Abstract
Breast cancer is the most common type of cancer in women globally. The overexpressed proteins, including EGFR, PI3K, AKT1, and CDK4, have a role in the growth of breast cancer cells. The 3D peptide structure of sea cucumber Cucumaria frondosa was modeled and then docked with EGFR, PI3K, AKT1, and CDK4 proteins using AutoDock Vina software. The docking result, which has the best binding affinity value, is continued with molecular dynamics simulation. The docking results showed that all peptides bind to the active sites of the four proteins. WPPNYQW and YDWRF peptides bind to proteins with lower binding affinity values than positive controls. The four proteins were in a stable state when complexed with the WPPNYQW peptide, which was seen from the RMSD and RMSF value. PI3K-YDWRF and AKT1-YDWRF complexes are stable, characterized by high RMSD values and increased volatility in several amino acids. WPPNYQW peptide has high potential as an antibreast cancer agent because it binds to the active sites of the four proteins with low binding affinity values and stable interactions. Meanwhile, the YDWRF peptide interacts with the four proteins with low binding affinity values, but the interaction is only stable on PI3K and AKT1 proteins.
Keywords: Sea cucumbers, peptide, EGFR, PI3K, AKT1, CDK4
Introduction
Breast cancer is the most common type of cancer in women worldwide, followed by cervical cancer and ovarian cancer. 1 In 2019, there were approximately 268 600 cases of breast cancer, and 82% of patients were found in women aged less than 50 years, with a mortality rate as high as 90%. 2 The highest mortality is found in developing countries due to the lack of resources for proper diagnosis and treatment. 3 In addition, cancer treatment methods, such as chemotherapy, also have many adverse side effects for the skin, hair, bone marrow, blood, gastrointestinal tract, and kidneys. They can even cause chronic side effects such as drug resistance, carcinogenicity, and infertility. 4 Therefore, an alternative breast cancer therapy that is cheaper and has minimal side effects is needed.
Marine life comprises thousands of bioactive metabolites to be explored for anticancer. 5 Sea cucumbers are marine invertebrates that have a high potential to be used as an antibreast cancer agent. Previous studies reported that sea cucumber (Holothuria tubulosa) coelom fluid extract could inhibit the growth of triple-negative breast cancer cells (MDA-MB231) by disrupting the cell cycle and inducing autophagy. 6 Sea cucumber extract TBL-12 can inhibit PCa cell (prostate cancer cell line) proliferation and metastasis and can induce apoptosis through the intrinsic apoptosis pathway. 7 Holothuria parva methanol extract induces apoptosis by releasing cytochrome c from mitochondria and by inducing caspase-3 activation in mouse hepatocellular carcinoma models. 8 The anticancer activity of sea cucumbers is thought to be caused by the presence of anticancer compounds such as holothurin A (HA), frondoside A, 24-dehydroechinoside A (DHEA), frondanol A5, okhtosides B, 9 colochiroside A, cucumarioside A2–2, ds-echinoside A, echinoside A, glycosides 1 & 2, intercedensides A, B, and C, philinopside A, philinopside E, stichoposide C, and stichoposide D. 10 In previous studies, these compounds were shown to have anticancer activity by various mechanisms. For example, frondoside A from Cucumaria frondosa can induce apoptosis by activating p53 and significantly increasing caspase-3/7 and caspase-9 activity in ER-MDA-MB-231 breast cancer cells. 11 Holothurin A and 24-glycosides isolated from Pearsonothuria graeffei inhibited HepG2s cell metastasis by suppressing Matrix Metalloproteinase-9 (MMP-9) and Nuclear Factor-kappaB (NF-kB) expression and by inducing Tissue Inhibitor of Metalloproteinase-1 (TIMP-1) expression. 12 However, there has been no research on peptides from sea cucumber as an anticancer agent. Peptides have several advantages as an alternative to anticancer drugs compared to compounds, eg, they have low toxicity, have multiple targets, and do not accumulate tissues because they are quickly metabolized. 13 Therefore, peptides can be used as an excellent alternative to anticancer drugs. In this study, peptides from sea cucumbers (C. frondosa) are tested to inhibit the proteins causing breast cancer growth and development, namely, Epidermal Growth Factor Receptor (EGFR), Phosphatidylinositol-3 kinase (PI3K), Protein Kinase B1 (AKT1), and Cyclin-Dependent Kinase 4 (CDK4).
EGFR, PI3K, AKT1, and CDK4 are proteins having a role in the growth of breast cancer cells. EGFR is a receptor family of tyrosine kinase receptors with a role in regulating cell growth and survival. 14 In an in vivo study, overexpression of EGFR led to the transformation of mouse cells into malignant cells and increased proliferation and resistance to apoptosis. 15 EGFR is overexpressed in about 14% of breast cancer cases due to the amplification of the EGFR gene. 16 One of the strategies to inhibit the activity of this protein is to inhibit its phosphorylation activity. Lapatinib and gefitinib are compounds that have been shown to inhibit EGFR phosphorylation activity.17,18 PI3K is a central protein in the PI3K/AKT/mTOR pathway regulating cell growth and proliferation. 19 In most breast cancer cases, the PI3K gene undergoes mutations or amplifications, resulting in the PI3K/AKT/mTOR pathway. 20 One strategy to inhibit the activity of this protein is to inhibit its phosphorylation activity using the wortmannin compound that binds to the ATP binding pocket of PI3K.21,22 Apart from PI3K, AKT1 also has a central role in the PI3K/AKT/mTOR pathway. AKT1 plays a role in cell proliferation, metabolism, and growth and is overexpressed and overactivated in various types of breast cancer cells.23,24 Therefore, different inhibitor compounds were developed to stop the activity of this protein. CDK4 plays a crucial role in breast cancer cell growth and is often overexpressed. 25 Cyclin D1 protein is also overexpressed in most breast cancer cells and requires CDK4 to induce breast cancer growth. Therefore, inhibiting CDK4 activity is one of the best strategies for inhibiting breast cancer growth.26,27
A number of researches have been done on the compounds from sea cucumbers that can potentially be a breast cancer drug. However, there is still no research on the peptide’s potential of sea cucumbers as antibreast cancer agents. This study presents the potential of sea cucumber peptides in inhibiting the growth of breast cancer cells by inhibiting the activity of the EGFR, PI3K, AKT1, and CDK4 proteins using a bioinformatics approach. This study aims to find and explain peptides’ potential in sea cucumber (C. frondosa) in inhibiting the activity of breast cancer-related proteins.
Methods
Breast cancer-related protein preparations
Breast cancer-related proteins were determined with the help of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (https://www.genome.jp/kegg/pathway.html). The 3D structure of the protein kinase domain EGFR (1XKK), 28 PI3K (1E90), 22 AKT1 (6HHF), 29 and CDK4 (2W9Z) 30 was obtained from the RCSB PDB database (https://www.rcsb.org/). Proteins were prepared by removing water molecules and ligand contaminants using BIOVIA Discovery Studio 2019 software (Dassault Systèmes BIOVIA, San Diego, California, USA).
Peptide preparation
The peptides contained in sea cucumbers (C. frondosa) are WPPNYQW, YDWRF, EMEWR, EEELAALVLDNG-SGMCK, KMLWK, MMSLHL, RMCCCSPLK, TEFHLL, VELWR, VMLGMLWTLLLR, WNWKL, and WNWKV. 31 Peptides structure were predicted using PEP-FOLD (webserver modeled://263bioserv.rpbs.univ-paris-diderot.fr /services/PEP-FOLD3/). The peptides were then prepared using PyRx 0.8 software 32 by minimizing the conformational energy.
Inhibitor preparation as positive controls
The 3D structure of the inhibitor compounds of each protein in the form of gefitinib 17 (CID: 123631), wortmannin 22 (CID: 312145), AZD5363 33 (CID: 25227436), and abemaciclib 34 (CID: 46220502) was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). The inhibitors were prepared using PyRx 0.8 software 32 by minimizing the conformational energy.
Molecular docking
Protein-ligand docking was performed using AutoDock Vina 35 software integrated with PyRx 0.8. 32 The docking result with a lower binding affinity value than the inhibitor was taken to be analyzed and continued with molecular dynamics simulations. Visualization of docking results was carried out using BIOVIA Discovery Studio 2019 by looking at the protein’s ligand-binding position.
Molecular dynamics simulation
The peptides with the lowest binding affinity value and inhibitor as a positive control continued with molecular dynamics simulation using Yet Another Scientific Artificial Reality Application (YASARA) software. 36 The parameters used were according to cell physiological conditions (37°C, 1 atm, pH 7.4, 0.9% salt content) for 20 ns, which are autosaved every 25 ps. 20 ns is the duration commonly used to determine the stability of the protein-ligand complex.37-39 The simulation was run using the md_run macro program, and the results were displayed using the md_analyze, md_analyzeres, and md_analyzebindenergy macro program.
Results
Molecular docking results
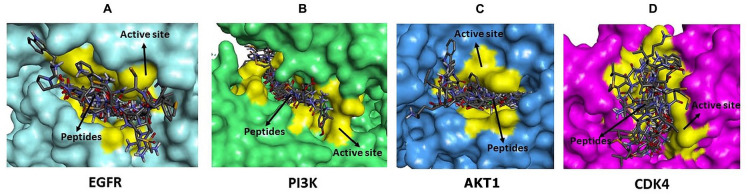
The molecular docking simulation aims to obtain peptides that bind to the protein’s active site and have a low binding affinity. The peptide has a high potential to inhibit the activity of the target protein. The results of protein-peptide docking show that all peptides (WPPNYQW, YDWRF, EMEWR, EEELAALVLDNGSGMCK, KMLWK, MMSLHL, RMCCCSPLK, TEFHLL, VELWR, VMLGMLWTLLLR, WNWKL, and WNWKV) interact on the active site of the protein (Figure 1). Besides, WPPNYQW and YDWRF peptides always have lower binding affinity than inhibitors (Table 1). This result shows that the two peptides have high potential as inhibitors of EGFR, PI3K, AKT1, and CDK4 proteins because they bind strongly to these proteins’ active sites, more potent than the inhibitors.
Figure 1.
All peptides bind to the active site of EGFR (A), PI3K (B), AKT1 (C), and CDK proteins (D). The active site of the protein is marked in yellow. The docking results showed that all peptides have potential as competitive inhibitors because they bind to the protein on the active site.
AKT1 indicates Protein Kinase B1; CDK4, Cyclin Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3K, Phosphatidylinositol-3 kinase.
Table 1.
Peptide docking results.
| Ligand | Binding affinity (kcal/mol) | |||
|---|---|---|---|---|
| EGFR | PI3K | Akt1 | CDK4 | |
| Inhibitor (positive control) | -8.2 | -9 | -9.8 | -9 |
| WPPNYQW | -9.5 | -10.6 | -11.3 | -9.4 |
| YDWRF | -9.3 | -9.4 | -10.7 | -9.3 |
| EMEWR | -8.3 | -8.2 | -9.4 | -7.4 |
| EEELAALVLDNGSGMCK | -6.1 | -6.8 | -6.1 | -5.7 |
| KMLWK | -7.8 | -7.5 | -8.9 | -6.7 |
| MMSLHL | -6.8 | -7.9 | -8.5 | -7.1 |
| RMCCCSPLK | -7.2 | -6.9 | -7.6 | -5.3 |
| TEFHLL | -8.3 | -8.4 | -9.5 | -7.6 |
| VELWR | -8.5 | -8.3 | -8.9 | -8 |
| VMLGMLWTLLLR | -4.7 | -5.3 | -8.1 | -8 |
| WNWKL | -8.7 | -8.9 | -8 | -9.2 |
| WNWKV | -9.5 | -9 | -8.5 | -8.9 |
Abbreviations: Akt1, Protein Kinase B1; CDK4, Cyclin-Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3 K, Phosphatidylinositol-3 kinase.
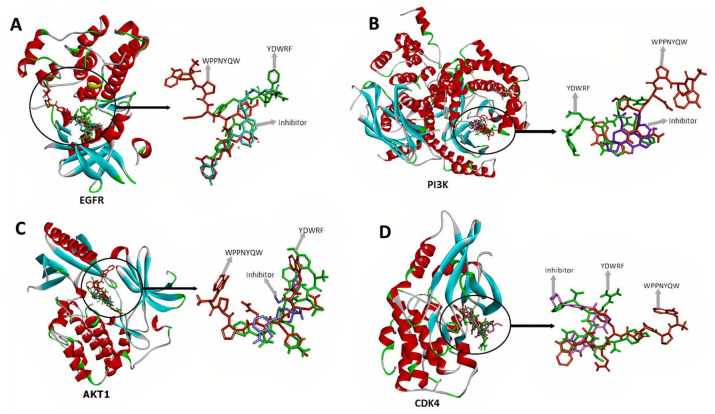
Based on docking results, the WPPNYQW and YDWRF peptides are shown to have high potential as EGFR, P13 K, AKT1, and CDK4 inhibitors because they bind to the ATP binding pocket and have lower binding affinity values than control inhibitors (Figure 2 and Table 2).
Figure 2.
WPPNYQW (orange) and YDWRF (green) peptides bind to EGFR (A), PI3K (B), AKT1 (C), and CDK4 (D) on the same side as the inhibitor. WPPNYQW and YDWRF peptides have a similar binding mode to the inhibitor indicating the peptides have the potential to act as protein inhibitors.
AKT1 indicates Protein Kinase B1; CDK4, Cyclin-Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3K, Phosphatidylinositol-3 kinase.
Table 2.
The interaction of WPPNYQW and YDWRF peptides with EGFR, PI3K, AKT1, and CDK4 proteins.
| Protein | Ligand | Binding affinity (kcal/mol) | Position of chemical bonds | |
|---|---|---|---|---|
| Hydrogen bond | Hydrofobic interaction | |||
| EGFR | Inhibitor (Gefitinib) | -8.2 | Arg841, Asn842, Asp855 | Ala743, Val726, Leu718, Lys745 |
| WPPNYQW | -9.5 | Ala722, Lys745, Met793, Asp837, Met793, Asp855, Ala743, Leu788, Gly721, Thr854, Leu718 | Met766, Phe856, Leu718, Trp880, Lys879, Leu858, Leu777 | |
| YDWRF | -9.3 | Cys797, Tyr998, Arg841, Asp800, Leu1001, Met1002, Val717, Phe795 | Lys745, Glu804, Cys797, Leu1001, met1002, Arg841, Val726, ala743 | |
| PI3K | Inhibitor (Wortmannin) | -9 | Lys883, Tyr867, Asp841, Asp950 | Trp812, Ile831, Ile881, Ile963, Ile879, Met953 |
| WPPNYQW | -10.6 | Lys807, Asn951, Lys833, Asp841, Asp964, Asp836, Asp841, Glu880, Tyr867, Val882 | Arg947, Trp812, Met953, ile881, Ala885, Leu1090 | |
| YDWRF | -9.4 | Asp950, Lys833, Thr886, Ala885, Val882, Asp950, Ser806 | Lys883, Trp812, Tyr867, Val822, Ile881, Met953, Ile963, Ile879 | |
| Akt1 | Inhibitor (AZD5363) | -9.8 | Gln79, Tyr272, Thr291 | Trp80, Val270, Lys268 |
| WPPNYQW | -11.3 | Gln79, Leu78, Asp274, Asn279, Tyr272, Ile290, Thr211 | Ala58, Trp80, Ile84, Ile186, Leu210, Lys268, Leu264 | |
| YDWRF | -10.7 | Thr82, Phe293, Gly294, Leu295, Glu85, Ile290, Thr211, Trp80 | Trp80, Tyr272, Cys296, Lys268, Val270 | |
| CDK4 | Inhibitor (Abemaciclib) | -9 | Gly15, Ile12, Asp99, Asp97, Thr102 | Val20, Leu147, His95, Ala10, Ile12 |
| WPPNYQW | -9.4 | Ile12, Val14, Ala16, Tyr17, Thr177, Glu144, Asp99, | Leu147, Trp179, Val176, Cys215, Ile12, Ala33, Ala157 | |
| YDWRF | -9.3 | Asp105, Val14, Thr102, Val96, Thr177, Asp99 | Ile12, Leu147, Val20, Ala33 | |
Abbreviations: Akt1, Protein Kinase B1; CDK4, Cyclin-Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3K, Phosphatidylinositol-3 kinase.
The docking results showed that all peptides bind to EGFR in the ATP binding pocket and gefitinib as an inhibitor. Besides, two peptides have a lower binding affinity than gefitinib, namely, WPPNYQW of -9.5 kcal/mol and YDWRF of -9.3 kcal/mol. These two peptides bind together with the amino acid residue Lys745, which plays an important role in binding ATP. 40 Therefore, these two peptides block ATP from binding to EGFR in the ATP binding pocket. Hydrogen bonds dominate protein-peptide interactions. WPPNYQW peptides form chemical interactions consisting of eleven hydrogen bonds (Ala722, Lys745, Met793, Asp837, Met793, Asp855, Ala743, Leu788, Gly721, Thr854, and Leu718) and seven hydrophobic interactions (Met766, Phe856, Leu718, Trp880, Lys879, Leu858, and Leu777). Meanwhile, YDWRF peptide forms eight hydrogen bonds (Cys797, Tyr998, Arg841, Asp800, Leu1001, Met1002, Val717, Phe795) and eight hydrophobic interactions (Lys745, Glu804, Cys797, Leu1001, Met1002, Arg841, Val726, and Ala743). The number of hydrogen bonds formed is predicted to have an essential role in the stability of protein-peptide interactions. 41 These results reveal that the WPPNYQW and YDWRF peptides have potential as EGFR inhibitors through the competitive ATP inhibitor mechanism (Figure 2A). The interaction of WPPNYQW and YDWRF peptides with EGFR have lower binding affinity values than interaction with inhibitors (Table 2).
The docking results showed that all peptides bind to the ATP binding pocket in the catalytic domain of the PI3K p110 subunit, and two peptides, WPPNYQW and YDWRF, have lower binding affinity values than wortmannin as inhibitors. The binding affinity value for the interaction between PI3K and WPPNYQW and YDWRF peptides is -10.6 kcal/mol and -9.4 kcal/mol, respectively. Hydrogen bonds dominate the interactions that form. The WPPNYQW peptide forms ten hydrogen bonds (Lys807, Asn951, Lys833, Asp841, Asp964, Asp836, Asp841, Glu880, Tyr867, and Val882) and six hydrophobic interactions (Arg947, Trp812, Met953, Ile881, Ala885, and Leu1090). Meanwhile, the YDWRF peptide forms seven hydrogen bonds (Asp950, Lys833, Thr886, Ala885, Val882, Asp950, and Ser806) and eight hydrophobic interactions (Lys883, Trp812, Tyr867, Val822, Ile881, Met953, Ile963, and Ile879). These two peptides have more hydrogen bonds than wortmannin as a positive control. Therefore, the WPPNYQW and YDWRF peptides have high potential as PI3K protein inhibitors through the competitive ATP inhibitor mechanism (Figure 2B and Table 2).
The docking results showed all the peptides were bound in the ATP binding pocket of AKT1 the same as AZD5363. However, two peptides, namely, WPPNYQW and YDWRF, have lower binding affinity than AZD5363, ie, -11.3 kcal/mol and -10.7 kcal/mol. The WPPNYQW peptide forms seven hydrogen bonds (Gln79, Leu78, Asp274, Asn279, Tyr272, Ile290, and Thr211) and seven hydrophobic interactions (Ala58, Trp80, Ile84, Ile186, Leu210, Lys268, and Leu264). Meanwhile, the YDWRF peptide forms eight hydrogen bonds (Thr82, Phe293, Gly294, Leu295, Glu85, Ile290, Thr211, and Trp80) and five hydrophobic interactions (Trp80, Tyr272, Cys296, Lys268, and Val270). The results reveal that WPPNYQW and YDWRF peptides have high potencial as an AKT1 inhibitor, even better than AZD5363 (Figure 2C and Table 2).
The docking results between CDK4 and peptides showed that the peptides bind to CDK4 in the ATP binding pocket with varying binding affinity values. Two peptides, namely, WPPNYQW and YDWRF, had lower binding affinities (WPPNYQW, -9.4 kcal/mol; YDWRF, -9.3 kcal/mol) than abemaciclib as positive controls. The WPPNYQW peptide forms seven hydrogen bonds (Val14, Ala16, Tyr17, Thr177, Glu144, Asp99, and Ile12) and seven hydrophobic interactions (Ile12, Val14, Ala16, Tyr17, Thr177, Glu144, and Asp99). Meanwhile, the YDWRF peptide forms six hydrogen bonds (Asp105, Val14, Thr102, Val96, Thr177, and Asp99) and four hydrophobic interactions (Ile12, Leu147, Val20, and Ala33). The number of chemical interactions that are formed will increase the stability of the protein–peptide interaction. Based on these docking results, the WPPNYQW and YDWRF peptides are shown to have high potential as CDK4 inhibitors because they bind to the ATP binding pocket and have lower binding affinity values than abemaciclib (Figure 2D and Table 2).
Molecular dynamics simulation results
Molecular dynamics simulations were carried out to analyze the structural stability and conformational fluctuations of the EGFR, PI3K, AKT1, and CDK4 proteins that interact with the WPPNYQW and YDWRF peptides. The molecular dynamics simulation result can be seen in Figure 3.
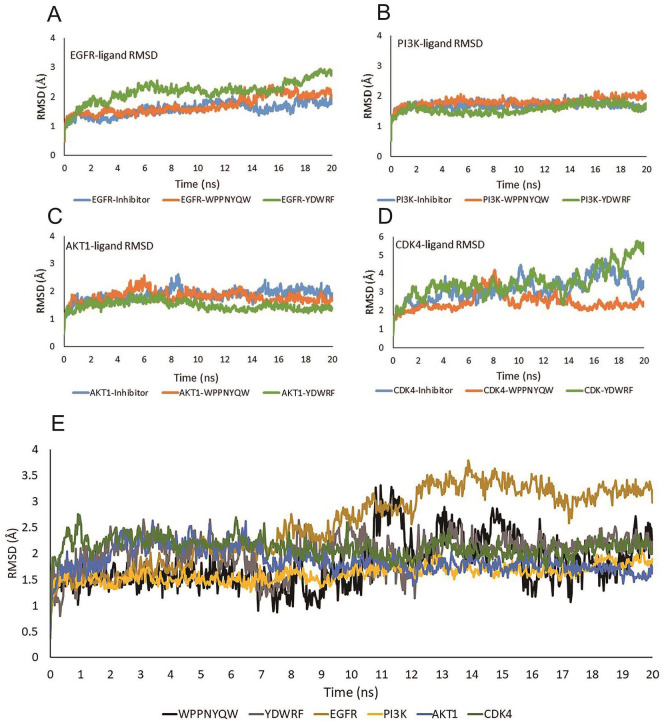
Figure 3.
The stability of protein-ligand complex interactions can be seen from RMSD values. (A) The RMSD values of the EGFR-inhibitor, EGFR-WPPYQW, and EGFR-YDWRF complexes tend to be stable. (B) The RMSD values of the PI3K-inhibitor, PI3K-WPPYQW, and PI3K-YDWRF complexes are stable during the simulation. (C) The RMSD values of the AKT1-inhibitor, AKT1-WPPNYQW, and AKT1-YDWRF complexes are stable during the simulation. (D) The RMSD value of the CDK4-WPPNYQW complex is more stable than the CDK4-inhibitor and CDK4-YDWRF. The CDK4-YDWRF complex tends to be unstable because of its high RMSD value. (E) RMSD peptides and target proteins.
AKT1 indicates Protein Kinase B1; CDK4, Cyclin-Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3K, Phosphatidylinositol-3 kinase; RMSD, root mean square deviation.
The EGFR, PI3K, and AKT1 proteins complexed with the WPPNYQW peptide had a stable RMSD value of less than 3 Å from the start to the end of the simulation, which is not much different from inhibitor compounds. Meanwhile, the RMSD CDK4 value complexed with inhibitor ligand and WPPNYQW peptide showed a different trend. CDK4 complexed with inhibitor (abemaciclib) had an unstable RMSD value during the simulation, which was more than 3 Å from the 4 ns time to the end of the simulation. The CDK4 complex with the WPPNYQW peptide was unstable at 6-8 ns time but returned to stability from 10 ns until the end of simulation. This result shows that CDK4 is more stable when interacting with the WPPNYQW peptide than the inhibitor (abemaciclib). Different results were shown when the four proteins interacted with the YDWRF peptide. PI3K and AKT1 proteins were stable from the beginning to the end of the simulation when complexed with the YDWRF peptide. Meanwhile, EGFR and CDK4 proteins were unstable when complexed with the YDWRF peptide characterized by high RMSD values (Figure 3).
EGFR in the form of a complex with peptides has a lower RMSD value than EGFR itself, which indicates that the EGFR complex with peptides is more stable (Figure 3A and E). The RMSD values of the PI3K and the PI3K-peptides complex tended to be the same during the simulation, indicating the complex’s stability (Figure 3B and E). The RMSD value of AKT1 with the AKT1-peptides complex was not much different during simulation, and even the AKT1-YDWRF was more stable because it had a lower RMSD value (Figure 3C and E). The CDK4-WPPNYQW RMSD value is not much different from CDK4. When interacted with YDWRF, CDK4 had a significant increase in the RMSD value, indicating a change in the structure of the CDK4 protein (Figure 3D and E).
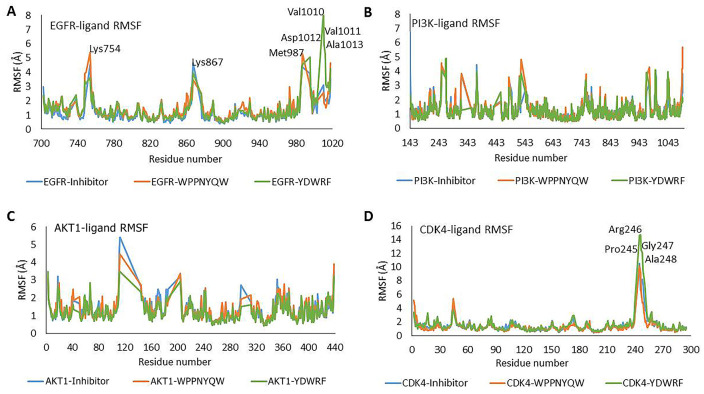
In EGFR complexed with peptides and inhibitors, several unstable amino acids are involved, including Lys754, Lys867, and Met987. However, amino acids are very unstable when EGFR is complexed with YDWRF, namely, Val1010, Val1011, Asp1012, and Ala1013. These residues cause the RMSD value of the EGFR-YDWRF complex to tend to be higher. Meanwhile, PI3K and AKT1 complexed with peptides showed RMSF values which were not significantly different than when the two proteins were complexed with an inhibitor. CDK4 complexed with YDWRF has very unstable residues, namely, Pro245, Arg246, Gly247, and Ala248 (Figure 4). These residues play a role in the structural instability of the CDK4-YDWRF protein, which can be seen from the high RMSD CDK4-YDWRF. From the RMSD and RMSF values of the protein-ligand complex, all proteins are found to be stable when are complexed with the WPPNYQW peptide. In contrast, the only proteins that were stable when interacted with YDWRF were PI3K and AKT1. EGFR and CDK4 are unstable because some residues have high flexibility when the protein is complexed with the YDWRF peptide. PI3K and AKT1 proteins were stable from the beginning to the end of the simulation when complexed with the YDWRF peptide. Meanwhile, EGFR and CDK4 proteins were unstable when complexed with the YDWRF peptide characterized by high RMSD values (Figure 3).
Figure 4.
The stability of each amino acid residue during the simulation can be seen from the RMSF value. (A) Val1010 and Val1011 residues of the EGFR-YDWRF complex have high flexibility. (B and C) The residues on the PI3K-Ligands and AKT1-Ligands complexes tend to be stable during simulation. (D) Pro245, Arg246, Gly247, and Ala248 residues of the CDK4-YDWRF complex have high flexibility. High flexibility indicates instability of residue.
AKT1 indicates Protein Kinase B1; CDK4, Cyclin-Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3 K, Phosphatidylinositol-3 kinase; RMSF, root mean square fluctuation.
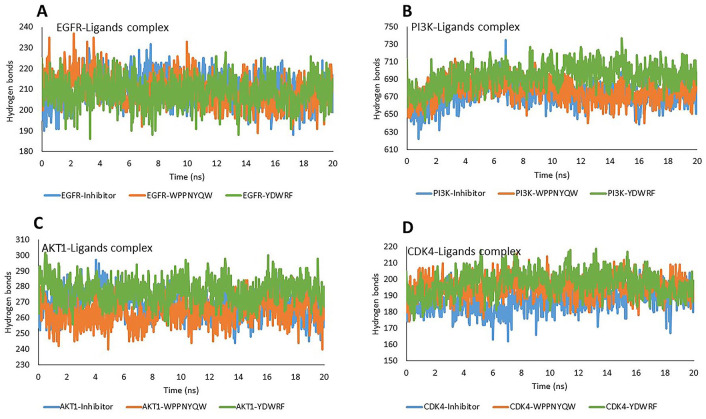
Hydrogen bonds have an important role in the stability of protein conformation and the stability of protein-ligand interactions. 42 The numbers of hydrogen bonds of the complexes during the simulation is shown in Figure 5. The numbers of hydrogen bonds complexed four proteins with the WPPNYQW and YDWRF peptides were not significantly different compared to the four proteins complexed with the inhibitors as positive controls. This indicates that the complex structures are stable.
Figure 5.
The stability of the complex structure can be seen from the number of hydrogen bonds during the simulation. The number of hydrogen bonds in EGFR-peptides (A), PI3K-peptides (B), AKT1-peptides (C), and CDK4-peptides (D) were not significantly different from proteins-inhibitors. These results indicate the structural stability of the protein-peptide complexes.
AKT1 indicates Protein Kinase B1; CDK4, Cyclin-Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3K, Phosphatidylinositol-3 kinase.
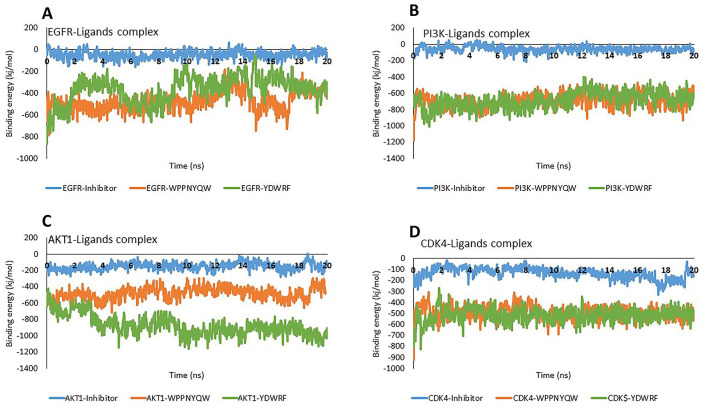
The molecular dynamic binding energy represents the stability of protein-ligand interactions during the simulation. 43 The simulation results show that all protein-peptide complexes have lower molecular dynamic binding energy values than protein-inhibitor complexes. Based on the molecular dynamic binding energy, the stability level of the protein-peptide interactions are still below the protein-inhibitor (Figure 6).
Figure 6.
The molecular dynamic binding energy represents the stability of the protein-ligand interaction during the simulation. The more positive the molecular dynamic binding energy value, the more stable the protein-ligand interaction. The molecular dynamic binding energy values of EGFR-peptides (A), PI3K-peptides (B), AKT1-peptides (C), and CDK4-peptides were lower than proteins-inhibitors.
AKT1 indicates Protein Kinase B1; CDK4, Cyclin-Dependent Kinase 4; EGFR, Epidermal Growth Factor Receptor; PI3K, Phosphatidylinositol-3 kinase.
Discussion
EGFR is a ligand epidermal growth factor (EGF) receptor, with an essential role in the growth and development of cancer cells. These receptors are overexpressed in various types of cancer cells, including breast cancer cells. 44 Activation of EGFR signaling is induced by the EGF ligand, causing EGFR to form dimers, and then tyrosine residues appear in the kinase domain, causing effector proteins to be recruited. 45 Phosphotyrosine motive in the kinase domain with ATP activates several pathways such as the KRAS/BRAF/MEK/ERK pathway, PI3K pathway, AKT pathway, and Signal Transducer and Activator of Transcription (STAT) pathway that stimulate proliferation, angiogenesis, migration, survival, and cell adhesion. 46 Therefore, one strategy to inhibit EGFR phosphorylation activity is to block the interaction between the tyrosine kinase EGFR domain and ATP. The ATP binding pocket is the region where the EGFR binds to ATP. This region is located near the C-helix and A-loop structures, to be precise around the amino acids Leu718, Val726, Gly745, Leu788, Gly796, Cys797, Leu844, and Asp855.47,48
This study reveals that the WPPNYQW and YDWRF peptides have potential as EGFR inhibitors through the competitive ATP inhibitor mechanism. Inhibition of EGFR activity can inhibit breast cancer cell growth through a variety of mechanisms. Inhibition of EGFR activity can reduce collagenase expression (MMP1) and Meiosis-specific Serine/Threonine Protein Kinase 1 (MEK1), which play a role in cancer cell invasion and metastasis.49,50 EGFR inactivation also induces apoptosis by decreasing the expression of antiapoptotic proteins such as Secreted Frizzled Related Protein 1 (SRFP1), Baculoviral IAP Repeat Containing 5 (BIRC5), and BAG Cochaperone 1 (BAG1). 51 EGFR inhibition can also lead to cell cycle arrest in cancer cells. 52 However, the use of EGFR inhibitors for cancer therapy as a single agent has a low success rate because other pathways play a role in cancer growth, such as the PI3K pathway. 53
PI3K is the central protein in the PI3K/AKT/mTOR pathway. PI3K is activated by receptor tyrosine kinases, such as EGFR. Once activated, the catalytic domain in the p110 subunit catalyzes the transfer of phosphate from ATP to Phosphatidylinositol Biphosphate (PIP2) to become Phosphatidylinositol triphosphate (PIP3). 54 PIP3 is what then activates AKT1 through the phosphoinositide-dependent kinase 1 (PDK1) protein recruit. 55 This pathway, called the master regulator for cancer, is responsible for regulating cell growth and proliferation. One strategy to inhibit this pathway is to inhibit the activity of the PI3K protein by blocking the binding between PI3K and ATP so that the activation mechanism of AKT1 cannot occur. 56 PI3K inhibitor compounds that target the ATP binding pocket will compete with ATP in binding to the PI3K. The ATP binding pocket protein PI3K is located between the residues of Ala805, Ser806, Trp812, Lys890, Asp950, and Asp964. 22
The docking results revealed that all peptides bind to the ATP binding pocket in the catalytic domain of the PI3K p110 subunit, and two peptides, WPPNYQW and YDWRF, have lower binding affinity values than wortmannin as inhibitors. That result shows that the WPPNYQW and YDWRF peptides have potential as PI3K inhibitors. The inhibition of PI3K activity can prevent the uncontrolled proliferation of cancer cells. 57 Inhibition of PI3K activity, can also suppress the expression of, ZAK, TACC1, ZFR, and ZNF565 genes, resulting in cell death. 58 In addition, PI3K inactivation also results in inhibition of Cyclin D1 activity, which plays an important role in proliferation and cycle regulation. PI3K inactivation can also induce apoptosis by increasing the amount of cleaved caspase-3 protein. 59
AKT is one of the central proteins in the PI3K/AKT/mTOR pathway, activated by PI3K. This protein is overexpressed in ER + or HER + breast cancer cells and plays an important role in proliferation, metabolism, antiapoptosis, and cell survival. 24 The activation stage of AKT1 is initiated by the PH domain that binds to PIP3 on the cell membrane previously phosphorylated by PI3K. The interaction between the AKT PH domain and PIP3 changes the AKT conformation so that it allows PDK1 to phosphorylate AKT1. After being phosphorylated, the ATP binding pocket in the kinase domain is exposed, and AKT1 will return to the cytoplasm to phosphorylate its target proteins. 60 AKT1 then binds to ATP and transfers phosphate from ATP to its target proteins to become the active protein product. Therefore, many AKT1 inhibitor compounds have been developed, which target ATP binding pockets that compete with ATP to bind to the ATP binding pocket so that AKT1 cannot phosphorylate the substrate. 61 One of them is AZD5363, which binds to AKT1 in the ATP binding pocket, thus preventing ATP from binding to AKT1. Consequently, there was no phosphorylation of the target protein from AKT1. 33
Inhibition of the activity of the AKT1 protein will induce apoptosis and cell cycle arrest in cancer cells. 62 The results reveal that WPPNYQW and YDWRF peptides have high potential as an AKT1 inhibitor, even better than AZD5363. AKT1, which is inhibited, will increase the activity of Checkpoint kinase 2 (Chk2) protein. This protein will degrade Cell Division Control Protein 2 (CDC2), resulting in cell arrest in the G2-M phase. 63 AKT1 inhibition can also inhibit the activity of proteins that play an important role in cell proliferation such, as Mammalian Target of Rapamycin (mTOR), IkB Kinase (IKK), and Cyclin D1, and increase the activity of antiproliferative proteins such as Glycogen Synthase Kinase 3 Betha (GSK3β) and Forkhead box protein O1 (FOXO1). 64
When overexpressed, CDK4 causes uncontrolled cell growth and proliferation. 65 In the G0/G1 phase, CDK4 binds to Cyclin D1 and then phosphorylates the retinoblastoma protein (Rb), thus inducing cells to enter the G1/S phase. 66 Previous studies reported that the CDK4/Cyclin D/Rb pathway causes the growth and development of various cancer types. One strategy to inhibit this pathway is by inhibiting the phosphorylation of Rb through the CDK4/Cyclin D complex. 34 Therefore, a CDK4 inhibitor was developed that targets the ATP binding pocket of CDK4 so that CDK4 cannot bind to ATP and phosphorylate Rb. This inhibitor binds to CDK4 in the ATP binding pocket located between the residues of His95, Val96, Asp97, Arg101, Thr102, and Glu144. 67 Some compounds with an activity as competitive ATP inhibitors include abemaciclib, palbociclib, and ribociclib have entered the clinical research stage. 34
The docking results between CDK4 and peptides showed that WPPNYQW and YDWRF peptides had lower binding affinity than abemaciclib as inhibitor. It indicates that both peptides have high potential as CDK4 inhibitors. The inhibition of CDK4 activity will stimulate cancer cells to undergo cell cycle arrest. When the CDK4/Cyclin D complex is active, Cyclin D-CDK4 will phosphorylate RB so that RB is released from E2F, and then E2F can bind to DNA as a transcription factor for genes that play an important role in the running of the cell cycle. 68 CDK4 inhibition will prevent Rb phosphorylation so that Rb will continue to bind to E2F (E2 Factor) protein, and E2F protein cannot bind to DNA as a transcription factor. 69 This interaction causes cell arrest in phase G1. 70
Molecular dynamics simulations were carried out to analyze the structural stability and conformational fluctuations of the EGFR, PI3K, AKT1, and CDK4 proteins that interact with the WPPNYQW and YDWRF peptides. Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), number of hydrogen bonds, and molecular dynamic binding energy analyses are also used to obtain important information regarding the stability and flexibility of protein-ligand complexes. High deviation and variability during the simulation indicate low stability. 71 Proteins stable during the simulation are marked with an RMSD value of not more than 3 Å. A value more than that indicates that the protein has undergone some structural changes. 72 RMSF represents a shift in each amino acid residue on a protein. Amino acids with a high RMSF value indicate that these amino acids are unstable or have high fluctuations during the simulation. 73 The stability of the RMSD and RMSF values is important to conclude a good binding affinity for protein-ligand interactions. 74 The conformational stability of the complex also can be determined from the number of hydrogen bonds during the simulation. Hydrogen bonds are required to form the secondary structures of proteins such as the α-helix and the ß-sheet. Hydrogen bonds make these structures very stable. 42 Hydrogen bonding also facilitates the stability of protein-ligand interactions. The more hydrogen bonds, the more stable the interaction. 41 The stability of the protein-ligand interaction during the simulation can be determined from the molecular dynamic binding energy value. The molecular dynamic binding energy is influenced by the value of the potential energy and the solvation energy of complexes, ligands, proteins, and ligands. The more positive the molecular dynamic binding energy value, the better the protein-ligand binding. 43
As the result of this study, the EGFR, PI3K, AKT1, and CDK4 protein were in a stable state when complexed with the WPPNYQW peptide, which was seen from the RMSD and RMSF value. PI3K-YDWRF and AKT1-YDWRF complexes are stable, characterized by high RMSD values and increased volatility in several amino acids.
Our result suggested that sea cucumber peptides namely WPPNYQW and YDWRF maybe are promising inhibitor candidates of key target proteins that highly expressed in breast cancer. Further research is needed to validate the in vitro and in vivo activity of the sea cucumber peptides in inhibit cancer progression.
Conclusion
In this study, two sea cucumber peptides had high potential as antibreast cancer agents, namely, WPPNYQW and YDWRF peptides. The two peptides bind to the active sites of EGFR, PI3K, AKT1, and CDK4 proteins with lower binding affinity values than inhibitors. The interactions between WPPNYQW peptide and the four proteins are stable. While the YDWRF peptide interacts stably with PI3K and AKT1 proteins, it is unstable when interacting with EGFR and CDK4 proteins. Therefore, the WPPNYQW peptide has more potential as an antibreast cancer agent than the YDWRF peptide.
Acknowledgments
The work was supported by the Ministry of Research and Technology/ National Research and Innovation Agency of the Republic of Indonesia under PDUPT scheme grant in 2020.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work was supported by the Ministry of Research and Technology/National Research and Innovation Agency of the Republic of Indonesia [8/E1/KPT/2020].
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: T.L.W.: Conceptualization, Supervision, Writing; Funding acquisition; H.R.: Project administration, Writing; N.W.: Conceptualization, Methodology, Formal analysis, Writing; M.H.W.: Investigation, Resources, Visualization, Writing.
Availability of Data and Materials: The data underlying this article are available in the article.
Ethical Approval: This article does not contain any studies with human participants or animals performed by any of the authors.
ORCID iD: Teresa Liliana Wargasetia  https://orcid.org/0000-0002-3433-3300
https://orcid.org/0000-0002-3433-3300
References
- 1. Mathur P, Sathishkumar K, Chaturvedi M, et al. Cancer statistics, 2020: report from national cancer registry programme, India. JCO Glob Oncol. 2020;6:1063-1075. doi: 10.1200/GO.20.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438-451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 3. Rivera-Franco MM, Leon-Rodriguez E. Delays in breast cancer detection and treatment in developing countries. Breast Cancer (Auckl). 2018;12:1178223417752677. doi: 10.1177/1178223417752677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schirrmacher V. From chemotherapy to biological therapy: a review of novel concepts to reduce the side effects of systemic cancer treatment (review). Int J Oncol. 2019;54:407-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wargasetia TL, Widodo N. The link of marine products with autophagy-associated cell death in cancer cell. Curr Pharmacol Rep. 2019;5:35-42. doi: 10.1007/s40495-019-00167-8. [DOI] [Google Scholar]
- 6. Luparello C, Ragona D, Asaro DML, et al. Cytotoxic potential of the coelomic fluid extracted from the sea cucumber holothuria tubulosa against triple-negative mda-mb231 breast cancer cells. Biology. 2019;8:76. doi: 10.3390/biology8040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuan L, Huang X, Zhou K, et al. Sea cucumber extract TBL-12 inhibits the proliferation, migration, and invasion of human prostate cancer cells through the p38 mitogen-activated protein kinase and intrinsic caspase apoptosis pathway. Prostate. 2019;79:826-839. doi: 10.1002/pros.23788. [DOI] [PubMed] [Google Scholar]
- 8. Seydi E, Motallebi A, Dastbaz M, et al. Selective toxicity of Persian Gulf sea cucumber (Holothuria parva) and sponge (Haliclona oculata) methanolic extracts on liver mitochondria isolated from an animal model of hepatocellular carcinoma. Hepat Mon. 2015;15:e33073. doi: 10.5812/hepatmon.33073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janakiram N, Mohammed A, Rao C. Sea cucumbers metabolites as potent anti-cancer agents. Mar Drugs. 2015;13:2909-2923. doi: 10.3390/md13052909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wargasetia TL, Widodo. Mechanisms of cancer cell killing by sea cucumber-derived compounds. Invest New Drugs. 2017;35:820-826. doi: 10.1007/s10637-017-0505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Al Marzouqi N, Iratni R, Nemmar A, et al. Frondoside A inhibits human breast cancer cell survival, migration, invasion and the growth of breast tumor xenografts. Eur J Pharmacol. 2011;668:25-34. doi: 10.1016/j.ejphar.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 12. Zhao Q, Xue Y, Liu Z, et al. Differential effects of sulfated triterpene glycosides, holothurin A1, and 24-dehydroechinoside A, on antimetastasic activity via regulation of the MMP-9 signal pathway. J Food Sci. 2010;75:H280-H288. doi: 10.1111/j.1750-3841.2010.01837.x. [DOI] [PubMed] [Google Scholar]
- 13. Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81:136-147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 14. Citri A, Yarden Y. EGF–ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505-516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 15. Rimawi MF, Shetty PB, Weiss HL, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer. 2010;116:1234-1242. doi: 10.1002/cncr.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al-Kuraya K, Schraml P, Torhorst J, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534-8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- 17. Brehmer D, Greff Z, Godl K, et al. Cellular targets of gefitinib. Cancer Res. 2005;65:379-382. [PubMed] [Google Scholar]
- 18. Wainberg ZA, Anghel A, Desai AJ, et al. Lapatinib, a dual EGFR and HER2 kinase inhibitor, selectively inhibits HER2-amplified human gastric cancer cells and is synergistic with trastuzumab in vitro and in vivo. Clin Cancer Res. 2010;16:1509-1519. doi: 10.1158/1078-0432.CCR-09-1112. [DOI] [PubMed] [Google Scholar]
- 19. Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. López-Knowles E, O’Toole SA, McNeil CM, et al. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality: PI3K pathway activation and breast cancer outcome. Int J Cancer. 2010;126:1121-1131. doi: 10.1002/ijc.24831. [DOI] [PubMed] [Google Scholar]
- 21. Abliz A, Deng W, Sun R, Guo W, Zhao L, Wang W. Wortmannin, PI3K/Akt signaling pathway inhibitor, attenuates thyroid injury associated with severe acute pancreatitis in rats. Int J Clin Exp Pathol. 2015;8:13821-13833. [PMC free article] [PubMed] [Google Scholar]
- 22. Walker EH, Pacold ME, Perisic O, et al. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, ly294002, quercetin, myricetin, and staurosporine. Mol Cell. 2000;6:909-919. [DOI] [PubMed] [Google Scholar]
- 23. Tokunaga E, Kimura Y, Oki E, et al. Akt is frequently activated in HER2/neu-positive breast cancers and associated with poor prognosis among hormone-treated patients. Int J Cancer. 2006;118:284-289. doi: 10.1002/ijc.21358. [DOI] [PubMed] [Google Scholar]
- 24. Yang Z-Y, Di M-Y, Yuan J-Q, et al. The prognostic value of phosphorylated Akt in breast cancer: a systematic review. Sci Rep. 2015;5:7758. doi: 10.1038/srep07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. An H-X, Beckmann MW, Reifenberger G, Bender HG, Niederacher D. Gene amplification and overexpression of CDK4 in sporadic breast carcinomas is associated with high tumor cell proliferation. Am J Pathol. 1999;154:113-118. doi: 10.1016/S0002-9440(10)65257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corona SP, Generali D. Abemaciclib: a CDK4/6 inhibitor for the treatment of HR+/HER2– advanced breast cancer. Drug Des Devel Ther. 2018;12:321-330. doi: 10.2147/DDDT.S137783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Q, Sicinska E, Geng Y, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23-32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 28. Wood ER, Truesdale AT, McDonald OB, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652-6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 29. Weisner J, Landel I, Reintjes C, et al. Preclinical efficacy of covalent-allosteric AKT inhibitor borussertib in combination with trametinib in KRAS-mutant pancreatic and colorectal cancer. Cancer Res. 2019;79:2367-2378. doi: 10.1158/0008-5472.CAN-18-2861. [DOI] [PubMed] [Google Scholar]
- 30. Day PJ, Cleasby A, Tickle IJ, et al. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci USA. 2009;106:4166-4170. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, He S, Bonneil Simpson ÉBK. Generation of antioxidative peptides from Atlantic sea cucumber using alcalase versus trypsin: in vitro activity, de novo sequencing, and in silico docking for in vivo function prediction. Food Chem. 2020;306:125581. doi: 10.1016/j.foodchem.2019.125581. [DOI] [PubMed] [Google Scholar]
- 32. Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. In: Hempel JE, Williams CH, Hong CC, eds. Chemical Biology: Methods in Molecular Biology. New York: Springer; 2015:243-250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y, Zheng Y, Faheem A, et al. A novel AKT inhibitor, AZD5363, inhibits phosphorylation of AKT downstream molecules, and activates phosphorylation of mTOR and SMG-1 dependent on the liver cancer cell type. Oncol Lett. 2016;11:1685-1692. doi: 10.3892/ol.2016.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016;6:353-367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krieger E, Vriend G. New ways to boost molecular dynamics simulations. J Comput Chem. 2015;36:996-1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bagheri M, Fatemi MH. Fluorescence spectroscopy, molecular docking and molecular dynamic simulation studies of HSA-Aflatoxin B1 and G1 interactions. J Lumin. 2018;202:345-353. doi: 10.1016/j.jlumin.2018.05.066. [DOI] [Google Scholar]
- 38. Fathima AJ, Murugaboopathi G, Selvam P. Pharmacophore mapping of ligand based virtual screening, molecular docking and molecular dynamic simulation studies for finding potent NS2B/NS3 protease inhibitors as potential anti-dengue drug compounds. Curr Bioinform. 2018;13:606-616. doi: 10.2174/1574893613666180118105659. [DOI] [Google Scholar]
- 39. Deeba F, Malik MdZ, Naqvi IH, et al. Potential entry inhibitors of the envelope protein (E2) of Chikungunya virus: in silico structural modeling, docking and molecular dynamic studies. Virusdisease. 2017;28:39-49. doi: 10.1007/s13337-016-0356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non–small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556-2568. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 41. Majewski M, Ruiz-Carmona S, Barril X. An investigation of structural stability in protein-ligand complexes reveals the balance between order and disorder. Commun Chem. 2019;2:110. doi: 10.1038/s42004-019-0205-5. [DOI] [Google Scholar]
- 42. Pace CN, Fu H, Lee Fryar K, et al. Contribution of hydrogen bonds to protein stability: hydrogen bonds and protein stability. Protein Sci. 2014;23:652-661. doi: 10.1002/pro.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen DE, Willick DL, Ruckel JB, Floriano WB. Principal component analysis of binding energies for single-point mutants of hT2R16 bound to an agonist correlate with experimental mutant cell response. J Comput Biol. 2015;22:37-53. doi: 10.1089/cmb.2014.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers. 2017;9:3-45. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arteaga CL, Baselga J. Tyrosine kinase inhibitors: why does the current process of clinical development not apply to them? Cancer Cell. 2004;5:525-531. doi: 10.1016/j.ccr.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 46. Twombly R. Failing survival advantage in crucial trial, future of Iressa is in Jeopardy. J Natl Cancer Inst. 2005;97:249-250. doi: 10.1093/jnci/97.4.249. [DOI] [PubMed] [Google Scholar]
- 47. Liao Q-H, Gao Q-Z, Wei J, Chou K-C. Docking and molecular dynamics study on the inhibitory activity of novel inhibitors on epidermal growth factor receptor (EGFR). Med Chem. 2011;7:24-31. doi: 10.2174/157340611794072698. [DOI] [PubMed] [Google Scholar]
- 48. Yun C-H, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070-2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brinckerhoff CE, Rutter JL, Benbow U. Interstitial collagenases as markers of tumor progression. Clin Cancer Res. 2000;6:4823-4830. [PubMed] [Google Scholar]
- 50. Pinkas J, Leder P. MEK1 signaling mediates transformation and metastasis of eph4 mammary epithelial cells independent of an epithelial to mesenchymal transition. Cancer Res. 2002;62:4781-4790. [PubMed] [Google Scholar]
- 51. Woodworth CD. Inhibition of the epidermal growth factor receptor increases expression of genes that stimulate inflammation, apoptosis, and cell attachment. Mol Cancer Ther. 2005;4:650-658. doi: 10.1158/1535-7163.MCT-04-0238. [DOI] [PubMed] [Google Scholar]
- 52. Huether A, Höpfner M, Sutter AP, Schuppan D, Scherübl H. Erlotinib induces cell cycle arrest and apoptosis in hepatocellular cancer cells and enhances chemosensitivity towards cytostatics. J Hepatol. 2005;43:661-669. doi: 10.1016/j.jhep.2005.02.040. [DOI] [PubMed] [Google Scholar]
- 53. Glaysher S, Bolton LM, Johnson P, et al. Targeting EGFR and PI3K pathways in ovarian cancer. Br J Cancer. 2013;109:1786-1794. doi: 10.1038/bjc.2013.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang M, Jang H, Nussinov R. Structural features that distinguish inactive and active PI3K lipid kinases. J Mol Biol. 2020;432:5849-5859. doi: 10.1016/j.jmb.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18:26. doi: 10.1186/s12943-019-0954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chang F, Lee JT, Navolanic PM, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590-603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- 58. Zwang Y, Jonas O, Chen C, et al. Synergistic interactions with PI3K inhibition that induce apoptosis. eLife. 2017;6:e24523. doi: 10.7554/eLife.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kunnimalaiyaan M, Ndiaye M, Chen H. Apoptosis-mediated medullary thyroid cancer growth suppression by the PI3K inhibitor LY294002. Surgery. 2006;140:1009-1014; discussion 1014–1015. doi: 10.1016/j.surg.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 60. Nitulescu GM, Margina D, Juzenas P, et al. Akt inhibitors in cancer treatment: the long journey from drug discovery to clinical use (review). Int J Oncol. 2016;48:869-885. doi: 10.3892/ijo.2015.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin K, Lin J, Wu W-I, et al. An ATP-site on-off switch that restricts phosphatase accessibility of Akt. Sci Signal. 2012;5:ra37. doi: 10.1126/scisignal.2002618. [DOI] [PubMed] [Google Scholar]
- 62. Kang D, Park W, Lee S, Kim J-H, Song JJ. Crosstalk from survival to necrotic death coexists in DU-145 cells by curcumin treatment. Cell Signal. 2013;25:1288-1300. doi: 10.1016/j.cellsig.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 63. Kuo P-L, Hsu Y-L, Cho C-Y. Plumbagin induces G2-M arrest and autophagy by inhibiting the AKT/mammalian target of rapamycin pathway in breast cancer cells. Mol Cancer Ther. 2006;5:3209-3221. doi: 10.1158/1535-7163.MCT-06-0478. [DOI] [PubMed] [Google Scholar]
- 64. Song M, Bode AM, Dong Z, Lee M-H. AKT as a therapeutic target for cancer. Cancer Res. 2019;79:1019-1031. doi: 10.1158/0008-5472.CAN-18-2738. [DOI] [PubMed] [Google Scholar]
- 65. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93-115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153-166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 67. Aubry C, Wilson AJ, Jenkins PR, et al. Design, synthesis and biological activity of new CDK4-specific inhibitors, based on fascaplysin. Org Biomol Chem. 2006;4:787. doi: 10.1039/b518019h. [DOI] [PubMed] [Google Scholar]
- 68. Bonelli M, La Monica S, Fumarola C, Alfieri R. Multiple effects of CDK4/6 inhibition in cancer: from cell cycle arrest to immunomodulation. Biochem Pharmacol. 2019;170:113676. doi: 10.1016/j.bcp.2019.113676. [DOI] [PubMed] [Google Scholar]
- 69. Pernas S, Tolaney SM, Winer EP, Goel S. CDK4/6 inhibition in breast cancer: current practice and future directions. Ther Adv Med Oncol. 2018;10:1758835918786451. doi: 10.1177/1758835918786451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Peurala E, Koivunen P, Haapasaari K-M, Bloigu R, Jukkola-Vuorinen A. The prognostic significance and value of cyclin D1, CDK4 and p16 in human breast cancer. Breast Cancer Res. 2013;15:2-10. doi: 10.1186/bcr3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. J Biomol Struct Dyn. 2020;7:1-14. doi: 10.1080/07391102.2020.1802347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Martínez L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE. 2015;10:e0119264. doi: 10.1371/journal.pone.0119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao Y, Zeng C, Massiah MA. Molecular dynamics simulation reveals insights into the mechanism of unfolding by the A130T/V mutations within the MID1 zinc-binding Bbox1 domain. PLoS ONE. 2015;10:e0124377. doi: 10.1371/journal.pone.0124377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dubey KD, Tiwari RK, Ojha RP. Recent advances in protein-ligand interactions: molecular dynamics simulations and binding free energy. Curr Comput Aided Drug Des. 2013;9:518-531. [DOI] [PubMed] [Google Scholar]