Abstract
Glycyrrhizin (GL) and Glycyrrhetic Acid 3-O-mono-β-D-glucuronide (GAMG) are the typical triterpenoid glycosides found in the root of licorice, a popular medicinal plant that exhibits diverse physiological effects and pharmacological manifestations. However, only few reports are available on the glycosylation enzymes involved in the biosynthesis of these valuable compounds with low conversion yield so far. In mammals, glycosyltransferases are involved in the phase II metabolism and may provide new solutions for us to engineer microbial strains to produce high valued compounds due to the substrate promiscuity of these glycosyltransferases. In this study, we mined the genomic databases of mammals and evaluated 22 candidate genes of O-glycosyltransferases by analyzing their catalytic potential for O-glycosylation of the native substrate, glycyrrhetinic acid (GA) for its glycodiversification. Out of 22 selected glycosyltransferases, only UGT1A1 exhibited high catalytic performance for biosynthesis of the key licorice compounds GL and GAMG. Molecular docking results proposed that the enzymatic activity of UGT1A1 was likely owing to the stable hydrogen bonding interactions and favorite conformations between the amino acid residues around substrate channels (P82~R85) and substrates. Furthermore, the complete biosynthesis pathway of GL was reconstructed in Saccharomyces cerevisiae for the first time, resulting in the production of 5.98 ± 0.47 mg/L and 2.31 ± 0.21 mg/L of GL and GAMG, respectively.
Keywords: O-glycosyltransferase, Homo sapiens, Glycyrrhizin (GL), Saccharomyces cerevisiae, Glycyrrhetic Acid 3-O-mono-β-D-glucuronide(GAMG)
Graphical abstract
1. Introduction
Triterpenoids are the secondary metabolites in many plant species that exhibit structural diversity and wide range of biological activities [1]. These are the chemical compounds of 30 carbon atoms constituted by six isoprene units (C5H8) [1] and often stored in the form of glycosylated triterpenoid saponins in plants. By glycosylation, one or more glycosyl groups are attached to the hydrophobic triterpenoid aglycones via glycosidic linkage by catalytic action of glycosyltransferase to produce triterpenoid saponins [[2], [3], [4]]. Modern pharmacological studies revealed that some triterpenoid saponins present significant physiological activities and high water solubility due to its nonpolar pentacyclic skeleton [5].
Glycyrrhizin (GL) is a distinctive pentacyclic triterpenoid saponin compound and is a key bioactive ingredient of Licorice, a traditional Chinese medicinal herb [6]. GL exhibits a wide spectrum of remarkable pharmacological activities,such as anti-inflammatory, anti-tumor, hepatoprotective and immunoregulation [7,8]. In addition to diverse pharmacological activities, GL also exhibits 170-fold more sweetness than that of sucrose rendering it to be widely used as a sweetener in the food industry [9]. Currently, GL is mainly extracted from licorice roots with the limitation of low yield due to the shortage of wild licorice resources making it difficult to meet the commercial needs [10]. Furthermore, prolonged growth period and high labor cost limit the production of this valuable compound at industrial scale.
Use of synthetic biology and metabolic engineering approaches in microbial cell factories has been a hot spot for microbial production of GL. Glycosyltransferases (GT) are categorized into 99 distinct families based on their substrate and catalytic specificity [11]. Family 1 glycosyltransferase, also known as UDP-glycosyltransferase (UGTs), catalyze the transfer of sugar moiety from UDP-sugar (sugar donor) to receptor aglycon, which regulates the biological activity, water solubility and stability of the receptor molecule [12]. UGTs play essential roles in glycodiversification of triterpenoid saponins by addition of monosaccharides to triterpene aglycones [13]. However, a few UGT enzymes have been identified to glycosylate triterpene aglycones so far and their substrate specificities and biochemical functions remained largely unknown.
Microbial cell factories exhibit several advantages over traditional extraction methods such as, shortened culture period, cheap carbon source and ability for large-scale fermentation, thus provide novel approaches to overcome the low yield of valuable natural products extracted from plants [14]. Analysis of metabolic pathways in plants, mining of key genetic elements and their optimization are the primary aspects to yield the plant derived natural products by microbial cell factories [15]. A stable and continuous production of GL requires the catalytic activity of the glycosyltransferases to transfer the glucuronic acid to the hydroxyl group at the 3-position of Glycyrrhetic acid (GA). Our previous research has already achieved the synthesis of GA in Saccharomyces cerevisiae [1]. To further elucidate the S. cerevisiae cell factory to biosynthesize the GL in vivo, limitation of mining the genes encoding UGTs to catalyze the GA to GL in plants has not been resolved. Currently, there are few reports on glycosyltransferases related to GL synthesis. In 2016, Xu et al. excavated a new glycosyltransferase in licorice, which could catalyze the GA to GL. However, no subsequent studies have been reported so far [16]. In 2018, He et al. mined the transcriptome data of G. uralensis and discovered a new glycosyltransferase UGT73F17, which can transfer the glucose moiety to the OH group at the 30-position of glycyrrhizin [17]. In 2019, Chen et al. mined a new glycosyltransferase to transfer one glucuronic acid to the OH group at the 3-position of GA, and produced Glycyrrhetic Acid 3-O-mono-β-D-glucuronide (GAMG) [18]. In another study, Nomura et al. reported that UGT73P12 catalyzes the second glucuronosylation as a final step of glycyrrhizin biosynthesis in G. uralensis [19].
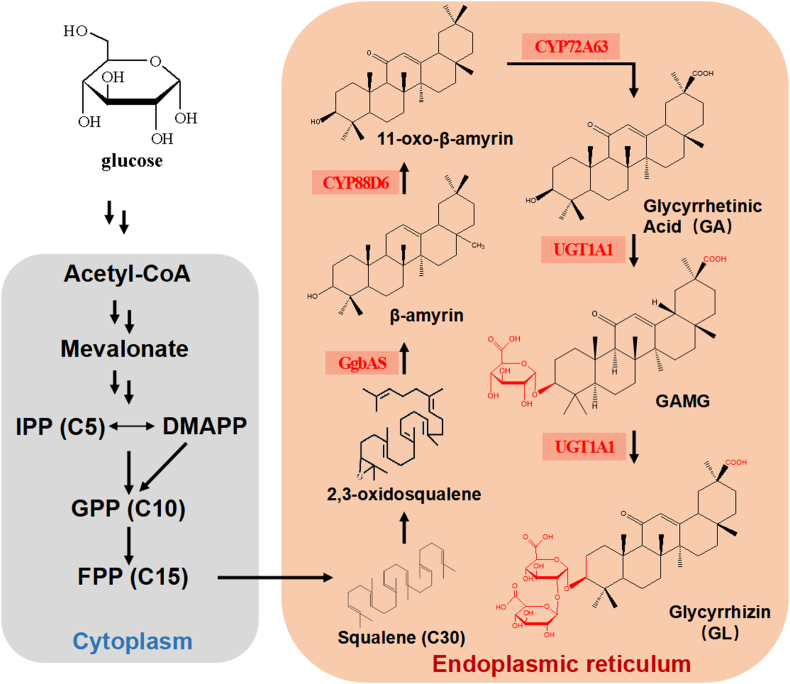
In this study, we mined and expressed 22 UGTs from phase II drug metabolism of mammals liver to evaluate their catalytic potential to glycosylate GA in vitro and constructed a novel platform to synthesize the high value plant triterpenoid saponins. Based on the in vitro results, UGT1A1 from Homo sapiens was expressed in S. cerevisiae to construct microbial cell factory to evaluate its catalytic potential to produce GL and GAMG in vivo with glucose as a substrate (Fig. 1). Results indicated that engineered strain produced 5.98 ± 0.47 mg/L and 2.31 ± 0.21 mg/L of GL and GAMG, respectively.
Fig. 1.
Biosynthesis pathway of Glycyrrhizin (GL)and Glycyrrhetic Acid 3-O-mono-β-D-glucuronide (GAMG) inS. cerevisiae
2. Material and methods
2.1. Strains, vectors and media
S. cerevisiae strain BY4741(MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0, SY022) (Invitrogen, Carlsbad, CA) was used for expression of candidate UGTs for enzyme assays in vitro. Engineered strains were cultured at 30 °C in SD medium lacking uracil with 20 g/L glucose. GA-producing strain (GA104), a derivative of S. cerevisiae INVSC1, was used as a platform strain for producing GA, and was cultured at 30 °C in SD medium with 20 g/L glucose. E. coli Top10 (Novagen, USA) competent cells were used for transformation and plasmid DNA extraction, and were cultivated at 37 °C in LB medium with 100 mg/L ampicillin or 100 mg/L kanamycin. The gene-accepting vectors HCKan-P, HCKan-O and HCKan-T for golden gate assembly were provided by Prof. Dai, Shenzhen Institutes of Advanced Technology. Plasmids used in this study are listed in (Supplementary Table 1). The genes and primers were synthesized by GENEWIZ (Suzhou, China).
2.2. DNA manipulation
For DNA manipulations, TIAN prep Mini Plasmid Kit (TIANGEN, China) and TIAN prep Yeast Plasmid DNA Kit (TIANGEN) were used to isolate plasmids from yeast and E.coli, respectively. Genomic DNA isolation was performed by using the TIAN amp Yeast DNA Kit (TIANGEN). Enzymes used for recombinant DNA cloning and Golden Gate Assembly were purchased from Thermo Scientific (Waltham, MA) and New England Biolabs (Ipswich, MA). Selected UGTs genes were synthesized by GENEWIZ, China. All these genes were codon optimized for S. cerevisiaeand cloned in pUC57 vector (GENEWIZ) followed by DNA amplification using their respective primers by PCR. PCR products were purified by TIAN quick Midi Purification Kit (TIANGEN) and UGTs genetic circuits were constructed with standard vector parts by employing the Golden Gate Assembly [20]. The candidate UGTs genes were ligated into the POT vectors along with TDH3 promotor upstream and SLM5t terminator downstream, followed by transformation into S. cerevisiae strain BY4741. Engineered strains were evaluated for expression of UGTs and their catalytic potential against GA glycosylation in vitro.
The UGT1A1 gene expressing cassette (in plasmid POT2) and HsUGDH gene expressing cassette (in plasmid POT3) were ligated with selection marker cassette FBA1p-KanMX-SLM5t (in plasmid POT1) onto the receiver plasmid K-3, using Golden Gate Assembly followed by integration into homologous HO-site of GA104 genome [21] (Supplementary Fig. 1). Primers used in the DNA assembly are summarized in (Supplementary Table 2).
2.3. Enzymatic assays for GA glycosylation in vitro
To screen the catalytic activities of selected UGTs, seed cultures of yeast strains harboring the POT plasmids with candidate UGTs were grown individually in 2 mL SD media lacking uracil at 30 °C in incubator shaker overnight and subjected to cultivation in 40 mL SD media without uracil in 250 mL Erlenmeyer flasks for 5 days. The yeast cells were harvested and cells were washed twice with double distilled deionized water followed by resuspension in 50 mM PBS buffer (pH 7.4). Cells were lysed by traditional glass beads method for 30 cycles at 4 °C in cell homogenizer. Cell lysates were collected by centrifugation at 12,000 rpm at 4 °C for 30 min. The glycosylation reactions were performed in 200 μL of reaction mixtures containing crude yeast cell lysates, 1 mM UDP-glucuronic acid (Sigma-Aldrich), and 100 μM GA (Sigma-Aldrich), followed by incubation at 37 °C for 2 h. All glycosylation reaction samples were prepared in triplicates and reactions were quenched by addition of equal volume of methanol and subjected to HPLC and LC-MS analysis.
2.4. Western blot analysis
Western Blot was used for evaluation of UGTs expression in S. cerevisiae. The seed culture was prepared as described in section 2.5. Briefly, a growing culture was inoculated into a 250 mL Erlenmeyer flask containing 40 mL of fresh YPD medium up to an OD600 of 0.1 prior to further cultivation at 30 °C for 5 days. Cells were harvested by centrifugation and the supernatant was discarded. Cells were washed 3 times in 100 mM of Tris-HCl (pH 7.0) buffer. The yeast cells were suspended in extraction buffer (100 mM of Tris-HCl, pH 7.0, 1 mM of DTT, 10 mM of MgCl2, 1 mM of EDTA) and disrupted by using ultrasonication (work 2 s, intermittent 2 s, duration 10 min, power 80 W) in an ice bath. The mixture was centrifuged at 8000 rpm and 4 °C for 5 min. Protein concentration in the supernatant was assayed using commercial kits (Jian Cheng Biotech Company, Nanjing, China) according to the manufacturers’ specifications and all samples were adjusted to the same protein concentration. For Western blot analysis, 10 μg solubilized protein samples were loaded into a 12% SDS polyacrylamide gel using an SDS-PAGE (Laemmli) Buffer System. After separation, the proteins were transferred to a nitrocellulose membrane (NCM, 0–45/SM pore size) by semi-dry electro-blotting (Pharmacia Nova-Blot electrophoretic transfer unit). Protein transfer was obtained by using 8 mA/sq. cm. of gel within 1 h. The membrane was incubated with 1% skim milk in TBST buffer at 4℃°C overnight and washed thrice with 10 mL TBST buffer for 5 min. The membranes were incubated with anti-myc antibody (Santa Cruz Biotechnology) at a 1:2000 dilution. Afterwards, the membranes were washed twice with 20 mL TBST buffer for 10 min, and incubated with 10 mL of TBST buffer containing 5 μL of anti-mouse IgG peroxidase-linked secondary antibody, anti-GAPDH (Zen Bioscience). After washing, ubiquitinated proteins were detected using ECL, Western lightening™ Chemiluminescence.
2.5. Shake-flask cultivation and metabolite extraction for GA glycodiversification in vivo
The engineered yeast strains were grown in YPD medium with 40 g/L glucose. Seed cultures were prepared by growing all strains individually in 15 mL culture tubes containing 2 mL medium at 30 °C and 220 rpm up to an OD600 of approximately 1.0. Flasks (250 mL) containing 30 mL medium were then inoculated to an OD600 of 0.1 using the resulting seed cultures. The strains were grown at 30 °C and 200 rpm for 6 days followed by cell lysis and resuspension of centrifuged pellets in ethyl acetate. Ethyl acetate was evaporated by employing vacuum concentrator system and subsequently, samples were analyzed by HPLC and LC-MS for presence of metabolites.
2.6. HPLC and LC-MS analysis
The HPLC analysis was performed by LC 10AD instrument (Shimadzu Corp., Kyoto, Japan). The chromatographic separations were carried out at UV detection wavelength 254 nm and 40 °C on a reverse-phase C18 column (5 mL; 250 × 4.6 mm; Shimadzu). A gradient elution method was employed with the mobile phase consisting a mixture of methanol and 0.6% acetic acid (84:16 v/v) at a flow rate of 1 mL/min. For LC-MS analysis, the product was evaporated by a vacuum concentrator system. The product was resuspended in 20 μL of methanol and injected into an LC-MS (LCMS-8040, Shimadzu Corp., Kyoto, Japan). For MS analysis, all spectra were obtained in positive mode over an m/z range of 100–1200; dry gas flow, 6.0 L min−1; dry temperature,180 °C; nebulizer pressure, 1 bar and probe voltage +4.5 kV.
2.7. Molecular docking
Molegro Virtual Docker and PyMOL v1.7.4.5. were used to calculate the best binding conformation of GL in the catalytic site of UGT1A1, and the structure of Sterol 3-beta-glucosyltransferase (ugt51) from Saccharomyces cerevisiae (SMTL ID: 5gl5.1) was used as a template to construct the 3D structures of UGT1A1 by homology modeling by employing the Swiss Model. The chem3D 16.0 was used to build the structures of GL for molecular docking.
3. Results and discussion
3.1. Candidate UGT genes screening and phylogenetic analysis
Construction of the S. cerevisiae cell factories to synthesize glycosylated derivatives of GA encounters the limitation of mining of UGT encoding genes from plants due to huge genomic databases. In order to avoid this limitation, previously reported already reported UGTs for glycosylation of other substrates may be evaluated for their glycosylation potential for GA glycodiversification. In this study, we focused on UGTs from Homo sapiens for their potential against the GA glycosylation. Although UGTs involved in phase II drug metabolism using UDP-glucuronic acid (UDPGA) as a sugar donor to make secondary metabolites, they can also catalyze a class of compounds with structural resemblance to GA to produce glucuronide derivatives [22]. As the structure of GA is similar to estrone (Supplementary Fig. 2) which can be glycosylated by UGT1A3 from Homo sapiens [23], we assumed that UGT1A3 may also glycosylate GA to GL.
We used UGT1A3 as a template and screened 10 more UGTs according to their amino acid sequence similarities (>80%) to that of UGT1A3 in NCBI (Supplementary Table 3). Researchers found that GA can induce protein transcription and expression of UGT1A family of phase II metabolizing enzymes in HepG2 cells [23]. Therefore, we further screened 11 UGTs from UGT1A family in NCBI, including UGT1A1, UGT1A4, UGT1A5, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT1A10, UGT2A1, UGT2A2 and UGT2A3. Hence, 22 candidate UGTs were evaluated for their catalytic potential to biosynthesize GL from GA. Their phylogenetic analysis has been shown in Supplementary Fig. 3.
3.2. Glycosylation of GA in vitro and the molecular docking to reveal the molecular basis of glycosylation activity
Initially, we tried to express all the 22 UGTs in E.coli expression system. The expression vectors, pET28a-UGTs were constructed and transformed into E.coli BL21 (DE3) competent cells. However, results showed that all UGTs could not be aptly expressed in E. coli and resulted in inclusion bodies, which is concordant with the previous report [24]. It is speculated that UGTs from Homo sapiens and mammals exhibit high glycosylation potential but there is no post translation glycosylation mechanism in E.coli.
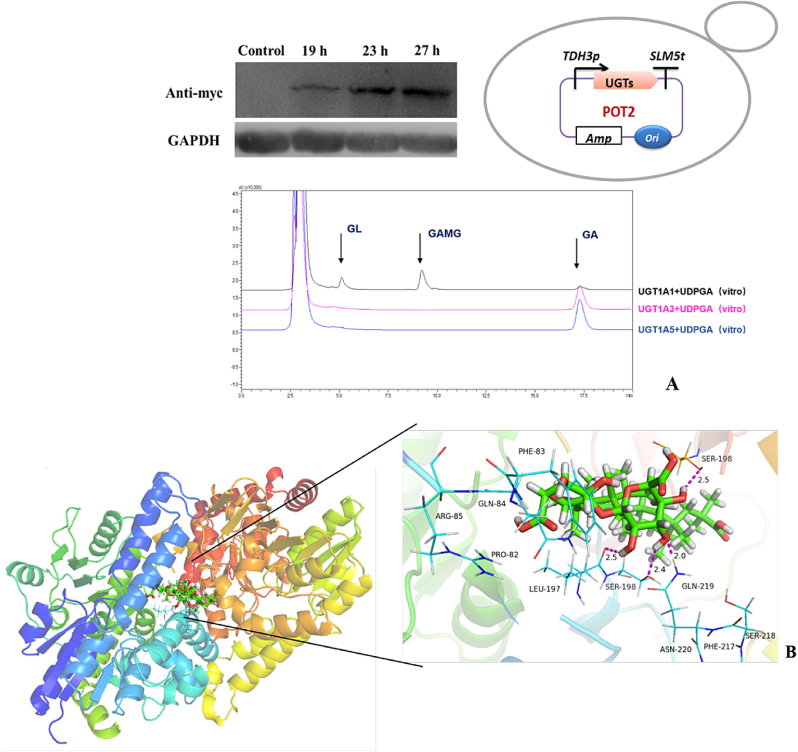
For heterologous expression of selected UGTs in S. cerevisiae, a promotor (TDH3p) and a terminator (SLM5t) were used to construct the expression vectors POT2-UGTs (Fig. 2A) followed by their transformation into S. cerevisiae BY4741. Strains were cultured and glycosylation reactions were performed by enzymatic assays of crude cell lysates with the UDPGA and GA. Results revealed that only UGT1A1 exhibits the catalytic potential to produce GL and GAMG by catalyzing the native substrate GA (Fig. 2A). The conversion rate of GA into GL and GAMG by bioactivity of UGT1A1 was roughly calculated to be 90.3%. While UGT1A1 may be the only active variant against GA glycosylation, there is no strong evidence that other UGTs do not have enzyme activity as screening from crude yeast lysate does not preclude poor expression.Western blot results also showed that UGT1A1 could be successfully expressed in S. cerevisiae (Fig. 2A).
Fig. 2.
Glycosylation of GA by UGT1A1 in vitro (A) Golden Gate Assembly of the UGTs, HPLC analysis of GA glycosylation assays in vitro and Western blot results of UGT1A1 expression. (B) Molecular docking of UGT1A1-GL complexes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In order to shed light on possible reasons for the novel catalytic activity of UGT1A1, we obtained the 3D structure of UGT1A1 and performed molecular docking of complexes UGT1A1-GL. Although, 22 candidate UGTs exhibited high sequence homology, but the enzymatic activity of UGT1A1 was likely owing to the stable hydrogen bonding interactions and favorite conformations between the amino acid residues around substrate channels and substrates. We chose three fragments located around the substrate channel of UGT1A1, having some sequence differences with other 21 UGTs (I215~N220,P82~R85 and S198~200) (Supplementary Fig. 4). We replaced the three corresponding regions of UGT1A3 and named the mutants M1, M2 and M3, respectively. Mutant strains were cultured and glycosylation reactions were performed by enzymatic assays of crude cell lysates with the UDPGA and GA. Results showed that only M2 exhibited the catalytic potential to produce GAMG by catalyzing the native substrates GA (Supplementary Fig. 5). To further confirm the role of pro82-arg85 in M2, a single point mutation was performed on pro82-arg85, and each amino acid was mutated to Alanine. Results of enzymatic catalysis in vitro showed that only P82A mutation, UGT1A3-M2 lost its catalytic activity (Supplementary Fig. 6), which proved that P82 played a key role in UGT1A1 catalyzed GA glycosylation (Fig. 2B).
3.3. Transformation of UDPG into UDPGA by HsUGDH in vivo
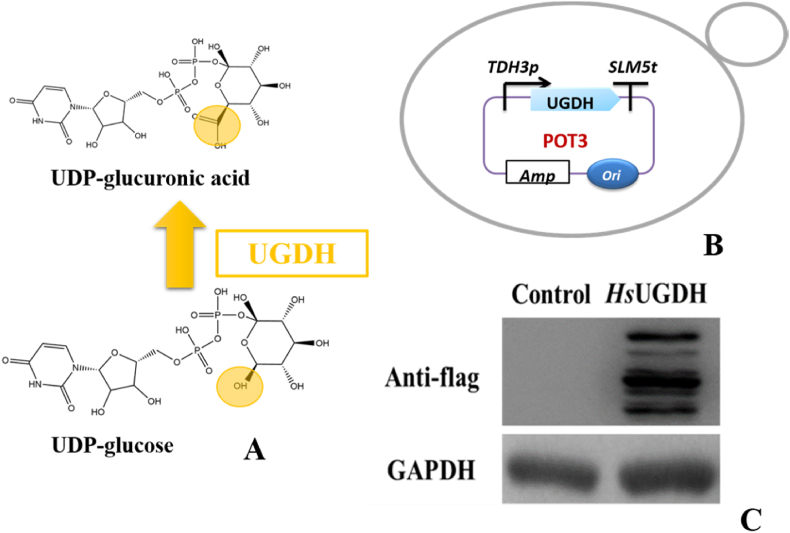
As the sugar donor of GL biosynthesis process, UDP-glucuronic acid (UDPGA) is vital for GL de novo biosynthesis in yeast cell factory. However, S. cerevisiae can only produce UDP-glucose (UDPG). In this regard, UDP glucose dehydrogenase (UGDH) from Homo sapiens was introduced in S. cerevisiae to produce UDPGA in vivo (Fig. 3A). To construct the vector harboring HsUGDH gene, TDH3p promoter and SLM5t terminator along with HsUGDH gene were cloned in POT3 vector by Golden gate assembly and the expression vector POT3-HsUGDH was transformed into S. cerevisiae BY4741 (Fig. 3B). After engineered strain was cultured in YPD medium for 72h, cells were harvested and samples were prepared for LC-MS analysis of metabolites. Results verified that HsUGDH could catalyze UDPG to produce UDPGA in S. cerevisiae while, the control strain with empty POT3 plasmid did not show any biotransformation of UDPG into UDPGA in vivo (Supplementary Fig. 7). Western blot analysis also showed that HsUGDH could be successfully expressed in S. cerevisiae (Fig. 3C).
Fig. 3.
Production of UDPGA in S. cerevisiae by HsUGDH. (A) Scheme of UDPG bioconversion into UDPGA (B) Description of POT3 vector harboring UGDH gene (C) Western blot results of HsUGDH expression.
3.4. Construction of S. cerevisiae cell factories to synthesize GL and GAMG in vivo
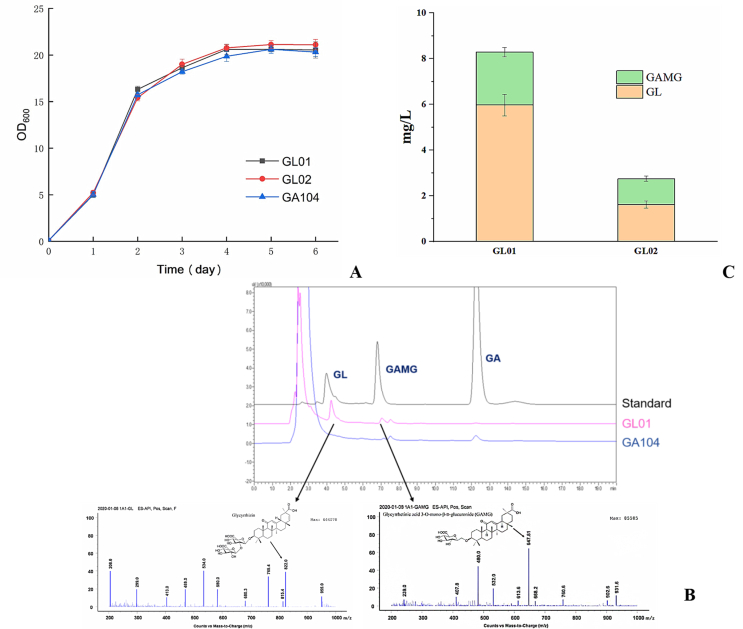
Based on the earlier experiments, HsUGDH and UGT1A1 were selected from Homo sapiens as the target genes to be integrated into the genome of GA producing strain GA104 (S. cerevisiae strain engineered by our lab to produce GA by using glucose as the substrate in vivo) [1]. The UGT1A1 gene cassette (in POT2) and HsUGDH gene cassette (in POT3) were ligated with selection marker cassette, FBA1p-KanMX-SLM5t (POT1) onto the receiver plasmid K-3 by using Golden Gate Assembly and subsequently integrated into homologous HO-site of GA104 genome and engineered strain was named as GL01. We also used the GuGT14 gene, which can transfer one glucuronic acid to the hydroxyl group at the 3-position of GA [18], and UGT73P12 gene, which can catalyze the second glucuronosylation as the final step of glycyrrhizin biosynthesis in G. uralensis [19] to construct another engineered strain (GL02 as a control strain). After GL01 and GL02 were cultured in YPD medium for 6 days (According to the results of the pre-experiment, a small amount of products could be detected after the fifth day of culture), samples were prepared and analyzed by LCMS for detection of target metabolites. Results showed that there was no significant difference in the growth curves of the three strains (Fig. 4A). The GA titer of GA producing chassis GA104 was 13.12±0.83 mg/L, and GA titers of GL01 and GL02 were 1.20±0.11 mg/L and 1.63±0.20mg/L, respectively. GL01 produced 5.98 ± 0.47 mg/L and 2.31 ± 0.21 mg/L of GL and GAMG, respectively while GL02 produced 1.62 ± 0.15 mg/L and 1.13 ± 0.11 mg/L of GL and GAMG, respectively (Fig. 4B). In addition, very minute quantity of GL and GAMG(no more than 10 μg/L)were detected in extracellular supernatant after centrifugation of the cell cultures, indicating the intracellular production of target metabolites. However, Parent strain (GA104) did not show any biotransformation of GA into GL and GAMG in vivo. The total triterpenoid saponin compounds produced by GL01 were 301% of the produced by GL02. These results proved that UGT1A1 from homo sapiens exhibits the catalytic potential for GA diversification in vivo due to its broad promiscuity and can be further elucidated for evaluation of its catalytic potential for production of glycosylated products of other substrates in microbial cell factories. Furthermore, UGT1A1 can comprehend the function of two UGTs together by its broad substrate promiscuity. It is expected to improve the activity of UGT1A1 through further rational design.
Fig. 4.
Construction of yeast cell factories for the production of GL and GAMG. (A) Growth curves of control and engineered strains (B) LCMS analysis of metabolites produced in vivo. (C) The total yield of GL and GAMG.
4. Conclusion
In this work, we constructed a novel platform to biosynthesize the high valued plant triterpenoid saponins by screening the UGTs from phase II drug metabolism of mammals due to their broad substrate promiscuity. This provided new insights and solutions for us to expand the range of UGTs substrates for their glycodiversification by glycosylation. We mined the genomic database of mammals and discovered 22 candidate genes for O-glycosyltransferase. The catalytic activities of the candidate UGTs were evaluated by catalyzing the native substrate, GA. As a result, UGT1A1 exhibited high performance for synthesizing the key licorice compounds, GL and GAMG. In addition, the complete pathways of GL biosynthesis were reconstituted in S. cerevisiae, resulting in the production of 5.98 ± 0.47 mg/L and 2.31 ± 0.21 mg/L of GL and GAMG, respectively. The construction of engineered S. cerevisiae to produce GL and GAMG by employing UGT1A1 is reported for the first time and its discovery for glycodiversification of GA makes it possible to construct yeast cell factories for the production of other valuable triterpenoid glycosides. Furthermore, this metabolic engineering approach of functional gene identification and strain engineering may serve as the basis for exploration of alternative ways for green microbial production of important natural products instead of chemical synthesis or extraction from plant sources.
CRediT authorship contribution statement
Ke Xu: Methodology, devised the processes for screening of candidate UGT genes, phylogenetic analysis, production of UDPGA in S. cerevisiae, Writing – review & editing. Yu-jia Zhao: devised the processes for screening of candidate UGT genes, Formal analysis, production of UDPGA in S. cerevisiae, Writing – review & editing. Nadeem Ahmad: Methodology. Jing-nan Wang: helped with DNA and genetic manipulations and fermentation experiments. Bo Lv: Methodology, Writing – review & editing. Ying Wang: helped with DNA and genetic manipulations and fermentation experiments. Jun Ge: Methodology, Supervision. Chun Li: Methodology, Writing – review & editing.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFA0901800), the Key Research and Development Program of Hebei Province (21374301D), the Natural Science Foundation of China (No.22078171), the Natural Science Foundation of Hebei Province (No.C2019105055) and the Scientific Research Foundation of Tangshan Normal University (No.2021B34).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.07.001.
Contributor Information
Bo Lv, Email: lv-b@bit.edu.cn.
Chun Li, Email: lichun@tsinghua.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu M., Wang C., Sun W. Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metab Eng. 2018;45:43–50. doi: 10.1016/j.ymben.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Vincken J.-P., Heng L., De Groot A. Saponins, classification and occurrence in the plant kingdom. 2007;68(3):275–297. doi: 10.1016/j.phytochem.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Sawai S., Saito K. Triterpenoid Biosynthesis and Engineering in Plants[J] Front Plant Sci. 2011;2(25):25. doi: 10.3389/fpls.2011.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y.-J., Li C. Biosynthesis of plant triterpenoid saponins in microbial cell factories. J Agric Food Chem. 2018;66(46):12155–12165. doi: 10.1021/acs.jafc.8b04657. [DOI] [PubMed] [Google Scholar]
- 5.He Y., Hu Z., Li A. Recent advances in biotransformation of saponins. Molecules. 2019;24(13):2365. doi: 10.3390/molecules24132365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nomura Y., Seki H., Suzuki T. Functional specialization of UDP‐glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019;99(6):1127–1143. doi: 10.1111/tpj.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen K., Yang R., Shen F.-Q. Current medicinal chemistry; 2019. Advances in pharmacological activities and mechanisms of glycyrrhizic acid [J] [DOI] [PubMed] [Google Scholar]
- 8.Lv B., Sun H., Huang S. Structure-guided engineering of the substrate specificity of a fungal β-glucuronidase toward triterpenoid saponins. J Biol Chem. 2018;293(2):433–443. doi: 10.1074/jbc.M117.801910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y., Lv B., Feng X. Perspective on biotransformation and de novo biosynthesis of licorice constituents. 2017;65(51):11147–11156. doi: 10.1021/acs.jafc.7b04470. [DOI] [PubMed] [Google Scholar]
- 10.Nose M., Yamanaka K., Hisaka S. Evaluation of the safety and efficacy of Glycyrrhiza uralensis root extracts produced using artificial hydroponic-field hybrid cultivation systems II: comparison of serum concentration of glycyrrhetinic acid serum concentration in mice. J Nat Med. 2019;73(3):661–666. doi: 10.1007/s11418-019-01312-9. [DOI] [PubMed] [Google Scholar]
- 11.Lairson L., Henrissat B., Davies G. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77 doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 12.Brazier-Hicks M., Gershater M., Dixon D. Substrate specificity and safener inducibility of the plant UDP‐glucose‐dependent family 1 glycosyltransferase super-family. Plant biotechnology journal. 2018;16(1):337–348. doi: 10.1111/pbi.12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahimi S., Kim J., Mijakovic I. Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol Adv. 2019;37(7) doi: 10.1016/j.biotechadv.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Zhao F., Bai P., Nan W. A modular engineering strategy for high‐level production of protopanaxadiol from ethanol by Saccharomyces cerevisiae. 2019;65(3):866–874. [Google Scholar]
- 15.Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164(6):1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Xu G., Cai W., Gao W. A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two‐step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. 2016;212(1):123–135. doi: 10.1111/nph.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J., Chen K., Hu Z.-M. UGT73F17, a new glycosyltransferase from Glycyrrhiza uralensis, catalyzes the regiospecific glycosylation of pentacyclic triterpenoids. 2018;54(62):8594–8597. doi: 10.1039/c8cc04215b. [DOI] [PubMed] [Google Scholar]
- 18.Chen K., Hu Z.M., Song W. Diversity of O-glycosyltransferases contributes to the biosynthesis of flavonoid and triterpenoid glycosides in Glycyrrhiza uralensis. ACS Synth Biol. 2019;8(8):1858–1866. doi: 10.1021/acssynbio.9b00171. [DOI] [PubMed] [Google Scholar]
- 19.Nomura Y., Seki H., Suzuki T. Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019;99(6):1127–1143. doi: 10.1111/tpj.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y., Dong J., Zhou T. YeastFab: the design and construction of standard biological parts for metabolic engineering in Saccharomyces cerevisiae. Nucleic Acids Res. 2015;43(13):e88. doi: 10.1093/nar/gkv464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H., Matsuda F., Yamada R. Cocktail delta-integration of xylose assimilation genes for efficient ethanol production from xylose in Saccharomyces cerevisiae. J Biosci Bioeng. 2013;116(3):333–336. doi: 10.1016/j.jbiosc.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Rowland A., Miners J.O., Mackenzie P.I. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol. 2013;45(6):1121–1132. doi: 10.1016/j.biocel.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Caillier B., Lepine J., Tojcic J. A pharmacogenomics study of the human estrogen glucuronosyltransferase UGT1A3. Pharmacogenetics Genom. 2007;17(7):481–495. doi: 10.1097/FPC.0b013e32806d87a4. [DOI] [PubMed] [Google Scholar]
- 24.Radominska-Pandya A., Bratton S.M., Redinbo M.R. The crystal structure of human UDP-glucuronosyltransferase 2B7 C-terminal end is the first mammalian UGT target to be revealed: the significance for human UGTs from both the 1A and 2B families. Drug Metab Rev. 2010;42(1):133–144. doi: 10.3109/03602530903209049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.