NEB | N0447L |
| Sodium acetate | Sigma-Aldrich | S2889 |
| Qubit dsDNA BR Assay Kit | Invitrogen | Q32853 |
| DNA Ladder GeneRuler DNA Ladder Mix | Thermo Scientific | SM0331 |
| Ethidium bromide | Sigma-Aldrich | 09-0617 |
| TAE 50× | Bio-Lab ltd | 20502375 |
| TWEEN® 20 | Sigma-Aldrich | P1379 |
| Experimental models: organisms/strains | ||
| Fresh mouse cortical tissue (from samples C57BL/6J mice) | The Jackson Laboratory | 000664 |
| Software and algorithms | ||
| nf-core ATAC-seq pipeline | (Ewels et al., 2020) | nf-core: nf-core: https://nf-co.re/atacseq Zenodo: https://doi.org/10.5281/zenodo.2634132 |
| distiller-nf Hi-C pipeline | GitHub: GitHub: Zenodo: https://doi.org/10.5281/zenodo.3350926 | |
| Oligonucleotides | ||
| ATAC seq Primer1: AATGATACGGCGACCACCGA GATCTACACTCGTCGGCAGCGTCAGATGTG |
Integrated DNA Technologies | N/A |
| ATAC seq Primer2_1: CAAGCAGAAGACGGCATA CGAGATTCGCCTTAGTCTCGTGGGCTCGGAGATGT |
Integrated DNA Technologies | N/A |
| ATAC seq Primer2_2: CAAGCAGAAGACGGCATA CGAGATAGCGTAGCGTCTCGTGGGCTCGGAGATGT |
Integrated DNA Technologies | N/A |
| Illumina Primer dir: 5′AATGATACGGCGA CCACCGAGAT 3′ |
Integrated DNA Technologies | N/A |
| Illumina Primer rev: 5′CAAGCAGAAGACGGCATACGA 3′ | Integrated DNA Technologies | N/A |
| Uni: AATGATACGGCGACCACCGAGATCTACACTC TTTCCCTACACGACGCTCTTCCGATCT |
Integrated DNA Technologies | N/A |
| Tru1: 5′ PO4 - GATCGGAAGAGCACACGTCTGAACTC CAGTCACATCACGATCTCGTATGCCGTCTTCTGCTTG |
Integrated DNA Technologies | N/A |
| Tru9: 5′ PO4 - GATCGGAAGAGCACACGTCTGAACT CCAGTCACGATCAGATCTCGTATGCCGTCTTCTGCTTG |
Integrated DNA Technologies | N/A |
| Other | ||
| 1.5 mL Eppendorf tube | Axygen | MCT-175-C |
| 50 mL Centrifuge tubes | Greiner | 227270 |
| 15 mL Centrifuge tubes | Greiner | 188261 |
| Falcon Round Bottom Polystyrene Test Tubes with Cell Strainer Cap | Falcon | 22719024 |
| Dounce homogenizer | Thomas Scientific | 3431D76 |
| Mechanical homogenizer | Kinematica AG | POLYTRON 2100 |
| Refrigerated centrifuge | Eppendorf | 5702R and 5810R |
| FACSAria cytometer equipped with a double argon (488 nm) and helium-neon (633 nm) laser | BD Biosciences | BD FACSAriaTM III Cell SorterModel number 648282-C1-010110-X-X-X |
| 30-kDa Amicon Ultra 0.5 Column | Millipore | UFC 503096 |
| Bioruptor® Plus Sonication Device | Bioruptor® | B01020001 |
| 1.5 mL TPX microtubes | Diagenode | C30010010 |
| Thermomixer | Fisher Scientific | FSGPD05 |
| 16-Tube SureBeads™ Magnetic Rack | Bio-Rad | 1614916 |
| DNA LoBind Tube 1.5 mL | Eppendorf | 022431021 |
| Corning® DeckWorks™ low binding tips | Corning | CLS4151 |
| CFX96 Touch Deep Well Real-Time PCR System | Bio-Rad | 1854095 |
| PCR machine (T100 Thermal Cycler) | Bio-Rad | 1861096 |
| Mini-Sub Cell GT Cell | Bio-Rad | 1704406 |
Alternatives: “The list of materials described in the key resources table are the ones tested in our laboratory. Equivalent chemicals, plastic and equipment from different suppliers may be suitable and should be tested by the users.
Step-by-step method details
Timing: 2 h 30 min for 1 mouse
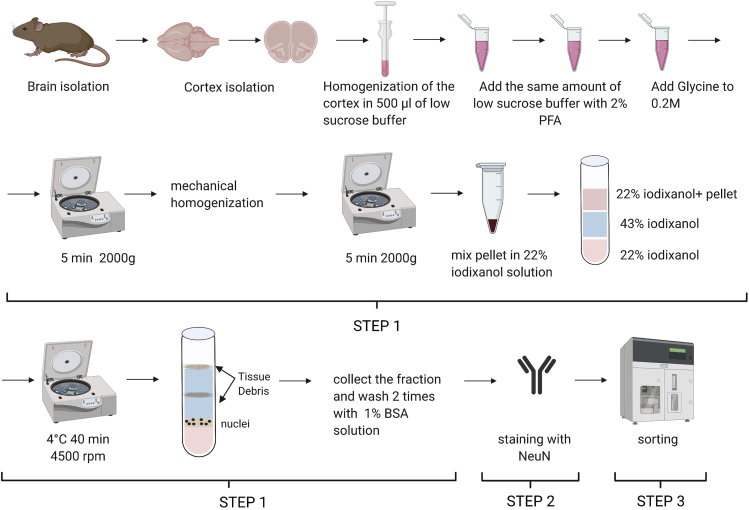
Isolation of nuclei from the mouse brain cortex (10 months old, C57/BL/6J) (Figure 1)
Note: This protocol describes the procedure for 1 mouse. Increase time by the number of mice to be processed.
-
1.
Mouse is sacrificed with isoflurane and perfused with cold PBS. Cortex is isolated on the ice
Note: We do not recommend keeping the isolated cortex more than 10–15 min on ice.
-
2.
Homogenize cortical tissue in 500 ul of ice-cold low sucrose buffer using Dounce homogenizer 10–15 times on ice.
Low sucrose buffer
| Reagent | Final concentration | Amount for 50 mL |
|---|---|---|
| 1 M Sucrose | 0,32 M | 16 mL |
| 1 M HEPES pH 8,0 | 10 mM | 500 μL |
| 1 M CaCl2 | 5 mM | 250 μL |
| 1 M Mg(CH3COO)2 | 3 mM | 150 μL |
| 0,5 M EDTA | 0,1 mM | 10 μL |
| 20% Triton X-100 | 0,1% | 250 μL |
| 1 M DTT | 1 mM | 50 μL |
| 100× PIC (Protease Inhibitor Cocktail) - add before use | Prepare 1× before homogenization | 500 μL |
| Ultra pure water | N/A | 32.29 mL |
Store at 4°C, and do not store more than 2 weeks.
-
3.
Measure the volume of homogenate and add the same amount of low sucrose buffer with 2% formaldehyde (to get in the end 1% formaldehyde low sucrose buffer). Mix well and leave for 10 min at 22°C temperature.
CRITICAL: Fixation time must be the same for all samples.
CAUTION: Working with fixation solutions should be done in a chemical hood.
-
4.
Add glycine to homogenate to get 0.2M glycine solution, mix gently and incubate for 10 min at 22°C temperature to quench the crosslinking reaction.
Note: In the case of multiple samples, it is important to keep them on ice after this step until the next one.
-
5.
Centrifuge the samples for 5 min at 2000 g at 4°C, discard the supernatant.
-
6.
Resuspend the pellet in 3 mL of low sucrose buffer with PIC.
-
7.
Homogenate each sample with the mechanical homogenizer (rotor/stator) at 15 rpm 4 times. Each time is about 6 s.
-
8.
Centrifuge the samples for 5 min at 2000 g at 4°C, discard the supernatant.
-
9.Prepare iodixanol gradient to isolate nuclei.
-
a.Iodixanol solution (5V Optiprep 60% iodixanol + 1V of dilution solution)
-
b.Dilution solution:
Reagent Final concentration Amount for 50 mL 1 M KCl 150 mM 7.5 mL 1 M MgCl2 30 mM 1.5 mL 1 М Tris-HCl pH 8,0 120 mM 6 mL Ultra pure water N/A 35 mL Store at 4°C, and do not store more than 1 month. -
c.Resuspension solution:
Reagent Final concentration Amount for 50 mL 1 M Sucrose 250 mM 4 mL 1 M KCl 25 mM 1.25 mL 1 M MgCl2 5 mM 0.25 mL 1 М Tris-HCl pH 8,0 20 mM 1 mL Ultra pure water N/A 43.5 mL Store at 4°C, and do not store more than 2 weeks. -
d.Prepare two gradient solutions: 22% and 43% iodixanol solutions by diluting iodixanol solution with a resuspension buffer. Keep on ice.
-
a.
-
10.
Resuspend pellet from step 8 in 2 mL of 22% iodixanol solution and prepare a gradient. Place in a 15 mL tube 1 mL of 43% iodixanol solution, then carefully add a layer of 1 mL of 22% iodixanol solution, on top add 2 mL of tissue homogenate in 22% iodixanol solution.
-
11.
Centrifuge at 2600 g for 40 min at 4°C on the bucket centrifuge.
CRITICAL: Usage of a bucket centrifuge is necessary
Note: You can increase the time of centrifugation if needed.
-
12.
Collect the nuclei interphase between 43% iodixanol solution and other layers.
-
13.
Dilute the nuclei interphase fraction with resuspension solution with 1% BSA (1:2)
-
14.
Centrifuge at 3200 g for 10 min at 4°C, discard the supernatant.
-
15.
Wash the pellet once with 1 mL of resuspension solution with 1% BSA
-
16.
Centrifuge at 3200 g for 5 min at 4°C.
Figure 1.
Schematic illustration of the preparation of cortical nuclei for ATAC-seq and Hi-C methods
FACS staining for sorting
Timing: 1 h15 min for 1 mouse
-
17.
Discard the liquid and resuspend the pellet in 200 μL PBST with 5% BSA and 3% goat serum.
CRITICAL: Take out 10 μL of pellet and dilute it in PBST with 5% BSA up to 500 μL. Use it for the non-stained control for FACS.
Pause point: Fixed nuclei can be stored at 4°C for up to 48 h.
-
18.
Add 4 μL of NeuN-PE antibody to 190 μL of resuspended pellet and incubate for 30 min on ice.
-
19.
Centrifuge at 3200 g for 5 min at 4°C, discard the supernatant.
-
20.
Resuspend the pellet in 200 μL PBST with 5% BSA.
-
21.
Centrifuge at 3200 g for 5 min at 4°C, discard the supernatant.
-
22.
Resuspend the pellet in 1 mL of PBST with 5% BSA
Sorting of immunolabeled nuclei
Timing: 45 min for 1 mouse
-
23.
Transfer the solution to Falcon™ Round-Bottom Polystyrene Test Tubes through Cell Strainer Snap Cap to filter the nuclei (35-μm cell strainer)
CRITICAL: Clumps and debris can clog the instrument fluidics
-
24.
Perform the fluorescence-activated sorting of fixed nuclei (Figure 2)
Note: We recommend using 70 μm nozzle for sorting of nuclei
-
25.
Collect the NeuN - positive population of nuclei (Figure 2)
-
26.
After FACS, the solution with nuclei was centrifuged for 10 min at 3200 g and 4°C.
-
27.
The pellet of fixed nuclei was resuspended in 50 ul of ultra-pure water. The pellet contained 1,3–1,5 m of neuron nuclei. The cell pellet was kept on ice.
Figure 2.
Representative gating strategy
Nuclei were immunostained with an antibody specific to NeuN, a nuclear membrane protein, filtered through a 35-μm cell strainer and sorted on a BD Biosciences FacsAria flow sorter. The right panels illustrate NeuN negative and positive populations. Positive NeuN-PE populations were used for downstream applications.
ATAC-seq on the fixed nuclei
Timing: ~8 h
Note: Increase time by the number of mice to be processed. Collected nuclei were used for downstream applications such as ATAC-seq (Figure 3).
Figure 3.
Schematic illustration of the ATAC-seq method
Tn5 transposase cuts the open chromatin and tags the adapters to it to generate DNA fragments. The chromatin is fragmented and tagged with sequencing adapters using the Tn5 transposase to generate the ATAC-Seq library.
100,000 nuclei were collected from the pellet (step 27) for the transposition reaction.
Note: The number of nuclei might be reduced to 50,000.
Transposition reaction mix
| Reagent | Final concentration | Amount for 50 μL |
|---|---|---|
| 2xTD buffer | 1× | 25 μL |
| TDE1 | N/A | 2,5 μL |
| Ultra pure water | N/A | up to 50 μL |
-
28.
Resuspend the pellet in the transposition reaction mix.
-
29.
Incubate the transposition reaction at 37°C for 30 min with shaking (1400 rpm).
-
30.
Add the equal volume of 2× reverse-crosslinked solution (up to 1×: 50 mM Tris-HCl pH8.0, 1mM EDTA, 0.1% SDS, 0.2 M NaCl, 2 μL Proteinase K).
CRITICAL: Since we perform ATAC-seq on fixed nuclei it’s crucial to do this step. It helps to achieve a good quality of ATAC-seq library.
-
31.
Incubate the mixture at 65°C without shaking for 4–5 h.
Pause point: This might be done for 16–18 h.
-
32.
Purify the transposed DNA using PCR Purification kit (we used NucleoSpin Gel and PCR Clean-up kit, Macherey-Nagel) and elute in 10 μL of 10 mM Tris-HCl pH 8.0.
-
33.
Amplify the transposed DNA fragments:
Amplification of transposed DNA fragments
| Reagent | Final concentration | Amount for 50 ul |
|---|---|---|
| Transposed DNA | N/A | 10 μL |
| PCR Primer 1 | 25 μM | 2,5 μL |
| Barcoded PCR Primer 2 | 25 μM | 2,5 μL |
| 2× PCR Master Mix | 1× PCR Master Mix | 25 μL |
| Ultra pure water | N/A | 10 μL |
Thermal cycle as follows:
| PCR cycling conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Pre-incubation | 72°C | 5 min | 1 |
| Initial denaturation | 98°C | 30 s | 1 |
| Denaturation | 98°C | 10 s | 5 |
| Annealing | 63°C | 30 s | |
| Extension | 72°C | 1 min | |
| Hold | 4°C | Forever | |
-
34.
Perform qPCR to choose an additional number of cycles.
| Reagent | Final concentration | Amount for 15 ul |
|---|---|---|
| Previously PCR-amplified DNA | N/A | 5 μL |
| PCR Primer 1 | 25 μM | 0,25 μL |
| Barcoded PCR Primer 2 | 25 μM | 0,25 μL |
| 100× SYBR GREEN I | 1× | 0,09 μL |
| 2× PCR Master Mix | 1× | 5 μL |
| Ultra pure water | N/A | 4,41 μL |
-
35.
Amplify the rest of the previously PCR-amplified DNA with the additional number of cycles you chose. We used 7 additional cycles.
-
36.
Check the library size by electrophoresis or other suitable methods.
-
37.
After PCR, we purified DNA using AMPure XP beads and selected fragments with up to 1 kb length for sequencing.
-
38.
Add 0.5 volumes of AMPureXP beads to the solution. Mix thoroughly by pipetting.
-
39.
Incubate for 10 min at 22°C. Mix from time to time.
-
40.
Separate the beads on a magnet for 2–3 min.
-
41.
Transfer the solution to a new tube.
-
42.
Add 1.3 volumes of AMPureXP beads to the solution. Mix thoroughly by pipetting.
-
43.
Incubate for 10 min at 22°C. Mix from time to time.
-
44.
Place the tube on a magnet for 2–3 min, discard the solution.
-
45.
Add 3 volumes of fresh 75% ethanol, wash the beads on the magnet. Discard the solution. Repeat this step again.
-
46.
Separate the beads on the magnet, discard the solution.
-
47.
Leave the beads on the magnet for 5–10 min to air-dry the beads from the ethanol.
-
48.
Remove the tube from the magnet.
-
49.
Add 10 μL of 10 mM Tris-HCl pH 8.0 to the beads to elute the DNA. Mix gently by pipetting. The volume of 10 mM Tris-HCl pH 8.0 depends on the volume of the initial sample and your needs, but the minimum is 10 μL.
-
50.
Incubate for 10 min at 22°C.
-
51.
Place the tube on the magnet for 2–3 min. Transfer the solution to a fresh tube.
Hi-C
Timing: ~4 days
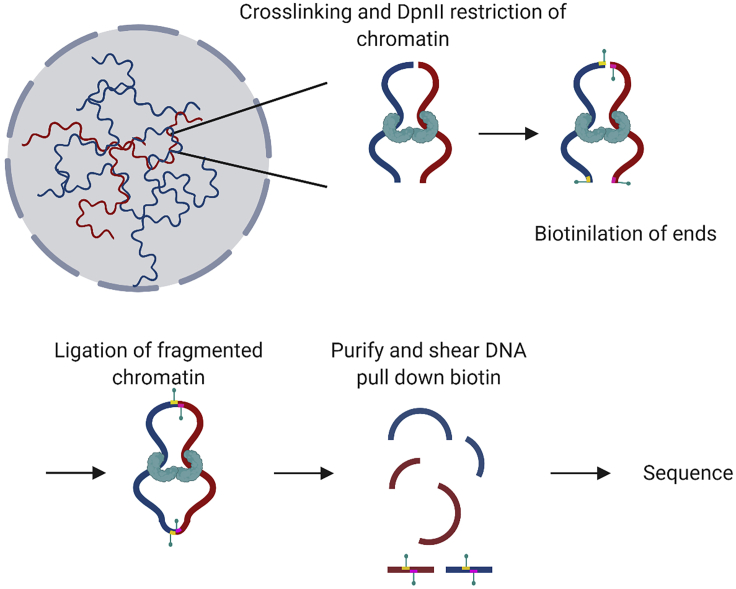
Collected nuclei were used for downstream applications such as Hi-C (Figure 4). Hi-C was performed as previously described (Belaghzal et al., 2017) with minor modifications.
-
52.
About 1,2–1,4 million of fixed neuron nuclei were collected from step 3.
-
53.
Add 11 μL of 10× DpnII buffer (up to 1.1×), 1,5 μL of 20% SDS (0,3% final) and ultra-pure water up to 100 μL total.
-
54.
Resuspend carefully.
-
55.
Incubate 1 h at 37°C with shaking 1400 rpm.
-
56.
Add 27 μL of 20% Triton-X100 (1,8% final) and 173 μL 1.1× DpnII buffer (300 μL total).
-
57.
Incubate 1 h at 37°C with shaking 1400 rpm.
-
58.
Take 15 μL of mixture for integrity control and place it at 4°C until step 7
CRITICAL: Integrity control is important to check stability of DNA before restriction.
-
59.
For DpnII restriction, add 4 μL (200 U) of DpnII, resuspend gently.
-
60.
Incubate 3–4 h at 37°C with shaking 1400 rpm.
Note: Incubation could be performed for 16–18 h
-
61.
Incubate 20 min at 65°C to inactivate DpnII.
-
62.
Take 15 μL of mixture for digestion control and place it at 4°C until step 7
CRITICAL: Digestion control is important and the effectiveness of restriction can impact the quality of the library.
-
63.
Centrifuge at 3200 g for 10 min at 20°C, discard the supernatant.
-
64.
Resuspend the pellet in 100 μL of 1× NEB2 buffer.
-
65.
Prepare the mixture for the biotinylation of DNA ends:
| Reagent | Final concentration | Amount for 1 sample (20 ul) |
|---|---|---|
| dCTP | 10 mM | 0.375 μL |
| dGTP | 10 mM | 0.375 μL |
| dTTP | 10 mM | 0.375 μL |
| biotin-14-dATP | 0.4 mM | 9.375 μL |
| Klenow DNA polymerase I | 5 U/μL | 2.5 μL |
| NEB2 Buffer 10× | 1× | 2 μL |
| Ultra pure water | N/A | 5 μL |
-
66.
Add 20 μL of the mixture to the 100 μL of sample and incubate 1.5 h at 37°C with shaking 1000 rpm.
-
67.
Centrifuge at 3200 g for 10 min at 20°C, discard the supernatant.
-
68.
For blunt-end ligation, add 196.5 μL of 1× T4 ligase buffer, 1 μL BSA, 2.5 μL (1000 U) of T4 ligase (NEB).
-
69.
Incubate at 20°C during 16–18 h with shaking 1400 rpm
Note: We recommend performing ligation for 16–18 h, but it’s possible to do it for 5–6 h.
-
70.
Reverse-crosslinking. Add 4 μL of Proteinase K and 10 μL 20% SDS to the samples.
-
71.
Add 2 μL of Proteinase K, 10 μL 20% SDS and 185 μL of 10 mM Tris-HCl pH 8.0 to the integrity and digestion controls.
-
72.
Incubate all the samples and controls at 65°C without shaking for 4–5 h.
Pause point: Incubation could be performed for 16–18 h.
-
73.
Cool down the samples at 20°C.
-
74.
Add PBS up to 300 μL.
-
75.
DNA purification. Add 1 volume (300 μL) of the mixture phenol:chloroform (1:1) to the samples, mix well.
-
76.
Centrifuge for 10 min at 20°C at the maximum speed.
-
77.
Transfer the aqueous phase to a new tube.
-
78.
Add 1 volume (300 μL) of the chloroform to the aqueous phase of each sample, mix thoroughly.
-
79.
Centrifuge for 10 min at 20°C at the maximum speed.
-
80.
Transfer the aqueous phase to a new tube.
-
81.
Add 1/9 of the aqueous phase volume (33.3 μL) of 3M sodium acetate solution (pH 5.0), 2 μL of glycogen (20 mg/mL), and 750 μL (2.5V) of 96% ethanol to the samples, mix well.
-
82.
Centrifuge for 15 min at the 4°C and maximum speed. Discard the supernatant.
-
83.
Add 500 μL of cold 75% ethanol to wash the pellet.
-
84.
Centrifuge for 15 min at the 4°C and maximum speed. Discard the supernatant.
-
85.
Air-dry the pellets.
-
86.
Dissolve the samples in 51 μL of 10 mM Tris-HCl pH 8.0 and controls in 20 μL of 10 mM Tris-HCl pH 8.0.
-
87.
Use 1 μL of each sample and 10 μL of each control to run 0.8% agarose gel. Check the quality of integrity control, restriction, and ligation.
-
88.
Removal of biotin from un-ligated ends.
Figure 4.
Schematic illustration of the Hi-C method
Prepare the mixture:
| Reagent | Final concentration | Amount for 1 sample (65 μL) |
|---|---|---|
| DNA sample | N/A | 50 μL |
| NEB2.1 Buffer 10× | 1× | 6.5 μL |
| dATP | 2.5 mM | 0.65 μL |
| dGTP | 2.5 mM | 0.65 μL |
| T4 DNA Polymerase | N/A | 2 μL |
| Ultra pure water | N/A | 5.2 μL |
-
89.
Incubate at 20°C for 4 h.
-
90.
Incubate at 75°C for 20 min to inactivate the enzyme.
-
91.
Cool down the samples and keep at 4°C.
Note: It is possible to perform steps 89–91 in a PCR machine for 16–18 h.
Pause point: Samples might be kept at 4°C for 16–18 h.
-
92.
DNA sonication. Add 150 μL of 2× Sonication buffer and 85 μL of ultra-pure water (300 total).
Sonication buffer 2×:
| Reagent | Final concentration | Amount for 150 μL |
|---|---|---|
| 1M Tris-HCl pH 8.0 | 50 mM | 7.5 μL |
| 0.5M EDTA | 20 mM | 6 μL |
| 20% SDS | 0.2% | 1.5 μL |
| Ultra pure water | N/A | 135 μL |
Store at 4°C, and do not store more than 2 weeks.
-
93.
Shear DNA to a size up to 1000 bp. Chromatin was solubilized by sonication for 20 cycles on the Diagenode Bioruptor with the following settings: high (H) power output, 30 s ON/30 s OFF pulses, +4°C water bath, no floating ice.
CRITICAL: It is important to use special Bioruptor tubes
-
94.
After sonication, transfer each sample to a 30-kDa Amicon Column and add 150 μL of 10 mM Tris-HCl pH 8.0 (450 μL total).
-
95.
Centrifuge for 5 min at 4°C and maximum speed.
-
96.
Discard the flow-through. Add 450 μL of 10 mM Tris-HCl pH 8.0 to the column.
-
97.
Centrifuge for 5 min at 4°C and maximum speed.
-
98.
Transfer the solution from the column to a new tube, measure the volume. Add 10 mM Tris-HCl pH 8.0 up to 50 μL.
-
99.
Add 100 μL (2 volumes) of AmpureXP beads to the solution. Mix thoroughly by pipetting.
-
100.
Incubate for 10 min at 22°C. Mix from time to time.
-
101.
Place the tube on a magnet for 2–3 min, then discard the solution.
-
102.
Add 150 μL (3 volumes) of fresh 75% ethanol, wash the beads on the magnet. Discard the solution. Repeat this step again.
-
103.
Separate the beads on the magnet, discard the solution.
-
104.
Leave the beads on the magnet for 5–10 min to air-dry the beads from the ethanol.
-
105.
Remove the tube from the magnet.
-
106.
Add 50 μL of 10 mM Tris-HCl pH 8.0 to the beads to elute the DNA. Mix gently by pipetting.
-
107.
Incubate for 10 min at 22°C.
-
108.
Place the tube on the magnet for 2–3 min. Transfer the solution to a fresh tube.
-
109.
Prepare the mixture for end repair:
| Reagent | Final concentration | Amount for 70 μL |
|---|---|---|
| DNA sample | N/A | 50 μL |
| 10×T4 ligase buffer | 1× | 7 μL |
| dNTP mix | 10 mM | 2.5 μL |
| T4 Polynucleotide kinase | 10 μ/μL | 2.5 μL |
| T4 DNA Polymerase | 3 μ/μL | 2.5 μL |
| Klenow DNA Polymerase I | 5 μ/μL | 0.5 μL |
| Ultra pure water | N/A | 5 μL |
-
110.
Incubate for 30 min at 20°C, then for 20 min at 75°C to deactivate the enzymes.
-
111.
Keep at 4°C until the next step.
Note: The incubations 110–111 might be performed in a PCR machine.
Pause point: Samples might be kept at 4°C for 16–18 h.
-
112.
Biotin pull-down with streptavidin-coated beads
CRITICAL: All the remaining steps must be done in DNA low-binding tubes using low-binding tips.
-
113.
Vortex the MyOne Streptavidin C1 beads and transfer 4 μL of bead solution to a low-binding tube.
-
114.
Resuspend the beads in 100 μL of Tween washing buffer (TWB) and incubate for 3 min at 20°C with shaking 900 rpm.
Tween washing buffer (TWB)
| Reagent | Final concentration | Amount for 1000 μL |
|---|---|---|
| 1M Tris-HCl pH 8.0 | 5 mM | 5 μL |
| 0.5M EDTA | 5 mM | 10 μL |
| 5M NaCl | 1M | 200 μL |
| 10% Tween | 0.05% | 5 μL |
| Ultra pure water | N/A | 780 μL |
Store at 4°C, and do not store more than 2 weeks.
-
115.
Separate the beads on the magnet, discard the supernatant.
-
116.
Resuspend the beads in 100 μL of TWB.
-
117.
Separate the beads on the magnet, discard the supernatant.
-
118.
Resuspend the beads in 100 μL of 2× Binding buffer (BB).
2× Binding buffer (BB):
| Reagent | Final concentration | Amount for 1000 μL |
|---|---|---|
| 1M Tris HCl pH 8.0 | 10 mM | 10 μL |
| 0.5M EDTA | 1 mM | 2 μL |
| 5M NaCl | 2 M | 400 μL |
| Ultra pure water | N/A | 588 μL |
Store at 4°C, and do not store more than 2 weeks.
-
119.
Add to the sample from step 111 (70 μL) 30 ul of TLE Buffer to 100 μL total.
TLE Buffer pH 8.0:
| Reagent | Final concentration | Amount for 1000 μL |
|---|---|---|
| 1M Tris HCl pH 8.0 | 10 mM | 10 μL |
| 0.5M EDTA | 0.1 mM | 2 μL |
| Ultra pure water | N/A | 988 μL |
Store at 4°C, and do not store more than 2 weeks.
-
120.
Mix the DNA sample (100 μL) and the beads in 2×BB from step 118 (100 μL).
-
121.
Incubate for 15 min in a shaker (1200 rpm).
-
122.
Separate the beads on the magnet, discard the supernatant.
-
123.
Resuspend the beads in 100 μL of 1× BB.
-
124.
Separate the beads on the magnet, discard the supernatant.
-
125.
Resuspend the beads in 100 μL of 1× TLE pH 8.0.
-
126.
Separate the beads on the magnet, discard the supernatant.
-
127.
Resuspend the beads in 25 μL of 1× TLE pH 8.0.
-
128.
Prepare the mixture for A-tailing
| Reagent | Final concentration | Amount for 50 μL |
|---|---|---|
| Streptavidin-coated beads | N/A | 25 μL |
| 10× NEBuffer 2.1 | 1× | 5 μL |
| dATP | 10 mM | 1 μL |
| Klenow fragment (3’–5’ exo-) | 5 U/ul | 5 μL |
| Ultra pure water | N/A | 14 μL |
-
129.
Incubate for 30 min at 20°C, then for 20 min at 65°C to deactivate the enzyme.
-
130.
Keep at 4°C until the next step.
Note: The incubations might be performed in a PCR machine.
Pause point: Samples might be kept at 4°C for 16–18 h.
-
131.
Separate the beads on the magnet, discard the supernatant.
-
132.
Resuspend the beads in 70 μL of 1× T4 ligation buffer.
-
133.
Separate the beads on the magnet, discard the supernatant.
-
134.
Resuspend the beads in the ligation mixture: 25 μL of 1.1× Quick ligase buffer +1 μL of Quick T4 ligase.
-
135.
Add 2.5 μL of 15 uM Illumina TruSeq adapter.
-
136.Adapters preparation
 CRITICAL: This step must be done in advance.Note: It is possible to use Illumina kit adapters
CRITICAL: This step must be done in advance.Note: It is possible to use Illumina kit adapters-
a.Prepare a mixture:
Reagent Final concentration Amount for 20 μL Adapter Uni 100 μM 3 μL Adapter Tru 100 μM 3 μL 10× NEBuffer 2 1× 2 μL Ultra pure water N/A 12 μL -
b.Set the settings of PCR machine to perform the annealing procedure as follows: ramp at 0.5°C/s from 25°C to 97.5°C hold at 97.5°C for 2.5 min drop at 0.5°C/s from 97.5°C to 4°C keep at 4°C
 CRITICAL: Keep adapters at −20°C.
CRITICAL: Keep adapters at −20°C.
-
a.
-
137.
Incubate for 2 h at 20°C.
-
138.
Incubate for 2 h at 20°C.
-
139.
Separate the beads on the magnet, discard the supernatant.
-
140.
Add 100 ul of TWB to the beads, mix carefully by pipetting.
-
141.
Incubate for 5 min at 20°C.
-
142.
Separate the beads on the magnet, discard the supernatant.
-
143.
Repeat 139–141 steps again.
-
144.
Resuspend the beads in 100 μL of 1× BB (Binding buffer).
-
145.
Separate the beads on the magnet, discard the supernatant.
-
146.
Resuspend the beads in 100 μL of 1× NEBuffer 2.1.
-
147.
Separate the beads on the magnet, discard the supernatant. Repeat 143–144 steps again.
-
148.
Resuspend the beads in 20 μL of 10 mM Tris-HCl pH 8.0 and transfer them to a new tube.
-
149.
Incubate the beads solution at 98°C for 10 min to remove the DNA from the beads. The incubation might be performed in a PCR machine. In that case, set the program: 98°C–10 min followed by 4°C - ∞.
CRITICAL: This step helps separate beads and leave just DNA in the solution. It's critical because beads prevent fluorescence during Real-Time PCR
-
150.
Place the tube on the magnet to separate the beads. Transfer the supernatant (DNA solution) to a new tube.
-
151.
Set up the Real-Time PCR to determine the optimal cycle for final amplification.
| Reagent | Final concentration | Amount for 15 μL |
|---|---|---|
| DNA | N/A | 1.5 μL |
| Illumina Primer dir (10 μM) | 1 μM | 1.5 μL |
| Illumina Primer rev (10 μM) | 1 μM | 1.5 μL |
| 2× NEB Next | 1× | 7.5 μL |
| SYBR Green 100× | 1× | 0.1 μL |
| Ultra pure water | N/A | 2.9 μL |
The thermal cycle as follows:
| PCR cycling conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial denaturation | 98°C | 30 s | 1 |
| Denaturation | 98°C | 10 s | 20 |
| Annealing | 65°C | 15 s | |
| Extension | 72°C | 20 s | |
| Hold | 4°C | forever | |
The optimal cycle is about ⅓ of the maximum fluorescence intensity. We chose 7 cycles.
-
152.
Perform the PCR reactions on the remaining DNA template with the chosen number of cycles and the same conditions. We performed 3 reactions with 5 μL of DNA template for each one.
| Reagent | Final concentration | Amount for 50 μL |
|---|---|---|
| DNA | N/A | 5 μL |
| Illumina Primer dir (10 μM) | 1 μM | 5 μL |
| Illumina Primer rev (1 0 μM) | 1 μM | 5 μL |
| 2× NEBNext | 1× | 25 μL |
| Ultra pure water | N/A | 10 μL |
-
153.
Pull 3 PCR reactions for each sample together.
-
154.
To remove primer dimers, purify the amplified Hi-C library using AMPure XP beads. Allow the AMPure mixture to come to 20°C and mix well before use.
-
155.
Add 270 μL (1.8 volumes) of AMPureXP beads to the solution. Mix thoroughly by pipetting.
-
156.
Incubate for 10 min at 20°C. Mix from time to time.
-
157.
Place the tube on a magnet for 2–3 min, discard the solution.
-
158.
Add 450 μL (3 volumes) of fresh 75% ethanol, wash the beads on the magnet. Discard the solution. Repeat this step again.
-
159.
Separate the beads on the magnet, discard the solution.
-
160.
Leave the beads on the magnet for 5–10 min to air-dry the beads from the ethanol.
-
161.
Remove the tube from the magnet.
-
162.
Add 30 μL of 10 mM Tris-HCl pH 8.0 to the beads to elute the DNA. Mix gently by pipetting.
-
163.
Incubate for 10 min at 20°C.
-
164.
Place the tube on the magnet for 2–3 min. Transfer the solution to a fresh tube.
-
165.
Quantify the amount of DNA in the Hi-C library fluorometrically with a Qubit dsDNA Broad Range kit according to the manufacturer's instructions.
-
166.
Take 1 μL of samples to check the quality running 1% agarose gel.
Expected outcomes
The protocol describes how to prepare both Hi-C and ATAC-seq libraries from fixed cortex tissue. The quality control of our libraries was performed by the GENEWIZ company. Figure 5 represents the library size distribution for ATAC-seq (A) and Hi-C (B). The ATAC-seq fragment size distribution has an expected profile. The first peak corresponds to the nucleosome-free library fragments, the second peak - mono-nucleosome fragments, the third peak - di-nucleosome fragments, the last peak - tri-nucleosome fragments. The average size of the ATAC-seq library fragments and Hi-C library is 497 bp and 557 bp, respectively.
Figure 5.
Size distribution of ATAC-seq and Hi-C libraries fragments
The average size for each library is indicated by a red dashed line (A, ATAC-seq library; B, Hi-C library). Peaks at 25 bp and 1500 bp are internal standards used for quantification determination with the High Sensitivity D1000 ScreenTape®.
ATACseq libraries were deep sequenced on the Illumina HiSeq by GENEWIZ (USA; www.GENEWIZ.com), resulting in 29–40 million 150-nt paired-end reads per sample. Sequencing reads were processed with the ATAC-seq nextflow pipeline (https://nf-co.re/atacseq) (Ewels et al., 2020) with default parameters, except for the peak calling option. The peaks were calculated by MACS2 (Zhang et al., 2008) with a narrow peak option and an initial threshold q-value of 0.05 as the cut-off. Reads were aligned on mm10 genome assembly. The number of mapped reads as well as the number of mitochondrial reads for each replicate is indicated in the Table 1 below. As expected, the number of mitochondrial reads is low (0.48%–0.86%).
Table 1.
Number of reads for ATAC-seq
| Sample | Number of read pairs |
Number of reads |
|
|---|---|---|---|
| Raw | Unique | Mitochondrial | |
| sample 1 | 40216981 | 27164935 | 191354 |
| sample 2 | 29099896 | 20843446 | 251066 |
The distribution of fragment sizes was plotted with Picard (CollectInsertSizeMetrics) inside the nf-core ATAC-seq pipeline (Figure 6). The fragment size distribution plot indicates a good quality of ATAC-seq data. The first peak corresponds to the nucleosome free region, the second – mononucleosome.
Figure 6.
Representable images of ATAC-seq and Hi-C analysis
(A) Insert size metrics of ATAC-seq data.
(B) ATAC-seq overage profile across 1726 kb region on chromosome 9.
(C) An example of Hi-C map for two 10-months-old C57/BL6 mice of 10 Mb region for chromosome 9 with 50 kb resolution
Hi-C libraries were deep sequenced on the Illumina NovaSeq by GENEWIZ (USA; www.GENEWIZ.com), resulting in 230.8–234.3 million 150-nt paired-end reads per sample. Sequencing reads were processed with the distiller-nf pipeline (https://github.com/open2c/distiller-nf). Reads were mapped on mm10 genome assembly with the default settings and with an option MAPQ30 filter. The number of uniquely mapped reads is indicated in the Table 2 below.
Table 2.
Number of paired reads for Hi-C samples
| Sample | Raw reads | Unique read pairs |
|---|---|---|
| sample 1 | 230844274 | 114504926 |
| sample 2 | 234327179 | 111373059 |
Limitations
Materials: In our protocol, we do not recommend substituting some reagents by other providers such asQubit ds; DNA BR Assay Kit; MyOne Strepavidin C1 beads; AMPure XP beads, Illumina Target DNA TDE1 Enzyme and Buffer Small kit, OptiPrep™ Density Gradient Medium.
Isolation of Nuclei and FACS Compared to whole cells, nuclei are generally more fragile and the possible loss of nuclei during the isolation procedure can be expected. Moreover, sorting also could be limited due to the number of samples to be processed in a short period of time.
Troubleshooting
Problem 1
The poor yield of nuclei after the iodixanol gradient (Isolation of nuclei from mice cortices) (step 12)
Potential solution
Make sure that the homogenate before the iodixanol gradient step does not contain any pieces of tissue. After the iodixanol gradient, take an interphase with nuclei carefully. The pipette tip position must be adjusted with the layer to maximize nuclei collection.
Problem 2
No interphase with nuclei observed after the iodixanol gradient (Isolation of nuclei from mice cortices) (step 12)
Carefully add the layer of nuclei on top of the gradient. For this step it is crucial to use a bucket rotor centrifuge instead of a fixed-angle rotor centrifuge.
Problem 3
Over-transposition or incomplete transposition (ATAC-seq) (step 28)
Potential solution
Optimization of nuclei number for transposition step is critical and directly impacts the library quality. We recommend calibrating the number of nuclei for your experiments. Using a large number of nuclei can result in incomplete transposition and bigger DNA fragments, while using a tiny number of nuclei can lead to over-transposition.
Problem 4
ATAC-seq fragment length distribution is small, around 200–300 bp (ATAC-seq) (step 29)
Optimization of the incubation time with transposase is required. In most cases it indicates that the transposase concentration is too high compared to nuclei number.
It can rarely happen in case of sample contamination. We recommend performing the sorting of nuclei in sterile conditions.
Problem 5
ATAC-seq fragment size plot does not have appropriate peaks (mono-, di-nucleosome) (ATAC-seq) (step 30)
Potential solution
Optimization of reverse-crosslinking reaction is required. Increasing the incubation time with proteinase K could be recommended.
Problem 6
Presence of high molecular DNA bands after the restriction step (Hi-C) (step 60)
Potential solution
Increasing the time of restriction could be required for this step. Also, this problem can occur if the permeabilization of nuclei was not complete. It could be useful to check the permeabilization of nuclei by a fluorescent microscope.
Problem 7
No signal or no amplification of Hi-C libraries by the real time PCR (Hi-C) (step 149)
Potential solution
This problem can occur if Streptavidin-coated beads are present in the solution. Remove streptavidin-coated beads from DNA by heating (Hi-C).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by lead contact Debra Toiber (toiber@bgu.ac.il)
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any datasets. Codes used for quantification in this study are available at nf-core: https://nf-co.re/atacseq, Zenodo: https://doi.org/10.5281/zenodo.3350926, GitHub: https://github.com/open2c/distiller-nf and Zenodo: https://doi.org/10.5281/zenodo.2634132.
Acknowledgments
This work was supported by the David and Inez Myers Foundation and ISF 188/17. The graphical abstract and Figures 1, 3, and 4 were created with BioRender.com.
Declaration of interests
The authors declare no competing interests.
References
- Belaghzal H., Dekker J., Johan H., Gibcus Hi-c 2.0: an optimized hi-c procedure for high-resolution genome-wide mapping of chromosome conformation. Methods. 2017;123:56–65. doi: 10.1016/j.ymeth.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Shen Y., Draper W., Buenrostro J.D., Litzenburger U., Cho S.W., Satpathy A.T., Carter A.C., Ghosh R.P., East-Seletsky A. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods. 2016;13:1013–1020. doi: 10.1038/nmeth.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso P., Nahnsen S. The nf-core framework for community-curated bioinformatics pipelines. Nat.Biotechnol. 2020;38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- Hempel C.M., Sugino K., Nelson S.B. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat. Protoc. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- Javier Rubio F., Li X., Liu Q., Cimbro R., Hope B. Fluorescence activated cell sorting (FACS) and gene expression analysis of Fos-expressing neurons from fresh and frozen rat brain tissue. J. Vis. Exp. 2014:54358. doi: 10.3791/54358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion-Poll L., Montalban E., Munier A., Hervé D., Girault J.A. Fluorescence-activated sorting of fixed nuclei: a general method for studying nuclei from specific cell populations that preserves post-translational modifications. Eur. J. Neurosci. 2014;39:1234–1244. doi: 10.1111/ejn.12506. [DOI] [PubMed] [Google Scholar]
- Policicchio S.S., Davies J.P., Chioza B., Burrage J., Mill J., Dempster E.L. Fluorescence-activated nuclei sorting (FANS) on human postmortem cortex tissue enabling the isolation of distinct neural cell populations for multiple omic profiling. protocols.io. 2020 [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any datasets. Codes used for quantification in this study are available at nf-core: https://nf-co.re/atacseq, Zenodo: https://doi.org/10.5281/zenodo.3350926, GitHub: https://github.com/open2c/distiller-nf and Zenodo: https://doi.org/10.5281/zenodo.2634132.