Abstract
Numerous physiological functions rely on distinguishing temperature by temperature-sensitive transient receptor potential channels (thermo-TRPs). While thermo-TRP function has been studied extensively, structural determination of their heat- and cold-activated states has remained a challenge. Here, we present cryo-EM structures of the nanodisc-reconstituted wild-type mouse TRPV3 in three distinct conformations: closed, heat-activated sensitized and open states. The heat-induced transformations of TRPV3 are accompanied by changes in the secondary structure of the S2–S3 linker, N- and C-termini and represent a conformational wave that links these parts of the protein to a lipid occupying the vanilloid binding site. State-dependent differences in the behaviour of bound lipids suggest their active role in thermo-TRP temperature-dependent gating. Our structural data supported by physiological recordings and molecular dynamics simulations provide an insight for understanding the molecular mechanism of temperature sensing.
Introduction
Temperature sensing is important for human physiology and pathophysiology and has been linked to ion channels, including temperature-activated TRP channels (thermo-TRPs)1–5. Ten closely-related channels constitute the group of thermo-TRPs, all of them being highly responsive to changes in temperature and covering the physiologically-relevant range of temperatures, from noxious cold (<15°C) to noxious heat (>43°C)6–10. A remarkable similarity of thermo-TRP structures1,11 suggests that these channels may share a common molecular mechanism of temperature sensing. Nevertheless, structural visualization of thermo-TRP opening by temperature had proven to be difficult and has only been successful using mutagenesis12. The usage of temperature-sensitizing mutations, however, may lead to significant alteration of TRP channel temperature activation, not necessarily recapitulating the process that naturally occurs in living cells. Therefore, to better understand the structural mechanisms of temperature sensing, there is a need to solve the missing strutures of wild-type thermo-TRPs in the heat-activated open states.
Here we embarked on structural and functional studies of TRPV3, a representative of vanilloid-subfamily temperature-sensitive TRP channels (thermo-TRPVs). TRPV3 is implicated in cutaneous sensation, including thermo-sensation, nociception and itch, in addition to the maintenance of the skin barrier and wound healing, hair growth, and embryonic development, while its dysfunction is associated with numerous human diseases, including a genodermatosis known as Olmsted syndrome, atopic dermatitis, rosacea and lung cancer13–15. We reconstituted wild-type mouse TRPV3 into lipid nanodiscs and structurally captured its temperature-activated open conducting conformation using cryo-EM. Our structural results combined with molecular dynamics (MD) simulations illustrate a critical role of lipids in TRPV3 activation and shed light on the mechanism of temperature sensing.
Results
Functional characterization and cryo-EM sample preparation
Compared to other representatives of thermo-TRPs that typically show time-dependent inactivation, TRPV3 exhibits a current increase (sensitization) in response to repetitive application of ligands or heat16–21 (Fig. 1a). Sensitization can also be achieved in response to prolonged exposure of TRPV3 to increasing temperature (Extended Data Fig. 1a), with a single TRPV3 channel showing no openings at low temperature and increasing probability of opening at high temperatures (Fig. 1b). Consistent with the previous estimates18,20,22, thermodynamic analysis of the temperature-induced single TRPV3 channel activity provided the high values of the temperature coefficient, Q10 = 27.0 ± 7.4 (n = 19; Extended Data Fig. 1b), an indicative of high sensitivity of thermo-TRPs to temperature, as well as transitional changes in enthalpy, ΔH = 91.2 ± 3.6 kcal/mol (n = 19), and entropy, ΔS = 287.5 ± 11.7 cal/mol • K (n = 19) (Extended Data Fig. 1c).
Figure 1. TRPV3 function and cryo-EM.
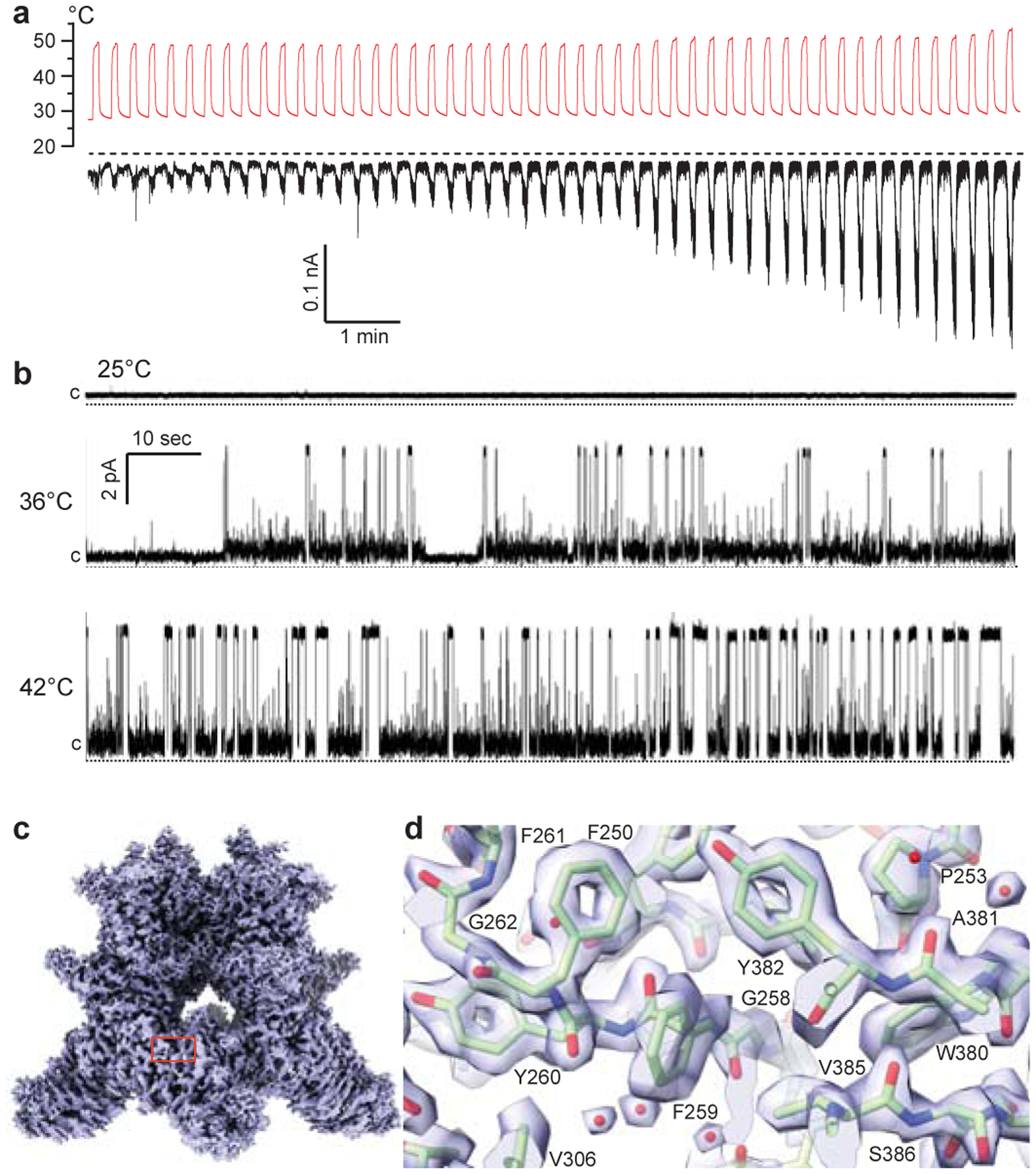
a, Whole-cell patch-clamp current (black) recorded from HEK-293T cell expressing wild-type mouse TRPV3 in response to repetitive applications of heat (red) at −70 mV membrane potential. The dashed line indicates zero current. b, Representative single-channel currents recorded at 25°C, 36°C and 42°C and 30 mV membrane potential from wild-type TRPV3 reconstituted into lipid bilayers. The single-channel conductance measured at 42°C, 146 ± 7 (n = 15)12, is in the range of conductances measured in experiments with cells (48 – 261 pS, see Methods). The dotted lines indicate zero current. c, 1.98-Å resolution cryo-EM map of wild-type TRPV3 reconstituted in MSP2N2 nanodiscs and incubated at 4°C. d, Close-up view of the map region indicated by red rectangle in c. Protein is shown in sticks. Red spheres represent water molecules.
To characterize temperature-induced TRPV3 activation structurally, we subjected the purified wild-type TRPV3 protein to cryo-EM. Previously we solved the structure of a detergent-solubilized mutant TRPV3 in the heat-activated open state but did not succeed with the wild-type channel12. This time, we adapted the strategy of recent high-resolution TRPV3 reconstruction23,24 and reconstituted our purified wild-type TRPV3 into lipid nanodiscs. At low temperature (4°C), the protein reconstituted into MSP2N2 nanodiscs produced a single three-dimensional (3D) class, with the cryo-EM map at 1.98-Å resolution (Extended Data Figs. 2–3, Table 1) and the core of the molecule reaching ~1.94-Å resolution, clearly demonstrating atomic details (Fig. 1c–d, Extended Data Fig. 4). The state of the corresponding structure is characterized by a non-conducting pore (Extended Data Fig. 5), I674 lining the lower gate and C-terminus forming a coil that wraps around the three-stranded β-sheet12,23–27, and will further be referred to as closed. To determine TRPV3 structures at high temperature, we tested different heat application protocols and found that repetitive cycles of heating and cooling showed better protein stability than protocols with prolonged exposure to heat (see Methods).
Table 1.
Cryo-EM data collection, refinement and validation statistics
| Closed MSP2N2 4°C (EMD-23853, PDB 7MIJ) |
Closed MSP2N2 42°C (EMD-23854, PDB 7MIK) |
Sensitized MSP2N2 42°C (EMD-23855, PDB 7MIL) |
Closed cNW11 4°C (EMD-23856, PDB 7MIM) |
Closed cNW11 42°C (EMD-23857, PDB 7MIM) |
Open cNW11 42°C (EMD-23858, PDB 7MIO) |
|
|---|---|---|---|---|---|---|
| Data collection and processing | ||||||
| Magnification | 105,000x | 105,000x | 105,000x | 81,000x | 105,000x | 105,000x |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 58 | 58.5 | 58.5 | 51.07 | 58 – 60 | 58 – 60 |
| Defocus range (μm) | −0.8 to −2.0 | −0.8 to −2.0 | −0.8 to −2.0 | −0.5 to −2.0 | −0.5 to −2.0 | −0.5 to −2.0 |
| Pixel size (Å) | 0.86 | 0.858 | 0.858 | 1.105 | 0.871 | 0.871 |
| Symmetry imposed | C4 | C4 | C4 | C4 | C4 | C4 |
| Initial particle images (no.) | 4,662,531 | 5,855,393 | 5,855,393 | 10,151,026 | 6,869,494 | 6,869,494 |
| Final particle images (no.) | 372,121 | 54,146 | 22,925 | 159,698 | 39,182 | 27,805 |
| Map resolution (Å) | 1.98 | 3.12 | 3.86 | 3.42 | 3.09 | 3.48 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 1.94–4.79 | 3.06–4.67 | 3.75–6.45 | 3.24–6.54 | 2.94–7.07 | 3.31–8.81 |
| Refinement | ||||||
| Initial model used (PDB code) | 6PVN | 6PVN | 6PVP | This study | This study | 6PVP |
| Model resolution (Å) | 1.98 | 3.12 | 3.86 | 3.42 | 3.09 | 3.48 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | 1.980–2.002 | 3.120–3.156 | 3.860–3.904 | 3.090–3.125 | 3.090–3.125 | 3.490–3,451 |
| Map sharpening B factor (Å2) | −42 | −114 | −137 | −151 | −50 | −80 |
| Model composition | ||||||
| Nonhydrogen atoms | 22,702 | 22,728 | 21,753 | 22,728 | 22,723 | 22,384 |
| Protein residues | 2,556 | 2,560 | 2,624 | 2,560 | 2,560 | 2,636 |
| Ligands | 2,202 | 1,876 | 365 | 1,876 | 1,875 | 1,457 |
| B factors (Å2) | ||||||
| Protein | 21.58 | 67.57 | 83.47 | 107.38 | 77.07 | 65.40 |
| Ligand | 47.58 | 30.50 | 78.61 | 40.05 | 85.22 | 70.34 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.004 | 0.006 | 0.005 | 0.009 | 0.006 | 0.008 |
| Bond angles (°) | 0.838 | 0.913 | 0.916 | 0.929 | 0.875 | 0.941 |
| Validation | ||||||
| MolProbity score | 1.25 | 1.49 | 1.40 | 1.50 | 1.54 | 1.50 |
| Clashscore | 3.13 | 4.36 | 2.86 | 3.91 | 4.27 | 4.27 |
| Poor rotamers (%) | 0.00 | 0.35 | 0.64 | 0.18 | 0.00 | 0.34 |
| Ramachandran plot | ||||||
| Favored (%) | 97.02 | 95.92 | 95.40 | 93.89 | 95.14 | 95.73 |
| Allowed (%) | 2.83 | 4.08 | 4.45 | 5.96 | 4.70 | 4.27 |
| Disallowed (%) | 0.16 | 0.00 | 0.15 | 0.16 | 0.16 | 0.00 |
Structures of TRPV3 activated by heat
We made cryo-EM samples of wild-type TRPV3 reconstituted into MSP2N2 nanodiscs after subjecting the protein to twenty to forty successive 30-sec heat pulses, by changing the temperature between 25°C and 42°C. Analysis of the collected cryo-EM data revealed two major high-resolution 3D classes (Extended Data Figs. 2–3), but neither one represented an open-pore structure (Extended Data Fig. 5). One class represented a structure in the closed conformation nearly identical to the closed state at 4°C. The other one yielded a structure with a slightly wider pore and α–helical C-terminus, the state of which will further be referred to as sensitized.
As an alternative strategy to determine the heat-activated open-state structure of TRPV3, we reconstituted the protein in cNW11 nanodiscs28. At 4°C, cNW11-reconstituted TRPV3 produced a single 3D class apparently representing the closed state, nearly identical to the closed states of TRPV3 reconstituted in MSP2N2 nanodiscs (Extended Data Fig. 5). Exposure of the protein reconstituted in cNW11 nanodiscs to twenty to forty 30-sec 42°C heat pulses resulted in two high-resolution 3D classes (Extended Data Figs. 2, 3 and 6, Movie 1). One class represented the closed state, which was similar to the other closed states (Extended Data Fig. 5), while the second class represented the open state with a wide pore (Fig. 2a–d), comparable in size to the pore of the previously solved open-state structures of mutant TRPV3 channels12,24,25,27. MD simulations (Extended Data Fig. 7a) confirmed that the closed state representing TRPV3 in cNW11 nanodiscs activated by heat was indeed not permeable to water or ions (Fig. 2e), while the open state conducted both (Fig. 2f).
Figure 2. TRPV3 structures and pore permeation at high temperature.
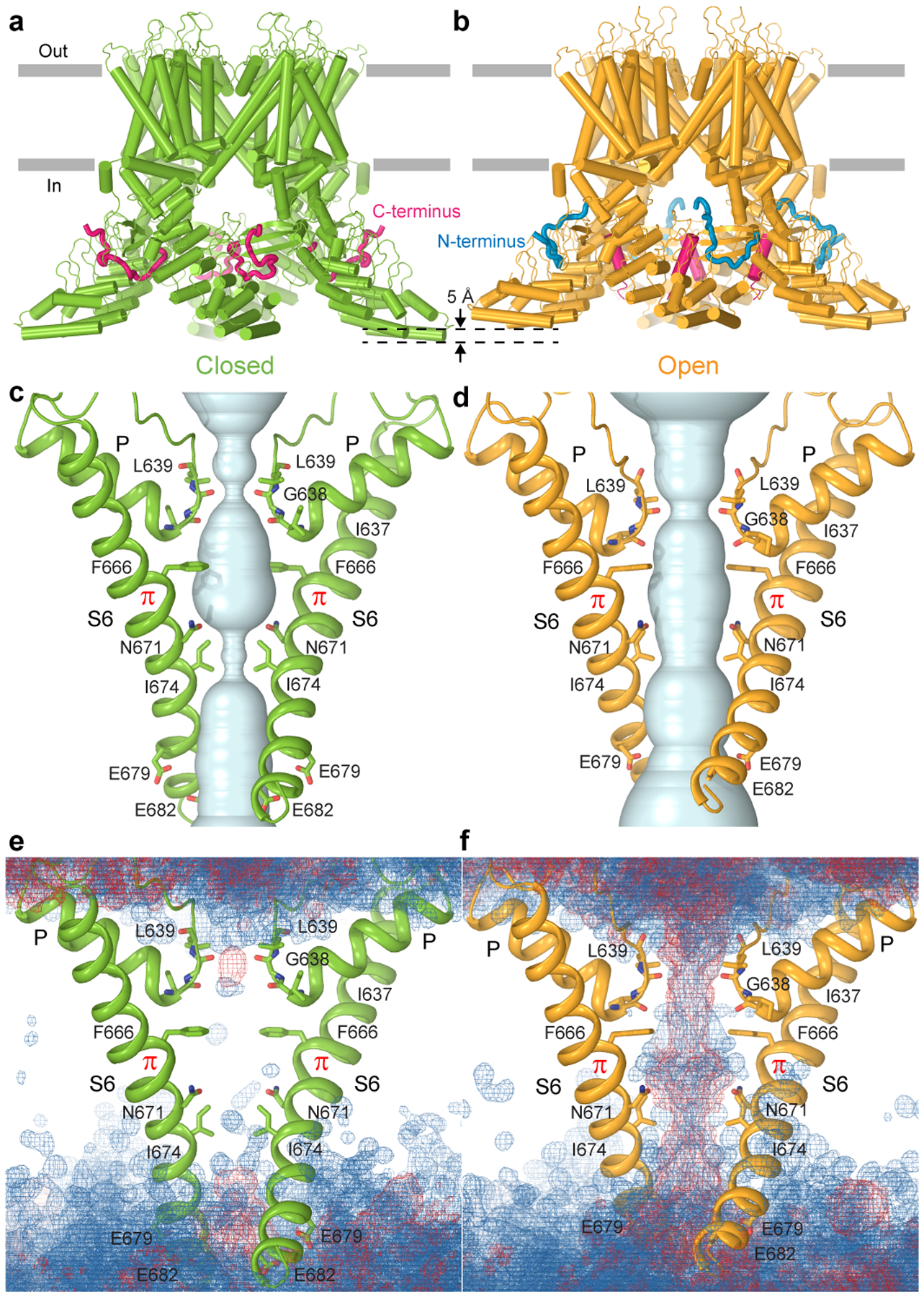
a-b, Closed-state (a) and open-state (b) structures of wild-type TRPV3 reconstituted in cNW11 nanodiscs and exposed to repetitive applications of heat. c-d, Pore-forming domains in the closed (c) and open (d) states with the residues lining the pore shown as sticks. Only two of four subunits are shown, with the front and back subunits omitted for clarity. The pore profiles are shown as space-filling models (light blue). e-f, Distribution of water (blue mesh) and ions (red mesh) during MD simulations of TRPV3 in the closed (e) and open (f) states, illustrating the lack and presence of pore permeation, respectively.
Why cNW11 but not MSP2N2 nanodiscs allow temperature-induced opening of TRPV3 remains unclear. It is possible that MSP2N2 nanodiscs create a tighter environment around the transmembrane domain region than in natural membranes. While this tight environment does not allow the channel to open completely, it allows it to “semi-open” and reveal the intermediate (sensitized) state of heat-induced activation. In contrast, cNW11 nanodiscs create a large ring around the transmembrane domain region, inside of which lipids move more freely and reproduce better the environment of natural membranes. In such environment, the channel can open in response to application of heat and its conformation resembles the open conformation of the heat-activated Y564A mutant (Extended Data Fig. 8). Similar to all previously solved open-state structures of mutant TRPV3 channels12,24,25,27, the pore’s narrow constriction at the gate region of the heat-activated open-state wild-type TRPV3 is formed by I674 (Fig. 2d, Extended Data Fig. 5c). Isoleucines 674 also form narrow constriction in the nanodisc-reconstituted closed and sensitized states (Fig. 2c, Extended Data Fig. 5c) and in all these structures, the pore-forming helix S6 contains the characteristic α-to-π transition23,24. In contrast, structures of detergent-solubilized or reconstituted in amphipols TRPV3 in the non-conducting closed state had the entire S6 α-helical12,25–27.
Distinct conformations of N- and C-termini
Apart from different pore sizes, the closed and temperature-activated open states demonstrate different conformations of the C-terminus. In the closed state, the C-terminus has an extended conformation and wraps around the three-stranded β-sheet (Fig. 2a). Similar to the closed states of the previously published TRPV3 structures12,23–25,27, the C-terminus in the extended conformation forms specific contacts with the ankyrin repeat domain (ARD) of the neighbouring subunit through residues conserved in the thermo-TRPVs but not in TRPV5/6 (Extended Data Fig. 9). These include electrostatic interactions of E751 and D752 with K169 and K174 as well as interactions of W739 and W742 with hydrophobic residues and R226 (Fig. 3a). The importance of these interactions for TRPV3 function has been previously confirmed by mutagenesis25,29. In the open state, the C-terminus forms an α-helix that lines the walls of the intracellular cavity (Fig. 2b). It participates in different contacts with the neighbouring subunit, including salt bridges between E308 and K743 as well as between R319 and E751 (Fig. 3b).
Figure 3. N- and C-termini.
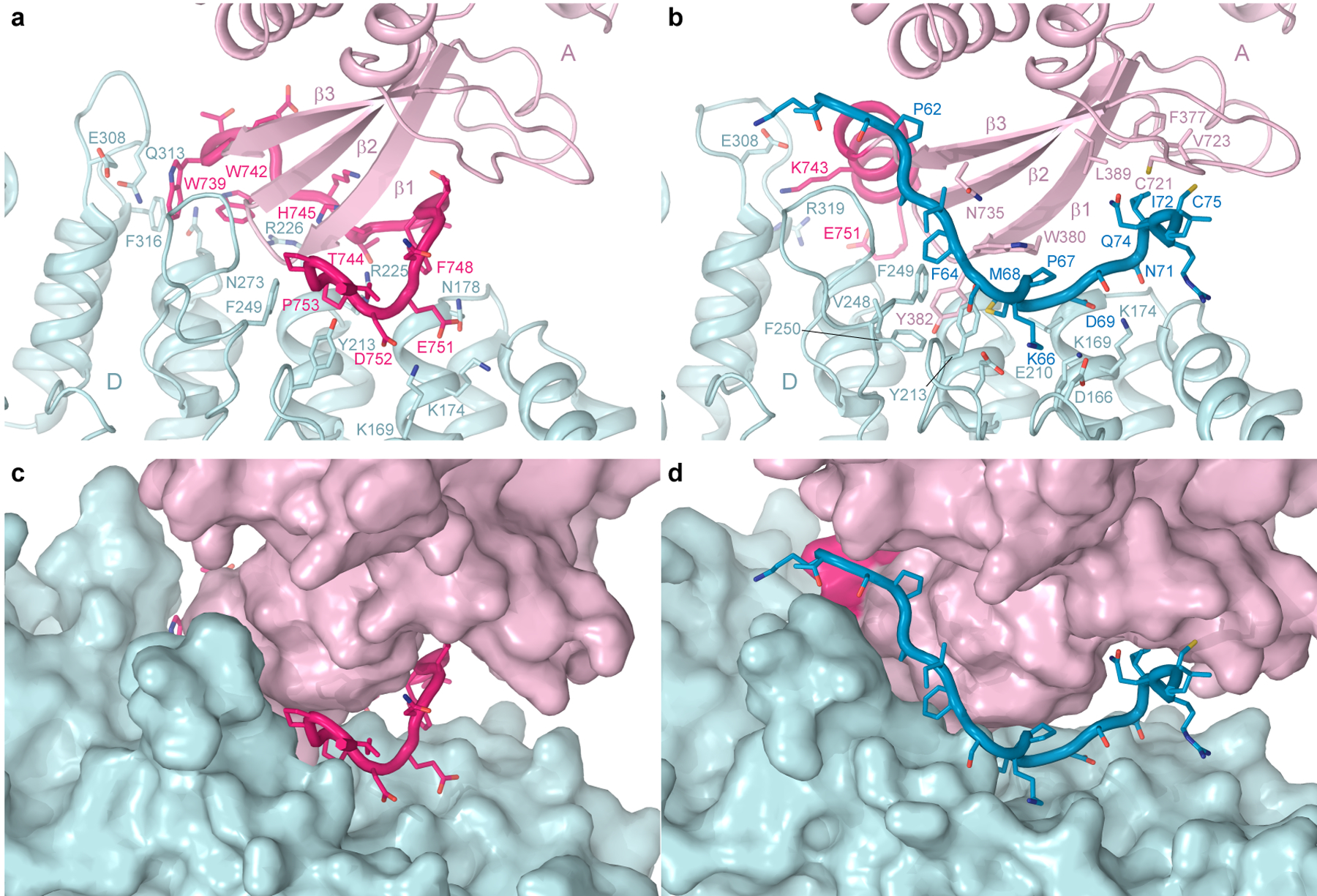
Interface between the neighbouring TRPV3 subunits (light pink and light blue) that connects elements of the intracellular skirt in the closed (a,c) and open (b,d) states, shown as cartoon (a-b) or surface (c-d), with the N-terminus shown in blue and C-terminus in hot pink and residues in stick representation.
A much more important role in strengthening the intracellular skirt intersubunit contacts in the open state, however, plays an extended polypeptide that snugly fits into the cleft formed between two subunits, only partially filled by the C-terminus in the closed state (Fig. 3c–d). It was previously suggested to represent a portion of the N-terminus that is unbound and not observed in the closed states of TRPV312,25. Because of the high quality of our cryo-EM reconstructions, we built a precise model of this polypeptide and confirmed that this is the K58-L76 region of the N-terminus. The amino acid sequence of this region is not highly conserved across thermo-TRPVs (Extended Data Fig. 10). Accordingly, numerous intra- and intersubunit interactions mediated by the N-terminal residues (Fig. 3b) are likely to be unique for TRPV3; they pull the three-stranded β-sheet and the C-terminal hook of one subunit towards the ARD of the neighbouring subunit (Fig. 3d) and make the cleft between them in the sensitized and open states narrower than in the closed state (Fig. 3c). As a result, the intracellular skirt of TRPV3 in the sensitized and open states rotates by ~8° and moves towards the transmembrane domain (Fig. 2a–b, Extended Data Fig. 5e–h).
Protein-lipid interactions
There are clearly distinguishable annular lipid densities, which look similar in the closed and open states on the extracellular side of the transmembrane domain but differently on its intracellular side (Fig. 4a–d). Indeed, a lipid that fits its head into a crevice between S2-S3 linker, S4, S4-S5 linker and TRP helix in the closed state (Fig. 4e), is not present in the open state due to closer positioning of the S4-S5 linker to S1-S4 as well as W521 and Q580 side chain rearrangements (Fig. 4f). In TRPV1, this site binds vanilloid ligands, like agonists resiniferatoxin (RTX) and capsaicin, competitive antagonist capsazepine, or phosphatidylinositol (PI) lipid30,31. In the closed state of TRPV3, this vanilloid site fits a two-tail lipid density with the head part being smaller than expected for PI but comparable in size to the head of phosphatidylcholine (PC).
Figure 4. Lipids.
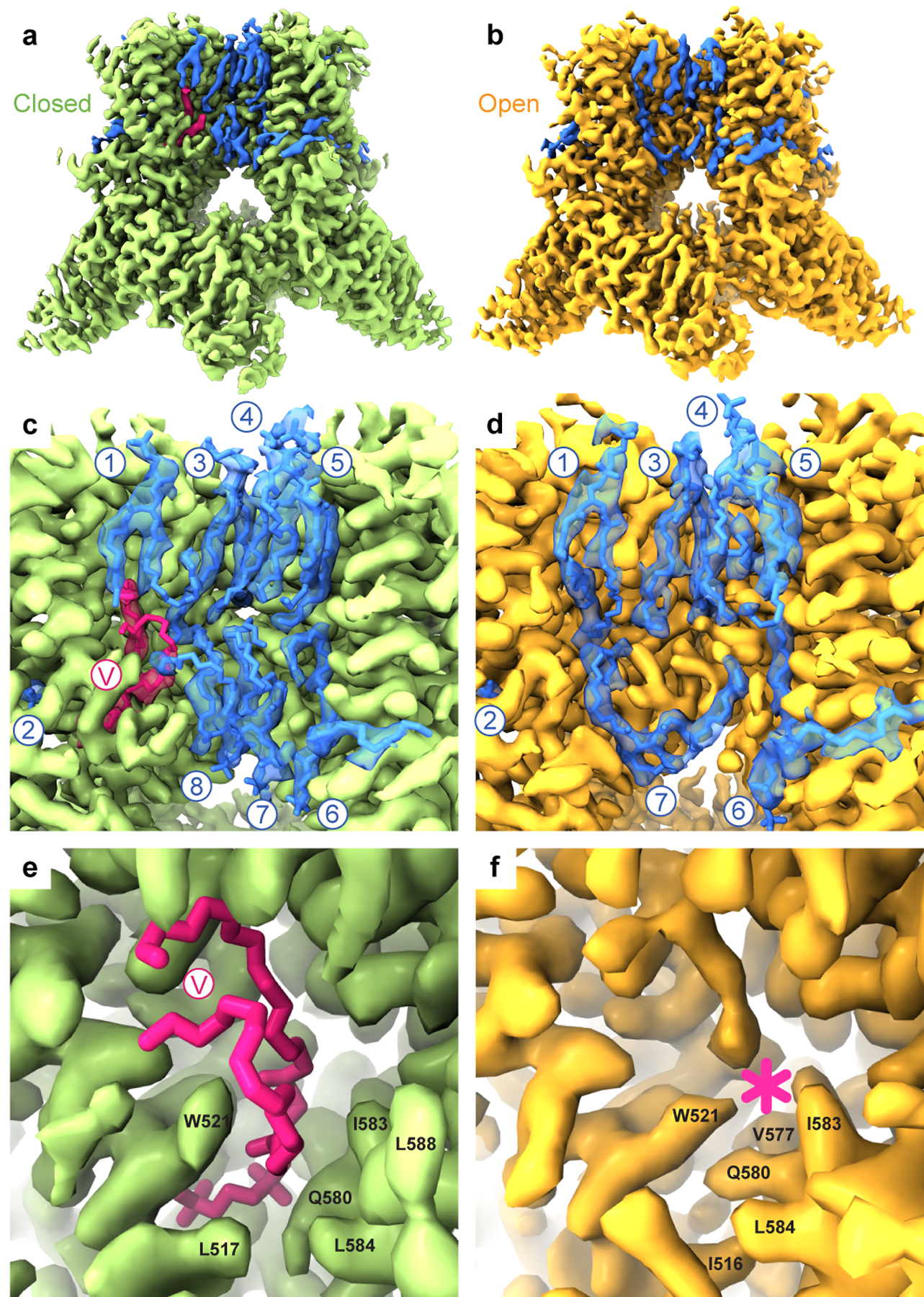
a-b, Cryo-EM density for cNW11-reconstituted wild-type TRPV3 in the closed (a) and heat-activated open (b) states, with the lipid densities coloured pink or blue. c-d, Close-up view of the membrane region, with the molecules of lipid shown in sticks and numbered. e-f, Close-up view of the vanilloid site (V), which harbours a phosphatidylcholine lipid in the closed state (sticks, e) and disappears in the open state (asterisk, f).
To get an insight into lipid binding specificity, we performed MD simulations of the closed-state structure by nesting different types of lipids in the vanilloid site (Extended Data Fig. 7b–g). All lipids, including cholesterol, PI, PC, phosphatidylserine (PS), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG) remained in the vanilloid pocket throughout the entire duration of 500 ns-long MD simulations. However, the MD-predicted density for cholesterol looked very different from the two-tails-and-one-head cryo-EM lipid density (Extended Data Fig. 7g), making it an unlikely candidate to occupy the vanilloid site of TRPV3. In contrast, the MD-predicted densities for all glycerophospholipids resembled the experimental cryo-EM density (Extended Data Fig. 7b–f). Nevertheless, promiscuity of the MD-predicted vanilloid site occupancy by glycerophospholipids suggests that the site has low lipid specificity. Together with high lipid mobility, this low specificity is a likely reason for poorer quality of lipid density compared to the protein density nearby. Accordingly, the drastic improvement in resolution of cryo-EM reconstruction from 3.09–3.86 Å to 1.98 Å does not rescue the poor quality of lipid density, which appears to be similar in all of our closed-state structures (Table 1).
Structural changes and conformational wave
Compared to the closed and open states that show striking differences in conductance, termini conformations and lipid binding, the sensitized state appears to be intermediate. We compared the corresponding structures to determine structural rearrangements accompanying the putative closed-to-sensitized and sensitized-to-open state transitions (Fig. 5). Calculation of the root-mean-square deviation (RMSD) values for Cα atoms with a short sliding window of 10 residues highlighted the structural regions that undergo drastic secondary structure rearrangements. For the putative closed-to-sensitized state transition, these include the S2-S3 linker that changes α-helicity between its C- and N-terminal portions, C-terminus that changes its coiled to α-helical conformation, and N-terminus, which is unfolded in the closed state and binds to the intersubunit skirt interface in the sensitized state (Fig. 5a). Strikingly, no such regions were identified for the sensitized-to-open state transition (Fig. 5b).
Figure 5. State-dependent structural changes.
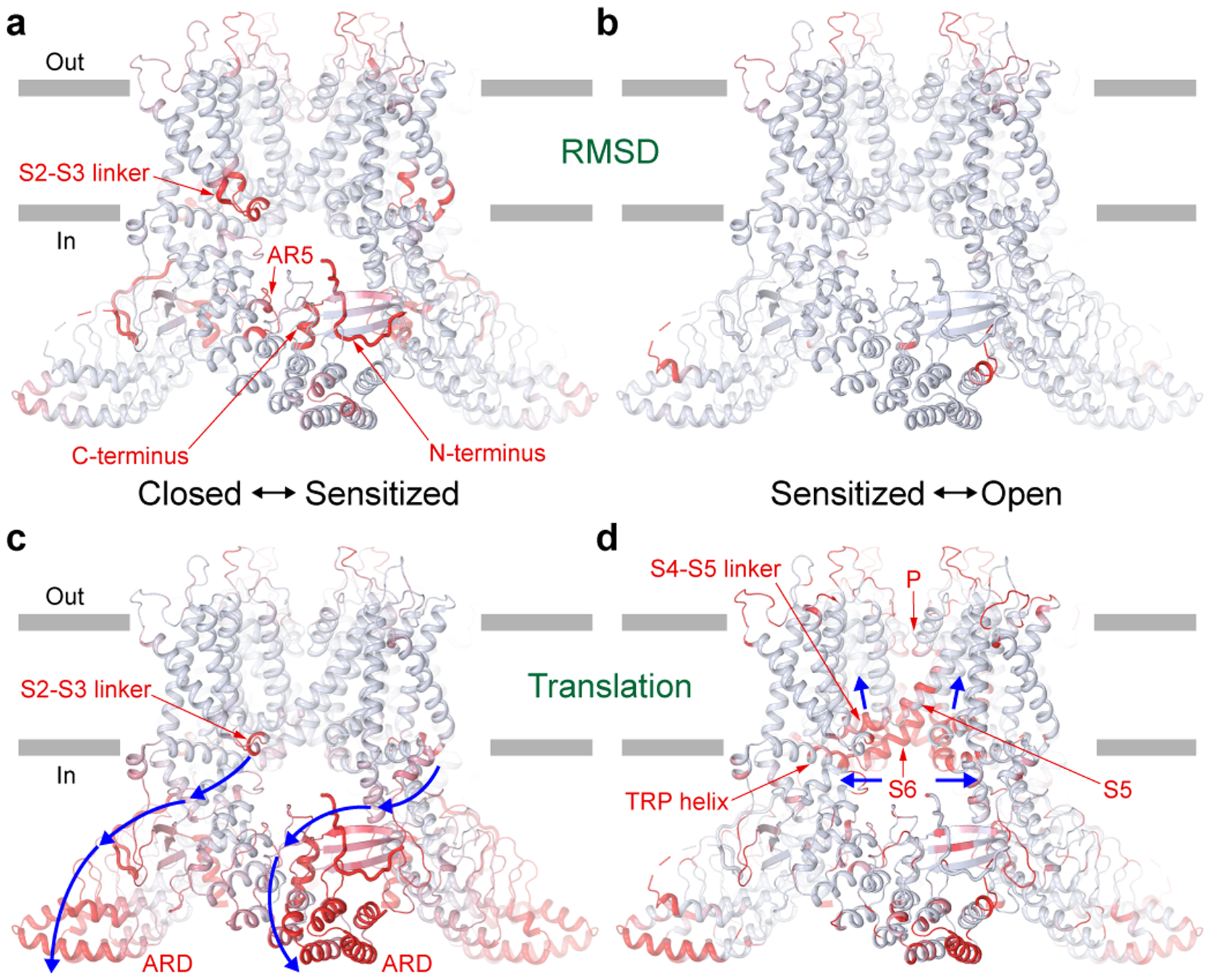
Light blue (no changes) to red (strong changes) gradient of RMSD (a-b) or translation (c-d) calculated between closed and sensitized (a, c) or sensitized and open (b, d) states and mapped on the sensitized state structure. RMSD values were calculated for Cα atoms along the entire TRPV3 sequence with a sliding window of 10 residues. Cα atom translations were calculated after aligning S1-S4 domains of the corresponding structures. Regions of the greatest structural changes are labelled. Blue arrows indicate the direction of the conformational wave accompanying closed to sensitized (c) and sensitized to open (d) state transitions.
As a measure of structural movement associated with the change in TRPV3 conformational state, we calculated Cα atom translations between the corresponding structures superposed based on the S1-S4 domains. For the putative closed-to-sensitized state transition, there is a cluster of interacting structural domains – the S2-S3 linker, linker domain, C-terminal hook, three-stranded β-sheet, N-terminus, C-terminus and ARD – that undergo significant translation (Fig. 5c). Most of them, except the S2-S3 linker and N- and C-termini (Fig. 5a), do not undergo drastic structural rearrangements but instead a rigid body motion only, and the concerted character of their motion resembles a wave of mutually dependent conformational changes that involves every contributing domain (Movie 1). Interestingly, the side chain size but not polarity or flexibility of N412, which resides at the interface between the S2-S3 loop and linker domain, was implicated in use-dependence of TRPV3 temperature sensitivity32.
For the sensitized-to-open state transition, the conformational changes have a different character. They are much more localized and involve the S4-S5 linker that pulls up upon channel opening, S5, S6 and the N-terminal portion of the TRP helix, which follow the S4-S5 linker, and pore-forming regions of the selectivity filter, which slightly move apart to increase channel conductance (Fig. 5d). Such different structural dynamics (Fig. 5c–d), together with a distinct character of structural rearrangements (Fig. 5a–b), highlight the different physical nature of the closed-to-sensitized and sensitized-to-open state transitions. Since heat exposure was necessary for the closed-to-sensitized state transition in MSP2N2 nanodiscs and closed-to-open state transition in cNW11 nanodiscs, the closed-to-sensitized state transition is expected to be temperature-dependent, while the sensitized-to-open state transition temperature-independent (Fig. 6).
Figure 6. Mechanism of TRPV3 temperature activation.
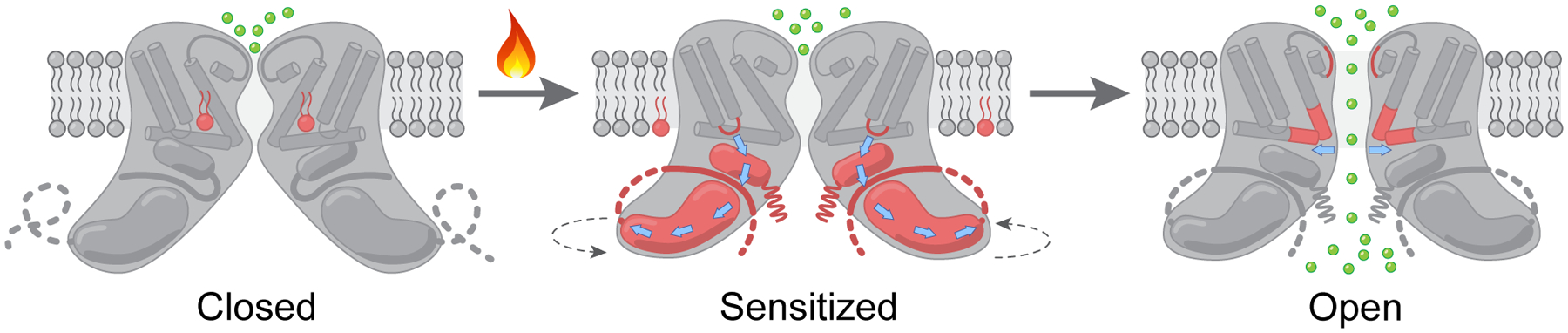
Heat-induced activation of TRPV3 occurs in two steps: sensitization and channel opening. Sensitization is highly temperature-sensitive and accompanied by withdrawal of the vanilloid-site lipid and a massive conformational wave that includes the S2-S3 loop, N- and C-termini changing their secondary structures and positioning. As a result of sensitization, the intracellular skirt of TRPV3 rotates by ~8° and moves towards the transmembrane domain. Channel opening is weakly temperature-sensitive and accompanied by more local structural changes that involve the TRP helix and pore-forming S6 and P-loop and results in pore widening sufficient for ion conductance.
Discussion
Molecular mechanism of temperature sensing
There are different competing hypotheses about the molecular mechanism of temperature sensing by TRP channels and ion channels in general. Since temperature sensitivity is an intrinsic property of the channel18,33, it can be localized to a specific domain (temperature sensor) or integrated over the entire protein34. Mutagenesis studies identified numerous temperature sensitivity-altering substitutions in different TRP channel domains, including the ARD-S1 linker domain, S1-S4 domain, pore domain, TRP helix and C-terminus32,35–46. It was therefore tempting to call each of these domains a temperature sensor. However, since all these domains together make up most of the TRP channel protein, the mutagenesis results rather favor the distributed temperature sensing.
On the other hand, comparison of the closed and heat-activated open states suggests that temperature-induced conformational changes involve several domains simultaneously, including the vanilloid site, S2-S3 and S4-S5 linkers, pore domain, TRP helix, linker domain, three-stranded β-sheet, loops of the ARD, N- and C-termini (Fig. 5, Extended Data Fig. 10). Changes in these domains, however, are mutually dependent and represent a conformational wave that propagates from one domain to another. The domino-like propagation of such conformational wave can be altered by obstacles at any location on its pathway, explaining why mutations all over the protein can affect temperature sensing32,35–45. The wave can potentially be triggered at a single location, which in turn can be considered a localized temperature sensor. The vanilloid site lipid that leaves its site with increasing temperature, possibly due to phase transitions in the membrane, can play a role of such sensor12,30 (Fig. 4). Alternatively, it could be the S2-S3 linker, N- or C-termini that drastically change their secondary structure between the closed and heat-activated states (Fig. 3 and 5, Extended Data Fig. 10).
The underlying energetics of temperature-associated conformational changes in the channel protein was proposed to be associated with changes in channel’s heat capacity47. For changes in heat capacity to be responsible for temperature activation of TRPV3, the estimated change in enthalpy associated with the temperature-induced activation of TRPV3, ΔH = 91.2 kcal/mol (22.8 kcal/mol per subunit, Extended Data Fig. 1), would need to be translated into 10–15 hydrophobic residues per subunit changing their local hydrophobic environment to hydrophilic one. Alternatively, the change in enthalpy can occur independently of changes in heat capacity, for example, due to formation or breakage of molecular bonds. Thus, in the presence of allosteric gating, the gating equilibrium constant as well as the strength- and temperature dependence of the coupling between gating and the temperature-sensitive transitions were proposed to determine temperature-dependent activity of TRP channels in heat capacity-independent manner48. There is a number of regions that can cumulatively account for energetics of temperature-associated conformational changes in the channel protein, including the lower pore with the surrounding intracellular pockets49 (Fig. 2e–f; Extended Data Fig. 5b), the vanilloid site (Fig. 4), but most importantly the S2-S3 linker and N- and C-termini that change their secondary structure between the closed and heat-activated open states (Fig. 5; Extended Data Fig. 10). These regions can potentially make their energetic contribution through short-living intermediate states that have not been captured structurally. It will therefore require additional efforts to estimate the role of each individual region and the physical nature of its contribution to energetics of temperature sensing.
In summary, while the details of the temperature sensing mechanism will need to be explored further, our structures of wild-type TRPV3 in the closed and temperature-activated sensitized and open states provide a molecular template for future studies.
Methods
Construct
The full-length mTRPV3 (residues 1–825) used in cryo-EM studies was cloned into a pEG BacMam vector50, with a C-terminal thrombin cleavage site (LVPRG), followed by a streptavidin affinity tag (WSHPQFEK), as was done previously12,27.
Expression and purification
TRPV3 construct was expressed and purified as previously described for mTRPV312,27 with minor modifications. Bacmids and baculoviruses were produced using a standard method50. Briefly, baculovirus was made in Sf9 cells for ~72 hours (Thermo Fisher Scientific, mycoplasma test negative, GIBCO #12659017) and was added to the suspension adapted HEK 293 cells lacking N-acetyl-glucosaminyltransferase I (GnTI–, mycoplasma test negative, ATCC #CRL-3022) that were maintained in Freestyle 293 media (Gibco-Life Technologies #12338–018) supplemented with 2% FBS at 37°C and 5% CO2. Twenty-four hours after transduction, 10 mM sodium butyrate was added to enhance protein expression, and the temperature was reduced to 30°C. Seventy-two hours after transduction, the cells were harvested by centrifugation at 5,471 g for 15 min using a Sorvall Evolution RC centrifuge (Thermo Fisher Scientific), washed in phosphate buffer saline (PBS, pH 8.0), and pelleted by centrifugation at 3,202 g for 10 min using an Eppendorf 5810 centrifuge. The cell pellet was resuspended in ice-cold lysis buffer, containing 20 mM Tris (pH 8.0), 150 mM NaCl, 0.8 μM Aprotinin, 4.3 μM Leupeptin, 2 μM Pepstatin A, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM β-mercaptoethanol (βME). Cells were subsequently lysed using a Misonix Sonicator with a preset program (6 cycles of 15 s “on” at the amplitude of 8 followed by 15 s “off”; this program was repeated three times for optimal cell lysis) under constant stirring on ice. Unbroken cells and cell debris were pelleted using an Eppendorf 5810 centrifuge at 3,202 g and 4°C for 10 min. The supernatant was subjected to ultracentrifugation in a Beckman Coulter ultracentrifuge using a Beckman Coulter Type 45Ti rotor at 186,000 g and 4°C for 1 hour to pellet the membranes. The membrane pellet was mechanically homogenized and solubilized in the lysis buffer supplemented with 2% (w/v) digitonin under stirring at 4°C for 2 hours. Insoluble material was removed by ultracentrifugation for 40 min in a Beckman Coulter Type 45Ti rotor at 186,000 g, and the supernatant was added to strep resin and rotated for 14–16 hours at 4°C. Next, the resin was washed with 10 column volumes of buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, 1 mM βME, 0.01% (w/v) glyco-diosgenin (GDN) and the protein was eluted with the same buffer supplemented with 2.5 mM D-desthiobiotin. The eluted protein was concentrated using a 100 kDa NMWL centrifugal filter (MilliporeSigma™ Amicon™) to 0.5 ml and then centrifuged in a Sorvall MTX 150 Micro-Ultracentrifuge (Thermo Fisher Scientific) using a S100AT4 rotor for 30 min at 66,000 g and 4°C before injecting into a size-exclusion chromatography (SEC) column. The protein was further purified using a Superose™ 6 10/300 GL SEC column attached to an AKTA FPLC (GE Healthcare) and equilibrated in 150 mM NaCl, 20 mM Tris, 1 mM βME, 0.01% GDN (pH 8.0). The tetrameric peak fractions were pooled and concentrated using 100 kDa NMWL centrifugal filter (MilliporeSigma™ Amicon™) to 2–4 mg/ml.
For TRPV3 reconstitution into MSP2N2 nanodiscs, the purified protein was mixed with MSP2N2 and soybean lipids (Soy polar extract, Avanti Polar Lipids, USA) at a molar ratio of 1:3:166 (monomer:MSP2N2:lipid). The MSP2N2 was stored in a buffer containing 150 mM NaCl and 20 mM Tris (pH 8.0). The lipids were dissolved to a concentration of 100 mg/ml in 150 mM NaCl, 20 mM Tris (pH 8.0) and subjected to 5–10 cycles of freezing in liquid nitrogen and thawing in a water bath sonicator. The nanodisc mixture (500 μl) was rocked at room temperature for 1 hour. Subsequently, 20 mg of Bio-beads SM2 (Bio-Rad) pre-wet in buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1 mM βME) was added to the nanodisc mixture and then it was rotated at 4°C. After one hour, additional 20 mg of Bio-beads SM2 was added and the resulting mixture was rotated at 4°C for ~14–20 hours. The Bio-beads SM2 were then removed by pipetting and mTRPV3 reconstituted in nanodiscs was purified from empty nanodiscs by SEC using Superose™ 6 10/300 GL SEC column equilibrated in 150 mM NaCl, 20 mM Tris (pH 8.0), 1 mM βME. The fractions of mTRPV3 reconstituted into nanodiscs were pooled and concentrated using 100 kDa NMWL centrifugal filter (MilliporeSigma™ Amicon™) to 2–3 mg/ml and 2–4 mM EGTA was added to the sample.
cNW11 circularized nanodiscs were prepared as described previously28 and stored (~2–3 mg/ml) before usage at −80°C in 20 mM Tris (pH 8.0) and 150 mM NaCl. Purified mTRPV3 was incorporated into cNW11 circularized nanodiscs using the same procedure described above for the MSP2N2 nanodiscs.
Cryo-EM sample preparation and data collection
Au/Au grids were prepared as described in the literature51. Briefly, grids were prepared by first coating C-flat (Protochips, Inc., Morrisville, NC) CF-1.2/1.3–2Au mesh holey carbon grids with ~60 nm gold using an Edwards Auto 306 evaporator. Subsequently, an Ar/O2 plasma treatment (4 min, 50 watts, 35.0 sccm Ar, 11.5 sccm O2) was used to remove the carbon with a Gatan Solarus (model 950) Advanced Plasma Cleaning System (Gatan, Pleasanton, CA, USA). The grids were again plasma treated (H2/O2, 25 s, 10 watts, 6.4 sccm H2, 27.5 sccm O2) prior to sample application to make their surfaces hydrophilic. These grids were used for MSP2N2-reconstituted specimens and specimens in detergent. For cNW11-reconstituted specimens, UltrAuFoil R 1.2/1.3, Au 300 grids were used. Prior to sample application and subsequent plunge-freezing, grids were plasma treated in a PELCO easiGlow glow discharge cleaning system (0.39 mBar, 15 mA, ‘glow’ 25 s, ‘hold’ 10 s).
Before subjecting TRPV3 protein to cryo-EM sample preparation, it was tested in different heat application protocols followed by tetrameric peak detection using Fluorescence-detection Size-Exclusion Chromatography (FSEC)52,53. Only those protocols were used, during which the tetrameric peak amplitude was not significantly reduced. For heat stimulation cryo-EM experiments the samples were pre-incubated in T100 Thermal Cycler (Bio-Rad) programmed to generate repetitive constant temperature steps (30 s at 25°C and 30 s at 42°C). After incubating the sample for 10 to 20 min using repetitive temperature pulses, 3 μl of the sample was immediately transferred to a Mark IV Vitrobot (Thermo Fisher Scientific) equilibrated at 42°C and 100% humidity, applied to a goal-coated side of the grid, and plunge-freezing was done in liquid ethane using a blot time of 3–5 s, a blot force of 3–5 and a wait time of 30 s. The samples without heat stimulation were prepared using the method described above, except for the samples were pre-incubated for 30 min on ice before plunge-freezing, and the temperature inside the Vitrobot was set to 4°C. The grids were stored in liquid nitrogen before imaging.
All images were collected on Titan Krios transmission electron microscopes (TEMs) (Thermo Fisher Scientific) operating at 300 kV and equipped with a post-column GIF Quantum energy filter and a Gatan K3 Summit direct electron detection (DED) camera (Gatan, Pleasanton, CA, USA). Images of frozen-hydrated particles of mTRPV3 in MSP2N2 nanodiscs (1.7 mg/ml) at 42°C were collected using Leginon54. Total number of 20,408 micrographs were collected in counting mode with an image pixel size of 0.858 Å and a defocus range of −0.8 to −2.0 μm. The total dose of ~58 e−Å−2 was attained by using a dose rate of ~16 e−pixel−1s−1 across 50 frames for 2.5 s total exposure time. Images of frozen-hydrated particles of mTRPV3 in MSP2N2 nanodiscs (1.6 mg/ml) at 4°C were collected using SerialEM. Total number of 9,702 micrographs were collected in counting mode with an image pixel size of 0.826 Å and a defocus range of −0.8 to −2.0 μm. The total dose of ~58 e−Å−2 was attained by using a dose rate of ~11.6 e−pixel−1s−1 across 50 frames for 2.5 s total exposure time.
Images of frozen-hydrated particles of mTRPV3 in cNW11 nanodiscs (2.4–3.2 mg/ml) at 42°C were collected in three sessions using Leginon or EPU software (Thermo Fisher). Total number of 18,667 micrographs were collected in counting mode with an image pixel size of 0.87 or 0.873 Å and a defocus range of −0.5 to −2.0 μm, and energy filter slit of 20 eV. The total dose of ~58–60 e−Å−2 was attained by using a dose rate of ~16 e−pixel−1s−1 across 50 frames for a 2.5 s total exposure time. Images of frozen-hydrated particles of mTRPV3 in cNW11 nanodiscs (2.4 mg/ml) at 4°C were collected using Leginon. Total number of 10,878 micrographs were collected in counting mode with an image pixel size of 1.083 Å and a defocus range of −0.8 to −2.0 μm, and energy filter slit of 20 eV. The total dose of 51.07 e−Å−2 was attained by using a dose rate of ~30 e−pixel−1s−1 across 40 frames for a 2.0 s total exposure time.
Image processing
All processing were completed in RELION55 and/or cryoSPARC56 (Table 1). The initial drift and beam-induced motion were corrected using MotionCor257 algorithm implemented in RELION55, and contrast transfer function (CTF) estimation was performed using Gctf58. Following CTF estimation, micrographs were manually inspected and those with outliers in defocus values, ice thickness, and astigmatism as well as micrographs with lower predicted CTF-correlated resolution (> 6 Å) were excluded from the rest of the processing pipeline (individually assessed for each parameter relative to overall distribution; no set threshold). Initial set of particles was picked using 4.48 Å cryo-EM map of mTRPV3 (EMD-20494)12 for 3D reference-based picking in RELION and further classified into 100 two-dimensional (2D) classes. A selection of 2D classes was used to generate templates to be used for the final round of template-based picking. Picked particles were further 2D- and 3D-classified in iterative classification and selection rounds with 20 Å lowpass-filtered map EMD-20494 as a starting reference model using RELION55 and/or cryoSPARC56. The reported resolutions of the final maps were estimated using the gold standard Fourier shell correlation in RELION55. The local resolution predictions were calculated in RELION, with the resolution range estimated by the gold standard FSC = 0.143 criterion55. Map versus model FSC curves with and without mask were calculated using Mtriage as part of Phenix package59. EM density visualization was done in UCSF Chimera60 and UCSF ChimeraX61.
As a representative of the image processing workflow, data for TRPV3 in cNW11 nanodiscs was processed as follows (see also Extended Data Fig. 6). Initially, 27119 particles were manually picked from 273 random picked micrographs in RELION55 previously published 4.48 Å cryo-EM map of mTRPV3 (EMD-20494)12 as s reference to generate 2D classes that were subsequently used as templates to automatically pick a total of 1,202,279, 2,886,493 and 2,780,712 particles from three separate collections. The extracted 4x-binned particle images were separately imported to cryoSPARC and subjected to several iterative rounds of 2D classification and heterogeneous refinement (3D classification). The particle images from the final 2D selections (131,394, 144,122 and 70,362, respectively) were imported separately back to RELION, re-extracted using the original box size of 256 pixels (without binning) and subjected to 3D auto-refinement with C1 symmetry using a box- and pixel-size corrected, previously published 4.48 Å cryo-EM map of mTRPV3 (EMD-20494)12 as a template volume. The aligned particles were CTF refined to estimate beamtilt, trefoil, 4th order aberrations, anisotropic magnification, and to fit per-particle defocus and per-micrograph astigmatism. The CTF-refined particles were further subjected to Bayesian polishing. Polished particles from three collections were joined and refined with the C1 symmetry to get a consensus 4.12-Å resolution map. Aligned particles from the previous step were subjected to 3D classification into eight classes with no symmetry imposed using the consensus map as a template. Particles from two best classes (36,446 and 39,172 in total) were refined with the C4 symmetry, with the respective map from last 3D classification step as a template. The particles form the class with an obvious closed pore was CTF-refined again as it was described previously and refined with the C4 symmetry using a soft mask covering the protein and nanodisc densities. The final post-processing with the soft mask resulted in a 3.09 Å map that represented the sensitized state of mTRPV3. The particles form the class with an obvious open pore was CTF-refined as was described previously and refined with the C4 symmetry using a soft mask covering the protein and nanodisc densities. To improve the quality of putative lipid densities, the refined map from the previous step was subjected to focussed 3D classification into four classes with a mask around transmembrane part of the protein. The total of 27,805 particles from the best class were refined with the C4 symmetry and postprocessed, which resulted in 3.48-Å resolution map. The final step of density-modification procedure62 was used to further enhance the quality of putative lipid densities that resulted in 3.45 Å map of mTRPV3 in the open state.
Model building
To build models of TRPV3 in Coot63, we used the previously published cryo-EM structures of TRPV3 as guides12,27. The models were tested for overfitting by shifting their coordinates by 0.5 Å (using Shake) in Phenix64, refining each shaken model against a corresponding unfiltered half map, and generating densities from the resulting models in Chimera. Structures were visualized, and figures were prepared in UCSF Chimera60, UCSF ChimeraX61, and Pymol65.
MD simulations
Structural models of TRPV3 in the closed, sensitized and open states were immersed into a fully hydrated lipid bilayer with the molecular composition close to composition of HEK 293 cell membrane66: 25% palmitoyloleoylphosphatidylcholine (PC), 15% palmitoyloleoylphosphatidylethanolamine (PE), 15% palmitoyloleoylphosphatidylserine (PS), 15% palmitoyloleoylphosphatidylglycerol (PG), 5% 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphoinositol (PI), 10% sphingomyelin (SMP) and 30% cholesterol (about 870 molecules in total) using the in-house IMPULSE software. About 180 sodium ions were added for electroneutrality. Because of the gap between the residues 76 and 113 in the open-state structure, the 58–76 segments were treated as independent polypeptides with added N-acetyl and methylamide terminal groups. Six different lipids (PC, PE, PS, PG, PI and cholesterol) were inserted into the vanilloid site of the closed-state structure using the cryo-EM structure-modeled lipid as a template (Fig. 4).
The simulated systems were first equilibrated in several stages: 5 × 104 steps of steepest descent minimization followed by heating from 5 to 315 K during 100 ps MD run and 10 ns of MD run at 315 K with fixed positions of the protein atoms to permit membrane relaxation after insertion of the protein. Then, 500-ns MD production runs were carried out for each system.
MD simulations were performed using the GROMACS 2020.4 package67, Amber99sd-ildn force field68, and TIP3P water model69. Simulations were carried out with an integration time of 2 fs, imposed 3D periodic boundary conditions, constant temperature (315 K) and pressure (1 bar). Electrostatic interactions were evaluated using the particle-mesh Ewald summation (real space cutoff 15 Å and 1.2 Å grid with fourth-order spline interpolation). A twin-range (15/15 Å) spherical cutoff function was employed to treat van der Waals interactions.
Conductance of water and Na+ ions through the selectivity filter and gate was calculated by explicitly accounting for water molecules (ions) consecutively passing through the two planes (in both directions) placed 7–8 Å above and below the residues G638 and I674 that form the narrow constrictions (Fig. 2c–d). Spatial distributions of water molecules and Na+ in the pore and lipids at the vanilloid site were calculated as their densities averaged over the MD trajectory at the time range of 100–500 ns.
Whole-cell patch-clamp recordings
Whole-cell patch-clamp recording were parformed as described previously45. HEK293T cells were cultured in Opti-MEM I medium (Invitrogen) supplemented with 5% fetal bovine serum (PAN-Biotech, Germany). The day before transfection, cells were plated in 24-well plates (2 × 105 cells per well) in 0.5 ml of medium and became confluent on the day of transfection. The cells were transiently co-transfected with 300 ng of plasmid encoding wild-type mouse TRPV3 (in pcDNA3 vector, kind gift of Prof. Michael Caterina, Johns Hopkins University School of Medicine, Baltimore, MD, USA), and with 200 ng of GFP plasmid (Takara, Japan) using the magnet-assisted transfection technique (IBA GmbH, Goettingen, Germany) and then plated on poly-L-lysine-coated glass coverslips. The presented data are representative of comparable results from 19 experiments with 6 independent transfections. The cells were used 24 h after transfection.
Whole-cell membrane currents were recorded by employing an Axopatch 200B amplifier and pCLAMP 10.2 software (Molecular Devices, Sunnyvale, USA). Patch electrodes were pulled from borosilicate glass capillary with a 1.5-mm outer diameter (Science Products GmbH, Germany). The tip of the pipette was heat-polished, and its resistance was 3–6 MΩ. Series resistance was compensated by 50–70% after compensation of fast and slow capacitance. Only one recording was performed on any one coverslip of cells to ensure that recordings were made from cells not previously exposed to heat.
The extracellular control bath solution for recording contained: 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM HEPES, 320 mOsm, adjusted to pH 7.4 with NaOH. The pipette solution contained 150 mM CsCl, 1 MgCl2, 10 HEPES and 5 mM EGTA (295 mOsm, pH 7.4 adjusted with CsOH). A system for rapid superfusion of the cultured cells was used for thermal stimulation and solution application70. Briefly, experimental solutions were driven by gravity from seven different barrels, through automatically controlled valves, to a manifold that consisted of fused silica tubes connected to a common outlet glass capillary. The lower part of the capillary was wrapped with densely coiled copper wire that heated the solution to a chosen final temperature. The volume of solution in the experimental dish and the immersion of the application capillary were maintained at a constant level and an internal table generated after balancing an electrical circuit of the system was utilized, which resulted in a good reproducibility of the heat stimuli. The average speed of temperature changes did not significantly differ throughout all experiments. Voltage commands for heat were generated from Digidata 1440A digitizer using pCLAMP 10.2 software (Molecular Devices, Sunnyvale, CA, USA). Temperature steps from room temperature to > 50°C with pulse duration of 5 s were applied at 5 or 10-s intervals. All chemicals were purchased from Sigma-Aldrich (Merck, Prague, Czech Republic).
Planar lipid bilayer recordings
Planar lipid bilayers measurements were performed as described previously71. Briefly, planar lipid bilayers were formed from a solution of synthetic 1-palmitoyl-2-oleoyl-glycero-3-phosphocoline (POPC) and 1-palmitoyl-2-oleoyl-glycero-3-phosphoethanolamine (POPE; Avanti Polar Lipids) at a 3:1 ratio in n-decane (Sigma-Aldrich). The solution was used to paint a bilayer in an aperture of ~250 μm diameter in a Delrin cup (Warner Instruments) between symmetric aqueous bathing solutions of 150 mM KCl, 0.02 mM MgCl2, and 20 mM HEPES (pH 7.2), in the presence of 4 mM EGTA. All reagents (Sigma-Aldrich) were ultrapure (>99%). Bilayer capacitances were in the range of 50–75 pF.
After the bilayers had formed, the TRPV3 protein was added by painting from the micellar solution of TRPV3 protein (20 ng/ml). Unitary currents were recorded using the Axopatch 200B patch-clamp amplifier (Molecular Devices). The trans solution (command voltage side) was connected to the CV 201A head-stage input, while the cis solution was held at a virtual ground via a pair of matched Ag-AgCl electrodes. Currents through the voltage-clamped bilayers (background conductance, <1 pS) were filtered at the amplifier output (low pass, −3 dB at 10 kHz, 8-pole Bessel response). Data were filtered at 100 Hz through an 8-pole Bessel filter (950 TAF; Frequency Devices) and digitized at 1 kHz with an analog-to-digital converter Digidata 1322A controlled by pClamp10.3 software (Molecular Devices). Single-channel conductance events, all-points histograms, open probability (Po), and other parameters were identified and analyzed with Clampfit10.3 software (Molecular Devices). The experiments were performed in the temperature range from 22 to 42°C. The single-channel conductance measured in our experiments (~146 pS at 42°C) was in the range of conductances (48 – 261 pS) measured in experiments with cells16,20,22,72–77. For the temperature-dependence measurements, the bilayer recording chamber was fitted onto a conductive stage containing a pyroelectric heater/cooler that was controlled by a temperature controller (CL-100; Warner Instruments). Deionized water was circulated through the stage and pumped into the system to remove the generated heat. The temperature of the bath was constantly monitored using a thermoelectric device in the cis chamber (the ground side) and was reliably controlled within ±0.5°C. The temperature coefficients (Q10) for Po were calculated using the following equation:
where X1 and X2 are Po values obtained at T1 and T2 temperatures measured in Kelvins.
Thermodynamics of the channel was determined from the Van’t Hoff plot obtained from the equilibrium constant, Keq, where Keq = Po/(1 – Po). The values for enthalpy and entropy changes were obtained from the linear fit of the plot ln(Keq) = −(ΔH/RT) + (ΔS/R), where R is the gas constant = 1.987 cal/mol • K. Statistical analysis was performed using Origin 9.0 (Microcal Software Inc.). Statistical significance was calculated using One-Way ANOVA followed by Fisher’s least significant difference test. All data are presented as mean ± SEM.
Data availability
All data needed to evaluate the conclusions of the paper are present in the paper or the Supplementary Materials. Cryo-EM density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession codes EMD-23853 (mTRPV3 closed, 4 °C, MSP2N2), EMD-23854 (mTRPV3 closed, 42 °C, MSP2N2), EMD-23855 (mTRPV3 sensitized, 42 °C, MSP2N2), EMD-23856 (mTRPV3 closed, 4 °C, cNW11), EMD-23857 (mTRPV3 closed, 42 °C, cNW11), and EMD-23858 (mTRPV3 open, 42 °C, cNW11) (see Table 1). The corresponding model coordinates have been deposited to the Protein Data Bank (PDB) under accession codes 7MIJ (mTRPV3 closed, 4 °C, MSP2N2), 7MIK (mTRPV3 closed, 42 °C, MSP2N2), 7MIL (mTRPV3 sensitized, 42 °C, MSP2N2), 7MIM (mTRPV3 closed, 4 °C, cNW11), 7MIN (mTRPV3 closed, 42 °C, cNW11), and 7MIO (mTRPV3 open, 42 °C, cNW11) (see Table 1). Source data are provided with this paper.
Extended Data
Extended Data Fig. 1. Temperature-dependent changes in currents and thermodynamics of TRPV3.
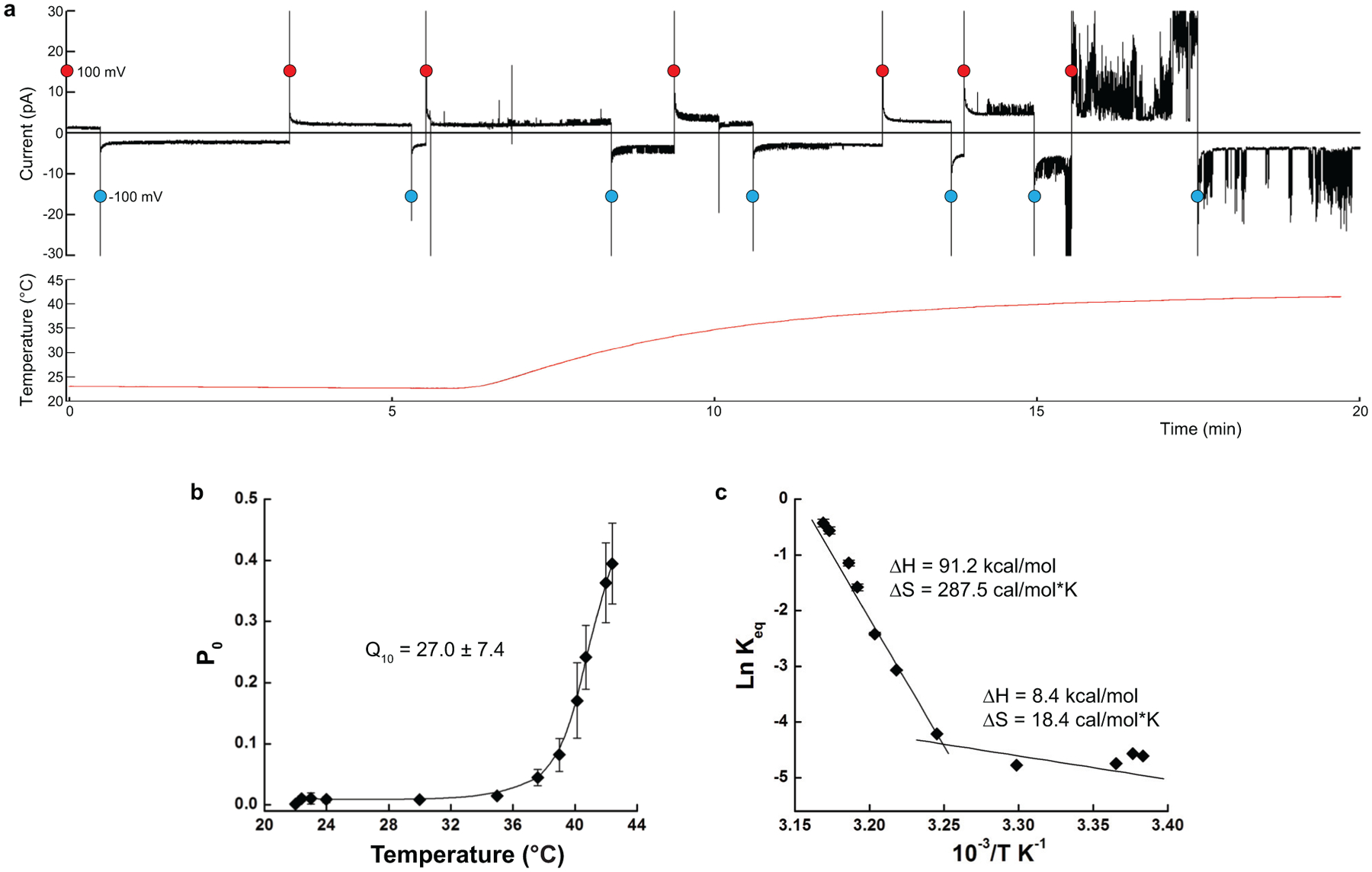
a, A representative continuous recording of current from multiple TRPV3 channels occasionally reconstituted into the synthetic lipid bilayer - (black) in response to the temperature ramp from 22 to 42°C (red), with the membrane potential alternating between +100 mV (red circles) and −100 mV (blue circles). Note the sharp increase in current activity in the 36–42°C temperature range. b, Temperature dependence of the open probability Po at +100 mV calculated using single-channel recordings (see examples in Fig. 1b; n = 19 independent experiments; 189,976 events were analyzed; the data includes the previously published12 and new recordings). Fitting of the data allows estimation of the temperature coefficient, Q10 = 27.0 ± 7.4 (n = 19 independent experiments). c, Van’t Hoff plot of the equilibrium constant Keq calculated using the values of Po (see Methods). Linear fits in the 22–36°C and 36–42°C temperature ranges provide the values of changes in enthalpy and entropy for the temperature-induced activation of TRPV3. Data in b and c are presented as mean values ± SEM. Source data for b and c are available online.
Extended Data Fig. 2. Characteristics of TRPV3 cryo-EM reconstructions.
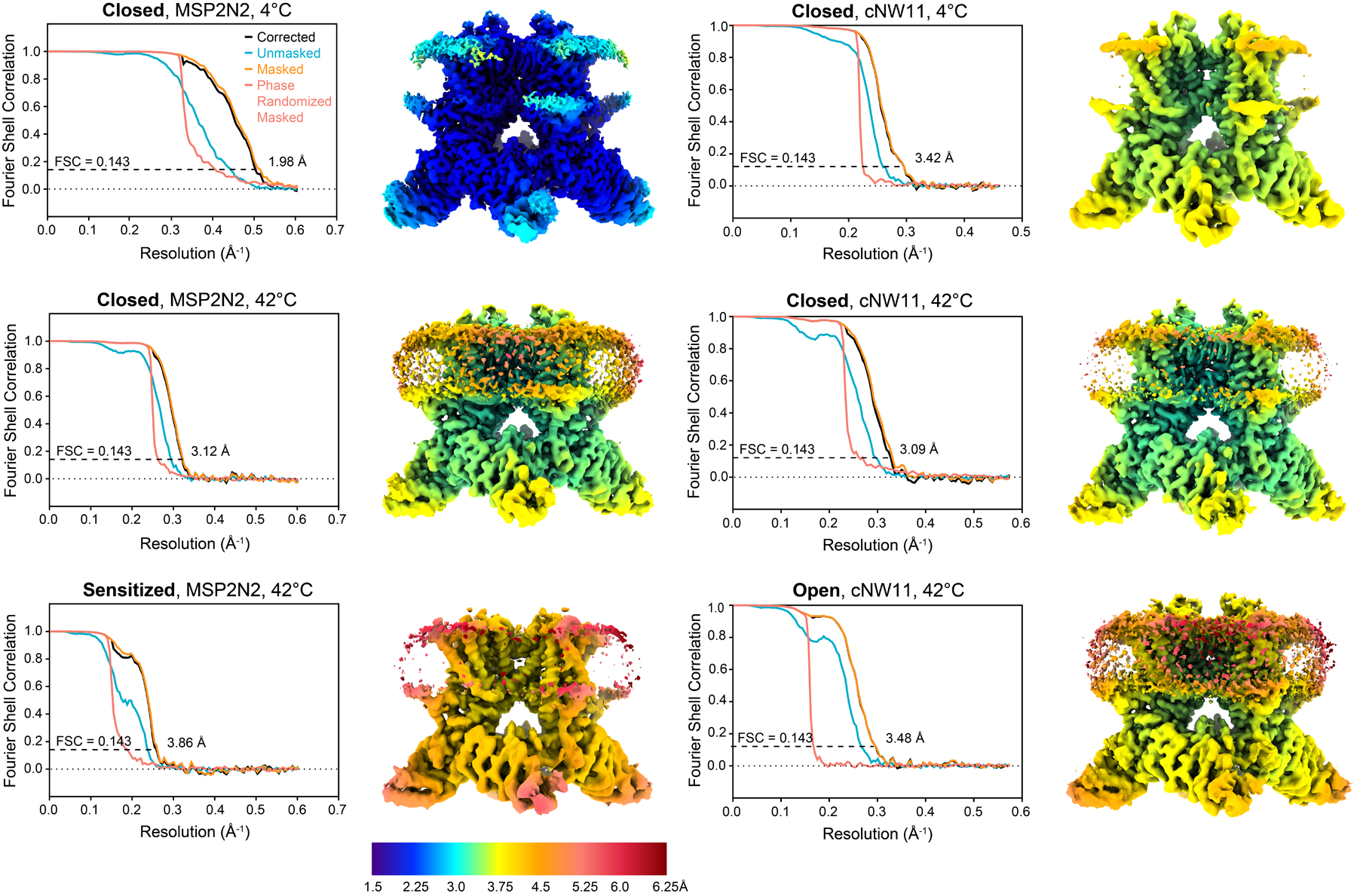
Plots show unmasked, masked and corrected FSC curves calculated between half maps, with the overall resolution estimated using the FSC = 0.143 criterion78. Cryo-EM maps are colored according to the local resolution estimation in Relion55.
Extended Data Fig. 3. Map versus model FSC curves.
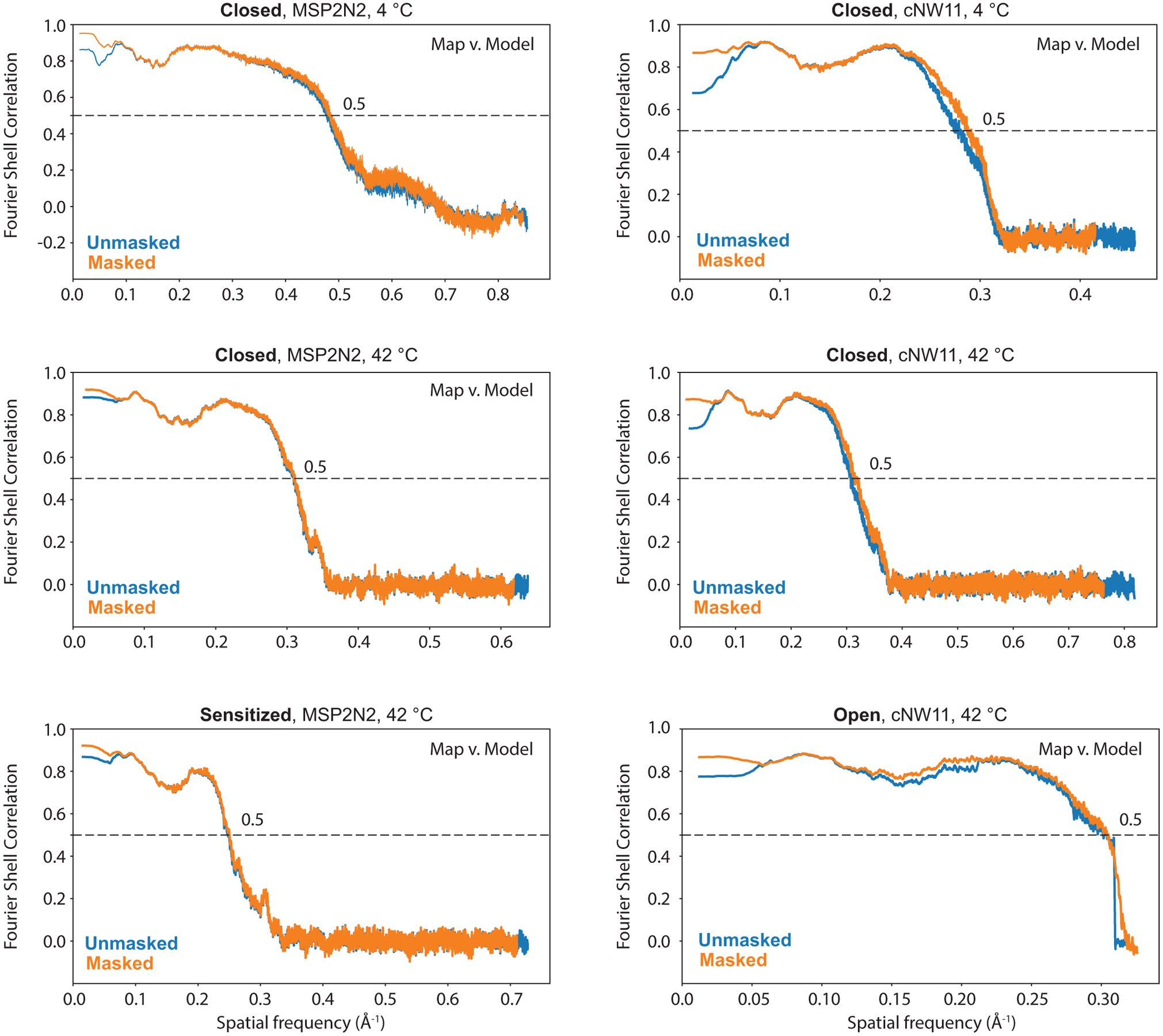
Map versus model FSC curves with and without mask were calculated using Mtriage as part of Phenix package59.
Extended Data Fig. 4. Cryo-EM density of TRPV3.
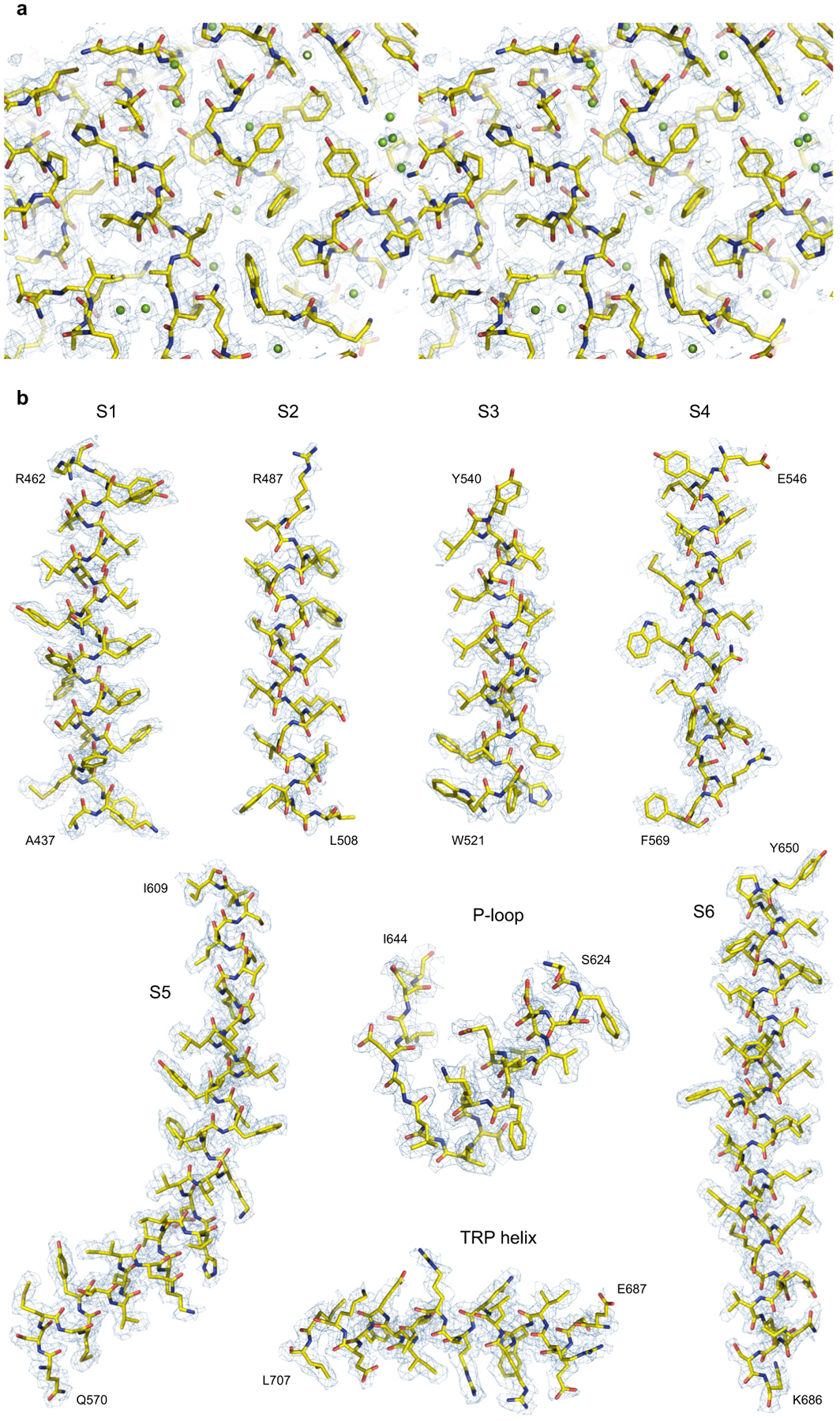
a, Stereo view of an ARD fragment of the 1.98-Å resolution cryo-EM map of TRPV3 reconstituted in MSP2N2 nanodiscs and incubated at 4°C. b, Fragments of the same map for the membrane segments and TRP helix.
Extended Data Fig. 5. Comparison of pore geometry and architecture of TRPV3 structures.
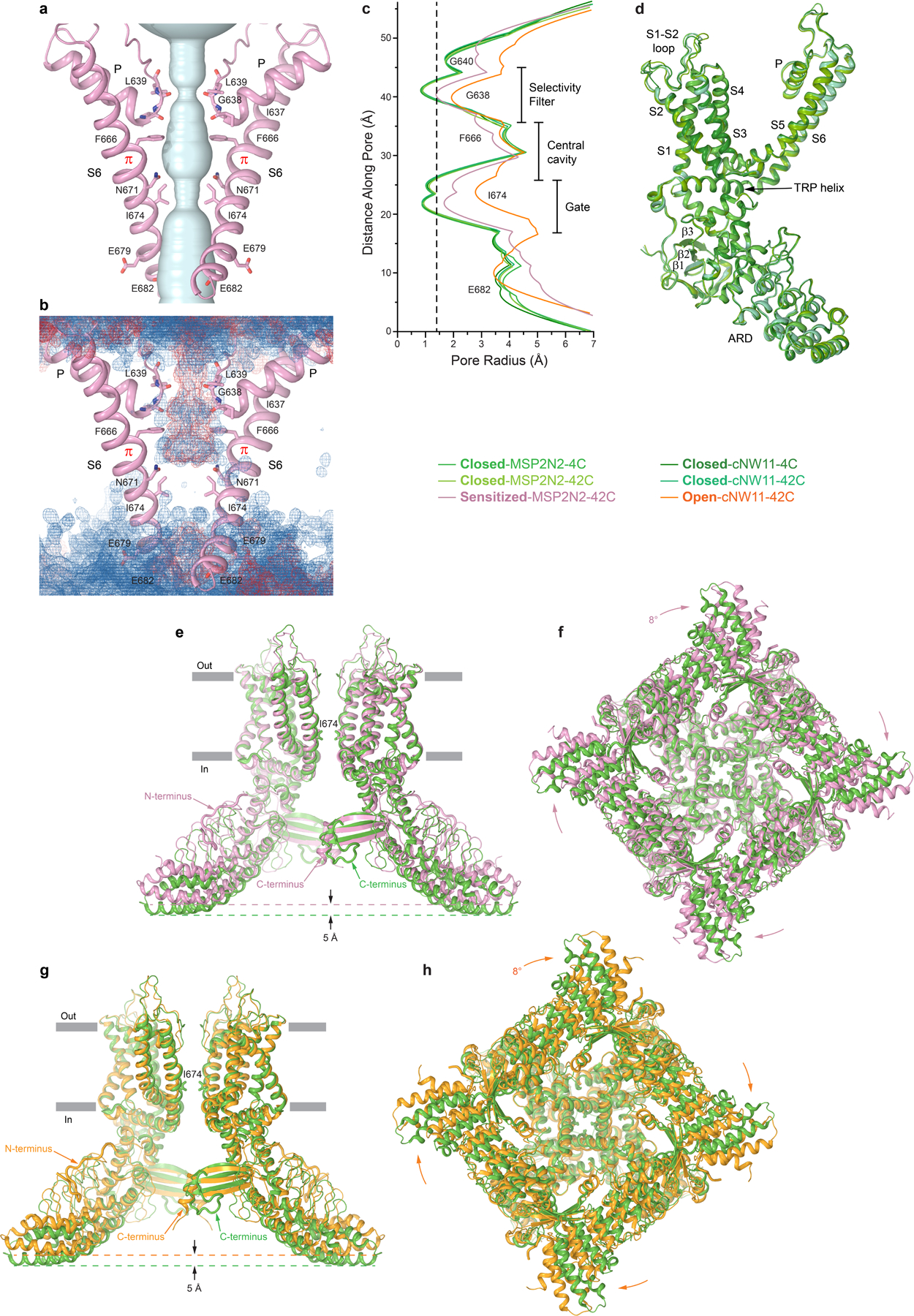
a, Pore-forming domains of TRPV3 in the sensitized state with the residues lining the pore shown as sticks. Only two of four subunits are shown, with the front and back subunits omitted for clarity. The pore profile is shown as a space-filling model (light blue). b, Distribution of water (blue mesh) and ions (red mesh) during MD simulations of TRPV3 in the sensitized state, illustrating the lack of pore permeation. c, The pore radius for different TRPV3 structures calculated using HOLE79. The vertical dashed line denotes the radius of a water molecule, 1.4 Å. d, Superposition of a single subunit from different closed-state structures of TRPV3, with the main structural elements labelled. The root-mean-square deviation (RMSD) calculated for each pair of these subunits ranges between 0.314 and 0.441 Å. e-f, Superposition of TRPV3 structures in the closed (MSP2N2, 4°C; green) and sensitized (pink) states (RMSD, 3.098 Å) viewed parallel to the membrane (e) or intracellularly (f). g-h, Superposition of TRPV3 structures in the closed (MSP2N2, 4°C; green) and open (orange) states (RMSD, 3.293 Å) viewed parallel to the membrane (g) or intracellularly (h). Only two of four subunits are shown in (e) and (g) with the front and back subunits omitted for clarity. Note (e-h) that TRPV3 in the sensitized and open states becomes shorter and its intracellular skirt undergoes a clockwise rotation when viewed intracellularly.
Extended Data Fig. 6. Overview of cryo-EM data collected for mTRPV3 in cNW11 nanodiscs at 42°C and 3D reconstruction workflow.
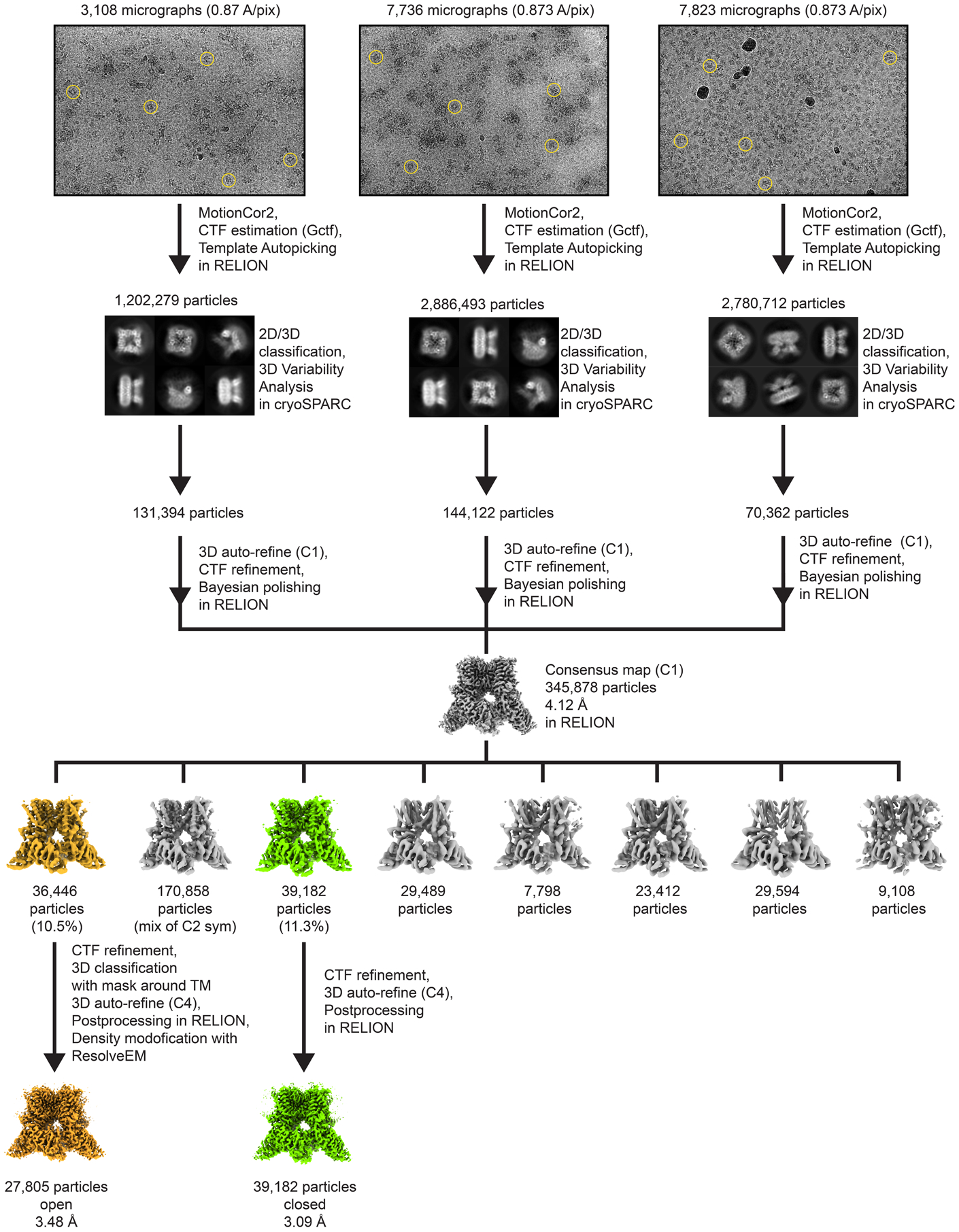
Representative micrographs with example particles circled in yellow and reference-free 2D class averages in different orientations are shown. Three datasets were collected and joined after particle clean-up. All processing steps were done in Relion, except the 2D/3D particle clean-up that was done in cryoSPARC.
Extended Data Fig. 7. Molecular dynamics simulations.
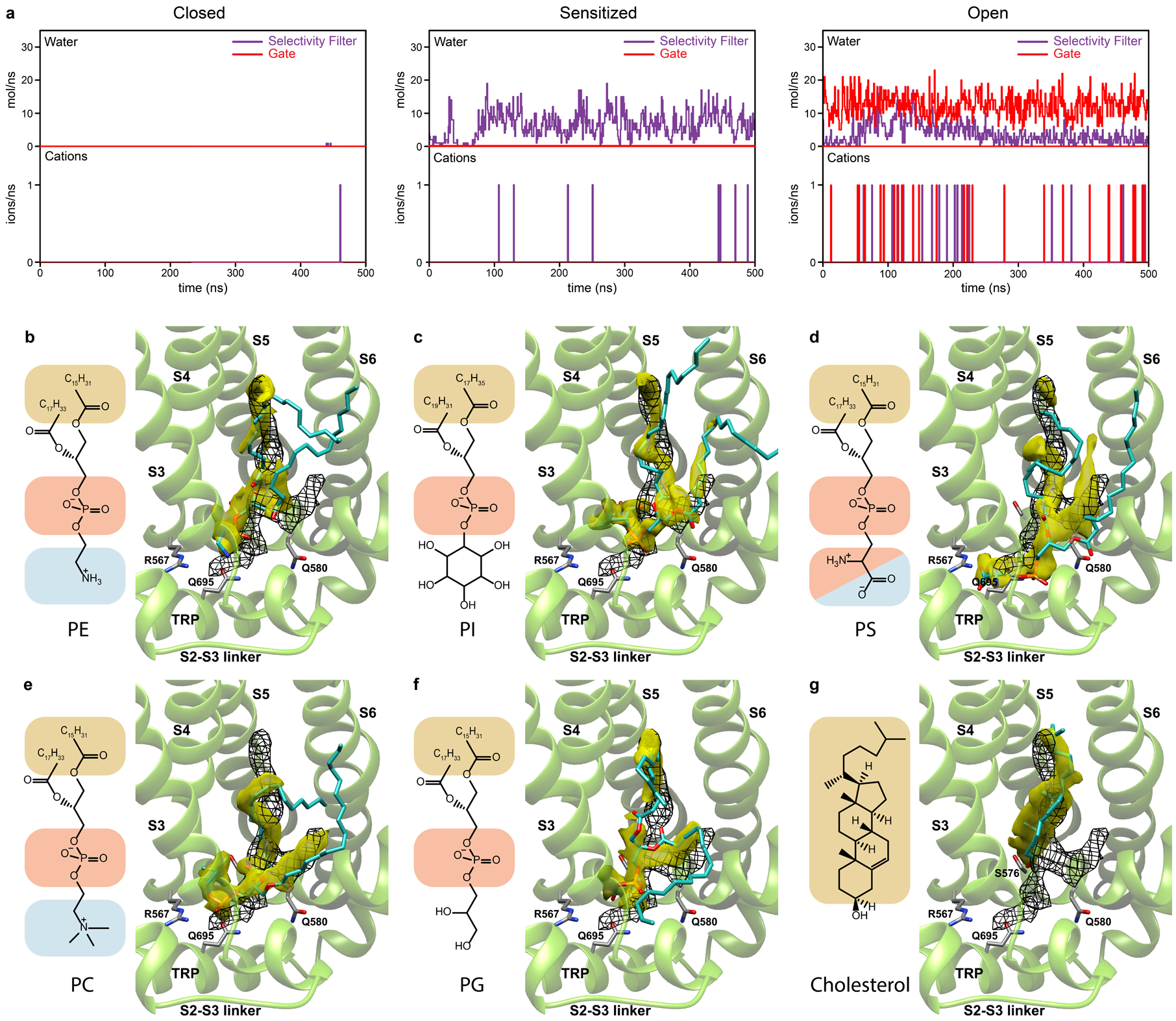
a, Conductance of water and Na+ ions through the selectivity filter and gate of the closed, sensitized and open TRPV3 plotted against the time course of MD simulation. Note that the closed state shows no permeation, the sensitized state permeates water through the selectivity filter only and the open state permeates water and Na+ ions through both selectivity filter and gate. b-g, Averaged MD density distributions (yellow) for non-hydrogen atoms of phosphatidylethanolamine (PE, b), phosphatidylinositol (PI, c), phosphatidylserine (PS, d), phosphatidylcholine (PC, e), phosphatidylglycerol (PG, f) and cholesterol (g) lipids nested in the vanilloid site of the closed TRPV3 at the beginning of 500-ns simulations. Black mesh shows cryo-EM density. MD snapshots of lipid molecules and residues coordinating their heads are shown in sticks. Chemical structures of the lipid molecules are shown to the left of each structural panel. The overlap of MD and EM densities are 26% for PE, 30% for PI, 36% for PS, 30% for PC, 33% for PG and 24% for cholesterol, calculated by multiplying the overlapping volume of MD and cryo-EM densities by two and dividing by their sum.
Extended Data Fig. 8. Comparison of open-state structures of wild-type TRPV3 and Y564A mutant.
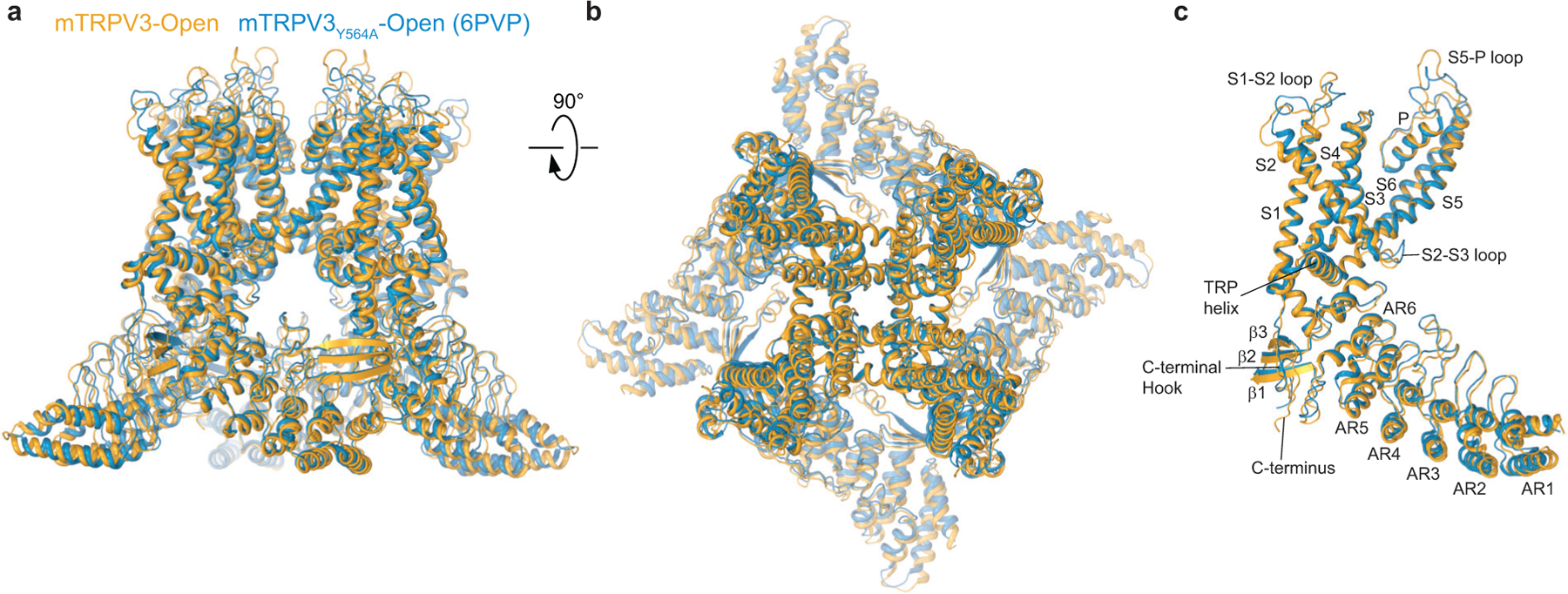
a-b, Overall superposition (RMSD, 2.131 Å) of the open-state structures of wild-type TRPV3 (orange) and previously published Y564A mutant12 (blue, PDB ID: 6PVP) viewed parallel to the membrane (a) and extracellularly (b). c, Single-subunit superposition based on the transmembrane domains (RMSD, 0.943 Å). Note, the most pronounced conformational differences are observed for the S1-S2, S2-S3, S5-P and ARD loops, while the transmembrane domains and TRP helices superpose closely.
Extended Data Fig. 9. Sequence alignment of mouse TRPV channels.
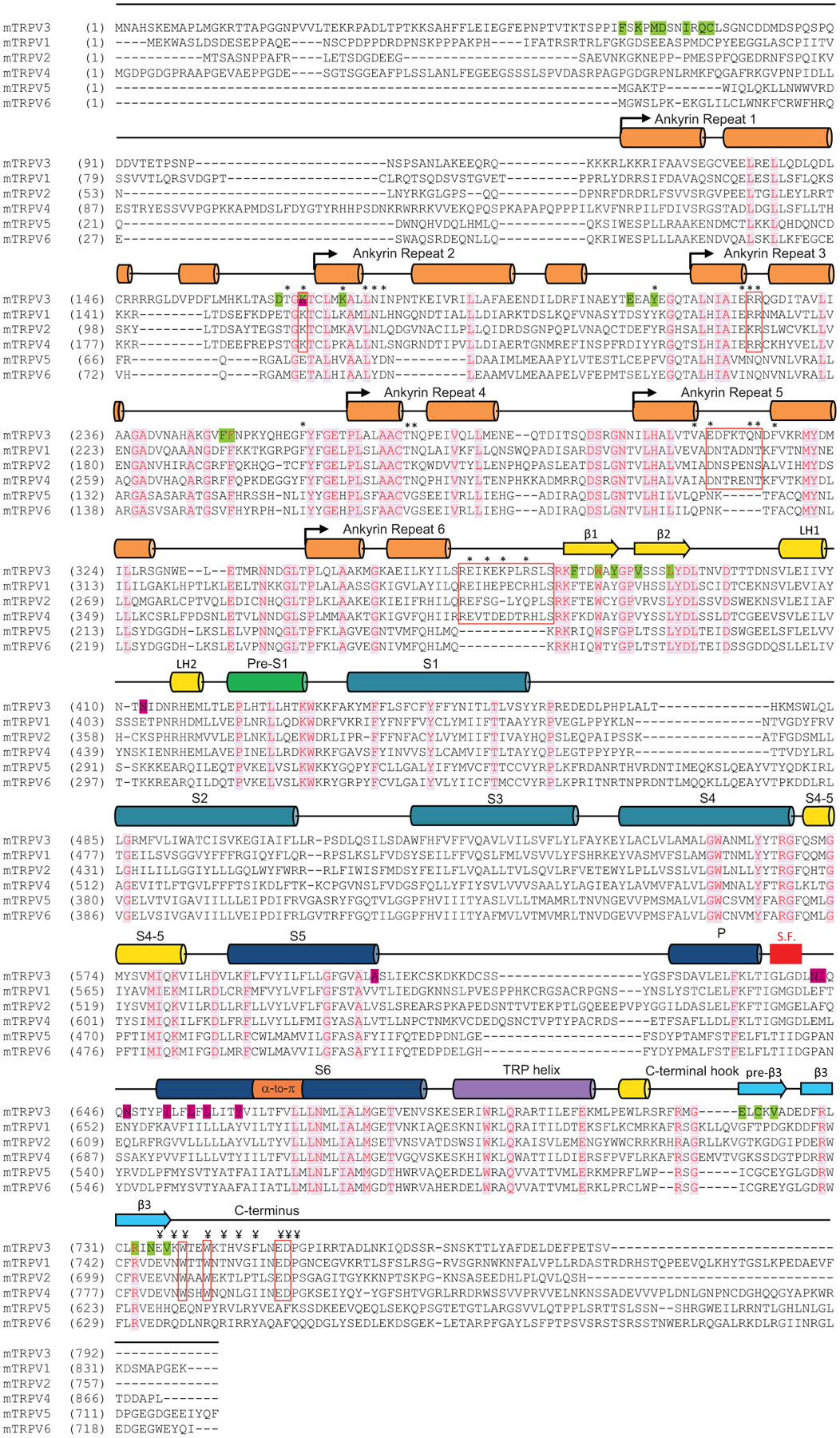
α helices and β strands are depicted above the sequences as cylinders and arrows, respectively. The * symbols indicate residues in the ARD and linker domain that interact with residues in the C-terminus (¥ symbols). Red rectangular outlines denote regions involved in the interaction of the C-terminus with the ARD and linker domain, including residues conserved in thermo-TRPVs, and the AR5 and linker domain loops, which are present in thermo-TRPVs and absent in TRPV5-6. The location of the selectivity filter (S.F.) is indicated by a red box. Identical residues are colored red and highlighted in light pink. Positions of the previously identified mutations in TRPV3 that are critical for thermal sensitivity are highlighted in dark pink.
Extended Data Fig. 10. Conformational changes accompanying temperature-induced opening of wild-type TRPV3.
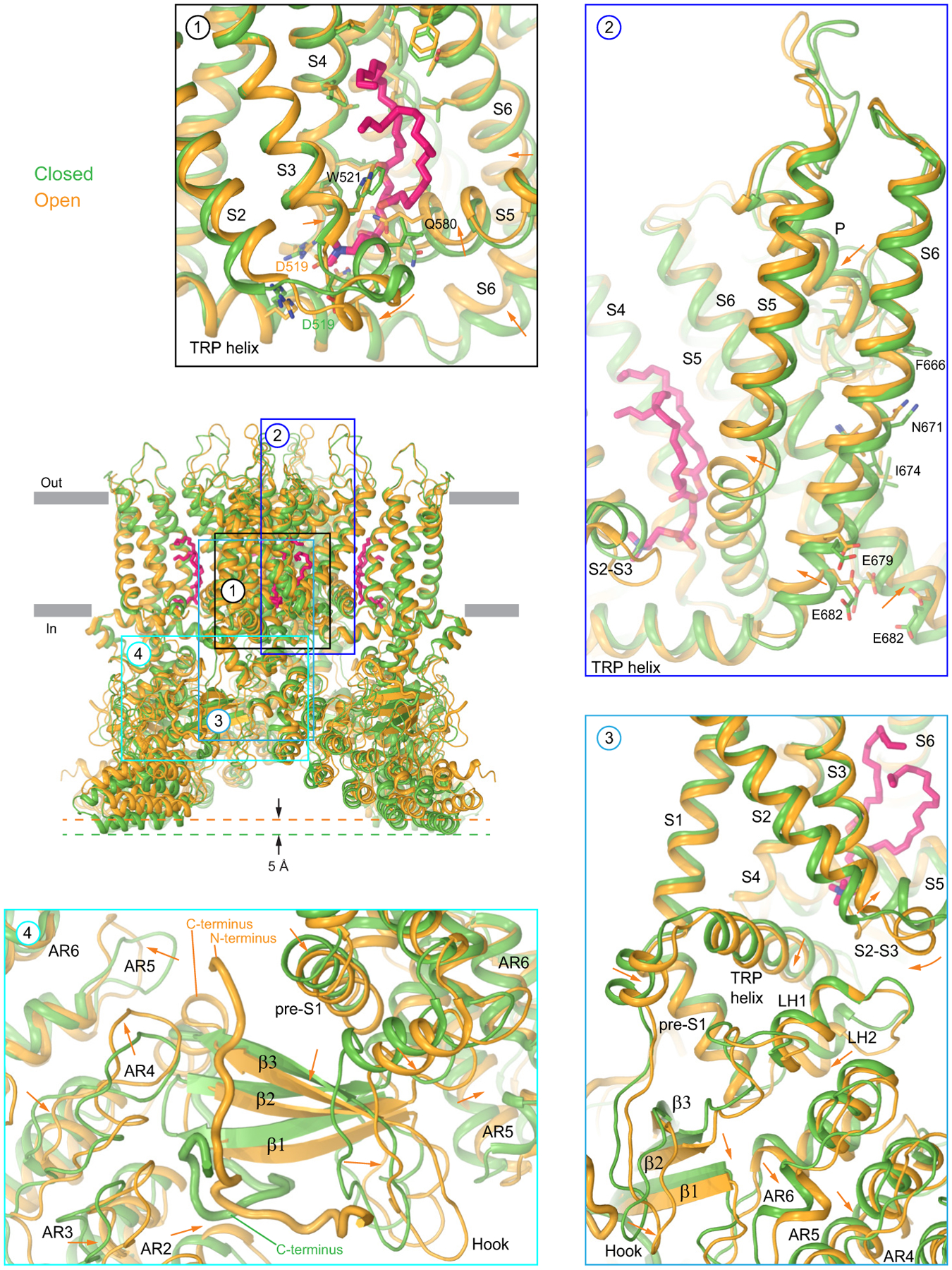
Superposition of the closed- and heat-activated open-state structures of TRPV3 (cNW11, 42°C) viewed parallel to the membrane is shown in the centre. Insets show select regions with the arrows indicating the displacement of domains in the open relative to the closed state. The lipid at the vanilloid site is shown in sticks (pink).
Supplementary Material
Acknowledgments
We thank Sean Mulligan (Pacific Northwest Center for Cryo-EM), Ulrich Baxa and Thomas Edwards (National Cancer Institute, The Frederick National Laboratory), R. Grassucci, Y.-C. Chi, Z. Zhang, C. Wang, J. Wang (Columbia University Cryo-Electron Microscopy Center), H. Kuang (New York Structural Biology Center/ National Center for CryoEM Access and Training) for help with microscope operation and data collection; personnel of the Supercomputer Center “Polytechnical” at the St. Petersburg Polytechnic University for access to the facility and D. Nolde for assistance with supercomputing. This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E. Some of this work was performed at the Columbia University Cryo-Electron Microscopy Center. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. Some of this work was performed at the National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539,) and by grants from the Simons Foundation (SF349247) and NY State Assembly Majority. MD simulations work was supported by the Russian Science Foundation (project #19-74-30014). Water and ions conductivity analysis was supported by the Russian Foundation for Basic Research (project #19-04-00350). Supercomputer calculations were supported within the framework of the HSE University Basic Research Program and the Russian Academic Excellence Project ”5-100”. Whole-cell patch-clamp recordings were supported by Czech Science Foundation (project #19-03777S). A.I.S. was supported by the NIH (R01 CA206573, R01 NS083660, R01 NS107253) and NSF (1818086).
Footnotes
Competing interests: The authors declare no competing interests.
References
- 1.Castillo K, Diaz-Franulic I, Canan J, Gonzalez-Nilo F & Latorre R Thermally activated TRP channels: molecular sensors for temperature detection. Phys Biol 15, 021001, doi: 10.1088/1478-3975/aa9a6f (2018). [DOI] [PubMed] [Google Scholar]
- 2.Voets T TRP channels and thermosensation. Handb Exp Pharmacol 223, 729–741, doi: 10.1007/978-3-319-05161-1_1 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Islas LD Thermal effects and sensitivity of biological membranes. Curr Top Membr 74, 1–17, doi: 10.1016/B978-0-12-800181-3.00001-4 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Lamas JA, Rueda-Ruzafa L & Herrera-Pérez S Ion Channels and Thermosensitivity: TRP, TREK, or Both? Int J Mol Sci 20, 2371, doi: 10.3390/ijms20102371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arrigoni C & Minor DL Global versus local mechanisms of temperature sensing in ion channels. Pflugers Arch 470, 733–744, doi: 10.1007/s00424-017-2102-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhaka A, Viswanath V & Patapoutian A Trp ion channels and temperature sensation. Annu Rev Neurosci 29, 135–161, doi: 10.1146/annurev.neuro.29.051605.112958 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Voets T, Talavera K, Owsianik G & Nilius B Sensing with TRP channels. Nat Chem Biol 1, 85–92, doi: 10.1038/nchembio0705-85 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Latorre R, Zaelzer C & Brauchi S Structure-functional intimacies of transient receptor potential channels. Q Rev Biophys 42, 201–246, doi: 10.1017/S0033583509990072 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Jordt SE, McKemy DD & Julius D Lessons from peppers and peppermint: the molecular logic of thermosensation. Curr Opin Neurobiol 13, 487–492, doi: 10.1016/s0959-4388(03)00101-6 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Clapham DE TRP channels as cellular sensors. Nature 426, 517–524, doi: 10.1038/nature02196 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Yuan P Structural biology of thermoTRPV channels. Cell Calcium 84, 102106, doi: 10.1016/j.ceca.2019.102106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh AK et al. Structural basis of temperature sensation by the TRP channel TRPV3. Nat Struct Mol Biol 26, 994–998, doi: 10.1038/s41594-019-0318-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilius B, Bíró T & Owsianik G TRPV3: time to decipher a poorly understood family member! J Physiol 592, 295–304, doi: 10.1113/jphysiol.2013.255968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang P & Zhu MX Trpv3. Handb Exp Pharmacol 222, 273–291, doi: 10.1007/978-3-642-54215-2_11 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Broad LM et al. TRPV3 in Drug Development. Pharmaceuticals (Basel) 9, 55, doi: 10.3390/ph9030055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung MK, Lee H, Mizuno A, Suzuki M & Caterina MJ 2-aminoethoxydiphenyl borate activates and sensitizes the heat-gated ion channel TRPV3. J Neurosci 24, 5177–5182, doi: 10.1523/JNEUROSCI.0934-04.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao R et al. Calcium plays a central role in the sensitization of TRPV3 channel to repetitive stimulations. J Biol Chem 283, 6162–6174, doi: 10.1074/jbc.M706535200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Yao J, Zhu MX & Qin F Hysteresis of gating underlines sensitization of TRPV3 channels. J Gen Physiol 138, 509–520, doi: 10.1085/jgp.201110689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moqrich A et al. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in the skin. Science 307, 1468–1472, doi: 10.1126/science.1108609 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Xu H et al. TRPV3 is a calcium-permeable temperature-sensitive cation channel. Nature 418, 181–186, doi: 10.1038/nature00882 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Peier AM et al. A heat-sensitive TRP channel expressed in keratinocytes. Science 296, 2046–2049, doi: 10.1126/science.1073140 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Smith GD et al. TRPV3 is a temperature-sensitive vanilloid receptor-like protein. Nature 418, 186–190, doi: 10.1038/nature00894 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Shimada H et al. The structure of lipid nanodisc-reconstituted TRPV3 reveals the gating mechanism. Nat Struct Mol Biol 27, 645–652, doi: 10.1038/s41594-020-0439-z (2020). [DOI] [PubMed] [Google Scholar]
- 24.Deng Z et al. Gating of human TRPV3 in a lipid bilayer. Nat Struct Mol Biol 27, 635–644, doi: 10.1038/s41594-020-0428-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zubcevic L, Borschel WF, Hsu AL, Borgnia MJ & Lee SY Regulatory switch at the cytoplasmic interface controls TRPV channel gating. Elife 8, 47746, doi: 10.7554/eLife.47746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zubcevic L et al. Conformational ensemble of the human TRPV3 ion channel. Nat Commun 9, 4773, doi: 10.1038/s41467-018-07117-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh AK, McGoldrick LL & Sobolevsky AI Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat Struct Mol Biol 25, 805–813, doi: 10.1038/s41594-018-0108-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasr ML et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods 14, 49–52, doi: 10.1038/nmeth.4079 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi DJ, Ye S, Cao X, Zhang R & Wang K Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 reveals unique conformation of finger 3 loop critical for channel function. Protein Cell 4, 942–950, doi: 10.1007/s13238-013-3091-0 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Cao E, Julius D & Cheng Y TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351, doi: 10.1038/nature17964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao E, Liao M, Cheng Y & Julius D TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118, doi: 10.1038/nature12823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu B & Qin F Single-residue molecular switch for high-temperature dependence of vanilloid receptor TRPV3. Proc Natl Acad Sci U S A 114, 1589–1594, doi: 10.1073/pnas.1615304114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao E, Cordero-Morales JF, Liu B, Qin F & Julius D TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron 77, 667–679, doi: 10.1016/j.neuron.2012.12.016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Q Temperature sensing by thermal TRP channels: thermodynamic basis and molecular insights. Curr Top Membr 74, 19–50, doi: 10.1016/B978-0-12-800181-3.00002-6 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Vlachová V et al. Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J Neurosci 23, 1340–1350 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao J, Liu B & Qin F Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc Natl Acad Sci U S A 108, 11109–11114, doi: 10.1073/pnas.1105196108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grandl J et al. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat Neurosci 13, 708–714 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grandl J et al. Pore region of TRPV3 ion channel is specifically required for heat activation. Nat Neurosci 11, 1007–1013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brauchi S, Orio P & Latorre R Clues to understanding cold sensation: thermodynamics and electrophysiological analysis of the cold receptor TRPM8. Proc Natl Acad Sci U S A 101, 15494–15499, doi: 10.1073/pnas.0406773101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brauchi S, Orta G, Salazar M, Rosenmann E & Latorre R A hot-sensing cold receptor: C-terminal domain determines thermosensation in transient receptor potential channels. J Neurosci 26, 4835–4840, doi: 10.1523/JNEUROSCI.5080-05.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F et al. Heat activation is intrinsic to the pore domain of TRPV1. Proc Natl Acad Sci U S A 115, E317–E324, doi: 10.1073/pnas.1717192115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voets T et al. The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature 430, 748–754, doi: 10.1038/nature02732 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Gregorio-Teruel L et al. The Integrity of the TRP Domain Is Pivotal for Correct TRPV1 Channel Gating.Biophys J 109, 529–541, doi: 10.1016/j.bpj.2015.06.039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SE, Patapoutian A & Grandl J Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations. PLoS One 8, e59593, doi: 10.1371/journal.pone.0059593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macikova L, Vyklicka L, Barvik I, Sobolevsky AI & Vlachova V Cytoplasmic Inter-Subunit Interface Controls Use-Dependence of Thermal Activation of TRPV3 Channel. Int J Mol Sci 20, 3990, doi: 10.3390/ijms20163990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laursen WJ, Schneider ER, Merriman DK, Bagriantsev SN & Gracheva EO Low-cost functional plasticity of TRPV1 supports heat tolerance in squirrels and camels. Proc Natl Acad Sci U S A 113, 11342–11347, doi: 10.1073/pnas.1604269113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clapham DE & Miller C A thermodynamic framework for understanding temperature sensing by transient receptor potential (TRP) channels. Proc Natl Acad Sci U S A 108, 19492–19497, doi: 10.1073/pnas.1117485108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jara-Oseguera A & Islas LD The role of allosteric coupling on thermal activation of thermo-TRP channels. Biophys J 104, 2160–2169, doi: 10.1016/j.bpj.2013.03.055 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasimova MA et al. A hypothetical molecular mechanism for TRPV1 activation that invokes rotation of an S6 asparagine. J Gen Physiol 150, 1554–1566, doi: 10.1085/jgp.201812124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goehring A et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc 9, 2574–2585, doi: 10.1038/nprot.2014.173nprot.2014.173 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russo CJ & Passmore LA Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380, doi: 10.1126/science.1259530346/6215/1377 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawate T & Gouaux E Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Hattori M, Hibbs RE & Gouaux E A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299, doi: 10.1016/j.str.2012.06.009S0969-2126(12)00231-6 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suloway C et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60, doi: 10.1016/j.jsb.2005.03.010 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530, doi: 10.1016/j.jsb.2012.09.006S1047-8477(12)00248-1 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296, doi: 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003S1047-8477(15)30100-3 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine PV et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840, doi: 10.1107/S2059798318009324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82, doi: 10.1002/pro.3943 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terwilliger TC, Ludtke SJ, Read RJ, Adams PD & Afonine PV Improvement of cryo-EM maps by density modification. Nat Methods 17, 923–927, doi: 10.1038/s41592-020-0914-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367, doi: 10.1107/S0907444912001308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA, USA, 2002). [Google Scholar]
- 66.Dawaliby R et al. Phosphatidylethanolamine Is a Key Regulator of Membrane Fluidity in Eukaryotic Cells. J Biol Chem 291, 3658–3667, doi: 10.1074/jbc.M115.706523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abraham MJ & Gready JE Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J Comput Chem 32, 2031–2040, doi: 10.1002/jcc.21773 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Lindorff-Larsen K et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958, doi: 10.1002/prot.22711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jorgensen WL & Tirado-Rives J Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci U S A 102, 6665–6670, doi: 10.1073/pnas.0408037102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dittert I et al. Improved superfusion technique for rapid cooling or heating of cultured cells under patch-clamp conditions. J Neurosci Methods 151, 178–185, doi: 10.1016/j.jneumeth.2005.07.005 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Cao C, Zakharian E, Borbiro I & Rohacs T Interplay between calmodulin and phosphatidylinositol 4,5-bisphosphate in Ca2+-induced inactivation of transient receptor potential vanilloid 6 channels. J Biol Chem 288, 5278–5290, doi: 10.1074/jbc.M112.409482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo J, Stewart R, Berdeaux R & Hu H Tonic inhibition of TRPV3 by Mg2+ in mouse epidermal keratinocytes. J Invest Dermatol 132, 2158–2165, doi: 10.1038/jid.2012.144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao X, Yang F, Zheng J & Wang K Intracellular proton-mediated activation of TRPV3 channels accounts for the exfoliation effect of alpha-hydroxyl acids on keratinocytes. J Biol Chem 287, 25905–25916, doi: 10.1074/jbc.M112.364869 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H et al. Mechanisms of proton inhibition and sensitization of the cation channel TRPV3. J Gen Physiol 153, e202012663, doi: 10.1085/jgp.202012663 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi SW et al. The novel high-frequency variant of TRPV3 p.A628T in East Asians showing faster sensitization in response to chemical agonists. Pflugers Arch 471, 1273–1289, doi: 10.1007/s00424-019-02309-9 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Doerner JF, Hatt H & Ramsey IS Voltage- and temperature-dependent activation of TRPV3 channels is potentiated by receptor-mediated PI(4,5)P2 hydrolysis. J Gen Physiol 137, 271–288, doi: 10.1085/jgp.200910388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schrapers KT, Sponder G, Liebe F, Liebe H & Stumpff F The bovine TRPV3 as a pathway for the uptake of Na(+), Ca(2+), and NH4(+). PLoS One 13, e0193519, doi: 10.1371/journal.pone.0193519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henderson R et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214, doi: 10.1016/j.str.2011.12.014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smart OS, Neduvelil JG, Wang X, Wallace BA & Samsom MS HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions of the paper are present in the paper or the Supplementary Materials. Cryo-EM density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession codes EMD-23853 (mTRPV3 closed, 4 °C, MSP2N2), EMD-23854 (mTRPV3 closed, 42 °C, MSP2N2), EMD-23855 (mTRPV3 sensitized, 42 °C, MSP2N2), EMD-23856 (mTRPV3 closed, 4 °C, cNW11), EMD-23857 (mTRPV3 closed, 42 °C, cNW11), and EMD-23858 (mTRPV3 open, 42 °C, cNW11) (see Table 1). The corresponding model coordinates have been deposited to the Protein Data Bank (PDB) under accession codes 7MIJ (mTRPV3 closed, 4 °C, MSP2N2), 7MIK (mTRPV3 closed, 42 °C, MSP2N2), 7MIL (mTRPV3 sensitized, 42 °C, MSP2N2), 7MIM (mTRPV3 closed, 4 °C, cNW11), 7MIN (mTRPV3 closed, 42 °C, cNW11), and 7MIO (mTRPV3 open, 42 °C, cNW11) (see Table 1). Source data are provided with this paper.


