Abstract
Although microRNA-155 (miR-155) is considered a pro-inflammatory mediator, cumulative evidence indicates that it also has anti-inflammatory effects in macrophages and dendritic cells. In this study, we identified the dramatic expression changes of more than half of potential miR-155-targeted genes upon lipopolysaccharide (LPS) stimulation; 223 genes were down-regulated and 85 genes were up-regulated, including suppressor of cytokine signaling 1 (SOCS1) and transforming growth factor-β-activated kinase 1-binding protein 2 (TAB2), two well-known genes involved in miR-155-mediated regulation of the Toll-like receptor 4 (TLR4) signaling pathway. We also found that miR-155 acted as an anti-inflammatory mediator in the initial stage of LPS-induced inflammatory response mainly through repressing TAB2 protein translation, and as a pro-inflammatory mediator by down-regulating SOCS1 in the later stage. Meanwhile, overexpression of TAB2 3' untranslated region (UTR) in macrophages promoted the development of endotoxin tolerance by competing for binding with miR-155, which resulted in an elevated expression level of SOCS1 protein. These findings provide new insights for understanding the regulatory mechanisms in fine-tuning of LPS-induced innate immune response.
Keywords: Toll-like receptor 4 (TLR4), Endotoxin tolerance, MicroRNA-155 (miR-155), Suppressor of cytokine signaling 1 (SOCS1), Transforming growth factor-β-activated kinase 1-binding protein 2 (TAB2)
1 Introduction
Toll-like receptor 4 (TLR4) is an important pattern recognition receptor (PRR) expressed on the cell surface for lipopolysaccharide (LPS) recognition and LPS-mediated inflammatory responses (Takeda et al., 2003; Wong et al., 2017). TLR4-triggered innate immune response is essential to eliminate pathogens and must be tightly regulated. Dysfunctional inflammatory responses result in failures of host defense against infection or extensive tissue damage (Roger et al., 2001; Martin et al., 2005; Freise et al., 2019). Recently, microRNAs (miRNAs) have been shown to play a pivotal role in regulating LPS-triggered innate immune response and the development of endotoxin tolerance (Rodriguez et al., 2007; Chen QY et al., 2012; Chen LQ et al., 2016; Zhang et al., 2019; Chen C et al., 2020). MicroRNA-155 (miR-155) has been identified as a key regulator of the TLR4 signaling pathway (Sayed and el Sayed, 2016; Sayed et al., 2018). There is currently plenty of evidence supporting the idea that miR-155, as a pro-inflammatory mediator, enhances the expression of pro-inflammatory cytokines such as interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) via suppressing the anti-inflammatory molecule suppressor of cytokine signaling 1 (SOCS1), a key negative regulator of LPS responsiveness (O'Connell et al., 2007; Gatto et al., 2008; El-Sahar et al., 2021). For example, Androulidaki et al. (2009) reported that the expression level of miR-155 was significantly elevated in AKT1-/- mouse macrophages upon LPS stimulation, which dampened the protein expression of its target gene SOCS1, leading to endotoxin tolerance that cannot be induced by LPS re-challenge in AKT1-/- macrophages. However, some studies have also shown that miR-155 suppresses TLR4 signaling adapters including myeloid differentiation factor 88 (MyD88) and transforming growth factor-β-activated kinase 1-binding protein 2 (TAB2), which are critical for TLR4 signaling cascades, and this supports an anti-inflammatory role for miR-155 (Ceppi et al., 2009; Tang et al., 2010; Xu et al., 2013; Sul et al., 2018).
We hypothesized that miR-155 might participate in regulation for the crucial homeostatic mechanism that prevents both the excessive innate immune response and immune suppression through interacting with different targets at different stages of the inflammatory response. miR-155 has been reported as a pro-inflammatory and anti-inflammatory mediator involved in regulating LPS-triggered immune response. In this study, we found that the profile of the miR-155-targeted gene pool in macrophages was dramatically changed upon LPS stimulation, and there were distinctly different expression patterns between TAB2 and SOCS1, the two well-known targets involved in miR-155-mediated regulation. We discovered that TAB2 messenger RNA (mRNA) was translationally repressed by miR-155 from the initial to the later stages of inflammation, but miR-155-mediated translational inhibition of SOCS1 mainly occurred in the later stage of inflammation. Moreover, we found that TAB2 3' untranslated region (UTR)-transfected RAW264.7 macrophages were more prone to developing endotoxin tolerance, accompanied by an elevated expression level of SOCS1 protein.
2 Results
2.1. Profile of the miR-155-targeted gene pool in macrophages in response to LPS stimulation
Given that miR-155 has been reported as both a pro-inflammatory regulator and an anti-inflammatory regulator in the innate immune response, we hypothesized that this inconsistency might be the result of the different content of the cellular miR-155-targeted gene pool depending on the functional status of innate immune cells. We performed high-throughput RNA sequencing of mouse peritoneal macrophages treated with or without LPS, and found that LPS caused dramatic expression changes to thousands of transcripts, including more than half of miR-155 potential target genes in macrophages (Fig. 1a, Table S1). Notably, among the altered transcripts, the expression levels of SOCS1 and TAB2, two well-known genes involved in regulating TLR4 signaling in response to LPS stimulation, were significantly enhanced. However, they had distinct expression patterns. SOCS1 mRNA expression was at a low endogenous basal level in resting macrophages but dramatically increased in LPS-triggered macrophages; in contrast; TAB2 had a high basal level but was mildly elevated upon LPS treatment (Fig. 1b, Table S2). Given that both SOCS1 and TAB2 contain a conserved 8-mer-binding site for miR-155 in their 3' UTRs, and earlier studies indicated that miR-155 could repress TAB2 and SOCS1 translation (O'Connell et al., 2007; Sul et al., 2018), we cloned both SOCS1 and TAB2 3' UTRs containing the conserved or mutant 8-mer miR-155-binding sites into the downstream luciferase gene, and then co-transfected RAW264.7 macrophages with miR-155 mimics. The miR-155 mimics significantly suppressed the expression of luciferase with both SOCS1 and TAB2 3' UTRs containing conserved seed sites, while mutation of the seed sequence for miR-155 abolished suppression (Figs. 1c and 1d). The above findings indicated that both the anti-inflammatory regulator SOCS1 and the TLR4 signaling adapter TAB2 were indeed miR-155 targets.
Fig.1. Profiles of microRNA-155 (miR-155)-targeted gene pool in macrophages treated with or without lipopolysaccharide (LPS). (a) Differential gene expression between control and LPS-triggered peritoneal macrophages displayed as a volcano plot with significance set at P<0.05. Genes with fold change of <0.5 are shown in blue, and genes with P<0.05 and fold change of >2 are shown in red. Analysis was performed in DESeq2 using a generalized linear model assuming negative binomial distributions. (b) Differential gene expression of 178 miR-155 potential target genes in macrophages displayed as a scatter plot. (c, d) RAW264.7 macrophages were co-transfected with wild-type or mutant transforming growth factor-β-activated kinase 1-binding protein 2 (TAB2) and suppressor of cytokine signaling 1 (SOCS1) 3' untranslated region (UTR) reporter plasmids, and pRL-TK-Renilla-luciferase plasmid, together with miR-155 mimics or scrambled control (Ctrl). After 24 h, firefly luciferase activity was measured and normalized by Renilla luciferase activity. Data are expressed as mean±standard deviation (n=3). * P<0.05.
2.2. Inconsistency between mRNA and protein levels in LPS-triggered macrophages caused by decreased translation efficiency of TAB2
Considering the distinct expression patterns of TAB2 and SOCS1 in response to LPS stimulation, we reasoned that miR-155 could interact with them in the different stages of the inflammatory response. To illustrate the influence of miR-155 on the translation of target genes, we first analyzed an external dataset (GSE99787) from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus, which contains RNA-seq and ribosome profiling data of macrophages treated with or without LPS. We found that the transcription and translation efficiencies of 178 potential miR-155-targeted genes significantly changed during LPS treatment (Figs. 2a and 2b, Table S3). Notably, TAB2 mRNA was transcriptionally induced from 2 to 6 h upon LPS stimulation (Fig. 2a). The translation efficiency of TAB2 mRNA temporarily increased from 0 to 1 h, and then significantly decreased from 2 to 6 h during LPS treatment (Figs. 2b and 2c), which indicated that TAB2 mRNA was translationally repressed from the early to the later stages of inflammation. The transcription of SOCS1 mRNA was induced from 0 to 6 h upon LPS stimulation, and its translation efficiency remained relatively stable from 0 to 4 h, and then decreased at 6 h post-stimulation (Fig. 2c), indicating that translation of SOCS1 mRNA might be repressed in the later stage of inflammation. We also evaluated the mRNA and protein levels of TAB2 and SOCS1 by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting, and found that all RNA expression levels of miR-155, TAB2, and SOCS1 were significantly up-regulated upon LPS stimulation (Fig. 2d). We observed general and significant associations between miR-155 and the two targets (Figs. 2e and 2f). However, unlike SOCS1 mRNA, for which the protein level consistently increased upon LPS stimulation at all five time points over 12 h, TAB2 protein expression markedly decreased during LPS stimulation, which appeared to be inconsistent with its increased mRNA levels (Fig. 2g).
Fig. 2. Decreased translation efficiency of TAB2 led to inconsistency between its messenger RNA (mRNA) and protein levels in lipopolysaccharide (LPS)-triggered macrophages. (a) Differential gene expression heatmap showing a subset of significantly differentially expressed potential miR-155-targeted genes at indicated time points. (b) Clustering analysis of the translation efficiency (TE) of the 178 potential miR-155-targeted genes differentially translated (empirical P<0.05) in the inflammatory response. Heatmap displays mean row-centered log2TE values at indicated time points. (c) Ribosome footprint reads were mapped to the genome and the number of footprints on the mRNAs for TAB2 and SOCS1 was visualized. (d) RAW264.7 cells were stimulated with 100 ng/mL LPS for the indicated time points. Expression of miR-155 was measured by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and normalized to the expression of U6. Expression of MyD88, TAB2, SOCS1, and SHIP1 mRNAs was measured by RT-qPCR and normalized to the expression of β-actin. (e, f) Correlations between miR-155 expression and its potential target genes (SOCS1 and TAB2) in RAW264.7 macrophages treated with LPS were assessed by means of Spearman’s Rho test. (g) Protein expression levels of SOCS1 and TAB2 upon LPS stimulation were measured at five time points by western blotting. TAB2, transforming growth factor-β-activated kinase 1-binding protein 2; SOCS1, suppressor of cytokine signaling 1; MyD88, myeloid differentiation factor 88; SHIP1, Src homology 2-containing inositol-5'-phosphatase 1.
2.3. Opposite effects of miR-155 in the early and later stages of inflammatory response in macrophages
To provide direct evidence supporting our hypothesis that miR155 exerts opposite effects by targeting TAB2 in the initial stage and SOCS1 in the later stage of inflammation, we analyzed the enrichment of target genes by using the biotinylated miR-155 pulldown assay. As shown in Fig. 3a, TAB2 mRNA significantly interacted with miR-155 compared to the scrambled control in resting macrophages. Meanwhile, the binding level was mildly elevated in LPS-triggered macrophages. Binding of miR-155 to SOCS1 mRNA was not observed in macrophages without LPS treatment, but significant, strong interaction between miR-155 and SOCS1 was identified at 6 h after LPS challenge. These findings were consistent with the distinct expression patterns of SOCS1 and TAB2 before and after LPS challenge, which suggested that miRNA passively interacted with targets depending on their relative abundance. To further investigate whether miR-155 exerts opposite effects in the early and later stages of inflammation, we transfected RAW264.7 macrophages with anti-miR-control or anti-miR-155 and treated them with phosphate-buffered saline (PBS) control or LPS. The mRNA levels of pro-inflammatory cytokines (TNF-α and IL-6) were measured at the indicated time points. As shown in Fig. 3b, inhibition of miR-155 slightly increased the mRNA expression levels of both TNF-α and IL-6 at 4 h after LPS stimulation but markedly decreased them at 12 h after LPS stimulation compared to the anti-miR-control-transfected cells. To validate these effects at the protein level, TNF-α and IL-6 production in the supernatant was measured at 4 h, 4‒12 h, and 12 h time points after LPS stimulation. For the 4‒12 h samples, RAW264.7 cells were primed with LPS for 4 h, followed by washing with PBS; cells were then incubated in fresh complete culture medium with the same concentration of LPS for another 8 h before the supernatant was collected. As expected, the same effects of anti-miR-155 were observed at the protein level for TNF-α and IL-6 (Fig. 3c). Moreover, as shown in Fig. 3d, TAB2 protein level was elevated from 0 to 12 h upon LPS stimulation in anti-miR-155-transfected cells, whereas SOCS1 protein expression was significantly enhanced at 6 h after LPS stimulation in anti-miR-155-transfected macrophages. These results indicated that the opposite effects of miR-155 in the early and later stages of inflammation were due to the altered main targets.
Fig. 3. MicroRNA-155 (miR-155) exerts opposite effects in the early and later stages of inflammatory response in macrophages. (a) Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of SOCS1 and TAB2 in the streptavidin-captured fractions from RAW264.7 cell lysates after transfection with 3'-end biotinylated miR-155 or scrambled control (Ctrl), followed by phosphate-buffered saline (PBS) or 100 ng/mL lipopolysaccharide (LPS) stimulation for 12 h. No template control (NTC) acted as negative control. (b) RAW264.7 cells were transfected with anti-miR-155 or inhibitor control for 24 h, followed by stimulation with 100 ng/mL LPS at the indicated time points. TAB2 and SOCS1 mRNA levels were measured by RT-qPCR and normalized to the expression of β-actin. (c) The supernatants were collected at the indicated time points and IL-6 and TNF-α protein levels were measured by enzyme-linked immunosorbent assay (ELISA). (d) Protein expression levels of SOCS1 and TAB2 were measured by western blotting. Data are expressed as mean±standard deviation (n=3). *P<0.05, **P<0.01. SOCS1, suppressor of cytokine signaling 1; TAB2, transforming growth factor-β-activated kinase 1-binding protein 2; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
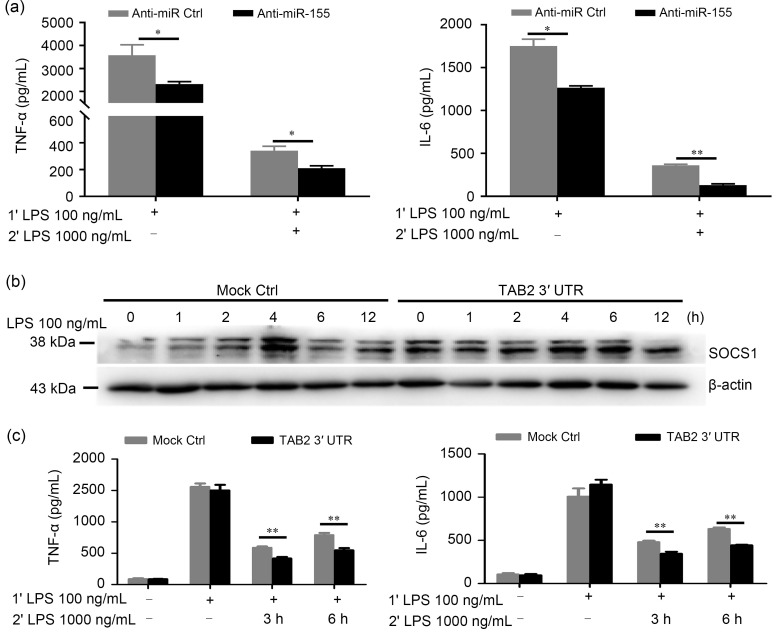
2.4. Elevated SOCS1 protein level and development of LPS desensitization in RAW264.7 cells caused by TAB2 3' UTR transfection
Considering that TAB2 mRNA had a high basal level and was elevated in LPS-triggered macrophages, but TAB2 protein expression was markedly decreased, and also that the ribosome-binding mRNA fragments of TAB2 stayed relatively stable during LPS stimulation, we decided to further investigate whether the increased TAB2 mRNA acts as a sponge in competing with SOCS1 for miR-155 instead of being recruited to the translation apparatus. Previous studies have reported that the up-regulation of SOCS1 acts as a hallmark of endotoxin-tolerant macrophages, so we first investigated whether LPS-mediated up-regulation of miR-155 was involved in negatively regulating the development of LPS desensitization. To induce LPS tolerance, RAW264.7 cells were primed with 100 ng/mL LPS for 18 h, followed by washing with PBS. Cells were incubated in fresh complete culture medium for 2 h before secondary LPS challenge (1000 ng/mL). The results showed that IL-6 and TNF-α protein levels in anti-miR-155-transfected RAW264.7 cell supernatant were significantly lower than those in anti-miR-control-transfected RAW264.7 cell supernatant (Fig. 4a), which is consistent with previous observations (Androulidaki et al., 2009) in murine bone marrow-derived macrophages (BMDMs). Given that overexpression or knockdown of TAB2 can change SOCS1 level through directly influencing nuclear factor-κB (NF-κB) signal strength downstream of TLR4, we transfected RAW264.7 cells with TAB2 3' UTR 24 h prior to LPS stimulation. As shown in Fig. 4b, RAW264.7 cells transfected with TAB2 3' UTR had significantly enhanced SOCS1 protein expression in response to LPS at indicated time points. Next, we detected the effect of TAB2 3' UTR transfection on the development of LPS desensitization and found that IL-6 and TNF-α protein levels were significantly decreased in TAB2 3' UTR-transfected RAW264.7 macrophages compared with mock control after the second LPS challenge (Fig. 4c). Taken together, these results suggested that TAB2 mRNA could compete for miR-155 and thus enhance SOCS1 protein expression.
Fig. 4. TAB2 3' untranslated region (UTR) transfection elevated SOCS1 protein levels and promoted development of lipopolysaccharide (LPS) desensitization in RAW264.7 cells. (a) An endotoxin tolerance model was developed in anti-miR-155 or anti-miR control (Ctrl)-transfected RAW264.7 cells. RAW264.7 cells were primed with 100 ng/mL LPS for 18 h, followed by washing with phosphate-buffered saline (PBS). Cells were incubated in fresh complete culture medium for 2 h before secondary challenge with 1000 ng/mL LPS. IL-6 and TNF-α production in supernatants from anti-miR-155- or miR-155 mimic-transfected RAW264.7 cells was measured by enzyme-linked immunosorbent assay (ELISA). (b) TAB2 3' UTR-transfected RAW264.7 cells and control cells were stimulated with LPS. SOCS1 protein level was determined by immunoblotting at the indicated time points. (c) IL-6 and TNF-α protein levels in supernatants were measured by ELISA. Data are expressed as mean±standard deviation (n=3). *P<0.05, **P<0.01. SOCS1, suppressor of cytokine signaling 1; TAB2, transforming growth factor-β-activated kinase 1-binding protein 2; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
3 Discussion
Since miRNAs emerged as an important regulator in biological processes, research on the topic has blossomed in recent years (Xu et al., 2020; Lin et al., 2021). It has been demonstrated that miRNAs participate in various biological processes such as cell proliferation and differentiation, development and apoptosis, and innate and adaptive immune responses (Zhang YY et al., 2016; Zhang M et al., 2018; Wan et al., 2019). It has also been found that miRNAs are deeply involved in regulating LPS-triggered immune responses (Guo and Zheng, 2020). Cumulative evidence indicates that miR-155 is one of the most important miRNAs and enables a robust inflammatory response (Piccinini and Midwood, 2012; Diwakar et al., 2019). However, some studies have shown that miR-155 suppresses TAB2 and thus inhibits TLR4 signals, supporting an anti-inflammatory role for miR-155. Taking into account the dynamic changes of miR-155 and its target genes in macrophages in response to LPS treatment, the conflicting conclusions from different research teams may be due to miR-155 interacting with different targets in different stages.
Recently, a new hypothesis termed "competing endogenous RNA (ceRNA)" was proposed, which suggests that mRNAs, transcribed pseudogenes, and long noncoding RNAs "talk" to each other using miRNA response elements (MREs) as letters of a new language (Salmena et al., 2011). Some experimental evidence supports this hypothesis. The tumor suppressor gene phosphatase and tension homolog (PTEN) and its pseudogene PTEN1, which share common miRNAs, are a pair of ceRNA, and overexpression of the PTENP1 3' UTR increased levels of PTEN and growth inhibition in a DICER-dependent manner. PTEN1 exhibits greater cancer-inhibiting ability than PTEN (Karreth et al., 2011). Long noncoding RNAs also function in muscle differentiation as a ceRNA. The transcripts of protein-coding RNA PTEN can crosstalk with its ceRNA by competing for common miRNAs. Depletion of its ceRNA transcripts did indeed result in a significant reduction in PTEN protein level (Tay et al., 2011). An abundance of studies have demonstrated that ceRNA regulation networks play an important role in many biological processes, especially in cancer development and differentiation (Bossi and Figueroa-Bossi, 2016; Nuzziello and Liguori, 2019). To our knowledge, there have been few reports about the signaling adaptors or negative regulators involved in the same signaling pathways which modulate each other via a ceRNA mechanism.
4 Conclusions
In the present study, we found that expression levels of miR-155 and its target genes SOCS1 and TAB2 mRNAs were significantly induced by LPS, but TAB2 protein expression markedly decreased, which could be reversed by knockdown of miR-155. Meanwhile, miR-155 knockdown promoted LPS-induced TNF-α and IL-6 production in the early stage of response to LPS, but significantly down-regulated them in the later stage and during an LPS re-challenge. Furthermore, we believe that transfecting noncoding TAB2 3' UTR into macrophages may significantly up-regulate the protein expression level of SOCS1 and promote the induction of endotoxin tolerance. Our results indicated that TAB2 mRNA may not only translate protein to maintain activation of TLR4 signaling, but also enhance SOCS1 protein expression upon LPS stimulation by competing with miR-155 to prevent an extensive immune response. This discovery could provide new insights for understanding the regulatory mechanisms of fine-tuning LPS-induced innate immune response.
Materials and methods
Detailed methods are provided in the electronic supplementary materials of this paper.
Supplementary information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 81701568, 81930041, 81571524, 81872248, and 91842103) and the Zhejiang Provincial Natural Science Foundation of China (Nos. Y15C080001 and Z19H100001). We would also like to thank the Zhejiang Provincial Key Laboratory for Immunity and Inflammatory Diseases for its support.
Author contributions
Yuhua LIU, Xiaopeng WAN, Yuan YUAN, Jingjing HUANG, Yijia JIANG, Kaiyue ZHAO, Yan WANG, and Yang LIU performed the experimental research and data analysis. Qingqing WANG and Hongchuan JIN contributed to the study design and data analysis. Yang LIU, Qingqing WANG, and Hongchuan JIN contributed to the writing and editing of the manuscript. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Yuhua LIU, Xiaopeng WAN, Yuan YUAN, Jingjing HUANG, Yijia JIANG, Kaiyue ZHAO, Yan WANG, Yang LIU, Qingqing WANG, and Hongchuan JIN declare that they have no conflict of interest.
This article does not contain any studies with human subjects performed by any of authors.
References
- Androulidaki A, Iliopoulos D, Arranz A, et al. , 2009. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity, 31(2): 220-231. 10.1016/j.immuni.2009.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Figueroa-Bossi N, 2016. Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Microbiol, 14(12): 775-784. 10.1038/nrmicro.2016.129 [DOI] [PubMed] [Google Scholar]
- Ceppi M, Pereira PM, Dunand-Sauthier I, et al. , 2009. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA, 106(8): 2735-2740. 10.1073/pnas.0811073106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu JM, Luo YP, 2020. MicroRNAs in tumor immunity: functional regulation in tumor-associated macrophages. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 21(1): 12-28. 10.1631/jzus.B1900452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Song YJ, He L, et al. , 2016. MicroRNA-223 promotes type I interferon production in antiviral innate immunity by targeting forkhead box protein O3 (FOXO3). J Biol Chem, 291(28): 14706-14716. 10.1074/jbc.M115.700252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, Wang H, Liu Y, et al. , 2012. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS ONE, 7(8): e42971. 10.1371/journal.pone.0042971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwakar BT, Yoast R, Nettleford S, et al. , 2019. Crth2 receptor signaling down‐regulates lipopolysaccharide‐induced NF‐αB activation in murine macrophages via changes in intracellular calcium. FASEB J, 33(11): 12838-12852. 10.1096/fj.201802608R [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sahar AE, Shiha NA, el Sayed NS, et al. , 2021. Alogliptin attenuates lipopolysaccharide-induced neuroinflammation in mice through modulation of TLR4/MYD88/NF-κB and miRNA-155/SOCS-1 signaling pathways. Int J Neuropsychopharmacol, 24(2): 158-169. 10.1093/ijnp/pyaa078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freise N, Burghard A, Ortkras T, et al. , 2019. Signaling mechanisms inducing hyporesponsiveness of phagocytes during systemic inflammation. Blood, 134(2): 134-146. 10.1182/blood.2019000320 [DOI] [PubMed] [Google Scholar]
- Gatto G, Rossi A, Rossi D, et al. , 2008. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-κB pathway. Nucleic Acids Res, 36(20): 6608-6619. 10.1093/nar/gkn666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zheng Y, 2020. Profiling of miRNAs in mouse peritoneal macrophages responding to Echinococcus multilocularis infection. Front Cell Infect Microbiol, 10: 132. 10.3389/fcimb.2020.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, et al. , 2011. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell, 147(2): 382-395. 10.1016/j.cell.2011.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Rehani K, Jope RS, et al. , 2005. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol, 6(8): 777-784. 10.1038/ni1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzziello N, Liguori M, 2019. The microRNA centrism in the orchestration of neuroinflammation in neurodegenerative diseases. Cells, 8(10): 1193. 10.3390/cells8101193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Taganov KD, Boldin MP, et al. , 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA, 104(5): 1604-1609. 10.1073/pnas.0610731104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinini AM, Midwood KS, 2012. Endogenous control of immunity against infection: tenascin-C regulates TLR4-mediated inflammation via microRNA-155. Cell Rep, 2(4): 914-926. 10.1016/j.celrep.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, et al. , 2007. Requirement of bic/microRNA-155 for normal immune function. Science, 316(5824): 608-611. 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger T, David J, Glauser MP, et al. , 2001. MIF regulates innate immune responses through modulation of Toll-like receptor 4. Nature, 414(6866): 920-924. 10.1038/414920a [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, et al. , 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146(3): 353-358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed AS, el Sayed NSED , 2016. Co-administration of 3-acetyl-11-keto-beta-boswellic acid potentiates the protective effect of celecoxib in lipopolysaccharide-induced cognitive impairment in mice: possible implication of anti-inflammatory and antiglutamatergic pathways. J Mol Neurosci, 59(1): 58-67. 10.1007/s12031-016-0734-7 [DOI] [PubMed] [Google Scholar]
- Sayed AS, Gomaa IEO, Bader M, et al. , 2018. Role of 3-acetyl-11-keto-beta-boswellic acid in counteracting LPS-induced neuroinflammation via modulation of miRNA-155. Mol Neurobiol, 55(7): 5798-5808. 10.1007/s12035-017-0801-2 [DOI] [PubMed] [Google Scholar]
- Sul OJ, Sung YB, Rajasekaran M, et al. , 2018. MicroRNA-155 induces autophagy in osteoclasts by targeting transforming growth factor β-activated kinase 1-binding protein 2 upon lipopolysaccharide stimulation. Bone, 116: 279-289. 10.1016/j.bone.2018.08.014 [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akira S, 2003. Toll-like receptors. Annu Rev Immunol, 21: 335-376. 10.1146/annurev.immunol.21.120601.141126 [DOI] [PubMed] [Google Scholar]
- Tang B, Xiao B, Liu Z, et al. , 2010. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett, 584(8): 1481-1486. 10.1016/j.febslet.2010.02.063 [DOI] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, et al. , 2011. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell, 147(2): 344-357. 10.1016/j.cell.2011.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan JH, Yang XY, Ren YP, et al. , 2019. Inhibition of miR-155 reduces impaired autophagy and improves prognosis in an experimental pancreatitis mouse model. Cell Death Dis, 10(4): 303. 10.1038/s41419-019-1545-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D, Nielsen TB, Bonomo RA, et al. , 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev, 30(1): 409-447. 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CL, Ren GW, Cao G, et al. , 2013. miR-155 regulates immune modulatory properties of mesenchymal stem cells by targeting TAK1-binding protein 2. J Biol Chem, 288(16): 11074-11079. 10.1074/jbc.M112.414862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu KJ, Jia QJ, et al. , 2020. Roles of miRNA and lncRNA in triple-negative breast cancer. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 21(9): 673-689. 10.1631/jzus.B1900709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu ZM, Yang YP, et al. , 2019. Catalpol ameliorates LPS-induced endometritis by inhibiting inflammation and TLR4/NF-κB signaling. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 20(10): 816-827. 10.1631/jzus.B1900071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Gillaspy AF, Gipson JR, et al. , 2018. Neuroinvasive Listeria monocytogenes infection triggers IFN-activation of microglia and upregulates microglial miR-155. Front Immunol, 9: 2751. 10.3389/fimmu.2018.02751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Zhang MY, Li XQ, et al. , 2016. Silencing microRNA-155 attenuates cardiac injury and dysfunction in viral myocarditis via promotion of M2 phenotype polarization of macrophages. Sci Rep, 6: 22613. 10.1038/srep22613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.