Abstract
At the very beginning of the new decade, the COVID-19 pandemic has badly hit modern human societies. SARS-CoV-2, the causative agent of COVID-19 acquiring mutations and circulating as new variants. Herein, we have found three new antiviral peptides (AVPs) against the SARS-CoV-2. These AVPs are analogous to the spike glycoprotein of the SARS-CoV-2. Antiviral peptides, i.e., Seq12, Seq12m, and Seq13m, can block the receptor-binding domain (RBD) of the SARS-CoV-2, which is necessary for communicating with the angiotensin-converting enzyme 2 (ACE2). Also, these AVPs sustain their antiviral properties, even after the insertion of 25 mutations in the RBD (Rosetta and FoldX based). Further, Seq12 and Seq12m showed negligible cytotoxicity. Besides, the binding free energies calculated using MM-PB/GBSA method are also in agreement with the molecular docking studies. The molecular interactions between AVPs and the viral membrane protein (M) also showed a favorable interaction suggesting it could inhibit the viral re-packaging process. In conclusion, this study suggests Seq12, Seq12m, and Seq13m could be helpful to fight against SARS-CoV-2. These AVPs could also aid virus diagnostic tools and nasal spray against SARS-CoV-2 in the future.
Keywords: SARS-CoV-2, spike glycoprotein, antiviral peptide, receptor-binding domain, mutation
Graphical abstract
1. Introduction
At present, the entire World is facing challenges to handle the COVID-19 pandemic [1]. SARS-CoV-2 is the causative agent of COVID-19, and it is new to the scientific community. Therefore, in-depth monitoring of all aspects of the COVID-19 is ongoing. For example, signs and symptoms [2], mode of transmission [3], WHO-solidarity trials [4], contact tracing by mobile apps such as “Arogya Setu” by India [5], CRISPR based rapid diagnostic of SARS-CoV-2 [6], and also monitoring daily cases by crowd-sourcing (https://www.covid19india.org/). Besides, recent reports suggest that repurposing known antiviral drugs [7], drugs [8], different phytochemicals [9] against COVID-19 could be fruitful. However, none of them has reached a final definitive clinical treatment for COVID-19. Indeed to address the urgent need for a safe and efficacious vaccine against the COVID-19 several vibrant initiatives have been started as never before. For example, vaccine manufacturing front-runner come-up with mRNA vaccines [10], viral vector vaccine [11], classical attenuated vaccine etc. [12]. However, reports showed COVID-19 vaccines would not be a silver bullet for the immunization of a community.
Furthermore, alternatives to traditional therapeutics would be necessary for the long run as before [13,14]. Moreover, SARS-CoV-2 acquires new mutations in its genome in a concise time frame [15], for instance, B.1.1.7, B.1.351, P.1, B.1.427, and B.1.429, etc. [16]. These SARS-CoV-2 variants are one of the main concerns for all anti-COVID-19 efforts [17]. SARS-CoV-2 encodes a spike glycoprotein that assembled as a trimer for function. The notorious entry of SARS-CoV-2 inside a human host is primarily mediated via a protein-protein interaction between angiotensin-converting enzyme 2 (ACE2) and the receptor-binding domain (RBD) of the SARS-CoV-2 [18]. Therefore, RBD holds importance as a potential pharmaceutical target. Herein, we made an effort to find out new antiviral peptides against SARS-CoV-2. In such an effort, we utilized Machine Learning and Supported Vector Machine for antiviral peptide predictions. After that, we opt for residue-resistant molecular docking, molecular dynamics simulations, and MM-PB/GBSA analysis to characterize the antiviral peptides and the molecular interactions between AVP-RBD complexes. Hydrogen bonds (H-bonds) are weak non-covalent interactions with a binding strength of one-tenth than a typical covalent bond [76]. However, H-bonds are essential for proteins and nucleic acids function [76,77]. And this study is no exception, as many H-bonds were formed during the molecular dynamics trajectory between the AVP-RBD complex and water. Best of our knowledge, this is the first report of antiviral peptides against the SARS-CoV-2 derived from its RBD of the spike glycoprotein.
2. Materials and methods
2.1. Data sampling and analysis
Nucleotide sequences of the SARS-CoV-2 were obtained from NCBI viruses [19]. The complete genome sequence of the SARS-CoV-2 strain Wuhan-Hu-1 (GenBank Sequence Accession: MN908947) was used as the query sequence. The viral spike glycoprotein (S-Protein) and membrane protein (M-Protein) (QHD43416.1) were studied using PSI-BLAST. Homology models of the M- and S-Protein were built using i-TASSER and SWISS-MODEL, respectively [20,21], followed by structure validation using PROCHECK [22].
2.2. Motif search
The aligned sequences of spike protein resulted from the PSI-BLAST were used to discover motifs using the MEME suite [23,24]. The primary query included a set of 69 protein sequences, between 16 to 1196 amino acids in length (average length 214.3 amino acids), with the following settings: Background: A order-0 background generated from the supplied sequences. Discovery Mode: Classic: optimize the E-value of the motif information content. Site Distribution: Zero or one occurrence (of a contributing motif site) per sequence. Motif Count: for 3 motifs. Motif Width: Between 15 wide and 47 wide (inclusive).
2.3. Antiviral property prediction
Identified motifs were checked and validated as antiviral peptide using a Machine Learning algorithm (MLA), and Supported Vector Machine (SVM), i.e., Meta-iAVP [25], and AVPred [26] with four prediction model settings viz., AVP motif, sequence alignment, composition analysis, physio-chemical properties (threshold value of 50). The theoretical half-maximum inhibitory concentration (IC50) value was calculated using AVP-IC50Pred [27].
2.4. Homology modeling and peptide characterizations
The predicted antiviral peptide sequences were used to build three-dimensional (3D) structures using the PEP-FOLD (v3.5) [28]. The structures obtained were then subject to energy minimization (GROMACS, v5.0) to remove unusual torsions and clashes from their structural geometry [29]. Properties of the predicted antiviral peptides were calculated using Innovagen peptide property calculator [30], ProtParam [31], AIPpred [53] AlgPred (Mapping of IgE epitopes and PID, MEME/MAST motif, Blast search on allergen representative peptides (ARPs)), [32], ToxinPred (SVM (TrEMBL) + Motif based and SVM (Swiss-Prot) based), [33], HemoPred, (SVM + Motif (HemoPI-2) based), IL4pred (Hybrid (SVM + motif) based), [34]. AVP-IC50Pred (Hybrid Model Features a: Composition (mono-di) + Physico + Secondary structure + Surface accessibility and Features b: Binary (N8/C8) + Physico + Secondary structure + Surface accessibility) was used by selecting a Machine Learning Techniques: Supported Vector Machine (SVMlight) Random Forest (R package), IBk (Weka), KStar (Weka), [27].
2.5. In silico mutant model of the receptor-binding domain
Crystal structure of the receptor-binding domain (6W41) of the SARS-CoV-2 was obtained from RCSB-PDB [35]. We manually select Chain C of the 6W41 as it is annotated as RBD for ACE2. The in silico multi-point mutant models were build using FireProt [36].
2.6. Molecular docking
HADDOCK-v2.2 was used for molecular docking between AVPs and receptor-binding domain (RBD) of the viral spike glycoprotein, viral membrane protein (M), RNA-dependent-RNA-polymerase (RdRp) [37]. Results were visualized using Discovery Studio Visualizer [38].
2.7. Molecular dynamics (MD) simulation
The MD-simulation of the antiviral peptides, i.e., Seq12, Seq12m, and Seq13m, were carried out for 50 ns using GROMACS (v2019) [39]. In addition, 100 ns MD-simulation studies of the AVP-RBD complexes were performed using NAMD as described previously [8,40]. In brief, input files were generated using the web-based server CHARMM-GUI (http://www.charmm-gui.org/). All systems were neutralized with KCl at 0.15 Molar, using the Monte-Carlo ion placing method. Systems were solvated using TIP3 water model, and CHARMM36m force field was used to assign charges. The MD simulations were performed in two stages. At the first stage of the MD run, the energy minimization was performed for 20,000 steps by the steepest descent method followed by restrained 5 ns-equilibration at NVT ensemble. We did not provide any trajectory analysis for this stage as it is a restrained system and would be a biased and unreliable examination in our study. At the second stage, unrestrained 100 ns production at 310 K was performed under the NPT ensemble. RMSD, RMSF, Radius of gyration (Rg), H-bonds, and SASA were analyzed using VMD. The average values of multiple replicated MD simulations were used for analysis. A detailed method of the MD simulation is described in the supplementary section.
2.7.1. Binding free energy calculation using MM-PB/GBSA method
The molecular mechanics Poisson-Boltzmann and Generalized Born and Surface Area continuum solvation (MM-PB/GBSA) methods were employed for the calculation of the binding free energy of the AVP-RBD/RBDm complexes. MM-PBSA calculations were performed using the Calculation of Free Energy (CaFE) tools [41]. In addition, MM-GBSA calculations all-atom MD simulations were performed using the AMBER 16 package with the FF99SB for the protein and peptide molecules. Entropy results (harmonic approximation) were calculated with NMODE analysis tool in AMBER 16 package [42,43]. Additionally, HawkDock was also used for MM-GBSA free energy calculations (per amino acid residues) [44]. The results were analyzed using GraphPad Prism 6 (San Diego, CA, USA). A detailed method is described in the supplementary section.
2.8. Characterization of immunogenic properties
Antiviral peptides were checked for their immunogenic properties using the IEDB epitope analysis tool, which includes sequential B-cell epitopes [45], T-cell (MHC-II), NetCTL-1.2 [46] and, mapped within the predicted epitopes.
3. Results and discussion
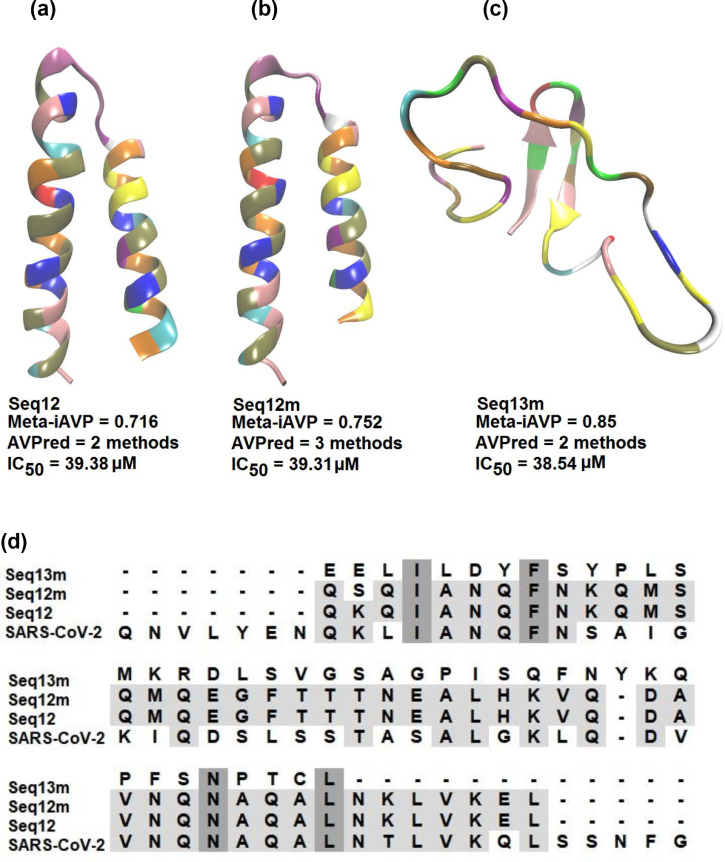
Genome sequences of the SARS-CoV-2 from the World's different continents were obtained from NCBI Viruses [19]. The nucleotide sequence of different genes enables researchers to look into the virus behavior from a distance. Moreover, bioinformatics tools accelerate new alternatives of therapeutics before expensive, extensive research work. An antiviral peptide is one of such new alternatives [47]. Besides, antiviral peptides are successful in combating the SARS-CoV and MERS-CoV [48], [49], [50]. Therefore, with a similar kind of anticipation for the SARS-CoV-2, we have found three new antiviral peptides against the SARS-CoV-2 (Fig. 1 a-c). Initially, the peptide sequences were interpreted as an antiviral peptide using AVPred [26] and Meta-iAVP [25]. The amino acid sequences of the predicted antiviral peptides were then mapped within the spike glycoprotein of the SARS-CoV-2. It is noteworthy that antiviral peptides Seq12, Seq12m, and Seq13m are analogous peptides of the spike glycoprotein (Fig. 1d). Fasta sequences of the antiviral peptides are available in the supplementary section, Fig. S1.
Fig. 1.
Peptide sequence mapping and three-dimensional structure of the AVPs. (a) Seq12, (b) Seq12m, (c) seq13m, (d) sequence mapping of the AVPs with the spike glycoprotein of the SARS-CoV-2 (Q913 – G972).
AVPred is an antiviral peptide prediction server that is based on amino acid sequence features viz., motifs, alignment, amino acid composition, and physicochemical properties [26]. Finally, the prediction of the antiviral peptide is made during 5-fold cross-validation using Supported Vector Machine (SVM). Antiviral peptides were predicted by the AVPred (based on the physicochemical model) can achieve up to 85% prediction accuracy with 0.70 Matthew's Correlation Coefficient (MCC). However, the experimental validation dataset shows 86% prediction accuracy with 0.71 MCC. Conversely, Meta-iAVP is based on a novel sequence-based meta-predictor with an effective features representation derived from Machine Learning Algorithms (MLA) and MLA types of features [25]. Interestingly, MLA and MLA types features have increased the overall prediction accuracy and MCC by 95.20%, and 0.90 respectively. At present more than 15 peptide-based drugs are in the pipeline of clinical trials [25]. However, a most worrying concern of an antiviral peptide is its immunogenic profile [51]. A low immunogenic profile is the desired characteristic of an antiviral peptide because a low immunogenic profile reduces the chances of elimination by the host defense system [52]. Therefore, to intensify the immunogenic profile of the AVPs, we have performed epitope prediction using IEDB tools. Soon after epitope prediction, AVP sequences were mapped within the aligned sequence of the epitopes (only most frequently human alleles were chosen).
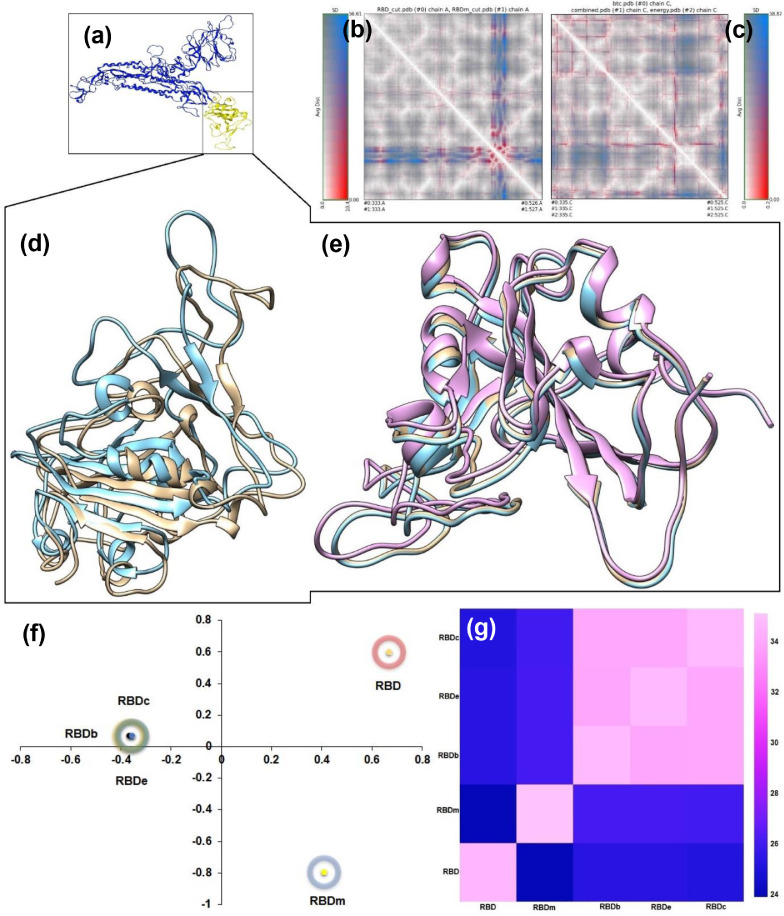
Results showed that the epitopes do not have significant similarities with the AVPs (Fig. S2). A few amino acid residues were apparently similar to the predicted epitopes. However, the binding postures (AVP-RBD) suggest these apparently similar amino acid residues were engaged with the AVP-RBD interactions. Further, the AVPs in this study can induce IL-4, an anti-inflammatory cytokine [53]. However, they are not capable of inducing IL-10 (a pro-inflammatory cytokine). Furthermore, AVPs were also non-allergenic as they do not have any known epitope for human IgE [32]. Other physicochemical properties viz., estimated half-life, instability index, water-solubility, theoretical IC50 values, etc., are summarized in Table 1 . It is noteworthy that Seq12 and Seq12m have excellent PROB Score compared to the previously reported antiviral peptides, i.e., P7, P8, P9, and P10 (Fig. 2 ). These P7, P8, P9, and P10 are actually human ACE2 peptide mimics, and they block SARS-CoV-2 pulmonary cell infections [54]. In addition, we have found that the predicted IC50 [27] values of these human ACE2 peptide mimics are 41.13, 41.38, 40.81, and 42.06 µM, respectively for P7, P8, P9, and P10 (Fig. 2). However, antiviral peptides viz., Seq12, Seq12m, and Seq13m in this study have predicted IC50 of only 39.38 µM, 39.31 µM, and 38.54 µM (Table 1), suggesting these AVPs might be promising too. Furthermore, other research groups have found antiviral peptides from a variety of different sources [47], such as Fibronectin by Beddingfield et al. [55], Aprotinin by Bestle et al. [56], Glycopeptide antibiotic by Zhang et al. [57], mouse β-defensin-4 by Zhao et al. [58], PCSK target motif by Cheng et al.[59], and from heptad repeats 1 and 2 (HR1 and HR2) in the S protein [60], [61], [62], [63]. However, in this study, Seq12, Seq12m, and Seq13m were derived from the RBD of the spike glycoprotein of the SARS-CoV-2. Besides, previous peer-reviewed works have failed to demonstrate how the studied peptides are effective against mutant RBD of the SARS-CoV-2. Here comes the uniqueness of this study, which suggests that Seq12, Seq12m, and Seq13m can sustain their properties with the mutant RBD model of the SARS-CoV-2 (RBDm). Moreover, a recent study on the classification of the circulating strains of novel coronavirus 2019 suggests there are A, B, and C types of coronavirus. A-type coronavirus is ancient/old, B-type is a bit new, meaning that it has acquired dozens of new mutations, and C-type is the current type, more contagious, and has more mutations [64]. The results depicted from this study suggest Seq12, Seq12m, and Seq13m might be effective to type A, B and C as well. As mentioned earlier, the interactions between RBD-ACE2 are essential for viral entry inside a human host cell [65]. A recent study has identified the molecular interactions between the RBD and ACE2 interactions [66], which include six aromatic amino acid residues viz., TYR449, TYR453, PHE456, PHE486, TYR489, and TYR505, five polar uncharged amino acid residues viz., ASN487, GLN493, GLN498, THR500, and ASN501. And three non-polar aliphatic amino acid residues, viz., LEU455, GLY496, and GLY502. We utilized this information in molecular docking studies to zoom in on the molecular interactions of the AVPs-RBD complexes. However, there is an increasing amount of fear of mutations in the RBD, which would probably make all anti-COVID-19 efforts nil [67]. Indeed, it is true that if mutations are incorporated into the RBD, then therapeutics have to evolve accordingly. Therefore, we have constructed a few in silico mutant models of the receptor-binding domain of the SARS-CoV-2, such as energy mutant (RBDe), evolutionary mutant (RBDb), and combined mutant (RBDc) (Fig. 3 ). The amino acid substitutions were incorporated based on Rosetta and FoldX suggestions [36] (Table 2 ). Additionally, the combined mutant model was again manually modified by following amino acid substitutions F486P, Y489F, Q493M, G496P, Q498L, T500Y, N501L, and Y505A. These manual amino acid substitutions have led to an improvement of the free energy of the combined mutant (Table 2) from - 27.63 kcal/mol (17 mutations) to -32.88 kcal/mol (25 mutations). It is noteworthy that N501Y is a common amino acid substitution among SARS-CoV-2 variants viz., B1.1.7, P.1, and B1.351, SARS-CoV-2 Variants of Concern [68]. However, according to FoldX and Rosetta, N501L (-0.997 kcal/mol) substitution is better than N501Y (-0.668 kcal/mol) (Fig. S3). Therefore, we speculate that Seq12, Seq12m, and Seq13m might also be effective for the B1.1.7, P.1, and B1.351 SARS-CoV-2 variants. However, detailed studies are required for further confirmation. Besides, we have found that among the critical RBD-ACE2 interacting amino acid residues [65], only N487 is conserved. Therefore mutation in the RBD of the SARS-CoV-2 is wide open. A detailed structural analysis of the receptor-binding domains (RBD) of the SARS-CoV-2 has been summarized in Fig. 3. The RRdisMap analysis showed that three-dimensional structures of the manual mutant of RBD (RBDm) and RBD could be overlapped. However, RBD and RBDm have an average distance of 51.74 (SD:1.486, at the selected site of the map), suggesting they are structurally different (Fig. S4). Furthermore, the RBDb/c/e are structurally distinguished from RBDm/RBD, and they cannot be overlapped with either RBD or RBDm. An average distance of the RBDb/c/e was 28.884 (SD:0.032, at the selected site of the map), suggesting they are structurally close to each other. In addition, the correspondence analysis also suggests RBDb/c/e are structural neighbors. On the other hand, RBD and RBDm are structurally distinguished from RBDb/c/e. The structural tree (Fig. S5) showed that RBDb, RBDc, and RBDe are orthologous. Conversely, RBD/RBDm are paralogous. Moreover, a comparison of all four mutant types of the RBD (by selecting the wild type RBD as a reference) revealed that all models have exactly the same RMSD value of 1.7 except the RBDe (RMSD = 1.5). Besides, RBDc is 96% identical with the wild type RBD, followed by RBDm (94%), RBDe (88%), and RBDb (84%). The Dali Z-scores are 25.3, 25.3, 25.2, and 23.9, respectively, for RBDm, RBDc, RBDe, and RBDb (Table 3 ).
Table 1.
Physicochemical and biological properties of the antiviral peptides
| Properties | Antiviral peptides |
||
|---|---|---|---|
| Seq12 | Seq12m | Seq13m | |
| APVpred1, 2, 3, 4, 5 | AVP3,5 | AVP3,4,5 | AVP4,5 |
| Meta-iAVP6 | 0.716 | 0.752 | 0.85 |
| PROB-HemoPI7 | 0.17 | 0.17 | 0.46 |
| Hydrophobicity | -0.23 | -0.21 | -0.10 |
| Steric hindrance | 0.69 | 0.67 | 0.65 |
| Solvation | 0.38 | 0.39 | 0.65 |
| Hydropathicity | -0.78 | -0.70 | -0.25 |
| Amphiphilicity | 0.72 | 0.62 | 0.73 |
| Hydrophilicity | 0.09 | 0.03 | -0.20 |
| Net hydrogen | 47.0 | 46.0 | 30.0 |
| Charge (pH = 7) | 1.1 | 0.0 | -1.0 |
| Isoelectric point (pI) | 8.71 | 7.10 | 4.79 |
| Molecular weight8 | 5302.68 | 5374.75 | 4657.83 |
| Instability index9 | 37.74s | 46.15u | 53.32u |
| Aliphatic index | 72.77 | 72.77 | 76.10 |
| GRAVY10 | -0.777 | -0.711 | -0.0246 |
| Estimated half life11 | 0.8 h | 0.8 h | 1 h |
| Water solubility | Good | Good | Poor |
| Epitope for IgE12 | Non-allergen | Non-allergen | Non-allergen |
| IL-4 inducer13 | Non-inducer* | Non-inducer* | Non-Inducer* |
| IL-10 inducer14 | Inducer | Inducer | Inducer |
| IC50 | 39.38 µM⁎⁎ | 39.31 µM⁎⁎ | 38.54 µM⁎⁎ |
AVPpred is available at http://crdd.osdd.net/servers/avppred.
Motif.
Align
Composition
Physicochemical composition.
Meta-iAVP is available at http://codes.bio/meta-iavp/.
Hemolytic prediction = PROB score HemoPI was calculated using HemoPI-1/1+motif and (SVM+Motif (HemoPI-2)).
Molecular weight in dalton.
Instability index : s = stable, u = unstable.
GRAVY = Grand average of hydropathy.
Predicted half-life in mammalian reticulocytes.
Calculated using AlgPred, it is available at http://crdd.osdd.net/raghava/algpred/.
IL-4 inducer predictions are made using the SVM method (SVM Threshold = 0.5). *However, the complete antiviral sequence contains two motifs to induce the IL-4 and hydride method (SVM+motif ) score is 1.04, suggesting it is an IL-4 inducer peptide.
L-10 inducing probability was calculated using RANDOM FOREST probability threshold = 0.5 and SVM method.
The half-maximal inhibitory concentration (IC50). It is a theoretical value.
Fig. 2.
Comparison of antiviral properties of Seq12, Seq12m, and Seq13m with other anti-SARS-CoV-2 peptides. *PROB score is the normalized SVM score and ranges between 0 and 1, i.e., 1 very likely to be hemolytic, 0 improbable to be hemolytic. **AVP-IC50Pred is a multiple machine learning technique based prediction of peptides' antiviral activity in terms of half-maximal inhibitory concentration (IC50).
Fig. 3.
Comparative structural analysis of the receptor-binding domain (RBD) of the SARS-CoV-2. (a) Wild type spike protein of SARS-CoV-2, RBD is highlighted in yellow (b) RRDis map of wild type and manual mutant model of the RBD (c) RRDis map of energy mutant, evolutionary mutant, combine mutant (d) overlapping three-dimensional structures of wild type RBD (gold) and manual mutant model of RBD (cyan), (e) overlapping three-dimensional structures of different mutant models of RBD, energy mutant (pink), evolutionary mutant (gold), combine mutant (cyan), (f) correspondence analysis of different structural models (based on Dali Z-scores), (g) heatmap of different mutant models of the RBD. *Corresponding analysis is a multidimensional scaling method. It positions data with the most similar structural neighborhoods near each other.
Table 2.
In silico mutant models of the receptor-binding domain of the SARS-CoV-2
| Amino acid residues (Wild type to mutant type) | FoldX (kcal/mol) |
Rosetta (kcal/mol) |
|
|---|---|---|---|
| Combined mutant -27.63 kcal/mol (17 mutations) | A348P | -1.66 | -2.34 |
| N354E | -0.45 | - | |
| A372T | 0.20 | - | |
| S373M | -2.30 | -2.82 | |
| T393F | -1.41 | -6.03 | |
| N394S | -0.63 | - | |
| S399M | -3.17 | -3.09 | |
| R403K | 0.48 | - | |
| K417V* | 0.21 | - | |
| N460K | -0.75 | - | |
| I468L | -0.40 | - | |
| T470Y | -2.10 | -2.19 | |
| S477G | -0.03 | - | |
| S494Y | -1.47 | -2.12 | |
| G502P | -1.94 | -2.61 | |
| V503P | -1.18 | -2.72 | |
| H519N | -0.02 | - | |
| Energy mutant: -25.41 kcal/mol (8 mutations) | A348P | -1.66 | -2.34 |
| S373M | -2.30 | -2.82 | |
| T393F | -1.41 | -6.03 | |
| S399M | -3.17 | -3.09 | |
| T470Y | -2.10 | -2.19 | |
| S494Y | -1.47 | -2.12 | |
| G502P | -1.94 | -2.61 | |
| V503P | -1.18 | -2.72 | |
| Evolution mutant: -6.29 kcal/mol (12 mutations) | A348P | -1.66 | -1.66 |
| N354E | -0.45 | -0.45 | |
| A372T | 0.20 | 0.20 | |
| T393S | 0.47 | 0.47 | |
| N394S | -0.63 | -0.63 | |
| R403K | 0.48 | 0.48 | |
| K417V* | 0.21 | 0.21 | |
| N460K | -0.75 | -0.75 | |
| I468L | -0.40 | -0.40 | |
| T470N | 0.06 | -0.06 | |
| S477G | -0.03 | -0.03 | |
| H519N | -0.02 | -0.02 |
In the manual mutant model of RBD (RBDm), F486P, Y489F, Q493M, G496P, Q498L, T500Y, N501L, and Y505A substitutions were manually incorporated. *Among SARS-CoV-2 variants, mutation at N501 is common in B1.1.7, P.1, B1.351, and at K417 in B.1.617.2.1
Table 3.
Structural analysis of mutant models of the receptor-binding domain (RBD) of the SARS-CoV-2
| Sl. No. | Mutant models of RBD | RMSDa | Percentage of identitiesb | Z-score* |
|---|---|---|---|---|
| 1. | RBDm | 1.7 | 94 | 25.3 |
| 2. | RBDc | 1.7 | 96 | 25.3 |
| 3. | RBDe | 1.5 | 88 | 25.2 |
| 4. | RBDb | 1.7 | 84 | 23.9 |
RMSD = root mean square deviations
Percentage of identities with the wild type RBD structure, RBDm = manual mutant, RBDc = combined model, RBDe = energy mutant, RBDb = evolutionary mutant model of the receptor-binding domain. *Dali Z-score.
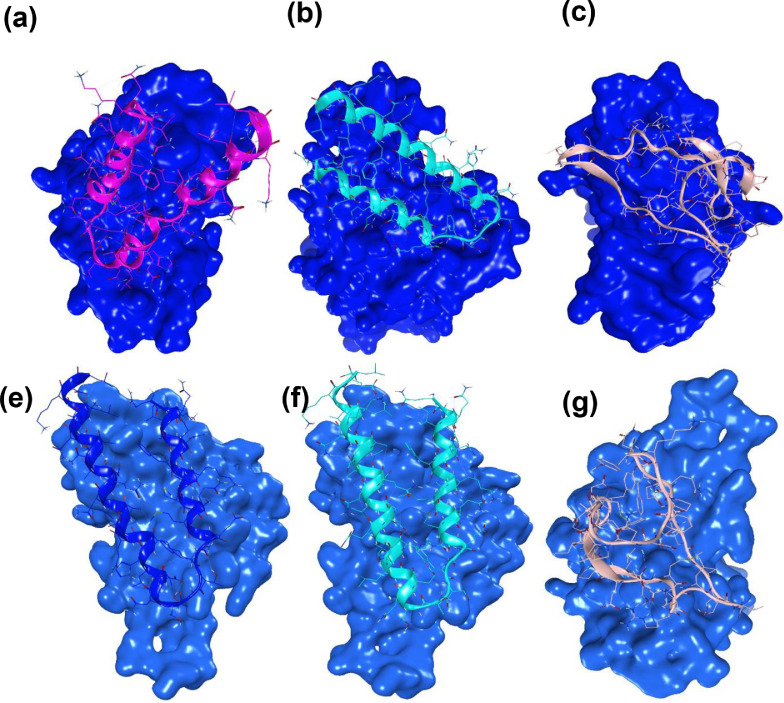
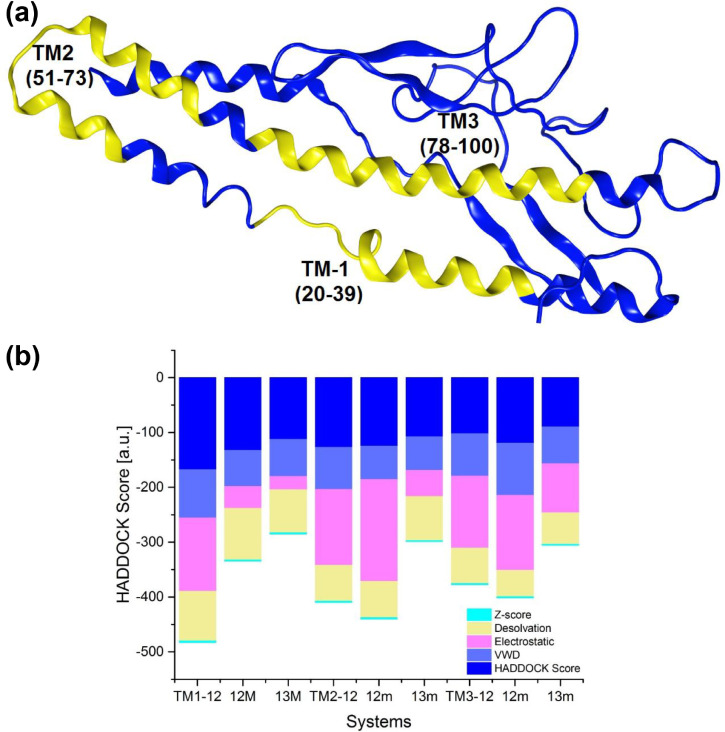
Results from molecular docking studies showed that AVP-RBD/RBDm complexes have thermodynamically favorable interactions (Table 4 ). Besides, Seq12, Seq12m, and Seq13m were engaged with nearly all critical amino acid residues of the RBD. Antiviral peptide of this study, i.e., Seq12, showed the best HADDOCK score of -111.2 kcal/mol followed by Seq13m (81.4 kcal/mol) and Seq12m (76.8 kcal/mol), suggesting AVPs were well-docked with the RBD (Fig. 4 ). Moreover, molecular docking results also suggest Seq12, Seq12m, and Seq13m retained the thermodynamically favorable binding properties with the mutant RBD (Fig. S6). Furthermore, Seq12 and Seq12m have negligible cytotoxicity (0.17; while 1.0 or higher is considered toxic and can rapture the human RBCs) (Fig. 2). These findings combinedly justify the novelty of the AVPs, and their possible therapeutic use in the future. SARS-CoV-2 has a structural membrane protein called Protein-M [69]. Protein-M is a tri-pass transmembrane protein (Fig. 5 ) which is essential for re-assembling viral structural units into a mature virus [70]. Therefore, protein-M also embraces importance as a potential therapeutic target. Results from our investigation indicate Seq12, Seq12m, and Seq13m are also participating in thermodynamically favorable interactions with the TM-1 region followed by TM-2 and TM-3, respectively (Fig. 5a). However, the molecular docking studies between the AVPs and active site of the viral RNA-dependent RNA polymerase (RdRp) were not thermodynamically favorable. Antiviral peptides usually rupture the viral capsid and eventually inhibit the viral replication cycles [69]. We speculate that the antiviral peptide, Seq12, Seq12m, and Seq13m could act as an anti-SARS-CoV-2 peptide by two possible mechanisms. Firstly, by inhibiting the RBD-ACE2 interaction. And secondly, by binding with M-protein followed by an eventual inhibition of the viral re-assembly/re-packaging.
Table 4.
HADDOCK docking properties of the AVP-RBD complexes
| Properties | Systems |
|||||
|---|---|---|---|---|---|---|
| Seq12 + RBD | Seq12 + RBDm3 | Seq12m + RBD | Seq12m + RBDm | Seq13m + RBD | Seq13m + RBDm | |
| HADDOCK Score | -111.2 | -92.8 | -76.8 | -76.7 | -81.4 | -41.7 |
| Cluster size | 104 | 36 | 13 | 27 | 40 | 14 |
| RMSD1 | 0. 5 | 0.4 | 2.6 | 4.7 | 0.5 | 3.5 |
| VDW2 energy | -68.8 | -71.3 | -71.9 | -78.3 | -75.2 | -63.4 |
| Electrostatic energy | -182.4 | -142.0 | -145.0 | -53.2 | -70.3 | -73.8 |
| Desolvation energy | -36.9 | -36.6 | -4.9 | -43.1 | -20.9 | -28.5 |
RMSD = Root mean square deviation from the overall lowest-energy structure. 2VDW = van der Waals energy. 3RBDm = mutant model of the receptor-binding domain of the SARS-CoV-2. Energy units are in kcal/mol.
Fig. 4.
The binding posture of the AVPs with the receptor-binding domain of the SARS-CoV-2. wild type receptor binding domain complex with (a) Seq12, (b) Seq12m, (c) Sqe13m, mutant receptor binding domain complex with (d) Seq12, (e) Seq12m, (f) Sqe13m.
Fig. 5.
Membrane protein (M) of the SARS-CoV-2. (a) predicted transmembrane domains of the M-protein are highlighted in yellow, (b) analyses of molecular docking results between the AVPs and the transmembrane domains.
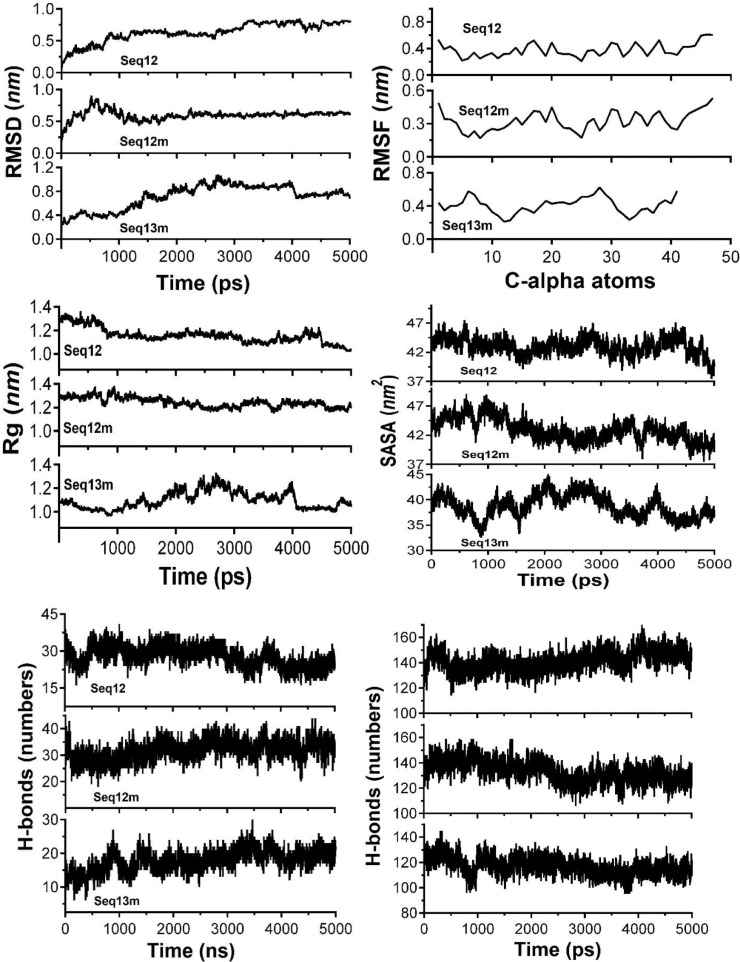
Results derived from MD-simulation of the studies of the Seq12, Seq12m, and Seq13m are summarised in Fig. 6 . The root mean square deviation (RMSD) of alpha carbon atoms of Seq12, Seq12m, and Seq13m are analyzed to detect their stabilities. It is observed from Fig. 6 that Seq12m has the lowest RMSD value than Seq12 and Seq13m, respectively. Antiviral peptides, namely Seq12, Se12m, and Seq13m, have ≈ 0.274 nm, ≈ 0.274 nm, and ≈ 0.286 nm of RMSD values at 1 ns. The highest RMSD values are respectively ≈ 0.852 nm, ≈ 0.908 nm, and ≈ 1.074 nm for Seq12, Seq12m, and Seq13m. A closer look at the RMSD plot suggests Seq12m has the highest fluctuation from 5 ns - 10 ns and was stabilized throughout the MD-trajectory. However, at the start, RMSD-fluctuation was lower in the case of Seq12, and the highest RMSD-fluctuation was observed between 30 ns - 40 ns. Quite similarly, the RMSD-fluctuation of Seq13m was highest between 20 ns -40 ns. Root means square fluctuation (RMSF) helps to understand the flexibility of each amino acid residue [71]. Seq13m is found to have the highest RMSF from the 5th to 7th and 25th to 30th position of the amino acid sequence. However, Seq12 and Seq12m showed a similar RMSF plot. As mentioned earlier, Seq12 and Seq12m have only one difference at the 2nd position of their sequences. This may be the reason for their similar RMSF value. However, a closer observation suggests Seq12 has a little higher RMSF value than Seq12m. The radius of gyration (Rg) represents the compactness of the molecular structure. The lower degree of fluctuation with its consistency through the simulation indicates the greater compactness and rigidity of a system [71]. Rg of Seq13m was fluctuated from 10 ns-40 ns and reached the highest value of 1.331 at ≈ 25 ns. However, Rg values of Seq12 and Seq12m are very similar to Seq13m at the beginning and remained the same up to 10 ns. Further, Rg fluctuations are prominent after 10 ns and sustain such dissimilarities throughout the trajectory. The lowest Rg values are ≈ 1.041 nm (50 ns), ≈ 1.380 nm (before 10 ns), and ≈ 0.944 nm (before 10 ns), respectively for Seq12, Seq12m, and Seq13m. In summary, Seq12 and Seq12m showed better stability throughout the complete trajectory. The number of inter water-peptide hydrogen bonds in the simulated systems were also compared. The highest number of hydrogen bonds are formed respectively by Seq12 (40 - 45 ns), Seq12m (15 ns), and Seq13m (before 5 ns).
Fig. 6.
Comparisons of 50 ns MD-Simulation results of the antiviral peptides Seq12, Seq12m, and Seq13m.
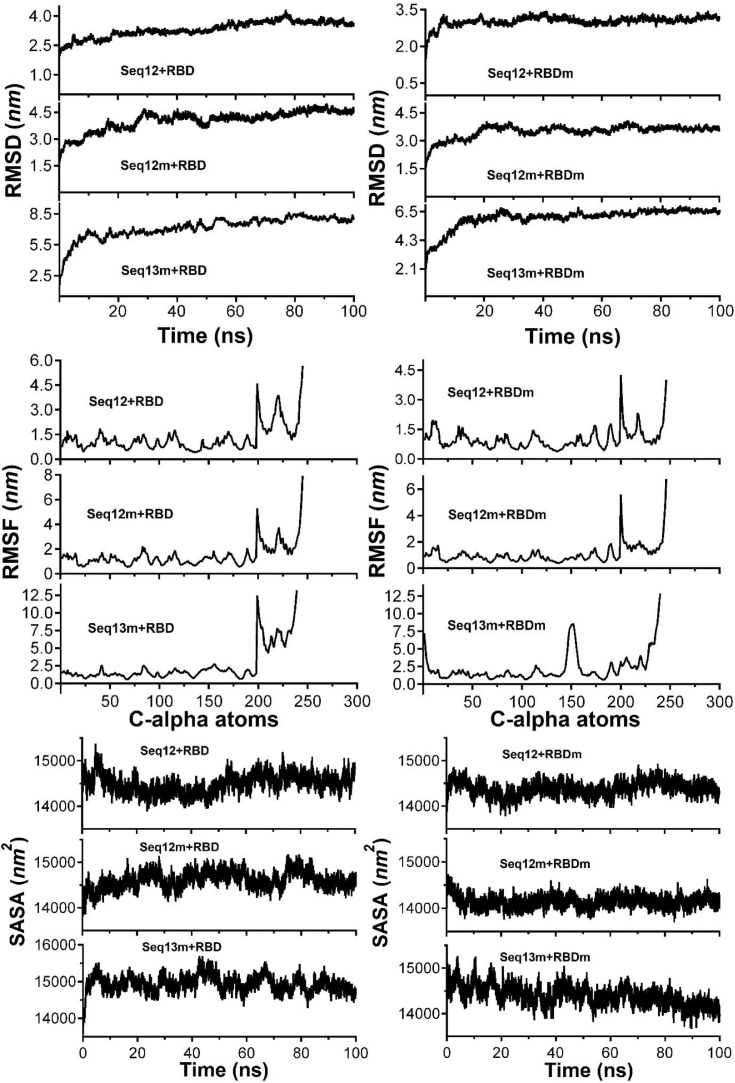
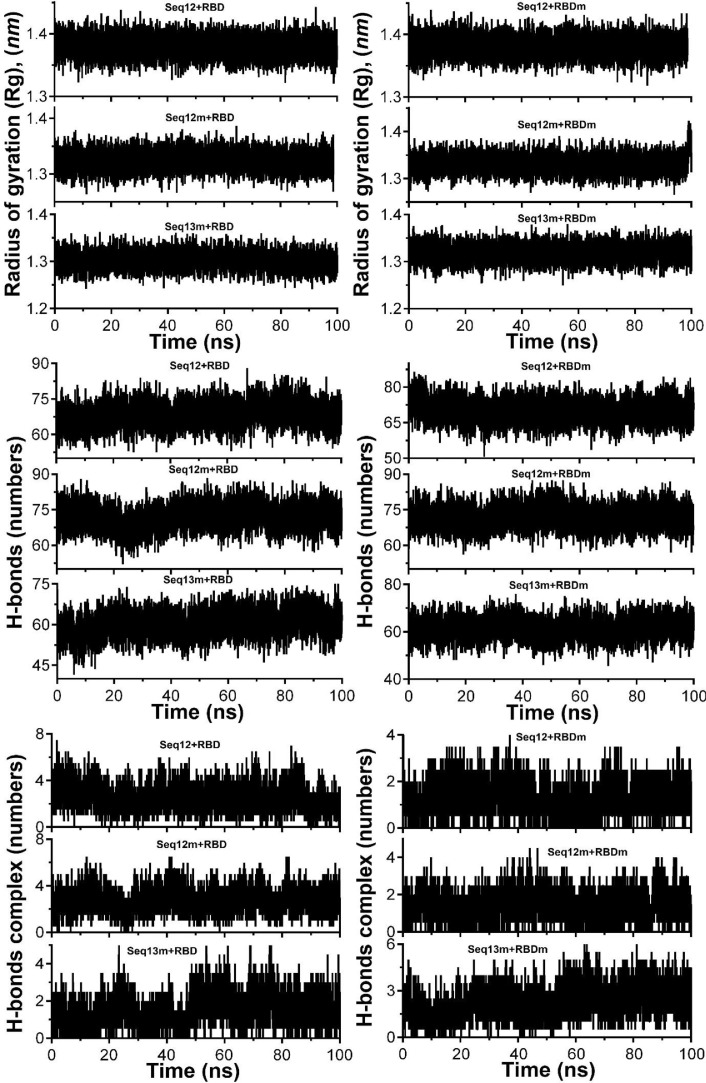
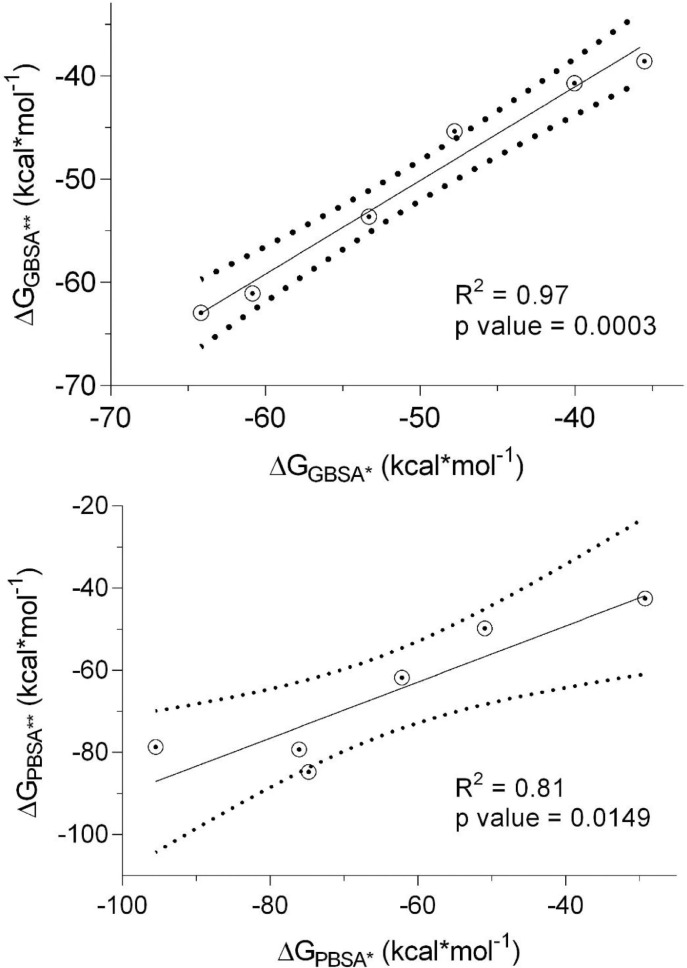
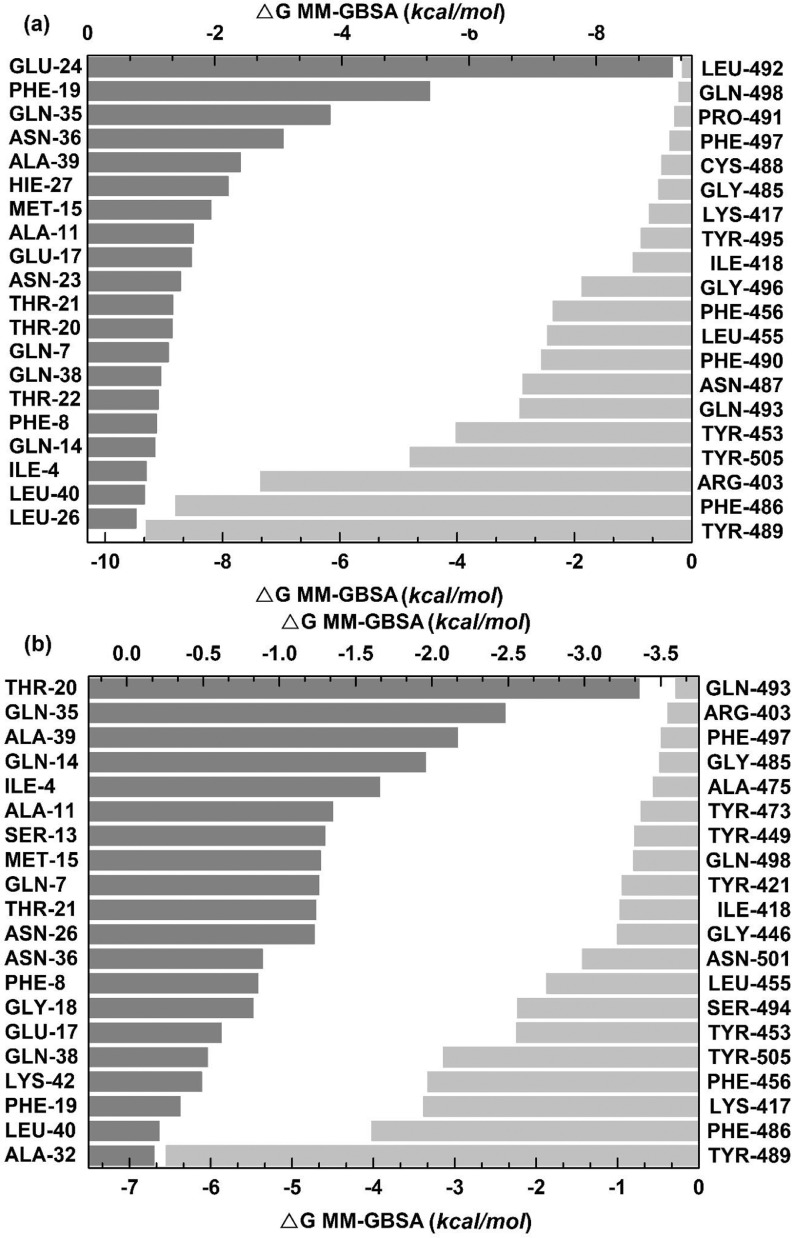
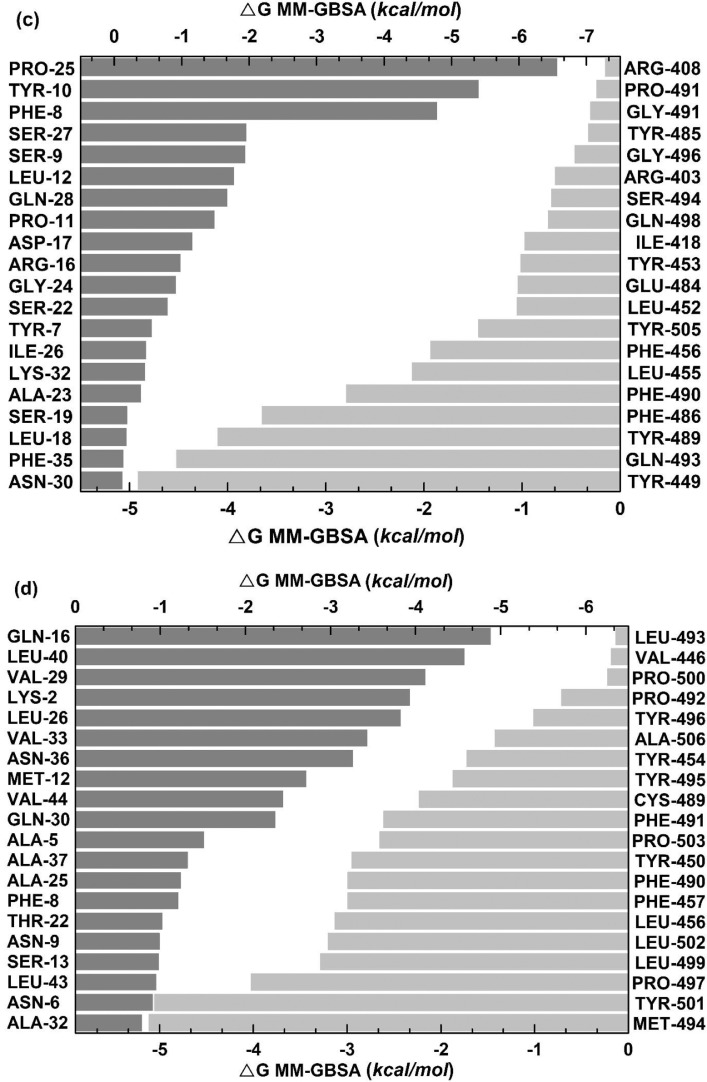
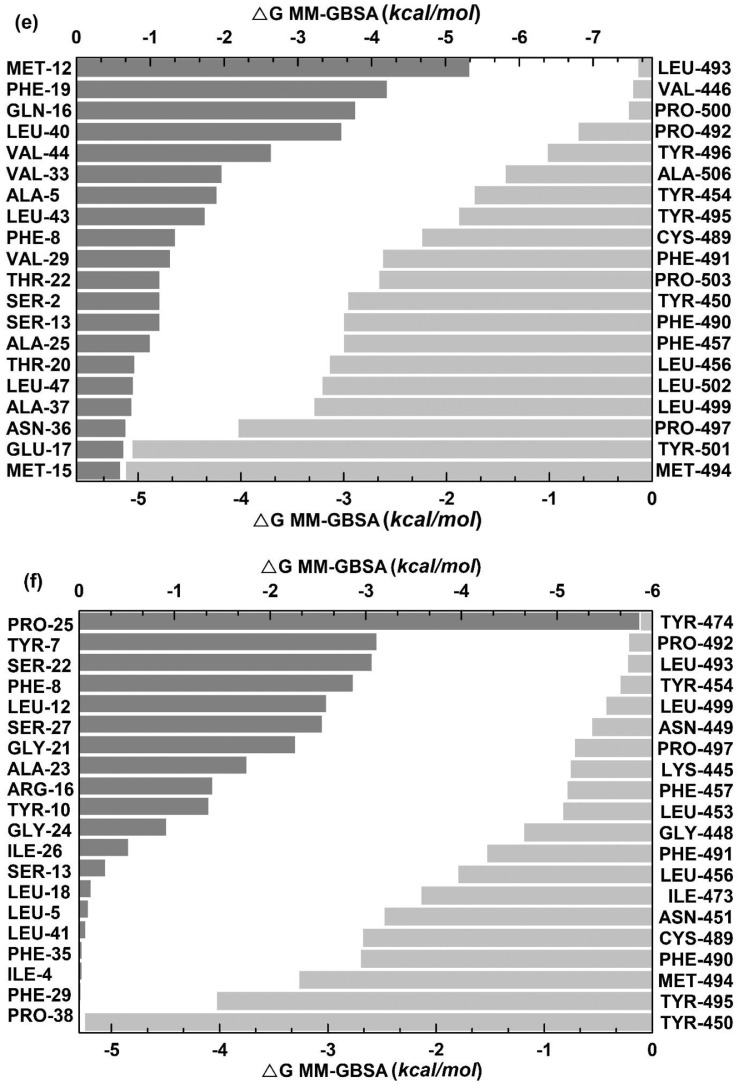
Furthermore, 100 ns scale MD-simulation studies of the AVP-RBD complexes are summarised in Fig. 7 . RMSD and RMSF were stable throughout all MD-trajectory and were most stable in the case of Seq12-RBD/RBDm, and Seq12m-RBD/RBDm complex. However, RMSD values of Seq13m-RBD/-RBDm complex were steadily increased during the initial 10 ns of the MD-trajectory and then stabilized throughout the remaining 90 ns of the MD trajectory. Similarly, the RMSF values fluctuate in the case of the Seq13m-RBD/-RBDm complex (Fig. 8 ). RMSF value is a measure of the average deviations of particular atoms or group of atoms from the initial reference structure [72]. Results showed that, in the case of the Seq13m-RBDm, RMSF fluctuation was high from about 138th -158th of the RBD. Similarly, in the case of Seq13m-RBD, the RMSF value highly fluctuate from about 200th to the rest of the C-alpha atoms (Fig. 8). However, only two crucial interacting amino acid residues, namely, PHE486 and TYR489, was remained within the fluctuating regions. And, the ΔGbind of Seq13m-RBDm and Seq13-RBD were also very good compared to the other RBD/RBDm-AVP complexes (Table 5 - 6), suggesting this minor structural fluctuation did not have negative impacts on binding. The radius of gyration (Rg) around the axis was stable (1.12-1.44 nm) for all AVP-RBD complexes. Besides, Rg of the AVP-RBD complexes were overlapping in the case of all AVP-RBD complexes except Seq12m-RBD. In addition, the solvent-accessible surface area (SASA) of the Seq13m-RBD/RBDm complexes were highest (15702.20 nm 2), and this value was lowest (13568.20 nm 2) for the Seq12m-RBDm complex. Moreover, the number of hydrogen bonds formed during the MD-simulation run was limited in the case of inter AVP-RBD/RBDm complex (0-8). However, more H-bonds (41-88) were formed between protein complex and water during the entire MD simulation run. The molecular mechanics Poisson-Boltzmann, and Generalized Born and Surface Area (MM-PB/GBSA) are popular methods for calculating binding free energy [73]. The binding free energy components of the AVP-RBD complexes were derived from MM-PB/GBSA method. Table 5 - 6 reports good predicted binding free energies of the AVP-RBD complexes. In particular, Seq13m-RBDm has the best ΔGPBSA (-88.44 kcal/mol), followed by Seq12m-RBD (-84.88 kcal/mol), and Seq12m-RBDm (-76.62 kcal/mol), etc. However, Seq12m-RBDm (-62.95 kcal/mol) showed best ΔGGBSA (calculated using AMBER 16) followed by Seq12m-RBD (-61.07 kcal/mol) and Seq12-RBDm (-53.64 kcal/mol), respectively. The “gas-phase” electrostatic energy (ΔEelec) is most important for complex formation [75], and the ΔEelec of the AVP-RBD complexes are -34.32, 0.56, -42.41, -12.33, -24.69, -40.48 kcal/mol for Seq12-RBD, Seq12-RBDm, Seq12m-RBDm, Seq13-RBD, and Seq13m-RBDm respectively. It is noteworthy that Seq12-RBDm has a positive ΔEelec value, but Seq12-RBDm also has a good binding free energy (-84.75 kcal/mol). Similarly, the van der Waals energy is a vital energy component for the ligand-protein interaction [75], dominating the AVP-RBD interactions (Table 5-6 ). The “gas-phase” van der Waals energies (ΔEvdw) are -55.44, -92.36, -65.07, -81.68. -41.09 and -93.84 kcal/mol respectively for Seq12-RBD, Seq12-RBDm, Seq12m-RBDm, Seq13-RBD, and Seq13m-RBDm. In addition, the non-polar solvation free energy (ΔGnpol) favors AVP-RBD complex formation, and the contribution of ΔG npol to the binding free energies are -62.98, -102.76, -74.02, -90.82, -46.46, and -104.74 kcal/mol, respectively for Seq12-RBD, Seq12-RBDm, Seq12m-RBDm, Seq13-RBD, and Seq13m-RBDm. Furthermore, the binding free energy components obtained from the MM-GBSA method are also in agreement with the binding free energy calculated using the MM-PBSA method. The linear regression fit between predicted ΔGPB/GBSA calculated for 50 ns and predicted ΔGPB/GBSA calculated for 100 ns is summarized in Fig. 9 . It showed that the R2 of the regression model of the ΔGGBSA was 0.97 (p = 0.0003), suggesting ΔGbind is significantly correlated with the model. In addition, Fig. 9a indicates that increasing the MD-Simulation time from 50 ns to 100 ns did not have an impact on the binding free energy. However, in the case of ΔGPBSA the R2 was 0.75 (p = 0.0256), although all variables were within 95% confidence intervals. This is because we have found that the ΔGPBSA calculated for 50 ns and 100 ns have high differences (Table S1). Furthermore, the MM-GBSA binding free energies per amino acid residues are summarized in Fig. 10 . It showed that TYR489 is a crucial amino acid residue of the RBD because TYR489 is a common contributor to the best binding free energies of Seq12-RBD (-9.30 kcal/mol), Seq12m-RBD (-6.55 kcal/mol), and Seq13m-RBD (-4.91 kcal/mol). On the other hand, GLU24 (-9.20 kcal/mol) of Seq12, THR20 (-3.36 kcal/mol) of Seq12m, and PRO25 (-6.56 kcal/mol) of Seq13m are critically important for the respective antiviral peptides for the same reasons. Furthermore, GLN498 (-0.22 kcal/mol) and GLN493 (-0.28 kcal/mol) are poor contributors for the AVP-RBD interactions, respectively for Seq12 and Seq12m. However, GLY496 poorly contributes to the Seq13m-RBD interaction but not for the Seq12-RBDm, Seq12m-RBDm, and Seq13m-RBDm interactions (Fig. 10). In the RBDm TYR501 (-6.32 kcal/mol), MET494 (-5.11 kcal/mol), and TYR-450 (-5.24 kcal/mol) contributes best binding free energy during interacting with Seq12, Seq12m, and Seq13m, respectively. It is noteworthy that in cases (RBD and RBDm), tyrosine contributes well binding free energy multiple times irrespective of its positions in the RBD. The biochemical and biophysical properties of tyrosine may be the reason for this phenomenon. However, tyrosine did not participate in the top twenty amino acid residues in the case of Seq12m-RBDm. On the contrary, LEU493 was a poor contributor to the binding free energies of Seq12/Seq12m-RBDm interactions. In addition, ASN449 was another poor contributor in the case of Seq13m-RBDm interaction. Overall, the molecular details of AVP-RBD/RBDm interactions calculated using MM-GBSA provide evidence for the fact that AVPs were occupied the crucial amino acid residues of the RBD, which are necessary for the RBD-ACE2 interactions [65].
Fig. 7.
Comparisons of the 100 ns MD-Simulation results of the AVP-RBD complexes.
Fig. 8.
Difference of RMSF values obtained from 100 ns MD-simulation of the AVP-RBD complexes. The major fluctuations of the C-alpha atoms are highlighted by black in the cartoon presentation of the RBD, from the 149th-158th amino acid residue (*) and from 200th to the rest of the amino acid residues (**).
Table 5.
Binding free energy (kcal/mol) resulting from MM-PBSA analysis of different AVP-RBD complexes
| Energy components* |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔEelec | ΔEvdw | ΔGPB | ΔGsa | ΔGgas | ΔGsol | ΔGpol | ΔGnpol | ΔGtotal* | |
| A | -34.32 | -55.44 | 46.67 | -7.53 | -89.76 | 39.13 | 12.35 | -62.98 | -50.62 |
| B | 0.56 | -92.36 | 17.31 | -10.39 | -91.80 | 6.91 | 17.87 | -102.76 | -84.88 |
| C | -42.41 | -65.07 | 56.69 | -8.94 | -107.49 | 47.75 | 14.27 | -74.02 | -59.74 |
| D | -12.33 | -81.68 | 26.52 | -9.13 | -94.01 | 17.39 | 14.19 | -90.82 | -76.62 |
| E | -24.69 | -41.09 | 34.51 | -5.36 | -65.78 | 29.14 | 9.8 | -46.46 | -36.64 |
| F | -40.48 | -93.84 | 56.78 | -10.89 | -134.33 | 45.89 | 16.3 | -104.74 | -88.44 |
A = Seq12+RBD, B = Seq12+RBDm, C = Seq12m+RBD, D = Seq12m+RBDm, E = Seq13m+RBD, F = Seq13m+RBDm. *MM-PBSA was calculated using CaFE tools with the 100 ns MD-trajectory generated using NAMD software.
Table 6.
Binding free energy (kcal/mol) resulting from MM-GBSA analysis of different AVP-RBD complexes
| Energy components* |
||||||||
|---|---|---|---|---|---|---|---|---|
| Vdw | EEL | EGB | ESURF | G gas | G solv | ΔGTotal | ΔGGBSA# | |
| A | -67.18 | -140.14 | 172.36 | -10.40 | -207.33 | 161.96 | -45.36 | -92.09 |
| B | -97.66 | -62.18 | 120.23 | -14.01 | -159.85 | 106.22 | -53.63 | -93.31 |
| C | -93.66 | -208.49 | 254.69 | -13.60 | -302.16 | 241.09 | -61.07 | -50.09 |
| D | -102.19 | -44.99 | 97.17 | -12.94 | -147.18 | 84.22 | -62.95 | -75.13 |
| E | -84.81 | -155.38 | 210.48 | -10.98 | -240.20 | 199.49 | -40.70 | -57.96 |
| F | -70.96 | -87.50 | 129.40 | -9.52 | -158.47 | 119.88 | -38.59 | -53.90 |
A = Seq12+RBD, B = Seq12+RBDm, C = Seq12m+RBD, D = Seq12m+RBDm, E = Seq13m+RBD, F = Seq13m+RBDm. ⁎MM-GBSA was calculated for 100 ns using Abmer 16 package. Generalized Born ESURF calculated using 'LCPO' surface areas. #ΔGGBSA was calculated using HawkDock.
Fig. 9.
Linear regression between predicted ΔGPB/GBSA calculated for 50 ns (*) and predicted ΔGPB/GBSA calculated for 100 ns (**). The dotted line around the solid line indicates 95% confidence intervals.
Fig. 10.
Distribution of binding free energies (MM-GBSA) per amino acid residues. (a) Seq12 + RBD, (b) Seq12m + RBD, (c) Seq13m + RBD, (d) Seq12 + RBDm, (e) Seq12m + RBDm, (f) Seq13m + RBDm. *Only top twenty participating amino acid are presented according to the ascending / desending order of energies (MM-GBSA, kcal/mol).
Entropy of a thermodynamic system is the measure of the degree of freedom of the system where the translational (ΔStr), rotational (ΔSrot), and vibrational (ΔSvib) entropic terms determine the total entropy of the system [74,75]. A change in total entropy correlates with the binding free energy, where the binding forces have to overcome the entropic effects during the binding process [75]. Therefore, a negative ΔS value indicates that the system becomes less disordered due to a decrease in the number of microstates for the protein-protein complexes [43]. In particular, the entropic values of the AVP-RBD systems are negative, which explains the low distortion of the system (Table 7 ). In addition, the Seq13m-RBDm complex has the highest ΔSvib value (-0.43 kcal/mol), contributing to the increase of the phase space and binding energy of the system.
Table 7.
Comparison of translational, vibrational, and rotational entropy among the AVP-RBD complexes calculated using the two-phase thermodynamic (2PT) model.
| Systems | ΔStr | ΔSrot | ΔSvib | Total ΔS¥ |
|---|---|---|---|---|
| Seq12 + RBD | -15.17 | -14.68 | -16.28 | -46.13 |
| Seq12 + RBDm$ | -15.17 | -14.71 | -13.44 | -43.33 |
| Sqe12m + RBD | -15.16 | -14.68 | -15.16 | -45.01 |
| Sqe12m + RBDm$ | -15.16 | -14.69 | -10.88 | -40.74 |
| Seq13m + RBD | -15.07 | -14.41 | -17.10 | -46.58 |
| Seq13m + RBDm$ | -15.07 | -14.33 | -0.43 | -29.85 |
Entropy results were (harmonic approximation) calculated with NMODE. All entropy results have units kcal/mol (Temperature is 298.15 K).
4. Conclusion
Antiviral peptides are a convenient alternative to conventional antiviral therapy. Now-a-days, AVPs are included in mainstream research against the SARS-CoV-2. In this study, peptide analogues of spike glycoprotein, i.e., Seq12, Seq12m, and Seq13m, showed antiviral properties against the SARS-CoV-2. These AVPs were derived from the RBD of the spike glycoprotein of the SARS-CoV-2. Seq12 and Seq12m showed negligible cytotoxicity, and they are also non-allergenic for humans. Besides, the predicted IC50 values of the Seq12, Seq12m, and Seq13m are better than the anti-SARS-CoV-2 peptide P7, P9, P9, and P10. Molecular dynamics simulation studies of the RBD/RBDm-Seq12 and RBD/RBDm-Seq12m showed stable RMSD and RMSF throughout the complete MD-trajectory. Furthermore, the binding free energies, van der Waals interaction patterns calculated using MM-PB/GBSA are also in agreement with the molecular docking studies. The molecular docking and molecular dynamics simulation studies suggest Seq12, Seq12m, and Seq13m can block RBD, which is necessary for communicating with ACE2. It is noteworthy that these AVPs can also block the mutant model of the RBD. The mutant model of the RBD contains 25 mutations, including N501, common in B1.1.7, P.1, B1.351, and K417 of B.1.617.2.1 variant of SARS-CoV-2. Furthermore, these AVPs can also interfere with viral membrane protein M. We speculate that these AVPs could eventually inhibit viral re-packaging cycles. Therefore, antiviral peptides, Seq12, Seq12m, and Seq13m could be helpful in the fight against the SARS-CoV-2. Moreover, in the future, these AVPs could also help to develop anti-SARS-CoV-2 nasal spray. However, more studies are required before any clinical or diagnostic use.
Author contributions
KD conceptualized the idea and method for finding new antiviral peptides against SARS-CoV-2. KD designed and executed most of the experiments, including but not limited to MD simulation, molecular docking. ADE and AA perform the MM-PBSA calculation and the MD simulation of the AVP-RBD complexes. SS performed the MM-GBSA and entropy calculations. WZ, OVM, SS, IK proofread and wrote part of the manuscript. KD wrote the manuscript.
Funding
This research received no external funding.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Data availability statement
The data presented in this study are available.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Council of Scientific and Industrial Research (CSIR), Govt. of India, New Delhi, India is sincerely acknowledged by K.D. for Senior Research Fellowship (SRF), sanction letter no. 09/599(0082)/2019-EMR-I.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.131113.
Appendix. Supplementary materials
References
- 1.Ting D.S.W., Carin L., Dzau V., Wong T.Y. Digital technology and COVID-19. Nat. Med. 2020;26(4):459–461. doi: 10.1038/s41591-020-0824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deeks J.J., Dinnes J., Takwoingi Y., Davenport C., Leeflang M.M., Spijker R., Hooft L., Van den Bruel A., Emperador D., Dittrich S. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews. 2020;(4) [Google Scholar]
- 3.Klompas M., Baker M.A., Rhee C. Airborne transmission of SARS-CoV-2: theoretical considerations and available evidence. JAMA. 2020 doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- 4.Pan H., Peto R., Karim Q.A., Alejandria M., Restrepo A.M.H., García C.H., Kieny M.P., Malekzadeh R., Murthy S., Preziosi M.-P. interim WHO SOLIDARITY trial results. MedRxiv; 2020. Repurposed antiviral drugs for COVID-19. [Google Scholar]
- 5.Kodali P.B., Hense S., Kopparty S., Kalapala G.R., Haloi B. How Indians responded to the Arogya Setu app? Indian J. Public Health. 2020;64(6):228. doi: 10.4103/ijph.IJPH_499_20. [DOI] [PubMed] [Google Scholar]
- 6.Azhar M., Phutela R., Ansari A.H., Sinha D., Sharma N., Kumar M., Aich M., Sharma S., Singhal K., Lad H. bioRxiv; 2020. Rapid, field-deployable nucleobase detection and identification using FnCas9. [Google Scholar]
- 7.K. Dutta, S. Shityakov, O. Morozova, I. Khalifa, J. Zhang, A. Panda, C. Ghosh, Beclabuvir can inhibit the RNA-dependent RNA polymerase of newly emerged novel coronavirus (SARS-CoV-2), (2020), doi: 10.20944/preprints202003.0395.v1. Available at: https://www.preprints.org/manuscript/202003.0395/v2. [DOI]
- 8.Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1758791. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalifa I., Zhu W., Mohammed H.H.H., Dutta K., Li C. Tannins inhibit SARS-CoV-2 through binding with catalytic dyad residues of 3CLpro: An in silico approach with 19 structural different hydrolyzable tannins. J. Food Biochem. 2020;44(10):e13432. doi: 10.1111/jfbc.13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahase E. Covid-19: Moderna vaccine is nearly 95% effective, trial involving high risk and elderly people shows. BMJ: British Medical Journal (Online) 2020:371. [Google Scholar]
- 11.Graham S.P., McLean R.K., Spencer A.J., Belij-Rammerstorfer S., Wright D., Ulaszewska M., Edwards J.C., Hayes J.W., Martini V., Thakur N. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. NPJ vaccines. 2020;5(1):1–6. doi: 10.1038/s41541-020-00221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020 doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boas L.C.P.V., Campos M.L., Berlanda R.L.A., de Carvalho Neves N., Franco O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019;76(18):3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahendran A.S.K., Lim Y.S., Fang C.-M., Loh H.-S., Le C.F. The Potential of Antiviral Peptides as COVID-19 Therapeutics. Frontiers in pharmacology. 2020;11:1475. doi: 10.3389/fphar.2020.575444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020 doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogels C.B., Breban M.I., Ott I.M., Alpert T., Petrone M.E., Watkins A.E., Kalinich C.C., Earnest R., Rothman J.E., Goes de Jesus J. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19(5) doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robson B. COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed Achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brister J.R., Ako-Adjei D., Bao Y., Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015;43(D1):D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26(2):283–291. [Google Scholar]
- 23.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(2):W202–W208. doi: 10.1093/nar/gkp335. suppl_. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaduangrat N., Nantasenamat C., Prachayasittikul V., Shoombuatong W. Meta-iAVP: A sequence-based meta-predictor for improving the prediction of antiviral peptides using effective feature representation. Int. J. Mol. Sci. 2019;20(22):5743. doi: 10.3390/ijms20225743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur N., Qureshi A., Kumar M. AVPpred: collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012;40(1):W199–W204. doi: 10.1093/nar/gks450. W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qureshi A., Tandon H., Kumar M. AVP-IC50Pred: Multiple machine learning techniques-based prediction of peptide antiviral activity in terms of half maximal inhibitory concentration (IC50) Pept. Sci. 2015;104(6):753–763. doi: 10.1002/bip.22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamiable A., Thévenet P., Rey J., Vavrusa M., Derreumaux P., Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44(W1):W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 30.Lear S., Cobb S.L. Pep-Calc. com: a set of web utilities for the calculation of peptide and peptoid properties and automatic mass spectral peak assignment. J. Comput. Aided Mol. Des. 2016;30(3):271–277. doi: 10.1007/s10822-016-9902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portal, ExPASy Bioinformatics Resource. “ProtParam tool.” (2011). https://web.expasy.org/protparam/.
- 32.Saha S., Raghava G. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34(2):W202–W209. doi: 10.1093/nar/gkl343. suppl_. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P., Consortium O.S.D.D. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Win T.S., Malik A.A., Prachayasittikul V., Wikberg J.E.S, Nantasenamat C., Shoombuatong W. HemoPred: a web server for predicting the hemolytic activity of peptides. Future medicinal chemistry. 2017;9(3):275–291. doi: 10.4155/fmc-2016-0188. [DOI] [PubMed] [Google Scholar]
- 35.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T., Lv H., Mok C.K., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musil M., Stourac J., Bendl J., Brezovsky J., Prokop Z., Zendulka J., Martinek T., Bednar D., Damborsky J. FireProt: web server for automated design of thermostable proteins. Nucleic Acids Res. 2017;45(W1):W393–W399. doi: 10.1093/nar/gkx285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Vries S.J., Van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5(5):883. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 38.BIOVIA D.S. Release 2017. Dassault Systèmes; San Diego: 2017. Discovery studio visualizer. 2016, to be found under http://accelrys. com/products/collaborative-science/biovia-discovery-studio/visualization-download. php (accessed Dec 12, 2017) [Google Scholar]
- 39.Dutta K., Nag K., Booth V., Smyth E., Dueck H., Fritzen-Garcia M., Ghosh C., Panda A.K. Paradoxical Bactericidal Effects of Hydrophobic Lung Surfactant Proteins and Their Peptide Mimics Using Liposome Molecular Trojan. J. Oleo Sci. 2018:ess18026. doi: 10.5650/jos.ess18026. [DOI] [PubMed] [Google Scholar]
- 40.Elmezayen A.D., Yelekçi K. Homology modeling and in silico design of novel and potential dual-acting inhibitors of human histone deacetylases HDAC5 and HDAC9 isozymes. J. Biomol. Struct. Dyn. 2020:1–19. doi: 10.1080/07391102.2020.1798812. [DOI] [PubMed] [Google Scholar]
- 41.Liu H., Hou T. CaFE: a tool for binding affinity prediction using end-point free energy methods. Bioinformatics. 2016;32(14):2216–2218. doi: 10.1093/bioinformatics/btw215. [DOI] [PubMed] [Google Scholar]
- 42.D. Case, D. Cerutti, T. Cheatham, T. Darden, R. Duke, T. Giese, H. Gohlke, A. Goetz, D. Greene, N. Homeyer, AMBER 2016 ( University of California ), San Francisco (2016).
- 43.Miller B.R., III, McGee Jr T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA. py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8(9):3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 44.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47(W1):W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(1):D405–D412. doi: 10.1093/nar/gku938. D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8(1):424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schütz D., Ruiz-Blanco Y.B., Münch J., Kirchhoff F., Sanchez-Garcia E., Müller J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug. Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21(2):131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K.-M., Chan C.C.-S., Leung H.-C., Fai N., Lin Y.-P. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashaolu T.J., Nawaz A., Walayat N., Khalifa I. Potential “biopeptidal” therapeutics for severe respiratory syndrome coronaviruses: a review of antiviral peptides, viral mechanisms, and prospective needs. Appl. Microbiol. Biotechnol. 2021:1–14. doi: 10.1007/s00253-021-11267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tourdot S., Oukka M., Manuguerra J.C., Magafa V., Vergnon I., Riche N., Bruley-Rosset M., Cordopatis P., Kosmatopoulos K. Chimeric peptides: a new approach to enhancing the immunogenicity of peptides with low MHC class I affinity: application in antiviral vaccination. J. Immunol. 1997;159(5):2391–2398. [PubMed] [Google Scholar]
- 52.Mahlapuu M., Björn C., Ekblom J. Antimicrobial peptides as therapeutic agents: opportunities and challenges. Crit. Rev. Biotechnol. 2020;40(7):978–992. doi: 10.1080/07388551.2020.1796576. [DOI] [PubMed] [Google Scholar]
- 53.Manavalan B., Shin T.H., Kim M.O., Lee G. AIPpred: sequence-based prediction of anti-inflammatory peptides using random forest. Frontiers in pharmacology. 2018;9:276. doi: 10.3389/fphar.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karoyan P., Vieillard V., Gómez-Morales L., Odile E., Guihot A., Luyt C.-E., Denis A., Grondin P., Lequin O. Human ACE2 peptide-mimics block SARS-CoV-2 pulmonary cells infection. Communications biology. 2021;4(1):1–9. doi: 10.1038/s42003-021-01736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beddingfield B.J., Iwanaga N., Chapagain P.P., Zheng W., Roy C.J., Hu T.Y., Kolls J.K., Bix G.J. The integrin binding peptide, ATN-161, as a novel therapy for SARS-CoV-2 infection. JACC: Basic to Translational Science. 2020 doi: 10.1016/j.jacbts.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bestle D., Heindl M.R., Limburg H., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life science alliance. 2020;3(9) doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang G., Pomplun S., Loftis A.R., Tan X., Loas A., Pentelute B.L. bioRxiv; 2020. Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 Spike RBD. [Google Scholar]
- 58.Zhao H., To K.K., Sze K.-H., Yung T.T.-M., Bian M., Lam H., Yeung M.L., Li C., Chu H., Yuen K.-Y. A broad-spectrum virus-and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-17986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Y.-W., Chao T.-L., Li C.-L., Chiu M.-F., Kao H.-C., Wang S.-H., Pang Y.-H., Lin C.-H., Tsai Y.-M., Lee W.-H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2) doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T.K., Wang Q., Du L., Tan W., Wilson I.A. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y., Yu D., Yan H., Chong H., He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 2020 doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ling R., Dai Y., Huang B., Huang W., Yu J., Lu X., Jiang Y. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;130 doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020;117(17):9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020:1–6. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 67.Abdullahi I.N., Emeribe A.U., Ajayi O.A., Oderinde B.S., Amadu D.O., Osuji A.I. Implications of SARS-CoV-2 genetic diversity and mutations on pathogenicity of COVID-19 and biomedical interventions. Journal of Taibah University Medical Sciences. 2020 doi: 10.1016/j.jtumed.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donal T. Skelly, Adam C. Harding, Javier Gilbert-Jaramillo et al. Vaccine-induced immunity provides more robust heterotypic immunity than natural infection to emerging SARS-CoV-2 variants of concern., 09 February 2021, PREPRINT (Version 1) available at Research Square [ 10.21203/rs.3.rs-226857/v1]. [DOI]
- 69.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. Sars-CoV-2 Envelope and Membrane proteins: differences from closely related proteins linked to cross-species transmission. Biomed. Res. Int. 2020 doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Islam R., Parves R., Paul A.S., Uddin N., Rahman M.S., Mamun A.A., Hossain M.N., Ali M.A., Halim M.A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–20. doi: 10.1080/07391102.2020.1761883. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martínez L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen F., Liu H., Sun H., Pan P., Li Y., Li D., Hou T. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein–protein binding free energies and re-rank binding poses generated by protein–protein docking. PCCP. 2016;18(32):22129–22139. doi: 10.1039/c6cp03670h. [DOI] [PubMed] [Google Scholar]
- 74.Brissaud J.-B. The meanings of entropy. Entropy. 2005;7(1):68–96. [Google Scholar]
- 75.Du X., Li Y., Xia Y.-L., Ai S.-M., Liang J., Sang P., Ji X.-L., Liu S.-Q. Insights into protein–ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17(2):144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.E. BAKER, 22.2. Hydrogen bonding in biological macromolecules, (2006). Available at: https://onlinelibrary.wiley.com/iucr/itc/Fa/ch22o2v0001/ch22o2.pdf.
- 77.G.A. Jeffrey, W. Saenger, Hydrogen bonding in biological structures, Springer Science & Business Media2012.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available.