Summary
HDV is a small, defective RNA virus that requires the HBsAg of HBV for its assembly, release, and transmission. Chronic HBV/HDV infection often has a severe clinical outcome and is difficult to treat. The important role of a robust virus-specific T cell response for natural viral control has been established for many other chronic viral infections, but the exact role of the T cell response in the control and progression of chronic HDV infection is far less clear. Several recent studies have characterised HDV-specific CD4+ and CD8+ T cell responses on a peptide level. This review comprehensively summarises all HDV-specific T cell epitopes described to date and describes our current knowledge of the role of T cells in HDV infection. While we now have better tools to study the adaptive anti-HDV-specific T cell response, further efforts are needed to define the HLA restriction of additional HDV-specific T cell epitopes, establish additional HDV-specific MHC tetramers, understand the degree of cross HDV genotype reactivity of individual epitopes and understand the correlation of the HBV- and HDV-specific T cell response, as well as the breadth and specificity of the intrahepatic HDV-specific T cell response.
Keywords: Hepatitis Delta, HBV, HDV, T cell, CD8+, CD4+, epitope, viral escape
Abbreviations: aa, amino acid(s); ADAR1, adenosine deaminases acting on RNA; ALT, alanine aminotransferase; AST, aspartate aminotransferase; cccDNA, covalently closed circular DNA; ELISpot, enzyme-linked immune spot assay; HDAg, hepatitis delta antigen; ICS, intracellular cytokine staining; IFN-, interferon-; L-HDAg, large hepatitis delta antigen; MAIT, mucosa-associated invariant T cells; NK cells, natural killer cells; NTCP, sodium taurocholate co-transporting polypeptide; PBMCs, peripheral blood mononuclear cells; PD-1, programmed cell death protein 1; Peg-IFN-α, pegylated interferon alpha; PTM, post-translational modification; S-HDAg, small hepatitis delta antigen; TCF, T cell-specific transcription factor; Th1, T helper 1; TNFα, tumour necrosis factor-α
Key points.
-
•
HDV causes severe hepatitis, often leading to hepatic complications and liver-related death; it is a major public health concern affecting 12 million patients worldwide, with few treatment options.
-
•
The virology and immunology of HDV infection, which is intricately connected with the concomitant HBV infection, is still not completely understood; it is only recently that several studies characterising the T cell response in patients with HDV have been published.
-
•
This review summarises our current knowledge on the virology and immunology of HDV infection, with a focus on the HDV-specific T cell response.
-
•
A comprehensive database of all HDV-specific CD4+ and CD8+ T cell epitopes published to date is presented.
-
•
Detailed functional and phenotypic studies on the peripheral and intrahepatic HDV-specific T cell response during future clinical trials are needed to understand the T cell corelates of HDV control.
Introduction
Apart from some initial immunological HDV studies in the 1990s, it was only more recently that several immunological research groups further characterised the HDV-specific T cell response using state of the art methods. In this comprehensive review, we summarise these studies and list all HDV-specific T cell epitopes identified in humans so far, with a focus on their potential significance as well as unresolved knowledge gaps. Detailed knowledge of the HDV-specific T cell epitope repertoire is needed to guide therapeutic vaccine design and to improve immune monitoring in future clinical trials.
Epidemiology
The global prevalence of HDV is estimated at 12 million individuals,1 while others have calculated that it affects 32–61 million,2,3 or even 62–72 million individuals.4 HDV is highly endemic in Africa, the Amazon Basin, Eastern and Mediterranean Europe, the Middle East, and parts of Asia,4 mostly coinciding with high numbers of chronic HBV infections in these areas. HDV genotype 1 has global prevalence, while genotypes 2-8 show distinct regional patterns.5 Genotypes 2 and 4 predominantly cause milder disease, while the South American genotype 3 causes more severe hepatitis.6 Genotypes 5-8 are mostly diagnosed in patients of African origin and have also been linked to milder disease,7 although the latter is a matter of debate.8 Untreated chronic HBV/HDV infection causes severe liver disease in many cases, with 50% of patients developing cirrhosis within 5-10 years.6 Whether HBV/HDV infection is associated with an increased risk of HCC per se,9 or whether this only occurs secondary to cirrhosis,10 is controversial.
HDV remains a high-priority public health concern11 for 3 main reasons: increasing immigration from HDV endemic areas towards the US and Northern Europe; high endemicity of HDV in low-income countries; ongoing outbreaks of HDV.5,6 HDV elimination thus depends on both specific therapies as well as preventive HBV vaccination.
HDV virology
HDV is an exceptionally small virus12 and considered defective due to its dependency on the HBV-derived HBsAg to form infectious virions. HDV cell entry is dependent on the interactions between HBsAg/heparan sulfate proteoglycans and sodium taurocholate co-transporting polypeptide (NTCP).13 HDV might also use envelope proteins of other viruses for transmission,14,15 however, the clinical significance is unknown.16 HDV contains a circular single-stranded RNA genome of 1.7 kb, encoding a single 214-amino acid (aa) peptide,5 the hepatitis delta antigen (HDAg), which exists in 2 variants, the small HDAg (S-HDAg) and the large HDAg (L-HDAg), which has 19 additional C-terminal aa.12 Approximately 200 of these molecules are included per virion,5,17 in the form of nucleosome-like ribonucleoproteins consisting of the HDAgs and viral RNA, which have essential roles in viral replication.18,19
HDV RNA is replicated and transcribed to host-like20,21 mRNA in a double rolling circle process by host DNA-dependent RNA polymerases, mainly RNA pol II, but possibly also RNA pol I and III.22,23 The small genome and the replication mode are unique among animal RNA viruses and more typical for plant viroids/virusoids.24,25 Other than these, HDV hijacks host enzymes and even forces a template shift from host DNA to viral RNA,25,26 probably using a histone H3 mimicry strategy.19
L-HDAg is translated after ADAR1 (adenosine deaminases acting on RNA 1)-mediated editing of a stop codon at the amber/w site (adenosine 1012) at a portion of the antigenomic RNA, effectively elongating the open reading frame by 19–20 aa.12,27,28 Thus, S-HDAg is the first of the 2 peptides and facilitates replication, while L-HDAg is translated at a later stage and inhibits replication to promote virion assembly.29
Post-translational modifications (PTMs) are essential for the function of HDAg.30 Namely serine-2, -123 and -177 are phosphorylated post-translationally, arginine-13 is methylated, and lysine-72 is acetylated. Cysteine-211, only found in the L-HDAg, is modified by isoprenylation/farnesylation. It has been reported that the acetylation of lysine-72 is required for the subcellular localisation of HDAg and RNA replication. Other important PTMs in this regard include the methylation of arginine-13 and the phosphorylation of serine-177 and -123. The farnesylation of cysteine-211 is required for virus assembly13,30,31 and to inhibit replication.32
Other functional domains include a coiled-coil domain that is important for self-dimerisation,23 a domain that determines the nuclear localisation of HDAg33 and the unique carboxyterminal region of L-HDAg with the nucleolar export signal.34 RNA-binding arginine-rich motifs of HDAg have been described, still, oligomerisation seems more important for the activating and inhibitory effects of S- and L-HDAg.25,35
Heterogeneity and viral evolution
HDV shows great genetic variance, a broad range of viral quasispecies exist within the same infected individual.5 The intragenotypic genetic variability of HDV genotype 1 is estimated to be 11.3–14.3%36 and, recently, a subclassification of genotypes was proposed.37
Like DNA polymerases, DNA-dependent RNA polymerase II is reported to have kinetic proofreading abilities, however, the template switch to RNA might cause a higher error rate.38 Additionally, stray ADAR1-mediated RNA editing might also contribute to sequence heterogeneity.39
The substitution rates range from 3.0∗10-2 to 3.0∗10-3 for the whole genome and 9.5∗10-3 to 1.2∗10-3 substitutions per site per year for the HDAg open reading frame (determined by next-generation sequencing),13 decaying over time towards a steady state.40 High evolution rates correlate with clinical flares, and evolution rates are only higher than other RNA viruses at the beginning of the infection, during adaptation to the host.41 Non-synonymous mutations happen relatively more often, likely as a result of selection of variants capable of immune escape.42
PTM sites and ribozymes seem rather conserved, while 10.6% of codons are under diversifying positive selection.43
A reduced in vitro sensitivity of HDV to interferon (IFN)-α during treatment has been reported,44 likely due to the selection of genetic variants that replicate despite IFN. Possibly, HDV is activating the IFN pathways itself to suppress HBV replication and increase RNA editing by the IFN-induced enzymes ADAR145,46 and apolipoprotein B mRNA editing enzyme (APOBEC).13,44
In conclusion, HDV shows higher initial mutation rates than other hepatotropic viruses, while a few conserved genomic regions are described. Mutations and quasispecies may contribute to immune escape and treatment failure.
HDV therapy
Until recently, the only recommended treatment option for chronically HBV/HDV-infected patients was a long-term (48 weeks) therapy with pegylated-IFN-α (peg-IFN-α).47 Only few patients respond to treatment and late relapses occur in almost half of responding patients after achieving a ‘sustained’ virological response.48 Only 11% of patients maintain a virological response after IFN-based regimens and late relapses after therapy discontinuation are not uncommon.49 Elongation of IFN therapy to 96 weeks is possible with acceptable safety in up to 80% of patients, leading to longer on-treatment suppression of HDV replication and amelioration of fibrosis.50 However, relapse rates are still high (around one-third of responders) and HBsAg clearance is not improved – even by addition of tenofovir disoproxil fumarate50 – which is possibly linked to almost undetectably low viraemia.51 These findings underscore that, while a sustained virological response by negative HCV PCR 12 weeks after therapy indicates cure from HCV, this concept cannot be extrapolated to HDV, where loss of HBsAg and seroconversion remain the best markers for cure of chronic HDV infection.49,52 Roche has officially stopped the production of peg-IFN-α which will only be available until the end of 2022.
The novel entry inhibitor bulevirtide targets host NTCP and has led to promising results in 2 phase II trials,[53], [54], [55] leading to its conditional marketing authorisation in Europe.56,57 Other novel anti-HDV agents are currently being investigated (reviewed in53 and 58). Pegylated-IFN-λ showed advantageous tolerability and comparable antiviral activity during 8 weeks of treatment compared to 48 weeks of peg-IFN-α in a randomised, open-label, multicentre study.53,59 Lonafarnib – an orphan drug for the rare genetic disorder progeria – inhibits the host enzyme farnesyltransferase and is being investigated for HDV therapy in multiple combinations.53 The HBsAg release inhibitor REP-2139 is another promising candidate for HDV treatment.60 So far, although paradigm-shifting, none of these novel therapeutic approaches is recommended in international treatment guidelines.47,61
Immunology of HDV infection
Humans are the only natural host of HDV and only chimpanzees62 and some non-primates such as Tupaia bengaleri63 can be infected with (human) HBV and co-/superinfected with HDV. Other mammals used for prospective studies on HDV are woodchucks, woolly monkeys, and bats.17 HDV-transgenic mice were used to demonstrate that HDV hepatotropism is only due to the entry restriction by HBsAg.17 HDAg/HBsAg-transgenic mice did not develop liver disease.64 In vitro, in transient transfection, there was no interference with the cell cycle or apoptosis, whereas in dividing cells a slight growth disadvantage could be observed.65 Accordingly, HDV may – to some degree – be cytopathic itself and drive histopathologic liver damage together with the immune response.66 In humanised uPA/SCID/beige mice, HDV monoinfection can persist intrahepatically for at least 6 weeks without HBsAg, while maintaining infectivity and the ability to convert to a productive co-infection after rescue by HBV infection.67
HDV and HBV interact in multiple ways. Although HDV is able to replicate without active HBV replication,67 HBsAg is needed for HDV to form infectious particles. Usually, HDV predominates over HBV and coinfected patients often only show low HBV viral loads, although this pattern might be reversed in some individuals68 or transiently during early treatment phases.69
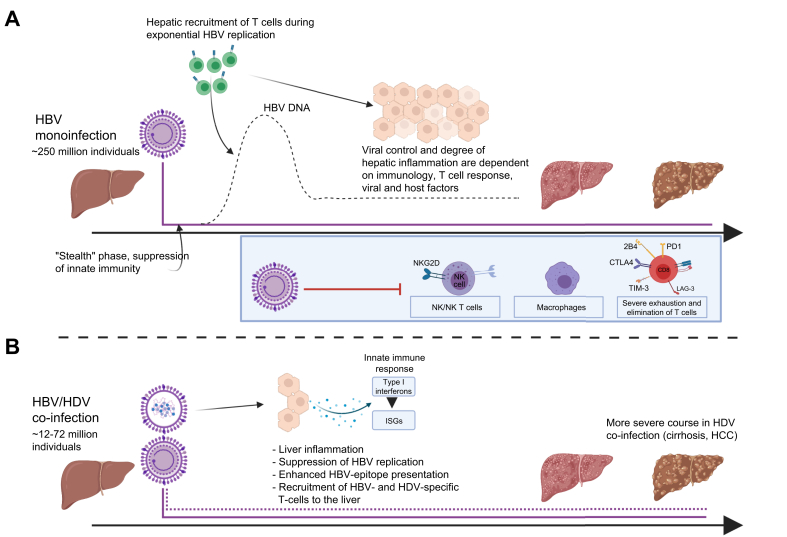
In vitro, upon superinfection there is a specific interference between the 2 viruses: HBV DNA, pregenomic RNA and HBeAg decrease while cccDNA and HBsAg stay constant.70 HDV infection is associated with a type-I IFN response and upregulation of IFN-induced genes.[70], [71], [72], [73] Upregulation of IFN-induced genes not only increases the HDV mutation rate, but also suppresses HBV replication,23 owing to decreased transcription of covalently closed circular DNA (cccDNA, without a decline in cccDNA abundance) as a result of IFN-α-induced epigenetic changes to the cccDNA.74,75 Notably, IFN partially suppresses HDV replication under certain circumstances (reviewed in detail12). HDV also enhances HBV epitope presentation, which could be one of the causes of the more severe liver pathology in HBV/HDV coinfection (Fig. 1).76
Fig. 1.
Immunological course of HBV monoinfection vs. HBV/HDV coinfection.
Note that chronic HBV inhibits various pathways of innate immunity and leads to different degrees of T cell exhaustion and deletion. Clearance of HBV is largely dependent on effective CD4+ and CD8+ T cell responses, as well as innate immune response, viral and host factors.117 Contrary, HDV activates pathways of innate immunity, thereby increasing type-I interferon (β and λ) responses and suppressing HBV replication. HBV epitope presentation and hepatotropic T cell recruitment is enhanced. Which immune responses are primarily required for HDV clearance is not well understood.12,74,76 ISG, interferon-stimulated genes; NKG2D, natural killer group 2 member D. Figure created with Biorender.com.
Few studies examined humoral immune responses to HDV. Anti-HDV antibodies are commonly generated,77 but probably unable to neutralise HDV.66,78 Anti-HDV IgM was traditionally used as clinical marker of disease activity before the establishment of standard pan-genotypic PCR assays.79,80
Chronically HDV-superinfected patients have higher serum type 1 to type 2 cytokine ratios, while HBV-monoinfected patients show elevated levels of both type 1 (tumour necrosis factor-α [TNFα], interleukin [IL]12, C-X-C motif chemokine ligand 9, IFN-γ) and type 2 (IL4, IL13, C-C motif chemokine ligand 26) cytokines.81 This predominance of type 1 responses, which mainly elicit cellular immune cascades, might explain the more aggressive course of disease in the case of HDV superinfection.81 Further, it has been shown that HDV strongly activates an IFN-β/λ response mainly through the pattern recognition receptor MDA5 (melanoma differentiation-associated protein 5) and that HDV can replicate in vivo despite this “interferon-activated state”.44
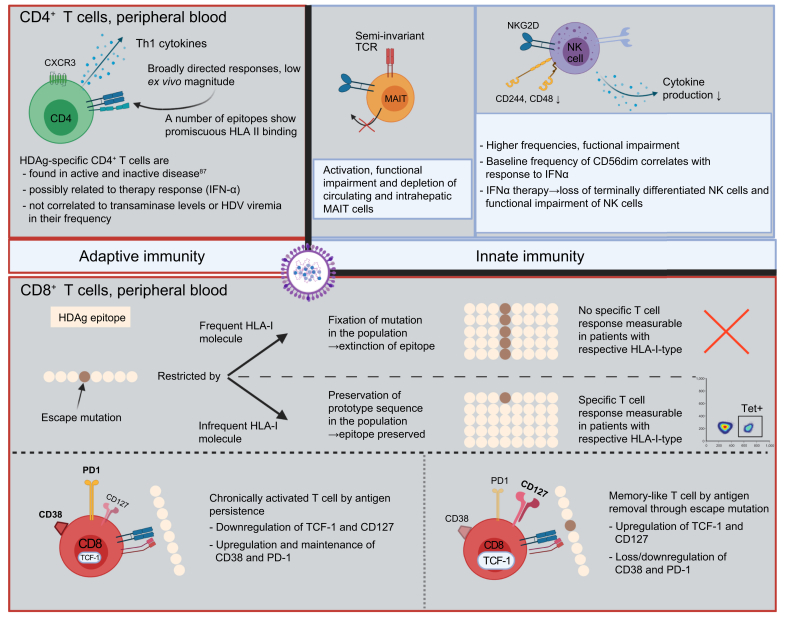
Functionally impaired CD56bright natural killer (NK) cells accumulate in viral hepatitis regardless of the virus itself (Fig. 2).82 The highest total frequencies of NK cells and CD56dim NK cells and the highest amounts of IFN-γ and TNF-α were found for HDV. However, phenotype and functional alterations were attributed primarily to the severity of infection rather than the virus itself.82 A higher frequency of CD56dim NK cells in HDV-infected patients is associated with better outcome after IFN-α treatment.83 IFN-α treatment seems to deplete terminally differentiated NK cells and cause functional impairment of NK cells.83
Fig. 2.
Key immunological findings in chronic HDV.
CXCR3, CXC-motif chemokine receptor 3; MAIT, mucosal associated invariant T cell; TCR, T cell receptor; TCF-1, T cell factor 1; PD-1, programmed cell death protein 1; Tet, HLA-I tetramer (loaded with HDAg-derived peptide). Based on [82], [83], [84],[86], [87], [88], [89],91,92. Figure created with Biorender.com
Intrahepatic and peripheral frequencies of mucosa-associated invariant T (MAIT) cells are reduced in patients with chronic HDV compared to healthy individuals and patients with HBV monoinfection of similar age.84 MAIT cells are also functionally impaired and exhibit an activated and exhausted compound phenotype of CD38hiPD-1hiCD28loCD127loPLZFloEomesloHelioslo .84
Similar to chronic HBV and HCV infection,85 it is assumed that specific T cell responses are needed for clearance of HDV and, at the same time, are drivers of histopathologic liver damage.78 There is great variability in the observed frequencies of HDV-specific T cell responses reported in the literature. Grabowski et al. describe HDV-specific cytokine responses after stimulation of peripheral blood mononuclear cells (PBMCs) with a peptide pool spanning the whole HDAg in 94% (16/17) of patients before the start of IFN treatment.86 Nisini et al. detected HDV-specific CD4+ T cell proliferation in 27% of patients after whole-antigen stimulation (8/30),87 whereas Landahl et al. detected CD4+ or CD8+ T cell responses in 53% of patients (17/32) by intracellular cytokine staining after in vitro expansion (ICS).88 Kefalakes et al. report an HDV-specific T cell response rate of 71% by IFN-γ ICS in a sub-cohort of 17 lonafarnib/ritonavir-treated patients after treatment discontinuation.89 Ex vivo frequencies of epitope-specific CD8+ T cells were either undetectable,90,91 detectable at very low frequencies after enrichment92 or reported at 0.013% of CD3+CD4- T cells,89 depending on the epitopes and multimers used.
Even less is known about the phenotype and the clinical correlate of the detection of a broad HDV-specific T cell response. Specific T cell responses have been linked to “inactive disease” – defined as normal alanine aminotransferase (ALT) levels for 1 year and negative anti-HDV IgM.87 A higher frequency of HDV-specific cytokine responses and a restoration of transiently diminished specific cytokine responses after peg-IFN-α treatment coincided with therapeutic response in the HIDIT-1 trial. However, this correlation between HDV-specific IFN-γ levels, ALT levels and HDV RNA was not significant.86 Correlations between elevated serum IL2 and IL12 levels and response to IFN treatment93 suggest that a T helper-1 (Th1)-polarised cellular immune response might be associated with viral clearance.
In contrast, Landahl et al. observed broad low-level HDV-specific T cell responses that did not correlate with HDV viral load, level of transaminases and presence or absence of HDV viraemia. There was also no difference between responses in spontaneous resolvers, treatment-induced PCR-negative patients and chronically viraemic patients, but there was a negative correlation between HBV viral load and number of responses.88 Interestingly, a correlation between activated HDV-specific T cells and aspartate aminotransferase (AST) levels was reported,89 suggesting that CD8+ T cells may contribute to liver damage in HDV infection. Furthermore, higher frequencies of IFN-producing CD8+ T cells were associated with lower viremia 4 weeks post treatment, emphasising their role in viral clearance.89 All in all, the exact interplay between specific T cell responses and treatment outcome or disease course remains unclear.
Cytokine secretion analyses suggest that HDV-specific CD4+ T cell clones belong either to Th1 or Th0 subsets with some cytotoxic capabilities.87 HDV-specific CD8+ T cells were described as non-terminally exhausted memory cells, with a lesser degree of CD8+ T cell exhaustion than in Epstein-Barr virus infection89,91 and as memory-like and “chronically activated, but not terminally differentiated”.89 Some of the epitope-specific memory-like CD8+ T cells were predominantly T cell-specific transcription factor 1 (TCF1)-positive, which was attributed to viral escape mutations resulting in loss of antigen recognition and a relative expansion of TCF1-positive memory-like cells in comparison to relatively diminishing amounts of exhausted effector cells.89,92 Interestingly, a quite similar CD127+PD1+TCF1+ population of epitope-specific CD8+ T cells, sharing characteristics of T cell memory and exhaustion, has been described in HCV.94 In HIV, TCF-1 expression maintains stem-cell like properties of virus-specific CD8+ positive T cells, thereby preventing exhaustion and is linked to the elite controller status.95 Significantly higher frequencies of perforin-positive CD4+ T cells could be observed in chronically HDV-infected patients compared to chronically HCV- or HBV-infected patients and correlated with elevated AST and decreased platelet count in one study.96 However, the antigen specificity of these perforin+CD4+ T cells was not assessed.
HDV-specific CD4+ T cell epitopes
In HCV infection, the detection of strong and long-lasting epitope-specific CD4+ T cells correlates with spontaneous viral clearance of acute HCV, while these responses are nearly absent in patients with chronic HCV.[97], [98], [99], [100] Significantly less is known about the role of CD4+ T cell responses in HBV monoinfection or HDV infection.
However, some studies have longitudinally characterised the HDV CD4+ T cell response during the primary HBV/HDV coinfection or acute HDV superinfection of HBsAg carriers using standardised CD4+ T cell assays (Fig. 3). So far, responses to 18 different HDAg-specific CD4+ T cell epitope specificities have been described in 2 studies,87,88 4 of these epitopes were detected in more than 1 tested individual. Landahl et al. identified 14 epitopes by in vitro enzyme-linked immune spot assay (ELISpot) using overlapping 20mer peptides spanning the whole L-HDAg, followed by ICS for IFN-γ in patients with positive ELISpots results.88 Nearly the whole HDAg was immunogenic for CD4+ responses, however, the hotspots were located towards the N-terminus. One of these, aa71-90, contains the nuclear localisation signal. The CD4+ T cell responses against the 2 N-terminal epitopes aa21-40 and 41-60 were also confirmed in a patient with acute HDV infection using direct ex vivo ELISpot assays.88
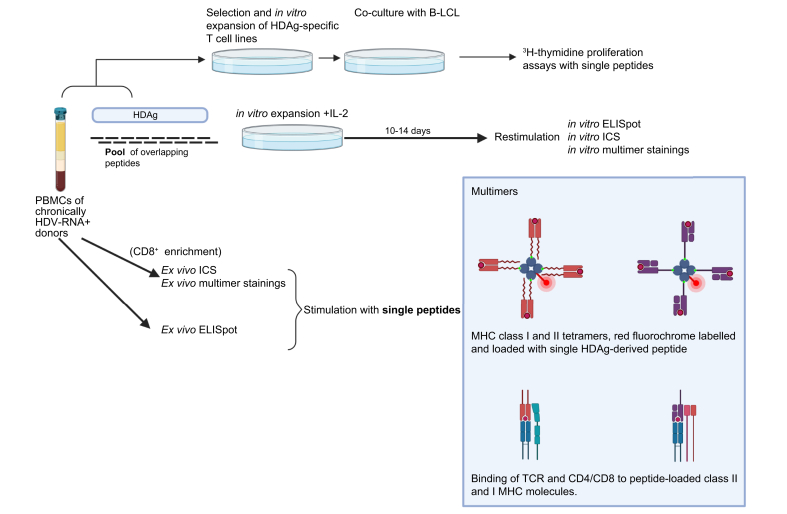
Fig. 3.
Simplified overview of commonly used methods of epitope detection/mapping.
B-LCL, B-lymphoblastoid cell line; ELISpot, enzyme-linked immune spot assay; ICS, intracellular cytokine staining; IL-2, interleukin-2. Based on [87], [88], [89], [90], [91], [92]. Figure created with Biorender.com
For assessment of the HLA restriction of the epitopes, Landahl et al. combined in silico predictions, HLA binding assays and HLA typing of responding patients to suggest likely HLA restrictions. Epitopes aa11-30 and 41-60 seem to bind to multiple HLA molecules in a rather promiscuous fashion. Truncating experiments suggest restriction by DRB1∗11:01 as described by Nisini et al. for an overlapping peptide (see below).
Interestingly, no difference was found in the detection rate, breadth or magnitude of the overall low-level CD4+ HDV-specific T cell responses between PCR-positive and negative patients.88
Nisini et al. tested pools of overlapping 16mer peptides spanning the whole L-HDAg on HDV-specific CD4+ T cell lines derived from 3 chronically HDV-infected patients with inactive disease (defined as normal ALT blood levels and undetectable anti-HDV IgM).87 Epitope-specific T cell proliferation was measured in a 3H-thymidine proliferation assay. The patients were preselected from a larger cohort of 30 patients based on their responsiveness to whole HDAg.
Four epitopes were identified by pool stimulations. Peptides were synthesised according to a genotype 1 sequence. However, the exact sequences of the tested peptides were not provided in the publication.87 HLA restrictions were studied by blocking experiments with anti-DR, anti-DP, and anti-DQ monoclonal antibodies. A B-lymphoblastoid cell line containing known haplotypes was then co-cultured with HDV-specific CD4+ T cell clones to assess their binding to certain HLA molecules.
The epitopes seem to be presented in conjunction with multiple MHC class II molecules, mostly DRB1 subtypes. Nisini et al. also described 13 additional epitopes in the study (with response rates ranging from 1 out of 7 to 3 out of 7 patients) by 3H thymidine proliferation of whole PBMCs derived from patients who responded to stimulation by whole HDAg. Given the stimulation mode, these are most likely CD4+ T cell epitopes, however, this has not been experimentally confirmed.87
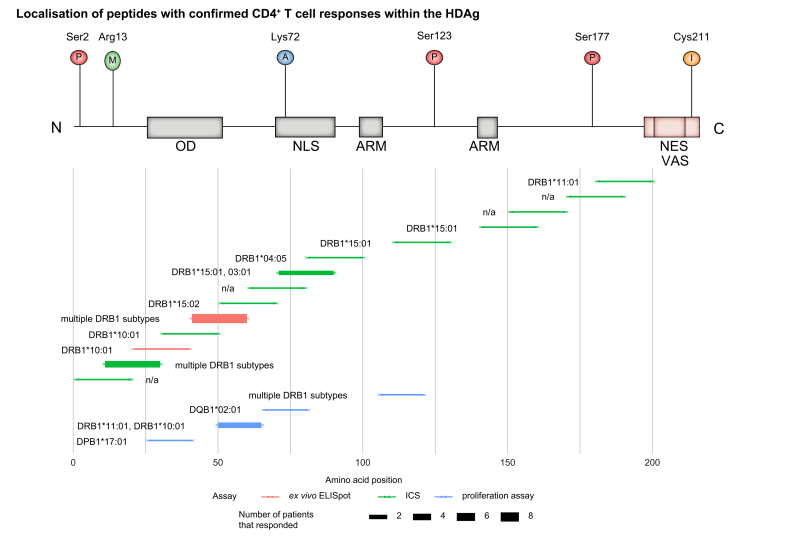
A schematic overview of the CD4+ T cell epitopes and their localisation within the HDAg is provided in Fig. 4.
Fig. 4.
Schematic overview of peptides eliciting CD4+ T cell responses in relation to the L-HDAg.
Bar thickness of lower plot represents number of responding patients, colour represents type of assay performed. Upper pictogram shows PTM sites and functional domains of HDAg, based on 17. ICS, intracellular cytokine staining; L-HDAg, large hepatitis delta antigen.
HDV-specific CD8+ T cell epitopes
It is generally thought that HBV-specific CD8+ cells are the main effector cells responsible for viral clearance during acute HBV infection.101 The role of CD8+ T cells in the different disease courses of patients with chronic HBV is less clear. CD8+ T cells cause liver injury and promote disease pathogenesis. A key event in the persistence of HBV is the exhaustion of virus-specific CD8+ T cells – indeed, diminished frequencies of functionally impaired HBV-specific CD8+ T cells expressing inhibitory receptors have been described. Immune checkpoint inhibition could restore antiviral CD8+ T cells responses.102
The role of the HDV-specific CD8+ T cell response in HDV resolution and pathogenesis remains unclear and most of the data are derived from animal models.66,103,104 Further, definition of the specificities of the anti-HDAg-specific T cells is an essential step towards understanding the heterogenous disease courses of HDV infection, non-responders, and paving the way for an immunotherapeutic approach.
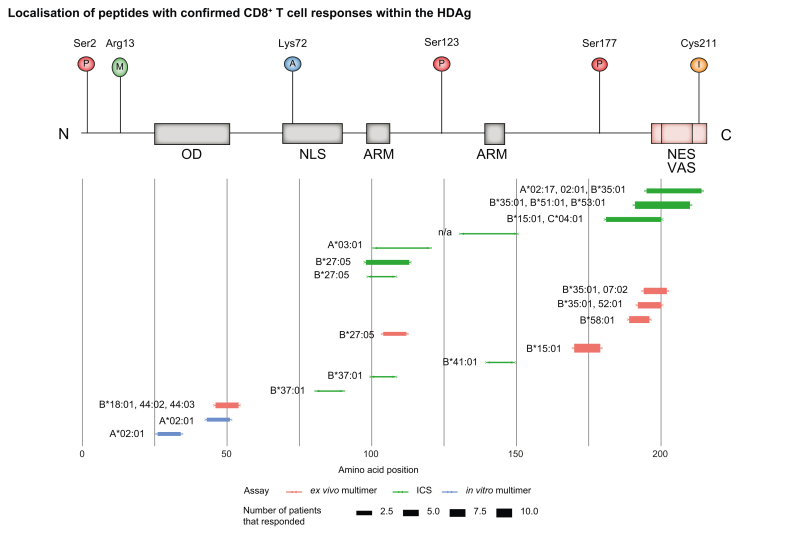
So far, T cell responses directed against 18 HDV-specific CD8+ T cell epitopes have been identified in 5 different studies, with partial overlap between the studied peptides.[88], [89], [90], [91], [92]
Huang et al. described the 2 CD8+ T cell epitopes that were restricted by HLA-A∗02:01 as predicted in silico. Out of 4 HLA-A∗02-positive patients, responses were detected in the 2 PCR-negative patients with normal ALT levels by ELISpot and HLA-A∗02:01 tetramer staining.90 However, these epitopes were not detectable in 4 European HLA-A∗02-positive patients by ICS,91 the overall response rate therefore being 2/8 for each epitope in a total of 2 studies.90,91 Possibly, different HLA-A∗02 subtypes in Asia and Europe may play a role in these discordant results.
Karimzadeh et al. analysed HLA-B∗27-restricted HDV-specific CD8+ T cell responses based on HLA-B∗27-associated sequence polymorphisms (indicating viral escape mutations within a putative HDV-specific CD8+ T cell epitope, see below). By IFNγ ICS, 2 epitopes were confirmed: aa99-108 and aa103/104-112. Three patients with resolved HDV infection responded to these 2 epitopes (1 patient for each epitope plus 1 patient who was only tested for the overlapping peptide containing both epitopes).
In 2019, Karimzadeh et al. expanded their viral sequence-based approach to all HLA class I alleles and described 5 additional HDV-specific CD8+ T cell epitopes by IFN-γ ICS of in vitro expanded CD8+ T cells from HLA-matched patients. Out of these HLA-B restricted novel epitopes, aa170-179 was additionally confirmed by direct ex vivo analysis after HLA-B∗15:01 tetramer enrichment.
Kefalakes et al. 2019 used overlapping peptides spanning the whole L-HDAg to detect HDV-specific CD8+ T cells. They found responses against a total of 6 HDV-specific CD8+ T cell epitopes, including the aforementioned HLA-B∗27 epitope aa104-11291 (with an arginine at the C-terminus), the HLA-B∗18 epitope aa46 - 5492 (additionally restricted by HLA-B∗44:02 and B∗44:03), and 4 additional novel epitopes.89 Most of these epitopes were further confirmed by direct ex vivo multimer staining. Response rates for these epitopes varied, however, responses clustered against epitopes located at the C-terminus of HDAg, which is unique to L-DHAg, with up to 8/17 patients responding to individual overlapping peptides, irrespective of the individual HLA types.
In addition to the identification and characterisation of HDV-specific CD4+ T cell epitopes (see above), Landahl et al. also found responses against 5 HDV-specific CD8+ T cell epitopes. However, fine-mapping and HLA class I restriction experiments were not performed in this study which focused on HDV-specific CD4+ T cell responses. In silico prediction, as well as HLA typing of responding patients, indicated that these responses were most likely restricted by B∗35:01, B∗51:01 and B∗53:01, indicating that the optimal epitope(s) may be identical to the 3 epitopes aa191–196, aa192–200, and aa194–202 identified by Kefalakes et al. in this viral region.89
Aa101-120 contains the B∗27 epitope aa104-112,89,91 interestingly, the responding patient in 88 was HLA-B∗27 negative. Other epitopes also overlap with those identified by the more CD8+ focused studies by Karimzadeh et al. and Kefalakes et al. (see Table 1 and Fig. 5).
Table 1.
Comprehensive overview of all described CD8+and CD4+T cell epitopes of the HDAg.
| Position | Sequence | Ref. | Response rate | Best assay | Assay details | HLA molecule | HLA assay | In silico prediction tool | Comments |
|---|---|---|---|---|---|---|---|---|---|
| CD8 | |||||||||
| 46-54 | DENPWLGNI | 89,92 | 4/24 | Ex vivo multimer | In vitro ICS (IFNγ) release92; ex vivo tetramer ICS89 | B∗18:01 (multimers); B∗44:02, B∗44:03 (HLA binding) | In silico, HLA-matched patients in ICS | ANN, netMHCpan (IEDB; SYFPEITHI; BIMAS | HLA B∗18:01 by multimers, others by binding assays |
| 81-90 | VDSGPRKRPL | 92 | 1/1 | ICS | In vitro ICS (IFNγ) | B∗37:01 | In silico, HLA-matched patients in ICS | ANN, netMHCpan (IEDB; SYFPEITHI; BIMAS | |
| 100-108 | QDHRRRKAL | 92 | 1/1 | ICS | In vitro ICS (IFNγ) | B∗37:01 | In silico, HLA-matched patients in ICS | ANN, netMHCpan (IEDB; SYFPEITHI; BIMAS | |
| 140-149 | RERRVAGPPV | 92 | 1/2 | ICS | In vitro ICS (IFNγ) | B∗41:01 | In silico, HLA-matched patients in ICS | ANN, netMHCpan (IEDB; SYFPEITHI; BIMAS | |
| 170-179 | SMQGVPESPF | 92 | 10/14 | Ex vivo multimer | In vitro ICS (IFNγ), ex vivo tetramer ICS (7/7) | B∗15:01 | Ex vivo tetramer ICS, In silico predicions | ANN, netMHCpan (IEDB; SYFPEITHI; BIMAS | |
| 192-200 | QGFPWDILF | 89 | 5/17 | Ex vivo multimer | In vitro ICS, ex vivo dextramer ICS | B∗35:01; B∗52:01 | HLA binding assays with radiolabelled HLA class I, dextramer | n.a. | HLA 35:01 is confirmed by multimer; QGFPWDMLF is also recognized and presented by both HLA-B∗ subtypes; QGFPWDLLF is presented by A∗02:05 und B∗52:01; aa193-200 GFPWDILF is presented by B∗35:01 |
| 194-202 | FPWDILFPA | 89 | 5/17 | Ex vivo multimer | In vitro ICS, ex vivo dextramer ICS | B∗35:01; B∗07:02 | HLA binding assays with radiolabelled HLA class I, dextramer | n.a. | Both HLA types by multimer; FPWDMLFPA also presented by both HLA types (binding assays), FPWDLLFPA: A∗02:05, B∗07:02 and B∗35:01 (binding assays) |
| 104-112 | RRKALENK/R | 89,92 | 2/17 | Ex vivo multimer | In vitro ICS, ex vivo pentamer ICS; ICS IFNγ release after restimulation on day 12 | B∗27:05 | HLA binding assays with radiolabelled HLA class I, pentamer | ANN, netMHCpan (IEDB; SYFPEITHI; BIMAS | 103-112 RRRKALENKK/R is also presented by HLA B∗27 and recognised by 1/7; escape mutation K106M |
| 189-196 | RGSQGFPW | 89 | 6/17 | Ex vivo multimer | In vitro ICS, ex vivo tetramer ICS | B∗58:01 | HLA binding assays with radiolabelled HLA class I, tetramer | n.a. | |
| 99-108 | RRDHRRRKAL | 91 | 1/7 | ICS | In vitro ICS (IFNγ) | B∗27:05/:02 | UV-mediated peptide exchange assay | IEDB and SYFPEITHI | Escape mutations R105K and K106M; RQDHRRRKAL, REDHRRRKAL, RKDHRRRKAL are also presented by B∗27:05 and recognised by one patient each |
| 98-113 | ERRDHRRRKALE | 91 | 3/8 | ICS | In vitro ICS (IFNγ) | B∗27:05 | In silico, HLA typing in responding patients | IEDB and SYFPEITHI | |
| 26-34 | KLEDLERDL | 90,91 | 2/8 | In vitro multimer | Cytotoxicity (mice) and tetramer staining; ELISPOT IFNγ release, tetramer qualitative binding (both after restimulation); In vitro ICS91 | A∗02:01 | MHC ligand assay, UV-mediatied peptide exchange assay | SYFPEITHI | |
| 43-51 | KLEDENPWL | 90,91 | 2/8 | In vitro multimer | Cytotoxicity (mice) and tetramer staining; ELISpot IFNγ release, tetramer qualitative binding (both after restimulation); In vitro ICS91 | A∗02:01 | MHC ligand assay, UV-mediatied peptide exchange assay | SYFPEITHI | Karimzadeh et al., found no responses in European cohort by ICS (0/4+2/4); only tested in HLA-A∗02:01 patients |
| 191-210 | GQGFPWDILFPS | 88 | 7/32 | ICS | In vitro ICS (IFNγ); ELISpot IFNγ | B∗35:01; B∗51:01; B∗53:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 101-120 | DHRRRKALENKR | 88 | 1/32 | ICS | In vitro ICS (IFNγ); ELISpot IFNγ | A∗03:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 131-150 | KRLTEEDERRER | 88 | 1/32 | ICS | In vitro ICS (IFNγ); ELISpot IFNγ | A∗02:02P/03:01P; B∗15:01P/41:01; C∗03:04/17:01P | HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 181-200 | RHGEGLGVRGG | 88 | 3/32 | ICS | In vitro ICS (IFNγ); ELISpot IFNγ | B∗15:01; C∗04:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 195-214 | PWDILFPSDPPF | 88 | 3/32 | ICS | In vitro ICS (IFNγ); ELISpot IFNγ | A∗02:17/02:01; B∗35:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| CD4 | |||||||||
| 26-41 | Data not provided | 87 | 1/3 | 3H thymidine proliferation | Epitope-specific 3H thymidine proliferation after cultivation+stim with HDAg and coculture of B-LCL as APCs | DPB1∗17:01 | Blocking experiments with MAbs; co-culture with B- LCL of known haplotypes | no in silico predictions used | |
| 50-65 | Data not provided | 87 | 3/3 | 3H thymidine proliferation | Epitope-specific 3H thymidine proliferation after cultivation+stim with HDAg and coculture of B-LCL as APCs | DRB1∗11:01; DRB1∗10:01 | Blocking experiments with MAbs; co-culture with B- LCL of known haplotypes | no in silico predictions used | |
| 66-81 | Data not provided | 87 | 1/3 | 3H thymidine proliferation | Epitope-specific 3H thymidine proliferation after cultivation+stim with HDAg and coculture of B-LCL as APCs | DQB1∗02:01 | Blocking experiments with MAbs; co-culture with B- LCL of known haplotypes | no in silico predictions used | |
| 106-121 | Data not provided | 87 | 1/3 | 3H thymidine proliferation | Epitope-specific 3H thymidine proliferation after cultivation+stim with HDAg and coculture of B-LCL as APCs | DRB1∗11:01; DRB1∗11:02; DRB1∗12:01; DRB1∗01:01; DRB1∗07:01; DRB1∗14:01; DRB5∗02:02 | Blocking experiments with MAbs; co-culture with B- LCL of known haplotypes | no in silico predictions used | |
| 11-30 | GGREEILEQWVN | 88 | 4/32 | ICS | In vitro (IFNγ) | DRB1∗08:02; DRB1∗10:01; DRB1∗14:01; DRB1∗15:01 | In silico predictions + MHC ligand assay | IEDB Consensus tool (ANN+SMM) | |
| 41-60 | IKKLEDENPWLG | 88 | 8/32 | Ex vivo ELISpot | in vitro ICS (IFNγ); ex vivo ELISpot IFNγ | DRB1∗10:01; DRB1∗11:01; DRB1∗08:02; DRB1∗13:02 | In silico predictions+MHC ligand assay | IEDB Consensus tool (ANN+SMM) | Confirmed by ex vivo ELISpot in acutely superinfected patient |
| 1-20 | MSRSESKKNRG | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗14:04/15:01; DQA1∗01:04/01:02; DGB1∗05:03/06:02P | HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 21-40 | VNGRKKLEELER | 88 | 1/32 | Ex vivo ELISpot | in vitro ICS (IFNγ); ex vivo ELISpot IFNγ | DRB1∗10:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | Confirmed by ex vivo ELISpot in acutely superinfected patient |
| 31-50 | ERDLRKIKKKIKK | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗10:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 51-70 | LGNIKGILGKKDK | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗15:02 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 61-80 | KDKDGEGAPPA | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗11:01P; DQA1∗05; DQB1∗03:01P | HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 71-90 | AKRARTDQMEID | 88 | 2/32 | ICS | in vitro ICS (IFNγ) | DRB1∗15:01/03:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 81-100 | IDSGPRKRPLRG | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗04:05 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 111-130 | KRKQLAGGGKSL | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗15:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 141-160 | ERRVAGPQVGG | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗15:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 151-170 | GVNPLEGGSRG | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗11:04/13:03; DQA1∗05; DQB1∗03:01P | HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 171-190 | MQGVPESPFTRH | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗11:04/13:03; DQA1∗05; DQB1∗03:01P | HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
| 181-200 | RHGEGLGVRGG | 88 | 1/32 | ICS | in vitro ICS (IFNγ) | DRB1∗11:01 | In silico, HLA typing in responding patients | IEDB Consensus tool (ANN+SMM) | |
HDAg, hepatitis delta antigen; ICS, intracellular cytokine staining.
Fig. 5.
Schematic overview of peptides eliciting CD8+ T cell responses in relation to the L-HDAg.
Bar thickness of lower plot represents number of responding patients, colour represents type of assay performed. Upper pictogram shows PTM sites and functional domains of HDAg, based on 17. ICS, intracellular cytokine staining; L-HDAg, large hepatitis delta antigen; PTM, post-translation modification.
In contrast to CD4+ epitopes that are distributed through the entire HDAg (with hotspots at the N-terminus) and can be presented by multiple HLA types, the known HDV-specific CD8+ T cell epitopes seem to cluster in a few distinct locations (mainly C-terminal) and are restricted mainly by HLA-B subtypes, including relatively infrequent subtypes.
A schematic overview of the CD8+ T cell epitopes and their localisation within the HDAg is provided in Fig. 5.
Viral escape from HDV-specific CD8+ T cell responses
Mutational viral escape is a major mechanism of virus-specific T cell failure in persistent viral infections. Viral escape was first described in the lymphocytic choriomeningitis virus mouse model,105 and has been best characterised in HIV and HCV infection in humans.106 While there is good evidence that viral escape from virus-specific CD8+ T cell responses impacts the outcome of infection, e.g. viral clearance vs. persistence, there is less evidence for a role of viral escape from virus-specific CD4+ T cell responses. Mutational escape can either occur at HLA binding anchors of epitopes (mostly aa residue 2 and the C-terminal aa residue), at the T cell receptor contact residues of the epitope (mostly the aa residues in the middle of the epitope), or even in the flanking regions of the epitope, interfering with proteasomal processing of the epitope.106 Since HDV has a relatively high mutation rate (see above), it is reasonable to argue that viral escape from virus-specific CD8+ T cell responses may also take place in persistent HDV infection. Indeed, a first study on this issue longitudinally sequenced HDV quasispecies in 4 HLA-A∗02+ patients with chronic HBV/HDV infection before and after hepatitis flares. They found evidence for selection pressure in the HLA-A∗02-restricted epitope aa43-51 KLEDDNPWL in 3/5 patients and in another predicted HLA-A∗02-restricted epitope aa114-122 QLSAGGKSL in 4/5 patients.107 However, functional T cell assays to confirm that the patients indeed targeted these epitopes were not performed and experimental evidence for the impact of the observed sequence mutations on recognition by epitope-specific CD8+ T cells or at least HLA-A∗02 binding was not supplied. Similarly, a more recent study performed a cross-sectional sequence analysis in 34 patients with chronic HBV/HDV infection and identified codons 136-159 to be under positive selection pressure, likely indicating CD8+ or B cell pressure.43 Unfortunately, HLA typing is not available. The HDAg region under positive selection pressure (aa136-159) overlaps with the HLA-B∗41-restricted epitope aa140-149 RERRVAGPPV, however, this CD8+ T cell epitope (restricted by a rather infrequent HLA class I allele) is unlikely to exert selection pressure at a population level. Due to the dominant role of the HLA class I type B∗27 in driving viral escape in HIV and HCV infection, the HLA-B∗27 background was also used in a pioneer study to functionally demonstrate viral escape from HDV-specific CD8+ T cell responses.91 Indeed, 2 predicted HLA-B∗27-restricted epitopes (aa99-108 RRDHRRRKAL and aa103/4-112 (R)RRKALENKK) were identified. Viral sequence analysis in 8 HLA-B∗27+ vs. 96 HLA-B∗27- patients demonstrated an enrichment of aa mutations in the epitope region in HLA-B∗27+ patients. These HLA-B∗27-associated viral sequence polymorphisms (also referred to as HLA-B∗27 footprints) indicated that viral escape occurs within these 2 HLA-B∗27-restricted HDV-specific CD8+ T cell epitopes. This was functionally confirmed by intracellular IFN-γ staining using wild-type vs. variant peptide, showing little cross-recognition of the variant peptide by wild-type-specific T cell lines.91 In a following multicentre, multinational study analysing HLA footprints for all HLA class I alleles present in a cohort of 104 patients with chronic HBV/HDV infection, a total of 21 HLA class I footprints were identified.92 Interestingly, these footprints were restricted by relatively infrequent HLA class I alleles, which might indicate that HDV has already adapted to its host’s HLA class I background at a population level, leading to the extinction of HDV-specific CD8+ T cell epitopes restricted by frequent HLA class I alleles. The most striking example of viral escape affected the HLA-B∗15:01-restricted HDV-specific CD8+ T cell epitope aa170-179 SMQGVPESPF: All 8 HLA-B∗15:01+ patients in the cohort displayed a viral sequence mutation at the N-terminal amino acid residue (S170N) that was detected in a minority of HLA-B∗15:01-negative patients only. This mutation impaired cross-recognition by the epitope-specific CD8+ T cell response. The loss of viral control in a patient with acute HDV superinfection coincided with the evolution of this escape mutation, indicating the biological significance of viral escape in HDV persistence. Virus-specific CD8+ T cells targeting this ‘escaped’ epitope displayed a memory-like phenotype (PD-1+CD127+TCF1+) and were thus not terminally exhausted. These data indicate that in parallel to the findings obtained in HCV infection,108,109 viral escape and terminal exhaustion are alternative and non-overlapping mechanisms of virus-specific T cell failure. Unfortunately, it was not possible to analyse the phenotype of HDV-specific CD8+ T cells targeting conserved (‘non-escaped’) epitopes in this study. However, Kefalakes et al.89 described viral sequence mutations in all 6 HDV-specific CD8+ T cell epitopes identified in 1-4 patients each and could confirm viral escape by HLA binding studies for 6 variant peptides and by functional T cell analysis for 4 variant peptides, respectively. Importantly, HDV-specific CD8+ T cells targeting escaped epitopes displayed a memory-like phenotype (PD-1+CD127+TCF+) without evidence of activation (CD38-), while HDV-specific CD8+ T cells targeting conserved epitopes had a ‘chronically activated’ phenotype (PD-1+CD127lowTCF1low, CD38+). These results further underline the complementary, non-overlapping roles of viral escape and terminal exhaustion in HDV-specific CD8+ T cell failure. In sum, these results highlight an important role of viral escape in HDV persistence and indicate that viral escape needs to be considered in vaccine design.
Discussion
Forty years after the discovery of HDV, its clinical peculiarities remain enigmatic. Clearly, the fate of the HDV infection is intricately intertwined with the replicative cycle and the clinical course of the HBV infection, since HBsAg loss and HBV seroconversion will ultimately terminate HDV propagation. While the current review focuses on the HDV-specific T cell response, the functional interactions between the 2 viruses and their respective effect on HBV- and HDV-specific adaptive and innate immunity must be studied in much more detail. For example, it is not clear why peg-IFN-α leads to relatively infrequent loss of the HBsAg in patients with HDV compared to those with HBV monoinfection. The immune correlates of spontaneous or therapy-induced control of HDV viraemia – apart from the known fact that HBsAg conversion itself can lead to clearance of HDV – and the role HDV-specific T cell responses play are poorly understood.
It is not clear whether immunological interventions in patients with HDV should a) aim at global blockade of co-inhibitory molecules on T cells, or b) be directly aimed at enhancing the HBV-specific immunity to achieve an HBV seroconversion; it is also not clear whether additionally targeting HDV antigens would synergistically help to achieve this aim. We also do not know whether halting HDV replication on its own – by therapeutic HDV vaccination or antiviral therapy103 – would be of significant benefit for chronically HBV/HDV-coinfected patients. Lastly, it is not currently clear whether a prophylactic HDV vaccine would be an epidemiologically useful tool to eradicate HDV infection.66,103
Nonetheless, it is essential to conduct further detailed longitudinal studies on the ex vivo phenotype and functionality of the HBV- and HDV-specific T cell response during therapeutic trials to understand the immunological correlates of HDV viral control and to achieve sustained virological responses in the majority of patients with HDV by application of antiviral and immunological combination therapies.
Only some of the recent studies included detailed T cell analysis and only 3 studies utilised ex vivo assays like MHC class I multimer stainings. Indeed, due to the generally low ex vivo frequencies of circulating HDV-specific T cells,[87], [88], [89] many researchers use peptide pool stimulation, measuring HDV-specific responses after in vitro expansion and re-stimulation. Epitope mapping by stimulating T cells with different pools of peptides spanning the whole antigen rather than testing each peptide individually greatly increases efficiency, while assay sensitivity may be slightly reduced, especially when using larger pool sizes.110 In vitro expansion and subsequent re-stimulation help to identify responses at low frequencies but carry the risk of altered T cell phenotypes and functionalities, limiting the comparability to the in vivo situation. Only 3 studies could detect T cell responses directed against 8 different peptide epitopes by direct ex vivo staining. Landahl et al. detected up to 800 HDV-specific T cells/106 PBMCs by ex vivo ELISpot in a patient with acute HDV88 and Kefalakes et al. performed ex vivo multimer stainings in 17 chronically HDV-infected patients after discontinuation of lonafarnib/ritonavir therapy.89 Karimzadeh et al. managed to detect responses against 2 epitopes ex vivo by bead-based CD8+T cell-enrichment, thereby increasing the assay sensitivity92 in HLA-matched patients. It is conceivable that a broad and strong specific response is induced in acutely infected patients, which diminishes as the infection persists. Analogously, suppression of viral replication by therapy could lead to partial recovery of exhausted specific memory cells, enabling them to initiate stronger, multi-specific responses upon re-stimulation ex vivo, similar to chronic HBV and HCV.85
HLA restriction is an additional insufficiently characterised aspect of HDV epitopes. None of the studies describing CD4+ T cell epitopes confirmed the HLA restriction experimentally, e.g. using multimer stainings, but rather relied on in silico predictions followed by antibody blocking and HLA-haplotype specific co-culturing of CD4+ T cells, or in vitro binding assays or HLA fine typing of responding patients. Regarding CD8+ T cell epitopes, 3 studies included multimer assays to confirm HLA restrictions, with 6 epitopes confirmed by direct ex vivo multimer staining.89,92
Another aspect which limits the current evidence base is the fact that only 4 studies mapped the whole HDAg for T cell epitopes.[87], [88], [89],91 Consequently, only a limited number of patients (8 in Nisini et al. 1997, 32 in Landahl et al. 2019, 4 in Karimzadeh et al. 2018, and 17 in Kefalakes et al. 2019) were included in complete mappings. Two studies – Kefalakes et al. 2019 and Karimzadeh et al. 2018 – focused on CD8+ T cell responses (including 21 patients in total). Landahl et al. 2019 aimed to measure both CD8+ and CD4+ T cell responses, although the peptide length of 20 aa is suboptimal for MHC class I presentation, which favour peptides of 8 to 10 aa,[111], [112], [113] and Nisini et al. only measured CD4+ responses. Karimzadeh et al. mapped HDAg for CD8+ T cell responses by IFN-γ ICS, a high-quality method for epitope mapping, although limited by the small number of patients. In this sense, a particular strength of the study conducted by Kefalakes et al. 2019 is the relatively large number of individual patients mapped by a high-quality method (namely ICS). Additionally, the authors were able to confirm epitopes and HLA binding by ex vivo multimer stainings of untreated HBV/HDV patients, and even to further characterise HDV-specific CD8+ T cells as discussed earlier in this review. Other studies performed in silico predictions of most probable epitopes and their HLA restrictions, which were subsequently experimentally confirmed. This approach carries the inherent risk of missing responses to epitopes with low binding affinities or restriction by uncommon HLA types. Usage of alternative binding pockets in MHC class I molecules and generally shallower (and thus more variable) binding pockets in MHC class II molecules further complicates this approach.114 Additionally, CD8+ T cell epitopes may span longer aa sequences than classically assumed, and thus be overlooked by in silico predictions presuming lengths of 8 to 10 aa.111
Of note, most HDV peptide sets used to experimentally screen for HDV-specific T cell responses are based on single genotype 1-based sequences and there is neither a consensus sequence available nor is there an understanding about the degree of cross-genotype reactivity of these epitopes. Optimally, these peptides should be based on, or compared with, autologous circulating HDV sequences to rule out T cell responses against suboptimal heterologous sequence variants.115,116
Furthermore, studies analysing the breadth, specificity, and functionality of the intrahepatic HDV-specific CD8+ T cell response have not yet been performed.
In summary, we have provided a detailed review of the current knowledge on HDV-specific T cells and a database of all human T cell epitopes of the hepatitis delta virus characterised to date. This evidence base will help to further elucidate the complicated immunology of this enigmatic viral infection that still has grave clinical implications for too many patients.
Financial support
DFG (German Research Foundation) grants to Julian Schulze zur Wiesch (SFB841 and SFB1328) and Christoph Neumann-Haefelin (TRR-179 “Determinants and dynamics of elimination versus persistence of hepatitis virus infection”, Project 02). German Center for Infection Research (DZIF) grants to Julian Schulze zur Wiesch and Christoph Neumann-Haefelin.
Authors’ contributions
Conception MK, JL and JSzW; Draft MK, JSzW and CNH; Proofread all authors, important contributions all authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgments
The authors thank Janna Heide for helpful advice on visualisation and tabular summary of epitopes.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100294.
Supplementary data
The following is the supplementary data to this article:
References
- 1.Stockdale A.J., Kreuels B., Henrion M.Y.R., Giorgi E., Kyomuhangi I., de Martel C. The global prevalence of hepatitis D virus infection: systematic review and meta-analysis. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao Z., Pan Q. Revisiting the estimation of hepatitis D global prevalence. J Hepatol. 2020;73(5):1279–1280. doi: 10.1016/j.jhep.2020.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Stockdale A.J., Kreuels B., Henrion M.R.Y., Giorgi E., Kyomuhangi I., Geretti A.M. Hepatitis D prevalence: problems with extrapolation to global population estimates. Gut. 2020;69(2):396–397. doi: 10.1136/gutjnl-2018-317874. [DOI] [PubMed] [Google Scholar]
- 4.Chen H.Y., Shen D.T., Ji D.Z., Han P.C., Zhang W.M., Ma J.F. Prevalence and burden of hepatitis D virus infection in the global population: a systematic review and meta-analysis. Gut. 2019;68(3):512–521. doi: 10.1136/gutjnl-2018-316601. [DOI] [PubMed] [Google Scholar]
- 5.Hughes S.A., Wedemeyer H., Harrison P.M. Hepatitis delta virus. Lancet. 2011;378(9785):73–85. doi: 10.1016/S0140-6736(10)61931-9. [DOI] [PubMed] [Google Scholar]
- 6.Farci P., Niro G.A. Clinical features of hepatitis D. Semin Liver Dis. 2012;32(3):228–236. doi: 10.1055/s-0032-1323628. [DOI] [PubMed] [Google Scholar]
- 7.Spaan M., Carey I., Bruce M., Shang D., Horner M., Dusheiko G. Hepatitis delta genotype 5 is associated with favourable disease outcome and better response to treatment compared to genotype 1. J Hepatol. 2020;72(6):1097–1104. doi: 10.1016/j.jhep.2019.12.028. [DOI] [PubMed] [Google Scholar]
- 8.Roulot D., Brichler S., Layese R., BenAbdesselam Z., Zoulim F., Thibault V. Origin, HDV genotype and persistent viremia determine outcome and treatment response in patients with chronic hepatitis Delta. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 9.Alfaiate D., Clement S., Gomes D., Goossens N., Negro F. Chronic hepatitis D and hepatocellular carcinoma: a systematic review and meta-analysis of observational studies. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Bockmann J.H., Grube M., Hamed V., von Felden J., Landahl J., Wehmeyer M. High rates of cirrhosis and severe clinical events in patients with HBV/HDV co-infection: longitudinal analysis of a German cohort. BMC Gastroenterol. 2020;20(1):24. doi: 10.1186/s12876-020-1168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization . 2017. Global hepatitis report 2017.https://apps.who.int/iris/bitstream/handle/10665/255016/9789241565455-eng.pdf Geneva. jsessionid=652F8A0ADA93EBC241298C96B2A25583?sequence=1 16/11/2020. [Google Scholar]
- 12.Zhang Z., Urban S. Interplay between hepatitis D virus and the interferon response. Viruses. 2020;12(11) doi: 10.3390/v12111334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabernero D., Cortese M.F., Buti M., Rodriguez-Frias F. HDV evolution-will viral resistance be an issue in HDV infection? Curr Opin Virol. 2018;32:100–107. doi: 10.1016/j.coviro.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Vargas J., Amirache F., Boson B., Mialon C., Freitas N., Sureau C. Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat Commun. 2019;10(1):2098. doi: 10.1038/s41467-019-10117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chemin I., Pujol F.H., Scholtes C., Loureiro C.L., Amirache F., Levrero M. Preliminary evidence for hepatitis delta virus exposure in patients who are apparently not infected with hepatitis B virus. Hepatology. 2021;73(2):861–864. doi: 10.1002/hep.31453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfluger L.S., Schulze Zur Wiesch J., Polywka S., Lutgehetmann M. Hepatitis delta virus propagation enabled by hepatitis C virus-Scientifically intriguing, but is it relevant to clinical practice? J Viral Hepat. 2020 doi: 10.1111/jvh.13385. [DOI] [PubMed] [Google Scholar]
- 17.Alfaiate D., Deny P., Durantel D. Hepatitis delta virus: from biological and medical aspects to current and investigational therapeutic options. Antivir Res. 2015;122:112–129. doi: 10.1016/j.antiviral.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Sureau C., Negro F. The hepatitis delta virus: replication and pathogenesis. J Hepatol. 2016;64(1 Suppl):S102–S116. doi: 10.1016/j.jhep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Abeywickrama-Samarakoon N., Cortay J.C., Sureau C., Muller S., Alfaiate D., Guerrieri F. Hepatitis Delta Virus histone mimicry drives the recruitment of chromatin remodelers for viral RNA replication. Nat Commun. 2020;11(1):419. doi: 10.1038/s41467-020-14299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudima S., Wu S.Y., Chiang C.M., Moraleda G., Taylor J. Origin of hepatitis delta virus mRNA. J Virol. 2000;74(16):7204–7210. doi: 10.1128/jvi.74.16.7204-7210.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh S.Y., Chao M., Coates L., Taylor J. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J Virol. 1990;64(7):3192–3198. doi: 10.1128/jvi.64.7.3192-3198.1990. https://www.ncbi.nlm.nih.gov/pubmed/1693700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greco-Stewart V., Pelchat M. Interaction of host cellular proteins with components of the hepatitis delta virus. Viruses. 2010;2(1):189–212. doi: 10.3390/v2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor J.M.P., R H., Farci P. Hepatitis D virus. In: Knipe D.M., Howley P.M., editors. Fields virology. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 2222–2241. [Google Scholar]
- 24.Tsagris E.M., Martinez de Alba A.E., Gozmanova M., Kalantidis K. Viroids. Cell Microbiol. 2008;10(11):2168–2179. doi: 10.1111/j.1462-5822.2008.01231.x. [DOI] [PubMed] [Google Scholar]
- 25.Taylor J.M. Hepatitis D virus replication. Cold Spring Harb Perspect Med. 2015;5(11) doi: 10.1101/cshperspect.a021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botelho-Souza L.F., Vasconcelos M.P.A., Dos Santos A.O., Salcedo J.M.V., Vieira D.S. Hepatitis delta: virological and clinical aspects. Virol J. 2017;14(1):177. doi: 10.1186/s12985-017-0845-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polson A.G., Bass B.L., Casey J.L. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380(6573):454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 28.Casey J.L. Control of ADAR1 editing of hepatitis delta virus RNAs. Curr Top Microbiol Immunol. 2012;353:123–143. doi: 10.1007/82_2011_146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao M., Hsieh S.Y., Taylor J. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64(10):5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. https://www.ncbi.nlm.nih.gov/pubmed/2398535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai M.M. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J Virol. 2005;79(13):7951–7958. doi: 10.1128/JVI.79.13.7951-7958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C.Z., Chen P.J., Lai M.M., Chen D.S. Isoprenylation of large hepatitis delta antigen is necessary but not sufficient for hepatitis delta virus assembly. Virology. 1994;199(1):169–175. doi: 10.1006/viro.1994.1109. [DOI] [PubMed] [Google Scholar]
- 32.Hwang S.B., Lai M.M. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J Virol. 1994;68(5):2958–2964. doi: 10.1128/jvi.68.5.2958-2964.1994. https://www.ncbi.nlm.nih.gov/pubmed/7512154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alves C., Freitas N., Cunha C. Characterization of the nuclear localization signal of the hepatitis delta virus antigen. Virology. 2008;370(1):12–21. doi: 10.1016/j.virol.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 34.Lee C.H., Chang S.C., Wu C.H., Chang M.F. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J Biol Chem. 2001;276(11):8142–8148. doi: 10.1074/jbc.M004477200. [DOI] [PubMed] [Google Scholar]
- 35.Xia Y.P., Lai M.M. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J Virol. 1992;66(11):6641–6648. doi: 10.1128/jvi.66.11.6641-6648.1992. https://www.ncbi.nlm.nih.gov/pubmed/1404608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Gal F., Brichler S., Drugan T., Alloui C., Roulot D., Pawlotsky J.M. Genetic diversity and worldwide distribution of the deltavirus genus: a study of 2,152 clinical strains. Hepatology. 2017;66(6):1826–1841. doi: 10.1002/hep.29574. [DOI] [PubMed] [Google Scholar]
- 37.Karimzadeh H., Usman Z., Frishman D., Roggendorf M. Genetic diversity of hepatitis D virus genotype-1 in Europe allows classification into subtypes. J Viral Hepat. 2019;26(7):900–910. doi: 10.1111/jvh.13086. [DOI] [PubMed] [Google Scholar]
- 38.Thomas M.J., Platas A.A., Hawley D.K. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93(4):627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 39.Chang J., Gudima S.O., Taylor J.M. Evolution of hepatitis delta virus RNA genome following long-term replication in cell culture. J Virol. 2005;79(21):13310–13316. doi: 10.1128/JVI.79.21.13310-13316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homs M., Rodriguez-Frias F., Gregori J., Ruiz A., Reimundo P., Casillas R. Evidence of an exponential decay pattern of the hepatitis delta virus evolution rate and fluctuations in quasispecies complexity in long-term studies of chronic delta infection. PloS One. 2016;11(6) doi: 10.1371/journal.pone.0158557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C.M., Bih F.Y., Chao Y.C., Govindarajan S., Lai M.M. Evolution of hepatitis delta virus RNA during chronic infection. Virology. 1992;188(1):265–273. doi: 10.1016/0042-6822(92)90756-f. [DOI] [PubMed] [Google Scholar]
- 42.Krushkal J., Li W.H. Substitution rates in hepatitis delta virus. J Mol Evol. 1995;41(6):721–726. doi: 10.1007/bf00173151. [DOI] [PubMed] [Google Scholar]
- 43.Shirvani-Dastgerdi E., Amini-Bavil-Olyaee S., Alavian S.M., Trautwein C., Tacke F. Comprehensive analysis of mutations in the hepatitis delta virus genome based on full-length sequencing in a nationwide cohort study and evolutionary pattern during disease progression. Clin Microbiol Infect. 2015;21(5):510 e11–23. doi: 10.1016/j.cmi.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z., Filzmayer C., Ni Y., Sultmann H., Mutz P., Hiet M.S. Hepatitis D virus replication is sensed by MDA5 and induces IFN-beta/lambda responses in hepatocytes. J Hepatol. 2018;69(1):25–35. doi: 10.1016/j.jhep.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 45.Hartwig D., Schoeneich L., Greeve J., Schutte C., Dorn I., Kirchner H. Interferon-alpha stimulation of liver cells enhances hepatitis delta virus RNA editing in early infection. J Hepatol. 2004;41(4):667–672. doi: 10.1016/j.jhep.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Hartwig D., Schutte C., Warnecke J., Dorn I., Hennig H., Kirchner H. The large form of ADAR 1 is responsible for enhanced hepatitis delta virus RNA editing in interferon-alpha-stimulated host cells. J Viral Hepat. 2006;13(3):150–157. doi: 10.1111/j.1365-2893.2005.00663.x. [DOI] [PubMed] [Google Scholar]
- 47.European Association for the Study of the Liver EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(2):370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 48.Heidrich B., Yurdaydin C., Kabacam G., Ratsch B.A., Zachou K., Bremer B. Late HDV RNA relapse after peginterferon alpha-based therapy of chronic hepatitis delta. Hepatology. 2014;60(1):87–97. doi: 10.1002/hep.27102. [DOI] [PubMed] [Google Scholar]
- 49.Wranke A., Hardtke S., Heidrich B., Dalekos G., Yalcin K., Tabak F. Ten-year follow-up of a randomized controlled clinical trial in chronic hepatitis delta. J Viral Hepat. 2020 doi: 10.1111/jvh.13366. [DOI] [PubMed] [Google Scholar]
- 50.Wedemeyer H., Yurdaydin C., Hardtke S., Caruntu F.A., Curescu M.G., Yalcin K. Peginterferon alfa-2a plus tenofovir disoproxil fumarate for hepatitis D (HIDIT-II): a randomised, placebo controlled, phase 2 trial. Lancet Infect Dis. 2019;19(3):275–286. doi: 10.1016/S1473-3099(18)30663-7. [DOI] [PubMed] [Google Scholar]
- 51.Bremer B., Anastasiou O., Hardtke S., Caruntu F.A., Curescu M.G., Yalcin K. Residual low HDV viremia is associated HDV RNA relapse after PEG-IFNa-based antiviral treatment of hepatitis delta: results from the HIDIT-II study. Liver Int. 2020 doi: 10.1111/liv.14740. [DOI] [PubMed] [Google Scholar]
- 52.Wranke A., Wedemeyer H. Antiviral therapy of hepatitis delta virus infection - progress and challenges towards cure. Curr Opin Virol. 2016;20:112–118. doi: 10.1016/j.coviro.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Koh C., Da B.L., Glenn J.S. HBV/HDV coinfection: a challenge for therapeutics. Clin Liver Dis. 2019;23(3):557–572. doi: 10.1016/j.cld.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogomolov P., Alexandrov A., Voronkova N., Macievich M., Kokina K., Petrachenkova M. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: first results of a phase Ib/IIa study. J Hepatol. 2016;65(3):490–498. doi: 10.1016/j.jhep.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Wedemeyer H., Bogomolov P., Blank A., Allweiss L., Dandri-Petersen M., Bremer B. GS-005 - final results of a multicenter, open-label phase 2b clinical trial to assess safety and efficacy of Myrcludex B in combination with Tenofovir in patients with chronic HBV/HDV co-infection. J Hepatol. 2018;68:S3. doi: 10.1016/S0168-8278(18)30224-1. [Oral presentation EASL 04/2018] [DOI] [Google Scholar]
- 56.European Medicines Agency . An overview of Hepcludex and why it is authorised in the EU. European Medicines Agency; The Netherlands: 2020. Hepcludex (bulevirtide) pp. 1–3.https://www.ema.europa.eu/en/medicines/human/EPAR/hepcludex 16.08.2020 [Google Scholar]
- 57.Kang C., Syed Y.Y. Bulevirtide: first approval. Drugs. 2020;80(15):1601–1605. doi: 10.1007/s40265-020-01400-1. [DOI] [PubMed] [Google Scholar]
- 58.Caviglia G.P., Rizzetto M. Treatment of hepatitis D, an unmet medical need. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.02.031. [DOI] [PubMed] [Google Scholar]
- 59.Etzion O., Hamid S.S., Lurie Y., Gane E., Bader N., Yardenia D. End of study results from LIMT HDV study: 36% durable virologic response at 24 weeks post-treatment with pegylated interferon lambda monotherapy in patients with chronic hepatitis delta virus infection. J Hepatol. 2019;70(1):e32. doi: 10.1016/S0618-8278(19)30058-1. [oral presentation]. [Oral presentation EASL 04/2019] [DOI] [Google Scholar]
- 60.Bazinet M., Pantea V., Cebotarescu V., Cojuhari L., Jimbei P., Albrecht J. Safety and efficacy of REP 2139 and pegylated interferon alfa-2a for treatment-naive patients with chronic hepatitis B virus and hepatitis D virus co-infection (REP 301 and REP 301-LTF): a non-randomised, open-label, phase 2 trial. Lancet Gastroenterol Hepatol. 2017;2(12):877–889. doi: 10.1016/S2468-1253(17)30288-1. [DOI] [PubMed] [Google Scholar]
- 61.Terrault N.A., Lok A.S.F., McMahon B.J., Chang K.M., Hwang J.P., Jonas M.M. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67(4):1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Engle R.E., De Battista D., Danoff E.J., Nguyen H., Chen Z., Lusso P. Distinct cytokine profiles correlate with disease severity and outcome in longitudinal studies of acute hepatitis B virus and hepatitis D virus infection in chimpanzees. mBio. 2020;11(6) doi: 10.1128/mBio.02580-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Y., Liu D., Feng D., Tang H., Li Y., You X. An animal study on transmission of hepatitis B virus through mosquitoes. Chin Med J (Engl) 1995;108(12):895–897. https://www.ncbi.nlm.nih.gov/pubmed/8728939 [PubMed] [Google Scholar]
- 64.Guilhot S., Huang S.N., Xia Y.P., La Monica N., Lai M.M., Chisari F.V. Expression of the hepatitis delta virus large and small antigens in transgenic mice. J Virol. 1994;68(2):1052–1058. doi: 10.1128/jvi.68.2.1052-1058.1994. https://www.ncbi.nlm.nih.gov/pubmed/8289334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D., Pearlberg J., Liu Y.T., Ganem D. Deleterious effects of hepatitis delta virus replication on host cell proliferation. J Virol. 2001;75(8):3600–3604. doi: 10.1128/JVI.75.8.3600-3604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fiedler M., Roggendorf M. Immunology of HDV infection. Curr Top Microbiol Immunol. 2006;307:187–209. doi: 10.1007/3-540-29802-9_10. [DOI] [PubMed] [Google Scholar]
- 67.Giersch K., Helbig M., Volz T., Allweiss L., Mancke L.V., Lohse A.W. Persistent hepatitis D virus mono-infection in humanized mice is efficiently converted by hepatitis B virus to a productive co-infection. J Hepatol. 2014;60(3):538–544. doi: 10.1016/j.jhep.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 68.Schaper M., Rodriguez-Frias F., Jardi R., Tabernero D., Homs M., Ruiz G. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J Hepatol. 2010;52(5):658–664. doi: 10.1016/j.jhep.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 69.Anastasiou O.E., Yurdaydin C., Maasoumy B., Hardtke S., Alexandru Caruntu F., Curescu M.G. A transient early HBV DNA increase during PEG-IFNalpha therapy of hepatitis D indicates loss of infected cells and is associated with HDV RNA and HBsAg reduction. J Viral Hepat. 2020 doi: 10.1111/jvh.13439. [DOI] [PubMed] [Google Scholar]
- 70.Alfaiate D., Lucifora J., Abeywickrama-Samarakoon N., Michelet M., Testoni B., Cortay J.C. HDV RNA replication is associated with HBV repression and interferon-stimulated genes induction in super-infected hepatocytes. Antivir Res. 2016;136:19–31. doi: 10.1016/j.antiviral.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 71.Giersch K., Allweiss L., Volz T., Helbig M., Bierwolf J., Lohse A.W. Hepatitis Delta co-infection in humanized mice leads to pronounced induction of innate immune responses in comparison to HBV mono-infection. J Hepatol. 2015;63(2):346–353. doi: 10.1016/j.jhep.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 72.He W., Ren B., Mao F., Jing Z., Li Y., Liu Y. Hepatitis D virus infection of mice expressing human sodium taurocholate co-transporting polypeptide. Plos Pathog. 2015;11(4) doi: 10.1371/journal.ppat.1004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suarez-Amaran L., Usai C., Di Scala M., Godoy C., Ni Y., Hommel M. A new HDV mouse model identifies mitochondrial antiviral signaling protein (MAVS) as a key player in IFN-beta induction. J Hepatol. 2017;67(4):669–679. doi: 10.1016/j.jhep.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 74.Belloni L., Allweiss L., Guerrieri F., Pediconi N., Volz T., Pollicino T. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122(2):529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tropberger P., Mercier A., Robinson M., Zhong W., Ganem D.E., Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci U S A. 2015;112(42):E5715–E5724. doi: 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tham C.Y.L., Kah J., Tan A.T., Volz T., Chia A., Giersch K. Hepatitis delta virus acts as an immunogenic adjuvant in hepatitis B virus-infected hepatocytes. Cell Rep Med. 2020;1(4):100060. doi: 10.1016/j.xcrm.2020.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rizzetto M., Shih J.W., Gocke D.J., Purcell R.H., Verme G., Gerin J.L. Incidence and significance of antibodies to delta antigen in hepatitis B virus infection. Lancet. 1979;2(8150):986–990. doi: 10.1016/s0140-6736(79)92561-3. [DOI] [PubMed] [Google Scholar]
- 78.Grabowski J., Wedemeyer H. Hepatitis delta: immunopathogenesis and clinical challenges. Dig Dis. 2010;28(1):133–138. doi: 10.1159/000282076. [DOI] [PubMed] [Google Scholar]
- 79.Ferns R.B., Nastouli E., Garson J.A. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J Virol Methods. 2012;179(1):189–194. doi: 10.1016/j.jviromet.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Le Gal F., Gordien E., Affolabi D., Hanslik T., Alloui C., Deny P. Quantification of hepatitis delta virus RNA in serum by consensus real-time PCR indicates different patterns of virological response to interferon therapy in chronically infected patients. J Clin Microbiol. 2005;43(5):2363–2369. doi: 10.1128/JCM.43.5.2363-2369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Townsend E.C., Zhang G.Y., Ali R., Firke M., Moon M.S., Han M.A.T. The balance of type 1 and type 2 immune responses in the contexts of hepatitis B infection and hepatitis D infection. J Gastroenterol Hepatol. 2019;34(4):764–775. doi: 10.1111/jgh.14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lunemann S., Malone D.F., Hengst J., Port K., Grabowski J., Deterding K. Compromised function of natural killer cells in acute and chronic viral hepatitis. J Infect Dis. 2014;209(9):1362–1373. doi: 10.1093/infdis/jit561. [DOI] [PubMed] [Google Scholar]
- 83.Lunemann S., Malone D.F., Grabowski J., Port K., Beziat V., Bremer B. Effects of HDV infection and pegylated interferon alpha treatment on the natural killer cell compartment in chronically infected individuals. Gut. 2015;64(3):469–482. doi: 10.1136/gutjnl-2014-306767. [DOI] [PubMed] [Google Scholar]
- 84.Dias J., Hengst J., Parrot T., Leeansyah E., Lunemann S., Malone D.F.G. Chronic hepatitis delta virus infection leads to functional impairment and severe loss of MAIT cells. J Hepatol. 2019;71(2):301–312. doi: 10.1016/j.jhep.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rehermann B., Thimme R. Insights from antiviral therapy into immune responses to hepatitis B and C virus infection. Gastroenterology. 2019;156(2):369–383. doi: 10.1053/j.gastro.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grabowski J., Yurdaydin C., Zachou K., Buggisch P., Hofmann W.P., Jaroszewicz J. Hepatitis D virus-specific cytokine responses in patients with chronic hepatitis delta before and during interferon alfa-treatment. Liver Int. 2011;31(9):1395–1405. doi: 10.1111/j.1478-3231.2011.02593.x. [DOI] [PubMed] [Google Scholar]
- 87.Nisini R., Paroli M., Accapezzato D., Bonino F., Rosina F., Santantonio T. Human CD4+ T-cell response to hepatitis delta virus: identification of multiple epitopes and characterization of T-helper cytokine profiles. J Virol. 1997;71(3):2241–2251. doi: 10.1128/jvi.71.3.2241-2251.1997. https://www.ncbi.nlm.nih.gov/pubmed/9032359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Landahl J., Bockmann J.H., Scheurich C., Ackermann C., Matzat V., Heide J. Detection of a broad range of low-level major histocompatibility complex class II-restricted, hepatitis delta virus (HDV)-Specific T-cell responses regardless of clinical status. J Infect Dis. 2019;219(4):568–577. doi: 10.1093/infdis/jiy549. [DOI] [PubMed] [Google Scholar]
- 89.Kefalakes H., Koh C., Sidney J., Amanakis G., Sette A., Heller T. Hepatitis D virus-specific CD8(+) T cells have a memory-like phenotype associated with viral immune escape in patients with chronic hepatitis D virus infection. Gastroenterology. 2019 doi: 10.1053/j.gastro.2019.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang Y.H., Tao M.H., Hu C.P., Syu W.J., Wu J.C. Identification of novel HLA-A∗0201-restricted CD8+ T-cell epitopes on hepatitis delta virus. J Gen Virol. 2004;85(Pt 10):3089–3098. doi: 10.1099/vir.0.80183-0. [DOI] [PubMed] [Google Scholar]
- 91.Karimzadeh H., Kiraithe M.M., Kosinska A.D., Glaser M., Fiedler M., Oberhardt V. Amino acid substitutions within HLA-B∗27-restricted T cell epitopes prevent recognition by hepatitis delta virus-specific CD8(+) T cells. J Virol. 2018;92(13) doi: 10.1128/JVI.01891-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karimzadeh H., Kiraithe M.M., Oberhardt V., Alizei E.S., Bockmann J., Zur Wiesch J.S. Mutations in hepatitis D virus allow it to escape detection by CD8+ T cells and evolve at the population level. Gastroenterology. 2019 doi: 10.1053/j.gastro.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nicolete L.D., Borzacov L.M., Vieira D.S., Nicolete R., Salcedo J.M. Correlation between TH1 response standard cytokines as biomarkers in patients with the delta virus in the western Brazilian Amazon. Mem Inst Oswaldo Cruz. 2016;111(4):275–276. doi: 10.1590/0074-02760160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wieland D., Kemming J., Schuch A., Emmerich F., Knolle P., Neumann-Haefelin C. TCF1(+) hepatitis C virus-specific CD8(+) T cells are maintained after cessation of chronic antigen stimulation. Nat Commun. 2017;8:15050. doi: 10.1038/ncomms15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rutishauser R.L., Deguit C.D.T., Hiatt J., Blaeschke F., Roth T.L., Wang L. TCF-1 regulates HIV-specific CD8+ T cell expansion capacity. JCI Insight. 2021;6(3) doi: 10.1172/jci.insight.136648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aslan N., Yurdaydin C., Wiegand J., Greten T., Ciner A., Meyer M.F. Cytotoxic CD4 T cells in viral hepatitis. J Viral Hepat. 2006;13(8):505–514. doi: 10.1111/j.1365-2893.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 97.Smith S., Honegger J.R., Walker C. T-cell immunity against the hepatitis C virus: a persistent research priority in an era of highly effective therapy. Cold Spring Harb Perspect Med. 2020 doi: 10.1101/cshperspect.a036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shoukry N.H., Cawthon A.G., Walker C.M. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391–424. doi: 10.1146/annurev.micro.58.030603.123836. [DOI] [PubMed] [Google Scholar]
- 99.Schulze zur Wiesch J., Lauer G.M., Day C.L., Kim A.Y., Ouchi K., Duncan J.E. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol. 2005;175(6):3603–3613. doi: 10.4049/jimmunol.175.6.3603. [DOI] [PubMed] [Google Scholar]
- 100.Binder B., Thimme R. CD4+ T cell responses in human viral infection: lessons from hepatitis C. J Clin Invest. 2020;130(2):595–597. doi: 10.1172/JCI133222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thimme R., Wieland S., Steiger C., Ghrayeb J., Reimann K.A., Purcell R.H. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77(1):68–76. doi: 10.1128/jvi.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ye B., Liu X., Li X., Kong H., Tian L., Chen Y. T-cell exhaustion in chronic hepatitis B infection: current knowledge and clinical significance. Cell Death Dis. 2015;6:e1694. doi: 10.1038/cddis.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roggendorf M. Perspectives for a vaccine against hepatitis delta virus. Semin Liver Dis. 2012;32(3):256–261. doi: 10.1055/s-0032-1323631. [DOI] [PubMed] [Google Scholar]
- 104.Burwitz B.J., Zhou Z., Li W. Animal models for the study of human hepatitis B and D virus infection: new insights and progress. Antivir Res. 2020;182:104898. doi: 10.1016/j.antiviral.2020.104898. [DOI] [PubMed] [Google Scholar]
- 105.Pircher H., Moskophidis D., Rohrer U., Burki K., Hengartner H., Zinkernagel R.M. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 1990;346(6285):629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 106.Timm J., Walker C.M. Mutational escape of CD8+ T cell epitopes: implications for prevention and therapy of persistent hepatitis virus infections. Med Microbiol Immunol. 2015;204(1):29–38. doi: 10.1007/s00430-014-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang S.Y., Wu J.C., Chiang T.Y., Huang Y.H., Su C.W., Sheen I.J. Positive selection of hepatitis delta antigen in chronic hepatitis D patients. J Virol. 2007;81(9):4438–4444. doi: 10.1128/JVI.02847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kasprowicz V., Kang Y.H., Lucas M., Schulze zur Wiesch J., Kuntzen T., Fleming V. Hepatitis C virus (HCV) sequence variation induces an HCV-specific T-cell phenotype analogous to spontaneous resolution. J Virol. 2010;84(3):1656–1663. doi: 10.1128/JVI.01499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bengsch B., Seigel B., Ruhl M., Timm J., Kuntz M., Blum H.E. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. Plos Pathog. 2010;6(6) doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fiore-Gartland A., Manso B.A., Friedrich D.P., Gabriel E.E., Finak G., Moodie Z. Pooled-peptide epitope mapping strategies are efficient and highly sensitive: an evaluation of methods for identifying human T cell epitope specificities in large-scale HIV vaccine efficacy trials. PloS One. 2016;11(2) doi: 10.1371/journal.pone.0147812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Burrows S.R., Rossjohn J., McCluskey J. Have we cut ourselves too short in mapping CTL epitopes? Trends Immunol. 2006;27(1):11–16. doi: 10.1016/j.it.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 112.Momburg F., Roelse J., Hammerling G.J., Neefjes J.J. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med. 1994;179(5):1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wenzel T., Eckerskorn C., Lottspeich F., Baumeister W. Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett. 1994;349(2):205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- 114.Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. 2017;2017:2680160. doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim A.Y., Lauer G.M., Ouchi K., Addo M.M., Lucas M., Schulze Zur Wiesch J. The magnitude and breadth of hepatitis C virus-specific CD8+ T cells depend on absolute CD4+ T-cell count in individuals coinfected with HIV-1. Blood. 2005;105(3):1170–1178. doi: 10.1182/blood-2004-06-2336. [DOI] [PubMed] [Google Scholar]
- 116.Altfeld M., Addo M.M., Shankarappa R., Lee P.K., Allen T.M., Yu X.G. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77(13):7330–7340. doi: 10.1128/jvi.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuen M.F., Chen D.S., Dusheiko G.M., Janssen H.L.A., Lau D.T.Y., Locarnini S.A. Hepatitis B virus infection. Nat Rev Dis Prim. 2018;4:18035. doi: 10.1038/nrdp.2018.35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.