Abstract
The whole mitochondrial genome sequence of Mactra quadrangularis (Reeve, 1854) was determined. It had a total length of 16,848 bp and it contained 12 protein coding genes, 2 ribosome RNA genes, and 22 transfer RNA genes. The base composition was 25.75% A, 20.82% G, 11.53% C, and 41.90% T, respectively. Furthermore, state codon of ND4 was ATT; ND1 and CYTB were ATA; COX1 was GTG; ND5, COX2, ND4L, ND6, ND2, COX3, ATP6, and ND3 were ATG. Phylogenetic analysis demonstrated that M. quadrangularis was most closely related to Mactra chinensis. The mitochondrial genome will provide reference for the further investigation and research of M. quadrangularis.
Keywords: Bivalvia, Venerida, phylogenetic analysis
As an important edible seashore clam and seafood resource in the coastal areas of China, Japan, and South Korea, the surf clam Mactra quadrangularis (Reeve, 1854), sometimes also referred to as Mactra veneriformis (Reeve, 1854), belongs to Bivalvia, Venerida, Mactridae, Mactra (Linnaeus, 1767) (Hou et al. 2006; Luan et al. 2011; Nie et al. 2013; Zhu et al. 2019). We reconstructed the complete mitochondrial genome of M. quadrangularis based on Illumina paired-end sequencing data.
Samples were collected at Lianyungang, Jiangsu province, China (34.9497°N, 119.1886°E). The specimen is stored at Laboratory of Marine Ecology, Ocean University of China (specimen code OUCMLE09529; chenguan1021@163.com). Total genomic DNA was extracted from muscular tissue according to the CTAB method as detailed in (Mirimin and Roodt-Wilding 2015). We generated 400 bp paired-end reads from total genomic DNA by whole genome shotgun (WGS) sequencing using the Illumina NovaSeq platform (GenomeAnalyzer, Illumina, San Diego, CA). De novo assembly was conducted using A5-miseq version 20150522 (Coil et al. 2014) and SPAdes version 3.9.0 (Bankevich et al. 2012; Choi et al. 2020). The annotation was performed using GeSeq software (Tillich et al. 2017).
The complete mitochondrial genome of M. quadrangularis was submitted to GenBank and the GenBank accession number was MW691169. The raw data of M. quadrangularis were submitted to Sequence Read Archive (SAR) and the accession number was SRR14764610. The complete mitochondrial genome was 16,848 bp in size. The A + T base content (67.65%) was higher than the G + C content (32.35%). The base composition of the complete mitochondrial genome of M. quadrangularis was 25.75% for A, 20.82% for G, 11.53% for C, and 41.90% for T, respectively. The newly sequenced mitochondrial genome encodes for a total of 36 genes, including 12 protein-coding genes (PCGs), 2 rRNA genes, and 22 tRNA genes. In PCGs of M. quadrangularis, ATP8 gene is missing. Of the 12PCGs, ND5, ND1, COX1, ND4L, COX3, and CYTB had TAA as stop codons; while the other PCGs had TAG as stop codons. The state codon of ND4 was ATT, that of ND1 and CYTB was ATA, that of COX1 was GTG, and that of ND5, COX2, ND4L, ND6, ND2, COX3, ATP6, and ND3 was ATG.
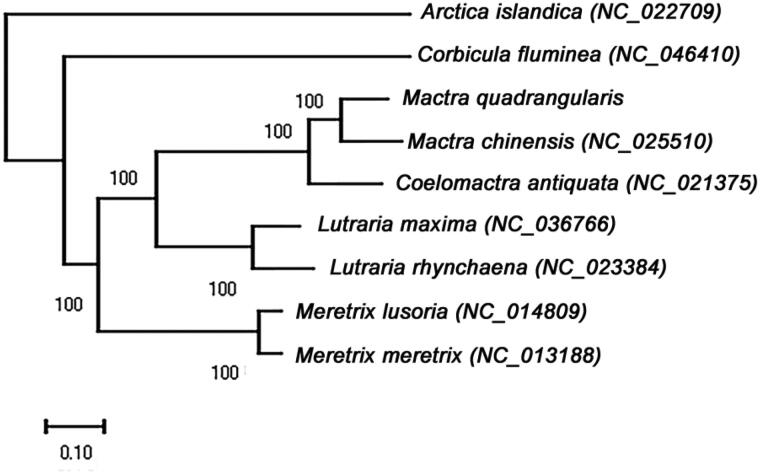
To investigate the phylogenetic relationships of M. quadrangularis, four complete mitochondrial genomes of Mactridae (Coelomactra antiquata [Spengler, 1802], Mactra chinensis [Philippi, 1846], Lutraria maxima [Jonas, 1844], and Lutraria rhynchaena [Jonas, 1844]) and two complete mitochondrial genomes of Veneridae (Meretrix [Linnaeus, 1758], Meretrix lusoria [Röding, 1798]), one mitochondrial genome of Arcticidae (Arctica islandica [Linnaeus, 1767]), as well as one mitochondrial genome of Cyrenidae (Corbicula fluminea [Müller, 1774]) were downloaded from GenBank and aligned using complete mitochondrial genome sequences (Figure 1). The maximum likelihood (ML) phylogeny was constructed based on the General Time Reversible + Invariant + gamma sites (GTR + I + G) model of nucleotide substitution with 1000 bootstrap replicates by Mega-X version 10.0.2 (Kumar et al. 2018). The ML tree analysis indicated that M. quadrangularis closely related to M. chinensis and Coelomactra antiquata (Figure 1). This published M. quadrangularis mitochondrial genome will provide evolutionary information in the Mactridae.
Figure 1.
Maximum likelihood (ML) phylogenetic tree based on 9 complete mitochondrial genome sequences of Venerida. ML bootstrap values are shown above nodes. All the sequences were downloaded from NCBI GenBank.
Funding Statement
This work was supported by the National Key R&D Program of China [No. 2019YFC1605704], the Key Laboratory of Marine Ecology and Environmental Science and Engineering, SOA [MESE-2019-02], and the National Natural Science Foundation of China [No. 61902368].
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW691169/, reference number MW691169. BioProject accession number was PRJNA736184 at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA736184. BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN19611295/ and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR14764610.
References
- Anton Bankevich, Sergey Nurk, Dmitry Antipov, Alexey A. Gurevich, Mikhail Dvorkin, Alexander S. Kulikov, Valery M. Lesin, Sergey I. Nikolenko, Son Pham, Andrey D. Prjibelski, Alexey V. Pyshkin, Alexander V. Sirotkin, Nikolay Vyahhi, Glenn Tesler, Max A. Alekseyev, Pavel A. Pevzner.. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beom-Soon Choi, Young Hwan Lee, Jin-Sol Lee, Erick O. Ogello, Hee-Jin Kim, Atsushi Hagiwara, Jae-Seong Lee. 2020. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus rubens (Rotifera, Brachionidae). Mitochondrial DNA B Resour. 5(1):5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Coil, Guillaume Jospin, Aaron E. Darling. 2014. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589. [DOI] [PubMed] [Google Scholar]
- Lin Hou, Hongli Lü, Xiangyang Zou, Xiangdong Bi, Deqin Yan, Chongbo He. 2006. Genetic characterizations of Mactra veneriformis (Bivalve) along the Chinese coast using ISSR-PCR markers. Aquaculture. 261(3):865–871. [Google Scholar]
- He-Mi Luan, Ling-Chong Wang, Hao Wu, Yan Jin, Jing Ji. 2011. Antioxidant activities and antioxidative components in the surf clam, Mactra veneriformis. Natural Product Research. 25(19):1838–1848. [DOI] [PubMed] [Google Scholar]
- Michael Tillich, Pascal Lehwark, Tommaso Pellizzer, Elena S. Ulbricht-Jones, Axel Fischer, Ralph Bock, Stephan Greiner. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Research. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L. Mirimin, R. Roodt-Wilding. 2015. Testing and validating a modified CTAB DNA extraction method to enable molecular parentage analysis of fertilized eggs and larvae of an emerging South African aquaculture species, the dusky kob Argyrosomus japonicus. Journal of Fish Biology. 86(3):1218–1223. [DOI] [PubMed] [Google Scholar]
- Hongtao Nie, Ning Li, Lingfeng Kong, Qi Li. 2013. Isolation and characterization of 18 polymorphic microsatellite loci in the surf clam (Mactra veneriformis). Conservation Genet Resour. 5(3):635–637. [Google Scholar]
- Sudhir Kumar, Glen Stecher, Michael Li, Christina Knyaz, Koichiro Tamura. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Zhu, Yanyan Yang, Hua Xu, Qing Wang, Yanyan Wei, Mingzhu Li, Fan Li, Yiqi Wang, Huawei Zhang, Yihao Liu, Xiaomeng Wang, Yan Fang. 2019. Bioaccumulation of decabromodiphenyl ether affects the antioxidant system in the clam Mactra veneriformis. Environmental Toxicology and Pharmacology. 68:19–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW691169/, reference number MW691169. BioProject accession number was PRJNA736184 at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA736184. BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN19611295/ and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR14764610.