Abstract
U2AF1 is involved in the recognition of the 3’ splice site during pre-mRNA splicing. Mutations in U2AF1 are frequently observed in myelodysplastic syndromes. However, the role of wild-type U2AF1 in normal hematopoiesis has remained elusive. Using a novel conditional U2af1 knockout allele, we have found that deletion of U2af1 results in profound defects in hematopoiesis characterized by pancytopenia, ablation of hematopoietic stem/progenitor cells (HSPC) leading to bone marrow failure and early lethality in mice. U2af1 deletion impairs HSPC function and repopulation capacity. U2af1 deletion also causes increased DNA damage and reduced survival in hematopoietic progenitors. RNA sequencing analysis reveals significant alterations in the expression of genes related to HSC maintenance, cell proliferation and DNA damage response-related pathways in U2af1-deficient HSPC. U2af1 deficiency also induces splicing alterations in genes important for HSPC function. This includes altered splicing and perturbed expression of Nfya and Pbx1 transcription factors in U2af1-deficient HSPC. Collectively, these results suggest an important role for U2af1 in the maintenance and function of HSPC in normal hematopoiesis. A better understanding of the normal function of U2AF1 in hematopoiesis is important for development of appropriate therapeutic approaches for U2AF1 mutant induced hematologic malignancies.
Keywords: Hematopoiesis, Hematopoietic stem cells, RNA splicing, U2AF1
Introduction
Somatic mutations in the RNA spliceosome genes (U2AF1, SRSF2, SF3B1 and ZRSR2) have been frequently observed in patients with myelodysplastic syndromes (MDS) [1–6]. These spliceosomal gene mutations are thought to play causal roles in clonal and aberrant hematopoiesis in MDS [1, 3, 5–7]. U2AF1, the gene encoding an RNA-binding protein, is involved in the recognition of the 3’ splice site required for the recruitment of the U2 snRNP during pre-mRNA splicing [1, 3, 8]. Mutations in U2AF1 have been identified in ~11% cases of MDS [1, 3]. U2AF1 mutations also have been associated with poor clinical outcomes in MDS and increased risk of transformation into secondary AML [9]. Most U2AF1 mutations are recurrent and found in two hotspots (S34 and Q157) within the first and second zinc finger domains [1, 3]. Several studies have suggested splicing changes in cells expressing mutant U2AF1 [10–14]. Retroviral expression of U2AF1 S34F mutant in human bone marrow CD34+ progenitors impairs erythroid differentiation and skews granulomonocytic differentiation toward granulocytes [14]. Transgenic and knock-in mice expressing U2AF1 S34F mutant exhibit aberrant hematopoiesis with MDS-like features [15, 16]. Although these studies have provided some insights into the effects of U2AF1 S34F mutant in the pathogenesis of MDS, the role of wild-type U2AF1 in regulating hematopoietic stem/progenitor cell (HSPC) function and normal hematopoiesis has remained unknown.
To determine the role of U2AF1 in normal hematopoiesis, we generated a conditional U2af1 knockout (floxed) allele. We found that deletion of U2af1 in mouse hematopoietic compartment leads to profound defects in hematopoiesis resulting in pancytopenia, bone marrow aplasia and early deaths in mice. U2af1 deficiency severely impairs HSPC function. Moreover, U2af1-deficient HSPC are unable to compete with wild-type HSPC in competitive repopulation assay. We also found the hematopoietic defects in U2af1-deficient mice is cell autonomous. Furthermore, U2af1 deficiency increases DNA damage and cell death in hematopoietic progenitor and precursor cells. Deletion of U2af1 induces splicing alterations of genes important for HSPC function. Overall, our results suggest a crucial role for U2af1 in the maintenance and function of HSPC in normal hematopoiesis.
Materials and Methods
Generation of conditional U2af1 knockout mice
To generate the conditional U2af1 knockout (floxed) allele, the exon 2 of U2af1 gene is targeted in embryonic stem (ES) cells by inserting a construct containing two LoxP sites between introns 1 and 2 of the U2af1 gene using homologous recombination. The U2af1 floxed allele can be deleted by the expression of Cre recombinase. Two correctly targeted ES clones were injected into C57BL/6 blastocysts. Both clones gave rise to germline transmission. The U2af1 floxed mice were crossed with the Mx1Cre mice [17] (Jackson Laboratory) to obtain Mx1Cre;U2af1fl/fl mice. To delete U2af1 in hematopoietic compartments, Cre expression was induced in Mx1Cre;U2af1fl/fl mice by intraperitoneal injection of 3 doses of 200 μg polyinosine-polycytosine (pI-pC) (GE healthcare) at 5 to 6 weeks after birth. All mice used in this study were on a C57BL/6 background. Littermate controls were used for all experiments, unless stated otherwise. Sequences of the genotyping primers are available in the supplemental Table 1. All animal studies were performed in accordance with the guidelines approved by the IACUC of the University of Virginia School of Medicine.
Cell lines and cell culture conditions
Human HEL and SET2 cells were grown in RPMI 1640 Media supplemented with 10% FBS and antibiotics (penicillin/streptomycin). Human MDS-L cells were grown in RPMI 1640 Media supplemented with 10% FBS, 50ng/ml human IL-3 and antibiotics (penicillin/streptomycin). Mouse BA/F3 cells were grown in RPMI 1640 Media supplemented with 10% FBS, 1 ng/ml mouse IL-3 and antibiotics (penicillin/streptomycin).
Plasmids
Glycerol stocks of pLKO.1 Human U2AF1 shRNAs (TRCN0000001156 and TRCN0000001158) and pLKO.1 mouse U2af1 shRNAs (TRCN0000123551 and TRCN0000123553) were purchased from Dharmacon (Horizon Discovery). HDVIR-Pbx1 construct was kindly provided by Dr. Licia Selleri (UCSF, USA). pSG5-Nfya construct was kindly provided by Dr. Roberto Mantovani (University of Milano, Italy). Nfya cDNA was sub-cloned into lentiviral pCDH vector at BamH1 and EcoR1 sites and confirmed by sequencing.
In vitro knockdown and apoptosis assays
For knockdown of U2AF1, cells were transduced with the lentiviruses expressing U2AF1 shRNAs or scramble (control) shRNA and infected cells were selected using puromycin (2μg/mL). Cell proliferation was assessed every day over 4 to 5 days by viable cell counts using trypan blue exclusion. Apoptosis assay was performed by Annexin V staining and evaluated by flow cytometry.
Bone marrow transplantation
For cell autonomous BM transplantation (BMT) assay, BM cells (1X106) from 6 weeks old Mx1Cre;U2af1fl/fl mice were injected into lethally irradiated (2X 550 cGy) C57BL/6 recipient mice. For competitive repopulation assay, BM cells from uninduced control (U2af1fl/fl; no cre) and Mx1Cre;U2af1fl/fl mice (CD45.2+) were mixed with CD45.1+ competitor BM cells at a ratio of 1:1 and then transplanted into lethally irradiated (CD45.1+) congenic mice. Four weeks after transplantation, recipients were injected with 3 doses of pI-pC. The chimerism in the BM of transplanted animals was assessed by CD45.2 and CD45.1 expression.
Colony-forming assays
BM (2 X104) cells were plated in duplicates in cytokine-supplemented complete methylcellulose medium (MethoCult M3434; StemCell Technologies). Burst forming units-erythroid (BFU-E), granulocyte-macrophage colony-forming units (CFU-GM) colonies were scored on day 7. To detect colony-forming units-megakaryocyte (CFU-Mk), BM cells were plated in collagen-based MegaCult medium (StemCell Technologies) in the presence of IL-3, IL-6, IL-11, and Tpo. CFU-Mk colonies were scored after 7–8 days according to manufacturer’s protocol. In some cases, BM cells from WT and U2af1cKO mice were transduced with empty pCDH lentiviral vector or lentivirus expressing NFYa or Pbx1. Infected cells were selected with 2.5μg/mL puromycin for 36 hours and plated in duplicates in methylcellulose medium containing complete cytokines or Epo (3U/ml). Myeloid progenitor colonies (CFU-GM, CFU-GEMM) and CFU-E colonies were counted.
Blood and tissue analysis
Peripheral blood counts were measured using Hemavet 950FS (Drew Scientific). For histologic analysis, mouse tissue specimens were fixed in 10% neutral buffered formalin and embedded in paraffin. Tissue sections (4 μm) were stained with hematoxylin and eosin (H&E).
Flow cytometry
Single-cell suspensions were prepared from BM and spleen, and red cells were lysed with red cell lysis solution. Cells were washed and resuspended in PBS plus 2% FBS and stained for 30 minutes on ice with directly conjugated (either PE or APC) monoclonal antibodies specific for Ter119, CD71, CD41, CD61, Mac-1, Gr-1, B220, or Tcrβ. For HSC/progenitor analysis, BM cells were stained for 60 minutes on ice with antibodies against c-Kit, Sca-1, Flk2 (CD135), CD34, CD16/32 (FcγR II/III), and antibodies against lineage (Lin) markers including CD3, CD4, CD8, CD19, B220, Gr-1, Ter119, and IL-7R (CD127). For competitive BM repopulation assays, PE-CD45.1 and FITC-CD45.2 conjugated antibodies were used for flow cytometry. FITC-conjugated anti-Annexin V and DAPI were used for apoptosis assays. All antibodies were purchased from eBioscience (Invitrogen) or BioLegend. Flow cytometry was performed with LSR Fortessa (BD Biosciences) and analyzed using FlowJo software (TreeStar, Ashland, OR).
Flow imaging
Image stream flow imaging was performed to asses DNA damage in the BM cells as previously described [18]. In short, single-cell suspensions were prepared from BM after the lysis of red blood cells. Cells were first stained with conjugated antibodies against surface markers (Percp Cy5.5-Gr-1 and PE-CD71) for 1 hour on ice. Cells were fixed using fixation/permeabilization buffer (eBioscience) and then stained with Alexa Fluor 647 conjugated phospho-H2AX antibodies for 1 hour at room temperature. Cells were then stained with DAPI for 5 minutes at room temperature. Flow imaging was performed using Amnis® ImageStream®X Mk II Imaging Flow Cytometer. Data were analyzed using IDEAS 6.0 software.
Immunoblotting
Mouse BM cells were lysed by direct boiling in 2x sample buffer. HEL, SET-2, MDS-L and BA/F3 cells with or without U2AF1 knockdown were lysed with RIPA lysis buffer containing protease inhibitors. Immunoblotting was performed using phospho-specific antibodies against p-CHK1, p-H2AX (Abclonal), or antibodies against total U2AF1 (cell signaling), Nfya, Pbx1, Meis1, Runx2, p15, (Abclonal), p16 (Thermo Scientific) and Flt3, Erk2, β-Actin (Santa Cruz Biotechnology).
Immunofluorescence
For detection of DNA damage, γ-H2AX staining was performed and for detection of R-loops, S9.6 antibody staining was performed as described below. Briefly, paraffin sections of BM were treated with 100% xylene for 5 min twice, 100% xylene and 100/% ethanol 1:1 ratio for 3 min, 100% ethanol for 5 min twice, 95% ethanol for 5 min, 70% ethanol for 5 min and 50% ethanol for 5 min and rinsed with cold water for 1 min followed by antigen retrieval with sodium citrate buffer at 100°C for 20 min. Blocking was performed with 10% goat serum for 2 hours. Phospho-serine 139 H2AX antibody (1:100 dilution) (Abclonal) and S9.6 antibody (1:75 dilution) (Kerafast) were used for primary staining. For secondary antibody staining, TRITC goat anti-rabbit antibody was used for γ-H2AX and TRITC goat anti-mouse antibody was used for S9.6 (1:200 dilutions) (Jackson Immunoresearch).
For R-loop detection in BM Lin- cells, the Lin- cells were first separated from the BM by using lineage depletion kit (Miltenyi Biotec). Lin- cells were then suspended in the pre-warmed 75 mM KCl solution in a drop-wise fashion and incubated for 15 min at 37°C. Freshly prepared ice-cold fixative solution [methanol:acetic acid(3:1)] were then added to cells drop wise and centrifugation was performed at 800 rpm for 5 min at 4°C. After centrifugation, the supernatant was aspirated down to 500 μl. Cells were added drop wise to 5 mL of fixative solution and incubated for 20 min on ice. After incubation, cells were washed once with fixative solution and then spotted onto slides using a Cytospin centrifuge followed by incubation at 65°C for 30 min. Fixed cells were blocked with 10% goat serum for 1 hour followed by incubation with S9.6 antibody (1:75 dilution) (Kerafast) overnight at 4°C. Secondary antibody staining was performed with TRITC goat anti-mouse antibody (1:200 dilution) (Jackson Immunoresearch). Mounting was done with vectashield mounting medium with DAPI (H-1200, Vector Labs). Fluorescence was visualized using Zeiss LAM 710 confocal microscope.
Real-time quantitative PCR
Total RNA was extracted from HEL cells and FACS sorted lineage-negative (Lin-) and Lin−Sca-1+c-kit+ (LSK) cells from the mice BM using RNeasy Mini kit (Qiagen) and cDNA samples were prepared by using QuantiTect Reverse Transcription kit (Qiagen). Real-time quantitative PCR (RT-qPCR) was performed in a Quantstudio3 machine (Applied Biosystems) using SYBR Green PCR master mix (Quatabio). The data were normalized to HPRT and fold changes in gene expression were determined by the ΔΔCt method. Sequences of the primers are available in the supplemental Table 2.
RNA-sequencing and splicing alterations
LSK cells from the BM of control and U2af1-deleted BMT mice, and lineage-negative (Lin-) cells from the BM of control and primary U2af1-deleted mice were first enriched using Lineage cell depletion kit from Miltenyi and then sorted using a FACS Aria II at 7 days after pI-pC induction. Total RNA was extracted from BM LSK and Lin- cells using RNeasy Mini kit (Qiagen). RNA sequencing was performed using Ultra II Directional RNA Library prep kit for Illumina (from NEB) and Hiseq next-generation sequencing instrument (Illumina). Gene set enrichment analysis was performed as described [19]. To identify splicing events, rMATS [20] was used. RT-PCR followed by gel electrophoresis was used to identify splicing events. Image J software was used to quantify the band intensity. Sequences of the primers are available in the supplemental Table 3. RNA-seq data from U2af1 cKO mice will be deposited to publicly available database (GEO). RNA-seq data for MP (Lin-c-kit+) cells from U2af1 S34F knock-in mice: GSE112174.
Statistical Analysis
Results are expressed as mean ± SEM, and statistical significance was determined by Student t-test. P <0.05 was considered statistically significant.
Data availability
The RNA-Sequencing datasets generated in this study are deposited to NCBI GEO database (GSE162888).
Results
Depletion of U2AF1 impairs cell growth and survival in hematopoietic cells
To assess the effects of U2AF1 depletion on hematopoietic cells, we performed lentiviral shRNA-mediated knock-down of U2AF1 in murine hematopoietic BA/F3 cell line as well as in human HEL and SET-2 leukemia cell lines and MDS patient-derived MDS-L cell line (Supplementary Fig. 1). The cells were transduced with lentiviral U2AF1 shRNA or control shRNA. Infected cells were selected with puromycin and cell proliferation was assessed using viable cell counts over a period of 4 to 5 days. We observed that knockdown of U2AF1 significantly inhibited the proliferation of BA/F3, HEL, SET-2 and MDS-L cells (Supplementary Fig. 1a-d). We also assessed the effects of U2AF1 knockdown on apoptosis in HEL, SET-2 and MDS-L cells using Annexin V staining and flow cytometry. We observed that knockdown of U2AF1 significantly increased apoptosis in HEL, SET-2 and MDS-L cells (Supplementary Fig. 1e-g). These data suggest an important role for U2AF1 in the growth and survival of hematopoietic cells.
Conditional deletion of U2af1 results in fatal bone marrow failure in mice
To elucidate the physiological role of U2af1 in normal hematopoiesis, we generated a new conditional U2af1 knockout (floxed) allele. We targeted the exon 2 of U2af1 by inserting a LoxP-flanked cassette, which can be excised by Cre-mediated recombination (Fig. 1a and Supplementary Fig. 2a, b). Two independent ES clones were injected into C57BL/6 blastocysts, and resulting chimeras were mated with C57BL/6 mice. Both clones gave rise to germ-line transmission. To determine the effects of U2af1 deficiency on HSPC and normal hematopoiesis, we crossed U2af1 floxed mouse with Mx1Cre transgenic mouse [17] and the expression of Cre recombinase was induced with polyinosine-polycytosine (pI-pC) injection at 5 to 6 weeks after birth. U2af1 was almost completely deleted in the bone marrow (BM) of Mx1Cre;U2af1fl/fl mice upon pI-pC induction (Fig. 1b and c). All induced Mx1Cre;U2af1fl/fl (hereafter U2af1 cKO) mice became moribund or died between 10–14 days after first pI-pC injection (Fig. 1d). Therefore, all mice analyses were done between 8–10 days after pI-pC induction unless otherwise specified. Mice with homozygous U2af1 deletion (U2af1 cKO) exhibited significantly reduced white blood cell (WBC), neutrophil, red blood cell (RBC), hemoglobin (Hb) and platelet counts in their peripheral blood compared with control animals (Fig. 1e-i). BM cellularity was significantly decreased in U2af1 cKO mice compared to control animals (Fig. 1j). Mice with heterozygous U2af1 deletion, however, did not exhibit any significant decrease in their peripheral blood counts or BM cellularity compared with control animals at this point of time (Supplementary Fig. 3a-g). Blood smears from U2af1 cKO mice exhibited anemia and pancytopenia (Fig. 1k). H&E staining of the BM sections from U2af1 cKO animals showed severe BM aplasia (Fig. 1k).
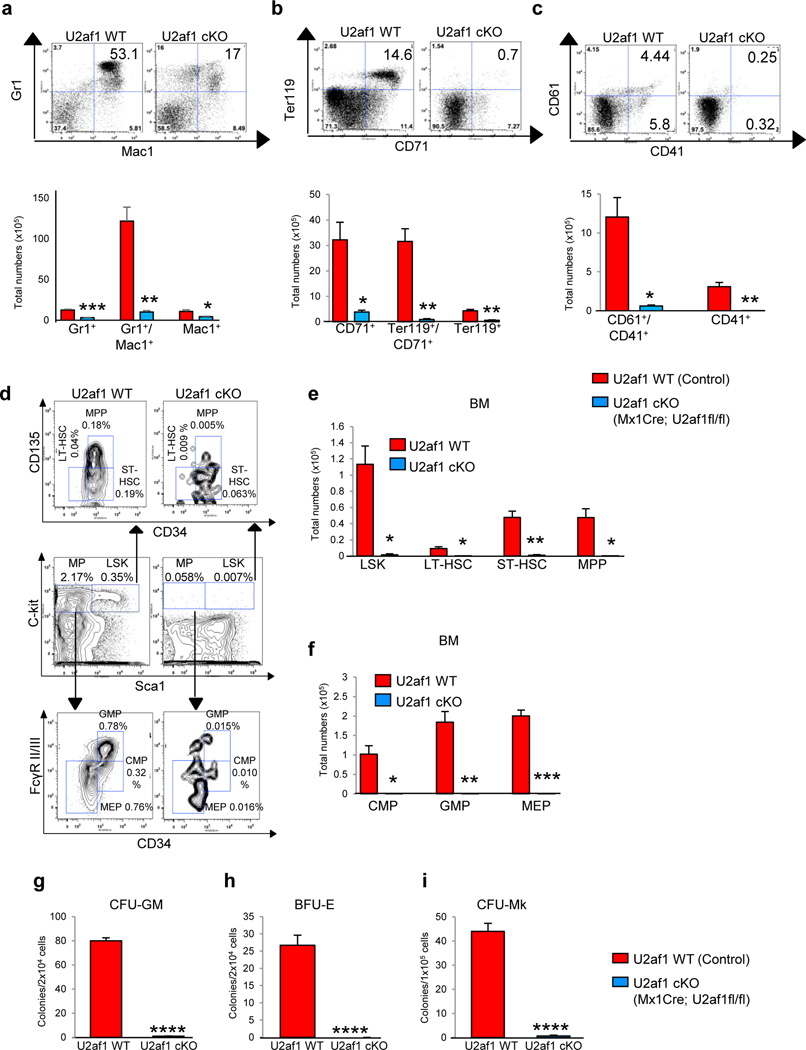
Figure 1. Conditional deletion of U2af1 in mouse hematopoietic compartments results in fatal BM failure.
(a) Gene-targeting strategy for the U2af1 gene is depicted. The exon 2 of U2af1 is targeted in ES cells by inserting two LoxP sites between introns 1 and 2 of U2af1 gene to create U2af1 floxed allele. The U2af1 floxed allele can be deleted by the expression of Cre recombinase. (b) Immunoblot analysis for U2af1 protein showed efficient deletion of U2af1 in the BM of Mx1Cre;U2af1fl/fl mice (U2af1 cKO) after induction with pI-pC. Erk2 served as loading control. (c) qRT-PCR analysis also showed markedly reduced expression of U2af1 mRNA in U2af1 cKO mice BM (n=6) (**p<0.005). (d) Kaplan-Meier analysis showed significant decrease in survival of U2af1-deleted mice compared to control mice. Conditional U2af1-deleted mice died or became moribund within 10 −14 days after pI-pC (n=15 each group). Peripheral blood (e) white blood cell (WBC), (f) neutrophil (NE), (g) Red blood cell (RBC), (h) hemoglobin (Hb) and (i) platelet counts were assessed at 10 days after pI-pC induction in control and U2af1 cKO mice (n=10–12). (j) BM cells were ablated in U2af1 deleted mice compare to control mice 10 days after pI-pC (n=10 to 12). All data are shown as mean ± SEM. Student t-test was used to compare between two groups of mice (**p<0.005, ***p<0.0005). (k) Representative peripheral blood smears (1000X) from control and U2af1 cKO mice at 11 days after pI-pC injection are depicted. The U2af1 cKO blood smear shows severe anemia. H&E staining of the BM sections (40X and 500X) demonstrate pancytopenia and severe aplasia in U2af1 cKO mice BM.
Flow cytometric analyses revealed a marked decrease in the frequencies as well as total numbers of myeloid (Mac-1/Gr-1), erythroid (Ter119/CD71) and megakaryocytic (CD61/CD41) precursors in the BM of U2af1 cKO mice compared with control animals (Fig. 2a-c and Supplementary Fig. 2c-e). However, mice with heterozygous U2af1 deletion did not exhibit any significant change in the frequencies of myeloid (Mac-1/Gr-1), erythroid (Ter119/CD71) and megakaryocytic (CD61/CD41) precursors in the BM compared with control animals at 4 weeks after pI-pC induction (Supplementary Fig. 3h- j). Together, these data suggest that homozygous deletion of U2af1 results in severe defects in hematopoietic development.
Figure 2. Effects of U2af1 deletion on hematopoietic stem/progenitor cells.
Representative dot plots and bar graphs of flow cytometric analysis show significant decrease in (a) myeloid Gr1+/Mac1+), (b) erythroid (CD71+/Ter119+) and (c) megakaryocytic (CD61+/CD41+) populations in U2af1 cKO mice BM as compared to control mice 10 days after pI-pC (n=8 to 10) (*p<0.05, **p<0.005, ***p<0.0005). (d) Representative contour plots of flow cytometric analysis of LSK (Lin-Sca-1+c-kit+), LT-HSC (Lin-Sca-1+c-kit+CD34-CD135-), ST-HSC (Lin-Sca-1+c-kit+CD34+CD135-), MPP (Lin-Sca-1+c-kit+CD34+CD135+), CMP (Lin-Sca-1-c-kit+CD34+FcγRII/IIlow), GMP (Lin-Sca-1-c-kit+CD34+FcγRII/IIhigh) and MEP (Lin-Sca-1-c-kit+CD34-FcγRII/III-) in the BM of control and U2af1 cKO mice BM at 10 days after pI-pC. (e) Total numbers of LSK, LT-HSC, ST-HSC and MPP in the BM of control and U2af1 cKO mice are shown in bar graphs as mean ± SEM (n=8 to 10). (f) Total numbers of CMP, GMP and MEP in the BM of control and U2af1 cKO mice are shown in bar graphs as mean ± SEM (n=8 to 10). (*p<0.05, **p<0.005, ***p<0.0005). (g-h) Hematopoietic progenitor colony assays. Total BM cells (2 × 104) from control WT and U2af1 cKO mice (n=6) were plated in methylcellulose medium (MethoCult 3434) with cytokines. CFU-GM and BFU-E colonies were scored 7 days after plating. (i) BM cells (1 × 105) from control and U2af1 cKO mice (n=6) were plated into collagen-based MegaCult medium supplemented with IL-3, IL-6, IL-11, and Tpo. Megakaryocytic (CFU-Mk) colonies were assessed after culturing for 8 days. All data are shown as mean ± SEM (****p<0.00005 by student t-test).
Effects of U2af1 deletion on hematopoietic stem/progenitor cells
The fatal BM failure in U2af1 cKO mice prompted us to examine the hematopoietic stem cells (HSC) and progenitor compartments in the BM of these animals. We observed a marked decrease in frequencies and absolute numbers of LSK and long-term hematopoietic stem cells (LT-HSC), short-term HSC (ST-HSC), and multipotential progenitors (MPP) as well as early progenitors including common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythroid progenitors (MEP) in the BM of U2af1 cKO mice, indicating a defect at the earliest stage of adult hematopoietic development (Fig. 2d-f and Supplementary Fig. 2f). Heterozygous U2af1 deleted mice, however, did not exhibit any significant alterations in HSC and progenitor compartments compared to control animals (Supplementary Fig. 3k). These findings suggest that dramatic reduction in HSC/progenitors may lead to multilineage cytopenia observed in U2af1 cKO mice.
Next, we performed hematopoietic progenitor colony (CFU) assays to assess HSC/progenitor differentiation. Consistent with the flow cytometric data, U2af1-deficient BM cells produced a markedly decreased numbers of myeloid (CFU-GM), erythroid (BFU-E), and megakaryocytic (CFU-Mk) colonies compared with control BM cells (Fig. 2g-i). Thus, loss of U2af1 causes significant defects in the differentiation potential of the HSPC.
BM failure observed in U2af1-deficient mice is cell autonomous
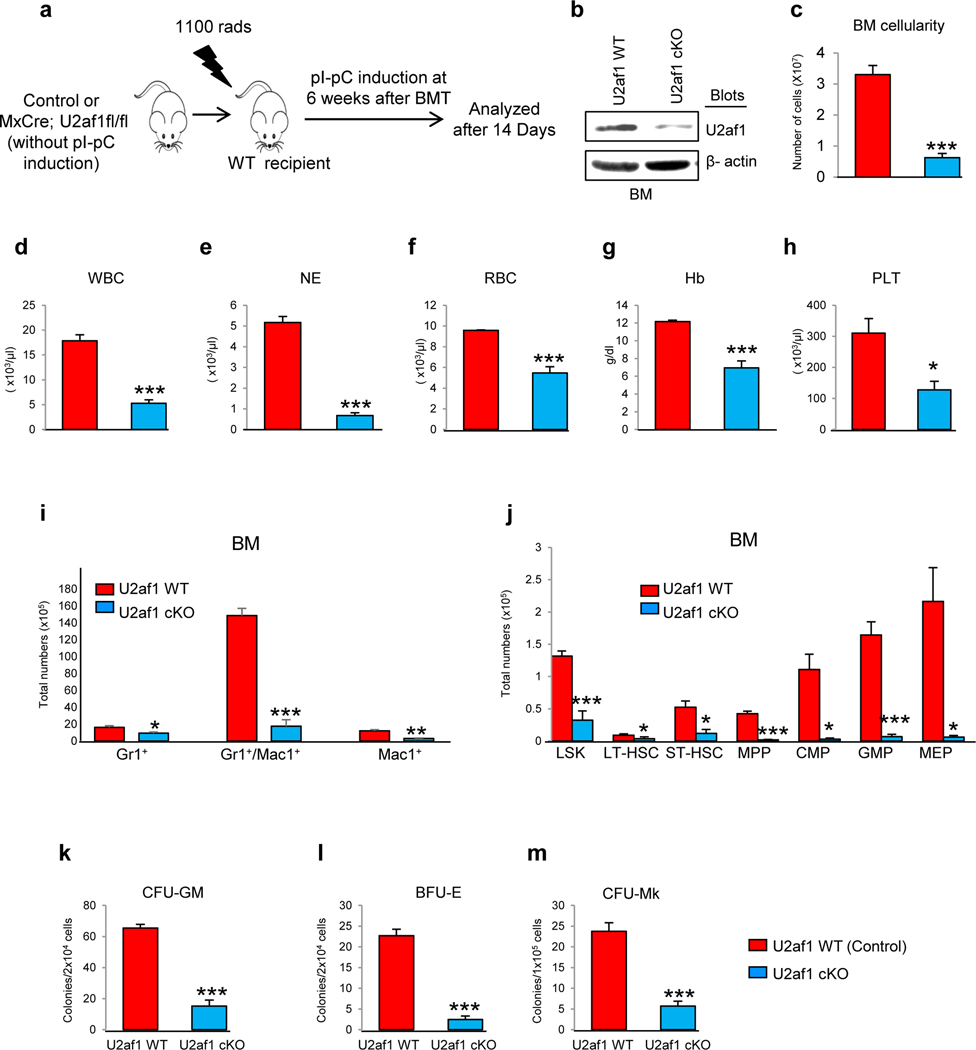
To determine whether the loss of HSC/progenitors in U2af1 cKO animals is cell autonomous, BM cells from uninduced control (U2af1fl/fl; no cre) and Mx1Cre;U2af1fl/fl mice were transplanted into lethally irradiated WT C57BL/6 mice. Six weeks after transplantation, recipients were injected with pI-pC to induce the deletion of U2af1 (Fig. 3a). All the recipients of U2af1-deficient BM became moribund within 14 days after pI-pC induction. Immunoblotting showed significant decrease in U2af1 protein levels upon pI-pC induction in the recipient animals (Fig. 3b). Deletion of U2af1 in the recipient animals resulted in marked decrease in BM cellularity and peripheral blood counts (pancytopenia) (Fig. 3c-h). U2af1 deletion also resulted in substantial decrease in HSC/progenitors as well as myeloid, erythroid and megakaryocytic precursors in the BM of recipient animals (Fig. 3i, j and Supplementary Fig. 4a, b). Hematopoietic progenitor colony assays also showed marked decrease in myeloid (CFU-GM), erythroid (BFU-E), and megakaryocytic (CFU-Mk) colonies upon U2af1 deletion in the recipient animals similar to that observed in the primary U2af1 cKO mice (Fig. 3k-m). These data strongly suggest that the hematopoietic defect in U2af1-deficient HSCs is cell intrinsic.
Figure 3. BM failure observed in U2af1 deficient mice is cell autonomous.
(a) Experimental design for cell autonomous bone marrow transplantation (BMT) assay. BM cells (1×106) were harvested from uninduced Mx1Cre;U2af1fl/fl and control mice, and transplanted into lethally irradiated wild type C57BL/6J recipient mice. Deletion of U2af1 was induced in donor-derived hematopoietic cells by injecting 3 doses of pI-pC at 6 weeks after BMT. Mice were analyzed 14 days after first pI-pC injection. (b) Immunoblot analysis showed efficient deletion of U2af1 in the BM of Mx1Cre;U2af1fl/fl recipient animals after induction with pI-pC. β-actin was used as a loading control. (c) BM cellularity was markedly reduced in U2af1-deficient mice compared to control mice 14 days after pI-pC (n=7 to 10). Peripheral blood (d) White blood cell (WBC), (e) neutrophil (NE), (f) Red blood cell (RBC), (g) hemoglobin (Hb) and (h) platelet (PLT) counts were assessed at 11 days after pI-pC induction in control and U2af1-deficient BMT mice (n=10 to 20). (i) Bar graphs of flow cytometric analysis show significant decrease in myeloid Gr1+/Mac1+) precursors in the BM of U2af1-deficient BMT mice as compared to control recipient mice at 14 days after pI-pC induction. (j) Bar graphs showing the total numbers of LSK, LT-HSC, ST-HSC, MPP, CMP, GMP and MEP in the BM of control and U2af1-deficient BMT mice. The data are presented as mean ± SEM (control, n=7; U2af1 cKO=10) (*p<0.05, **p<0.005, ***p<0.0005). (k-l) BM cells (2 × 104) from control and U2af1-deficient BMT mice (n=5) were plated in methylcellulose medium supplemented with cytokines. CFU-GM and BFU-E colonies were scored 7 days after plating. (m) BM cells (1 × 105) from control and U2af1-deficient BMT mice (n=5) were plated into collagen-based MegaCult medium supplemented with IL-3, IL-6, IL-11, and Tpo. CFU-Mk colonies were assessed after culturing for 8 days. All data are shown as mean ± SEM (***p<0.0005).
U2af1 deficiency impairs repopulation capacity of HSC
We performed competitive repopulation assays to further evaluate the ability of U2af1-deficient HSCs in hematopoietic reconstitution. BM cells from uninduced control (U2af1fl/fl; no cre) and Mx1Cre; U2af1fl/fl mice (CD45.2+) were mixed with CD45.1+ competitor BM cells at a ratio of 1:1 and then transplanted into lethally irradiated congenic recipient animals (CD45.1+) (Fig. 4a). Four weeks after transplantation (to allow the establishment of steady-state hematopoiesis), recipients were injected with 3 doses of pI-pC. Chimerism analysis in peripheral blood (PB) of transplanted animals revealed that BM cells from U2af1-deficient mice were completely unable to compete with WT BM cells. Within 2 weeks after pI-pC induction, the percentages of mutant (U2af1-deficient) donor-derived (CD45.2+) myeloid, B and T cells were significantly lower than those derived from control (U2af1fl/fl) BM donor (Fig. 4b-e) in the peripheral blood. At 16 weeks after pI-pC induction, mutant donor derived (CD45.2+) myeloid, B and T cells were almost abolished in the peripheral blood as well as in the BM of the recipient animals (Fig. 4b-g and Supplementary Fig. 5a-d). At 16 weeks after pI-pC induction, Mx1Cre; U2af1fl/fl mice BM-derived (CD45.2+) LSK cells were almost undetectable in the recipient animals, whereas ~50% LSK cells in recipients that had received control (U2af1fl/fl; no cre) BM were CD45.2+ (Fig. 4h). These data strongly suggest that U2af1 deficiency contributes to severe functional impairment in HSCs. Due to severe BM failure observed in both primary as well as in BMT mice, we were not able to perform any secondary transplantation to further test the self-renewal capacity of U2af1-deficient HSCs.
Figure 4. Defective stem cell function in U2af1-deficient mice.
(a) Competitive reconstitution assay. BM cells (5×105) from uninduced CD45.2+ Mx1Cre;U2af1fl/fl (U2af1 cKO) or littermate control mice were mixed with CD45.1+ wild type mice BM (5×105) at a 1:1 ratio and transplanted into lethally irradiated CD45.1+ recipient mice. Four weeks after BMT, recipient mice were treated with 3 doses of pI-pC to induce U2af1 deletion after hematopoietic reconstitution. The recipient mice were analyzed at 16 weeks after pI-pC injection. (b) The percentages of donor-derived CD45.2+ cells, (c) percentages of CD45.2+ Gr-1+ myeloid cells (d) percentages of CD45.2+ B220+ B cells and (e) percentages of CD45.2+ Tcrβ+ T cells in the peripheral blood of recipient animals at 2 and 16 weeks after pI-pC induction are shown in bar graphs. (f) Representative contour plots of flow cytometric analysis and the percentages of donor-derived CD45.2+ total BM cells, (g) percentages of CD45.2+ Gr-1+ myeloid cells and (h) percentages of CD45.2+ LSK cells in the BM of recipient animals at 16 weeks after pI-pC induction are shown; bar graphs represent mean ± SEM. (Control, n=10; U2af1 cKO=10). Student t-test was used to compare between two groups of mice (**p<0.005, ***p<0.0005 and ****p<0.00005).
Loss of U2af1 increases cell death and DNA damage in hematopoietic progenitors
Since U2af1 deletion resulted in rapid decrease in hematopoietic cells of multiple lineages in the BM, we asked whether deletion of U2af1 insulted the genome and induced apoptosis in hematopoietic progenitor and precursors cells. We performed Annexin V and DAPI staining to determine the apoptotic cells in the BM. We observed significant increase in apoptotic cells in the total BM as wells as in c-kit+, Gr1+, Ter119+ and CD41+ cells suggesting that hematopoietic progenitors and precursors of multiple cell lineages underwent apoptosis upon U2af1 deletion (Fig. 5a). Histone-H2AX is rapidly phosphorylated on serine 139 (γ-H2AX) in response to DNA damage. We performed γ-H2AX assay using ImageStreamX imaging flow cytometry[18] to evaluate DNA damage in total BM, Gr1+ (myeloid) and CD71+ (erythroid) cells in control and U2af1 cKO mice. ImageStreamX imaging flow cytometry technique allows quantitative measurement of γ-H2AX fluorescence intensity in individual cells [18]. Indeed, we observed markedly elevated levels of γ-H2AX in total BM, Gr1+ and CD71+ cells from U2af1 cKO mice compared with control mice (Fig. 5b and Supplementary Fig. 6a and b). Imaging flow cytometry also showed marked shift in the γ-H2AX fluorescence intensity in U2af1 cKO BM cells (Fig. 5c). Quantification using imaging flow cytometry showed significantly increased percentage of γ-H2AX positive cells in the total BM as well as myeloid and erythroid cells from U2af1 cKO mice compared with control mice (Fig. 5d).
Figure 5. Deletion of U2af1 leads to DNA damage and apoptosis in hematopoietic cells.
(a) Annexin V/DAPI staining was performed on hematopoietic progenitors and precursor cells from U2af1 cKO and control mice BM at 8 days after pI-pC induction and apoptosis was measured by flow cytometry. Bar graphs showing increased percentage of apoptotic cells in the total BM, c-Kit+ progenitors, Gr1+ myeloid, CD71+ erythroid and CD41+ megakaryocyte cells in U2af1 cKO mice compared to control mice. Data are presented as mean ± SEM (control n=6; U2af1 cKO n=6) (***p<0.0005, ****p<0.00005). (b) Representative images from the Image stream flow imaging show increased γ-H2AX+ cells in the BM of U2af1 cKO mice compared with WT control mice. γ-H2AX (red) and DAPI (blue). (c) Image stream flow imaging show marked shift in the fluorescence intensity of γ-H2AX+ cells suggesting increased DNA damage in U2af1 cKO BM. (d) Bar graphs showing quantification of γ-H2AX+ cells in the total BM, Gr1+ myeloid and CD71+ erythroid cells in control and U2af1 cKO mice BM at 8 days after pI-pC administration. Data are presented as mean ± SEM (control n=6; U2af1 cKO n=6) (*p<0.05, **p<0.005, ****p<0.00005). (e) Representative immunofluorescence images of BM sections showing increased γ-H2AX (serine 139) staining in U2af1 cKO mice BM as compared to control mice BM. γ-H2AX (green) and DAPI (blue) Scale bars, 10μm. (f) Immunoblot analyses showing increased phosphorylation of H2AX at serine 139 (γ-H2AX) and CHK1 at serine 345, and increased ubiquitination of H2A at lysine 119 (H2AK119Ub) in U2af1 cKO mice BM cells compared with WT control BM cells. β-Actin was used as a loading control. (g) Immunoblot analyses showing increased phosphorylation of H2AX at serine 139 (γ-H2AX) and CHK1 at serine 345, and increased ubiquitination of H2AK119 (H2AK119Ub) in HEL cells upon U2AF1 knockdown. β-Actin was used as a loading control. (h) Representative immunofluorescence images of BM sections showing increased R-loop signals by S9.6 staining in U2af1 cKO mice BM as compared to control mice BM. S9.6 (green) and DAPI (blue) Scale bars, 15μm. (i) R-loop signals detected by immunofluorescence staining with S9.6 in BM Lin- cells from control and U2af1 cKO mice at 7 days after pI-pC induction. S9.6 (green) and DAPI (blue) Scale bars, 15μm. (j) Quantification of S9.6+ cells from control and U2af1 cKO mice BM Lin- cells is presented in bar graphs. Data are presented as mean ± SEM (control n=3; U2af1 cKO n=3; more than 100 cells were analyzed) (***p<0.0005).
Immunofluorescence staining of the BM sections exhibited increased γ-H2AX positive cells in the BM sections from U2af1 cKO mice compared with control mice (Fig. 5e). Immunoblotting analysis also showed increased histone-H2AX phosphorylation at Ser139 (γ-H2AX) in the BM of U2af1 cKO mice (Fig. 5f). In addition, we observed increased Chk1 phosphorylation at serine 345, a hallmark for activation of the ATR pathway, and increased histone H2A K119 ubiquitination (H2AK119Ub), a marker for DNA damage response, in the BM of U2af1 cKO mice (Fig. 5f). We further validated these results by knocking down U2AF1 in HEL cells. Indeed, we observed significantly increased histone-H2AX phosphorylation (γ-H2AX), Chk1 phosphorylation and H2AK119 ubiquitination upon U2AF1 knockdown in HEL cells (Fig. 5g). Therefore, loss of U2af1 causes insult to the genome and induces DNA damage and increased cell death.
Previous studies showed that spliceosome mutations augment R-loop formation, resulting in increased DNA damage in MDS cells [21, 22]. We performed S9.6 immunostaining to assess R-loop in total BM and Lin- cells. We observed significantly increased R-loops in total BM and Lin- cells from U2af1 cKO mice compared with control mice (Fig. 5h, i). Quantification of the S9.6+ cells also showed elevated levels of R-loop formation in U2af1 cKO BM Lin- cells as compared to control BM Lin- cells (Fig. 5j). Thus, loss of U2af1 promotes R-loop formation and increased DNA damage in hematopoietic progenitors.
U2af1 deletion impairs expression of genes responsible for HSC maintenance
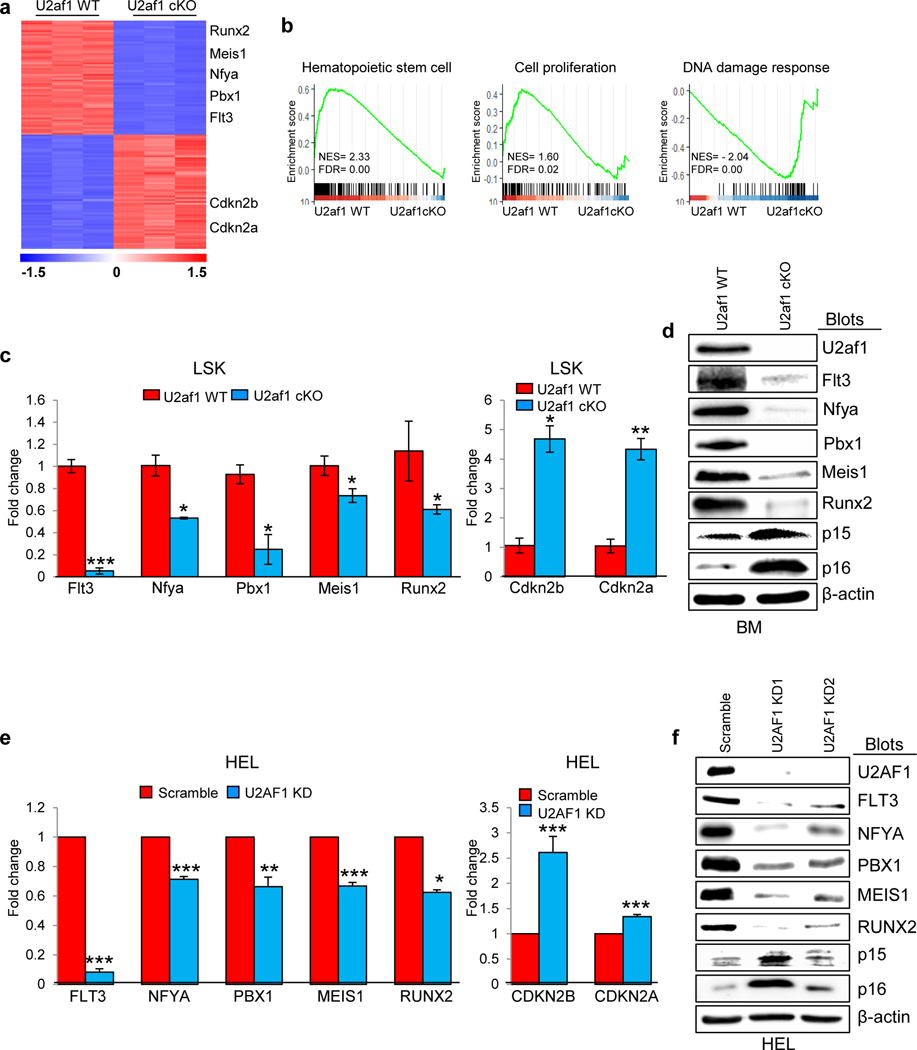
To gain insights into severe hematopoietic defects observed in U2af1-deficient mice, we performed transcriptome profiling through RNA sequencing of sorted LSK cells from the BM of U2af1 wild type (control) and U2af1-deleted (U2af1 cKO) BMT mice. Three independent sets of LSK cells were sorted from control and U2af1-deficient mice at 7 days after pI-pC administration. We also performed RNA sequencing on sorted lineage-negative (Lin-) BM progenitors from primary U2af1-deleted (U2af1 cKO) and control mice since the number of LSK cells in the primary U2af1 cKO mice was extremely low within 7 days after pI-pC induction. Heat map showing significant changes in gene expression profiles between U2af1-deleted LSK and Lin- cells compared with U2af1 WT (control) LSK and Lin- cells (Fig. 6a and Supplementary Fig. 7a, b). We also compared our U2af1 cKO LSK RNA-seq data with publicly available RNA-seq data (GSE112174) from U2af1 S34F knock-in mice LK cells and found that expression of 1,007 genes were commonly altered in both U2af1 cKO LSK and U2af1 S34F mutant LK cells (Supplementary Fig. 7c).
Figure 6. Effect of U2af1 deletion on gene expression profile in HSPC.
(a) Heat map showing significantly up-regulated and down-regulated (p-adj <0.05; fold change >1.5 fold and <−1.5 fold) genes in U2af1-deficient LSK cells compared with control LSK cells in BMT mice. (b) Gene-set enrichment analyses show significant alterations of genes related to hematopoietic stem cells, cell proliferation and DNA damage response pathways in U2af1-deficient LSK cells compared with control LSK cells. (c) Relative expression of Flt3, Nfya, Pbx1, Meis1, Runx2, Cdkn2a (p16), and Cdkn2b (p15) mRNA was determined in control WT and U2af1-deficient LSK cells by RT-qPCR and normalized with Hprt expression. (d) Immunoblot analyses show decreased protein levels of Flt3, Nfya, Pbx1, Meis1 and Runx2 and increased levels of p15 and p16 in U2af1 cKO BM compared with control BM. (e) Validation of U2AF1 targets using lentiviral U2AF1 knockdown in HEL cells. Relative expression of FLT3, NFYA, PBX1, MEIS1, RUNX2, CDKN2A (p16), and CDKN2B (p15) was assessed by RT-qPCR and normalized by HPRT expression. Data are shown in bar graphs as mean ± SEM (n =4; *p<0.05, **p<0.005, ***p<0.0005). (f) Immunoblot analyses show decreased protein levels of FLT3, NFYA, PBX1, MEIS1 and RUNX2 and increased levels of p15 and p16 in U2AF1 knockdown HEL cells. β-Actin was used as a loading control.
Gene Set Enrichment Analysis (GSEA) [19] of RNA sequencing data revealed significant downregulation of HSC-specific genes in U2af1-deficient LSK (Fig. 6b). GSEA also revealed enrichment for cell proliferation and DNA damage response-related genes in U2af1-deficient LSK and Lin- cells (Fig. 6b and Supplementary Fig. 7d), consistent with decreased proliferation and increased DNA-damage and apoptosis observed in U2af1-deficient hematopoietic cells and progenitors. We found that expression of Flt3, Nfya, Pbx1, Meis1 and Runx2 was significantly downregulated in U2af1-deficient LSK and/or Lin- cells compared with control LSK or Lin- cells (Fig. 6a and Supplementary Fig. 7a). Nfya and Pbx1 are transcription factors that are known to play important roles in the survival and function of HSC [23–25]. We also observed increased expression of cell cycle regulators Cdkn2b (p15) and Cdkn2a (p16) in U2af1-deficient LSK and/or Lin- cells compared with control LSK or Lin- cells (Fig. 6a and Supplementary Fig. 7a).
RT-qPCR analysis also confirmed that expression of Flt3, Nfya, Pbx1, Meis1 and Runx1 was significantly reduced whereas expression of Cdkn2b (p15) and Cdkn2a (p16) was significantly increased in U2af1-deficient LSK cells compared with control LSK (Fig. 6c). Expression of these genes was similarly affected in U2af1-deficient Lin- cells (Supplementary Fig. 7e). We also observed decreased protein expression of Flt3, Nfya, Pbx1, Meis1 and Runx2 and increased expression of Cdkn2b (p15) and Cdkn2a (p16) in the BM of U2af1 cKO mice compared with control mice (Fig. 6d).
We further validated these U2af1 target genes using lentiviral shRNA-mediated knockdown of U2AF1 in human erythroleukemia (HEL) cell line. We observed that knockdown of U2AF1 in HEL cells resulted in significantly decreased expression of FLT3, NFYA, PBX1, MEIS1 and RUNX2 (Fig. 6e). Conversely, knockdown of U2AF1 resulted in significantly increased expression of CDKN2A (p16) and CDKN2B (p15) transcripts (Fig. 6e). Protein expression levels for these targets were similarly altered upon U2AF1 knockdown in HEL cells (Fig. 6f), suggesting that these are bona fide targets of U2AF1.
Effects of U2af1 deletion on RNA splicing
Since U2AF1 is involved in recognition of the 3’ splice site required for the recruitment of the U2 snRNP during pre-mRNA splicing [8], we determined the effects of U2af1 deletion on RNA splicing in U2af1-deficient LSK and Lin- cells. Changes in RNA splicing were identified using rMATS [20] analysis of the RNA-seq data from control and U2af1-deficient LSK and Lin- cells. Alteration in exon skipping/inclusion (cassette exons) was the most common type of aberrant splicing event induced by U2af1 deletion in LSK cells (Fig. 7a and b). Similar to U2af1-deficient LSK cells, exon skipping/inclusion (cassette exons) was the most frequent aberrant splicing event observed in U2af1-deleted Lin- cells (Supplementary Fig. 8a and b). Interestingly, we observed significant changes in gene expression as well as splicing alterations in Nfya and Pbx1 transcripts in both LSK and Lin- cells (Fig. 6a and c; Fig. 7c; and Supplementary Fig. 7a and e; Fig. 8d). We further compared the alternatively spliced genes between U2af1-deficient LSK and U2af1 S34F mutant LK cells (Supplementary Fig. 8e). We identified 348 gene transcripts had commonly altered splicing events between these two groups (Supplementary Fig. 8e). Next, we assessed the motif preferences around 3’ splice sites of cassette exons in U2af1-deficient LSK and compared with U2af1 S34F mutant LK cells. We observed that the cassette exons in U2af1 S34F mutant mice LK cells exhibited different sequence preferences (C/A >> T/C) at the −3 position (flanking the AG) of the 3’ splice site in U2af1 S34F mutant mice LK cells (Supplementary Fig. 8f). In contrast, the cassette exons that were differentially spliced (increased inclusion/exclusion) in U2af1-deleted LSK cells did not exhibit sequence-specific changes at the −3 position of the 3’ splice site (Supplementary Fig. 8f).
Figure 7. Effect of U2af1 deletion on RNA splicing.
(a) Number and type of alternative splicing events in U2af1-deficient LSK cells compared with control LSK cells. (b) Inclusion levels of cassette exons (exon skipping/inclusion) in U2af1-deficient LSK cells compared with control LSK cells are depicted. Red and blue colored dots represent individual cassette exons that are significantly more included (red) or excluded/skipped (blue) in U2af1-deficient versus control LSK cells (>5% inclusion level differences and FDR <0.05). (c) Venn diagrams comparing gene expression changes and splicing events in U2af1-deficient LSK cells compared with control LSK cells. (d-e) Sashimi plots, RT-PCR and gel electrophoresis analyses confirmed altered splicing events in Nfya and Pbx1. % of long isoform was significantly decreased in U2af1 cKO LSK cells compared with control LSK cells for both Nfya and Pbx1(*p<0.05, **p<0.005). (f-g) Ectopic expression of Nfya or Pbx1 by lentiviral transduction partially rescues the defects in hematopoietic progenitor colony formation in U2af1-deficient BM progenitors. Transduced cells were selected using puromycin and plated in methylcellulose medium containing complete cytokines or Epo (3U/ml). Myeloid progenitor colonies (CFU-GM and CFU-GEMM) and erythroid (CFU-E) colonies were counted. Results are expressed as percentage of controls (n=4; **p<0.005, ***p<0.0005).
We identified altered splicing events in Nfya and Pbx1 transcripts in U2af1-deficient hematopoietic progenitors (Fig. 7d and e). Sashimi plot from our RNA-seq data clearly showed that U2af1 deletion is associated with altered pre-mRNA splicing of Nfya, with decreased usage of exon 3 (skipping of exon 3) (Fig. 7d). RT-PCR followed by gel electrophoresis showed significant decrease in the Nfya long isoform (including exon 2, 3 and 4) in U2af1 cKO LSK cells compared with control LSK cells suggesting exon 3 is skipped due to U2af1 deletion (Fig. 7d). We also confirmed that exon 7 of Pbx1 is skipped in U2af1 cKO LSK cells (Fig. 7e).
Previous reports have suggested important roles for Nfya and Pbx1 in the maintenance and function of HSC [23–25]. Since deficiency of U2af1 resulted in both decreased expression and splicing alterations of Nfya and Pbx1 transcription factors (Fig. 6a, c; Fig. 7d, e), we asked if enforced expression of Nfya or Pbx1 could rescue the defects in U2af1-deficient HSPC using hematopoietic progenitor colony formation assay. Lentiviral expression of Nfya or Pbx1 alone significantly increased myeloid progenitor (CFU-GM, CFU-GEMM) and erythroid (CFU-E) colony formation in U2af1-deficient BM progenitors compared with vector expression (Fig. 7f, g) although neither Nfya nor Pbx1 alone is sufficient to make complete rescue. Notably, expression of Nfya resulted in larger number of myeloid progenitor and erythroid colonies than Pbx1 in U2af1-deficient HSPC, suggesting more important contribution of Nfya in HSPC function. Overall, these results suggest that aberrant RNA splicing associated with perturbed expression of Nfya and Pbx1 may contribute to hematopoietic defects in U2af1-deficient mice.
Discussion
Here we report the development of a novel conditional U2af1 knockout mouse to elucidate the role of U2af1 in normal adult hematopoiesis. Our studies show that U2af1 deletion results in pancytopenia, ablation of hematopoietic stem/progenitor cells (HSPC) with a dramatic decrease in bone marrow cellularity and early lethality in mice. Loss of U2af1 also causes profound defect in HSC repopulation capacity as well as increased cell death in hematopoietic progenitors. Overall, our results suggest an essential role for U2af1 in normal hematopoiesis.
HSC functional analyses using non-competitive and competitive BM transplantation assays reveal that U2af1-deficient HSCs are severely defective in self-renewal capacity and they are unable to maintain long-term hematopoiesis. There was marked reduction of HSC, progenitors and all types of blood and BM cell precursors within 2 weeks after U2af1 deletion (by pI-pC administration) in the transplanted animals. Also, U2af1-deficient HSCs were unable to compete with WT HSCs and there was rapid loss of hematopoietic progenitors/precursors derived from the U2af1-deficient HSCs. These data clearly suggest a critical requirement for U2af1 in HSC maintenance and function.
RNA-seq analyses also reveal that HSC maintenance programs are perturbed in the absence of U2af1. In addition, cell proliferation-related and DNA damage response genes are significantly altered in U2af1-deficient HSPC. We found that expression of Flt3, Nfya, Pbx1, Mesi1 and Runx2 is significantly downregulated whereas expression of Cdkn2b (p15) and Cdkn2a (p16) is significantly increased in U2af1-deficient LSK and Lin- cells (Figure 6). Increased expression of Cdkn2b (p15) and/or Cdkn2a (p16) can lead to cell cycle arrest [26]. Consistent with this, we observed significantly reduced cell growth associated with increased expression of CDKN2B (p15) and CDKN2A (p16) in HEL cells upon U2AF1 knockdown (Fig. 6e, f and Supplementary Fig. 1b). We also observed splicing alterations in Nfya and Pbx1 transcripts in U2af1-deficient hematopoietic progenitors. Nfya and Pbx1 transcription factors have been suggested to play important roles in HSC survival and self-renewal [23–25]. Nfya-deficient mice exhibit rapid decrease in peripheral blood cell counts and BM failure [24]. Furthermore, loss of Nfya results in increased apoptosis in BM cells and impairment in HSC repopulation ability [24] similar to that observed with U2af1 deficiency. Conditional inactivation of Pbx1 also results in progressive loss of HSC/progenitors, decrease in BM cellularity and impairment of HSC self-renewal [25]. Furthermore, we show that ectopic expression of Nfya and Pbx1 significantly rescue hematopoietic progenitor colony formation in U2af1-deficient BM HSPC. It also has been reported that Flt3 promotes expansion of early hematopoietic progenitors [27], whereas Runx2 regulates differentiation of hematopoietic progenitors [28]. Meis1 is also suggested to play an important role in the maintenance and function of HSPC [29, 30]. Thus, perturbation of multiple genes related to HSC maintenance and function may contribute to severe HSPC defects in U2af1-deficient mice.
Somatic U2AF1 mutations have been associated with MDS [1, 3–6]. Knock-in mice expressing MDS-associated U2af1 S34F mutant exhibit macrocytic anemia and reduction in the HSC/progenitors [16]. Also, U2af1 S34F mutant expressing HSCs exhibit competitive disadvantage in repopulations assays [16]. Our current work reveals that loss of U2af1 causes more severe defects in HSC repopulation than observed with U2af1 S34F mutant expression [16]. Moreover, U2af1 deletion induces significant apoptosis in hematopoietic progenitors and multilineage precursors in the BM of U2af1 cKO mice. Further, we show that U2af1 deficiency leads to accumulation of R-loop and increased DNA damage in BM progenitor and precursor cells. We also have observed increased H2K119Ub, a marker for DNA damage response, and increased phosphorylation of CHK1 (S345), a downstream target of ATR pathway activation. It has been suggested that expression of U2AF1 S34F mutant induces DNA damage and ATR-CHK1 pathway activation [21, 22]. However, U2af1 S34F mutant expression does not shorten the lifespan in mice [16]. In contrast, deletion of U2af1 causes severe pancytopenia and early lethality in mice. The rapid depletion of HSPC, BM aplasia and pancytopenia observed in U2af1 cKO mice could possibly be due to markedly increased apoptosis caused by U2af1 deficiency. Thus, disruption of U2af1 results in more severe hematopoietic phenotype than that observed in U2af1 S34F knock-in mice [16].
In conclusion, this study provides important new understanding on the role of wild-type U2af1 in hematopoietic stem cell/progenitor function and normal hematopoietic development. We show that U2af1 plays an essential role in the maintenance and normal function of HSPC. Our inducible U2af1 knockout mouse will be valuable for further understanding the role of U2af1 in different hematopoietic lineages and other tissue development. Recently, therapies targeting spliceosome proteins have been developed and undergoing pre-clinical and clinical investigations in various hematologic malignancies [31–35]. Since U2af1 deletion leads to fatal hematopoietic defects, it is very important to carefully examine the long-term effect of spliceosome inhibitors on hematopoiesis. A better understanding of the role of U2AF1 in normal hematopoiesis will allow us to delineate its contribution in hematologic diseases and facilitate the development of appropriate therapeutic approaches for U2AF1 mutant induced hematologic malignancies.
Supplementary Material
Acknowledgements
We thank Dr. Roberto Mantovani (University of Milan) and Dr. Licia Selleri (University of California San Francisco) for NF-Ya and Pbx1 expression constructs, and Ms. Julia Dreksler for assistance with generating U2AF1 knockdown cell lines. We also thank the flow cytometry and microscopy core facilities at the University of Virginia for assistance with FACS sorting and confocal microscopy. Flow cytometry and microscopy cores are supported by the UVA Cancer Center through NCI P30CA044578 grant. This work was supported in part by US National Institute of Health (NIH) grants R01 HL095685 (G.M.), R35 GM133712 (C.Z.) and a start-up fund from the Department of Biochemistry and Molecular Genetics of the University of Virginia (G.M.).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent Pathway Mutations of Splicing Machinery in Myelodysplasia. Nature, 2011. 478(7367):64–69. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 Mutation in Myelodysplasia With Ring Sideroblasts. N Engl J Med, 2011. 365(15): 1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet., 2012. 44(1): 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thol F, Kade S, Schlarmann C, Löffeld P, Morgan M, Krauter J, et al. Frequency and Prognostic Impact of Mutations in SRSF2, U2AF1, and ZRSR2 in Patients With Myelodysplastic Syndromes. Blood, 2012. 119(15): 3578–84. [DOI] [PubMed] [Google Scholar]
- 5.Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, et al. Groupe Francophone des Myélodysplasies., Mutations Affecting mRNA Splicing Define Distinct Clinical Phenotypes and Correlate With Patient Outcome in Myelodysplastic Syndromes. Blood, 2012. 119(14): 3211–18. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa S. Genetics of MDS. Blood, 2019. 133(10):1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue D, Bradley R, Abdel-Wahab O. Spliceosomal Gene Mutations in Myelodysplasia: Molecular Links to Clonal Abnormalities of Hematopoiesis. Genes Dev, 2016. 30(9):989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S, Romfo CM, Nilsen TW, Green MR. Functional Recognition of the 3’ Splice Site AG by the Splicing Factor U2AF35. Nature, 1999. 402(6763):832–35. [DOI] [PubMed] [Google Scholar]
- 9.Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al.Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood, 2012. 119(14): 3203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Przychodzen B, Jerez A, Guinta K, Sekeres MA, Padgett R, Maciejewski JP, et al. Patterns of Missplicing Due to Somatic U2AF1 Mutations in Myeloid Neoplasms. Blood, 2013. 122(6): 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, et al. U2AF1 Mutations Alter Splice Site Recognition in Hematological Malignancies. Genome Res, 2015. 25(1):14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks AN, Choi P, de Waal L, Sharifnia T, Imielinski M, Saksena G, et al. A Pan-Cancer Analysis of Transcriptome Changes Associated With Somatic Mutations in U2AF1 Reveals Commonly Altered Splicing Events. PLoS One., 2014. 9(1):e87361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia, 2015. 29(4):909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yip BH, Steeples V, Repapi E, Armstrong RN, Llorian M, Roy S, et al. The U2AF1S34F Mutation Induces Lineage-Specific Splicing Alterations in Myelodysplastic Syndromes. J Clin Invest, 2017. 127(6): 2206–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer Cell, 2015. 27(5):631–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fei DL, Zhen T, Durham B, Ferrarone J, Zhang T, Garrett L, et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1. Proc Natl Acad Sci U S A, 2018. 115(44): E10437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science, 1995. 269(5229):1427–29. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y, Wang Q, Shuryak I, Brenner DJ, Turner HC. Development of a High-Throughput γ-H2AX Assay Based on Imaging Flow Cytometry. Radiat Oncol., 2019. 14(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A., 2005. 102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A, 2014. 111(51):E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Chen J, Huang YJ, Gu Y, Qiu J, Qian H, et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol Cell., 2018. 69(3):412–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen HD, Leon W, Li W, Reddy PNG, Sullivan JD, Walter MJ, et al. Spliceosome Mutations Induce R Loop-Associated Sensitivity to ATR Inhibition in Myelodysplastic Syndromes. Cancer Res., 2018. 78(18):5363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. NF-Ya Activates Multiple Hematopoietic Stem Cell (HSC) Regulatory Genes and Promotes HSC Self-Renewal. Proc Natl Acad Sci U S A, 2005. 102(33):11728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bungartz G, Land H, Scadden DT, Emerson SG. NF-Y Is Necessary for Hematopoietic Stem Cell Proliferation and Survival. Blood, 2012. 119(6):1380–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ficara F, Murphy MJ, Lin M, Cleary ML.Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell, 2008. 2(5):484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev., 1999. 13(12):1501–12. [DOI] [PubMed] [Google Scholar]
- 27.Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR. Targeted Disruption of the flk2/flt3 Gene Leads to Deficiencies in Primitive Hematopoietic Progenitors. Immunity, 1995. 3(1):147–61. [DOI] [PubMed] [Google Scholar]
- 28.Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 Induces Acute Myeloid Leukemia in Cooperation With Cbfbeta-SMMHC in Mice. Blood, 2009. 113(14):3323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, Copeland NG, et al. Meis1 Preserves Hematopoietic Stem Cells in Mice by Limiting Oxidative Stress. Blood, 2012. 120(25):4973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ariki R, Morikawa S, Mabuchi Y, Suzuki S, Nakatake M, Yoshioka K, et al. Homeodomain Transcription Factor Meis1 Is a Critical Regulator of Adult Bone Marrow Hematopoiesis. PLoS One, 2014. 9(2):e87646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folco EG, Coli KE, Reed R. The Anti-Tumor Drug E7107 Reveals an Essential Role for SF3b in Remodeling U2 snRNP to Expose the Branch Point-Binding Region. Genes Dev, 2011. 25(5):440–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eskens FA, Ramos F, Burger H, O’Brien JP, Piera A, de Jonge MJ, et al. Phase I Pharmacokinetic and Pharmacodynamic Study of the First-In-Class Spliceosome Inhibitor E7107 in Patients With Advanced Solid Tumors. Clin Cancer Res, 2013. 19(22):6296–04. [DOI] [PubMed] [Google Scholar]
- 33.Lee SC-W, Abdel-Wahab O. Therapeutic Targeting of Splicing in Cancer. Nat Med, 2016. 22(9):976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirai CL, White BS, Tripathi M, Tapia R, Ley JN, Ndonwi M, et al. Mutant U2AF1-expressing Cells Are Sensitive to Pharmacological Modulation of the Spliceosome. Nat Commun., 2017. 8:14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seiler M, Yoshimi A, Darman R, Chan B, Keaney G, Thomas M, et al. H3B-8800, an Orally Available Small-Molecule Splicing Modulator, Induces Lethality in Spliceosome-Mutant Cancers. Nat Med., 2018. 24(4):497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Sequencing datasets generated in this study are deposited to NCBI GEO database (GSE162888).