Abstract
The occurrence of the COVID-19 pandemic caused by the SARS-CoV-2 virus since the end of 2019 has significantly affected the entire world. Now SARS-CoV-2 diagnostic tests are not only required for screening of suspected infected people for their medical treatment, but have also become a routine diagnosis for all people at a place where new cases have emerged in order to control spread of the disease from that region. For these reasons, sensitive methods for detection of SARS-CoV-2 are highly needed in order to avoid undetected infections. In addition, sample pooling that uses pooled specimens has been routinely employed as a time- and cost-effective strategy for community monitoring of SARS-CoV-2. In this regard, the content of each viral RNA sample of an individual will be further diluted in detection; therefore, higher detection sensitivity would be rather preferred. Among nucleic acid-based detection methods, isothermal nucleic acid amplifications are considered quite promising because they typically take less time to complete the test (even less than 20 min) without the need of thermal cycles. Hence, it does not necessitate the use of highly costly real-time PCR machines. According to recently published isothermal nucleic acid amplification methods, the reverse transcription recombinase polymerase amplification (RT-RPA) approach shows outstanding sensitivity with up to single-copy sensitivity in a test reaction. This chapter will mainly focus on how to employ RT-RPA technology to sensitively detect SARS-CoV-2 RNA. Besides, recently published RT-RPA based detection methods will be summarized and compared regarding their detection parameters and the primers and probes being used. In addition, we will also highlight the key considerations on how to design an ultrasensitive RT-RPA assay and the precautions needed to conduct the assay. Moreover, based on our recent report, we will also detail the methods we developed to detect SARS-CoV-2 RNA using modified RT-RPA, or RT-ERA, with single-copy sensitivity and the possible extensions beyond this method.
Keywords: RT-RPA, SARS-CoV-2 RNA, Exo probe, Nfo probe, Lateral flow, Single-copy sensitivity
1. Introduction
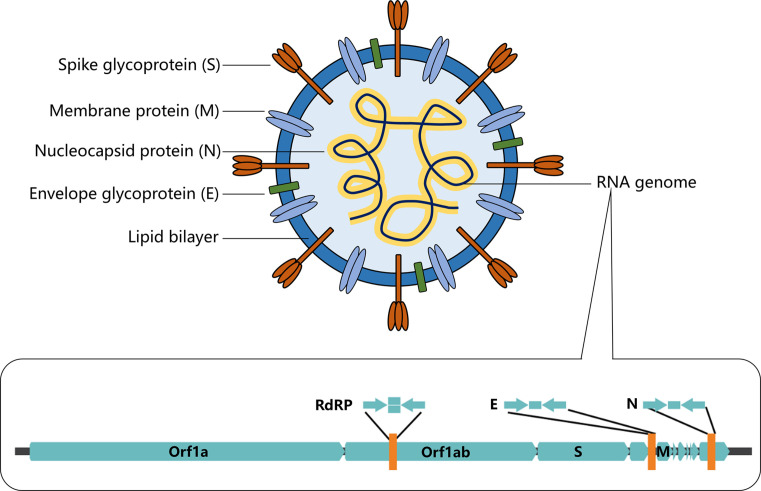
Since December 2019, an outbreak of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus occurred and soon spread to the entire world (Wang, Horby, Hayden, & Gao, 2020). The COVID-19 pandemic has led to a dramatic loss of human life worldwide and presents an unprecedented challenge to publish health and food systems; the global economic growth was largely slowed down and social activities were greatly disrupted. As of July 1, 2021, COVID-19 has infected over 182 million people worldwide with over 3.9 million reported casualties. COVID-19 is caused by an RNA virus named severe acute respiratory syndrome coronavirus 2 (Chen et al., 2020). SARS-CoV-2 virus is a coronavirus belonging to the beta family of coronaviruses that also includes SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus OC43 (HCoV-OC3), and human coronavirus HKU1 (HCoV-HKU1). These beta-viruses are enveloped, positive-sense, single-stranded RNA viruses of zoonotic origin. As for SARS-CoV-2, this virus contains four structural proteins, namely the envelope protein (E), spike protein (S), membrane protein (M), and the nucleocapsid (N) protein (Fig. 1 ). The S, M, and E proteins form the envelope of the virus while the N protein is associated with the single-stranded RNA genome forming nucleocapsid inside the envelope.
Fig. 1.
Schematic illustration of the structure of coronavirus SARS-CoV-2 and its single-stranded RNA genome. The sequence information of the single-stranded RNA genome of SARS-CoV-2 serves as the basis for the development of nucleic acid-based diagnosis.
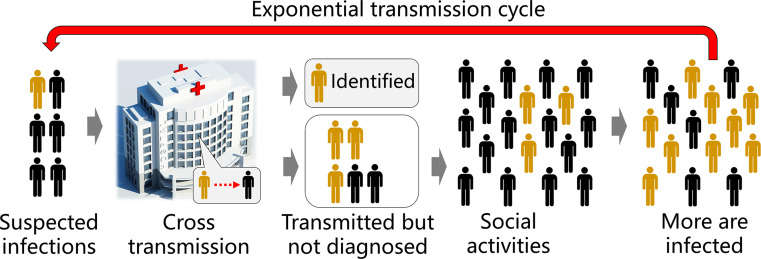
According to initial studies, this disease caused by SARS-CoV-2 showed a high transmissibility with a basic reproduction number R0 = 1.4–5.5 (WHO, 2020; Zhao et al., 2020), likely to be below 5 and above 3 (Chen, 2020) and therefore it is highly desired to perform early testing of SARS-CoV-2 for timely screening of infected individuals and to stop the spread of this disease from a location to its surroundings. Because detection sensitivity is the key to reduce false negative results, a detection method with a high level of sensitivity can minimize non-diagnosed infected individuals and reduce the chance of further cross transmission (Fig. 2 ).
Fig. 2.
Sensitive detection methods will be highly beneficial to prevent the spread of COVID-19. High sensitivity of diagnostic methods ensures low false negative results and therefore reduces the latent cross transmission of non-diagnosed but actually infected individuals to their closely contacted people.
2. General approaches for the detection of SARS-CoV-2
There are several different types of diagnostic approaches used in the current pandemic, which can be mainly classified into the following three categories: (i) computed tomography (CT) chest scan, (ii) antigen-antibody interaction-based serological tests, and (iii) nucleic acid (NA)-based tests (Kilic, Weissleder, & Lee, 2020; Qin, Peng, Baravik, & Liu, 2020). The CT chest scan (Ai et al., 2020) detects the pathological change of the respiratory system including the lung by visualization of the transmission of the X-ray through the chest. Typically, a brighter image that reveals a low transmission of the X-ray through the chest could potentially indicate the infection by SARS-CoV-2. However, the specificity of CT chest scan is low because the symptomatic features of COVID-19 CT scan are similar to those of other types of viral pneumonia; and hence this method is generally considered as an auxiliary method for SARS-CoV-2 diagnosis. Serological testing is based on the detection of antibodies generated by the immune system which can be used to confirm the infection of SARS-CoV-2 (Amanat et al., 2020; Udugama et al., 2020). However, it typically takes 1–2 weeks before the body can generate detectable antibodies for serological testing; as a result, this approach is not well suited for early-stage diagnosis. Other issues associated with serological tests are the high variability and sometimes low sensitivity and specificity (Tang et al., 2020). Nucleic acid-based detection is the detection of the RNA genomic materials of the SARS-CoV-2, usually using nucleic acid amplification approaches, which has been considered as the gold standard for SARS-CoV-2 viral detection.
Reverse transcription-polymerase chain amplification (RT-PCR) has been widely used for the detection of viral RNA for many years. RT-PCR requires a first reverse transcription to produce a cDNA from the viral RNA, and then amplification of cDNA via polymerase chain reaction. During amplification, a fluorescent probe, either a fluorogenic dye (e.g. SYBR safe) or a rationally designed probe (e.g. TagMan probe) is included in the PCR reaction which will respond to the amplified DNA to produce fluorescent signals. As a result, the amplification could be detected in real-time via fluorescence using a real-time quantitative PCR machine. A drawback of RT-PCR diagnosis is the high cost of the PCR machine and the expertise required to design the program and to conduct the analysis. In fact, RT-PCR is among the first reported and approved SARS-CoV-2 detection methods (Chu et al., 2020; Corman et al., 2020). At the earliest time after the COVID-19 outbreak, Drosten et al. reported the detection of SARS-CoV-2 by real-time RT-PCR method. The PCR primers and probes were designed for the detection of RdRp gene, E-gene and N-gene. Although single-copy sensitivity was not achieved, this RT-PCR assay was found to be still quite sensitive with 5.2 copies per reaction at 95% detection probability for E-gene and 3.8 copies per reaction at 95% detection probability for RdRp gene using non-clinical samples; the sensitivity for the N-gene is less and hence was not subjected to intensive evaluation (Corman et al., 2020). Another RT-PCR based approach for the molecular diagnosis of SARS-CoV-2 introduced in the very beginning of the pandemic detects the ORF1b and N-gene of SARS-CoV-2 with a sensitivity up to ≤ 10 copies per reaction using none-RNA testing sample (Chu et al., 2020).
3. Isothermal amplification methods for sensitive detection of SARS-CoV-2
Isothermal nucleic acid amplification approaches are emerging as new promising methods for detection of viral RNA (James & Alawneh, 2020; Zhao, Chen, Li, Wang, & Fan, 2015). In isothermal nucleic acid amplification, no thermal cycles are needed and therefore, exempted from using sophisticated thermocyclers. The respective detection device is hence simpler, more portable and meet the requirement of so-called point of care (POC) testing (Dinnes et al., 2020; Dohla et al., 2020). Sometimes, lateral flow strips can also be used to facilitate the diagnosis so that the testing can be applied in a POC, or even home-based fashion. Further, isothermal amplification approaches can achieve a very high sensitivity, with up to single-copy sensitivity per reaction. The ultrahigh sensitivity makes these approaches suitable for early-stage diagnosis of virus infections. Therefore, isothermal nucleic acid amplification has been considered as a promising method for the detection of virus infection and could compensate with the currently widely used RT-PCR methods.
Indeed, as for SARS-CoV-2, isothermal amplification detection methods have already been considered as highly promising detection tools (Shen et al., 2020). Early development of isothermal amplification techniques include LAMP (loop-mediated isothermal amplification) (Notomi et al., 2000; Wong, Othman, Lau, Radu, & Chee, 2018), NASBA (nucleic acid sequence-based amplification) (Compton, 1991), HDA (helicase-dependent amplification) (Vincent, Xu, & Kong, 2004), EXPAR (exponential amplification reaction of nucleic acids), and SDA (strand-displacement amplification) (Walker et al., 1992). Among these isothermal detection methods, LAMP coupled with reverse transcription, i.e. RT-LAMP, seems to be the most popular in SARS-CoV-2 analysis (Kashir & Yaqinuddin, 2020). In RT-LAMP, four or six primers that target 6–8 regions in the genome are designed in combination with the use of Bsm DNA polymerase. Along with the proceeding LAMP reaction, pairs of primers generate a dumbbell-shaped DNA structure, which functions as the LAMP initiator. In this method, around 109 DNA copies can be generated within an hour and the reaction takes place at constant temperature in the range of 60–65 °C. Since magnesium pyrophosphate is generated as a byproduct during LAMP, metal-sensitive indicators or pH-sensitive dyes can be employed for visual detection. An advantage of the RT-LAMP approach is that regular primers are used which do not necessitate the design of specially functionalized primers or probes; hence this feature is helpful to reduce the cost of a RT-LAMP reaction. On the other hand, LAMP, NASBA and HDA methods do not require thermocycle machines for amplification; however, specifically designed heating devices are still needed. In addition, LAMP requires the use of 4–6 primers, which makes primer design more complicated than other methods. Furthermore, LAMP, NASBA or HDA typically needs at least 60 min to complete the amplification reaction, which is longer than RT-RPA.
Recently some more contemporary isothermal amplification methods have been introduced, for example, NEAR, DETECR, STOP and so on. NEAR (nicking enzyme amplification reaction) or nicking enzyme-assisted amplification (NEAA) uses not only strand-displacement DNA polymerase (e.g. Bst polymerase), but also nicking endonuclease enzymes to exponentially amplify short oligonucleotides (Wang et al., 2018). Thousands of copies of DNA fragment can be produced from only one restriction site making this approach a unique technique with rather high amplification efficiency. On the other hand, a drawback of NEAR is the formation of non-specific products which limits detection sensitivity. DETECTR (DNA endonuclease-targeted CRISPR trans reporter) method (Chen et al., 2018) is associated with CRISPR-based detection approaches that involves the use of the genome editing Cas12a enzymes. DETECTR uses a crRNA-Cas12a complex to recognize amplified DNA targets and binding of the crRNA-Cas12a complex to target DNA induced discrimination cleavage of non-target FQ-DNA reporters. Similar CRISPR-based approaches include SHERLOCK detection (Gootenberg et al., 2017) in one-pot (STOP) that uses Cas13a enzyme.
4. Sensitive detection of SARS-CoV-2 via RT-RPA
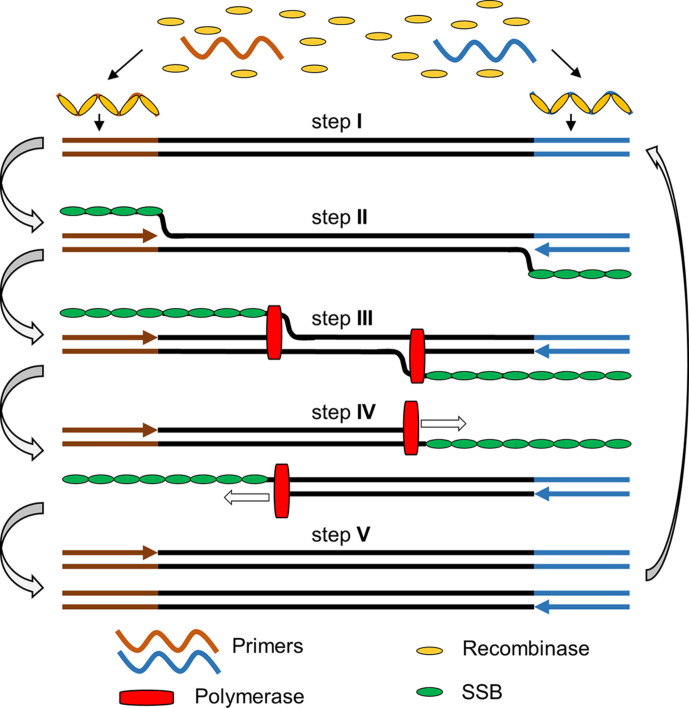
Reverse transcription recombinase polymerase amplification (RT-RPA) is a widely recognized isothermal amplification assay for the amplification of RNA, which combines reverse transcription that converts RNA to cDNA, and recombinase polymerase amplification (RPA) that amplifies cDNA under isothermal conditions. RPA sometimes is also called ERA (enzymatic recombinase amplification) as a modified/improved version of RPA according to the commercially supplied kit from the company GenDx (http://gendx.cn), or RAA (recombinase aided amplification) (Wu et al., 2020; Xue et al., 2020). RPA is a molecular technology introduced by Piepenburg, Williams, Stemple, and Armes (2006) using proteins involved in cellular DNA synthesis, recombination and repair (Piepenburg et al., 2006). The RPA process starts when a recombinase protein binds to primers in the presence of ATP and a crowding agent (e.g. a high molecular weight polyethylene glycol), forming a recombinase-primer complex (step I). The complex then interrogates double-stranded DNA seeking a homologous sequence and facilitates strand hybridization with the primer at the cognate site (step II & III). In the meanwhile, single-stranded binding protein (SSB) is added in the reaction mixture to stabilize the dissociated single-stranded DNA and prevent the ejection of the inserted primer by branch migration, and facilitate the amplification to proceed at room temperature. The DNA polymerase (e.g. Bsu) will bind to the 3′ end of the primer to elongate the primer in the presence of dNTPs (step IV & V). Cyclic repetition of this process results in the exponential amplification of a DNA (Fig. 3 ) (Piepenburg et al., 2006).
Fig. 3.
The general principle of recombinase polymerase amplification. Step I: recombinase and primer form complexes and target homologous DNA; step II: strand exchange forms a D-loop; herein, the brownish and blue-coloured arrows refer to the forward and reverse primers, respectively, upon annealing with their templates; step III: polymerase initiates synthesis; step IV: parental strands separate & synthesis continues; herein, the hollow boxed arrows refer to the directions of the polymerization reaction; step V: two duplexes form. Abbreviation(s): SSB, single-stranded DNA binding protein.
In order to employ RPA for the amplification of RNA, additional reverse transcriptase was added in order to convert RNA to cDNA for subsequent RPA amplification. In addition, since RNA is a much less stable species compared to DNA and is highly prone to degradation by the ubiquitously existing RNase, RNase inhibitor protein was added into the RT-RPA reaction. In order to facilely detect the amplification product, a probe and a nuclease was used. Moreover, creatine kinase, phosphocreatine and ATP were needed to generate energy for the reaction. Therefore, a practical RT-RPA based detection reaction requires at least seven enzymes/proteins including: (i) strand-displacing DNA polymerase, (ii) recombinase, (iii) recombination-mediator protein (RMP), (iv) single-strand DNA binding protein, (v) RNase inhibitor, (vi) creatine kinase, and (vii) nuclease; in addition, multiple necessary reagents are also required like dNTPs, creatine, the crowding agent—polyethylene glycol, the activator Mg(OAc)2, the forward and reverse primers, and a probe. For the most reliable test, viral RNA samples need to be purified via a standard RNA purification process rather than using a non-purified viral sample. The list of proteins, enzymes, reagents necessary for conducting an RT-RPA reaction with their recommended concentrations are summarized in Table 1 (Chen et al., 2020). The concentrations of primers, probes, RNase inhibitor and nucleases used in our hand for exo-probe, nfo-probe and multiplexing RT-RPA reaction have also been given (Table 1).
Table 1.
List of concentrations of primers, probes, nucleases, inhibitors and Mg(OAc)2 that need to be additionally supplied in a RT-RPA or RT-ERA reaction kit for viral RNA detection.
| Components | Concentrations used in our study | Recommended ranges by other studies | |
|---|---|---|---|
| Exo-probe detection | Fw & Rw primers | 500 nM | 420 nM, can be varied in the range of 150–600 nM |
| Exo probe | 150 nM | In the range of 50–150 nM | |
| Exonuclease III | 100 U in 50 μL | – | |
| Nfo-probe LFD | Fw & Rw primers | 400 nM | 420 nM, can be varied in 150–600 nM range |
| Nfo probe | 120 nM | In the range of 50–150 nM | |
| Endonuclease IV | 10 U in 50 μL | – | |
| Duplexing | Rw & Fw Primers | 100 nM | Subjected to testing and optimization |
| Exo probes | 30 nM | ||
| Exonuclease III | 100 U in 50 μL | ||
| Other necessary reagents | RNase A inhibitor | 5 U in 50 μL | – |
| Mg(OAc)2 | 2 μL | Standard 14 mM, can be varied in the range of 12–30 mM |
Note: Other components include (i) RPA enzymes (120 ng·μL− 1 T4 UvsX recombinase, 60 ng·μL− 1 T4 UvsY recombination-mediator protein, 600 ng·μL− 1 T4 gp32 single-stranded binding protein, 30 ng·μL− 1 Bsu or 8.6–12.8 μg Sau DNA strand-displacing polymerase), (ii) energy-supply system constituents (50 mM phosphocreatine, 100 ng·μL− 1 creatine kinase, and 3 mM ATP), (iii) 200 μM each dNTPs, (iv) buffering constituents (typically pH 7.9 50 mM Tris/100 mM KOAc), (v) reducing agent 2 mM DTT, (vi) crowding reagent (5% carbowax 20M), and (vii) reverse transcriptase are generally included in the commercially available RT-RPA or RT-ERA kit with fixed concentration without the need for further adjustment (Li, Macdonald, & von Stetten, 2020).
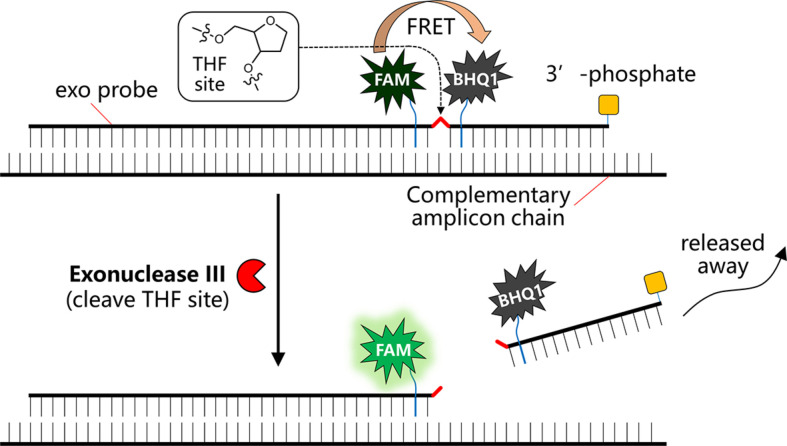
For the detection of viral RNA using RT-RPA, multiple detecting formats are available. These formats include exo probe, nfo probe, fpg probe, digital (Shen et al., 2011), nesting, microfluidics (Lutz et al., 2010), solid phase, template generation, electrochemistry, colorimetric or SNP detection, and so on (Daher, Stewart, Boissinot, & Bergeron, 2016; Li et al., 2020). Among all these, it seems that using exo probe for fluorescence detection (Behrmann et al., 2020; Tu et al., 2020) and using nfo probe coupled with lateral flow strip detection (Zheng et al., 2021) are the most popular and hence we will mainly focus on these two formats for the detection of SARS-CoV-2 viral RNA by RT-RPA. Herein, either approach has its own advantages. For the detection of RNA using exo probe, first forward and reverse primers that are around 30–35 nucleotides long are designed; the melting temperature of an oligonucleotide is normally not a critical factor for the performance as a primer. The primer pairs allow the amplification of a relatively short length DNA amplicon ideally around 100–200 bp long that is most suited for RPA amplification. In addition to the primer pair, an exo probe was designed for the fluorogenic detection of the amplicon in the presence of exonuclease III (exo enzyme). The exo probe is a modified oligonucleotide featuring an abasic nucleotide analogue—tetrahydrofuran residue (THF, sometimes referred to as “dSpacer”) that is prone to cleavage by exo enzyme when the nucleotide is in a double-stranded state. In addition, the exo probe is usually around 46–52 nucleotides long, with at least 30 nucleotides located 5′- to the THF site, and a further at least 15-bp long nucleotide located 3′- to the THF site. A blocking group, such as a phosphate, a C3-spacer, a biotin-TEG or an amine, is situated at its 3′ side so that the exo probe cannot act as a primer and block the probe from polymerase extension. The entire exo probe is able to specifically hybridize with one chain of the amplicon (THF needs to be counted as one nucleotide residue). In order to allow this probe to detect the amplicon in a fluorogenic way, exo probe features a flanking dT-fluorophore (e.g. fluorescein) on one side of the THF residue and a flanking corresponding dT-quencher group (typically a suitable Black Hole Quencher (BHQ)) on the other side of the THF site. In such a way, the dT-fluorophore is temporally quenched by its FRET quencher (e.g. BHQ1) and the probe will be in a none or weakly fluorescent state. Once RPA reaction produces an amplicon that can hybridize with the exo probe, the exo probe is converted from single-stranded state to double-stranded state that allows the THF residue to be readily cleaved by the exo III enzyme. Hence the quencher moiety is separated from the probe and the fluorescence of the dT-fluorophore is restored (Fig. 4 ). Exo probe allows sensitive detection in RT-RPA based on the enhancement of fluorescence.
Fig. 4.
Schematic view of the working principle of a typical exo probe for RT-RPA detection of viral RNA.
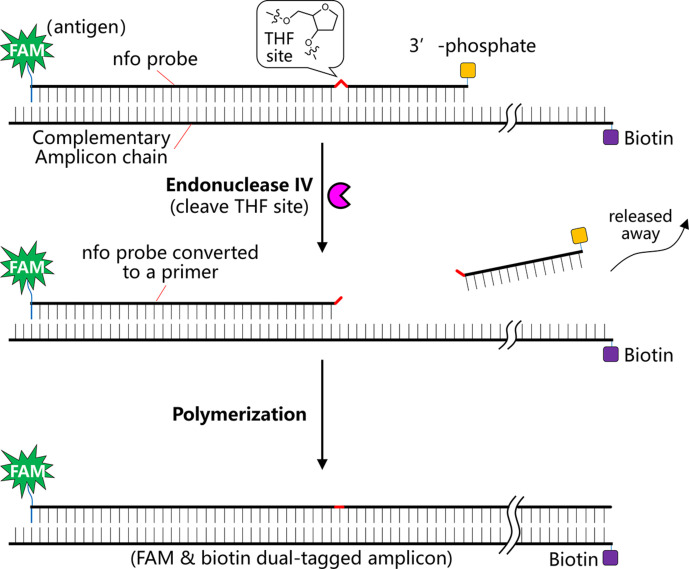
In order to make the sensitive detection approach more field-deployable with minimal instrumentation, nfo probe could be designed in combination with the use of lateral flow strips for direct visual detection. In this regard, nfo probe can hybridize with the amplicon to give a bifunctional amplicon, typically with a biotin antigenic tag at one end and a FAM antigenic tag at the other end. Similar to the exo probe, a nfo probe is around 46–52 nucleotides long, features an abasic THF residue in between, at least 30 nucleotides 5′ to the THF site, at least a further 15 nucleotides located 3′ to the THF side, and a blocking group (e.g. a phosphate) at the 3′ side to block the probe from polymerization. In contrast to exo probe, the nfo probe does not necessitate any modified dT-fluorophore nor dT-quencher, but requires an affinity biotin or FAM antigenic tag at the 5′ side of the probe. In addition, the primer with reverse direction to the nfo probe should also be modified with a different affinity tag (e.g. a FAM tag or a biotin tag) complementary to the nfo probe at the 5′ end. Once the nfo probe hybridizes with the amplicon generated in the RPA amplification reaction, the nfo probe turns to a double-stranded state, rendering its internal THF residue susceptible to cleavage by the endonuclease IV, i.e. the nfo enzyme. Then the ~ 15 bp oligonucleotide 5′ to the THF side is released, converting the probe to a primer and initializing polymerization in the RT-RPA reaction. In the end, a bifunctional amplicon featuring a FAM moiety at one end and a biotin moiety at the other end is generated, which allows easy visual detection using lateral flow strips (Fig. 5 ). Since RT-RPA reaction is highly viscous, a further dilution of the RT-RPA reaction solution is necessary prior to lateral flow strip analysis.
Fig. 5.
Schematic view of the working mechanism of a nfo probe in RT-RPA for detection of viral RNA.
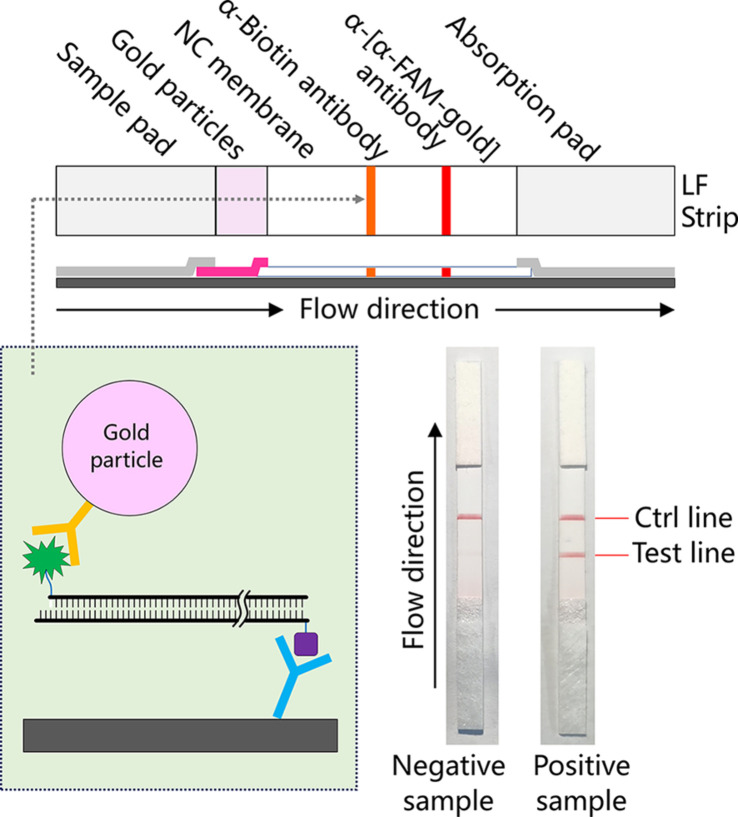
In the lateral flow (LF) test using a lateral flow strip, the bifunctional amplicon acts as a “bridge” that connects the gold nanoparticle and the antibody immobilized in the test line so that the test line turns into a red colour, the colour of the gold nanoparticle (Fig. 6 ). In brief, the lateral flow strip contains two lines. The first line which is closer to the absorption pad side is immobilized with α-biotin antibody. The second control line is fixed with an antibody against α-FAM antibody (i.e. α-[α-FAM-gold] antibody). A colloidal gold particle region (the gold particle is coated with α-FAM antibody) is located below the two lines and above the sample pad. When the sample pad of the test strip is inserted into a diluted RT-RPA reaction solution, the colloidal gold particle will flow along with the eluent and sequentially cross the test and the control line. Once the double-tagged amplicon is presented in the RT-RPA reaction, it will bridge the gold particle and the α-biotin antibody in the test line, turning the test line to a red colour. Meanwhile the control line that contains α-[α-FAM-gold] antibody will bind the α-FAM antibody-coated gold particle, suggesting that the LF strip is effective (Fig. 6).
Fig. 6.
Schematic view of the working mechanism of lateral flow detection (LFD) for the detection of bifunctional amplicon, i.e. amplicon with two antigenic labels, produced in RT-RPA reaction.
5. General considerations for designing an ultrasensitive RT-RPA assay
RT-RPA reaction contains multiple proteins/enzymes, ingredients, primers, and a probe. In addition, multiple factors can also affect the performance of RT-RPA reaction. Therefore, in order to achieve an optimal performance in RT-RPA reaction for the diagnosis of viral RNA, several points need to be taken into consideration. Herein, we will mainly discuss the most important factors that could help the setup of an optimal RT-RPA reaction for RNA detection. These factors include: (i) primer design, (ii) probe design, and (iii) temperature.
5.1. Primer design
For either exo probe or nfo probe used in RT-RPA test, the optimal primer length is 30–35 nucleotides, which is usually longer than typical PCR primers. Regarding the design of a primer pair for RPA, the melting temperature of an oligonucleotide is not the critical factor for its performance as a primer, but rather its length. In addition, the optimal length of a primer can also be temperature dependent as RPA reaction can still take place above or below 37 °C. Oligonucleotides shorter than 30 bp may still function albeit at a typically slower reaction rate. Longer oligonucleotides may also be used as a primer to increase the RPA kinetics; yet longer primer may increase the possibility of secondary-structure formation that could lead to the so-called primer noise. Moreover, since RPA reaction is best suited for relatively short length DNA amplification compared to PCR methods, it is advised that the primer pair amplify an amplicon with a length ideally between 100 and 200 bp, not exceeding 500 bp. Amplification of a short length of DNA is also beneficial to achieve an increased product/noise ratio because shorter products will be generated in a shorter period of time and the longer an amplicon is, the more likely primer noise will outcompete target amplification. In fact, short amplicon is an important criterion for designing ultrasensitive RT-RPA assays. In order to select an appropriate primer pair annealing to an appropriate region within a nucleic acid target, it is advised that the target region is characterized by relatively “average” nucleotide sequence composition with a GC content between 40% and 60%, featuring repetitive sequences, and with minimal direct/inverted repeats, palindromes, etc. More importantly, when RT-RPA is used for viral nucleic acid detection, the specificity of the primers toward the nucleic acid target of the virus should be carefully evaluated. This can be tested, via, e.g. BLAST, to ensure that the designed primers indeed specifically detect certain species, but not any other species. In order to screen a best Fw and Rw primer pair, different Fw primer and Rw primers could be paired and evaluated.
5.2. Probe design
Both exo and nfo probes should be 46–52 nucleotides long, featuring at least 30 bp located 5′ to the THF site, and at least a further 15 bp located 3′ to THF residue appended with a blocking group at the 3′ group. The 3′ block group should always be added in order to prevent polymerization from the probe. Care should be taken to avoid the occurrence of primer-probe dimers, although the primer in the same direction as the probe could partially overlap at its 5′ part. In addition, secondary structures in the probe should be avoided to prevent the probe folding back on itself. For the design of either exo probe or nfo probe, it needs to be kept in mind that the THF site occupies one base site, meaning THF needs to replace an existing base rather than being added as an additional base. For the design of the exo probe, the most important factor is to identify a pair of dT residues in close proximity to each other, typically with only 1–5 interval nucleotides. If the distance between the dT-fluorophore and dT-quencher is too large, quenching could be poor. For the design of a nfo probe, a 5′-antigenic label needs to be appended in order to pair with the antigenic label on the opposing primer.
5.3. Reaction temperature
Most commercially available RPA kits are designed for being conducted at a temperature range of 37–42 °C. If the temperature is higher, enzymes gradually lose activity inhibiting amplification. When the reaction temperature is lower, the RPA may also work excellently but at a reduced reaction rate. However, the problem is that the consumption of the reagents in the RPA reaction still proceeds quickly even at a reduced temperature. The result is fuel “burn-out” before RPA amplification produces a reasonable amount of amplicon for detection.
6. Comparative reviews of recently published RT-RPA assays for SARS-CoV-2 detection
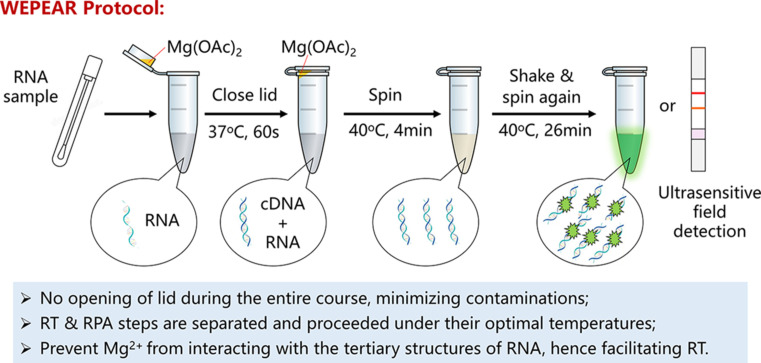
Until now, there have been multiple reports about the detection of SARS-CoV-2 viral RNA using RT-RPA approach following our initial report in March 2020 and May 2020 (Abd El Wahed et al., 2021; Arizti-Sanz et al., 2020; Behrmann et al., 2020; Qian et al., 2020; Tu et al., 2020; Wang, Cai, et al., 2020; Xia and Chen, 2020a, Xia and Chen, 2020b; Xiong et al., 2020; Xue et al., 2020; Zheng et al., 2021). We introduced an WEPEAR (whole-course encapsulated procedure for exponential amplification from RNA) that seamlessly combines the reverse transcription (optimally at 37 °C, in the absence of Mg(OAc)2 catalyst) and the RPA reaction (optimally at 40 °C, in the presence of Mg(OAc)2 catalyst) at their own optimal conditions (Fig. 7 ) (Xia and Chen, 2020a, Xia and Chen, 2020b). Because the optimal temperature used for performing reverse transcription and RPA reaction is different (although a bit close), direct combination of reverse transcription and RPA reactions in one tube would compromise the entire amplification, although possible. In addition, the Mg2 + ion used as a catalyst in the PRA reaction can complex with the RNA to inhibit the transcription step. Hence it is advantageous to physically separate the reverse transcription step and the PRA amplification step. On the other hand, however, it is also highly desirable to perform reverse transcription and RPA reactions in one-tube without re-opening the lid again after introducing the detection sample. In this case, it can simplify the testing procedure and reduce possible contamination during the amplification course, such as aerosol contamination from the air. The WEPEAR protocol readily solved this problem by seamlessly combining the RT step and the RPA step in one-pot without opening the lid at all during the entire detection course (Fig. 7).
Fig. 7.
The WEPEAR protocol seamlessly combines reverse transcription and RPA reactions in one tube allowing both steps to be conducted under their own optimal temperatures and prevents the Mg2 + used in the RPA reaction from interacting with the RNA sample and inhibiting reverse transcription.
In WEPEAR protocol, the RNA sample is added into the reaction solution in a vial containing all necessary reagents for both RT and RPA reaction aside from the magnesium activator. In the meanwhile, a small volume of the magnesium acetate solution catalyst is loaded onto the lid of the reaction vial; due to the surface tension force, the small volume of the magnesium solution will still sit inside of the inner lid after gently closing the lid. The reaction vial is first warmed to 37 °C for 60 s to finish the reverse transcription process. Afterwards, the reaction vial is subjected to brief centrifugation, vortex, centrifugation again to mix the magnesium catalyst into the reaction mixture and initialization of the RPA reaction. Finally, the reaction vial is warmed to 40 °C for 4 min as the pre-reaction step, vortexed, centrifuged again, and warmed to 40 °C for another 26 min to complete the entire RPA reaction. Herein, vortexing and centrifuging the reaction again after 4 min is beneficial to enhance the sensitivity because RT-RPA reaction mixture is quite viscous and the internal vortex step could help disperse the initially amplified amplicon in the entire reaction mixture for a more efficient amplification (Fig. 7).
In the WEPEAR-powered RT-PRA approach, two exo probes were designed for the detection of N-gene and S-gene. Both probes show high sensitivity for the detection of SARS-CoV-2 RNA. The exo probe for N-gene can detect four copies of RNA in a reaction while the exo probe for S-gene is able to achieve single-copy sensitivity in the RT-RPA reaction.
As has been mentioned above, false negative results that are usually caused by lack of sufficient sensitivity in the detection method should be avoided. False positive results also need to be minimized for a reliable diagnostic approach. Herein, if two genes could be simultaneously detected, many false positive results will be excluded. In our recent report, the exo probe for N-gene is green-colour emissive and the exo probe for S-gene is red-colour emissive. Hence, it makes it possible to multiplex the RT-RPA reaction for simultaneous dual-gene detection. We achieved this simply by reducing the concentrations of both primers and probes to around one fifth of their original concentration. Real-time dual-gene detection using green and red channels was readily achieved using an advanced real-time PCR machine. This result suggested that rational adjustment/reduction of the primer and probe concentrations is an efficient way to establish a successful multiplexing RT-RPA reaction.
In addition to the exo probes, nfo probe for both N- and S-gene were also designed for lateral flow test without the need of a real-time PCR machine, nor a blue light imaging plate. Hence, nfo probe combined with lateral flow strips provide a more field-deployable way for conducting SARS-CoV-2 detection. Household detection using this LFD method is also possible simply using a daily used thermos cup to warm up the reaction vial. Herein, ultrasensitivity has been achieved for both nfo probes. The sensitivity tested using in vitro transcribed RNA sample reached four-copy per reaction.
Among other RT-RPA methods for SARS-CoV-2 detection (Table 2 ), enhanced RT-RPA has been introduced which is named eRPA protocol (Qian et al., 2020). Internally quenched (exo-IQ) probe was designed for the detection of SARS-CoV-2 N gene (Behrmann et al., 2020). Also, all the required detection setup and reagents could be included in a suitcase called a “Suitcase Lab” for deployable detection of SARS-CoV-2 (Behrmann et al., 2020). Moreover, RT-RPA can be coupled with gene editing enzymes, e.g. Cas12a or Cas13a for viral RNA detection (Arizti-Sanz et al., 2020; Xiong et al., 2020). RT-RPA has the following advantages over RT-PCR: (1) the amplification proceeds faster which can be accomplished within 20–30 min; (2) RT-RPA proceeds at a constant temperature without the need of thermocyclers, and often gives higher sensitivity. For example, there are already several reports achieving single-copy detection sensitivity for the detection of SARS-CoV-2 using RT-RPA (Xia and Chen, 2020a, Xia and Chen, 2020b; Xue et al., 2020). A comparison of the recently reported RT-RPA detection against SARS-CoV-2 RNA is summarized in Table 2.
Table 2.
Summarization of recently published RT-RPA methods for sensitive detection of SARS-CoV-2 (as of February 2021).
| Entry | Gene | Type | Sequence & structure | Duration | Sensitivity | Specificity | Readout | References | Date |
|---|---|---|---|---|---|---|---|---|---|
| 1 and 2 | N | Fw | TTTGGTGGACCCTCAGATTCAACTGGCAGTAAC | 1 min (37 °C) + 30 min (40 °C) with internal vortex | ≤ 4-copy/reaction (50 μL) | Specific over SARS-CoV | Green emission | Xia and Chen (2020a) and Xia and Chen (2020b) | March 23, 2020 |
| Rw | GAATTTAAGGTCTTCCTTGCCATGTTGAGTGAG | ||||||||
| Exo-probe | TATTATTGGGTAAACCTTGGGGCCGACGTTGTT/i6FAMdT/T/idSp/A/iBHQ1dT/CGCGCCCCACTG-Phosphate | ||||||||
| N | Fw | TTTGGTGGACCCTCAGATTCAACTGGCAGTAAC | 1 min (37 °C) + 30 min (40 °C) with internal vortex + 2 min LF test | ≤ 4-copy/reaction (50 μL) | Specific over MERS-CoV but not SARS-CoV | LFD | May 28, 2020 | ||
| Rw | Biotin-GAATTTAAGGTCTTCCTTGCCATGTTGAGTGAG | ||||||||
| Nfo-probe | 6-FAM-GCGATCAAAACAACGTCGGCCCCAAGGTTTACC/idSp/AATAATACTGCGTCT-Phosphate | ||||||||
| S | Fw | GTCTCTAGTCAGTGTGTTAATCTTACAACCAGAAC | 1 min (37 °C) + 30 min (40 °C) with internal vortex | 1-copy/reaction (50 μL) | Specific over SARS-CoV | Red emission | Xia and Chen (2020b) | May 28, 2020 | |
| Rw | CATTGGAAAAGAAAGGTAAGAACAAGTCCTGAG | ||||||||
| Exo-probe | CCTGCATACACTAATTCTTTCACACGTGGTG/iTAMdT/T/idSp/A/iBHQ2dT/TACCCTGACAAAGTT-Phosphate | ||||||||
| S | Fw | GTCTCTAGTCAGTGTGTTAATCTTACAACCAGAAC | 1 min (37 °C) + 30 min (40 °C) with internal vortex + 2 min LF test | ≤ 4-copy/reaction (50 μL) | Specific over SARS-CoV, MERS-CoV | LFD | |||
| Rw | Biotin-CATTGGAAAAGAAAGGTAAGAACAAGTCCTGAG | ||||||||
| Nfo-probe | 6-FAM-CCTGCATACACTAATTCTTTCACACGTGGTG/idSp/TTATTACCCTGACAA-Phosphate | ||||||||
| 3 | N | Fw | CCTCTTCTCGTTCCTCATCACGTAGTCGCAAC | 21 min (42 °C) + internal vortex | 10-copy/reaction (~ 57 μL) | Specific over SARS-CoV, MERS-CoV, OC43/229E/NL63 | Green emission | Behrmann et al. (2020) | May 8, 2020 |
| Rw | AGTGACAGTTTGGCCTTGTTGTTGTTGGCCTT | ||||||||
| Exo-Probe | CCTGCTAGAATGGCTGGCAATGGCGGTGA/idFAMdT/idSp/C/iBMNQ535dT/TGCTCTTGCTTTGC-C3 | ||||||||
| 4 | S | Fw | CTTCAACCTAGGACTTTTCTATTAAAATATAATG | 4 min + 20 min (39 °C) | 10-copy/reaction (50 μL) | Specific over OC43/229E/NL63/HKU1 | Green emission | Xue et al. (2020) | May 22, 2020 |
| Rw | GTTGGTTGGACTCTAAAGTTAGAAGTTTGATAG | ||||||||
| Exo-probe | CCATTACAGATGCTGTAGACTGTGCACTTGACCC/iFAMdT/C/idSp/C/iBHQ1dT/CAGAAACAAAGTGTACG-Phosphate | ||||||||
| Orf1ab | Fw | TACGCCAAGCTTTGTTAAAAACAGTACAATTCTG | 4 min + 20 min (39 °C) | 1-copy/reaction (50 μL) | Green emission | ||||
| Rw | GGCATTAACAATGAATAATAAGAATCTACAACAGG | ||||||||
| Exo-probe | TTGTTGGTGTACTGACATTAGATAATCAAGATC/iFAMdT/C/idSp/A/iBHQ1dT/GGTAACTGGTATGATTTCG-Phosphate | ||||||||
| 5 | N | Fw | CAGTTCAAGAAATTCAACTCCAGGCAGCAGTAG | 7 min (39 °C) + 20 min (39 °C) | 10-copy/reaction (50 μL) | Specific over OC43/229E/NL63/HKU1, influenza A/B; unknown for SARS-CoV | Green emission | Wu et al. (2020) | July 29, 2020 |
| Rw | CAGTTTGGCCTTGTTGTTGTTGGCCTTTAC | ||||||||
| Exo-probe | CAGACATTTTGCTCTCAAGCTGGTTCAATC/iFAMdT/idSp/iBHQ1dT/CAAGCAGCAGCAAAG-C3 | ||||||||
| 6 | S | Fw | TCTTGTTTTATTGCCACTAGTCTCTAGTCAGT | 25 min (42 °C) + 3 min (90 °C) + 3 min (RT) + 3 min LF test | 3-copy/reaction (50 μL) SuperScript IV & RNase H added |
Specific over SARS-CoV, MERS-CoV, 229E/HKU1 | LFD | Qian et al. (2020) | November 20, 2020 |
| Rw | FAM-GAATGTAAAACTGAGGATCTGAAAACTTTG | ||||||||
| Nfo-probe | Biotin-TGCATACACTAATTCTTTCACACGTGGT | ||||||||
| 7 | Orf1ab | Fw | CCAAGGTAAACCTTTGGAATTTGGTGCCAC | ≥ 50 min | 10-copy/μL input | Specific over SARS-CoV | Green emission or LFD | Arizti-Sanz et al. (2020) | November 20, 2020 |
| Rw | ACTATCATCATCTAACCAATCTTCTTCTTG | ||||||||
| Cas13a crRNA | CUCUUCUUCAGGUUGAAGAGCAGCAGAA | ||||||||
| FQ/FB reporter |
6-FAM-rUrUrUrUrUrUrUrUrUrUrUrUrUrU-Biotin 6-FAM-rUrUrUrUrUrUrU-IABkFQ |
||||||||
| 8 | Orf1ab | Fw | TTGCCTGGCACGATATTACGCACAACTAATGGT | 20 min (42 °C) + up to 120 min (37 °C) for Cas12a detection | 1-copy/reaction (12.5 μL) | Specific over MERS-CoV, SARS-CoV, OC43/HKU1 | Green emission or LFD | Xiong et al. (2020) | December 15, 2020 |
| Rw | CAAGCTGATGTTGCAAAGTCAGTGTACTCTAT | ||||||||
| Cas12a crRNA | UAAUUUCUACUAAGUGUAGAUgugcaguugguaacaucuguuac | ||||||||
| FQ/FB reporter |
FAM-TTATT-BHQ1 FAM-TTATT-Biotin |
||||||||
| N | Fw | CTTCCTCAAGGAACAACATTGCCAAAAGGCT | 20 min (42 °C) + up to 120 min (37 °C) for Cas12a detection | 1–10-copy/reaction (12.5 μL) | Specific over SARS-CoV, MERS-CoV, OC43/HKU1 | Green emission or LFD | |||
| Rw | TCTAGCAGGAGAAGTTCCCCTACTGCTGCCTGG | ||||||||
| Cas12a crRNA | UAAUUUCUACUAAGUGUAGAUuugaacuguugcgacuacgugau | ||||||||
| FQ/FB reporter |
FAM-TTATT-BHQ1 FAM-TTATT-Biotin |
||||||||
| 9 | RdRp | Fw | TATGCCATTAGTGCAAAGAATAGAGCTCGCAC | 15 min | 2-copy/reaction (50 μL) | NOT specific over SARS-CoV | Green emission | Abd El Wahed et al. (2021) | January 20, 2021 |
| Rw | CAACCACCATAGAATTTGCTTGTTCCAATTAC | ||||||||
| Exo-probe | TCCTCTAGTGGCGGCTATTGATTTCAATAA/iBHQ1dT/idSp/iFAMdT/TTGATGAAACTGTCTATTG-Phosphate | ||||||||
| E | Fw | GAAGAGACAGGTACGTTAATAGTTAATAGCGTA | 15 min | 15-copy/reaction (50 μL) | NOT specific over SARS-CoV | Green emission | |||
| Rw | AAAAAGAAGGTTTTACAAGACTCACGTTAACsA | ||||||||
| Exo-probe | ATCGAAGCGCAGTAAGGATGGCTAG/iBHQ1dT/idSp/iFAMdT/AACTAGCAAGAATAC-Phosphate | ||||||||
| N | Fw | CCTCTTCTCGTTCCTCATCACGTAGTCGCAAC | 15 min | 15-copy/reaction (50 μL) | Specific over SARS-CoV | Green emission | |||
| Rw | AGTGACAGTTTGGCCTTGTTGTTGTTGGCCTT | ||||||||
| Exo-probe | TAGAATGGCTGGCAATGGCGGTGATGCTGC/iBHQ1dT/idSp/iFAMdT/TGCTTTGCTGCTGCTT-Phosphate | ||||||||
| 10 | N | Fw | CTAACAAAGACGGCATCATATGGGTT | 30 min (39 °C) + LFD | 10-copy/reaction (50 μL) | Specific over SARS-CoV, OC43/HKU1/NL63/229E, HBV human influenza A/B, RSV A/B, | LFD | Zheng et al. (2021) | February 1, 2021 |
| Rw | FITC-GGCCTTTACCAGACATTTTGCTCTCA | ||||||||
| Nfo-probe | Biotin-CTCTTCTCGTTCCTCATCACGTAGTCGCAAC/dSp/GTTCAAGAAATTCA-Phosphate |
Abbreviations: FITC, fluorescein isothiocyanate; FAM, fluorescein; 6-FAM, 6-carboxyfluorescein; C3, C3-blocking group; idFAMdT, internal FAM modified dT nucleotide; id6FAMdT, internal 6-FAM modified dT nucleotide; idTAMdT, internal TAMRA modified dT nucleotide; idSp, internal tetrahydrofuran (THF) spacer; idBHQ1, internal BHQ1 quencher modified dT nucleotide; idBHQ2, internal BHQ2 quencher modified dT nucleotide; iBMNQ535dT, internal BMNQ535 quencher modified dT nucleotide; s, phosphorothioate backbone.
7. Methods section
In the following, we will detail the methods using the WEPEAR protocol for ultrasensitive field-deployable detection of SARS-CoV-2 RNA.
Methods section (All Using Basic RT-ERA Kit).
8. Before you begin
Timing: few days
Design and customer synthesis of exo probes, nfo probes, and respective primers (few days);
Reconstitution of the obtained exo probes, nfo probes and respective primers to 10 μM as stock solutions; store at − 40 °C and thaw them before use;
Preparation of 5 U/μL murine RNase inhibitor in glycerol containing solution and store at − 40 °C before use;
Make exonuclease III at 200 U/μL and endonuclease IV at 10 U/μL; store at − 40 °C before use;
Preparation of viral RNA sample using a qualified RNA extraction kit.
9. Key resources table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Bacterial and virus strains | ||
| DH5α E. coli | HaiGene | Cat# K10119 |
| Recombinant proteins | ||
| Exonuclease III (exo III) | Thermo Scientific | Cat# EN0191 |
| Murine RNase inhibitor | Vazyme | Cat# R301-01 |
| Endonuclease IV (nfo enzyme) | Vazyme | Cat# R301-01 |
| NdeI | NEB | Cat# R0111S |
| HindIII | NEB | Cat# R3104S |
| Critical commercial assays | ||
| T7 high yield transcription kit | Vazyme Biotech Co., Ltd. | Cat# TR101-01 |
| Basic RT-ERA reaction kit | GenDx Biotech Co., Ltd. | Cat# KS102 |
| FastPure gel DNA extraction mini kit | Vazyme Biotech Co., Ltd. | Cat# DC301 |
| Express RNA purification kit | GenDx Biotech Co., Ltd | Cat# NR202-50T |
| Oligonucleotides | ||
| N-gene Exo Fw: tttggtggaccctcagattcaactggcagtaac | LoGenBio | N.A. |
| N-gene Exo Rw: gaatttaaggtcttccttgccatgttgagtgag | LoGenBio | N.A. |
| N-gene Exo probe: tattattgggtaaaccttggggccgacgttgtt/i6FAMdT/t/idSp/a/iBHQ1dT/cgcgccccactg-Phosphate | LoGenBio | N.A. |
| N-gene Nfo Fw: tttggtggaccctcagattcaactggcagtaac | LoGenBio | N.A. |
| N-gene Nfo Rw: tttggtggaccctcagattcaactggcagtaac | LoGenBio | N.A. |
| N-gene Nfo probe: 6-FAM-gcgatcaaaacaacgtcggccccaaggtttacc/idSp/aataatactgcgtct-Phosphate | LoGenBio | N.A. |
| S-gene Exo Fw: gtctctagtcagtgtgttaatcttacaaccagaac | LoGenBio | N.A. |
| S-gene Exo Rw: cattggaaaagaaaggtaagaacaagtcctgag | LoGenBio | N.A. |
| S-gene Exo probe: cctgcatacactaattctttcacacgtggtg/iTAMdT/t/idSp/A/iBHQ2dT/taccctgacaaagtt-Phosphate | LoGenBio | N.A. |
| S-gene Nfo Fw: gtctctagtcagtgtgttaatcttacaaccagaac | LoGenBio | N.A. |
| S-gene Nfo Rw: Biotin-cattggaaaagaaaggtaagaacaagtcctgag | LoGenBio | N.A. |
| S-gene Nfo probe: 6-FAM-cctgcatacactaattctttcacacgtggtg/idSp/ttattaccctgacaa-Phosphate | LoGenBio | N.A. |
| Recombinant DNA | ||
| SARS-CoV-2 N-gene plasmid | LoGenBio | N.A. |
| SARS-CoV-2 S-gene plasmid | LoGenBio | N.A. |
| SARS-CoV N-gene plasmid | LoGenBio | N.A. |
| SARS-CoV S-gene plasmid | LoGenBio | N.A. |
| MERS-CoV N-gene plasmid | LoGenBio | N.A. |
| MERS-CoV S-gene plasmid | LoGenBio | N.A. |
| Software and algorithms | ||
| SnapGene viewer | SnapGene | www.snapgene.com/snapgene-viewer |
| BLAST | NCBI | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| Image J | National Institutes of Health (NIH) | https://imagej.net/Downloads |
| Other | ||
| HybriDetect LF strips | Amplification Future | Cat# WLF8201 |
| DL5000 DNA marker | Vazyme | Cat# MD102 |
| 100 bp DNA ladder | Vazyme | Cat# MD104 |
| Agarose | Biosharp | Cat# BS081-100g |
| 50 × TAE buffer | Alphabio | Cat# A1558 |
| SYBR safe DNA gel stain | APExBio | Cat# A8743 |
| DNA sample loading buffer | Alphabio | Cat# A1550 |
10. Materials and equipment
-
•
Mini centrifuge (DLAB, D1008)
-
•
PCR machine (MIULAB, PR-96E)
-
•
Real-time PCR machine (Applied Biosystem, QuantStudio 6Flex)
-
•
Fluorescence spectrometer (Molecular Devices, SpectraMax i3x)
-
•
Pipettes (Eppendorf, 2.5, 10, 20, 200, 1000 μL)
-
•
Horizontal gel electrophoresis kit (LIUYI, DYCP-31C)
-
•
Heating block (DLAB, Mini HCL100)
-
•
Mini blue and white light imaging plate (LIUYI, WD-9403 ×)
11. Step-by-step method details
11.1. Detection of SARS-CoV-2 N- or S-gene using exo probes and primers
Timing: around 31 min
-
1.
For a 50 μL reaction system, 2.5 μL of exo forward primer (10 μM), 2.5 μL of exo reverse primer (10 μM), 0.75 μL of exo FRET probe (10 μM), 1 μL of 5 U/μL of murine RNase inhibitor, 1 μL of RNA sample, 20 μL of DI-H2O, and 0.5 μL of 200 U/μL exonuclease III were added into 20 μL dissolving buffer (DA solution).
-
2.
The resultant 48 μL of reaction solution was added into the PCR tube containing dry powder that includes all necessary enzymes and ingredients for RT-ERA to occur, and then gently pipette to mix the reaction mixture.
-
3.
Meanwhile, 2 μL of Mg(OAc)2 solution as the ERA activator was loaded into the lid of the PCR tube, then close the lid gently.
-
4.
Afterwards, the reaction vial was incubated in a 37 °C water bath for 60 s to allow solely transcription to take place.
-
5.
Centrifuge the tube to mix the Mg2 + activator with the RT-ERA reaction mixture, shake and centrifuge again to collect all liquids to the bottom of the PCR tube.
-
6.
Heat the PCR tube at 40 °C for 4 min using a heating block or PCR machine or water inside a thermos cup to allow pre-reaction.
-
7.
The PCR tube was shaken and spun again, and incubated at 40 °C for another 26 min to compete the RT-ERA reaction.
-
8.
Visualize the fluorescence signal by putting the PCR tube on top of a mini blue-light plate and capture the fluorescence image using a smartphone camera; alternatively, the fluorescence intensity can be quantified using Molecular Devices SpectraMax i3x with a 96-well plate.
Important notes:
-
(i)
The area around PCR machine where RT-ERA amplification is conducted can produce invisible aerosols in the air that contain the target amplicon. Hence the aerosols could potentially contaminate the surrounding areas. As a result, the area for setting up the RT-ERA reaction and the area for conducting the RT-ERA amplification should be strictly separated in order to prevent the contamination of the RT-ERA reaction solution by aerosols in the air.
-
(ii)
Since RNase ubiquitously exists which is detrimental for RNA samples, the whole working area should be carefully cleaned using DEPEC water and the equipment used (e.g. mini-centrifuge and pipettes) should also be swiped using DEPEC water prior to use; in addition, all consumables, e.g. tips, should be autoclaved after spraying with DEPEC water prior to use.
-
(iii)
Always wear appropriate personal protective equipment (PPE) including at least a lab coat, goggles, gloves, and a mask; wearing of a mask is required in order to prevent any foams/droplets that contain RNase and other contaminant to be expelled from the mouth and nose contaminating the RT-ERA reaction.
-
(iv)
Be highly cautious to avoid any part of your skin from touching the working bench, equipment and consumables when performing the SARS-CoV-2 detection because skin surfaces have a lot of secretions (like oil) that contain RNase and other contaminates.
-
(v)
Due to the ultrahigh sensitivity of this RT-ERA test, both positive control and negative control samples must be included in every test in order to exclude possible false positive and false negative results.
11.2. Detection of SARS-CoV-2 N- or S-gene using nfo probes and lateral flow strips
Timing: around 31 min + around 3 min of LFD
-
1.
For a 50 μL reaction system, 2 μL of nfo forward primer (10 μM), 2 μL of nfo reverse primer (10 μM), 0.6 μL of nfo probe (10 μM), 1 μL of 5 U/μL of murine RNase inhibitor, 1 μL of RNA sample, 21 μL of DI-H2O, and 1 μL of 10 U/μL endonuclease IV (nfo enzyme) were added into 20 μL dissolving buffer (DA solution).
-
2.
The resultant 48 μL of reaction solution were added into a PCR tube containing dry powder that includes all necessary enzymes and ingredients for RT-ERA to occur, and then gently pipette to mix the reaction mixture.
-
3.
Meanwhile, 2 μL of Mg(OAc)2 solution as the ERA activator was loaded into the lid of the PCR tube, and then the lid was closed gently.
-
4.
Afterwards, the reaction vial was incubated in a 37 °C water bath for 60 s to allow solely transcription to take place.
-
5.
Centrifuge the tube to mix the Mg2 + activator with the RT-ERA reaction mixture, shake and centrifuge again to collect all liquids to the bottom of the PCR tube.
-
6.
Warm the PCR tube at 40 °C for 4 min using a heating block, a PCR machine or water bath inside a thermos cup to allow for pre-reaction.
-
7.
The PCR tube was shaken and centrifuge again, and incubate at 40 °C for another 26 min to complete the RT-ERA reaction.
-
8.
10 μL of nfo reaction solution was diluted into 200 μL by DI-H2O in a 1.5 mL centrifuge tube.
-
9.
The sample pad end of the HybriDetect LF strip was dipped into the diluted reaction solution.
-
10.
Read the detection result after 2–3 min that if both the test line and control line turn red, SARS-CoV-2 viral RNA is detected.
Important notes:
-
(i)
The area around the PCR machine where RT-ERA amplification is conducted can produce invisible aerosols in the air that contain the target amplicon. Hence the aerosols could potentially contaminate the surrounding areas. As a result, the area for setting up the RT-ERA reaction and the area for conducting the RT-ERA amplification should be strictly separated in order to prevent the contamination of the RT-ERA reaction solution by aerosols in the air.
-
(ii)
Since RNase ubiquitously exists, which is detrimental for RNA samples, the whole working area should be carefully cleaned using DEPEC water and the equipment used (e.g. mini-centrifuge and pipettes) should also be swiped using DEPEC water prior to use; in addition all consumables, e.g. tips, should be autoclaved after spraying with DEPEC water prior to use.
-
(iii)
Always wear appropriate PPE including at least a lab coat, goggles, gloves, and mask; wearing of a mask is required in order to prevent any foams/droplets that contain RNase and other contaminants to be expelled from the mouth and nose contaminating the RT-ERA reaction.
-
(iv)
Be highly cautious to avoid any part of your skin from touching the working bench, equipment and consumables when performing the SARS-CoV-2 detection because skin surfaces have a lot of secretions (like oil) that contain RNase and other contaminates.
-
(v)
Due to the ultrahigh sensitivity of this RT-ERA test, both a positive control and a negative control samples must be included in every test in order to exclude possible false positive and false negative results.
-
(vi)
After the LF strip is dipped into the diluted RT-ERA reaction, the results should be read within the initial few minutes to get a reliable readout because the test line may still gradually, although slowly, develop some light red colour after an extended duration even for a blank or negative control sample.
11.3. Optional steps
Timing: < 1 min
-
1.
For single-copy sensitivity assay, more internal vortex steps are advised, such as at 3rd, 6th and 9th min
11.4. Simultaneous dual N- and S-gene detection using paired exo probe and primers for N-gene and exo probe and primers for S-gene
Timing: > 30 min
-
1.
For a 50 μL reaction system, 0.5 μL of exo forward primers (10 μM) for N- and S- gene (final 100 nM), 0.5 μL of exo reverse primers (10 μM) for N- and S-genes (final 100 nM), 0.3 μL of exo probes (each 5 μM) for N- and S-genes (final 30 nM for each exo probe), 1 μL of 5 U/μL of murine RNase inhibitor, 1 μL RNA sample, 23 μL of DI-H2O, and 0.5 μL of 200 U/μL exonuclease III were added into 20 μL of DA solution.
-
2.
The resultant ~ 48 μL of reaction solution was added into the PCR tube containing dry powder that includes all necessary enzymes and ingredients for RT-ERA to occur; and then gently pipette to mix the reaction mixture.
-
3.
Transfer the PCR mixture to a 0.2 mL 8-strip PCR tubes equipped with transparent and flat lids.
-
4.
Meanwhile, 2 μL of Mg(OAc)2 solution as the ERA activator was loaded into the lid of the PCR tube strip, and then close all lids gently.
-
5.
Afterwards, the PCR tube strip was incubated in a 37 °C water bath for 60 s to allow solely the transcription to take place.
-
6.
Centrifuge the PCR tube strip to mix the Mg(OAc)2 activator into the RT-ERA reaction mixture, shake and centrifuge again.
-
7.
Load the PCR strips into Applied Biosystem QuantStudioTM 6Flex real-time PCR machine according to the instructions of the manufacturer.
-
8.
Record the fluorescence increase of both fluorescein and TAMRA channels.
12. Summary
In this chapter, we have described nucleic acid-based approaches for sensitive detection of SARS-CoV-2 with a particular focus on the recombinase polymerase amplification that belongs to the isothermal amplification category. Sensitivity is the key to avoid false negative results and would be highly beneficial to prevent the spread of a pandemic. Some recent reports have already pointed out the necessity to avoid insensitive methods, e.g. some serological tests, for practical SARS-CoV-2 diagnosis. According to recently published papers, more than one RT-RPA-based reports have achieved single-copy sensitivity, the highest sensitivity in diagnosis. This is generally higher than the widely used RT-PCR methods and is clearly higher than other testing approaches. In order to achieve a high sensitivity, primers and probe should be rationally designed in order to achieve the best performance. Specificity should also be taken into account when designing primers and probes for sensitive viral RNA detection. The region within an RNA genome for RT-RPA amplification also needs to be carefully taken into account. Unlike the direct detection of DNA using RPA, the detection of RNA using RPA requires an additional reverse transcription step. However, the reaction condition of reverse transcription is typically not fully consistent with the conditions used in RPA; and moreover, some reagents used in RPA (e.g. the catalyst Mg2 +) can potentially inhibit reverse transcription. In order to make the two steps fully compatible, whole-course encapsulated procedure for exponential amplification from RNA (WEPEAR) protocol has been introduced for “sample-in, results-out” one tube detection of viral RNA sample (Xia and Chen, 2020a, Xia and Chen, 2020b).
RT-RPA has several distinct advantages; however, it also has some limitations. RT-RPA is performed under a constant temperature, in the range between room temperature to 42 °C without the need for thermocyclers. As a result, highly costly real-time PCR machine is not compulsory for performing the detection. RT-RPA could be coupled with either fluorogenic detection using a blue light imaging plate, or lateral flow strips without any particular imaging devices. All these features make RT-RPA highly field-deployable and would be rather suited for grassroots clinics. In fact, we have shown that RT-PRA coupled with LF test using thermos cups as the heating device can be conducted in a house-hold fashion (Xia and Chen, 2020a, Xia and Chen, 2020b). Another key advantage of RT-RPA is the fast detection rate with a typical sample-to-readout time in the range of 20–30 min, and even within 20 min. This is clearly shorter than the RT-LAMP method (usually > 60 min) and RT-PCR methods (typically > 2 h). On the other hand, the RT-RPA reaction contains multiple components, including several enzymes, proteins, energy-supply system, crowding agents, aside from primers and probes. This makes a standard RT-RPA reaction relatively more expensive. However, given that no sophisticated instrumentation is required and that diagnosis time is much shorter, the general cost for performing an RPA assay could still be reduced to a comparative level to other nucleic acid-based detection approaches as suggested by some previous evaluations (Londono, Harmon, & Polston, 2016). Along with the further development of RT-RPA, such as the digital version, microfluidic version and so on, and the fact that there are several unique advantages of RT-RPA (e.g. ultrahigh sensitivity), we predict RT-RPA approach will show great potential for sensitive and field-deployable viral RNA detection in the future.
Acknowledgement
We thank the research fund from Harbin Institute of Technology, and National Natural Science Foundation of China (grant No. 32071410 to X.C.) for the support of this work.
References
- Abd El Wahed A., Patel P., Maier M., Pietsch C., Ruster D., Bohlken-Fascher S., et al. Suitcase lab for rapid detection of SARS-CoV-2 based on recombinase polymerase amplification assay. Analytical Chemistry. 2021;93(4):2627–2634. doi: 10.1021/acs.analchem.0c04779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nature Medicine. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nature Communications. 2020;11(1):5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann O., Bachmann I., Spiegel M., Schramm M., Abd El Wahed A., Dobler G., et al. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (Exo-IQ) Clinical Chemistry. 2020;66(8):1047–1054. doi: 10.1093/clinchem/hvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes and Infection. 2020;22(2):69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.J., Liu W.Y., Zhang Q., Xu K., Ye G.M., Wu W.C., et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerging Microbes & Infections. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y.Z., et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clinical Chemistry. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nucleic-acid sequence-based amplification. Nature. 1991;350(6313):91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher R.K., Stewart G., Boissinot M., Bergeron M.G. Recombinase polymerase amplification for diagnostic applications. Clinical Chemistry. 2016;62(7):947–958. doi: 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database of Systematic Reviews. 2020;8:1–129. doi: 10.1002/14651858.CD14013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohla M., Boesecke C., Schulte B., Diegmann C., Sib E., Richter E., et al. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health. 2020;182:170–172. doi: 10.1016/j.puhe.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A.S., Alawneh J.I. COVID-19 infection diagnosis: Potential impact of isothermal amplification technology to reduce community transmission of SARS-CoV-2. Diagnostics. 2020;10(6):399. doi: 10.3390/diagnostics10060399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashir J., Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Medical Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: Current status and challenges. iScience. 2020;23(8):101406. doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Macdonald J., von Stetten F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification (vol 144, pg 31, 2019) Analyst. 2020;145(5):1950–1960. doi: 10.1039/c9an90127b. [DOI] [PubMed] [Google Scholar]
- Londono M.A., Harmon C.L., Polston J.E. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virology Journal. 2016;13:48. doi: 10.1186/s12985-016-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Muller C., et al. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA) Lab on a Chip. 2010;10(7):887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28(12) doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biology. 2006;4(7):1115–1121. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Boswell S.A., Chidley C., Lu Z.X., Pettit M.E., Gaudio B.L., et al. An enhanced isothermal amplification assay for viral detection. Nature Communications. 2020;11(1):5920. doi: 10.1038/s41467-020-19258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z., Peng R., Baravik I.K., Liu X.Y. Perspective fighting COVID-19: Integrated micro- and nanosystems for viral infection diagnostics. Matter. 2020;3(3):628–651. doi: 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Davydova E.K., Du W.B., Kreutz J.E., Piepenburg O., Ismagilov R.F. Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Analytical Chemistry. 2011;83(9):3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.Z., Zhou Y., Ye J.W., Al-Maskri A.A.A., Kang Y., Zeng S., et al. Recent advances and perspectives of nucleic acid detection for coronavirus. Journal of Pharmaceutical Analysis. 2020;10(2):97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W., et al. Clinical performance of two SARS-CoV-2 serologic assays. Clinical Chemistry. 2020;66(8):1055–1062. doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu F., Yang X.T., Xu S.K., Chen D.J., Zhou L., Ge X.N., et al. Development of a fluorescent probe-based real-time reverse transcription recombinase-aided amplification assay for the rapid detection of classical swine fever virus. Transboundary and Emerging Diseases. 2020 doi: 10.1111/tbed.13849. [DOI] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., et al. Diagnosing COVID-19: The disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Vincent M., Xu Y., Kong H.M. Helicase-dependent isothermal DNA amplification. EMBO Reports. 2004;5(8):795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G.T., Fraiser M.S., Schram J.L., Little M.C., Nadeau J.G., Malinowski D.P. Strand displacement amplification—An isothermal, in vitro DNA amplification technique. Nucleic Acids Research. 1992;20(7):1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai K., He X., Shen X., Wang J., Liu J., et al. Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clinical Microbiology and Infection. 2020;26(8):1076–1081. doi: 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern (vol 395, pg 470, 2020) Lancet. 2020;395(10223):496. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Qian C., Wu H., Qian W.J., Wang R., Wu J. Technical aspects of nicking enzyme assisted amplification. Analyst. 2018;143(6):1444–1453. doi: 10.1039/c7an02037f. [DOI] [PubMed] [Google Scholar]
- WHO . Online Post; 2020. Statement on the first meeting of the international health regulations (2005) Emergency committee regarding the outbreak of novel coronavirus (2019-nCoV) [Google Scholar]
- Wong Y.P., Othman S., Lau Y.L., Radu S., Chee H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. Journal of Applied Microbiology. 2018;124(3):626–643. doi: 10.1111/jam.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Ge Y.Y., Zhao K.C., Zhu X.J., Chen Y., Wu B., et al. A reverse-transcription recombinase-aided amplification assay for the rapid detection of N gene of severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) Virology. 2020;549:1–4. doi: 10.1016/j.virol.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Chen X. Ultrasensitive and whole-course encapsulated field detection of 2019-nCoV gene applying exponential amplification from RNA combined with chemical probes. ChemRixv. 2020 doi: 10.26434/chemrxiv.12012789. [DOI] [Google Scholar]
- Xia S., Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discovery. 2020;6(1):37. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong D., Dai W.J., Gong J.J., Li G.D., Liu N.S., Wu W., et al. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biology. 2020;18(12) doi: 10.1371/journal.pbio.3000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G.H., Li S.L., Zhang W.W., Du B., Cui J.H., Yan C., et al. Reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2) Analytical Chemistry. 2020;92(14):9699–9705. doi: 10.1021/acs.analchem.0c01032. [DOI] [PubMed] [Google Scholar]
- Zhao Y.X., Chen F., Li Q., Wang L.H., Fan C.H. Isothermal amplification of nucleic acids. Chemical Reviews. 2015;115(22):12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- Zhao S., Lin Q.Y., Ran J.J., Musa S.S., Yang G.P., Wang W.M., et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. International Journal of Infectious Diseases. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.Z., Chen J.T., Li J., Wu X.J., Wen J.Z., Liu X.Z., et al. Reverse transcription recombinase-aided amplification assay with lateral flow dipstick assay for rapid detection of 2019 novel coronavirus. Frontiers in Cellular and Infection Microbiology. 2021;11:613304. doi: 10.3389/fcimb.2021.613304. [DOI] [PMC free article] [PubMed] [Google Scholar]