Abstract
Aim of the study
Liver regeneration is one of the essential fields of regenerative medicine as a branch of tissue engineering and molecular biology that draws global researchers’ attention. This study aims to conduct a systematic review and meta-analysis on the high-throughput gene expression microarray dataset of liver regeneration on the NCBI-GEO database to identify the significant genes and signaling pathways and confirm the genes from literature studies on associated diseases.
Material and methods
We thoroughly searched the NCBI-GEO database to retrieve and screen the GEO microarray datasets’ contents. Due to the inclusion of different species in eligible GEO datasets in the meta-analysis, the list of significant genes for the random-effects model were identified. Moreover, we carried out detailed gene analyses for three main gene ontology components and the KEGG signaling pathway. Furthermore, we investigated the possibility of genes’ association with liver cancer through the Kaplan-Meier plot.
Results
The random-effects model from six eligible GEO datasets identified 71 genes with eight down-regulated and 63 up-regulated genes. The target genes are involved in various cellular functions such as cell proliferation, cell death, and cell cycle control. Finally, we noted that 58 out of 71 genes are associated with different types of diseases related explicitly to other liver and inflammation diseases.
Conclusions
The current study assessed various GEO datasets at the early stages of liver regeneration with promising results. The present systematic review and meta-analysis results are beneficial for future novel drug design and discovery specifically for patients in the liver transplantation process.
Keywords: liver regeneration, meta-analysis, systematic review, systems biology, gene
Introduction
The liver is one of the magic metabolic organs in the live body. It is mainly composed of hepatocytes in hemostasis and normalizes different metabolisms ranging from carbohydrates to lipids [1-3]. Several factors can cause liver injury, such as accidental or intentional ingestion of toxins, surgery, or transplantation processes [4, 5]. The liver is known to have a high capacity in the regeneration of its damaged tissues compared to other body organs [6]. Additionally, regenerative medicine, as one of the interdisciplinary translational science fields attracting the attention of the world’s research community, uses tissue engineering and stem cell therapy techniques in replacing the injured organ with an artificial organ, which is in a direct relationship with the immune system [3, 7]. Through different original or review literature articles, the molecular and functional mechanisms have been explicitly investigated on animal models (e.g., rats and mice) and Homo sapiens to identify the significant genes and signaling pathways involved in liver regeneration [6, 8].
The processes of development, repair, and regeneration of the liver are vital and complex in several aspects deserving to be studied in molecular mechanisms and epigenetics [9, 10]. Moreover, various therapeutics for liver regeneration have been proposed, including cyclosporine A, tri-iodothyronine, mesenchymal stem cell infusion, transforming growth factor β (TGF-β), interleukin (IL)-1, nuclear factor κB (NF-κB), tumor necrosis factor α (TNF-α), IL-6, glutamine, and amino acids, with simulating and inhibiting effects [10]. Despite its high regeneration capacity, it may be affected by severe and advanced liver diseases. That sometimes can be repaired using liver progenitor cell-driven liver regeneration; however, this approach may not be useful in progressive conditions and needs careful and promotional considerations for future designing and developing tactics [11].
The reports showed that the pattern of gene expression levels in liver regeneration after an injury is unique. Different samples are affected by a specific gene with its interconnection with other genes. The other gene expression pattern types can also affect one molecule on other regenerated liver cells, considering the ethanol-treated and parenchyma liver cells. Additionally, the miRNA pattern changes, for example, on male rat miRNA microarray profiles for a 24-hour duration have similar characteristics. Finally, these types of pattern changes can also represent differences in cytokine levels [e.g., interferon γ (IFN-γ) in aged humans].
Hence, we conducted a systematic review and metaanalysis approach to investigate the molecular and functional analyses of liver regeneration of publicly available microarray datasets for humans, mice, and rats. However, some studies in the literature have performed the same analysis on only one type of species. The current research output will present the significant essential genes, gene ontology, and KEGG signaling pathway analyses in liver transplantation. Finally, the identified genes can help develop novel therapeutics and drug design and discovery approaches relating to specifically liver regeneration failures.
Material and methods
Microarray profiling datasets
We searched the National Center for Biotechnology Information for retrieving the target GEO datasets by the Boolean query “hepatectomy OR hepatic systematically” AND “liver regeneration”. The included datasets’ eligibility was determined based on their titles and contents by having all types of species, liver biopsy tissues, expression types, and platforms with 1-hour duration without age restriction. We omitted those GEO datasets not satisfying the inclusion criterion from further analysis and utilized the included ones for other processes.
Meta-analysis approach
The ExAtlas online web server is a comprehensive and useful tool for performing meta-analysis on GEO datasets on various platforms with a user-friendly interface to import and utilize the preprocessing steps (i.e., normalization and single-factor ANOVA) in the background for comparing the paired samples based on p-values and F-statistics. Then, the ExAtlas web server processed the imported datasets for the data quality, in which it discarded those samples with a standard deviation (STD) of more than 0.3. Moreover, from conducting the meta-analysis, the final step, the false discovery rate (FDR ≤ 0.05), and fold change ≤ 1.5 were important sets of parameters. The Benjamini-Hochberg procedure was the method used to decrease the significant number of false-positive genes.
Gene ontology and KEGG enrichment analyses of significant genes
DAVID v. 6.8 (Database for Annotation, Visualization, and Integrated Discovery) website was used to analyze three gene ontology (GO) domains (cellular component, biological process, and molecular function) and involved several Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways [12, 13]. Moreover, the p-value term for the genes to be considered significant were set to be less than 0.05.
Kaplan-Meier analysis
To further analyze the obtained significant genes from the meta-analysis approach, the Kaplan-Meier methodology was applicable to investigate survival rates of the input genes in liver cancer versus a living period. This process was necessary because the liver regeneration process had an indirect relationship with liver diseases such as liver cancer [14, 15]. To this end, we used the mRNA liver cancer section of kmplot.com to determine the overall survival (OS; n = 364), recurrence-free survival (RFS; n = 316), progression-free survival (PFS; n = 370), and disease-specific survival (DSS; n = 362). We used the website’s features inducing pathology, patients, and risk factors as default by enabling the plot option to generate high-quality TIFF image output files. Moreover, the significant genes involved in liver cancer in terms of OS, RFS, PFS, and DSS) were indicated by p-values less than 0.05.
Results
An overview of the systematic review of the NCBI-GEO database is available in Figure 1. The process is generally useful in applying the inclusion and exclusion criteria for identifying the final eligible GEO datasets. The systematic review procedure revealed 190 datasets based on the Boolean query from which 11 datasets remained after a critical inspection of the titles and contents. On the other hand, we examined the datasets in the ExAtlas online web server for their availability for further analysis. The outcomes showed that only 6 out of 11 GEO datasets had the required information. The remaining five datasets had platforms and experiment types related to being not RNAs. At the end, six GEO datasets were identified including GSE12720 (n_control = 21 and n_regeneration = 21), GSE15239 (n_control = 2 and n_regeneration = 2), GSE20427 (n_control = 3 and n_regeneration = 3), GSE33785 (n_control = 6 and n_regeneration = 6 with various diets including ethanol, carbohydrates, and high fat), GSE34057 (n_control = 2 and n_regeneration = 2 with diets including ethanol and carbohydrate), and GSE6998 (n_control = 2 and n_regeneration = 2) for 1 hour after the liver transplantation period on liver biopsy tissues. The detailed information for the six datasets is available in Table 1. In the next step, after the successful import of six datasets on Homo sapiens, rats, and mice as well as data analysis quality analysis with no outliers, the meta-analysis approach resulted in seventy-one genes. The criteria parameters were FC ≤ 1.5 (those genes with FC larger than 1.5) and FDR ≤ 0.05 for an overall number of 72 samples for both control (n = 36) and liver transplantation (n = 36).
Fig. 1.

The systematic review flowchart for applying including and excluding criteria to retrieve the final eligible GEO datasets
Table 1.
Detailed information on six GEO datasets eligible for meta-analysis
| GEO | Published (PMID) | Species | Platform | Experiment type | Sample type | Source name |
|---|---|---|---|---|---|---|
| GSE12720 | 19353763 | Homo sapiens | GPL570 | Expression profiling by array | RNA | Liver biopsy |
| GSE15239 | N/A | Homo sapiens | GPL570 | Expression profiling by array | RNA | Liver |
| GSE20427 | 20564353 | Mus musculus | GPL81 GPL1261 | Expression profiling by array | RNA | Liver |
| GSE33785 | 22790595 27012785 | Rattus norvegicus | GPL6247 | Expression profiling by array | RNA | Chronic ethanol-fed liver Fed carbohydrate liver Fed high fat liver |
| GSE34057 | 22823254 | Rattus norvegicus | GPL14889 | Non-coding RNA profiling by array | RNA | Ethanol-fed liver Ethanol-fed liver |
| GSE6998 | 17227769 | Mus musculus | GPL1261 | Expression profiling by array | RNA | Hepatectomy |
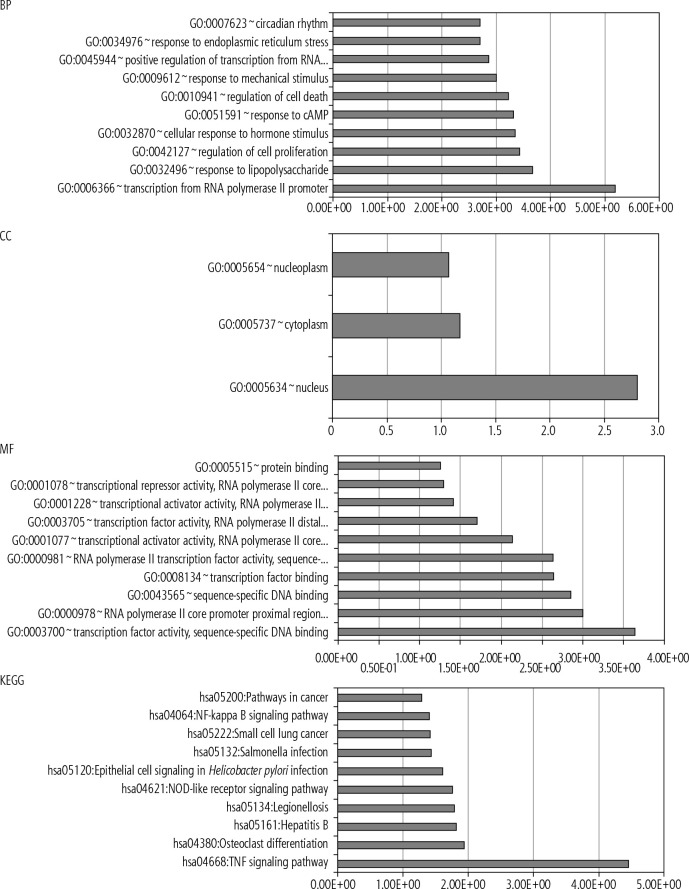
Additionally, significant genes among the three species were eligible for analysis in DAVID’s bioinformatics web server. A portion of the input genes enriched the BP (e.g., GO:0006366~transcription from RNA polymerase II promoter, GO:0032496~response to lipopolysaccharide), CC (e.g., GO:0005634~nucleus), MF (e.g., GO:0003700~transcription factor activity, sequence-specific DNA binding, GO:0000978~RNA polymerase II core promoter proximal region sequence-specific DNA binding), and KEGG signaling pathways (e.g., hsa04668: TNF signaling pathway, hsa04380: Osteoclast differentiation).
Finally, the KM plot analysis for the seventy-one genes revealed that only 24 out of 56 genes for OS, 17 out of 56 genes for RFS, 24 out of 56 genes for PFS, and 26 out of 56 genes for DSS are significantly useful for liver cancer (shown in Table 2 and Fig. 2). However, the KM plot website did not analyze fifteen genes as they were not available in its database. That is to say that the inhibition of liver cancer may happen after successful liver transplantation.
Table 2.
Output of Kaplan-Meier plot for overall survival (OS), recurrence-free survival (RFS), progression-free survival (PFS), and disease-specific survival (DSS) rates. The genes with significant p-values and no data are highlighted in bold and gray, respectively
| Gene ID | OS | RFS | PFS | DSS |
|---|---|---|---|---|
| ZBED5 | 0.003 | 0.0691 | 0.0045 | 0.0009 |
| LDLRAD4 | No data | No data | No data | No data |
| LRRC37A2 | 0.1425 | 0.0692 | 0.0143 | 0.1632 |
| ADORA2A-AS1 | No data | No data | No data | No data |
| RTP4 | 0.0219 | 0.021 | 0.0063 | 0.037 |
| DLEU1 | 0.2554 | 0.0108 | 0.0168 | 0.0503 |
| FLJ37453 | 0.2374 | 0.2816 | 0.3419 | 0.2479 |
| SMIM10L2A | No data | No data | No data | No data |
| BTG2 | 0.3459 | 0.0008 | 0.015 | 0.2117 |
| MYC | 0.0883 | 0.1173 | 0.1623 | 0.1357 |
| SIK1 | 3.7e-5 | 0.1546 | 0.1058 | 0.0012 |
| JUN | 0.3599 | 0.1405 | 0.2655 | 0.2081 |
| MIR6732 | No data | No data | No data | No data |
| CXCL8 | No data | No data | No data | No data |
| NOCT | No data | No data | No data | No data |
| KLF6 | 0.0782 | 0.1276 | 0.1145 | 0.1881 |
| CCN1 | No data | No data | No data | No data |
| NFIL3 | 0.1694 | 0.203 | 0.2328 | 0.2092 |
| GDF15 | 0.0769 | 0.0534 | 0.0648 | 0.2222 |
| SGK1 | 0.0116 | 0.0731 | 0.0343 | 0.0313 |
| ARL5B | 0.0578 | 0.0694 | 0.0102 | 0.1265 |
| SLC38A2 | 0.1796 | 0.2052 | 0.1286 | 0.1061 |
| BIRC3 | 0.4525 | 0.2439 | 0.2981 | 0.3399 |
| GADD45B | 0.0005 | 0.0231 | 0.0043 | 0.0005 |
| SDS | 0.0169 | 0.0792 | 0.0065 | 0.0427 |
| TSC22D2 | 0.128 | 0.099 | 0.1211 | 0.0324 |
| ZNF331 | 0.0264 | 0.2766 | 0.1639 | 0.1927 |
| VNN3 | 0.1836 | 0.0104 | 0.0246 | 0.0599 |
| MAFF | 0.0583 | 0.4564 | 0.2274 | 0.2622 |
| CSRNP1 | 0.0071 | 0.005 | 0.0007 | 0.026 |
| JUNB | 0.1361 | 0.0103 | 0.004 | 0.0447 |
| ZFP36 | 0.0796 | 0.0571 | 0.0872 | 0.0441 |
| S100A9 | 0.0001 | 0.1028 | 0.057 | 0.0005 |
| THBD | 0.0007 | 0.0035 | 0.0027 | 0.0014 |
| BHLHE40 | 0.1656 | 0.1846 | 0.2219 | 0.1462 |
| SDC4 | 0.0027 | 0.0177 | 0.013 | 0.0358 |
| TCIM | No data | No data | No data | No data |
| IFRD1 | 0.0997 | 0.083 | 0.1972 | 0.0727 |
| PTP4A1 | 0.002 | 0.2971 | 0.0712 | 0.093 |
| CEBPD | 0.0022 | 0.009 | 0.0066 | 0.0004 |
| SLC2A14 | 0.0611 | 0.2228 | 0.339 | 0.1177 |
| WSB1 | 0.0209 | 0.017 | 0.0007 | 0.0022 |
| CCNL1 | 0.2028 | 0.2989 | 0.1965 | 0.0541 |
| ELL2 | 0.0227 | 0.0068 | 0.0223 | 0.0849 |
| PIM3 | 0.2517 | 0.1098 | 0.0192 | 0.1525 |
| RETREG1 | No data | No data | No data | No data |
| GPCPD1 | 0.0062 | 0.1136 | 0.0876 | 0.0062 |
| ALAS1 | 6.8e-5 | 0.0555 | 0.0112 | 0.0015 |
| NNMT | 0.085 | 0.0488 | 0.0702 | 0.0391 |
| JUND | 0.0017 | 0.2282 | 0.0537 | 0.0007 |
| NRBF2 | 0.1633 | 0.1174 | 0.0641 | 0.1889 |
| MAFK | 0.3297 | 0.2452 | 0.217 | 0.3273 |
| TIMM23B | No data | No data | No data | No data |
| LOC100996792 | No data | No data | No data | No data |
| MT1M | 0.2659 | 0.0536 | 0.0177 | 0.3193 |
| SLC25A33 | 0.3775 | 0.0462 | 0.0585 | 0.284 |
| CEMIP2 | No data | No data | No data | No data |
| USP36 | 0.0423 | 0.0165 | 0.0007 | 0.0394 |
| CEBPB | 0.0087 | 0.1607 | 0.0647 | 0.0083 |
| PNRC1 | 0.0273 | 0.2189 | 0.2985 | 0.0847 |
| MIR636 | No data | No data | No data | No data |
| CXCL3 | 0.0013 | 0.1966 | 0.0297 | 0.0128 |
| CHAC1 | 0.0569 | 0.0704 | 0.1669 | 0.1139 |
| NFKBIA | 0.0656 | 0.0216 | 0.081 | 0.035 |
| AKAP12 | 0.0676 | 0.2141 | 0.0991 | 0.0259 |
| DNAJB4 | 0.0617 | 0.1582 | 0.2692 | 0.029 |
| HBG1 | 0.0004 | 0.0004 | 0.0009 | 0.0009 |
| MIR21 | No data | No data | No data | No data |
| BCL2L11 | 0.0289 | 0.0748 | 0.0269 | 0.0691 |
| BTG3 | 0.0967 | 0.2024 | 0.1592 | 0.066 |
| PRR26 | No data | No data | No data | No data |
Fig. 2.
DAVID’s webserver results demonstrated for biological processes (BP), cellular components (CC), molecular functions (MF), and the KEGG signaling pathway enrichment analysis
Discussion
We performed a systematic review and meta-analysis and a systems biology approach on six GEO datasets that remained from screening and analyzing the NCBI-GEO database contents. The outcomes will be beneficial to thoroughly study liver regeneration and the common genes involved in three species (i.e., Homo sapiens, mice, and rats). Whether they are present in genetics or signaling pathways involved in liver regeneration, mechanisms are paramount to effectively utilize them in clinical experiences. The above issue has also been frequently investigated in both sciences, including biological and clinical fields, considering its complicated structure to find better novel drug design and discovery tactics. The researchers reported that the time required for liver regeneration is twelve weeks. However, in all of the available GEO datasets it was far less than the reported completion of the liver regeneration period (e.g., 0.5, 1, 1.5, 2, 6, 36, and 72 hours) from which for the meta-analysis, the one-hour samples were in common among the GEO datasets [16]. It is worth mentioning that complex process of liver regeneration is composed of three phases: priming, proliferative, and termination phases [17]. In the priming stage, main cytokine-associated pathways such as TNF-α and IL-6 are involved. In the proliferative stage, the complete mitogens epidermal growth factor (EGF), transforming growth factor α (TGF-α), and heparin-binding EGF-like growth factor (HB-EGF) can play significant roles. Meanwhile, the terminative process of liver regeneration has remained unclear. However, some studies have shown that TGF-β family members such as TGF-β1 are enough to end the liver regeneration after 48 h following partial hepatectomy (about two-thirds of the liver) [18]. This period could be enough for obtaining the information to understand the process of liver regeneration in a time-series analysis. Moreover, liver regeneration’s early proliferation phase is critical to capturing significant transcriptomic signatures [19].
The meta-analysis results reveal the significant common gene biomarkers among three species playing a vital role in their liver regeneration at the early phase.
We also analyzed the possible direct relationships among genes and their involved diseases using a gene-disease network approach to further discuss the outcomes. DisGeNET (v7.0), a publicly available database web interface, provided a diverse update (June 2020) and curated data bank for retrieving information on genes linked to human diseases using 1,134,942 gene-disease associations [20-22]. We used the “disgenet2r” R package to accurately automate the retrieval procedure of the gene-disease associations (GDAs) using the “gene2disease” command through a working Internet connection [22]. This section provides the required data to discuss the related diseases with the identified significant human genes. Among the reported genes of different illnesses, 58 out of 71 genes were directly consistent with various conditions. These conditions included liver, infection, bile, biliary, blood, measurement, bleed, mental, seizure, cirrhosis, behavior, metabolic, cirrus, cardiologic variants, edema, coronary, vascular, sepsis, and inflammatory. A detailed summary of these GDAs as a collective set of .csv files is available as supplementary material.
As a complex process in Homo sapiens, rats, and mice, liver regeneration needs a more clinical and experimental research to understand various biological and molecular aspects of the process for long periods. The liver regeneration core depends explicitly on the cell-cycle proliferation, which activates multiple signaling pathways, including NF-κB, MAPK, PI3K/AKT, Wnt, JAK/STAT, cellular senescence, and TNF signaling pathway [23, 24]. In this regard, the total of fourteen identified associated genes with these signaling pathways, playing an important role in the other liver regeneration phases, are summarized in Table 3. In contrast to the abovementioned accelerating factors, a diverse range of active proliferative-inhibiting liver regeneration pathways are mainly associated with cytokines, oncogenes, gene expression regulation, miRNAs, protein modification, and hormone, metabolism, and pathological factors [25]. The decrease and increase of inhibitor promoters’ expression levels may affect different stages of liver regeneration, namely “hepatostat”, which requires more studies on inhibitory factors than promoting aspects of liver regeneration [24, 25]. Finally, because cyclin-dependent kinase (Cdks)/cyclin complexes regulate cell-cycle progression, it can be deduced that the main factors, whether they are inhibiting or promoting the process of liver regeneration, occur in the cell cycle [26]. Researchers have reported that the Cdk1 gene contributes to metabolic pathway functions such as cell division and DNA repair [27, 28].
Table 3.
Identified associated genes with involved signaling pathways of liver regeneration
| Pathways | Gene terms |
|---|---|
| NF-κB | CXCL8, BIRC3, GADD45B, CXCL3, NFKBIA |
| MAPK | GADD45B, JUND |
| PI3K/AKT | MYC, SGK1, BCL2L11 |
| Wnt | MYC, JUN |
| JAK/STAT | MYC |
| Cellular senescence | MYC, CXCL8, GADD45B, ZFP36 |
| TNF signaling pathway | CEBPB, CXCL3, NFKBIA, BIRC3, JUNB |
Furthermore, Schmidt and Dalhof reported that α-fetoprotein (AFP) in acute liver failure (ALF) patients could be regarded as a functional marker of hepatocyte proliferation [29]. One of the treatments that can be clinically applied for successful liver surgery is platelet therapy (e.g., thrombopoietin receptor agonists, artificial platelets [30], and freeze-dried platelets [31]). It has a positive protective role for hepatocytes resulting in liver regeneration promotion [23]. Finally, although gene therapy through activating autophagy using mesenchymal stem cells (MSC) transplantation can promote liver regeneration, autophagy suppression may result in functional failure of the liver [32, 33].
However, one of the main limitations of the current systematic review and meta-analysis is the availability of a limited number of GEO datasets for liver regeneration, considering those tracked for extended periods (e.g., six weeks). The next limitation is related to the “experiment type”, not supported in ExAtlas due to the microarray platform and non-RNA samples (e.g., genomic in GSE74273). This systematic review and meta-analysis provide in-depth information on liver regeneration’s biological and molecular processes common among three species. This study paves the way for future research on liver-related diseases such as acute and chronic liver failure, injury, and transplantation.
Conclusions
Regeneration medicine is a vital field contributing several advantages to the liver regeneration process. We carried out a systematic review and meta-analysis to understand further its biological and functional mechanisms for the process’s importance and complexity. Through a systematic review on the NCBI-GEO database, six datasets were eligible for metaanalysis; their organisms include Mus musculus, Rattus norvegicus, and Homo sapiens. The meta-analysis results identified seventy-one significant genes common to three types of organisms. Among them, 27/56, 24/56, 26/56, and 17/56 genes were directly relative to liver cancer for PFS, OS, DSS, and RFS, considering the KM plot (i.e., fifteen genes were not available for KM plot analysis in liver cancer). Moreover, the enrichment analysis of KEGG signaling pathways showed that TNF, NOD-like receptor, and NF-κB signaling pathways were important in liver transplantation. Finally, the involved genes common to three species can predict future drug design and discovery to expedite the liver regeneration process.
Acknowledgements
This is to acknowledge the equal contributions of the two first authors. Hence, the two first authors should be considered as joint co-first authors.
Disclosure
The authors declare no conflict of interest.
References
- 1.Valizadeh A, Majidinia M, Samadi Kafil H, et al. The roles of signaling pathways in liver repair and regeneration. J Cell Physiol 2019; 234: 14966-14974. [DOI] [PubMed] [Google Scholar]
- 2.Xing X, Li M, Guo Z, et al. Expression profiles of genes associated with mitochondria-mediated apoptosis and their roles in liver regeneration. Genet Mol Res 2016; 15: 1-13. [DOI] [PubMed] [Google Scholar]
- 3.Yin L, Wang Y, Guo X, et al. Comparison of gene expression in liver regeneration and hepatocellular carcinoma formation. Cancer Manag Res 2018; 10: 5691-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horn KD, Wax P, Schneider SM, et al. Biomarkers of liver regeneration allow early prediction of hepatic recovery after acute necrosis. Am J Clin Pathol 1999; 112: 351-357. [DOI] [PubMed] [Google Scholar]
- 5.Clemens MM, McGill MR, Apte U. Mechanisms and biomarkers of liver regeneration after drug-induced liver injury. Adv Pharmacol 2019; 85: 241-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kojima H, Nakamura K, Kupiec-Weglinski JW. Therapeutic targets for liver regeneration after acute severe injury: a preclinical overview. Expert Opin Ther Targets 2020; 24: 13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sergeeva O, Sviridov E, Zatsepin T. Noncoding RNA in liver regeneration–from molecular mechanisms to clinical implications. Semin Liver Dis 2020; 40: 070-083. [DOI] [PubMed] [Google Scholar]
- 8.Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Comprehensive Physiol 2013; 3: 485-513. [DOI] [PubMed] [Google Scholar]
- 9.Macchi F, Sadler KC. Unraveling the epigenetic basis of liver development, regeneration and disease. Trends Genet 2020; 36: 587-597. [DOI] [PubMed] [Google Scholar]
- 10.Hyslip J, Martins P. Liver repair and regeneration in transplant: state of the art. Curr Transplant Rep 2020; 1-9. [Google Scholar]
- 11.So J, Kim A, Lee SH, et al. Liver progenitor cell-driven liver regeneration. Exp Mol Med 2020; 52: 1230-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li HM, Ye ZH. Microenvironment of liver regeneration in liver cancer. Chin J Integr Med 2017; 23: 555-560. [DOI] [PubMed] [Google Scholar]
- 15.Guest RV, Boulter L, Dwyer BJ, et al. Understanding liver regeneration to bring new insights to the mechanisms driving cholangiocarcinoma. NPJ Regen Med 2017; 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olthoff KM, Smith AR, Abecassis M, et al. Defining long-term outcomes with living donor liver transplantation in North America. Ann Surg 2015; 262: 465-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao Y, Wang M, Chen E, et al. Liver regeneration: analysis of the main relevant signaling molecules. Mediators Inflamm 2017; 2017: 4256352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueda S, Yamanoi A, Hishikawa Y, et al. Transforming growth factor-β1 released from the spleen exerts a growth inhibitory effect on liver regeneration in rats. Lab Invest 2003; 83: 1595-1603. [DOI] [PubMed] [Google Scholar]
- 19.Colak D, Al-Harazi O, Mustafa OM, et al. RNA-Seq transcriptome profiling in three liver regeneration models in rats: comparative analysis of partial hepatectomy, ALLPS, and PVL. Sci Rep 2020; 10: 1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piñero J, Queralt-Rosinach N, Bravo À, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015; 2015: bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2016; 45: D833-D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020; 48: D845-D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi K, Liang C, Oda T, et al. Platelet and liver regeneration after liver surgery. Surg Today 2020; 50: 974-983. [DOI] [PubMed] [Google Scholar]
- 24.Zhu C, Dong B, Sun L, et al. Cell sources and influencing factors of liver regeneration: a review. Med Sci Monit 2020; 26: e929129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M, Chen P. Proliferation-inhibiting pathways in liver regeneration (Review). Mol Med Rep 2017; 16: 23-35. [DOI] [PubMed] [Google Scholar]
- 26.Nevzorova YA, Trautwein C. Chapter 11. Regulation of cell cycle during liver regeneration. In: Apte U (Ed.). Liver regeneration. Academic Press, Boston: 2015; 153-166. [Google Scholar]
- 27.Caldez MJ, Van Hul N, Koh HWL, et al. Metabolic remodeling during liver regeneration. Dev Cell 2018; 47: 425-438.e425. [DOI] [PubMed] [Google Scholar]
- 28.Yalcin A, Clem BF, Imbert-Fernandez Y, et al. 6-Phosphofructo-2-kinase (PFKFB3) promotes cell cycle progression and suppresses apoptosis via Cdk1-mediated phosphorylation of p27. Cell Death Dis 2014; 5: e1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt LE, Dalhoff K. Alpha-fetoprotein is a predictor of outcome in acetaminophen-induced liver injury. Hepatology 2005; 41: 26-31. [DOI] [PubMed] [Google Scholar]
- 30.Karagiannis P, Eto K. Manipulating megakaryocytes to manufacture platelets ex vivo. J Thromb Haemost 2015; 13 Suppl 1: S47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horimizu M, Kawase T, Nakajima Y, et al. An improved freeze-dried PRP-coated biodegradable material suitable for connective tissue regenerative therapy. Cryobiology 2013; 66: 223-232. [DOI] [PubMed] [Google Scholar]
- 32.Amiri F, Molaei S, Bahadori M, et al. Autophagy-modulated human bone marrow-derived mesenchymal stem cells accelerate liver restoration in mouse models of acute liver failure. Iran Biomed J 2016; 20: 135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chien WS, Chen YH, Chiang PC, et al. Suppression of autophagy in rat liver at late stage of polymicrobial sepsis. Shock 2011; 35: 506-511. [DOI] [PubMed] [Google Scholar]