Abstract
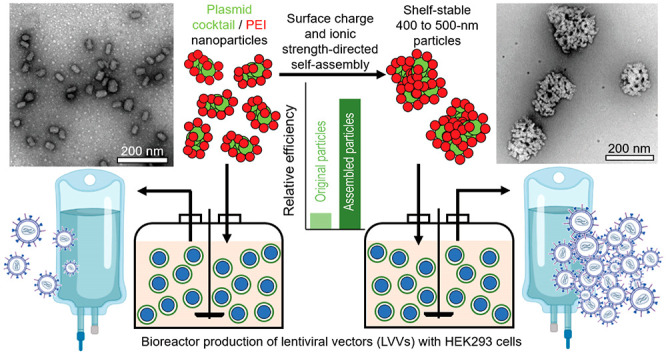
Polyelectrolyte complex particles assembled from plasmid DNA (pDNA) and poly(ethylenimine) (PEI) have been widely used to produce lentiviral vectors (LVVs) for gene therapy. The current batch-mode preparation for pDNA/PEI particles presents limited reproducibility in large-scale LVV manufacturing processes, leading to challenges in tightly controlling particle stability, transfection outcomes, and LVV production yield. Here we identified the size of pDNA/PEI particles as a key determinant for a high transfection efficiency with an optimal size of 400–500 nm, due to a cellular-uptake-related mechanism. We developed a kinetics-based approach to assemble size-controlled and shelf-stable particles using preassembled nanoparticles as building blocks and demonstrated production scalability on a scale of at least 100 mL. The preservation of colloidal stability and transfection efficiency was benchmarked against particles generated using an industry standard protocol. This particle manufacturing method effectively streamlines the viral manufacturing process and improves the production quality and consistency.
Keywords: plasmid DNA, poly(ethylenimine), transfection, lentiviral vector production, particle size, kinetic growth
Introduction
Gene therapy has become an increasingly valuable modality for treating congenital and acquired conditions and for prophylactic and treatment vaccines. Many of these therapies include the use of vectorized viruses based on lentivirus (LVVs)1 and adeno-associated virus (AAVs),2 such as ABECMA approved by the U.S. FDA for engineering CAR-T cells and ZYNTEGLO approved by the E.U. EMA for modifying hematopoietic stem cells. One of the most common methods to produce LVVs is transient transfection and vector assembly in production cell lines such as HEK293 cells.3 Benchmark transfection vehicles include calcium phosphate,4 lipofectamine,5 and poly(ethylenimine) (PEI).6 In a typical transfection procedure using PEI, solutions of pDNAs and PEI are mixed to form a polyelectrolyte complex particle suspension and incubated in a batch mode for a few minutes to 1 h, before being added to the cell culture. The pDNA/PEI particles facilitate cellular uptake,7,8 endosomal escape,9 nuclear transport,10 and transgene expression of pDNAs encoding LVV components. This widely adopted production method of pDNA/PEI particles delivers high levels of transfection efficiency and meets the critical criteria for LVV production. However, the batch-mode mixing process, conducted immediately before transfection, lacks adequate engineering control to ensure uniform assembly during the complexation process and is prone to operator-dependent variability. The objective of this study is to develop an engineering approach to produce shelf-stable pDNA/PEI particles in a highly scalable and consistent fashion to ensure high transfection efficiency with ease-of-use features. Such a platform is essential to ensure the consistent yield and quality of LVVs to deliver the intended therapeutic efficacy.11
Despite the widespread use of pDNA/PEI nanoparticles for transfecting various types of cells, the relationship between particle size and transfection efficiency has been poorly understood; particularly, transfection activities for particles of sizes beyond 200 nm have been scarcely reported.12−15 Here we provide the first direct correlation of the transfection efficiency of pDNA/PEI particles within a wide size range of 60–1000 nm, reporting an optimal particle size of 400–500 nm in both adherent and suspension cultures.
Existing methods for the controlled assembly of pDNA/PEI nanoparticles target a size range of 30–200 nm,16,17 which are suboptimal for in vitro transfection in LVV production cell lines. There has been no specific effort to date for controlling particle size and uniformity in this range while adequate particle stability is maintained. Conventional batch mixing methods (e.g., pipet mixing or dropwise addition) offer limited control of assembly kinetics, resulting in particles with poor uniformity and a high degree of instability. Here we devised a scalable method to produce pDNA/PEI particles at any size between 60 and 1000 nm by a bottom-up assembly strategy through reversible tuning of the particle surface charge. Using this particle series, we revealed that the rate-limiting step in the intracellular delivery process is size-dependent cellular uptake. We also demonstrated the superior shelf stability of the ready-to-use particles with preservation of physical properties and transfection efficiencies in bench-scale bioreactor LVV production systems.
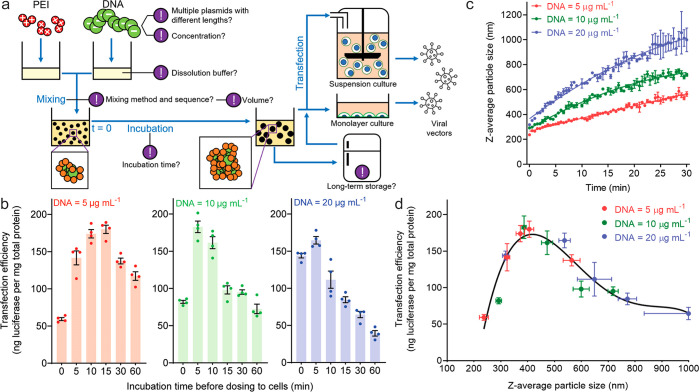
Particle-Size-Dependent Transfection Efficiency
In a typical transfection process for LVV production, multiple pDNAs encoding different viral components are used. In this study, we used a 3-pDNA mixture with weight ratios of 10% (4.4 kb, noncoding), 45% (6.8 kb, gWiz-Luc luciferase reporter), and 45% (9.6 kb, noncoding) in Opti-MEM medium to mimic an LVV plasmid cocktail. On a 200 μL scale preparation, we used a 10 s vortex as the mixing method after pipetting 100 μL of PEI solution into 100 μL of pDNA solution (5, 10, or 20 μg/mL) at a nitrogen to phosphate (N/P) ratio of 5.5, followed by incubation at room temperature for 10 s to 60 min before transfection tests in HEK293T cells (Figure 1a). When the pDNA dose remained constant, i.e., 0.1 μg per 104 cells at 1 μg/mL, the transfection efficiency showed a bell-shaped relationship with incubation time, and the peak occurred at different incubation times for different DNA concentrations in particle preparation (Figure 1b). We next monitored the particle size as a function of incubation time upon mixing of pDNA and PEI solutions by dynamic light scattering (DLS) and found that the size increased in a highly predictable manner during incubation (Figure 1c). When the transfection efficiency was plotted against the z-average particle size as shown in in Figure 1d, a strong correlation emerged with a curve fitting all data points collected under different pDNA concentrations and different incubation times during particle preparation. This demonstrated the dominant effect of pDNA/PEI particle size on transfection efficiency with an optimal size of 400–500 nm for the highest transfection efficiency.
Figure 1.
Size of pDNA/PEI particles dictates their transfection efficiency in LVV production cells. (a) Schematic of preparation of pDNA/PEI particles and transfection process for production of LVVs. Each exclamation mark indicates a critical process control parameter influencing transfection outcomes. (b) Transfection efficiencies, characterized as transgene expression levels of the luciferase reporter, in a monolayer culture of HEK293T cells as a function of pDNA concentration at the mixing step and incubation time (10 s to 60 min) before dosage. For the group of mixing at a DNA concentration of 5, 10, or 20 μg/mL, the particles were diluted 5, 10, or 20 times, respectively, to 1 μg/mL to dose cells. (c) Growth in the average size (z-average diameter given by dynamic light scattering, DLS) of pDNA/PEI particles following mixing of pDNA and PEI solutions in Opti-MEM. The growth kinetics is dependent on the concentration of pDNA. The error bars were derived from three independent experiments, demonstrating reproducibility and predictability under the experimental conditions used. (d) Direct correlation between transfection efficiency and the z-average particle size based on data points from all experiments from (b) and (c) with varying pDNA concentrations and incubation times.
Production of Sub-Micrometer pDNA/PEI Particles with Controlled Size
The above finding motivated us to develop a method for producing shelf-stable pDNA/PEI particles with a controlled size of 400–500 nm. Discrete size control for pDNA/PEI particles was achieved in the sub-100 nm range using the flash nanocomplexation (FNC) technique,16,17 but controlling the size in the sub-micrometer range has been particularly challenging. Previous work from our laboratory17 and the Kataoka group18 demonstrated that one pDNA/polycation nanoparticle that consists of a single pDNA molecule is only 20–50 nm in size, indicating that a 400–500 nm particle will need nearly thousands of copies of pDNA to be constructed (Figure S1). During the particle assembly process, negatively charged pDNA collapses into a condensed state upon charge neutralization with positively charged PEI.19 The assembly kinetics is extremely fast with a time scale of tens of milliseconds,17 whereas the diffusion rates of pDNA and the complexes are substantially slower.20 It may only be possible to generate a 400–500 nm size in a single complexation step using a DNA concentration much higher than 5 mg/mL by our estimation,17 which would be difficult to handle or scale up due to high solution viscosity.
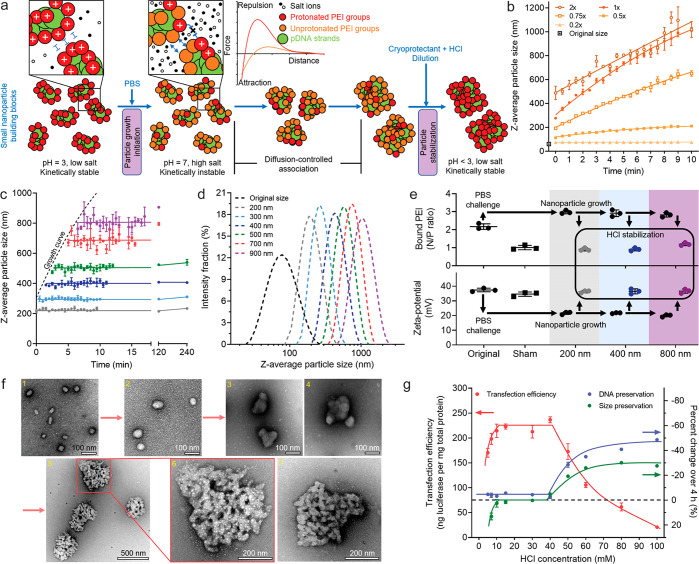
To address this challenge, we developed a bottom-up assembly strategy based on the surface charge tunability and controllable assembly kinetics of pDNA/PEI particles (Figure 2a). First, we prepared uniform small nanoparticles under low-salt (conductivity ∼0.4 mS/cm) and low-pH (∼3) conditions. Over 80% of the secondary nitrogen groups in PEI are protonated (i.e., positively charged) at pH 3, rendering a high surface density of positive charge,21 such that the individual nanoparticles are sufficiently stable against aggregation. When the pH is switched to 7, the nanoparticle surface becomes sufficiently deprotonated. Together with the shortening of the Debye length associated with the residual surface charges by salt-induced charge screening, the medium condition change triggers particle association and size growth. It is important to note that the ionic strength needs to be controlled at a level that does not induce dissociation of the pDNA/PEI complexes.22 The particle size growth is primarily driven by van der Waals interactions, with the rate being controlled by particle concentration and ionic strength of the medium. It can be effectively quenched by reversing the pH to 3 to reprotonate the particle surfaces and by dilution to reduce the ionic strength, thus re-establishing the long-range Debye screening effect.
Figure 2.
Process for production of size-controlled pDNA/PEI particles in the range of 60–1000 nm through controlling the assembly kinetics and surface charge. (a) Schematic of the stepwise kinetic growth and quenching. (b) Predicable size growth induced under different concentrations of PBS. (c) The particle size growth was arrested by dilution with 20 mM HCl in 19% (w/w) trehalose solution at different time points along the growth curve in 1× PBS. (d) The z-average diameter distributions measured by DLS of a series of stabilized particles with distinct sizes. (e) The ζ potential, and bound PEI content (measured by N/P ratio) changed along with the growth and stabilization steps. The particles in the sham control were treated with premixed 1× PBS and 20 mM HCl solutions, and the size stayed unchanged (66 nm) after the treatment. (f) TEM images of the particles obtained under the conditions of (1) original 66 nm nanoparticles as the building blocks, (2) stabilized particles with an average size of 120 nm, (3, 4) stabilized particles with an average size of 180 nm, (5, 6) stabilized 400 nm particles with an enlarged view of one of the particles, and (7) another enlarged 400 nm particle with less salt precipitation (white speckles) in the negatively stained region. (g) Effect of HCl concentration used in the quenching step on size stability, DNA protection, and transfection efficiency of the 400 nm particles. Note that the percentage axis is inverted to spread data points, showing that a high HCl concentration resulted in size shrinkage and loss of pDNA. In (b) and (c) the error bars were derived from three independent experiments, demonstrating the predictability and reproducibility of the process. In (g) the error bars were derived from three replicates within a single experiment.
The building block pDNA/PEI nanoparticles were prepared using the FNC technique16,17 in a confined impinging jet (CIJ) mixer.23,24 With an input pDNA concentration of 100 μg/mL, the nanoparticles had an average size of 66.0 ± 1.0 nm as measured by DLS (Figure 2b and Figure S2a). The nanoparticle suspension was challenged by mixing it with an equal volume of PBS, which initiated size growth with a rate dependent on the PBS concentration (Figure 2b). The buffering component of PBS was important to confer the pH change and maintain particle uniformity, while the salt components determined the growth kinetics (Figure S3). Assembling with 1× PBS and quenching the growth by mixing the particle suspension with an equal volume of 20 mM HCl in 19% w/w trehalose (a cryoprotectant) at different time points along its growth curve successfully stabilized the average particle sizes at 200, 300, 400, 500, 700, and 900 nm (Figure 2c) with a high degree of uniformity (Figure 2d; polydispersity index values are reported in Figure S3b). All particles with average sizes of less than 500 nm were stable at ambient temperature for at least 4 h in the final solution (Figure 2c).
The proposed size control mechanism was verified by ζ-potential measurements through phase analysis light scattering (PALS) and PEI composition assessment25 of the growing and stabilized particles (Figure 2e). Upon challenge by 1× PBS, the ζ potential dropped from +37 to +20 mV, which was sufficient to overcome the potential energy barrier at the appropriate ionic strength in the medium. The potential recovered after the addition of a stabilization medium. Transmission electron microscopy (TEM) analysis confirmed DLS measurements showing the nature of association of small individual nanoparticles (Figures 2f-2, and S2). The stabilized 400 nm particles appeared as uniform agglomerate constructs (Figures 2f-, 6, 7, and Figure S2). The HCl concentration used for the growth arrest is close to that required to fully protonate the buffering salts added during the growth phase. Lower or higher concentrations resulted in ineffective stabilization or particle shrinkage and DNA degradation, respectively (Figure 2g).
Transfection Efficiencies of Size-Controlled pDNA/PEI Particles
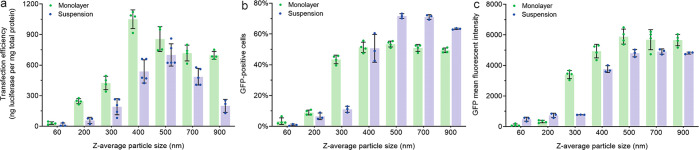
The concentration of stabilized particles containing luciferase or GFP reporter pDNAs was diluted to 1 μg of pDNA/mL to effectively limit further size growth under the transfection conditions in the physiological medium (Figure S4). The luciferase activity analysis verified the optimal size of 400 nm in a monolayer culture, as observed in Figure 1d, and indicated an optimal size of 500 nm in a suspension culture (Figure 3a). The GFP expression analysis revealed a substantial efficiency jump for particle sizes between 200 and 300 nm in the monolayer culture and between 300 and 400 nm in the suspension culture (Figure 3b,c). The trends in luciferase and GFP expression levels agreed well for particles with average sizes from 60 to 500 nm, but differed for particles over 500 nm, presumably due to different expression kinetics, transgene product stability, and assessment methods of the two reporters. Clearly, particles smaller than 200 nm were far less effective in transfection even though they may eventually grow in the transfection medium to a larger size within 4 h (Figure S4). This trend was also consistently observed in the suspension culture, even though the particles were not removed from the culture during the entire transfection period of 48 h. These results demonstrated the importance of controlling the particle size before dosing to the cells.
Figure 3.
Transfection efficiencies of stabilized particles with controlled sizes ranging from 60 to 1000 nm. (a) Efficiency of transgene expression of luciferase as a reporter. (b, c) The efficiency of transgene expression of GFP is shown in (b) for the percentage of GFP-positive cells and (c) for the mean fluorescent intensity in the population of GFP-positive cells. For a monolayer culture of HEK293T cells, the cells were harvested and lysed at 24 h post-transfection, and the error bars present the standard deviation from four replicates in a single experiment; for the suspension culture of HEK293F cells, the cells were harvested and lysed at 48 h post-transfection, and the error bars present the standard deviation of three or four independent experiments (each was conducted in a single well of a 12-well plate).
Intracellular Trafficking of pDNA/PEI Particles
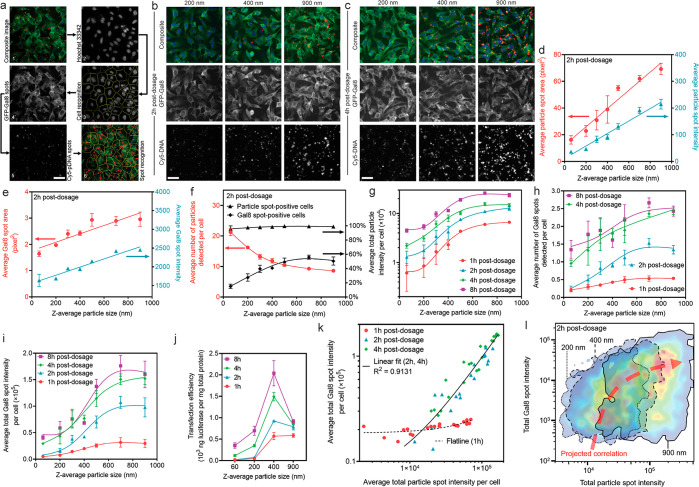
To assess cellular uptake7,8,26,27 and endosomal escape,9 which are two major intracellular transport barriers for cell transfection,28 we labeled pDNAs with Cy5 dye29 and used a genetically modified B16F10 cell line that expresses galectin-8 (Gal8) fused with GFP as the assessment tools. The Gal8 protein that distributed throughout the cytosol binds to the cell membrane glycans exposed upon damage of endosomal vesicles, which subsequently aggregate and form GFP spots (Figure 4a).30 In this study, the Cy5 and GFP-Gal8 spots were quantitatively analyzed by Cellomics high-content analysis (HCA) on fixed cells after treatment with particles for 1, 2, 4, or 8 h (Figures S7 and S9). The analysis showed a clear increasing trend in the average particle spot area and intensity as the particle size increased (Figure 4b–d and Figures S5 and S6). Larger particles induced Gal8 spots with higher average area and intensity (Figure 4e), indicating a higher degree of endosomal membrane disruption and formation of larger endocytic vesicles prior to escape events. The drop in particle spot number per cell as the particle size increased reflected a decreasing particle number concentration because of an increasing pDNA payload per particle (Figure 4f). Nonetheless, the fewer larger particles yielded much higher efficiency in endosomal escape, presented as a higher percentage of Gal8 spot-positive cells (Figure 4f). There was a sharp increase in the total uptake amount measured by average total particle intensity per cell and by the amount of intracellular pDNA per cell (Figure S8) at all time points as the average particle size increased from 60 to 500 nm, though a gradual increase was observed as the average particle size increased further to 900 nm (Figure 4g). In addition to reporting the overall endosomal escape indicator of average Gal8 spot number per cell31,32 (Figure 4h), the average total Gal8 spot intensity per cell gave a better assessment of the overall endosomal escape level (Figure 4i), considering the differences observed in Gal8 spot area and intensity (Figure 4e). In addition, the kinetics of uptake and endosomal escape matched the kinetics of luciferase expression following particle incubation with different durations (Figure 4j). The reasons for a drop in transfection efficiency mediated by particles with an average size larger than 400 nm (HEK293T) or 500 nm (HEK293F), even though they showed higher degrees of particle uptake and endosomal escape, remain to be elucidated in future studies, although it was not due to a change in cellular metabolism activities (Figure S10).
Figure 4.
Quantitative Cellomics high-content analysis (HCA) of cellular uptake and endosomal escape by particles with different sizes. (a) The image analysis modality to analyze fixed cells directly in the tissue culture plates. Representative images are shown in (b) at 2 h and (c) at 4 h after incubation with particles of different sizes. Quantitative results are presented in terms of (d) particle spot characteristics (area and intensity), directly suggesting successful size control during particle–cell interactions. (e) Gal8 spot characteristics (area and intensity) indicating the formation of larger endosomal vesicles by larger particles. (f) Frequency of detected particles and Gal8 spots in cells at 2 h. (g) Average total particle intensity per cell at all time points as a representative measure of total particle uptake quantity. (h) Average number of Gal8 spots per cell at all time points as an indication of endosomal escape level, serving as a predictive index for transfection efficiency according to previous reports using this assay. (i) Quantitative measure of overall endosomal escape degree, i.e., average total Gal8 spot intensity per cell, due to different Gal8 spot characteristics observed for different particle sizes. (j) Transfection efficiencies (luciferase reporter expression level) as a result of incubation with particles at different sizes for different periods of time, which correlated well with the trends of total cellular uptake and endosomal escape levels. (k) Regardless of the particle size, fitting the overall endosomal escape level (Y axis) of all plate well-averaged data points against the overall cellular uptake level (X axis) shows a strong positive correlation at 2–4 h postdosage. In the figure, n = 21 wells for the fitted line of 1 h and n = 42 wells for the fitted line of 2 and 4 h. (l) Fitting the endosomal escape level in a single cell (Y axis) of all cells assessed in the same well of the group of 200 nm (n = 5400 cells), 400 nm (n = 4693 cells), and 900 nm (n = 4336 cells), against the cellular uptake level in the same single cell (X axis), shows a strong positive correlation. The figure was generated by overlapping the FlowJo-generated pseudocolor heat maps showing the cell distribution density with an arbitrary correlation curve plotted. In (a–c), all figures share the same scale bar of 50 μm.
The relationship between cellular uptake and endosomal escape was further evaluated on the basis of a plate-well-average (Figure 4k) or single-cell-level (Figure 4l) analysis. Most endosomal escape events of the particles took place after 1 h of incubation. A linear regression correlated all data points of the plate-well-average readings at 2 and 4 h postdosing, regardless of the particle size (Figure 4k). There was a positive correlation that lines all the areas with the highest cell density on the heat maps across different particle sizes on the single-cell level (Figure 4l and Figure S11). These analyses demonstrated that the endosomal escape degree scaled with the cellular uptake level and that the size-dependent transfection efficiency was strongly influenced by cellular uptake. Results from the HEK293F cell suspension culture presented a similar trend of cellular uptake (Figure S12), which also similarly correlated with the transfection efficiency (Figure 3).
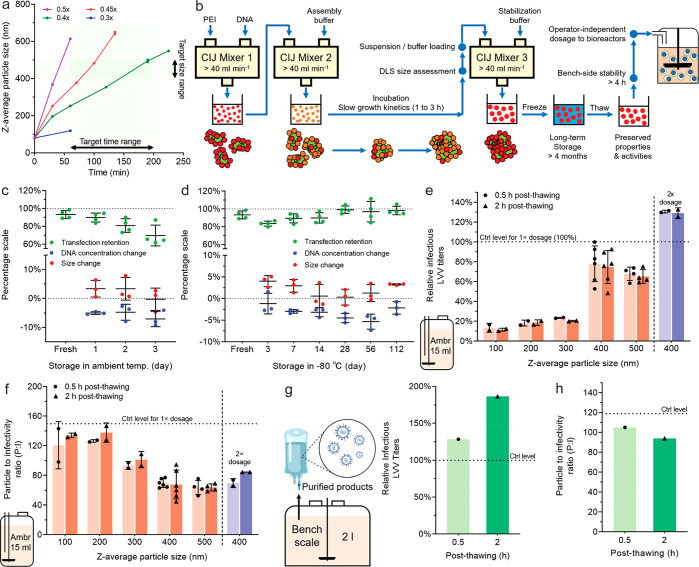
Scalable Production of Sub-Micrometer Particles for LVV Production
The particle assembly process could be scaled up by implementing the two mixing steps (particle growth and stabilization) with relatively high flow rates (e.g., 40 mL/min) in CIJ devices. The pDNA concentration was doubled to reduce the volume handled (Figure S13). The ionic strength of the medium for the particle growth step was optimized to control the particle growth duration between 1 and 3 h (Figure 5a). As streamlined in Figure 5b, given the extended particle growth time, the time required to collect growing particles out of the CIJ mixer #2 and to load solutions into the CIJ mixer #3 was rendered insignificant. This whole process was easier to implement and more reproducible. The lowered ionic strength of the assembly buffer and consequently the lower amount of acid required to revert the pH for the stabilization step provided additional benefits to maintaining the colloidal stability of the particles and preservation of transfection efficiency in storage. The particles were stable in the suspension at ambient temperature for 2 days (Figure 5c) and ofr at least 4 months when they were stored at −80 °C (Figure 5d).
Figure 5.
Scale-up production of pDNA/PEI particles with controlled sizes and validation of transfection efficiency for LVV production in bioreactors. (a) Tunable particle size growth kinetics as a function of ionic strength of the particle growth medium (i.e., PBS concentration, 0.3×, 0.4×, 0.45×, and 0.5× of the full ionic strength). (b) Schematic of the scale-up production process enabled by conducting the mixing steps in CIJ mixers at a flow rate of higher than 40 mL/min. (c) Stability of the 400 nm particles at ambient temperature. (d) Stability of the 400 nm particles at different time points during storage at −80 °C. Particle suspension samples were thawed at ambient temperature before testing. (e, f) Effect of pDNA/PEI particle size on the infectious titers (e) and P:I ratios (f) of the LVVs produced in the 15 mL small bioreactors (ambr 15). A 1× dosage level represents 1 μg of pDNA/mL in the suspension cultures. The data power of each size group differed in the experimental design and is fully indicated by the individual data points shown in the figures. (g, h) The infectious titers (g) and P:I ratios (h) of LVVs produced in a 2 L bioreactor using the 400 nm pDNA/PEI particles, at a dosage level of 1 μg of pDNA/mL. n = 1 bioreactor for each condition. In (e–h), the control level represents the optimal results from the standardized in-house procedures to prepare pDNA/PEI particles manually immediately before the transfection experiments.
Using this protocol, we prepared 100, 200, 300, 400, and 500 nm particles at a final concentration of 50 μg of pDNA/mL in a 20 mL batch size (Table S1). The samples were frozen to −80 °C and shipped on dry ice to bluebird bio, Inc., to test for LVV production in stirred-tank bioreactors (STR). The initial evaluation was conducted in an in-house-developed screening model on the ambr 15 system at a 15 mL scale. The thawed particle suspensions were dosed into the vessels using a liquid handler. LVV titers obtained from the culture supernatants increased with particle size from 100 to 400 nm and then slightly decreased for the 500 nm particles, which was consistent with the reporter transfection results (Figure 5e). The 400 nm particles, whether used at 30 min or 2 h after thawing, produced titers (78%) comparable to those of an internal control that represents the highest level achieved by the freshly prepared particles using the standard method (Figure 1a). The high stability at ambient temperature for 2 h after thawing and nearly 70% increase in LVV titer at a 2× DNA dose were features unattainable with particles prepared using the standard method (Figure 5e). In addition, the comparable titers by the 400 nm particles were achieved by producing a fewer number of LVVs, indicated by a lower particle to infectivity (P:I) ratio (i.e., higher infectivity) in comparison to the standard control (Figure 5f).
We next increased the production scale of the 400 nm particles (50 μg of pDNA/mL) from 20 mL to 70 and 100 mL (Table S2) and tested the particles in a 2 L benchtop single-use STR system. The thawed particles were added to the culture using a peristaltic pump with a 0.5 or 2 h stand after thawing. The infectious titers of the purified LVV preparations from these stabilized 400 nm particles were superior (128% and 187%, n = 1 bioreactor for each) to the highest level achieved by the standard protocol (Figure 5g), and a lower P:I ratio was also verified at this production scale (Figure 5h). These results suggest that the 400 nm particles might have improved the coexpression of multiple plasmids essential for LVV production and assembly.
Conclusions
This study revealed the key insight that the transfection efficiency in LVV production cell lines critically depends on the size of pDNA/PEI particles and identified 400–500 nm as the optimal size range. A stepwise process was designed on the basis of surface charge inversion and conditioning of ionic strength, and pDNA/PEI particles with an average size of 200–1000 nm were prepared with a high degree of size control. The prepared particles exhibited excellent stability in suspension at ambient temperature for transfection operations and at −80 °C for long-term storage. A manufacturing process was developed on the basis of the FNC process with a tailored assembly kinetics to accommodate the mixing procedure. This process was fully characterized at a scale up to 100 mL, although further scale-up is intrinsically straightforward due to its continuous flow setup. The optimal transfection activity and stability of the 400 nm pDNA/PEI particle formulation were validated in the production of LVVs using premanufactured, cryostored, transported, and thawed particles, showing performance matching that of the particles produced using the industry standard in a bioreactor setting. This new scalable manufacturing method has high translational potential that can be easily extended to the production of a wide range of gene therapy vectors with improved productivity and quality control.
Acknowledgments
This work was supported by bluebird bio, Inc. and the National Institutes of Health (NIBIB EB018358 to H.-Q.M.). The authors thank Calvin Chang, Wentao Wang, and Prof. Jamie B. Spangler from the JHMI Translational Tissue Engineering Center (TTEC) for assistance with the suspension culture of HEK293F cells, Stipe Iveljic and Rich Middlestadt from the JHU Whiting School of Engineering Machine Shop for manufacturing of the CIJ devices, Hanhvy Bui from the JHU Integrated Imaging Center (IIC) for assistance with flow cytometry assessments, Professor Emeritus Michael McCaffery from the JHU IIC for assistance with transmission electron microscopy, and Dr. Lesley Y. Chan from bluebird bio, Inc., for scientific input.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c01421.
Methods, correlation of the pDNA payload with the size of pDNA/PEI particles, transmission electron microscopy (TEM) images of particles at the original or stabilized grown sizes, effect of ionic strength and pH of the particle growth medium on growth kinetics and uniformity, the limited particle size change in the transfection medium, representative Cellomics images of B16F10-Gal8-GFP cells incubated with Cy5-pDNA NPs for different durations, confocal laser scanning microscopy images of B16F10-Gal8-GFP cells incubated with Cy5-pDNA particles for 4 h, complete data set of the particle cellular uptake assessed by Cellomics, verification of the particle cellular uptake by pDNAs labeled with tritium, complete data set of the particle-induced endosomal escape assessed by Cellomics, metabolic activities of cells incubated with particles of different sizes, positive scaling of endosomal escape and cellular uptake on a single-cell level, cellular uptake in a suspension culture of HEK293F cells, scaling the size growth process at a DNA concentration of 200 μg/mL, quality control parameters of particles shipped to bluebird bio for testing of lentiviral vector production (ambr 15 scale), quality control parameters of particles shipped to bluebird bio for testing of lentiviral vector production (bench scale), references for methods, and supplementary figures (PDF)
Author Contributions
● Y.H. and Y.Z. contributed equally.
Author Contributions
Y.H., N.D.S., and H.-Q.M. conceived of and designed the study. Y.H. and Y.Z. performed the experiments on particle size control, transfection with reporter plasmids, BS intracellular trafficking mechanisms, with help on data collection and analysis from M.P., E.L., and J.R.S. The cell line and imaging algorithms for assessing intracellular trafficking were developed by D.R.W., C.A.B., D.J.Z., and J.J.G. Validation experiments in bioreactors and characterizations of LVVs produced were carried out by N.D.S. The manuscript was written by Y.H. and H.-Q.M., with revisions by D.R.W., N.D.S., and S.K.R. and inputs from all the other authors.
The authors declare the following competing financial interest(s): Y.H., Y.Z., and H.-Q.M. are inventors on patents and patent applications filed through and managed by Johns Hopkins Technology Ventures. N.D.S. is a Scientist in Process Development at bluebird bio, Inc. and holds company stocks. S.K.R. and H.-Q.M. are co-founders and equity holders of SpaceTime Therapeutics LLC.
Supplementary Material
References
- Milone M. C.; O’Doherty U. Clinical use of lentiviral vectors. Leukemia 2018, 32, 1529–1541. 10.1038/s41375-018-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Tai P. W. L.; Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discovery 2019, 18, 358–378. 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merten O.-W.; Hebben M.; Bovolenta C. Production of lentiviral vectors. Mol. Ther.--Methods Clin. Dev. 2016, 3, 16017. 10.1038/mtm.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear W. S.; Nolan G. P.; Scott M. L.; Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 8392–8396. 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby B.; et al. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 2004, 33, 95–103. 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- Boussif O.; et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 7297–7301. 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mislick K. A.; Baldeschwieler J. D. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 12349–12354. 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J.; Bragonzi A.; Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 2005, 12, 468–474. 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Bus T.; Traeger A.; Schubert U. S. The great escape: how cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B 2018, 6, 6904–6918. 10.1039/C8TB00967H. [DOI] [PubMed] [Google Scholar]
- Pollard H.; et al. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998, 273, 7507–7511. 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- van der Loo J. C. M.; Wright J. F. Progress and challenges in viral vector manufacturing. Hum. Mol. Genet. 2016, 25, R42–R52. 10.1093/hmg/ddv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogris M.; et al. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998, 5, 1425–1433. 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- Zhang W.; et al. Nano-structural effects on gene transfection: large, botryoid-shaped nanoparticles enhance DNA delivery via macropinocytosis and effective dissociation. Theranostics 2019, 9, 1580–1598. 10.7150/thno.30302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.; et al. Size-dependent gene delivery of amine-modified silica nanoparticles. Nano Res. 2016, 9, 291–305. 10.1007/s12274-015-0909-5. [DOI] [Google Scholar]
- Hines E.; et al. Rambutan-like silica nanoparticles at tailored particle sizes for plasmid DNA delivery. J. Mater. Sci. 2021, 56, 5830–5844. 10.1007/s10853-020-05660-w. [DOI] [Google Scholar]
- Santos J. L.; et al. Continuous production of discrete plasmid DNA-polycation nanoparticles using flash nanocomplexation. Small 2016, 12, 6214–6222. 10.1002/smll.201601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; et al. Kinetic control in assembly of plasmid DNA/polycation complex nanoparticles. ACS Nano 2019, 13, 10161–10178. 10.1021/acsnano.9b03334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tockary T. A.; et al. Single-stranded DNA-packaged polyplex micelle as adeno-associated-virus-inspired compact vector to systemically target stroma-rich pancreatic cancer. ACS Nano 2019, 13, 12732–12742. 10.1021/acsnano.9b04676. [DOI] [PubMed] [Google Scholar]
- Osada K.; et al. Quantized folding of plasmid DNA condensed with block catiomer into characteristic rod structures promoting transgene efficacy. J. Am. Chem. Soc. 2010, 132, 12343–12348. 10.1021/ja102739b. [DOI] [PubMed] [Google Scholar]
- Robertson R. M.; Laib S.; Smith D. E. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 7310–7314. 10.1073/pnas.0601903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K. A.; et al. Unusual salt and pH induced changes in polyethylenimine solutions. PLoS One 2016, 11, e0158147 10.1371/journal.pone.0158147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertschinger M.; et al. Disassembly of polyethylenimine-DNA particles in vitro: implications for polyethylenimine-mediated DNA delivery. J. Controlled Release 2006, 116, 96–104. 10.1016/j.jconrel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Johnson B. K.; Prud’homme R. K. Chemical processing and micromixing in confined impinging jets. AIChE J. 2003, 49, 2264–2282. 10.1002/aic.690490905. [DOI] [Google Scholar]
- Hao Y.; Seo J.-H.; Hu Y.; Mao H.-Q.; Mittal R. Flow physics and mixing quality in a confined impinging jet mixer. AIP Adv. 2020, 10, 045105. 10.1063/5.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertschinger M.; Chaboche S.; Jordan M.; Wurm F. M. A spectrophotometric assay for the quantification of polyethylenimine in DNA nanoparticles. Anal. Biochem. 2004, 334, 196–198. 10.1016/j.ab.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Rejman J.; Oberle V.; Zuhorn I. S.; Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. 10.1042/bj20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopatz I.; Remy J.-S.; Behr J.-P. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J. Gene Med. 2004, 6, 769–776. 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- Lächelt U.; Wagner E. Nucleic acid therapeutics using polyplexes: a journey of 50 years (and beyond). Chem. Rev. 2015, 115, 11043–11078. 10.1021/cr5006793. [DOI] [PubMed] [Google Scholar]
- Wilson D. R.; et al. A triple-fluorophore-labeled nucleic acid pH nanosensor to investigate non-viral gene delivery. Mol. Ther. 2017, 25, 1697–1709. 10.1016/j.ymthe.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston T. L. M.; Wandel M. P.; von Muhlinen N.; Foeglein Á.; Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012, 482, 414–418. 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchrist K. V.; et al. Gal8 Visualization of endosome disruption predicts carrier-mediated biologic drug intracellular bioavailability. ACS Nano 2019, 13, 1136–1152. 10.1021/acsnano.8b05482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y.; et al. Carboxylated branched poly(β-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019, 5, eaay3255 10.1126/sciadv.aay3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.