Abstract
Metformin, a first-line drug for type 2 diabetes mellitus, has been recognized as a potential anti-tumor agent in recent years. Epigallocatechin-3-gallate (EGCG), as the dominant catechin in green tea, is another promising adjuvant agent for tumor prevention. In the present work, the potential effect of EGCG on the anti-tumor efficacy of metformin in a mouse melanoma cell line (B16F10) was investigated. Results indicated that EGCG and metformin exhibited a synergistic effect on cell viability, migration, and proliferation, as well as signal transducer and activator of transcription 3/nuclear factor-κB (STAT3/NF-κB) pathway signaling and the production of inflammation cytokines. Meanwhile, the combination showed an antagonistic effect on cell apoptosis and oxidative stress levels. The combination of EGCG and metformin also differentially affected the nucleus (synergism) and cytoplasm (antagonism) of B16F10 cells. Our findings provide new insight into the potential effects of EGCG on the anti-tumor efficacy of metformin in melanoma cells.
Keywords: Epigallocatechin-3-gallate (EGCG), Metformin, Melanoma, Anti-tumor efficacy
1 Introduction
Metformin is the main biguanide that is widely used throughout the world to treat diabetes (Nicolucci et al., 2019; Ahmad et al., 2020). More recently, researchers have been gaining interest in the potential use of metformin in treating or preventing various cancers (Chae et al., 2016; Kim et al., 2020). A variety of studies have shown that metformin could potentially inhibit the development of diverse cancers such as renal cancer (Liu et al., 2019; Pasha et al., 2019), ovarian cancer (Tang et al., 2018; Hart et al., 2019; Ma et al., 2019; Zou et al., 2019), oesophageal carcinoma (Wang et al., 2019), gastric cancer (Courtois et al., 2019; Lu et al., 2019), melanoma (de Souza Neto et al., 2017; Li et al., 2018), pancreatic cancer (Chen et al., 2017; Suzuki et al., 2019), and breast cancer (Zordoky et al., 2014; Lord et al., 2018). The principal mechanisms of the action of metformin are its inhibition of circulating insulin and mammalian target of rapamycin complex 1 (mTORC1), as well as activation of the AMP-activated protein kinase (AMPK) pathway (Jaune and Rocchi, 2018). In addition, other studies have shown the anti-tumor synergy between metformin and some natural products including aloin (Sun et al., 2020), epigallocatechin-3-gallate (EGCG) (Yu et al., 2017), and vemurafenib (Niehr et al., 2011).
EGCG, the main antioxidant extracted from green tea leaves, is a natural product that has been used for thousands of years to treat human diseases in China. In recent decades, studies have demonstrated the role of EGCG in cancer prevention and tumor growth reduction both in vivo and in vitro, with some clinical trials showing promising results (Yang et al., 2009; Fujiki et al., 2015). Some studies have identified the specific interactions between EGCG and other anti-cancer compounds; for example, EGCG can enhance the anti-cancer effects of vitamin A (Lee et al., 2010), vorinostat, interferon-α, hinokitiol, and gemcitabine (Kim et al., 2004; Lee et al., 2010; Nihal et al., 2010; Chen et al., 2014; Wei et al., 2019a, 2019b).
One type of cancer, which has been receiving increasing attention, is cutaneous malignant melanoma (CMM), which occurs when cells in the skin develop abnormally. CMM is one of the most severe and lethal types of skin cancer (Zou et al., 2019). CMM accounts for 4% to 10% of all skin malignancies, but it is linked to 75% of skin cancer deaths, and melanoma patients have a poor prognosis with a five-year survival rate of only 17% (Hartman and Lin, 2019; Iglesias-Pena et al., 2019). Many reports have described the inhibition of melanoma development by metformin treatment in vivo and in vitro (Janjetovic et al., 2011; Tomic et al., 2011; Cerezo et al., 2013; Zhang et al., 2015; de Souza Neto et al., 2017; Yu et al., 2019). Other studies have investigated potential inhibitory effects of EGCG in melanoma development (Taniguchi et al., 1992; Caltagirone et al., 2000; Wu et al., 2008; Nihal et al., 2009; Ravindranath et al., 2009; Shen et al., 2009; Ellis et al., 2011; Watanabe et al., 2012; Yamada et al., 2016; Zhang et al., 2016). In tandem with these studies, possible synergistic anti-tumor effects between EGCG and metformin were revealed in previous studies. For instance, Sabry et al. (2019) reported that combined treatment with EGCG and metformin exhibited a highly significant effect against hepatocellular carcinoma, pancreatic cancer (Hodges et al., 2015), and non-small cell lung cancer (Yu et al., 2017).
Despite the evidence provided in the aforementioned studies, we still know little about the potential of combining metformin and EGCG in treating CMM. The goals of the present work were to identify whether metformin could be effectively used in combination with EGCG to retard growth of the highly metastatic mouse melanoma cell line (B16F10), and to further dissect the underlying mechanisms of action of EGCG and metformin in melanoma development using advanced technologies such as Raman spectroscopy.
2 Materials and methods
2.1. Chemicals
EGCG (purity of >98%) was purchased from Chengdu Must Biotechnology Co., Ltd. (Chengdu, China). Anti-nuclear factor-κB (anti-NF-κB), anti-signal transducer and activator of transcription 3 (anti-STAT3), anti-β actin (Proteintech Group, USA), anti-phospho-NF-κB, and anti-phospho-STAT3 (Cell Signaling Technology, USA) were the primary antibodies used in western blot.
2.2. Cell line
Mouse melanoma cell line B16F10 cells were purchased from the American Type Culture Collection (ATCC, USA). B16F10 cells were kept in a 37 ℃ incubator with 5% CO2 and maintained in Roswell Park Memorial Institute-1640 (RPMI-1640) medium with 4 mmol/L glutamine, 10% (volume fraction) fetal bovine serum (FBS), 100 U/mL streptomycin, and penicillin (all from Gibco, CA, USA).
2.3. Cell proliferation assay
Cell counting kit-8 (CCK-8; Biosharp, China) was used to investigate cell proliferation according to a previously published protocol (Xu et al., 2020). Briefly, a total of 1×104 B16F10 cells suspended in 100 μL of RPMI-1640 medium were seeded into 96-well plates and incubated overnight. The following day, cells were treated with metformin at doses of 0, 1, 5, 10, 20, 40, 80, and 100 mmol/L or EGCG at doses of 0, 10, 50, 100, 200, 400, and 800 μmol/L. CCK-8 reagent was added to each well at 10 μL, and the absorbance at 450 nm was recorded after 1 h of incubation in the dark.
2.4. Wound healing assay
B16F10 cells were cultured overnight in six-well plates. On the second day, a microtube tip was used to directly scratch the canter of each well. The cells were washed with phosphate-buffered saline (PBS; Gibco), and then treated with 100 μmol/L EGCG, 1 mmol/L metformin, and either 100 μmol/L EGCG plus 1 mmol/L metformin (EM1) or 100 μmol/L EGCG plus 5 mmol/L metformin (EM5) for 48 h. Images of the wound surfaces were taken at 0, 24, and 48 h.
2.5. Flow cytometry analysis
We analyzed apoptosis and the cell cycle distribution of B16F10 cells by flow cytometry according to the protocol described by Xu et al. (2020). Briefly, 5×105 cells were seeded into six-well plates. After 24 h of different treatments, cell apoptosis rate was analyzed by employing propidium iodide (PI) and Annexin V-fluorescein isothiocyanate (FITC) staining reagents. The cell cycle distribution was analyzed using PI. ACEA NovoCyteTM (Biosciences, San Diego, USA) equipment was used for flow cytometry. Flowjo (Version 10.0, Stanford, USA) software was used to analyze the results.
2.6. Immunohistochemistry staining
B16F10 cells were seeded with a density of 1×105 cells/mL onto circular microscope cover slip (NEST Biotechnology Co., Ltd., Wuxi, China) pre-placed in a six-well plate and were cultured for 24 h. Following different treatments for 24 h, cells were rinsed with PBS and were fixed in 4% (volume fraction) formalin. Immunohistochemistry staining was then performed at the histology platform at Zhejiang University (reagents were purchased from Beijing Zhongshan Jinqiao Biotechnology Company, Beijing, China).
2.7. ELISA
For enzyme-linked immunosorbent assay (ELISA), 5×105 cells/mL were seeded into six-well plates, and then followed with different treatments for 24 h. Cell supernatants were collected after centrifugation. The productions of inflammatory cytokines including interleukin-6 (IL-6), IL-10, and tumor necrosis factor-α (TNF-α), as well as the levels of antioxidant enzymes (superoxide dismutase (SOD), catalase (CAT), and glutathione (GSH)) were detected by different ELISA kits (Kenuodibio, China) following the manufacturer's protocols.
2.8. qRT-PCR
Total RNA was extracted from the cells using TRIzol reagent (TaKaRa, Japan). Reverse transcriptase (Toyobo, Japan) was used to generate the first strand of complementary DNA (cDNA) from total RNA. SYBR Green reagents (TaKaRa, Japan) were applied in the quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) following the manufacturer's protocol. Table S1 lists the sequences of all primers used in this study.
2.9. Western blot analysis
Cold RIPA buffer (Solarbio Science & Technology, Beijing, China), which was added with phenylmethanesulfonyl fluoride (PMSF; Solarbio Science & Technology) and phosphatase inhibitor (Solarbio Science & Technology) at 1:100 (volume ratio), was used to lyse cells. The cell lysates were centrifuged for 10 min at 12 000g to remove the precipitate. The BCA protein detection kit (Beyotime Biotechnology, Shanghai, China) was used to measure protein concentration. The same amount of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Then, the membranes were blocked with 5% (50 mg/mL) skim milk in Tris-buffered saline+Tween (TBST) at room temperature for 1 h before being incubated with primary antibody overnight at 4 ℃. After incubation, the membranes were washed three times with TBST, and then incubated with horse radish peroxidase (HRP)-labelled goat anti-rabbit IgG antibody (Servicebio, Wuhan, China) for 1 h at room temperature.
2.10. Raman spectroscopy
2.10.1. Cell culture
Following the method of Xu et al. (2019), B16F10 cells were seeded with a density of 5×104 cells/mL onto circular glass cover slip (NEST) pre-placed in a six-well plate, cultured overnight, and then received different treatments for 24 h. Cells were then rinsed three times in PBS and fixed in 75% ethanol for 30 min. Finally, cells were stored at 4 ℃ ready for measurement.
2.10.2. Single-cell Raman spectra
Single-cell Raman spectra were carried out using a Raman spectroscopy system (Renishaw plc, Gloucestershire, UK) supplied with a 50× microscope objective. Measurements were performed with 514 nm excitation wavelength and the collected effective spot diameter was about 3 nm. Under a laser power of 10 mW, the Raman spectra of the samples were obtained within an integration time of 10 s. Three sampling locations were randomly selected within each cell and the mean values of the three spectra collected represented this cell. In each treatment group, six cells were randomly selected according to the area division, and were considered to represent the group. MatLab (MathWorks Inc., USA) was used for data processing. All measurements were pre-processed by noise filtering, cosmic ray removal, baseline subtraction, and principal component analysis (PCA).
2.11. Statistical analyses
To determine whether EGCG and metformin had a synergistic effect, we introduced a theoretical value (T-EM) and compared it to the actual experimental value (EM), where EM value equals the effect value of EGCG plus the effect value of metformin in experiment. If the EM was higher than the T-EM, the two compounds were considered to have a synergistic effect. GraphPad Prism (Version 6.0; San Diego, CA, USA) and SPSS software (Version 20.0; Stanford, USA) were applied for statistical analyses. All data are shown as mean±standard deviation (SD). The comparison of the mean values for the biochemical data of each group was performed by one-way analysis of variance (ANOVA) and Duncan's multiple comparison test. An unpaired two-tailed t-test was used to analyze the differences between T-EM and EM. P value of <0.05 was considered statistically significant.
3 Result
3.1. Effects of EGCG and metformin on cell viability and migration ability of B16F10 cells
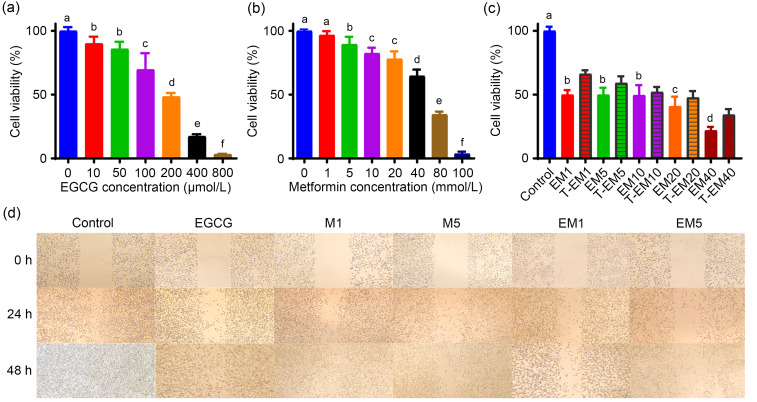
To illustrate potential synergistic effects of EGCG and metformin on the cell growth of B16F10 cells, we first investigated the suitable treatment doses of EGCG and metformin. B16F10 cells were treated with various doses of EGCG (0, 10, 50, 100, 200, 400, and 800 µmol/L) or metformin (0, 1, 5, 10, 20, 40, 80, and 100 mmol/L) alone. Interestingly, EGCG and metformin both reduced cell viability in a dose-dependent manner but the cells were more sensitive to EGCG than to metformin (Figs. 1a and 1b). The concentration of EGCG for the combination experiment was selected at 30% inhibition concentration (IC30; 100 µmol/L). To evaluate the combined effect of the two drugs, we compared the EM and T-EM. As shown in Fig. 1c, the EMs of these three combinations (100 μmol/L EGCG plus 1 mmol/L metformin (EM1), 100 μmol/L EGCG plus 5 mmol/L metformin (EM5), and 100 μmol/L EGCG plus 40 mmol/L metformin (EM40)) exhibited higher inhibitory effects on cell growth than their T-EMs, indicating a synergistic effect of EGCG and metformin on cell viability using these three combinations. Since the combination of the two drugs has shown obvious effects at low concentrations, we selected EM1 and EM5 combinations for subsequent experiments.
Fig. 1. Impact of EGCG on the inhibitory effects of metformin on the growth and migration of B16F10 cells. Cell viability analysis of B16F10 cells following treatments with EGCG (10‒800 µmol/L) (a), metformin (1‒100 mmol/L) (b), or both (c) for 24 h. Control: culture solution; EM1: 100 μmol/L EGCG plus 1 mmol/L metformin; EM5: 100 μmol/L EGCG plus 5 mmol/L metformin; EM10: 100 μmol/L EGCG plus 10 mmol/L metformin; EM20: 100 μmol/L EGCG plus 20 mmol/L metformin; EM40: 100 μmol/L EGCG plus 40 mmol/L metformin. The corresponding theoretical value is shown as T-EM. (d) Wound-healing assay images of B16F10 cells from different groups. Images were visualized at 0, 24, and 48 h. Data are presented as mean±standard deviation (SD) and are representative of at least three independent experiments. Different letters among groups indicate significantly different means at P<0.05. EGCG: epigallocatechin-3-gallate; M1: 1 mmol/L metformin; M5: 5 mmol/L metformin.
The wound healing experiment was carried out to evaluate the effects of EGCG and metformin on cell migration. The results showed that the inhibition rate of the EGCG treatment group (100 μmol/L) was higher than that of the 1 mmol/L metformin (M1) or 5 mmol/L metformin (M5) groups. Moreover, the combination groups of EM1 and EM5 showed higher inhibitory effects on cell migration than any other single treatment group, suggesting a synergistic effect of EGCG and metformin on cell migration (Fig. 1d).
3.2. Effects of EGCG and metformin on cell apoptosis and proliferation of B16F10 cells
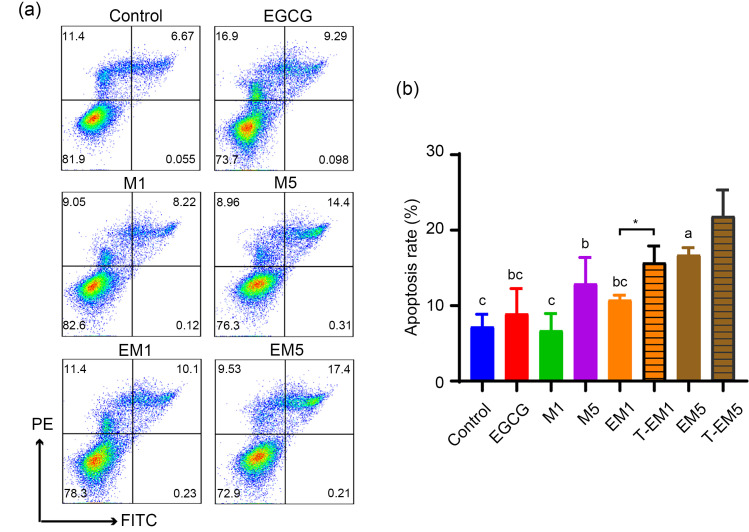
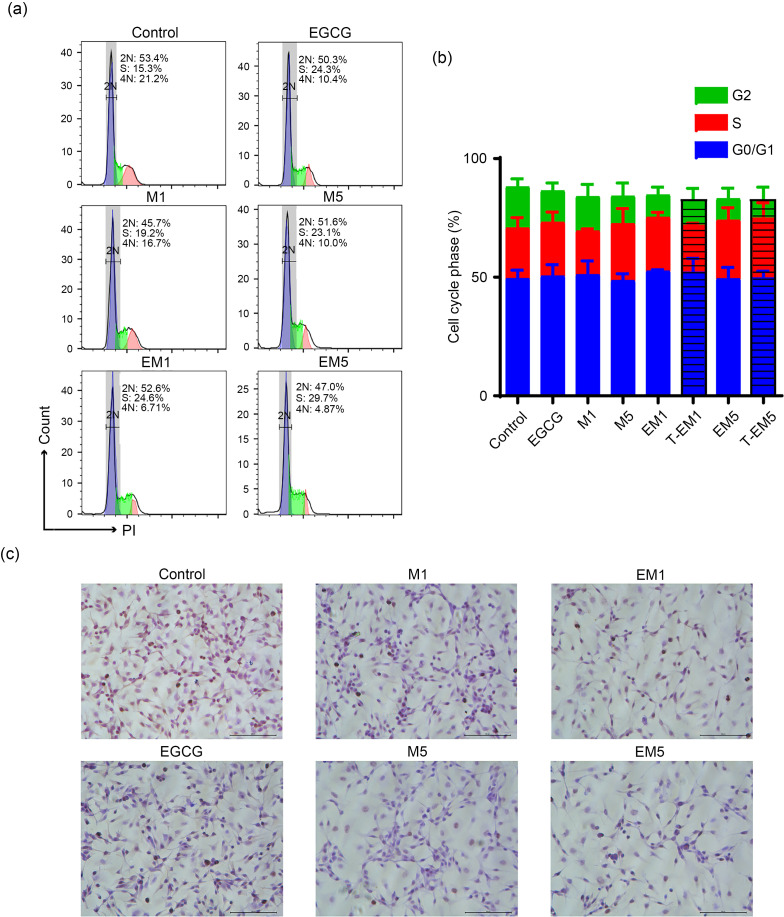
To further verify the effects of EGCG and metformin on different cellular processes of B16F10 cells, we carried out cell apoptosis and proliferation assays. As shown in Figs. 2a and 2b, the proportion of apoptotic cells in the EM5 group was significantly higher than that in the control group (P<0.05). The apoptosis rates of the EM1 and EM5 groups were also higher than those of the single drug treatment groups, but lower than those of T-EM groups. This suggested that a combination treatment of EGCG and metformin may not induce cell apoptosis in B16F10 cells. Figs. 3a and 3b show the effects of EGCG and metformin on cell cycle distribution. The EM1 and EM5 treatment groups showed slight synthesis/Gap2 (S/G2) phase blocks after 24-h treatment, but the differences were not significant when compared with T-EM groups (P>0.05). Furthermore, we determined the combined effect of EGCG and metformin on cell proliferation using immunohistochemistry staining of Ki-67, which is a marker of proliferating cells. Intriguingly, the numbers of Ki-67+ cells in the EM1 and EM5 groups were significantly less than that in other treatment groups (P<0.05; Fig. 3c).
Fig. 2. Impact of EGCG on the pro-apoptotic effects of metformin on B16F10 cells. Representative flow cytometric plots (a) and statistical chart (b) of apoptosis of control B16F10 cells and cells exposed to various treatments for 24 h. Data are presented as mean±standard deviation (SD) and are representative of at least three independent experiments. Different letters among groups indicate significantly different means at P<0.05. EGCG: epigallocatechin-3-gallate; FITC: fluorescein isothiocyanate; PE: phycoerythrin; M1: 1 mmol/L metformin; M5: 5 mmol/L metformin; EM1: 100 μmol/L EGCG plus 1 mmol/L metformin; EM5: 100 μmol/L EGCG plus 5 mmol/L metformin; T-EM1: theoretical value of EM1; T-EM5: theoretical value of EM5.
Fig. 3. Combined effects of EGCG and metformin on cell cycle arrest of B16F10 cells. Representative cell cycle analysis images (a) and statistical chart (b) of B16F10 cells treated with EGCG, metformin, or both for 24 h. There was no significant difference in the data. (c) Representative Ki-67 staining of B16F10 cells. Scale bars=100 μm. Data are presented as mean±standard deviation (SD) and are representative of at least three independent experiments. EGCG: epigallocatechin-3-gallate; M1: 1 mmol/L metformin; M5: 5 mmol/L metformin; EM1: 100 μmol/L EGCG plus 1 mmol/L metformin; EM5: 100 μmol/L EGCG plus 5 mmol/L metformin; T-EM1: theoretical value of EM1; T-EM5: theoretical value of EM5.
In summary, the results showed that metformin and EGCG had an obvious synergistic effect in inhibiting cell proliferation but had a certain antagonistic effect in inducing cell apoptosis, with no significant changes in cell cycle distribution.
3.3. Effects of EGCG and metformin on inflammatory cytokine secretion in B16F10 cells
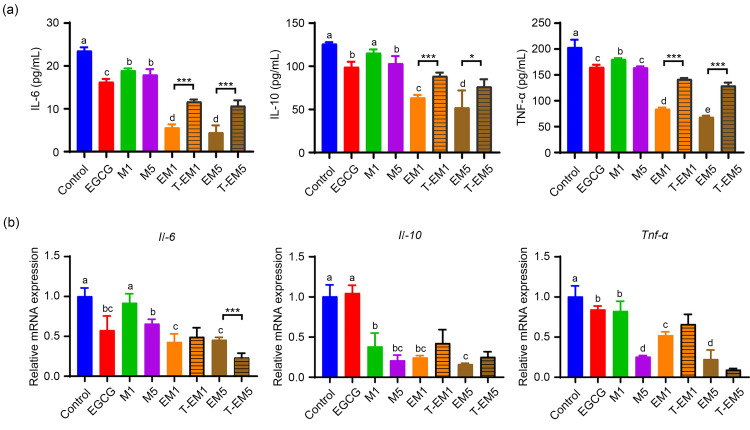
The production of inflammatory cytokines affects the tumor microenvironment in a variety of ways. To further confirm whether the combination of EGCG and metformin could impact the secretion of inflammatory cytokines, the levels of IL-6, IL-10, and TNF-α in the cell supernatant were determined by ELISA. Both EGCG and metformin treatments reduced the levels of these three inflammatory cytokines, while the combination treatment with EGCG and metformin showed significantly decreased levels of IL-6, IL-10, and TNF-α (P<0.05; Fig. 4a), suggesting a synergistic effect of EGCG and metformin on cytokine production. At the messenger RNA (mRNA) level, the EM1 group showed a more significant synergistic inhibition of inflammatory cytokines than the EM5 group (Fig. 4b). In general, EGCG and metformin synergistically inhibited the expression and secretion of inflammatory cytokines in B16F10 melanoma cells, suggesting that the regulation of inflammatory cytokines may be an important mechanism to inhibit the growth and migration of these cells.
Fig. 4. Effects of EGCG on metformin-induced suppression of inflammatory cytokine expression and secretion in B16F10 cells. (a) Levels of IL-6, IL-10, and TNF-α in the cell supernatant of the control and other treated groups were measured by ELISA. (b) Il-6, Il-10, and Tnf-α mRNA expression in B16F10 cells from the different treatment groups. Data are presented as mean±standard deviation (SD) and are representative of at least three independent experiments. Different letters among groups indicate significantly different means at P<0.05. IL-6: interleukin-6; TNF-α: tumor necrosis factor-α; EGCG: epigallocatechin-3-gallate; M1: 1 mmol/L metformin; M5: 5 mmol/L metformin; EM1: 100 μmol/L EGCG plus 1 mmol/L metformin; EM5: 100 μmol/L EGCG plus 5 mmol/L metformin; T-EM1: theoretical value of EM1; T-EM5: theoretical value of EM5; ELISA: enzyme-linked immunosorbent assay.
3.4. Effects of EGCG and metformin on the level of oxidative stress in B16F10 cells
The expression of inflammatory cytokines is often accompanied by oxidative stress, and the induction of oxidative stress plays an important role in the action of many anticancer drugs (Sun et al., 2011; Klaunig, 2018). Adaptation to oxidative stress is beneficial to cancer cells, while over-exposure of cells to reactive oxygen species (ROS) can induce apoptosis of cancer cells, transform the redox state to the oxidized state, and regulate the immune system to cause tumor degeneration (Chikara et al., 2018; Klaunig, 2018; Farhood et al., 2019). Therefore, we explored whether EGCG and metformin could regulate the oxidation state of B16F10 cells.
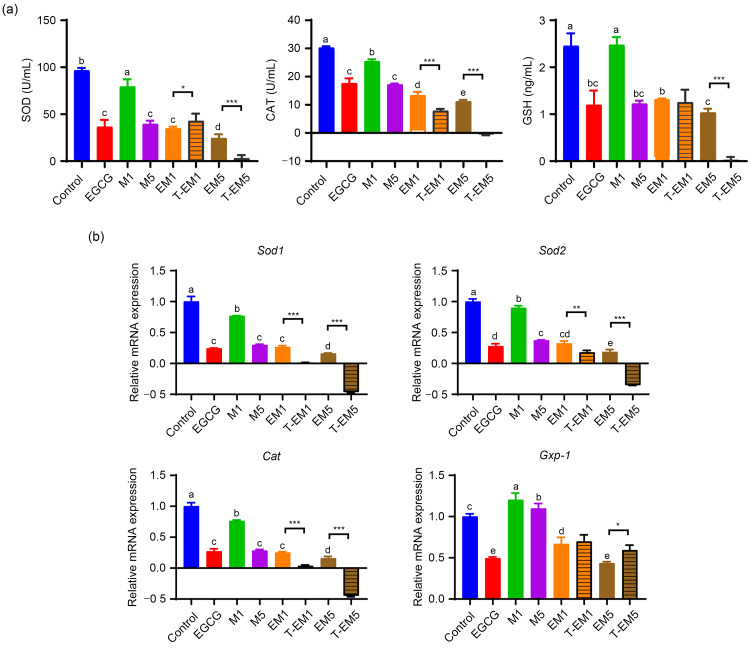
As shown in Fig. 5a, 100 μmol/L EGCG treatment exhibited similar SOD, CAT, and GSH levels to M5, but both treatments showed significantly reduced levels when compared to the control group or M1 treatment group (P<0.05). Additionally, the combination treatment of EM5 displayed the lowest productions of SOD, CAT, and GSH, indicating that it further reduced antioxidant enzyme levels. Based on the comparison of EM and T-EM, the EM5 combination showed a synergistic effect on the secretion of SOD, CAT, and GSH (Fig. 5a). Similar results were obtained when we investigated the mRNA expression of Sod1, Sod2, and Cat (Fig. 5b). In contrast, the mRNA expression of Gxp-1 in the EM5 group was significantly reduced when compared with T-EM5 (P<0.05).
Fig. 5. Effects of EGCG and metformin on oxidative stress levels in B16F10 cells. (a) Levels of SOD, CAT, and GSH in the cell supernatant treated for 24 h with EGCG, metformin, or both were measured by ELISA. (b) Sod1, Sod2, Cat, and Gpx-1 mRNA expression in B16F10 cells in the different treatment groups. Data are presented as mean±standard deviation (SD) and are representative of at least three independent experiments. Different letters among groups indicate significantly different means at P<0.05. * P<0.05, ** P<0.01, and *** P<0.001 for T-EM vs. EM. EGCG: epigallocatechin-3-gallate; M1: 1 mmol/L metformin; M5: 5 mmol/L metformin; EM1: 100 μmol/L EGCG plus 1 mmol/L metformin; EM5: 100 μmol/L EGCG plus 5 mmol/L metformin; T-EM1: theoretical value of EM1; T-EM5: theoretical value of EM5; SOD: superoxide dismutase; CAT: catalase; GSH: glutathione; ELISA: enzyme-linked immunosorbent assay.
In general, EGCG and metformin showed an antagonistic effect on the oxidative stress level of B16F10 cells, but the oxidative stress level of the combined group was still higher than that of the single treatment group. Interestingly, we found that the oxidative stress level of the EM group was similar to that of the EGCG group in both protein and mRNA expression, especially in the low-concentration metformin group (EM1 group), indicating that the combined effect of the two is likely to be mainly driven by EGCG.
3.5. Effects of EGCG and metformin on STAT3 and NF-κB p65 signaling pathways in B16F10 cells
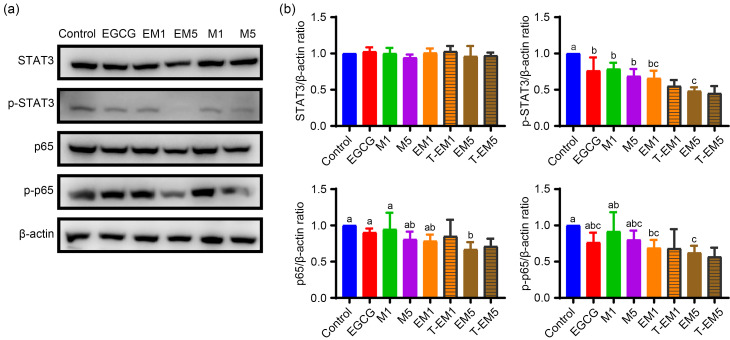
STAT3 (Yu et al., 2009; Chai et al., 2016) and NF-κB (Li et al., 2015; Taniguchi and Karin, 2018) are highly regulated transcription factors that are overactive in many tumors. Therefore, we further investigated the underlying signaling pathways involved in the EGCG and metformin treatments in B16F10 cells. Western blot analysis (Figs. 6a and 6b) showed that there was no change in the total protein level of STAT3 among the groups, and the phosphorylation level, EM or T-EM values were not significantly different (P>0.05), indicating that EGCG and metformin had a certain synergistic effect in inhibiting the STAT3 phosphorylation of B16F10 cells, but the synergistic effect was not obvious. For the NF-κB p65 signaling pathway, the p65 total levels or phosphorylation levels of the EGCG and EM groups were not significantly different from the control group (P>0.05), but the phosphorylation levels of the EM groups showed a significant inhibitory effect (P<0.05), and the EM and T-EM showed no differences, indicating synergistic inhibition of p65 activation. In summary, the combination of EGCG and metformin could synergistically inhibit the phosphorylation levels of the STAT3 and NF-κB p65 signaling pathways, thereby regulating the cellular process of B16F10 cells.
Fig. 6. EGCG exacerbated the inhibitory effects of metformin on the STAT3 and NF-κB p65 signaling pathways in B16F10 cells. (a) Immunoblot analysis of phosphorylated (p-) and total STAT3 and NF-κB p65 protein levels in lysates of B16F10 cells exposed to various treatments and control cells. (b) Protein expression was normalized to that of β-actin. Data are presented as mean±standard deviation (SD) and are representative of at least three independent experiments. Different letters among groups indicate significantly different means at P<0.05. EGCG: epigallocatechin-3-gallate; M1: 1 mmol/L metformin; M5: 5 mmol/L metformin; EM1: 100 μmol/L EGCG plus 1 mmol/L metformin; EM5: 100 μmol/L EGCG plus 5 mmol/L metformin; T-EM1: theoretical value of EM1; T-EM5: theoretical value of EM5; STAT3: signal transducer and activator of transcription 3; NF-κB: nuclear factor-κB.
3.6. Effects of EGCG and metformin on the biochemical composition of B16F10 cells at the subcellular level
Raman spectroscopy is a fast, environmentally friendly, and simple detection method that can evaluate the molecular composition of biological samples at the subcellular level (Heath et al., 2016; Mignolet et al., 2018; Xu et al., 2019). Therefore, we used Raman spectroscopy to further identify the effects of EGCG and metformin on the biochemical components of B16F10 cells at the subcellular level.
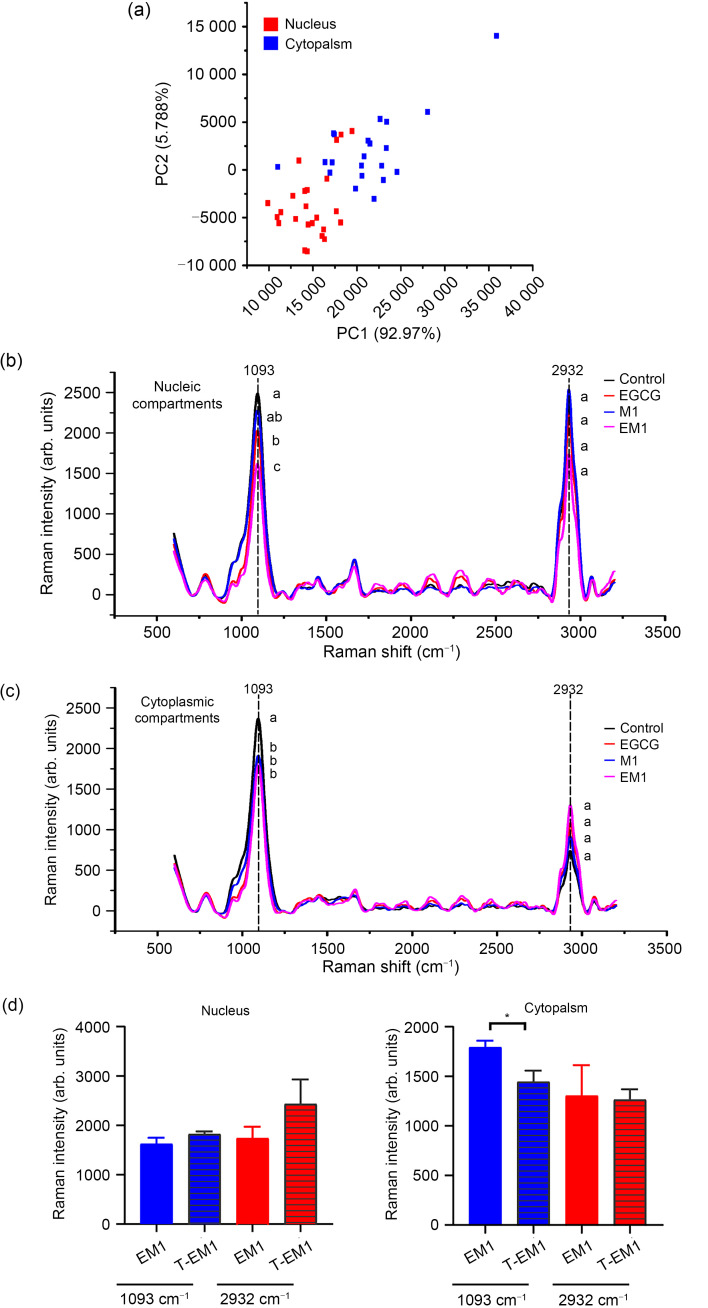
First, the location of the nucleus was determined by crystal violet staining. As shown in Fig. S1, the spectra of the nucleus and cytoplasm of the B16F10 cell were obtained and subjected to PCA. As shown in Fig. 7a, the nuclear and cytoplasmic spectra of B16F10 cells could be distinguished by first principal component (PC1), indicating that the chemical composition of the two was different.
Fig. 7. Effects of EGCG and metformin on nucleic acids, lipids, and proteins in the B16F10 cell nucleus and cytoplasm. (a) The PCA score plot of the PC2 versus the PC1. Raman spectra from the nucleus (b) and cytoplasm (c) of B16F10 cells in the control, EGCG, metformin, and combination-treated groups. (d) Statistical analysis of the Raman intensity at 1093 and 2932 cm -1. Data are presented as mean±standard deviation (SD) and are representative of at least three independent experiments. Different letters among groups indicate significantly different means at P<0.05. * P<0.05 for theoretical value vs. experimental value. EGCG: epigallocatechin-3-gallate; M1: 1 mmol/L metformin; EM1: 100 μmol/L EGCG plus 1 mmol/L metformin; T-EM1: theoretical value of EM1; PCA: principal component analysis; PC2: the second principal component; PC1: the first principal component; arb.: arbitrary.
We obtained the Raman bands of B16F10 cells by treating them with different compounds (EGCG, M1, and EM1) for 24 h, and used the untreated group as a control. The results were obtained at both the nuclear (Fig. 7b) and the cytoplasmic (Fig. 7c) levels. The comparison showed that there were two obvious peaks at bands 1093 and 2932 cm-1, which were respectively related to the symmetric extension of phosphate (PO2 -) of nucleic acids and the asymmetric extension of CH3 of lipids and proteins (Movasaghi et al., 2007).
At the nuclear level, the Raman intensity of each treatment group at the 1093 and 2932 cm-1 bands was lower than that of the control group; there was a significant difference at 1093 cm-1 (P<0.05), but no significant difference at 2932 cm-1 (P>0.05) (Fig. 7b). Based on the comparative analysis of EM1 and T-EM1 at the nucleus level (Fig. 7d), EGCG and metformin synergistically destroyed the nuclear structure of B16F10 cells, which was mainly reflected in the destruction of the nucleic acid structure of the cell nucleus.
Cytoplasmic spectrum changes are shown in Fig. 7c. Compared with the control group, each treatment group induced a decrease at 1093 cm-1. The intensity of the EM1 group was slightly lower than that of the EGCG and M1 groups, but the effect was weaker than that of the T-EM1 (Fig. 7d). The Raman band observed at 2932 cm-1 showed the opposite trend. The Raman intensity of each treatment group was higher than that of the control group and generally similar, although the EM1 group had a slightly larger increase. The results showed that compared with single administration, combined administration of EGCG and metformin showed antagonistic effects to some extent, although lipids and proteins may be further accumulated and the nucleic acid structure of B16F10 cytoplasm destroyed.
In summary, EGCG and metformin showed a synergistic effect on the destruction of the nuclear nucleic acid structure of B16F10 cells, but this effect was not shown in the cytoplasm. Interfering with the transcription of DNA damage can lead to apoptosis and senescence (Wolters and Schumacher, 2013), which suggests that changes in nucleic acid may be related to the apoptosis of B16F10 cells induced by EGCG and metformin.
4 Discussion
The incidence of melanoma has gradually increased in the past few decades, and because of its high degrees of malignancy, rapid development, and extremely poor prognosis, it is becoming the most fatal disease of the skin (Miller et al., 2019; Siegel et al., 2019; Zou et al., 2019). Both EGCG and metformin have been shown to have potential anti-cancer effects, but different tumor cells exhibited different sensitivities to these two compounds (Fujiki et al., 2015; de Souza Neto et al., 2017; Hart et al., 2019; Liu et al., 2019). This study evaluated the anti-tumor activity of EGCG and metformin in B16F10 cells to explore their roles in the treatment of melanoma. Our results showed that EGCG and metformin have significant synergistic effects on inhibiting the growth and migration of B16F10 cells and inhibiting the levels of inflammatory factors and the STAT3/NF-κB pathway; they have a certain antagonistic effect on the levels of cell apoptosis, but the effect of the combination is superior to that of the individual drugs.
The relationship between inflammation and tumor development is a long-standing scientific problem (Balkwill and Mantovani, 2001). Inflammation is known as the eighth major biological feature of malignant tumors, and it plays an important role in tumor development, invasion, and metastasis (Hanahan and Weinberg, 2011). Inflammatory cytokines such as IL-1, IL-6, IL-10, IL-12, TNF-α, and transforming growth factor-β (TGF-β) are abundant in tumor microenvironments. Under the stimulation of inflammatory mediators, tumor cells can maintain their own growth, invasion, and migration, and recruit inflammatory cells by releasing various inflammatory cytokines to amplify the inflammatory effect (Hanahan and Weinberg, 2011). In addition, inflammatory cytokines can also mediate complex pathways, such as by activating NF-κB, STAT3, and other cell signaling pathways, thereby inducing the expression of a variety of pro-inflammatory cytokines, chemokines, and angiogenic factors, and accelerating the development of tumors (Karin, 2006; Yu et al., 2007). Our experimental results also confirmed this point. EGCG and metformin synergistically inhibited the phosphorylation levels of NF-κB p65 and STAT3 signaling pathways, thereby inhibiting the transcription and expression of pro-inflammatory genes. This led to significant decreases in IL-6, IL-10, and TNF-α levels, thus inhibiting the growth and migration of B16F10 cells. Based on these findings, we suggest that the combination of EGCG and metformin could play a potential role in tumor progression mediated by inflammatory cytokines, and has the potential for development into anti-tumor drugs targeting inflammatory cytokines.
The process of apoptosis is essential to maintain the physiological balance between cell death and cell growth (Sabry et al., 2019). Our experiment explored the effects of EGCG and metformin on apoptosis of B16F10 cells through mediation of oxidative stress. An increase in ROS is related to abnormal growth of cancer cells and reflects the destruction of redox homeostasis, leading to a state called oxidative stress (Toyokuni et al., 1995). Excessive ROS can be toxic to cells, causing oxidative damage to lipids, proteins, and DNA. This leads to the death of malignant cells, thereby limiting cancer progression (Perry et al., 2000; Fruehauf and Meyskens, 2007; Schumacher et al., 2008). The increase in ROS production is largely due to the decrease in ROS scavenging ability. ROS scavenging systems include SOD (SOD1, SOD2, and SOD3), GSH peroxidase, peroxide redoxin, glutaraldehyde, thioredoxin, and CAT (Trachootham et al., 2009). Our results showed that both EGCG and metformin treatment had a significant inhibitory effect on antioxidant enzyme (SOD, CAT, and GSH) activity. Although the combined effect of the two was stronger than that of the single treatment group, it did not show a synergistic effect. Interestingly, we found that the combined effect of the two was likely to be largely driven by EGCG. Raman spectroscopy results showed that EGCG and metformin synergistically destroyed the nuclear nucleic acid structure of B16F10 cells, which may be partly due to the increase of ROS. Therefore, EGCG and metformin may damage the nuclear nucleic acid structure by increasing ROS, thereby interfering with transcriptional DNA damage and leading to cell apoptosis and senescence. Existing studies have shown that in order to maximize ROS-mediated cell death mechanism as a therapeutic strategy, drugs that induce ROS production can be combined with compounds that inhibit the antioxidant capacity of cells. For example, the combination of ROS generator arsenic trioxide and SOD inhibitor 2-methoxyestradiol (2-ME) was effective in treating primary chronic lymphocytic leukemia (CLL) cells (Zhou et al., 2003). Although the combination of EGCG and metformin did not show a significant synergistic inhibitory effect on antioxidant enzyme activity in this experiment, EGCG was found to be a promising compound for inhibiting the antioxidant capacity of cells. Future studies could try to combine EGCG and drugs that induce ROS production to promote tumor cell apoptosis through oxidative stress.
5 Conclusions
In summary, the work presented here suggests that EGCG exerts a partially additive effect when combined with metformin in melanoma cancer cells by inhibiting cell growth, cytokine levels, and the STAT3/NF-κB pathway. On the other hand, the two compounds possess antagonistic effects on apoptosis and anti-enzyme secretion. To further confirm their potential combined effects on melanoma, more studies are necessary to define the pharmacodynamic interaction between EGCG and metformin.
Supplementary information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. U19A2034). We thank Dr. Le YING (Hudson Institute of Medical Research, Australia) for English-language editing.
Author contributions
An'an XU performed the experimental research and data analysis, wrote and edited the manuscript. Jeehyun LEE and Yueling ZHAO performed the experimental research and data analysis. Yuefei WANG contributed to the study design. Xiaoli LI contributed to the data analysis. Ping XU contributed to the study design, writing and editing of the manuscript. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
An'an XU, Jeehyun LEE, Yueling ZHAO, Yuefei WANG, Xiaoli LI, and Ping XU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Ahmad E, Sargeant JA, Zaccardi F, et al. , 2020. Where does metformin stand in modern day management of type 2 diabetes? Pharmaceuticals, 13(12): 427. 10.3390/ph13120427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F, Mantovani A, 2001. Inflammation and cancer: back to Virchow? Lancet, 357(9255): 539-545. 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- Caltagirone S, Rossi C, Poggi A, et al. , 2000. Flavonoids apigenin and quercetin inhibit melanoma growth and metastatic potential. Int J Cancer, 87(4): 595-600. [DOI] [PubMed] [Google Scholar]
- Cerezo M, Tichet M, Abbe P, et al. , 2013. Metformin blocks melanoma invasion and metastasis development in AMPK/p53-dependent manner. Mol Cancer Ther, 12(8): 1605-1615. 10.1158/1535-7163.MCT-12-1226-T [DOI] [PubMed] [Google Scholar]
- Chae YK, Arya A, Malecek MK, et al. , 2016. Repurposing metformin for cancer treatment: current clinical studies. Oncotarget, 7(26): 40767-40780. 10.18632/oncotarget.8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai EZP, Shanmugam MK, Arfuso F, et al. , 2016. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol Ther, 162: 86-97. 10.1016/j.pharmthera.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Chen K, Qian WK, Jiang ZD, et al. , 2017. Metformin suppresses cancer initiation and progression in genetic mouse models of pancreatic cancer. Mol Cancer, 16: 131. 10.1186/s12943-017-0701-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SN, Zhu XM, Lai XF, et al. , 2014. Combined cancer therapy with non-conventional drugs: all roads lead to AMPK. Mini Rev Med Chem, 14(8): 642-654. 10.2174/1389557514666140820104444 [DOI] [PubMed] [Google Scholar]
- Chikara S, Nagaprashantha LD, Singhal J, et al. , 2018. Oxidative stress and dietary phytochemicals: role in cancer chemoprevention and treatment. Cancer Lett, 413: 122-134. 10.1016/j.canlet.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Courtois S, Lehours P, Bessède E, 2019. The therapeutic potential of metformin in gastric cancer. Gastric Cancer, 22(4): 653-662. 10.1007/s10120-019-00952-w [DOI] [PubMed] [Google Scholar]
- de Souza Neto FP, Bernardes SS, Marinello PC, et al. , 2017. Metformin: oxidative and proliferative parameters in-vitro and in-vivo models of murine melanoma. Melanoma Res, 27(6): 536-544. 10.1097/CMR.0000000000000391 [DOI] [PubMed] [Google Scholar]
- Ellis LZ, Liu WM, Luo YC, et al. , 2011. Green tea polyphenol epigallocatechin-3-gallate suppresses melanoma growth by inhibiting inflammasome and IL-1β secretion. Biochem Biophys Res Commun, 414(3): 551-556. 10.1016/j.bbrc.2011.09.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhood B, Najafi M, Salehi E, et al. , 2019. Disruption of the redox balance with either oxidative or anti-oxidative overloading as a promising target for cancer therapy. J Cell Biochem, 120(1): 71-76. 10.1002/jcb.27594 [DOI] [PubMed] [Google Scholar]
- Fruehauf JP, Meyskens FL, 2007. Reactive oxygen species: a breath of life or death? Clin Cancer Res, 13(3): 789-794. 10.1158/1078-0432.CCR-06-2082 [DOI] [PubMed] [Google Scholar]
- Fujiki H, Sueoka E, Watanabe T, et al. , 2015. Primary cancer prevention by green tea, and tertiary cancer prevention by the combination of green tea catechins and anticancer compounds. J Cancer Prev, 20(1): 1-4. 10.15430/JCP.2015.20.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA, 2011. Hallmarks of cancer: the next generation. Cell, 144(5): 646-674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hart PC, Chiyoda T, Liu XJ, et al. , 2019. SPHK1 is a novel target of metformin in ovarian cancer. Mol Cancer Res, 17(4): 870-881. 10.1158/1541-7786.MCR-18-0409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman RI, Lin JY, 2019. Cutaneous melanoma—a review in detection, staging, and management. Hematol Oncol Clin North Am, 33(1): 25-38. 10.1016/j.hoc.2018.09.005 [DOI] [PubMed] [Google Scholar]
- Heath JR, Ribas A, Mischel PS, 2016. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov, 15(3): 204-216. 10.1038/nrd.2015.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges V, Tucci M, Benghuzzi H, 2015. The effects of metformin and EGCG on PANC-1 cell survival. Biomed Sci Instrum, 51: 393-399. [PubMed] [Google Scholar]
- Iglesias-Pena N, Paradela S, Tejera-Vaquerizo A, et al. , 2019. Cutaneous melanoma in the elderly: review of a growing problem. Actas Dermosifiliogr, 110(6): 434-447. 10.1016/j.ad.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Janjetovic K, Harhaji-Trajkovic L, Misirkic-Marjanovic M, et al. , 2011. In vitro and in vivo anti-melanoma action of metformin. Eur J Pharmacol, 668(3): 373-382. 10.1016/j.ejphar.2011.07.004 [DOI] [PubMed] [Google Scholar]
- Jaune E, Rocchi S, 2018. Metformin: focus on melanoma. Front Endocrinol, 9: 472. 10.3389/fendo.2018.00472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, 2006. Nuclear factor-κB in cancer development and progression. Nature, 441(7092): 431-436. 10.1038/nature04870 [DOI] [PubMed] [Google Scholar]
- Kim DS, Park SH, Kwon SB, et al. , 2004. (-)-Epigallocatechin-3-gallate and hinokitiol reduce melanin synthesis via decreased MITF production. Arch Pharm Res, 27(3): 334-339. 10.1007/bf02980069 [DOI] [PubMed] [Google Scholar]
- Kim K, Yang WH, Jung YS, et al. , 2020. A new aspect of an old friend: the beneficial effect of metformin on anti-tumor immunity. BMB Rep, 53(10): 512-520. 10.5483/BMBRep.2020.53.10.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaunig JE, 2018. Oxidative stress and cancer. Curr Pharm Des, 24(40): 4771-4778. 10.2174/1381612825666190215121712 [DOI] [PubMed] [Google Scholar]
- Lee JH, Kishikawa M, Kumazoe M, et al. , 2010. Vitamin A enhances antitumor effect of a green tea polyphenol on melanoma by upregulating the polyphenol sensing molecule 67-kDa laminin receptor. PLoS ONE, 5(6): e11051. 10.1371/journal.pone.0011051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhang JW, Arfuso F, et al. , 2015. NF-κB in cancer therapy. Arch Toxicol, 89(5): 711-731. 10.1007/s00204-015-1470-4 [DOI] [PubMed] [Google Scholar]
- Li K, Zhang TT, Wang F, et al. , 2018. Metformin suppresses melanoma progression by inhibiting KAT5-mediated SMAD3 acetylation, transcriptional activity and TRIB3 expression. Oncogene, 37(22): 2967-2981. 10.1038/s41388-018-0172-9 [DOI] [PubMed] [Google Scholar]
- Liu MH, Zhang Z, Wang H, et al. , 2019. Activation of AMPK by metformin promotes renal cancer cell proliferation under glucose deprivation through its interaction with PKM2. Int J Biol Sci, 15(3): 617-627. 10.7150/ijbs.29689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord SR, Cheng WC, Liu D, et al. , 2018. Integrated pharmacodynamic analysis identifies two metabolic adaption pathways to metformin in breast cancer. Cell Metab, 28(5): 679-688.e4. 10.1016/j.cmet.2018.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Chiang JH, Tsai FJ, et al. , 2019. Metformin triggers the intrinsic apoptotic response in human AGS gastric adenocarcinoma cells by activating AMPK and suppressing mTOR/AKT signaling. Int J Oncol, 54(4): 1271-1281. 10.3892/ijo.2019.4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LW, Wei JW, Wan JH, et al. , 2019. Low glucose and metformin-induced apoptosis of human ovarian cancer cells is connected to ASK1 via mitochondrial and endoplasmic reticulum stress-associated pathways. J Exp Clin Cancer Res, 38(1): 77. 10.1186/s13046-019-1090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet A, Wood BR, Goormaghtigh E, 2018. Intracellular investigation on the differential effects of 4 polyphenols on MCF-7 breast cancer cells by Raman imaging. Analyst, 143: 258-269. 10.1039/c7an01460k [DOI] [PubMed] [Google Scholar]
- Miller KD, Nogueira L, Mariotto AB, et al. , 2019. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin, 69(5): 363-385. 10.3322/caac.21565 [DOI] [PubMed] [Google Scholar]
- Movasaghi Z, Rehman S, Rehman IU, 2007. Raman spectroscopy of biological tissues. Appl Spectrosc Rev, 42(5): 493-541. 10.1080/05704920701551530 [DOI] [Google Scholar]
- Nicolucci A, Charbonnel B, Gomes MB, et al. , 2019. Treatment patterns and associated factors in 14 668 people with type 2 diabetes initiating a second-line therapy: results from the global DISCOVER study programme. Diabetes, Obes Metab, 21(11): 2474-2485. 10.1111/dom.13830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehr F, von Euw E, Attar N, et al. , 2011. Combination therapy with vemurafenib (PLX4032/RG7204) and metformin in melanoma cell lines with distinct driver mutations. J Transl Med, 9: 76. 10.1186/1479-5876-9-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihal M, Ahsan H, Siddiqui IA, et al. , 2009. (-)-Epigallocatechin-3-gallate (EGCG) sensitizes melanoma cells to interferon induced growth inhibition in a mouse model of human melanoma. Cell Cycle, 8(13): 2057-2063. 10.4161/cc.8.13.8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihal M, Roelke CT, Wood GS, 2010. Anti-melanoma effects of vorinostat in combination with polyphenolic antioxidant (-)-epigallocatechin-3-gallate (EGCG). Pharm Res, 27(6): 1103-1114. 10.1007/s11095-010-0054-5 [DOI] [PubMed] [Google Scholar]
- Pasha M, Sivaraman SK, Frantz R, et al. , 2019. Metformin induces different responses in clear cell renal cell carcinoma Caki cell lines. Biomolecules, 9(3): 113. 10.3390/biom9030113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G, Raina AK, Nunomura A, et al. , 2000. How important is oxidative damage? Lessons from Alzheimer’s disease. Free Radical Bio Med, 28(5): 831-834. 10.1016/S0891-5849(00)00158-1 [DOI] [PubMed] [Google Scholar]
- Ravindranath MH, Ramasamy V, Moon S, et al. , 2009. Differential growth suppression of human melanoma cells by tea (Camellia sinensis) epicatechins (ECG, EGC and EGCG). Evid Based Complement Alternat Med, 6(4): 523-530. 10.1093/ecam/nem140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabry D, Abdelaleem OO, el Amin Ali AM, et al. , 2019. Anti-proliferative and anti-apoptotic potential effects of epigallocatechin-3-gallate and/or metformin on hepatocellular carcinoma cells: in vitro study. Mol Biol Rep, 46(2): 2039-2047. 10.1007/s11033-019-04653-6 [DOI] [PubMed] [Google Scholar]
- Schumacher B, Garinis GA, Hoeijmakers JHJ, 2008. Age to survive: DNA damage and aging. Trends Genet, 24(2): 77-85. 10.1016/j.tig.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Shen Q, Tian F, Jiang P, et al. , 2009. EGCG enhances TRAIL-mediated apoptosis in human melanoma A375 cell line. J Huazhong Univ Sci Technol Med Sci, 29: 771. 10.1007/s11596-009-0620-4 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2019. Cancer statistics, 2019. CA Cancer J Clin, 69(1): 7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- Sun RJ, Zhai RR, Ma CL, et al. , 2020. Combination of aloin and metformin enhances the antitumor effect by inhibiting the growth and invasion and inducing apoptosis and autophagy in hepatocellular carcinoma through PI3K/AKT/mTOR pathway. Cancer Med, 9(3): 1141-1151. 10.1002/cam4.2723 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun Y, Huang LQ, Mackenzie GG, et al. , 2011. Oxidative stress mediates through apoptosis the anticancer effect of phospho-nonsteroidal anti-inflammatory drugs: implications for the role of oxidative stress in the action of anticancer agents. J Pharmacol Exp Ther, 338(3): 775-783. 10.1124/jpet.111.183533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Takeuchi O, Suzuki Y, et al. , 2019. Mechanisms of metformin’s anti-tumor activity against gemcitabine-resistant pancreatic adenocarcinoma. Int J Oncol, 54(2): 764-772. 10.3892/ijo.2018.4662 [DOI] [PubMed] [Google Scholar]
- Tang GJ, Guo JF, Zhu YP, et al. , 2018. Metformin inhibits ovarian cancer via decreasing H3K27 trimethylation. Int J Oncol, 52(6): 1899-1911. 10.3892/ijo.2018.4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Karin M, 2018. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol, 18(5): 309-324. 10.1038/nri.2017.142 [DOI] [PubMed] [Google Scholar]
- Taniguchi S, Fujiki H, Kobayashi H, et al. , 1992. Effect of (-)-epigallocatechin gallate, the main constituent of green tea, on lung metastasis with mouse B16 melanoma cell lines. Cancer Lett, 65(1): 51-54. 10.1016/0304-3835(92)90212-e [DOI] [PubMed] [Google Scholar]
- Tomic T, Botton T, Cerezo M, et al. , 2011. Metformin inhibits melanoma development through autophagy and apoptosis mechanisms. Cell Death Dis, 2(9): e199. 10.1038/cddis.2011.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokuni S, Okamoto K, Yodoi J, et al. , 1995. Persistent oxidative stress in cancer. FEBS Lett, 358(1): 1-3. 10.1016/0014-5793(94)01368-B [DOI] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P, 2009. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov, 8(7): 579-591. 10.1038/nrd2803 [DOI] [PubMed] [Google Scholar]
- Wang L, Li K, Lin XJ, et al. , 2019. Metformin induces human esophageal carcinoma cell pyroptosis by targeting the miR-497/PELP1 axis. Cancer Lett, 450: 22-31. 10.1016/j.canlet.2019.02.014 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Kuramochi H, Takahashi A, et al. , 2012. Higher cell stiffness indicating lower metastatic potential in B16 melanoma cell variants and in (-)-epigallocatechin gallate-treated cells. J Cancer Res Clin Oncol, 138(5): 859-866. 10.1007/s00432-012-1159-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R, Penso NEC, Hackman RM, et al. , 2019a. Epigallocatechin-3-gallate (EGCG) suppresses pancreatic cancer cell growth, invasion, and migration partly through the inhibition of Akt pathway and epithelial-mesenchymal transition: enhanced efficacy when combined with gemcitabine. Nutrients, 11(8): 1856. 10.3390/nu11081856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R, Hackman RM, Wang YF, et al. , 2019b. Targeting glycolysis with epigallocatechin-3-gallate enhances the efficacy of chemotherapeutics in pancreatic cancer cells and xenografts. Cancers, 11(10): 1496. 10.3390/cancers11101496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters S, Schumacher B, 2013. Genome maintenance and transcription integrity in aging and disease. Front Genet, 4: 19. 10.3389/fgene.2013.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Lin Y, Liu HJ, et al. , 2008. Inhibition of invasion and up-regulation of E-cadherin expression in human malignant melanoma cell line A375 by (-)-epigallocatechin-3-gallate. J Huazhong Univ Sci Technolog Med Sci, 28(3): 356-359. 10.1007/s11596-008-0330-3 [DOI] [PubMed] [Google Scholar]
- Xu N, Zhu PP, Liang J, et al. , 2019. Label-free Raman spectroscopy monitoring of cytotoxic response induced by a telomerase inhibitor. Sens Actuat B Chem, 293: 1-10. 10.1016/j.snb.2019.03.146 [DOI] [Google Scholar]
- Xu P, Yan F, Zhao YL, et al. , 2020. Green tea polyphenol EGCG attenuates MDSCs-mediated immunosuppression through canonical and non-canonical pathways in a 4T1 murine breast cancer model. Nutrients, 12(4): 1042. 10.3390/nu12041042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Tsukamoto S, Huang YH, et al. , 2016. Epigallocatechin-3-O-gallate up-regulates microRNA-let-7b expression by activating 67-kDa laminin receptor signaling in melanoma cells. Sci Rep, 6: 19225. 10.1038/srep19225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Wang X, Lu G, et al. , 2009. Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nat Rev Cancer, 9(6): 429-439. 10.1038/nrc2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CX, Jiao Y, Xue J, et al. , 2017. Metformin sensitizes non-small cell lung cancer cells to an epigallocatechin-3-gallate (EGCG) treatment by suppressing the Nrf2/HO-1 signaling pathway. Int J Biol Sci, 13(12): 1560-1569. 10.7150/ijbs.18830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D, 2007. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol, 7(1): 41-51. 10.1038/nri1995 [DOI] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R, 2009. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer, 9(11): 798-809. 10.1038/nrc2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zhou W, Wang HM, et al. , 2019. Transdermal metformin hydrochloride-loaded cubic phases: in silico formulation optimization, preparation, properties, and application for local treatment of melanoma. Drug Deliv, 26(1): 376-383. 10.1080/10717544.2019.1587046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JL, Lei Z, Huang ZN, et al. , 2016. Epigallocatechin-3-gallate (EGCG) suppresses melanoma cell growth and metastasis by targeting TRAF6 activity. Oncotarget, 7(48): 79557-79571. 10.18632/oncotarget.12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YP, Peng GY, Hsueh EC, 2015. Abstract 1196: enhancement of anti-melanoma effect of BRAF and MEK inhibition bymetformin. Cancer Res, 75(15): 1196. 10.1158/1538-7445.Am2015-1196 [DOI] [Google Scholar]
- Zhou Y, Hileman EO, Plunkett W, et al. , 2003. Free radical stress in chronic lymphocytic leukemia cells and its role in cellular sensitivity to ROS-generating anticancer agents. Blood, 101(10): 4098-4104. 10.1182/blood-2002-08-2512 [DOI] [PubMed] [Google Scholar]
- Zordoky BNM, Bark D, Soltys CL, et al. , 2014. The anti-proliferative effect of metformin in triple-negative MDA-MB-231 breast cancer cells is highly dependent on glucose concentration: implications for cancer therapy and prevention. Biochim Biophys Acta, 1840(6): 1943-1957. 10.1016/j.bbagen.2014.01.023 [DOI] [PubMed] [Google Scholar]
- Zou G, Bai J, Li DD, et al. , 2019. Effect of metformin on the proliferation, apoptosis, invasion and autophagy of ovarian cancer cells. Exp Ther Med, 18(3): 2086-2094. 10.3892/etm.2019.7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.