Abstract
High-throughput screening (HTS) and computational technologies have emerged as important tools for chemical hazard identification. The US Environmental Protection Agency (EPA) launched the Toxicity ForeCaster (ToxCast™) Program, which has screened thousands of chemicals in hundreds of mammalian-based HTS assays for biological activity. The data are being used to prioritize toxicity testing on those chemicals likely to lead to adverse effects. To use HTS assays in predicting hazard to both humans and wildlife, it is necessary to understand how broadly these data may be extrapolated across species. The US EPA Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS; https://seqapass.epa.gov/seqapass/) tool was used to assess conservation of the 484 protein targets represented in the suite of ToxCast™ assays and other HTS assays. To demonstrate the utility of the SeqAPASS data for guiding extrapolation, case studies were developed which focused on targets of interest to the US Endocrine Disruptor Screening Program and the Organisation for Economic Cooperation and Development. These case studies provide a line of evidence for conservation of endocrine targets across vertebrate species, with few exceptions, and demonstrate the utility of SeqAPASS for defining the taxonomic domain of applicability for HTS results and identifying organisms for suitable follow-up toxicity tests.
Graphical Abstract
Introduction
The international adoption of 21st century toxicity testing practices, aimed at increasing efficiencies in chemical safety assessments and reducing reliance on whole animal studies has led to significant advancements in cell-based and computational approaches.1 High-throughput screening (HTS) assays allow for rapid screening of hundreds to thousands of chemicals, using cell-free assays, primary cells, immortalized cell lines, and even small scale in vivo models to prioritize which chemicals are of most concern and therefore may require in vivo toxicity testing. For example, the United States Environmental Protection Agency (US EPA) Endocrine Disruptor Screening Program (EDSP) and more broadly, the Organization of Economic Cooperation and Development (OECD; http://oe.cd/endocrine-disrupters) have both recently described frameworks to screen for endocrine active chemicals using computational models informed by HTS data for prioritization of thousands of chemicals for follow-up testing.2,3,4,5 When used for prioritization, HTS assays can significantly reduce the number of animals used for testing and reduce the costs associated with whole organism tests. Further, with advances in technology, many of these assays can be conducted using robotics and multi-well plates, of 96-wells or greater, which increase efficiencies in the chemical testing pipeline.
One example of such an effort includes the US EPA ToxCast™ screening and prioritization program, along with the collaborative multi-agency Tox21 effort, which has led the development and employment of rapid HTS assays for the assessment of large numbers of chemicals and the utilization of generated data to characterize bioactivity to focus future testing.6,7,8 Since initiation of the ToxCast™ program in 2007, over 4,000 chemicals have been screened in at least a subset of the bioassay platforms.7 The suite of ToxCast™ bioassays were developed to have extensive biological coverage with molecular targets spanning all major protein superfamily groups, including many known to be chemical targets.6
Species coverage
Traditionally, HTS assays were developed for screening chemicals for adverse impacts to human health and therefore primarily utilize mammalian-based cell lines, primary cells, and gene sequences.6 The available HTS assays with mammalian models were incorporated in the suite of ToxCast™ assays including human (Homo sapiens), cattle (Bos taurus), chimpanzee (Pan troglodytes), domestic guinea pig (Cavia porcellus), rabbit (Oryctolagus cuniculus), rat (Rattus norvegicus), house mouse (Mus musculus), pig (Sus scrofa), and sheep (Ovis aries). More recently, HTS assays have been developed and some incorporated into ToxCast™ to evaluate chemical effects on alternative vertebrate species using an African clawed frog (Xenopus laevis)-based thyroid pathway screening assay and a zebrafish (Danio rerio) development assay.9,10 With the original intention of ToxCast™/Tox21 assays to identify and prioritize chemicals that have the greatest likelihood to produce adverse effects in humans, and the subsequent realization that such data may be useful for protecting wildlife, a challenge emerged to understand whether the predominantly mammalian-based prioritization approach reasonably reflects potential impacts on other species. Therefore, understanding the domain of applicability across species for the HTS data is important to taking full advantage of both the existing and new data not only in support of the EDSP and OECD efforts, but also in support of other hazard identification efforts for non-mammalian species.
A tool that can aid in addressing this species-extrapolation challenge is the US EPA Sequence Alignment to Predict Across Species Susceptibility tool (SeqAPASS v3.0; https://seqapass.epa.gov/seqapass/).11 The SeqAPASS tool can be used to evaluate protein sequence similarity to predict chemical susceptibility across species. The underlying assumption for such protein-based species comparisons is that the greater the similarity between a chemical’s protein target in a known sensitive species to a protein in other species, the greater likelihood that the chemical would also interact with this protein in the other species. This concept of linking evolutionary conservation of protein targets to cross species chemical sensitivity was first described in 2008.12 The publicly available SeqAPASS tool has since expanded upon and advanced this concept into practice allowing for the comparison of any species to all other species with sequence information available to computationally predict chemical susceptibility. Therefore, the SeqAPASS output provides evidence as to whether a known protein target is present or absent in another species based on similarity, which is useful for laying the initial foundation for species extrapolation. Because the SeqAPASS tool takes advantage of the National Center for Biotechnology Information (NCBI) protein database, program outputs include similarity evaluations and conservation predictions for hundreds or even thousands of vertebrates, invertebrates, plants, bacteria, and viruses.
A tiered framework was proposed by Ankley et al.13 to evaluate biological pathway conservation across taxa as a means to understand whether current HTS approaches would be considered protective of non-mammalian species in the context of the EDSP. The tiered approach was previously applied to explore the taxonomic applicability domain of human estrogen receptor α (ERα)-based HTS assays.13 Since perturbation or modulation of estrogenic pathways are relatively well studied in a variety of species (e.g., 14,15,16), a robust evaluation of conservation at all tiers (i.e., computational, in vitro, and in vivo studies) of the taxonomic relevance framework was feasible for ERα. However, cross-species data related to perturbation of other molecular targets are often much more limited. Therefore, for a majority of chemical targets, the most practical and rapid means to evaluate the potential for chemical-protein target interactions across species will be through computational evaluation of protein sequence and structural conservation using SeqAPASS.
To initiate a SeqAPASS query in the context of HTS assays, two key pieces of information are necessary. Specifically, the model organism from which the cell, protein, or gene was derived for a given assay is needed, and the gene for the protein target measured in the assay must be known. Depending on the objective of the analysis and how well the protein has been characterized, as well as the level of detail known about the chemical-protein interaction, the SeqAPASS tool allows for evaluation of protein similarity at three different levels of sequence specificity.11 Level 1 facilitates comparative cross-species evaluation of the full primary amino acid sequence, Level 2 focuses the comparative analysis on the functional domain(s) in the protein target, and Level 3 compares individual amino acid residues across species.11,17 Level 3 queries are typically based on information obtained from the published literature regarding which specific amino acids are critical for the interaction of a specific chemical, or group of chemicals, with the protein of interest. Level 3 evaluations may be applied to compare known amino acid residues that are important for the specific action of that chemical. 11,17 Advancing through each level of the SeqAPASS evaluation adds additional evidence for protein similarity, with Level 3 providing the highest degree of taxonomic resolution for predictions of protein conservation and chemical susceptibility. However, in conducting the SeqAPASS evaluation to determine whether the assay target is conserved in other species, Level 1 and 2 data are generally sufficient to provide an initial line of evidence that can inform further toxicity testing (e.g., selection of relevant species to test) and extrapolation of screening data when considering all chemicals screened (as opposed to an individual chemical).
As a starting point in determining the taxonomic domain of applicability for mammalian-based HTS screening data, SeqAPASS evaluations comparing primary amino acid sequences (Level 1) and functional domains (Level 2) were performed for all molecular targets associated with the ToxCast™ assays and made publicly available. Since it was not reasonable to discuss all SeqAPASS results for these targets, data for two ToxCast™ assay groups are summarized. In addition, case studies were developed for hypothalamic-pituitary-gonadal/-thyroidmolecular targets relevant to the EDSP and OECD. These case studies examined conservation of the androgen receptor (AR), enzymes involved in the sex steroid synthesis (i.e., steroidogenesis), and proteins found in the thyroid axis. These case studies are used as examples to demonstrate the practical application of this approach to a current regulatory challenge.
Material and Methods
Identifying Protein Targets from ToxCast™/Tox21 HTS Assays
ToxCast™ data were downloaded from https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data, and filtered to identify all unique protein identifications (i.e., protein accessions) associated with HTS assay endpoints (Supplemental Data, Materials and Methods). This protein information, which included the protein accession and model species represented in the ToxCast™ assay, was used as a query protein sequence in the SeqAPASS analysis (Supplemental Data, Workbook S1).
Additional assay targets were identified as new HTS assays have been, and are being, developed to support the EDSP and OECD to more thoroughly screen for chemicals that may disrupt the thyroid axis and steroidogenesis. Since the summary data that was made public in October 2015, six thyroid related HTS assays and the high-throughput H295R assay (HT-H295R; a steroidogenesis assay) were developed to support EDSP and OECD efforts. The data for the HT-H295R assay and the thyroid peroxidase (TPO) assay are available through the ToxCast™ dashboards and available for public download. Chemical screening results from the other thyroid-related HTS assays can be obtained from the published literature, but has yet to be incorporated in the ToxCast™ dashboard.18,19,9,20,21
SeqAPASS Level 1: Primary Amino Acid Sequence Comparisons of HTS Targets
A total of 484 unique assay targets available in the ToxCast™ suite were evaluated using SeqAPASS v3.0, Data Version 3 (Supplemental Data, Table S1). The output from SeqAPASS provides a prediction of relative intrinsic susceptibility across species based on similarity of the hit proteins to the query protein (Supplemental Data, Table S2). In the context of evaluating HTS assays, a susceptibility prediction of “yes” in the SeqAPASS output indicates the protein target is conserved in that species whose sequence aligned with the query species (Supplemental Data, Materials and Methods).
SeqAPASS Level 2: Functional Domain Comparisons of HTS Targets
For each protein accession submitted in SeqAPASS Level 1, functional domains (e.g., ligand binding domains, DNA binding domains, zinc finger domains) categorized as “specific hits” in the NCBI Conserved Domain Database (CDD; https://www.ncbi.nlm.nih.gov/cdd/) were identified (0 to 17 domains) and submitted as SeqAPASS Level 2 queries (Supplemental Data, Materials and Methods).11,22, 23 Overall, functional domains were evaluated for 452 of the 484 proteins.
Processing SeqAPASS Data to Evaluate the Taxonomic Domain of Applicability for HTS Data
All generated SeqAPASS Level 1 and Level 2 data were saved in a .zip file using the Save Reports function described in the SeqAPASSv3.0 user guide.22 Briefly, this save function allows the user to download and save multiple reports at one time (Supplemental Data, Materials and Methods). All Level 1 and Level 2 data from the current evaluation can be accessed via SeqAPASS and through the US EPA CompTox Chemistry Dashboard (https://comptox.epa.gov/dashboard/) (Supplemental Data, Tables S3–S4; Supplemental Data, Zip file S1). The CompTox Chemistry Dashboard is a resource that provides access to chemical information, including HTS bioassay data.24 Therefore, links from the dashboard to the SeqAPASS tool, and specifically to SeqAPASS data for HTS assay targets, are available as a means to efficiently access chemical and effects data. As new “Data Versions” are released in the SeqAPASS tool (See SeqAPASS About page), the SeqAPASS data on the CompTox Chemistry Dashboard will be updated for all bioassay targets, allowing access to the most current protein data for cross-species extrapolation.
To broadly describe the results of the SeqAPASS data, Level 1 and 2 data outputs with default settings (i.e., primary reports; E-value <0.01; common domains ≥1; default 1st local minimum was used to set the susceptibility cut-off)25,11 were collected as a multi-dataset download (SeqAPASS User Guide22) for each protein target. Protein targets were then grouped according to their intended target family (Supplemental Data, Materials and Methods). The target families included: 1) cell adhesion molecules; 2) cytochrome P450 enzymes (i.e., cyp); 3) cytokine; 4) DNA binding; 5) esterase 6) G-protein-coupled receptors (i.e., gpcr); 7) growth factor; 8) hydrolase; 9) ion channel; 10) kinase; 11) lyase; 12) methyltransferase; 13) nuclear receptor; 14) oxidoreductase; 15) phosphatase; 16) protease; 17) protease inhibitor; and 18) transporter. Of the 484 proteins corresponding to ToxCast™ assays and evaluated in SeqAPASS, 346 were placed into these groups (Supplemental Data, Table S1). The remaining proteins were not placed in a group and were categorized as miscellaneous proteins. Conservation was broadly described within a defined assay group (except for miscellaneous proteins), deeming protein groups either broadly conserved (e.g., vertebrates and invertebrates, plants) or narrowly conserved (e.g., vertebrates only) (Supplemental Data, Zip File S1). Synthesis of the SeqAPASS Level 1 results for two of the ToxCast™ assay groups (G protein coupled receptor and growth factor) are described below to illustrate the information available for all assay groups. Level 2 SeqAPASS data are described for an individual protein, human transforming growth factor beta 1 (found in assay group growth factor), although Level 2 data were produced for all proteins evaluated (Supplemental Data, Zip File S1).
Case Studies Focused on Endocrine Targets
Androgen Receptor
Eleven ToxCast™ assays have been developed to detect chemicals that perturb the AR, most of which utilize the human AR gene or protein. However, AR genes from the chimpanzee and Norway rat are also represented in the suite of assays (Table 1). Therefore, to evaluate conservation of the AR across species, proteins for all three species were queried using SeqAPASS. Level 1 analysis comparing primary amino acid sequences and Level 2 analyses focused on the ligand binding domain were performed (Table 1; Supplemental Data, Figure S1a–d).
Table 1.
| Assay Name | Assay Target | Model organism | SeqAPASS Query Accession | Domain evaluated (NCBI Accession) |
|---|---|---|---|---|
| ATG_TRANS | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| NVS_NR_hAR | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| OT_AR_ARELUC_AG_1440 | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| OT_AR_ARSRC1_0480 | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| OT_AR_ARSRC1_0960 | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| TOX21_AR_BLA_Agonist | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| TOX21_AR_BLA_Antagonist | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| TOX21_AR_LUC_MDAKB2_Agonist | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| TOX21_AR_LUC_MDAKB2_Antagonist | Androgen receptor, AR | Human (Homo sapiens) | P10275 | Ligand binding domain (cd07073) |
| NVS_NR_cAR | Androgen receptor, AR | Chimpanzee (Pan troglodytes) | O97775 | Ligand binding domain (cd07073) |
| NVS_NR_rAR | Androgen receptor, AR | Norway rat (Rattus norvegicus) | P15207 | Ligand binding domain (cd07073) |
| HT-H295R | Cholesterol side-chain cleavage enzyme; CYP11A1 | Human (Homo sapiens) | P05108 | NA |
| HT-H295R | Steroid 17α-hydroxylase/17,20 Lyase; CYP17A1 | Human (Homo sapiens) | P05093 | NA |
| HT-H295R | 3β-hydroxysteroid dehydrogenase; HSD3B2 | Human (Homo sapiens) | P26439 | NA |
| HT-H295R | Aromatase; CYP19A1 | Human (Homo sapiens) | P11511 | NA |
| NVS_ADME_hCYP19A1 | Aromatase; CYP19A1 | Human (Homo sapiens) | P11511 | NA |
| TOX21_Aromatase_Inhibition | Aromatase; CYP19A1 | Human (Homo sapiens) | P11511 | NA |
| HT-H295R | 17β-hydroxysteroid dehydrogenase type1; 17βHSD1 | Human (Homo sapiens) | P14061 | NA |
| HT-H295R | 17β-hydroxysteroid dehydrogenase type3; 17βHSD3 | Human (Homo sapiens) | P37058 | NA |
| HT-H295R | Steroid 21-hydroxylase; CYP21A2 | Human (Homo sapiens) | AFK10138 | NA |
| HT-H295R | Steroid 11-beta-hydroxylase; CYP11B1 | Human (Homo sapiens) | AAA35741 | NA |
| ATG_THRa1_TRANS | Thyroid hormone receptor alpha; THRα | Human (Homo sapiens) | P10827 | Ligand binding domain (cd06935) |
| NVS_NR_hTRa_Antagonist | Thyroid hormone receptor alpha; THRα | Human (Homo sapiens) | P10827 | Ligand binding domain (cd06935) |
| TOX21_TR_LUC_GH3_Agonist | Thyroid hormone receptor alpha; THRα | Human (Homo sapiens) | P10827 | Ligand binding domain (cd06935) |
| ATG_THRb_TRANS2 | Thyroid hormone receptor beta; THRβ | Human (Homo sapiens) | P10828 | Ligand binding domain (cd06935) |
| NCCT_TPO_AUR | Thyroid peroxidase, TPO | Norway rat (Rattus norvegicus) | P14650 | Thyroid _peroxidase (cd09825) |
| NCCT_TPO_GUA | Thyroid peroxidase, TPO | Pig (Sus scrofa) | P09933 | Thyroid _peroxidase (cd09825) |
| hNIS-HEK293T-EPA | Sodium/iodide cotransporter, NIS | Human (Homo sapiens) | Q92911 | NA |
| HTS assay in development | Type I iodothyronine deiodinase, DIO1 | Human (Homo sapiens) | P49895 | NA |
| HTS assay in development | Type II iodothyronine deiodinase, DIO2 | Human (Homo sapiens) | Q92813 | NA |
| HTS assay in development | Type III iodothyronine deiodinase, DIO3 | Human (Homo sapiens) | P55073 | NA |
| HTS assay in development | Thyroid stimulating hormone receptor, TSHR | Human (Homo sapiens) | P16473 | Peptide binding pocket (cd15964) |
| HTS assay in development | Iodotyrosine deiodinase, IYD | Human (Homo sapiens) | Q6PHW0 | Iodotyrosine_dehalogenase domain (cd02144) |
Assay name key: Attagene = ATG; NovaScreen = NVS; Odyssey-Thera = OT; Tox21 platform = TOX21; Steroidogenesis Assay = HT-H295R; Radioactive iodide uptake assay (RAIU) assay = hNIS; high-throughput screening = HTS
Steroidogenesis
The SeqAPASS tool was used to evaluate cross-species conservation of steroidogenic enzymes by comparing primary amino acid sequences representing several steroidogenic genes: human cholesterol side-chain cleavage enzyme (CYP11A1), steroid 17-alpha-hydroxylase/17,20 lyase (CYP17A1), 3β-hydroxysteroid dehydrogenase (HSD3B2), aromatase (CYP19A1), 17β-hydroxysteroid dehydrogenase type 1 (17βHSD1), 17β-hydroxysteroid dehydrogenase type 3 (17βHSD3), 21-hydroxylase (CYP21A2), and steroid 11-beta-hydroxylase (CYP11B1) to similar proteins in other species (Table 1). Only Level 1 SeqAPASS evaluations were carried out for the CYPs, as the domain identified as a specific hit in the NCBI conserved domain database (pfam00067) was shown to encompass almost the entire protein sequence for each of the CYPs. Therefore, Level 2 evaluations of functional domain(s) were not used to describe the data for steroidogenic CYPs as they did not add greater taxonomic resolution to the SeqAPASS prediction of conservation.
Thyroid axis
The ToxCast™/Tox21 suite contains seven assays relevant to the thyroid axis, three of which were developed to detect chemicals that act on human thyroid hormone receptor alpha (THRα), one for human thyroid hormone receptor beta (THRβ), one each for Norway rat and pig thyroid peroxidase (TPO) and one for the human sodium/iodide cotransporter (NIS).21,19,18 In addition to the assays already incorporated into ToxCast™, SeqAPASS analyses were also performed for additional thyroid axis targets for which HTS assays are being developed (i.e., human type I (DIO1), II (DIO2), and III (DIO3) deiodinases, thyroid stimulating hormone receptor (TSHR), and iodotyrosine deiodinase (IYD);9 Personal communication S. Degitz, April 2018). Each target was evaluated with SeqAPASS using default settings (Table 1). Relevant domains were either not identified or encompassed nearly the full length of the full protein sequence for DIO1, DIO2, and DIO3 in NCBI. Because the primary reports for SeqAPASS Level 1 identify hits with at least one common domain to the query protein, very few protein hits were found for DIO1, DIO2, and DIO3. Therefore, for these proteins the full reports (as opposed to the default primary reports) were used to describe the SeqAPASS data (see SeqAPASS user guide for detailed description of full report22).
Results and Discussion
SeqAPASS results can be used as an initial line of evidence for defining the taxonomic domain of applicability for HTS assays, with the recognition that the tool specifically identifies a protein conservation across species. Such evidence of protein conservation indicates that chemicals identified as modulators of a particular protein (or gene) in an assay developed with a specific model organism are likely to act as such in other organisms that have the same protein. This information can be useful for both extrapolation of HTS results to other taxa or in identifying organism for which assay development might be warranted due to unique biology not covered by current HTS assays. The level of detail used in the SeqAPASS evaluation is dependent on the purpose. The intent of the present study was to provide an initial understanding of how broadly the HTS data may be reasonably extrapolated across species. All 484 proteins corresponding to ToxCast™ assay gene targets were evaluated. Consequently, it is not feasible to discuss the results for every target in detail. Instead, two protein groups were selected for discussion, G protein coupled receptor and growth factor. Description of these data are intended to provide an illustration of the available data and methods for application.
SeqAPASS to Evaluate Select ToxCast™ Assay Groups
G protein-coupled receptors are transmembrane cell surface receptors that bind extracellular substances and generate intracellular signals.26 There are 79 GPCR-related HTS assays for these receptors in the ToxCast™ suite, representing 71 unique mammalian protein targets (Supplemental Data, Table S1). The SeqAPASS evaluations of several of these (e.g., human Beta-2 adrenergic receptor, Norway rat neurotensin receptor type 1A, human D(1A) dopamine receptor, cattle adenosine receptor), provided evidence of conservation across both vertebrates and invertebrates. At least 62 of the 71 GPCRs were conserved among vertebrates including Mammalia, Testudines, Aves, Crocodylia, Lepidosauria, Amphibia, Actinopteri, Coelacanthiformes, and Chondrichthyes. Between 28 and 41 gpcr were conserved across invertebrate taxon such as Insecta, Merostomata (horseshoe crab, sea scorpions), Arachnida, Branchiopoda (water fleas), Bivalvia, Malacostraca, and Gastropoda. Flowering plants were rather limited in their similarity to the mammalian GPCRs with only the Liliopsida taxonomic group shown to have a glutamate receptor aligning with the Norway rat gamma-aminobutyric acid (GABA) B receptor 1.
There is only one growth factor protein represented in ToxCast™, human transforming growth factor beta 1 (TGF-β), which is involved in intercellular signaling during development.27 Based on Level 1 SeqAPASS analysis, this protein was only found in vertebrates and Trematoda (i.e., parasitic flatworms). Two domains for human TGF-β were identified as specific hits in the NCBI conserved domain database and evaluated using SeqAPASS Level 2. The first domain is from the protein family TGF-β propeptide, while the second is a disulphide-linked homo- or heterodimer. Both domains are conserved across vertebrates and Trematoda, providing additional evidence of conservation.
Based on SeqAPASS evaluation of proteins in all other assay groups, conservation was broadly identified across vertebrates, invertebrates, and even plants. As such, the initial SeqAPASS data (using default settings) provides a line of evidence that screening results generated from mammalian-based ToxCast™ assays are more broadly applicable beyond the model organism used in the assay itself. These data are now available through both SeqAPASS and the CompTox Chemistry Dashboard (Supplemental Data, Figures S1 and S2). Below we present three illustrative case examples of how SeqAPASS data can inform the EDSP and OECD for understanding the taxonomic domain of applicability for available and developing HTS assays.
Protecting Human Health and Wildlife from Endocrine Disruption
The EDSP and OECD are exploring the utility of HTS data for screening to identify endocrine active chemicals for additional, higher-tier, toxicity testing.3,28 Assays included in the ToxCast™ HTS screening battery suitable for this purpose capture aspects of estrogen, androgen, or thyroid hormone signaling pathways, as well as steroidogenesis. Empirical studies that evaluate conservation of key protein components of these pathways across taxonomic groups are relatively limited, so a simple pragmatic approach is needed to provide an initial understanding of the taxonomic domain of applicability for the HTS results. We previously described use of SeqAPASS to evaluate cross-species conservation of one key endocrine target, the estrogen receptor.13 Herein, three additional SeqAPASS case studies have been developed, focusing on the AR, thyroid axis, and steroidogenic enzymes. In addition to defining the taxonomic domain of applicability of the HTS data, the results of these SeqAPASS analyses could be used to focus higher-tiered toxicity testing on those species that are most likely susceptible to chemicals that can act on relevant protein targets in these endocrine pathways. Additionally, the results from SeqAPASS may inform the development of additional HTS assays to capture unique biology of a particular taxonomic group.
The Taxonomic Domain of Applicability for AR HTS Assays
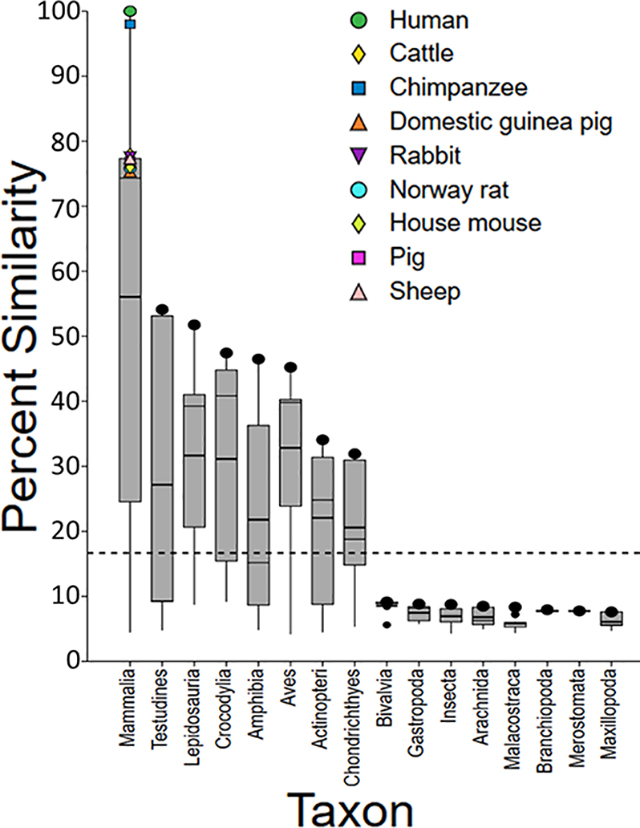
Eleven HTS assays using human, chimpanzee, and Norway rat ARs are included in the ToxCast™ suite to screen for endocrine disrupting chemicals (Table 1).29 Because human, chimpanzee, and Norway rat SeqAPASS evaluations yielded the same susceptibility predictions across species, in the following analysis we focus on the human AR as a comparative probe sequence. However, these data are representative of the other species (both chimpanzee and rat SeqAPASS data are provided in Supplemental Data, Figure S1a–d). The primary amino acid sequence comparison shows that the human AR and specifically the ligand binding domain is conserved in vertebrate taxa including Mammalia, Testudines, Lepidosauria, Crocodylia, Amphibia, Aves, Coelacanthiformes, Actinopteri, Chondrichthyes, Ceratodontimorpha (lungfish), Cladistia (eel shaped bony fish), Myxiniformes (hagfish) and Petromyzontiformes (lamprey) (Figure 1a and b; Supplemental Data, Workbook S2). However, upon further evaluation of the SeqAPASS data, it was noted that the proteins aligning with the human AR ligand binding domain from Myxiniformes and Petromyzontiformes were annotated as nuclear receptors, but not specifically AR (e.g., steroid receptor 2, ER1 or partial sequences for the corticoid receptor, retinoic acid receptor 2 and retinoic acid receptor 4). These results are consistent with previously published evolutionary evidence from genome mapping and phylogenetic analyses demonstrating that orthologs of AR are present in tetrapods, teleosts, and sharks; whereas, AR is not present in lamprey.30,31,32,33 Overall, these data provide a line of evidence that ToxCast™ assay results with human, chimpanzee, or Norway rat ARs are likely applicable to a majority of vertebrate species due to the presence of a similar molecular (i.e., protein) target.
Figure 1.
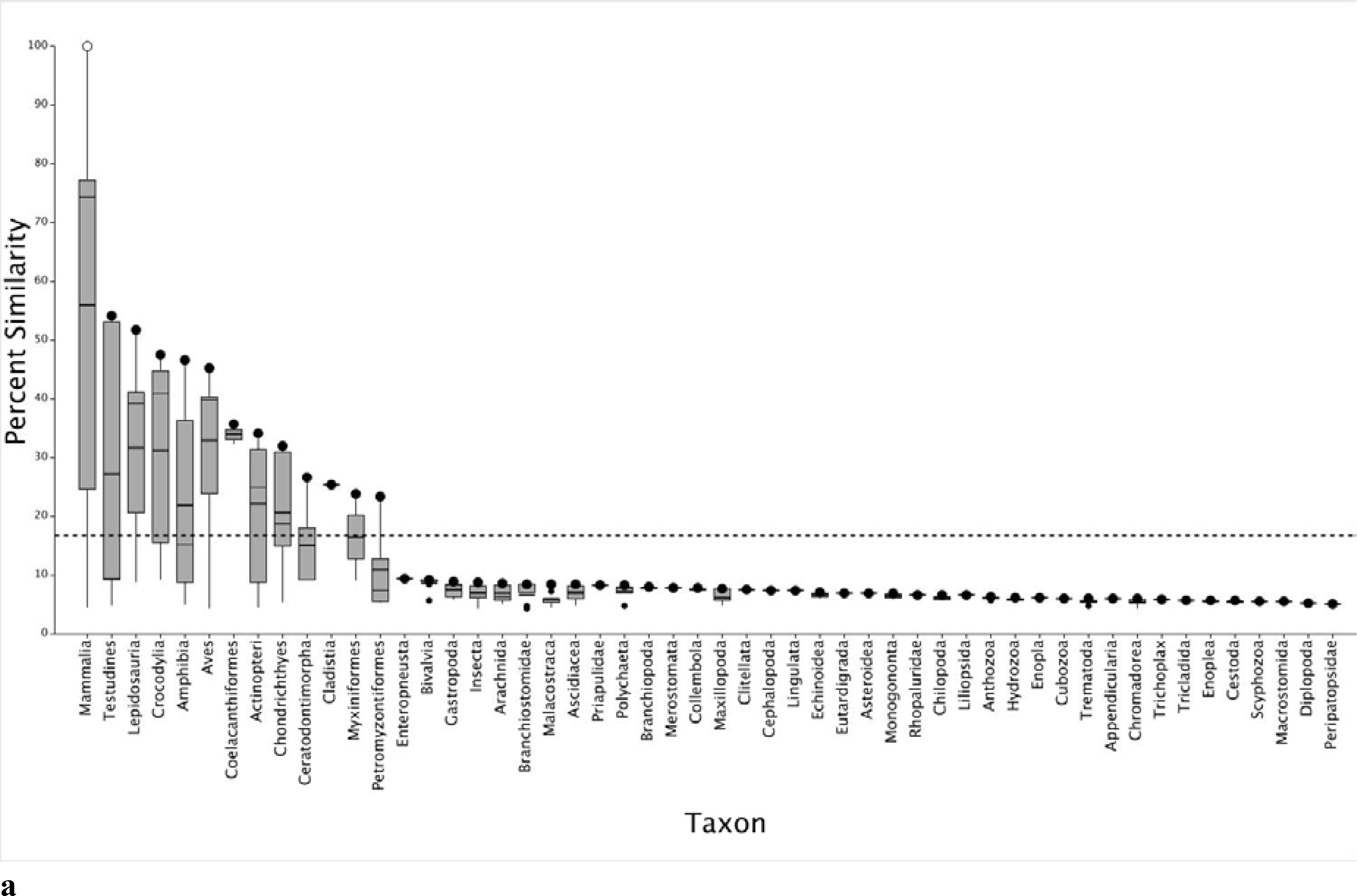
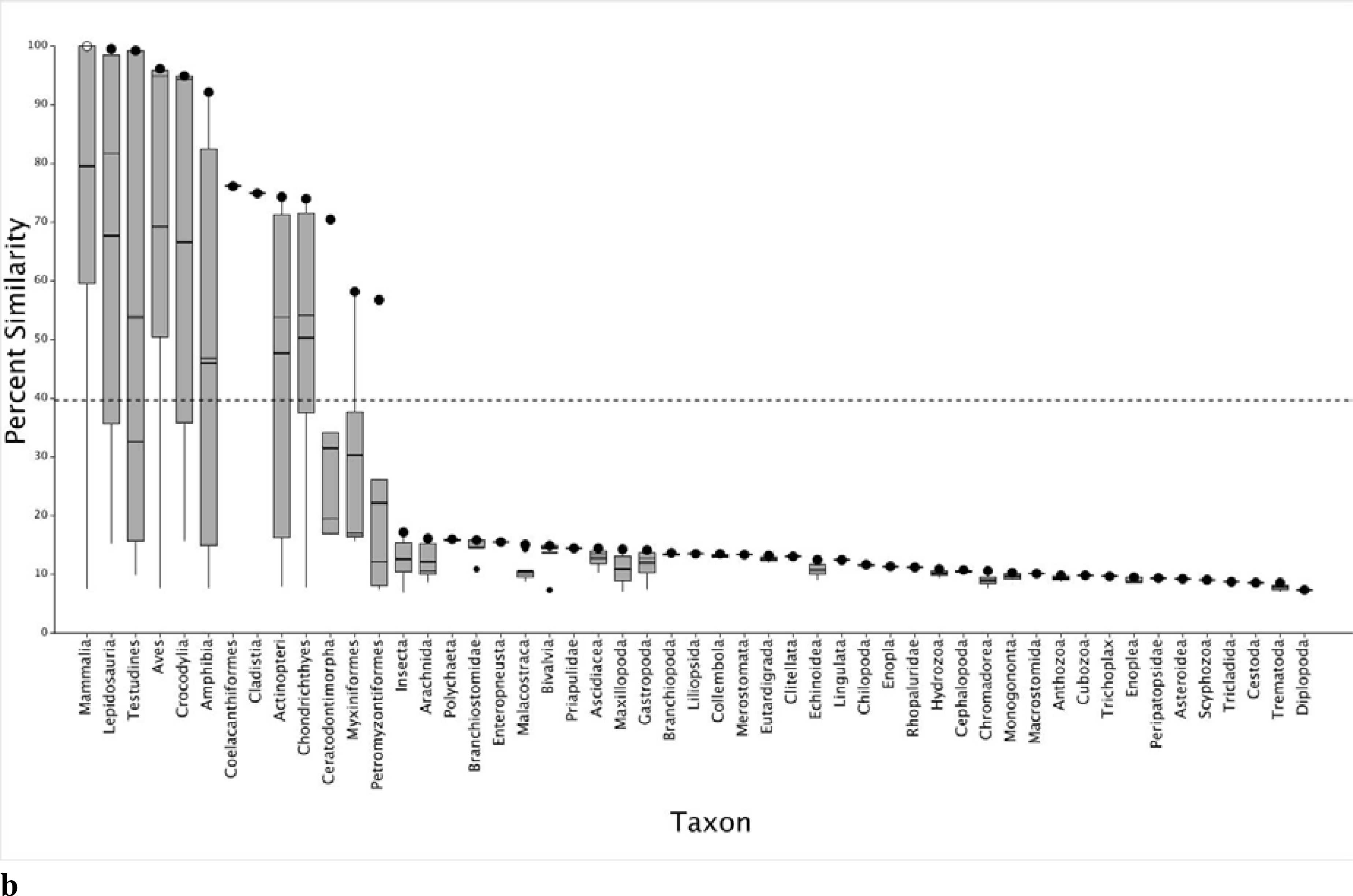
Boxplots depicting SeqAPASS (v3.0) data illustrating the percent similarity across species compared to human (Homo sapiens) androgen receptor (AR), examining the primary amino acid sequences (a) and the ligand binding domain (b). In each plot, the o represents the human AR and • represents the species with the highest percent similarity within the specified taxonomic group. The top and bottom of each box represent the 75th and 25th percentiles, respectively. The top and bottom whiskers extend up to 1.5 times the interquartile range. The mean and median values for each taxonomic group are represented by horizontal thick and thin black lines on the box, respectively. The dashed line indicates the cut-off for predictions of intrinsic susceptibility. Species from taxonomic groups that cross the susceptibility cut-off or those found above the cut-off are predicted as likely to be susceptible to chemicals that act on the query species protein target. The protein target is therefore conserved in species predicted to be susceptible.
The Taxonomic Domain of Applicability for HTS Assays Evaluating Steroidogenesis
Several enzymes are involved in converting cholesterol to steroid hormones, including corticosteroids and sex steroids. Sex hormones serve key functions in immunosuppression, blood pressure, sexual differentiation, reproduction, and fertility. Steroidogenic enzymes feature many structurally-related CYPs. However, the coverage of all steroidogenic enzymes in the suite of ToxCast™ assays is minimal, with only human CYP19A1 (i.e., aromatase) represented as a discrete target in the HTS assays. To address this gap, the EDSP is evaluating the utility of a HTS assay that quantitatively measures steroid hormone production in the human adrenocortical H295R cell line.34 This assay measures 11-deoxycortisol, deoxycorticosterone, cortisol, corticosterone, 17α-hydroxyprogesterone, 17α-hydroxypregnenolone, progesterone, pregnenolone, dehydroepiandrosterone (DHEA), androstenedione, testosterone, estrone, and estradiol in control and chemically-treated cells (Supplemental Data, Figure S2). Therefore, data from the HT-H295R assay provides profiles of steroid synthesis that can serve as indirect measures of chemical perturbation of enzyme activity/function.
As an initial step in determining the taxonomic domain of applicability of the HT-H295R assay results, it is important to understand whether other species also have similar steroidogenic enzymes to humans. To this end, several key enzymes were queried using the SeqAPASS tool (Table 1; Supplemental Data, Workbook S2). The human CYP11B1 primary amino acid sequence was conserved across vertebrate taxa, with the exception of Chondrichthyes, Aves, and Petromyzontiformes (Table 2; Supplemental Data, Figure S3a). From these results, it could be hypothesized that the biosynthesis of corticosterone may be different for sharks, skates, rays, birds, and lamprey than other vertebrates. Consistent with SeqAPASS results, CYP11B has not been identified in any species of Chondrichthyes.35 In fact, Chondrichthyes have been shown to convert 11-deoxycorticosterone to a unique steroid, 1α-hydroxycorticosterone, which has been reported to function as a corticosteroid.36 The enzyme responsible for the formation of 1α-hydroxycorticosterone in Chondrichthyes has yet to be identified.35 Also consistent with SeqAPASS results, CYP11B gene was reported to be found only in the kiwi but not in other birds.37 The Brown kiwi (Apteryx spp.) was present in the SeqAPASS data, having a predicted protein sequence for CYP27C isoform X1 aligning with the human CYP11B1 with 19.7 percent similarity. It has been reported that lamprey species do not have a CYP11B ortholog, which is consistent with their lack of serum cortisol and corticosterone.38,39
Table 2.
SeqAPASS Level 1 predictions of conservation of mammalian steroidogenic targets across taxa
| Query Protein | CYP11A1 | CYP17A1 | HSD3B2 | CYP19A1 | 17βHSD1 | 17βHSD3 | CYP21A2 | CYP11B1 | |
|---|---|---|---|---|---|---|---|---|---|
| bSeqAPASS Cut-off | 32.92 | 30.95 | 16.16 | 46.79 | 38.91 | 38.40 | 33.59 | 37.60 | |
| Vertebrates | Mammalia | Yes (115 of 127) |
Yes (113 of 153) |
Yes (117 of 118) |
Yes (111 of 121) |
Yes (110 of 116) |
Yes (112 of 113) |
Yes (96 of 142) |
Yes (108 of 127) |
| Actinopteri | Yes (58 of 74) |
Yes (63 of 118) |
Yes (59 of 59) |
Yes (137 of 147) |
Yes (19 of 49) |
Yes (42 of 49) |
Yes (20 of 118) |
Yes (38 of 76) |
|
| Amphibia | Yes (8 of 8) |
Yes (5 of 6) |
Yes (4 of 4) |
Yes (8 of 10) |
Yes (3 of 3) |
Yes (3 of 4) |
Yes (3 of 6) |
Yes (6 of 8) |
|
| Aves | Yes (45 of 65) |
Yes (61 of 71) |
Yes (73 of 73) |
Yes (65 of 67) |
Yes (16 of 66) |
Yes (45 of 66) |
Yes (4 of 71) |
No (0 of 70) |
|
| Chondrichthyes | Yes (4 of 5) |
Yes (3 of 4) |
Yes (4 of 4) |
Yes (4 of 5) |
No (0 of 2) |
Yes (2 of 2) |
Yes (1 of 4) |
No (0 of 5) |
|
| Coelacanthiformes | Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
- | Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
|
| Crocodylia | Yes (2 of 4) |
Yes (4 of 4) |
Yes (4 of 4) |
Yes (4 of 4) |
Yes (3 of 3) |
Yes (4 of 4) |
No (0 of 4) |
Yes (1 of 3) |
|
| Lepidosauria | Yes (4 of 8) |
Yes (7 of 8) |
Yes (7 of 7) |
Yes (6 of 6) |
No (0 of 7) |
Yes (7 of 7) |
Yes (4 of 8) |
Yes (5 of 8) |
|
| Myxiniformes | Yes (1 of 1) |
- | - | - | - | - | - | No (0 of 1) |
|
| Testudines | Yes (3 of 3) |
Yes (4 of 6) |
Yes (3 of 3) |
Yes (7 of 7) |
Yes (2 of 3) |
Yes (3 of 3) |
No (0 of 6) |
Yes (1 of 4) |
|
| aInvertebrates | Anthozoa | No (0 of 5) |
Yes (3 of 5) |
Yes (3 of 4) |
No (0 of 5) |
No (0 of 5) |
No (0 of 5) |
No (0 of 5) |
No (0 of 5) |
| Arachnidia | No (0 of 15) |
No (0 of 11) |
Yes (8 of 9) |
No (0 of 15) |
No (0 of 2) |
No (0 of 14) |
No (0 of 10) |
No (0 of 15) |
|
| Bivalvia | No (0 of 8) |
No (0 of 7) |
Yes (4 of 4) |
No (0 of 10) |
No (0 of 1) |
No (0 of 4) |
No (0 of 7) |
No (0 of 5) |
|
| Branchiopoda | No (0 of 2) |
No (0 of 2) |
Yes (1 of 2) |
No (0 of 2) |
- | No (0 of 2) |
No (0 of 2) |
No (0 of 2) |
|
| Branchiostomidae | No (0 of 2) |
Yes (2 of 2) |
Yes (2 of 2) |
No (0 of 2) |
- | No (0 of 2) |
No (0 of 2) |
No (0 of 2) |
|
| Gastropoda | No (0 of 5) |
No (0 of 3) |
Yes (2 of 3) |
No (0 of 6) |
No (0 of 1) |
No (0 of 5) |
No (0 of 3) |
No (0 of 8) |
|
| Insecta | No (0 of 176) |
No (0 of 137) |
Yes (8 of 56) |
No (0 of 172) |
No (0 of 4) |
No (0 of 117) |
No (0 of 135) |
No (0 of 189) |
|
Invertebrate taxon represent a subset of those found to be conserved for CYP17A1 and HSD3B2. Those invertebrate taxa shown represent common taxonomic groups (Full taxon list in Supplemental Data, Workbook S2). Invertebrates were shown to be conserve for only CYP17A1 and HSD3B2.
The SeqAPASS cut-offs define those species with conserved proteins found above the cut-off and those species less likely to be conserved below the cutoff (Details in Supplemental Data, Table S3)
Table cells indicate whether the protein is conserved compared to the human query protein, “Yes,” or less likely to be conserved, “No.” In parentheses are the number of species found above the SeqAPASS cut-off of the total number of species aligning with the mammalian target for that taxonomic group. Dashed line indicates that no protein sequences aligned with the query sequence for that taxonomic group.
Abbreviations and query protein accessions: CYP11A1= human cholesterol side-chain cleavage enzyme (P05108.2); CYP17A1 = human steroid 17-alpha-hydroxylase/17,20 lyase (P05093.1); HSD3B2 = human 3 beta-hydroxysteroid dehydrogenase/Delta 5-->4-isomerase type 2 (P26439.2); CYP19A1 = aromatase (P11511.3); 17βHSD1 = human estradiol 17-beta-dehydrogenase 1 (P14061.3); 17βHSD3 = human testosterone 17-beta-dehydrogenase 3 (P37058.2); CYP21A2 = human steroid 21-hydroxylase (AFK10138.1); CYP11B1 = human 11-beta-hydroxylase (AAA35741.1).
Steroid 21-hydroxylase catalyzes the conversion of progesterone to 11-deoxycorticosterone and 17α-OH-progesterone to 11-deoxycortisol. Comparative evaluation of human CYP21A2 with SeqAPASS provided evidence that the enzyme was conserved in a majority of vertebrate taxa evaluated with the exception of Crocodylia and Testudines (Table 2; Supplemental Data, Figure S3b). In these taxa a putative “PREDICTED: steroid 17-alpha-hydroxylase/17,20 lyase” aligned most closely with the human enzyme.
There is a group of fourteen human enzymes known as 17β-hydroxysteroid dehydrogenases which perform various reactions within the steroidogenesis pathway. However, due to the well-established roles of 17βHSD1 in the interconversion of estrone and estradiol and 17βHSD3 in the reduction of DHEA, 5α-androstanedione, and androsterone to precursors of testosterone and dihydrotestosterone, these two enzymes were the focus of the SeqAPASS evaluation (Table 1).40 For 17βHSD1, conservation was limited to vertebrates with the exception of Lepidosauria and Chondrichthyes (both had retinol dehydrogenase 8-like align with human 17βHSD1) (Table 2; Supplemental Data, Figure S3c). Results of a Baker et al.38 study of the evolution of enzymes involved in adrenal and sex steroid biosynthesis support SeqAPASS results, and found that Chondrichthyes do not contain an ortholog to human17βHSD1.
SeqAPASS analysis of human CYP11A1, CYP19A, and 17βHSD3 provided evidence for conservation of these enzymes across all vertebrates, but not invertebrates (Table 2; Supplemental Data, Figure S3d–f). However, human CYP17A1, the enzyme responsible for converting pregnenolone and progesterone to 17-alpha-hydroxylated products and further to dehydroepiandrosterone and androstenedione was broadly conserved across both vertebrate and invertebrate taxa (Table 2; Supplemental Data, Figure S3g). The same was true for HSD3B, the enzyme that catalyzes the conversion of pregnenolone to progesterone, 17α-hydroxypregnenolone to 17α-hydroxyprogesterone, and DHEA to androstenedione (Table 2; Supplemental Data, Figure S3h).
Overall, the SeqAPASS analyses of enzymes involved in steroidogenesis suggest that results from the human cell-based HT-H295R assay may be broadly extrapolated to other vertebrates, but not invertebrates (Supplemental Data, Workbook S2). However, caution should be applied in extrapolating screening results involving glucocorticoid synthesis to Chondrichthyes, Aves, and Petromyzontiformes, as enzymes involved in biosynthesis may differ from humans in these taxa. Additionally, the apparent lack of 17βHSD1 in Lepidosauria and Chondrichthyes indicate that estrone and 17β-estradiol profiles measured by the HT-H295R assay may not be reflective of potential impacts to species in these groups.
The Taxonomic Domain of Applicability for HTS Assays Evaluating Thyroid Axis Disruption
The thyroid axis is essential in vertebrate development, growth, and neurogenesis.41 There are several proteins that are critical to the normal function of the human thyroid axis (Supplemental Data, Figure S4). Accordingly, several HTS assays are under development or already being used in the EDSP to screen for chemicals that act on key proteins in the thyroid axis that could, if disrupted by a chemical stressor, lead to adverse health effects.
The current ToxCast™ suite includes assays for three thyroid axis targets: THRα, THRβ, and TPO. The THRα and THRβ bind triiodotyrosine (T3) and regulate subsequent thyroid hormone responsive gene transcription.42 Thyroid peroxidase is an enzyme present at the apical cell surface of thyrocytes that oxidizes iodide for incorporation into thyroglobulin and production of thyroid hormones, thyroxine (T4) and T3.43 In addition to these relatively well-established potential thyroid targets, there are other proteins in the axis that could be subject to chemical perturbation and hence, would be desirable to capture with HTS assays. For example, a 96-well plate, stable human cell-based (sodium/iodide symporter-expressing HEK293T cell line), radioactive iodide uptake (RAIU) assay was recently developed to screen for inhibitors of sodium/iodide symporter (NIS)-mediated iodide uptake.18 The NIS utilizes a Na+ gradient to allow for iodide uptake into the thyroid cell and is the first rate-limiting step for synthesis of T3 and T4 (Supplemental Data, Figure S4).18
Additional HTS assays are being developed to screen for chemicals that disrupt human DIO1, DIO2, and DIO3, TSHR, and IYD.20,9 To provide insights as to how broadly results from currently available and developing HTS assays may reasonably be extrapolated across species, SeqAPASS was used to evaluate sequence similarity of all assay targets used or being developed to screen the thyroid axis (Table 1; Supplemental Data, Workbook S2).
Levels 1 and 2 evaluations of human THRα and THRβ and their respective ligand binding domains, showed that these receptors are well conserved across vertebrates, with the exception of Ceratodontimorpha (lungfish). Conservation of THRβ, but not THRα is also found for several invertebrate taxa, including Polychaeta (sandworms), Gastropoda, Lingulata (lampshells), Bivalvia, Enteropneusta (Acorn worms), Asteroidea (starfish), Branchiostomidae (lancelet) and Ascidiacea (sea squirts, tunicate) (Table 3; Supplemental Data, Figure S5a–d). These SeqAPASS results are concordant with results presented by Holzer et al.44 describing the evolution of thyroid hormone signaling across species.
Table 3.
SeqAPASS level 1 and 2 predictions of conservation of mammalian thyroid targets across taxa
| Query Protein | THRα | THRβ | TPO | NIS | DIO1 | DIO2 | DIO3 | TSHR | IYD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SeqAPASS Level | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | |
| bSeqAPASS Cutoff | 34.53 | 70.03 | 33.15 | 28.04 | 38.75 | 54.65 | 44.54 | 25.83 | 40.00 | 16.77 | 48.42 | 82.57 | 24.53 | 33.62 | |
| Vertebrates | Mammalia | Yes (113 of 121) |
Yes (115 of 162) |
Yes (115 of 167) |
Yes (113 of 152) |
Yes (94 of 117) |
Yes (84 of 141) |
Yes (100 of 110) |
Yes (126 of 127) |
Yes (123 of 127) |
Yes (127 of 127) |
Yes (114 of 157) |
Yes (113 of 181) |
Yes (106 of 106) |
Yes (111 of 111) |
| Actinopteri | Yes (68 of 121) |
Yes (71 of 127) |
Yes (71 of 120) |
Yes (78 of 117) |
Yes (17 of 53) |
Yes (19 of 63) |
Yes (48 of 63) |
Yes (62 of 70) |
Yes (52 of 70) |
Yes (69 of 70) |
Yes (52 of 1409) |
Yes (1 of 2169) |
Yes (45 of 45) |
Yes (45 of 45) |
|
| Amphibia | Yes (12 of 24) |
Yes (13 of 29) |
Yes (15 of 23) |
Yes (28 of 28) |
Yes (3 of 3) |
Yes (3 of 3) |
Yes (3 of 3) |
Yes (4 of 10) |
Yes (4 of 10) |
Yes (7 of 10) |
Yes (2 of 16) |
Yes (3 of 16) |
Yes (3 of 3) |
Yes (3 of 3) |
|
| Aves | Yes (68 of 80) |
Yes (68 of 75) |
Yes (69 of 80) |
Yes (75 of 80) |
Yes (58 of 68) |
Yes (58 of 68) |
Yes (21 of 67) |
Yes (70 of 70) |
Yes (63 of 70) |
Yes (70 of 70) |
Yes (67 of 108) |
Yes (68 of 137) |
Yes (66 of 137) |
Yes (66 of 68) |
|
| Ceratodontimorpha | No (0 of 2) |
No (0 of 2) |
No (0 of 2) |
No (0 of 2) |
- | - | - | Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
No (0 of 1) |
No (0 of 4) |
- | - | |
| Chondrichthyes | Yes (3 of 6) |
Yes (3 of 6) |
Yes (3 of 7) |
Yes (5 of 7) |
Yes (2 of 2) |
Yes (2 of 3) |
Yes (2 of 4) |
Yes (3 of 3) |
Yes (3 of 3) |
Yes (3 of 3) |
Yes (1 of 9) |
Yes (1 of 11) |
Yes (2 of 2) |
Yes (2 of 2) |
|
| Coelacanthiformes | Yes (1 of 2) |
Yes (1 of 2) |
Yes (1 of 2) |
Yes (1 of 1) |
No (0 of 1) |
No (0 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
|
| Crocodylia | Yes (4 of 6) |
Yes (4 of 5) |
Yes (4 of 6) |
Yes (4 of 5) |
Yes (4 of 4) |
Yes (4 of 4) |
Yes (3 of 4) |
Yes (4 of 4) |
Yes (4 of 4) |
Yes (4 of 4) |
Yes (4 of 4) |
Yes (4 of 4) |
Yes (3 of 3) |
Yes (4 of 4) |
|
| Lepidosauria | Yes (8 of 15) |
Yes (8 of 13) |
Yes (8 of 15) |
Yes (9 of 11) |
Yes (6 of 6) |
Yes (6 of 7) |
Yes (4 of 6) |
Yes (9 of 9) |
Yes (7 of 9) |
Yes (8 of 9) |
Yes (6 of 341) |
Yes (7 of 422) |
Yes (6 of 6) |
Yes (6 of 6) |
|
| Myxiniformes |
cYes (0 of 1) |
cYes (0 of 2) |
No (0 of 1) |
Yes (1 of 2) |
No (0 of 1) |
No (0 of 1) |
- | - | - | - | No (0 of 1) |
No (0 of 1) |
- | - | |
| Petromyzontiformes | Yes (1 of 4) |
Yes (1 of 4) |
Yes (1 of 4) |
Yes (4 of 4) |
- | No (0 of 1) |
- | Yes (1 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
No (0 of 5) |
No (0 of 5) |
- | - | |
| Testudines | Yes (3 of 9) |
Yes (3 of 5) |
Yes (3 of 9) |
Yes (3 of 4) |
Yes (3 of 3) |
Yes (3 of 3) |
Yes (2 of 3) |
Yes (3 of 3) |
Yes (3 of 3) |
Yes (3 of 3) |
Yes (3 of 4) |
Yes (3 of 26) |
Yes (3 of 3) |
Yes (3 of 3) |
|
| aInvertebrates | Ascidiacea | No (0 of 5) |
No (0 of 5) |
Yes (1 of 5) |
Yes (2 of 5) |
No (0 of 2) |
Yes (1 of 2) |
No (0 of 1) |
Yes (2 of 2) |
No (0 of 2) |
Yes (2 of 2) |
No (0 of 1) |
No (0 of 1) |
- | - |
| Asteroidea | No (0 of 1) |
No (0 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 2) |
No (0 of 3) |
Yes (1 of 1) |
Yes (1 of 1) |
|
| Bivalvia | No (0 of 10) |
No (0 of 10) |
Yes (2 of 10) |
Yes (3 of 10) |
No (0 of 6) |
No (0 of 8) |
No (0 of 4) |
Yes (4 of 5) |
No (0 of 5) |
Yes (4 of 5) |
No (0 of 4) |
No (0 of 5) |
- | - | |
| Branchiostomidae | No (0 of 4) |
No (0 of 4) |
Yes (3 of 4) |
Yes (4 of 4) |
No (0 of 3) |
No (0 of 4) |
No (0 of 2) |
Yes (2 of 2) |
No (0 of 2) |
Yes (2 of 2) |
No (0 of 4) |
No (0 of 4) |
Yes (2 of 2) |
Yes (2 of 2) |
|
| Dictyosteliida | - | - | - | - | - | - | - | No (0 of 6) |
No (0 of 6) |
Yes (1 of 5) |
- | - | - | - | |
| Enteropneusta | No (0 of 1) |
No (0 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
- | Yes (1 of 1) |
|
| Gastropoda | No (0 of 9) |
No (0 of 9) |
Yes (2 of 2) |
Yes (8 of 9) |
No (0 of 3) |
No (0 of 4) |
No (0 of 3) |
No (0 of 2) |
No (0 of 2) |
No (0 of 2) |
No (0 of 5) |
No (0 of 5) |
- | - | |
| Lingulata | No (0 of 1) |
No (0 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
Yes (1 of 1) |
No (0 of 1) |
Yes (1 of 1) |
cYes (0 of 1) |
cYes (0 of 1) |
Yes (1 of 1) |
Yes (1 of 1) |
|
| Polychaeta | No (0 of 2) |
No (0 of 1) |
Yes (1 of 2) |
Yes (1 of 1) |
No (0 of 5) |
No (0 of 10) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 1) |
No (0 of 2) |
No (0 of 2) |
Yes (1 of 1) |
Yes (1 of 1) |
|
Invertebrate taxon represent a subset of those found to be conserved for THRβ, TPO, NIS, DIO1, DIO3 and IYD. Those invertebrate taxa shown represent common taxonomic groups (Full taxon list in Supplemental Data, Workbook S2).
The SeqAPASS cut-offs define those species with conserved proteins found above the cut-off and those species less likely to be conserved below the cutoff (Details in Supplemental Data, Table S3)
Predicted as conserved “Yes,” because species in Myxiniformes for THRα and in Lingulata for TSHR are ortholog candidates, although the taxonomic group is below the cutoff.
Table cells indicate whether the protein is conserved compared to the human query protein, “Yes,” or less likely to be conserved, “No.” In parentheses are the number of species found above the SeqAPASS cut-off of the total number of species aligning with the mammalian target for that taxonomic group. Dashed line indicates that no protein sequences aligned with the query sequence for that taxonomic group.
Abbreviations: THRα = human thyroid hormone receptor alpha (P10827.1; domain cd06935); THRβ = human thyroid hormone receptor beta (P10828.2; domain cd06935); TPO = rat thyroid peroxidase (P14650.1; domain cd09825); NIS = human sodium/iodide cotransporter (Q92911.1); DIO1 = human type I iodothyronine deiodinase (P49895.3); DIO2 = human type II iodothyronine deiodinase (Q92813.4); DIO3 = human type III iodothyronine deiodinase (P55073.4); TSHR = human thyroid-stimulating hormone receptor (P16473.2; domain cd15964); and IYD = iodotyrosine deiodinase 1 (Q6PHW0.2; domain cd02144).
Norway rat and pig TPO sequences and their peroxidase functional domains, which contain the heme binding site and putative substrate binding site, were conserved across vertebrates, as well as Ascidiacea (sea squirts, tunicate) taxa (Table 3; Supplemental Data, Figure S5e–h). Conversely, probable orthologs of human NIS were restricted to vertebrates only (Table 3; Supplemental Data, Figure S5i).
The deiodinase enzymes activate and deactivate thyroid hormones. The SeqAPASS results for DIO1, DIO2, and DIO3 demonstrate conservation of these enzymes across vertebrate species, including Ceratodontimorpha and Coelacanthiformes, and Petromyzontiformes (Table 3; Supplemental Data, Figure S5j–l). DIO1and DIO3 are also found in invertebrate species, whereas DIO2 iodothyronine deiodinase is not. DIO3 has the broadest conservation across invertebrates and is even found in Dictyosteliida (slime molds) taxa.
Through binding of thyroid stimulating hormone, the TSHR signaling modulates thyroid function by regulating genes coding for enzymes involved in iodide metabolism, such as TPO and NIS. The human TSHR is well conserved across vertebrates with the exception of Ceratodontimorpha, Myxiniformes and Petromyzontiformes (Table 3; Supplemental Data, Figure S5m). Further, SeqAPASS results yielded the same conclusion upon evaluation of the TSHR functional domain containing the putative peptide ligand binding pocket (Table 3; Supplemental Data, Figure S5n).
The final protein evaluated using SeqAPASS was the human IYD, involved in the recycling of iodide. This protein had ortholog candidates across vertebrates and invertebrates, displaying conservation in all but one taxon, Trichomonadida (anaerobic protists), based on the primary amino acid sequence that aligned with human (Table 3; Supplemental Data, Figure S5o). However, upon evaluation of the functional domain (positions 93–285) which contains the putative dimer interface, additional taxa were identified that were not conserved compared to the human iodotyrosine deiodinase (Supplemental Data, Figure S5p).
Overall, the protein targets associated with ToxCast™ assays intended to screen for thyroid axis disruption were conserved across all vertebrate species, in some cases identifying conservation in more ancient fish species such as hagfish, Coelacanth, lamprey, and lungfish (Supplemental Data, Workbook S2). Therefore, it would be anticipated that screening results identifying chemical disruption via HTS assays for the human thyroid axis targets could be reasonably extrapolated across vertebrate species. Additionally, THRβ, TPO, IYD, DIO1, and DIO3 are present in certain invertebrates, suggesting perhaps applicability of these assays to an even broader array of taxa. However, additional research is needed to understand the actual role of these proteins in invertebrate species.
The overall objectives of this work were to evaluate mammalian-based HTS assay targets for protein similarity using the SeqAPASS tool, make the resultant data available for other researchers and regulators (via SeqAPASS and the CompTox Chemistry Dashboard), and demonstrate the utility of the analysis as applied to a current regulatory challenge, screening of endocrine-active chemicals. This study illustrates that HTS targets relevant to EDSP and OECD are likely to be broadly applicable across most vertebrate taxa and some targets may be applicable to certain invertebrates. Ultimately, application of the SeqAPASS tool can lay the foundation for future work integrating in vitro and in silico approaches to support more rapid evaluations of chemical safety for the protection of human health and wildlife.
Supplementary Material
Acknowledgements –
We thank Dr. M. Hornung for providing thoughtful review comments on an earlier version of the paper. This manuscript has been reviewed in accordance with the requirements of the US EPA Office of Research and Development; however, the recommendations made herein do not represent US EPA policy. Mention of products or trade names does not indicate endorsement by the US EPA.
Footnotes
Supporting Information
Supplemental data are provided that includes detailed information on the SeqAPASS analyses for all ToxCast targets and data visualization to accompany the SeqAPASS results.
References
- (1).National Research Council. (2007). Toxicity Testing in the 21st Century: A Vision and Strategy. The National Academies Press, Washington, DC. [Google Scholar]
- (2).Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia M, Huang R, Rotroff DM, Filer DL and Houck KA (2015). Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol Sci 148(1), 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Browne P, Noyes PD, Casey WM and Dix DJ (2017). Application of Adverse Outcome Pathways to US EPA’s Endocrine Disruptor Screening Program. Environmental health Perspectives, 96001, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Pinto CL, Markey K, Dix D, and Browne P (2018). Identification of candidate reference chemicals for in vitro steroidogenesis assays. Toxicology in Vitro 47, 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).US Environmental Protection Agency. Endocrine disruptor screening program. [cited 2018 February 15]. Available from: http://www.epa.gov/endo/
- (6).Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, Reif D, Richard A, Rotroff D, Sipes N and Dix D (2012). Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chemical research in toxicology 25(7), 1287–1302. [DOI] [PubMed] [Google Scholar]
- (7).Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF, Knudsen TB, Kancherla J, Mansouri K, Patlewicz G, Williams AJ, Little SB, Crofton KM and Thomas RS (2016). ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chemical research in toxicology 29(8), 1225–1251. [DOI] [PubMed] [Google Scholar]
- (8).Tice RR, Austin CP, Kavlock RJ and Bucher JR, (2013). Improving the human hazard characterization of chemicals: a Tox21 update. Environmental health perspectives, 121 (7), 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hornung MW, Korte JJ, Olker JH, Denny JS, Knutsen C, Hartig PC, Cardon MC, and Degitz SJ (2017). Screening the Toxcast Phase 1 Chemical Library For Inhibition of Deiodinase Type 1 Activity. Toxicol Sci 162 (2), 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Truong L, Reif DM, St Mary L, Geier MC, Truong HD, and Tanguay RL (2014). Multidimensional in vivo hazard assessment using zebrafish. Toxicol Sci 137(1), 212–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).LaLone CA, Villeneuve DL, Lyons D, Helgen HW, Robinson SL, Swintek JA, Saari TW and Ankley GT (2016). Editor’s Highlight: Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS): A Web-Based Tool for Addressing the Challenges of Cross-Species Extrapolation of Chemical Toxicity. Toxicol Sci 153 (2), 228–45. [DOI] [PubMed] [Google Scholar]
- (12).Gunnarsson L, Jauhiainen A, Kristiansson E, Nerman O, and Larsson DGJ (2008). Evolutionary Conservation of Human Drug Targets in Organisms use dfor Environmental Risk Assessments. Environmental Science & Technology 42(15), 5807–5813. [DOI] [PubMed] [Google Scholar]
- (13).Ankley GT, LaLone CA, Gray LE, Villeneuve DL and Hornung MW (2016). Evaluation of the scientific underpinnings for identifying estrogenic chemicals in nonmammalian taxa using mammalian test systems. Environ Toxicol Chem 35 (11), 2806–2816. [DOI] [PubMed] [Google Scholar]
- (14).Kohno S, Bernhard MC, Katsu Y, Zhu J, Bryan TA, Doheny BM, Iguchi T and Guillette LJ Jr (2015). Estrogen receptor 1 (ESR1; ERα), not ESR2 (ERβ), modulates estrogen-induced sex reversal in the American alligator, a species with temperature-dependent sex determination. Endocrinology, 156(5), 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tohyama S, Miyagawa S, Lange A, Ogino Y, Mizutani T, Ihara M, Tanaka H, Tatarazako N, Kobayashi T, Tyler CR and Iguchi T (2016). Evolution of estrogen receptors in ray-finned fish and their comparative responses to estrogenic substances. The Journal of steroid biochemistry and molecular biology, 158, 189–197. [DOI] [PubMed] [Google Scholar]
- (16).Ogino Y, Tohyama S, Kohno S, Toyota K, Yamada G, Yatsu R, Kobayashi T, Tatarazako N, Sato T, Matsubara H, Lange A, Tyler CR, Katsu Y, Iguchi T, and Miyagawa S (2018). Functional distinctions associated with the diversity of sex steroid hormone receptors ESR and AR. The Journal of steroid biochemistry and molecular biology. In Press. 10.1016/j.jsbmb.2018.06.002 [DOI] [PubMed] [Google Scholar]
- (17).Doering J, Lee S, Kristiansen K, Evenseth L, Barron M, Sylte I and LaLone C (2018) In silico site-directed mutagenesis informs species-specific predictions of chemical susceptibility derived from the Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) tool. Toxicol Sci 10.1093/toxsci/kfy186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hallinger DR, Murr AS, Buckalew AR, Simmons SO, Stoker TE and Laws SC (2017). Development of a screening approach to detect thyroid disrupting chemicals that inhibit the human sodium iodide symporter (NIS). Toxicology in Vitro. 40, 66–78. [DOI] [PubMed] [Google Scholar]
- (19).Paul KB, Hedge JM, Macherla C, Filer DL, Burgess E, Simmons SO, Crofton KM and Hornung MW (2013). Cross-species analysis of thyroperoxidase inhibition by xenobiotics demonstrates conservation of response between pig and rat. Toxicology. 312, 97–107. [DOI] [PubMed] [Google Scholar]
- (20).Titus S, Neumann S, Zheng W, Southall N, Michael S, Klumpp C, Yasgar A, Shinn P, Thomas CJ, Inglese J and Gershengorn MC (2008). Quantitative high-throughput screening using a live-cell cAMP assay identifies small-molecule agonists of the TSH receptor. Journal of biomolecular screening. 13(2), 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Freitas J, Miller N, Mengeling BJ, Xia M, Huang R, Houck K, Rietjens IM, Furlow JD and Murk AJ (2014). Identification of thyroid hormone receptor active compounds using a quantitative high-throughput screening platform. Current chemical genomics and translational medicine. 8, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).LaLone CA (2018). Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) Version 3.0 User Guide. https://cfpub.epa.gov/si/si_public_record_report.cfm?dirEntryId=339888. US EPA Office of Research and Development, Washington, DC, EPA/600/R-18/062. [Google Scholar]
- (23).Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C and Bryant SH (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS and Richard AM, 2017. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. Journal of Cheminformatics. 9(1), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).LaLone CA, Villeneuve DL, Burgoon LD, Russom CL, Helgen HW, Berninger JP, Tietge JE, Severson MN, Cavallin JE and Ankley GT (2013). Molecular target sequence similarity as a basis for species extrapolation to assess the ecological risk of chemicals with known modes of action. Aquatic toxicology, 144, 141–154. [DOI] [PubMed] [Google Scholar]
- (26).Marinissen MJ and Gutkind JS (2001). G-protein-coupled receptors and signaling networks: emerging paradigms. Trends in pharmacological sciences, 22(7), 368–376. [DOI] [PubMed] [Google Scholar]
- (27).Massague J (1990). The transforming growth factor-beta family. Annual review of cell biology. 6 (1), 597–641. [DOI] [PubMed] [Google Scholar]
- (28).Browne P, Judson RS, Casey WM, Kleinstreuer NC, and Thomas RS (2015). Screening chemicals for estrogen receptor bioactivity using a computational model. Environmental science & technology. 49 (14), 8804–8814. [DOI] [PubMed] [Google Scholar]
- (29).Kleinstreuer NC, Ceger P, Watt ED, Martin M, Houck K, Browne P, Thomas RS, Casey WM, Dix DJ, Allen D and Sakamuru S, (2016). Development and validation of a computational model for androgen receptor activity. Chemical research in toxicology. 30 (4), 946–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Colombe L, Fostier A, Bury N, Pakdel F, Guiguen Y (2000). Steroids. 65, 319–328. [DOI] [PubMed] [Google Scholar]
- (31).Escriva H, Safi R, Hanni C, Langloi MC, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V (1997). Proc. Natl. Acad. Sci. USA. 94, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Colbert EH, Morales M (1991). Evolution of the Vertebrates: A history of the Backboned Animals Through Time (Wiley-Liss, New York: ), 4th Ed. [Google Scholar]
- (33).Thornton JW (2001). Proc. Natl. Acad. Sci. USA. 98, 5671–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Karmaus AL, Toole CM, Filer DL, Lewis KC and Martin MT (2016). High-throughput screening of chemical effects on steroidogenesis using H295R human adrenocortical carcinoma cells. Toxicol Sci. 150 (2), 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Anderson WG (2012). The endocrinology of 1α-hydroxycorticosterone in elasmobranch fish: a review. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 162(2), 73–80. [DOI] [PubMed] [Google Scholar]
- (36).Nunez S and Trant JM (1999). Regulation of interrenal gland steroidogenesis in the Atlantic stingray (Dasyatis sabina). Journal of Experimental Zoology Part A: Ecological Genetics and Physiology. 284 (5), 517–525. [PubMed] [Google Scholar]
- (37).Nelson DR (2017). Cytochrome P450 diversity in the tree of life. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 1866 (1), 141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Baker ME, Nelson DR, and Studer RA (2015). Origin of the response to adrenal and sex steroids: Roles of promiscuity and co-evolution of enzymes and steroid receptors. The Journal of steroid biochemistry and molecular biology, 151, 12–24. [DOI] [PubMed] [Google Scholar]
- (39).Close DA, Yun SS, McCormick SD, Wildbill AJ, & Li W (2010). 11-Deoxycortisol is a corticosteroid hormone in the lamprey. Proceedings of the National Academy of Sciences, 107 (31), 13942–13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Miller WL, and Auchus RJ (2011). The Molecular Biology, Biochemistry, and Physiology of Human Steroidogenesis and Its Disorders. Endocrine Reviews, 32(1), 81–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gauthier K, Chassande O, Plateroti M, Roux JP, Legrand C, Pain B, Rousset B, Weiss R, Trouillas J and Samarut J (1999). Different functions for the thyroid hormone receptors TRα and TRβ in the control of thyroid hormone production and post‐natal development. The EMBO journal. 18(3), pp.623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bassett JD, Harvey CB and Williams GR (2003). Mechanisms of thyroid hormone receptor-specific nuclear and extra nuclear actions. Molecular and cellular endocrinology. 213(1), 1–11. [DOI] [PubMed] [Google Scholar]
- (43).Ruf J and Carayon P (2006). Structural and functional aspects of thyroid peroxidase. Archives of biochemistry and biophysics. 445 (2), 269–277. [DOI] [PubMed] [Google Scholar]
- (44).Holzer G and Laudet V (2017). New Insights into Vertebrate Thyroid Hormone Receptor Evolution. Nuclear Receptor Research, 4, 1–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


