Abstract
Codon usage bias, the preference for certain synonymous codons, is found in all genomes. Although synonymous mutations were previously thought to be silent, a large body of evidence has demonstrated that codon usage can play major roles in determining gene expression levels and protein structures. Codon usage influences translation elongation speed and regulates translation efficiency and accuracy. Adaptation of codon usage to tRNA expression determines the proteome landscape. In addition, codon usage biases result in nonuniform ribosome decoding rates on mRNAs, which in turn influence the cotranslational protein folding process that is critical for protein function in diverse biological processes. Conserved genome-wide correlations have also been found between codon usage and protein structures. Furthermore, codon usage is a major determinant of mRNA levels through translation-dependent effects on mRNA decay and translation-independent effects on transcriptional and posttranscriptional processes. Here, we discuss the multifaceted roles and mechanisms of codon usage in different gene regulatory processes.
Keywords: codon usage, translation elongation, translation efficiency, cotranslational protein folding, mRNA decay, transcription, chromatin structure
INTRODUCTION
The central dogma of molecular biology describes the flow of genetic information from DNA to RNA to protein. Transcription of DNA generates mRNA, and the coding region of the mRNA is translated to protein by ribosomes. The amino acid sequence of the protein is determined by the triplet genetic codons, which are individually deciphered through the interaction of amino acid–specific tRNAs with the ribosome to direct the synthesis of a protein with a defined structure. Studies of the regulation of this information pathway, which results in the production of the optimal amount of active protein, have been focused on transcriptional and posttranscriptional controls that determine mRNA levels, as well as posttranslational mechanisms that regulate protein structure, activity, and stability. The level of transcription is mostly thought to be determined by promoter strength and upstream regulatory elements, whereas posttranslational modifications and protein degradation processes determine functional protein levels.
The degeneracy of genetic codons, predominantly due to flexibility in the identity of the third nucleotide—in what is known as the wobble position—allows 18 of the 20 standard amino acids to be encoded by two to six synonymous codons. Although the genetic code is almost identical in all organisms, the preferential use of certain synonymous codons, a phenomenon called codon usage bias, has been found in all genomes evaluated (1–3). The preferentially used synonymous codons are called preferred or optimal codons (i.e., codons with high optimality), and others are referred to as nonoptimal or rare codons. Different organisms have different codon usage biases. Mammals have a bias for C/G at wobble positions, whereas budding yeast prefer A/U codons, likely due to different mutation biases in each organism (3, 4). Within each genome, genes have a wide range of codon usage bias distributions. Some genes exhibit strong codon usage bias, but others have weak or even opposite biases (5). Furthermore, different regions of the same open reading frame (ORF) can have different codon usage biases. Because synonymous codon mutations do not change protein sequences, synonymous codons were previously considered to be redundant, and their mutations were regarded as silent mutations. However, accumulating genetic, biochemical, molecular, and bioinformatic evidence now demonstrates that gene codon usage acts as an important layer of genetic information that influences protein expression levels and structure (3, 6–8). Although codon optimization for heterologous gene expression has been widely and successfully used in biotechnology and biological studies (8), the mechanisms that lead to enhancement of gene expression when codons are optimized are not clear. Understanding the roles and mechanisms by which codon usage regulates the flow of genetic information will have major impacts on our understanding of gene regulatory mechanisms.
In diverse eukaryotes and prokaryotes, highly expressed, protein-encoding genes, such as ribosomal protein genes, are strongly enriched with optimal codons. Genome codon usage bias often correlates with levels of cognate tRNAs or with corresponding tRNA gene copy numbers (1, 9–12), suggesting that different synonymous codons may be decoded with different efficiencies to influence the translation elongation process. Genome-wide correlations between codon usage bias and protein levels have also been observed (13), suggesting that codon usage can determine protein expression levels. Indeed, codon optimization of endogenous genes in many prokaryotic and eukaryotic organisms leads to upregulation of protein production, sometimes by many hundred fold (5, 13–18). Moreover, codon usage is now known to influence multiple steps during the translation process by regulating elongation speed, translation efficiency, initiation, and termination, as well as accuracy (Table 1). Genetic and biochemical evidence also shows that codon usage affects cotranslational protein folding due to its effect on translation elongation speed. In addition to these effects on translation, codon usage is now known to play a major role in determining mRNA levels due to its translation-dependent effect on RNA stability. Furthermore, translation-independent effects of codon usage due to differences in nucleotide sequences can play important roles in transcription, chromatin structure, splicing, mRNA structure, transcriptional termination, and mRNA export (Table 1). Therefore, codon usage represents a previously unappreciated code that has multiple functions during the gene expression process. Here, we provide a systematic review of different functions and mechanisms of codon usage that are involved in gene expression, protein synthesis, and cotranslational protein folding with an emphasis on recent experimental advances.
Table 1.
Translation-dependent and translation-independent effects of codon usage
| Translation-dependent cell function | Effect of codon usage |
|---|---|
| Translation elongation | Optimal codons speed up the translation elongation rate, while rare codons slow it down, due to differential expression levels of corresponding tRNAs. |
| Translation efficiency | Codon usage regulates protein synthesis by affecting the amount of protein produced per mRNA in a given time. |
| Translation initiation | Rare codons can cause ribosome stalling and may inhibit translation initiation, whereas optimal codons may result in rapid liberation of start codons and high initiation rates. |
| Translation fidelity | Codon usage regulates the fidelity of amino acid incorporation and maintenance of the translation reading frame. |
| Premature termination | Rare codons can cause ribosome stalling and promote eRF1-mediated premature translation termination at sense codons. |
| tRNA expression, modification, and charging | Differential tRNA expression levels and tRNA modification and charging levels determine codon usage-biased mRNA translation in different tissues and cell types. |
| Cotranslational protein folding | The effect of codon usage on translation elongation speed affects the time available for the cotranslational protein folding process, thus affecting protein structure and function. |
| Cotranslational mRNA decay | Rare codons can cause ribosome stalling and promote translation-dependent mRNA decay mediated by the CCR4-NOT complex. |
| Translation-independent cell function | Effect of codon usage |
| Transcription | Codon optimality determines gene transcription levels from fungi to mammalian cells. |
| Chromatin structure | Codon composition affects chromatin structures by affecting transcription activation and suppression-related histone modification marks. |
| Transcription termination | Rare codons promote premature transcription termination through the formation of noncanonical poly(A) signals within open reading frames. |
| mRNA structure | Codon usage changes can result in mRNA structure changes, which may influence RNA stability and translation. |
| Splicing | Codon composition can influence splice site and exonic splicing enhancer and silencer sequences. |
| mRNA localization/transport | Codon usage profiles can influence mRNA cellular transport and localization. |
| mRNA toxicity | mRNAs with certain codon usage profiles can cause cellular toxicity in Escherischia coli. |
CODON USAGE REGULATES TRANSLATION ELONGATION SPEED
During the translation elongation cycle, the search for and selection of the cognate tRNA that recognizes the corresponding codon is the rate-limiting step, whereas the transpeptidation and ribosome translocation steps are usually fast. Early studies in Escherichia coli based on reporter genes and biochemical assays suggested that the rate of synthesis of heterologous protein can be determined by codon usage and cognate tRNA concentrations (19, 20). This effect was proposed to be due to the role of codon usage in regulating mRNA translation elongation speed; rare codons may take longer to be recognized because their corresponding tRNAs are present at low concentrations (1, 3, 21). However, most of the early experimental studies concerning the role of codon usage in the control of elongation speed relied on indirect measurements and protein overexpression systems, which led to conflicting conclusions (19, 22–24).
The development of ribosome profiling provided a powerful molecular tool to study ribosome translation dynamics across the genome with codon-level resolution in many organisms (25, 26). Because ribosome occupancies at individual codons should in principle inversely correlate with translation elongation speed, it was assumed that ribosome profiling would provide a conclusive genome-wide assessment of the role of codon usage in elongation speed. However, multiple early analyses of ribosome profiling data obtained in both prokaryotic and eukaryotic organisms found no correlation between codon usage and ribosome occupancy, which led to the early conclusion that codon usage does not play a significant role in modulating translation elongation kinetics (26–28). Later reanalyses of the same ribosome profiling data by a different method, however, revealed a correlation between tRNA concentration and the putative codon-decoding rate (29).
Despite its codon-level resolution, ribosome profiling experiments are influenced by experimental conditions, cloning and sequencing biases, methods of bioinformatic analysis, and experimental noise (30). These factors could impair the sensitivity of ribosome profiling for determining codon decoding rates. To resolve the effects of codon usage on codon decoding rate, we used cell-free translation systems made from Neurospora and Drosophila cells to directly compare the translation elongation speeds of mRNA templates encoding luciferase with different codon usage profiles (31, 32). Because luciferase folding is cotranslational in these systems, the time when the luciferase activity first appears reflects the translation elongation speed. A comparison of the time of first appearance of the luciferase signal clearly demonstrated, in both systems, that optimal codons increase the speed of translation elongation, whereas rare codons slow down translation elongation (Figure 1a). Therefore, the effect of codon usage on elongation rate is conserved from fungi to animals. Consistent with these results, codon optimality was also found to affect ribosome traffic on mRNAs. Rare codons cause ribosome pausing and accumulation of intermediate nascent peptides at the expected positions during translation in manners both dependent on and independent of amino acid context (31–34). Furthermore, by designing mRNAs with high signal to noise ratios, the effect of codon usage bias in ribosome profiling experiments could also be clearly seen both in vitro and in vivo without the need for sophisticated bioinformatic analyses, indicating the limited sensitivity of the ribosome profiling performed in previous analyses (31).
Figure 1.
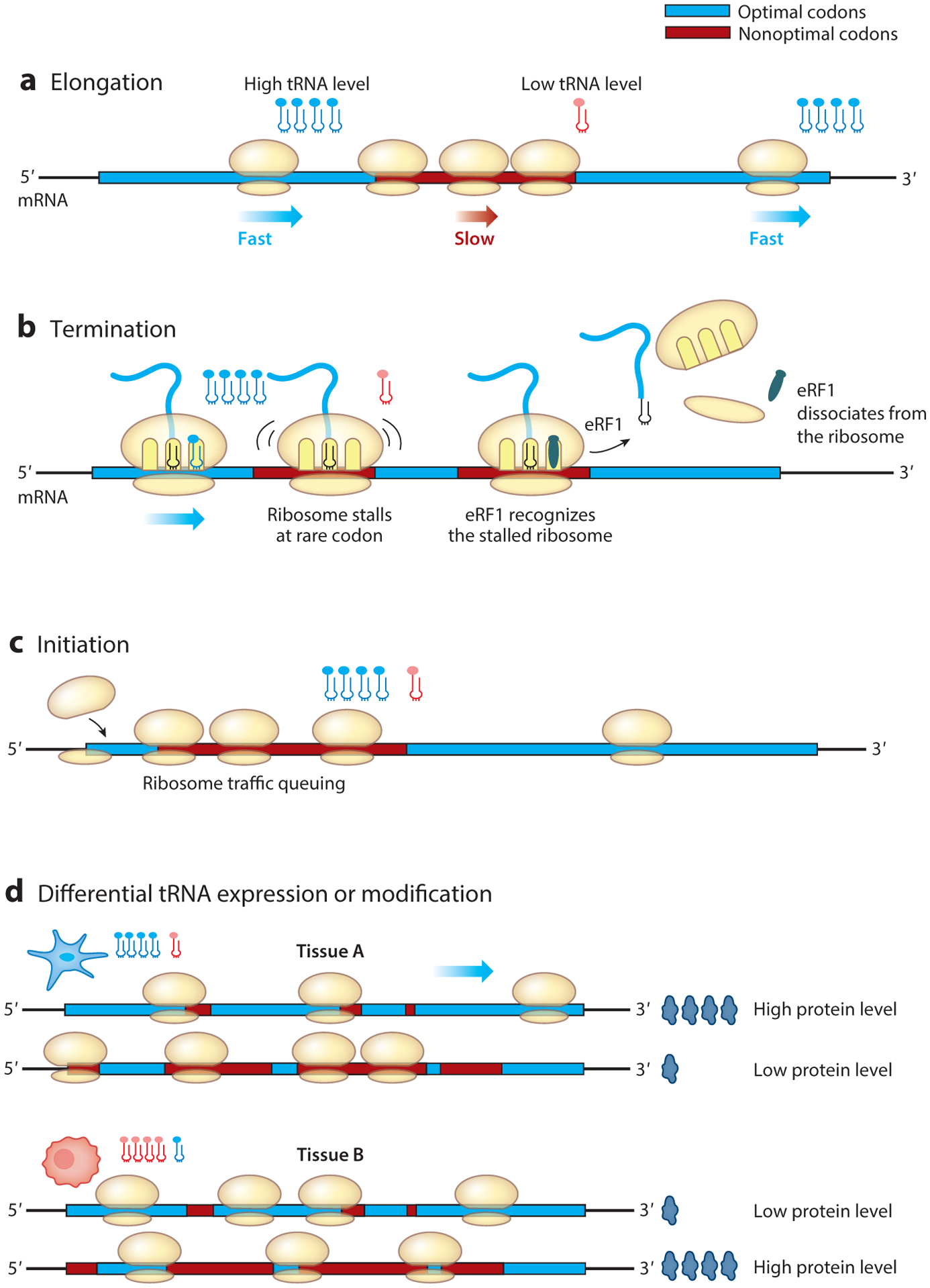
(a) Codon usage affects the speed of translation elongation: Optimal codons (light blue) speed up elongation, while nonoptimal codons (red) slow it down. (b) Codon usage regulates translation efficiency by affecting premature translation termination. Nonoptimal codons result in the ribosome stalling with an empty A-site, which can be recognized by the termination factor eRF1 to trigger premature termination. (c) Rare codon clusters downstream of the start codon can result in ribosome queuing, which may inhibit translation initiation. (d) Differential tRNA expression or modification levels in different cell types can result in the presence of different tRNA pools, influencing the cell type–specific translation of mRNAs.
Using improved ribosome profiling methods, genome-wide correlations between codon optimality and the ribosome decoding rate were further demonstrated in multiple organisms (31, 33, 35–37): Optimal codons are decoded faster by the ribosome than are rare codons. These results demonstrated that the failure to detect correlations between codon usage and ribosome occupancy in the early studies was due to the lack of sensitivity and technical issues associated with the early ribosome-profiling experiments. More recently, single-molecule imaging of mRNA translation further confirmed the role of codon usage in elongation speed in vivo (38). Together, these studies firmly established a role for codon usage in regulating the translation elongation rate and provided a mechanistic explanation for many codon usage–dependent translation effects.
CODON-USAGE EFFECTS ON TRANSLATION EFFICIENCY
Because of the correlation between strong codon usage biases and highly expressed proteins, codon usage was proposed to regulate protein synthesis by affecting translation efficiency (i.e., the amount of protein produced per mRNA in a given time). Although this notion was consistent with observations that manipulation of gene codon usage had robust effects on protein expression levels and that protein levels and observed codon usage were correlated genome wide in different organisms (7, 8, 13, 17, 39), most of the early studies did not take into account potential changes in mRNA level. We now know that codon usage can influence mRNA decay and transcription (see the section titled Codon Usage Is a Major Determinant of mRNA Levels). For actively translated mRNAs, translation efficiency is mainly determined by the number of ribosomes loaded at the 5′ end of the mRNA, and translation initiation—rather than elongation—was previously thought to be the main rate-limiting step and primary determinant of translation efficiency (35, 40). Further complicating the issue, codon usage–dependent mRNA sequence changes can alter mRNA structures, which can also affect the translation initiation rate (41).
Some proteome-wide analyses based on absolute protein levels, synthesis rate, or ribosome profiling found weak or no correlation between gene codon usage bias and predicted translation efficiency (42–44). Such whole-proteome analyses, however, are complicated by the fact that protein levels, mRNA levels, and ribosome occupancy can be influenced by multiple uncontrollable factors, such as analysis method and mRNA and protein stability, which can reduce their sensitivity. Despite these issues, accumulating cellular and biochemical evidence based on reporter genes now indicates that codon usage does regulate translation efficiency. First, in vitro translation assays using cell lysates showed that rare codons cause ribosome stalling, the accumulation of intermediate nascent peptides, and a reduction in full-length translation products (18, 33). Second, when a self-cleaving StopGo peptide was introduced into the middle of an eGFP-Luc translational fusion gene, the codon usage of the downstream Luc reporter gene had a major effect on its translation efficiency in vivo in both Neurospora and Drosophila cells (33). Third, codon usage optimization shifted mRNAs into high-polysome fractions in mammalian cells and resulted in dramatic increases in protein levels (17, 18). Fourth, differences in tRNA expression profiles in different growth conditions and tissues are correlated with protein expression (45, 46). Similarly, mutations that impair normal modification of tRNAs that result in codon-dependent reprograming of elongation kinetics also cause codon usage–dependent changes in protein levels in different organisms (47–50). Together, these results suggest that codon usage bias influences mRNA translation efficiency.
Ribosome Stalling and Premature Translation Termination
Rare codons in Neurospora and Drosophila and the multiple CGA codon repeats in a Saccharomyces cerevisiae strain that lacks tRNASer(UCG) were found to result in strong ribosome pausing or stalling in in vitro translation systems (33, 51, 52). In addition, ribosomal stalling can cause premature translation termination, thus reducing translation efficiency. In E. coli, translation of the rare CGA codon is inefficient, and repeats of CGA suppress reporter gene expression (53). Amino acid starvation of E. coli also results in translational pausing and premature termination that is dependent on tRNA aminoacylation, leading to a global reduction of protein synthesis (54). These results indicate that premature termination induced by rare codons is a conserved mechanism that regulates translation efficiency in both prokaryotic and eukaryotic organisms. In Neurospora, the premature termination caused by rare codons is mediated by the eukaryotic translation termination factor eRF1, which recognizes ribosomes stalled on rare sense codons (33). Silencing of eRF1 expression in Neurospora results in reduced rare codon–dependent ribosome stalling and upregulated expression of proteins encoded by mRNAs enriched with rare codons. In S. cerevisiae, the rare CGA codon can also be decoded as a stop codon by eRF1 and eRF3 (52). Together, these results suggest that translation termination factors can compete with canonical tRNAs for the empty A-sites of ribosomes stalled on rare sense codons to trigger premature translation termination (Figure 1b).
Translation Initiation
Although a universal role for codon usage in translation initiation is still not clear, ribosome stalling or collision that occurs near the AUG start codon and results in ribosome queuing likely affects the translation initiation efficiency (55, 56) (Figure 1c). In S. cerevisiae, analyses of the expression of CFLuc reporter genes with different codon usage profiles suggested that codon usage–dependent ribosome movement downstream of the start codon regulates translation initiation: Rare codons caused ribosome stalling and inhibited translation initiation, whereas optimal codons resulted in rapid liberation of start codons and high initiation rates (57). A study in E. coli, however, suggested that ribosome collisions affect translation by acting as a timer for translational quality control pathways (58). In mice, a mutation in a central nervous system–specific tRNA causes the loss of GTPBP2, a ribosome recycling protein, and results in ribosome stalling and widespread neurodegeneration. This mutation also results in elevated phosphorylation of eIF2α, an important regulator of translation initiation, due to activation of the eIF2α kinase GCN2 (59), suggesting that ribosome stalling may be sensed by GCN2 to provide feedback on translation initiation.
Previous analyses of prokaryotic and eukaryotic ribosome profiling results revealed a shared feature of highly expressed genes: enrichment of rare codons in the region 30 to 50 codons downstream of the start codon. This region is called the ramp sequence and serves to slow ribosome movement (43). The presence of the ramp was first proposed to reduce ribosome traffic jams to allow efficient translation of highly expressed genes but was later proposed to be the result of selection for reduced mRNA secondary structures, which would facilitate efficient translation initiation (41, 44, 60). The presence of the ramp may also be due to a data analysis bias: Shorter genes have higher translation initiation rates than longer genes, and technical issues with early ribosome profiling results selected for shorter genes (60). It was further shown that avoiding intramolecular base pairing between the Shine-Dalgarno sequence and the coding region of mRNA, rather than rare codons, can promote translation initiation in E. coli (61). Thus, whether codon usage broadly influences translation initiation efficiency due to its role in translation elongation is still unclear.
Codon Pair Bias and Codon Context
Like codon usage, codon context (i.e., the codon neighboring a specific codon) is not random. This phenomenon, called codon pair bias or dicodon bias, has been found in Eukarya, Archaea, and Bacteria and is different from the predicted usage bias of single codons (62–64). Although codon pair biases in different organisms are influenced by mutational biases (65, 66), evidence suggests that functional selection at the level of translation plays an important role (64). Similar to codon usage bias, codon pair bias is proposed to influence translation efficiency due to its effect on the translation elongation process (67, 68). In addition, codon context has been suggested to influence translation elongation speed in different organisms (23, 33, 67). Multiple studies suggest that interaction between tRNAs at the A- and P-sites of the ribosome, the codon-anticodon interaction, and the size of the tRNAs can all influence the decoding efficiency of the codon pair by the ribosome and, thus, translation efficiency (68–70). In yeast, some codon pairs potently inhibit translation (69). This effect is attributed to wobble-base pairing and is different from individual codon effects.
Because of the effect of codon pair bias on translation efficiency, codon optimization that takes codon pair bias into account has been successfully used in gene design to optimize protein expression (8, 71). Codon-pair deoptimization has been proposed as a strategy to attenuate viruses by reducing translation efficiency (72). This approach has been attempted in vaccine development for multiple viruses, including influenza virus and Zika virus (73). It should be noted that the viral attenuation that results from codon-pair deoptimization was later proposed to be mainly due to attenuation of viral replication caused by an increase in CpG and UpA dinucleotides rather than attenuation of translation (74).
TRANSLATION FIDELITY
In addition to translation efficiency, codon usage has also been implicated in translation fidelity, including correct decoding and maintenance of the correct translation reading frame. The miscoding rate for each codon, as measured using different assays in different organisms, varies many fold (75, 76). Early insights into contributions to translation accuracy were based on studies using reporter genes that measured miscoding rates for selected codons and amino acids (77, 78). Although tRNA levels were believed to underlie the effects of codon usage bias, evidence confirming that differential tRNA levels determine accuracy was lacking. Rather, early studies indicated that decoding accuracy is dependent on individual tRNAs and their inherent properties when interacting with codons and ribosomes and showed that this is also influenced by codon context (77, 79). Since error-prone tRNAs and codons tend to be underrepresented in the genome, translation accuracy is proposed to be a selective pressure that shapes codon usage. Consistent with this hypothesis, computational analysis of sequence conservation suggests that evolutionary selection for codon usage reduces missense and nonsense mutations in E. coli (80, 81). In addition, conserved patterns of covariation between sequence evolution, codon usage, and tRNA levels in different organisms led to the proposal that codon usage is under selective pressure to limit protein mistranslation and nonsense mutations (81, 82).
It is still not clear whether preferred codons increase or decrease decoding accuracy. Some studies suggest that codon usage was optimized during evolution to maximize both elongation rate and accuracy (83, 84), whereas other studies provide evidence that there is a trade-off between speed and accuracy, as slow decoding might promote fidelity checks during decoding (85, 86). Recently, a methodology for systematically detecting amino acid substitution errors in entire proteomes was developed (87). Analyses of E. coli and yeast proteomes using this method showed that most amino acid substitutions result from codon-anticodon mispairing at sites that are evolving rapidly. Ribosome profiling results suggest that errors occur at sites where ribosome velocity is higher (87), suggesting that a trade-off between speed and accuracy does exist. Further studies are still needed to determine how codon optimality broadly affects translation fidelity.
Codon usage is also under selection to minimize frameshifting events during translation, as frameshifts usually lead to early translation termination and truncated protein products (88). Although there is no clear experimental data showing a correlation between codon usage and frameshifting events, cognate tRNA levels are indeed key factors in determining the frameshifting efficiency at frameshift-prone codons (89), suggesting that ribosome stalling promotes frameshifting events. In particular, the combination of strong mRNA secondary structure and slippery sequences that allow ribosomes to easily move back and forth can cause frameshifting with high frequency (90). The classic example is the AAA codon, which is prone to frameshifting due to the rarity of cognate tRNAs and the tendency of tRNAs to mismatch to the adjacent nucleotide (91). Consecutive AAA codons are often found in programmed frameshift sites but are rare in other coding sequences.
tRNA MEDIATES CODON-USAGE EFFECTS ON TRANSLATION
Correlation Between Codon Usage and tRNA Abundance
The decoding rate of a codon can be affected by tRNA concentration, tRNA charging level, and codon-anticodon wobble decoding. Soon after the discovery of nonuniform usage of synonymous codons, codon usage frequency was found to correlate with cognate tRNA levels and tRNA copy numbers (1, 9–12, 92, 93). As a result, it is generally thought that tRNA concentration is the primary factor that determines the decoding rate. The tRNA adaptation index and its variants, which take into account the tRNA concentration (based on tRNA gene-copy numbers) and the efficiencies of codon-anticodon pairing, are commonly used to quantify gene codon usage bias in different organisms (94, 95). Since tRNA concentrations normally correlate with tRNA gene copy numbers, tRNA repertoires and codon usage are believed to have coevolved to achieve balance between supply and demand during translation, thus optimizing translation efficiency (4, 94, 96). Consistent with this model, recent high-throughput studies based on ribosome profiling experiments have confirmed genome-wide correlations among codon usage, tRNA concentration, and codon decoding time (29, 31, 33, 35, 37).
Further supporting the role of tRNA concentration in mediating the codon usage effect, mutations that alter tRNA levels have been shown to cause predicted changes in protein synthesis rates and gene expression. Overexpression of cognate tRNAs also overcomes the inhibitory effects of rare codons (11, 97, 98). The arginine CGA codon is the rarest codon for arginine in S. cerevisiae, and it has a strong inhibitory effect on translation due to inefficient wobble decoding by the isoacceptor tRNA (98). Overexpression of the cognate tRNA for the CGA codon suppresses the inhibitory effect of the rare CGA codon. Changing gene codon usage to match host codon usage or overexpressing certain cognate tRNAs of rare codons promotes the production of heterologous proteins in many systems (8).
The connection between codon usage and the makeup of cellular tRNA pools plays an important role in determining global translation levels and the proteomic landscape in cells. In E. coli, the simultaneous mutation of abundant codons to their rare synonymous counterparts in several highly expressed genes reduced the global translation efficiency and cellular fitness (99). These phenotypes were rescued by increasing the supply of the tRNA for the mutated codon, leading to the proposal that the codon usage bias of highly expressed genes was due to selective pressure to maintain the efficiency of global protein translation. In Neurospora, we showed that depletion of the tRNA modification enzyme adenosine deaminase (ADAT) altered relative tRNA levels for different synonymous codons in cells and caused global reprogramming of codon usage–dependent translation kinetics and global codon usage–biased protein level changes (50). These results suggest that the adaptation of codon usage to the tRNA pool is important for determining the global translation landscape.
Regulation of tRNA Expression
Recent studies suggest that both the tRNA pool and mRNA transcript levels contribute to the translation-dependent function of codon usage. tRNA expression was previously thought to be less regulated than expression of mRNA; however, accumulating evidence indicates that control of tRNA expression plays an important role in defining the translational landscape in cells. Depending on the demand for mRNA translation, cells can adjust their tRNA supply to achieve optimal gene expression. tRNA transcription levels, charging levels, and anticodon modifications are fine-tuned to regulate protein expression under different nutrient and growth conditions and in different developmental stages.
In E. coli, differential tRNA expression was detected at different growth rates that roughly correlated with the codon usage frequencies in the mRNA pools under different conditions, suggesting that tRNA expression was modulated to allow optimal cell growth (100, 101). Consistent with this model, the aminoacylation levels of tRNAs were found to differ depending on the growth media used and that different tRNA levels differentially regulate the translation of subsets of mRNAs (102, 103). In S. cerevisiae, different stresses were found to induce different patterns of up- or downregulation of tRNA pools (104, 105). In Caenorhabditis elegans, identical tRNA genes can have very different expression levels in different tissues in a time-specific manner (106).
Analysis of genome-wide tRNA levels in eukaryotes is challenging due to high gene copy numbers, sequence similarities, strong secondary structures, and extensive posttranscriptional modifications. A microarray-based method to quantify a subset of tRNAs representing all 20 amino acids revealed that tRNA levels within the total cellular RNA vary widely among different human tissues (45), suggesting that tRNA levels contribute to tissue-specific gene expression (Figure 1d). Highlighting the importance of tRNA regulation in human tissues, genes involved in cell-autonomous functions and those involved in differentiation have different codon usage patterns (107, 108). Importantly, tRNAs that are upregulated in proliferating cells are typically repressed in differentiated cells and have anticodons corresponding to the codon usage profile of proliferation-related genes (107). In contrast, the tRNAs elevated in differentiated cells are often repressed in proliferating cells. These data suggest that different translational programs based on different tRNA pools contribute to tissue-specific gene expression in mammals. In different human cell lines, we previously showed that the same codon usage manipulation of the oncogene KRAS, which encodes an oncogenic Ras GTPase family member, has different effects on its protein and mRNA expression levels (18). Similarly, codon optimization of the Adh gene in Drosophila elevated alcohol dehydrogenase activity in larvae but not adults (16). However, the codon-dependent translational efficiency was reported to be stable in different mammalian cell types, and the codon-usage differences in mammalian transcriptomes could be explained by mutational biases caused by GC-content differences (109).
The cellular tRNA pool is regulated at multiple levels from tRNA abundance to tRNA charging to tRNA modifications (104, 110–113). tRNA expression levels are mainly regulated at the transcriptional level through interactions between RNA polymerase III and transcription factors (107, 114). For example, the transcription factor SOX4 directly regulates the transcription of a subset of tRNA genes (115). Thus, transcriptional regulation appears to coordinate gene codon usage and translation efficiency to allow proper cell growth and differentiation in diverse tissues and cell types. Dysregulation of tRNA expression can lead to disease, as the upregulation of specific tRNAs drives cancer-specific gene expression and promotes breast cancer metastasis (46). Together, these results suggest that codon usage and tissue- or cell type–specific tRNA expression may be important for tissue- and cell type–specific gene expression.
tRNA Charging and tRNA Modifications
Like cellular tRNA concentration, tRNA charging and tRNA modifications can also influence codon usage–dependent effects on decoding speed and translation efficiency. Selective charging of tRNAs can be induced by amino acid starvation, and the effects can be profound since various isoacceptor tRNAs compete for the same amino acids (116, 117). When amino acid concentration becomes growth limiting in E. coli, the differential charging of tRNAs maintains the expression of essential genes due to their preference for optimal codon usage, leading to the promotion of cell survival and suppression of the expression of genes with rare codons (110, 116, 118). In addition, the codon optimality of genes contributes to differential mRNA translation in response to amino acid starvation, indicating the physiological significance of codon usage bias in the cellular stress response (110). In mammalian cells, amino acid starvation also results in differential tRNA charging and codon-specific ribosome pausing on mRNA, which can affect translation efficiency (119).
tRNAs have the most diverse and abundant nucleoside modifications of all types of RNA. These modifications are important for tRNA structure, function, and stability, and some modifications have been shown to affect codon-biased protein expression. There are 61 codons in the genome, but no organism expresses all of the 61 corresponding tRNAs, and modified tRNAs are needed to decode many codons. tRNA modifications that affect wobble decoding shape the codon usage effect on translation. Isoacceptor tRNAs with modified anticodons are used to decode multiple synonymous codons, but the different affinities of the Watson-Crick and non-Watson-Crick base pairings result in different decoding efficiencies that can have major effects on translation rate (120, 121). For instance, G:C base pairs are more stable than G:U base pairs, and the I:A base pair is even less stable (53, 120). Based on these observations, a model was proposed whereby two tRNA wobble codon modifications—A34 to I34 (A-to-I) editing mediated by tRNA-dependent ADATs in bacterial and eukaryotic cells and the uridine to xo5U34 modification mediated by tRNA-dependent uridine methyltransferases in bacteria—explain the correlation between codon usage and tRNA gene frequency (122). These wobble-position modifications expand the base-pairing ability of the wobble codon: I34 can wobble pair with A, C, and U, and xo5U34 can pair with A, G, and U.
In Neurospora, silencing of ADAT expression abolishes most of the I34 modifications of tRNAs, resulting in large tRNA pool changes (decreases in ADAT-related INN tRNAs, where N can be any nucleotide, and increases in ANN tRNAs) and reprogramming of translation elongation kinetics for ADAT-related codons in a codon usage–dependent manner (50). ADAT silencing also causes genome-wide codon usage–biased ribosome pausing on mRNAs and changes to the proteome landscape. During human embryonic stem cell differentiation, self-renewing cells are dependent on the I34 tRNA modification to optimize translation, and human pluripotent embryonic stem cells have higher levels of I34 tRNA modifications and higher ADAT protein levels than differentiated cells, suggesting that the dynamic I34 modification may contribute to differential codon usage–dependent translation with effects on mammalian cell growth and differentiation (123).
Mutations that disrupt modification of U34 in S. cerevisiae and C. elegans cells cause ribosome pausing at their cognate codons, resulting in proteotoxic stress and the accumulation of endogenous protein aggregates (124, 125). In mice with a mutation in IKBKAP (also known as ELP1), a gene encoding the Elongator complex involved in U34 modification, the level of mcm5s2U34 tRNA modification is reduced, resulting in codon-biased gene misregulation (49). This codon-biased effect results in the developmental and neurodegenerative phenotypes of human familial dysautonomia. Thus, both I34 and U34 modifications are conserved tRNA modifications important for codon usage–dependent translation.
Other types of tRNA modification also contribute to the effects of codon usage on translation. In yeast, hydrogen peroxide treatment induces Trm4 methyltransferase–dependent increases in levels of tRNALeu(CAA) with a 5-methyl C34 modification, which results in selective translation of mRNA enriched in the corresponding UUG codon (47). In Mycobacterium bovis, exposure to hypoxia alters the modifications of many tRNAs, which may promote translation of codon-biased transcripts that enhance the expression of stress-response proteins (48). Together, these studies demonstrate that both tRNA charging and tRNA modifications play critical roles in driving codon usage–dependent translation in diverse organisms.
CODON USAGE REGULATES COTRANSLATIONAL PROTEIN FOLDING AND PROTEIN FUNCTION
Although amino acid sequences determine protein structure, folding of most proteins occurs cotranslationally in vivo. Folding of a nascent peptide, which is synthesized from the N-terminal to the C-terminal end, is a modular process. After the newly synthesized nascent chains emerge from the ribosome, their folding is mediated by their interactions with the ribosome, protein chaperones, and other folding catalysts (126–129). Translation kinetics were predicted to influence cotranslational protein folding (130–132). Supporting this notion, it was shown that changes in the elongation rate affect folding of overexpressed proteins in E. coli (21, 34). Heterologous protein expression often leads to protein misfolding and aggregation, a phenomenon that can be corrected by culturing cells at a low temperature, which is presumably due to the enhancement of correct folding caused by the slower translation elongation rate. Because of the role of codon usage in translation kinetics, it is expected to influence protein folding by affecting the time available for cotranslational folding events (Figure 2).
Figure 2.
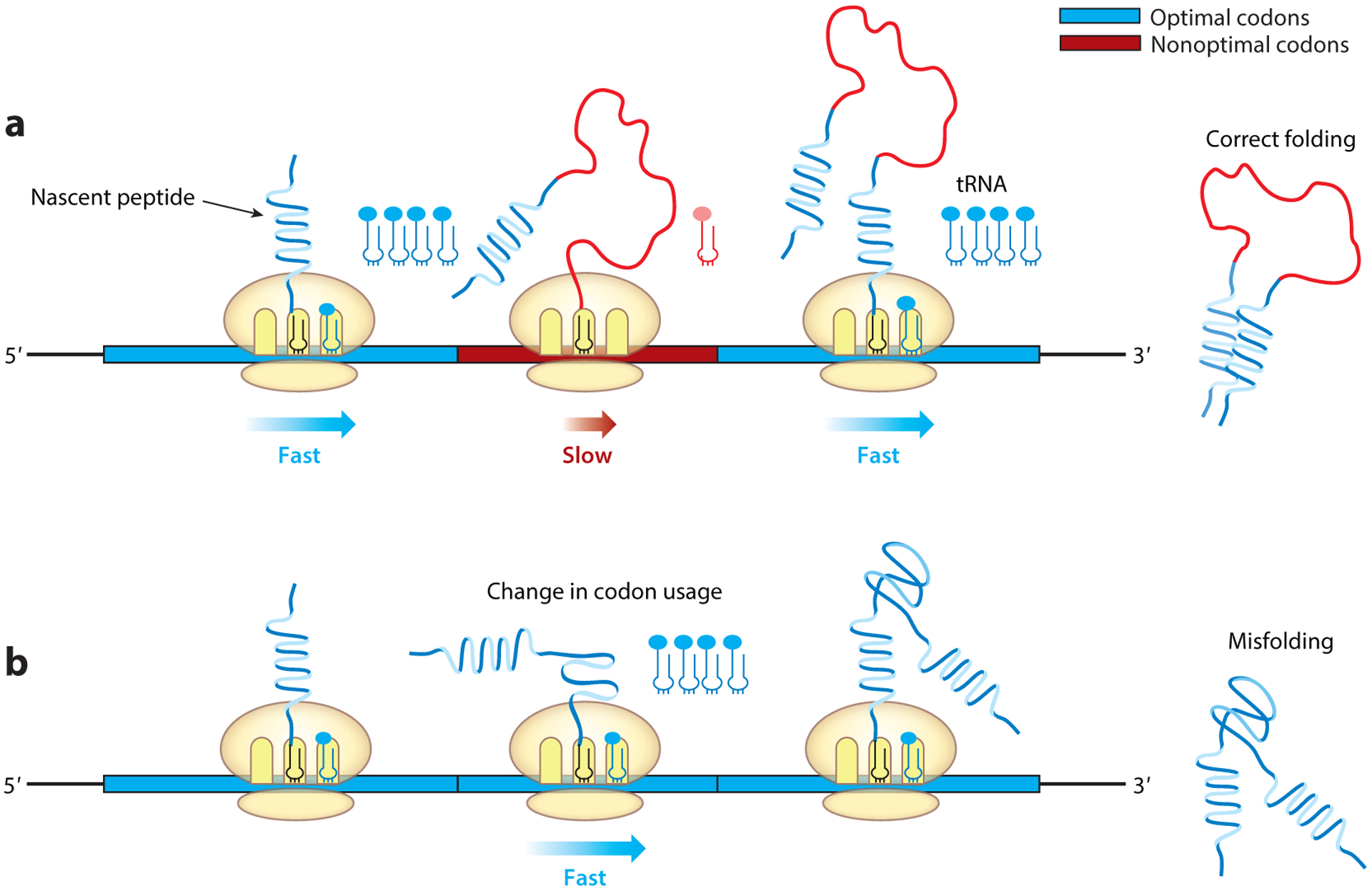
Codon usage affects cotranslational protein folding. (a) Cotranslational folding of different domains of the nascent peptide is adapted to the elongation kinetics defined by codon usage to allow optimal protein folding. (b) Changes to codon usage alter elongation kinetics, which can increase protein misfolding.
Codon Usage Affects Protein Folding in E. coli
The role of codon usage in protein folding was first investigated in E. coli by examining the activity and folding of overexpressed proteins. Analyses of proteins overexpressed in E. coli showed that changing rare codons to preferred ones caused modest decreases in protein activity or solubility (133, 134) or increased the amount of insoluble and aggregated protein (134). In addition, codon substitutions to preferred codons impaired the folding of the multidomain protein SufI in vitro and in E. coli cells (34). In contrast, codon optimization, with synonymous codons chosen to mimic ribosome movement in E. coli, improved the specific activity of firefly luciferase overexpressed in E. coli (11). Furthermore, codon substitutions influenced the folding of an artificially designed fluorescent YKB (yellow-black-blue) protein expressed in E. coli (135). Importantly, the effect of codon usage on the protein folding of mammalian gamma-B crystallin overexpressed in E. coli was later directly confirmed by real-time FRET (fluorescence resonance energy transfer) analysis and NMR (nuclear magnetic resonance) spectroscopy (136). More recently, overexpression of chloramphenicol acetyltransferase in E. coli showed that various synonymous codon substitutions affect protein folding and protein activity that support cell growth (137). It should be noted that studies in E. coli relied on protein overexpression and, therefore, may not reflect the physiological role of codon usage in protein folding.
Codon Usage–Mediated Protein Folding in Eukaryotes and Its Physiological Roles
A study of the human MDR1 gene, which encodes a protein involved in multidrug resistance, showed that a synonymous single-nucleotide polymorphism (SNP) can cause altered drug and inhibitor interactions and sensitivity to protease digestion of MDR1 protein transiently overexpressed in human cells; this result provided the first suggestion of a role for codon usage in eukaryotic protein folding (138). Later, by studying the Neurospora circadian clock gene frequency (frq), which has an ORF enriched for nonoptimal codons, we demonstrated genetically that codon usage influences eukaryotic protein folding and function in vivo (5). Codon optimization of regions of frq enriched for rare codons led to complete abolishment of circadian rhythms; impaired FRQ activity; and altered protein stability, phosphorylation profiles, and protease sensitivity (5, 12). Also, importantly, the codon optimization of different regions of frq resulted in phenotypes that could be predicted by the effects of localized codon usage on protein structure (5). Unlike previous studies on codon usage, the expression of frq was under the control of its own promoter and regulatory context and did not rely on protein overexpression, thus demonstrating the physiological role of codon usage. Similar to results in Neurospora, codon optimization of a component of the Drosophila circadian clock encoded by the Period (Per) gene, which, although not homologous to frq, is also enriched for rare codons, abolished the circadian locomotor rhythm due to severe impairment of PER activity and reduction of PER phosphorylation in a location- and codon usage–dependent manner (139). The altered structure of PER from fly tissues was evident from altered trypsin sensitivity and changes in thermal denaturation and aggregation temperatures and was not due to protein overexpression. By performing in vitro translation assays in the Neurospora and Drosophila cell-free translation systems, we provided further support for the role of codon usage in cotranslational protein folding because codon usage changes had dramatic effects on luciferase folding and specific activity that were consistent with effects on translation elongation speed (31, 32). Even though the codon-optimized Luc mRNA was translated more rapidly and produced more full-length protein, its light emission was dramatically reduced compared to that of the protein made from the wild-type mRNA. These results demonstrate that the physiological role of codon usage in cotranslational protein folding is conserved from fungi to animals and that protein misfolding is increased when codon usage is not adapted to the cotranslational folding kinetics in vivo (Figure 2).
Despite the dramatic effect of codon usage on protein function in both the Neurospora and Drosophila clock genes, in the cyanobacterium Synechococcus, codon optimization of the circadian clock genes kaiB and kaiC, which are not sequence homologs of the eukaryotic clock genes, produced a very different outcome (15). The Synechococcus clock actually became more robust upon codon optimization due to an increase in Kai protein levels, even under conditions in which the clock is not normally functional, and this resulted in impaired cell growth (15). The different outcomes of codon optimization for these clock genes suggest that sensitivity to codon usage–mediated cotranslational folding effects is protein dependent.
Cotranslational folding plays an important role in the function of the conductance regulator CFTR, the protein that is mutated in cystic fibrosis patients (140). Codon optimization of a region of the NBD1 domain, which is enriched with nonoptimal codons, results in aggregation of full-length CFTR, suggesting that translation elongation kinetics modulated by codon usage are important for cotranslational folding of different CFTR protein regions in human cells (140). Furthermore, a synonymous SNP in the CFTR gene created a codon that is recognized by a tRNA present at low levels in human bronchial epithelial cells, resulting in reduced CFTR protein stability and channel activity (97). Overexpression of this tRNA in HeLa cells rescued the phenotype resulting from this SNP, suggesting that CFTR cotranslational folding is influenced by the alteration of translation elongation kinetics caused by the SNP (97). In cancer cells, a short stretch of rare codons within certain regions of the gene encoding the transcription factor ZEB2 results in translational pausing and compromises its protein production, likely by influencing its folding (141). We and others showed that the codon usage of the oncogene KRAS, which is rich in rare codons, regulates the expression and protein structure of KRAS protein in human cells (17, 18). More recently, codon usage optimization of coagulation factor IX and a single SNP of ADAMTS13 has been shown to affect protein folding or activity (142, 143). Together, these studies established that codon usage has a universal role in cotranslational protein folding in prokaryotes and eukaryotes.
The Codon Usage Code for Cotranslational Protein Folding
The Synechococcus circadian clock protein KaiC is a highly structured protein, as shown by a high-resolution crystal structure. In contrast, the Neurospora and Drosophila clock proteins, FRQ and PER, encode large proteins that are mostly made of predicted intrinsically disordered domains (5, 12, 15, 139). The different circadian phenotypes that resulted from codon optimization of kaiC versus frq and Per suggest that codon usage affects the cotranslational folding of different types of protein structures differently. This conclusion is further supported by the differing effects of codon optimization of different regions of the frq ORF: Codon optimization of the putative intrinsically disordered regions impairs clock function, whereas codon optimization of predicted structured domains has little or no effect (12). Moreover, codon optimization of an unstructured loop region that is highly conserved among luciferase homologs causes the greatest reduction of luciferase-specific activity (31). Together, these results suggest that cotranslational folding of well-structured protein domains is less sensitive to codon usage changes, whereas less stably structured or intrinsically disordered domains are more sensitive.
Because codon usage influences the time available for cotranslational folding, it is likely that well-structured domains fold readily and are insensitive to changes in translation kinetics. Conversely, less structured domains, structures that are difficult to fold, or intrinsically disordered domain–containing regions may require more time for proper folding of the entire protein; rapid elongation in these regions likely interferes with the cotranslational folding process and increases the chance of protein misfolding (Figure 2). Although intrinsically disordered domains are not predicted to form stable structures by themselves, some may form structures due to interactions with other regions of the protein or protein partners.
Bioinformatics analyses have uncovered correlations between codon usage and secondary and tertiary protein structures (12, 95, 144–149). Consistent with a role for nonoptimal codons in predicted unstructured or weakly structured domains, multiple genome-wide studies have shown that domains predicted to be unstructured are enriched for rare codons, whereas predicted α-helical regions in proteins from E. coli, Neurospora, yeast, C. elegans, and Drosophila are encoded by common codons (12, 95, 144). In addition, analyzing proteins with known structures found that buried residues are usually encoded by common codons, especially in highly expressed proteins (150). Structurally buried residues in highly expressed proteins are mostly found within well-structured protein domains.
The relationship between codon usage and protein structure, however, is not a simple one, perhaps reflecting the complex nature of cotranslational folding processes for diverse protein structure variations. Rare codons were found to be enriched at domain boundaries in a number of studies (34, 144, 149), but some studies found only weak or no correlation (147, 151). Analysis of homologous coding sequences in different organisms revealed that although many rare codon clusters are conserved, most of these clusters are found within conserved protein domains (148). Therefore, the role of codon usage in different proteins and structure types is likely to be specific to the given protein and dependent on its cotranslational folding kinetics.
Other cotranslational folding–related processes can also be regulated by codon usage. Rare synonymous codons in the 5′ end of the γ -actin gene affect cotranslational arginylation, which regulates the expression of the protein by influencing its ubiquitination (152). In addition, the 5′ ends of the coding sequences of secreted proteins are enriched in rare codons, which may promote cotranslational translocation and increase the secretion efficiency of these proteins (153). Moreover, rare codons downstream of the region encoding the signal recognition particle (SRP) binding site in some yeast genes encoding membrane proteins are proposed to promote protein translocation by regulating the interaction between nascent polypeptides and SRP (154). A pair of rare codons in the gene encoding the urea transporter is important for the production and localization of the UreA protein in Aspergillus nidulans (155). Together, these studies suggest that codon usage has diverse physiological roles in regulating cotranslational folding–related processes.
CODON USAGE IS A MAJOR DETERMINANT OF mRNA LEVELS
Codon usage was previously thought to mediate its effect on protein expression mostly due to its role in translation. Recent evidence, however, demonstrates that the codon usage effect on mRNA levels is a major, and in some cases the main, mechanism by which codon usage affects protein expression. Multiple early studies showed that codon optimization or deoptimization of specific genes had positive or negative effects on the mRNA levels of specific genes (156, 157). In addition, genome-wide expression profiling studies uncovered positive correlations between codon usage and mRNA levels in both prokaryotic and eukaryotic organisms (13, 39, 158, 159). Recent studies showed that codon usage acts as an important posttranscriptional determinant of mRNA levels by affecting cotranslational mRNA decay. Unexpectedly, accumulating evidence has now demonstrated that codon usage can play a major role in determining mRNA levels through its transcriptional and posttranscriptional effects in a translation-independent manner.
mRNA Decay
The connection between translation and mRNA decay has long been known, and mRNA can be stabilized after translation inhibition (160). Most previous mechanistic studies on the effects of translation on mRNA stability have focused on nonsense-mediated decay (the degradation of mutant mRNAs with premature stop codons), no-go decay (the degradation of mRNAs with stalled ribosomes), and nonstop decay pathways (the degradation of mRNAs lacking stop codons) (161). Although these studies demonstrated the importance of translation in mRNA decay pathways, these surveillance mechanisms are normally activated when translation is abnormally halted or stalled. Recent establishment of the link between gene codon usage and mRNA stability in yeast, E. coli, Drosophila, zebrafish, and mammalian cells (32, 162–169) has demonstrated codon usage as a universal mechanism contributing to mRNA stability.
Although early genome-wide studies using classic codon bias measurements failed to reveal a clear correlation between codon usage and mRNA stability (170), Presnyak and colleagues (162) assessed the contribution of individual codons to genome-wide mRNA half-lives in budding yeast and found that stable and unstable transcripts are enriched for different sets of codons, which largely coincide with codon optimality. The importance of codon optimality for determining mRNA stability in a translation-dependent manner was confirmed by using reporter genes with modified codon optimality. The DEAD-box helicase Dhh1 was identified as a critical mediator of the codon usage effect on mRNA stability (171). Dhh1 interacts with the ribosome and preferentially binds to transcripts enriched in rare codons. It is hypothesized that Dhh1 preferentially binds to slow-moving ribosomes and targets transcripts enriched in rare codons for degradation. mRNA deadenylation is also affected by codon usage: The poly(A) tails of transcripts with different codon usage biases are differentially protected by poly(A) binding proteins, leading to different efficiencies of deadenylation by the CCR4-NOT deadenylation complex (165, 172). In human cells, the importance of codon optimality and translation elongation speed in translation-dependent mRNA decay was demonstrated by independent studies using ORFeome libraries, which allowed mRNA decay rates to be determined independently of 5′ and 3′ regulatory sequences (166, 168).
Recently, a structure-based study provided important insights into how codon optimality modulates cotranslational mRNA decay in budding yeast by demonstrating how the interaction between the CCR4-NOT complex and the ribosome monitors translation kinetics (173) (Figure 3a). Cryo–electron microscopy structures revealed that the N-terminal domain of NOT5, a subunit of the CCR4-NOT complex, specifically interacts with the ribosome E-site when the ribosome A-site is empty. The association of Dhh1 with ribosomes is also dependent on the interaction of NOT5 with the E-site, suggesting that the Dhh1-promoted decapping process is downstream of deadenylation. Ribosome profiling revealed that less optimal codons were enriched at the A-site when CCR4-NOT was bound to ribosomes (173). Thus, the interaction between the CCR4-NOT complex and the ribosome can monitor codon usage–dependent translation elongation kinetics and regulate cotranslational mRNA decay.
Figure 3.
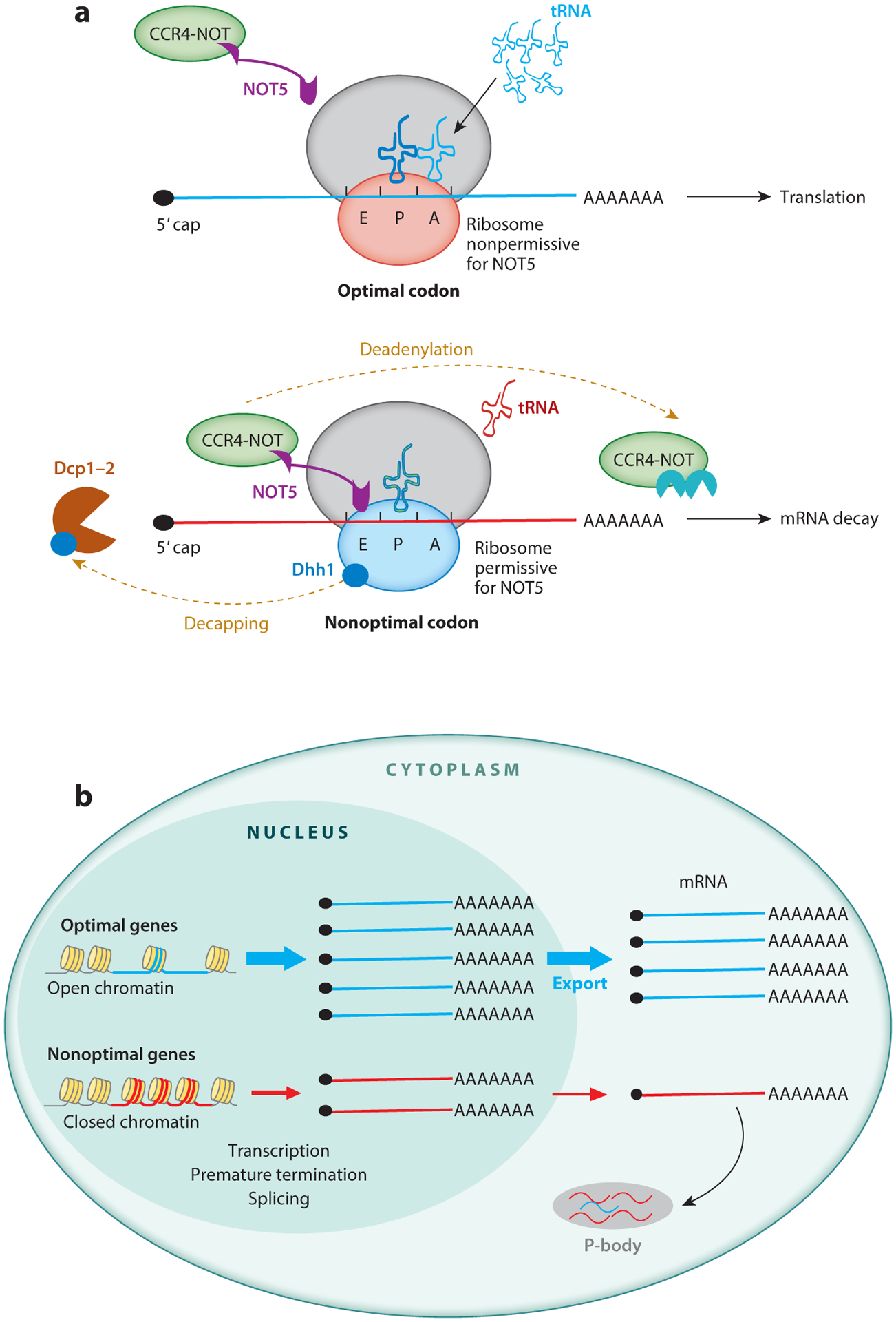
Transcriptional and posttranscriptional codon usage effects on mRNA in eukaryotes. (a) A model explaining how codon usage regulates cotranslational mRNA decay. (top) Optimal codons allow the ribosome A-site to be quickly recognized by cognate tRNA, leading to a ribosome conformation that does not permit the association of NOT5 at the ribosome E-site and leads to continued mRNA translation. (bottom) Nonoptimal codons result in a ribosome with an empty A-site, leading to a ribosome conformation that allows NOT5 to interact at the E-site and recruits the CCR4-NOT complex to mediate mRNA decay from the 3′ end. (b) Translation-independent effects of codon usage on mRNA levels. Genes with strong codon usage biases (indicated by blue lines) are associated with open chromatin and high transcription levels. Genes enriched for rare codons (indicated by red lines) promote the formation of closed chromatin, resulting in suppressed transcription and increased premature transcription termination. Codon optimality may also influence the efficiency of mRNA transport and localization.
Although most studies of mRNA decay have focused on the translation-dependent effects of codon usage, a recent study in which translation was inhibited by cycloheximide suggested that codon usage might also affect mRNA decay in a translation-independent manner (167). It should be noted, however, that the use of a global translation inhibitor may induce unintended secondary effects and that the translation-independent effect has not been observed in other studies (162, 168).
Transcription
The effect of codon usage on mRNA levels often cannot be explained by the effect on mRNA half-lives alone. For example, codon optimization had little effect on mRNA decay for reporter genes in Neurospora (13), and deletion of the dhh1 homolog did not affect the genome-wide correlation between codon usage and mRNA levels (F. Zhao & Y. Liu, unpublished data). In human cells, high GC content within the ORF, which usually correlates with optimal codons, increases mRNA and protein levels independently from mRNA decay (174). Moreover, codon optimization of the ORF of the gene encoding Toll-like receptor 7 was found to increase its protein and mRNA levels mainly due to enhanced transcription, with only modest effects on translation and mRNA stability (175).
In Neurospora, we showed that codon usage is strongly correlated with mRNA levels genome wide (13). Using a series of endogenous and exogenous reporter genes, codon optimization was found to have dramatic effects on mRNA levels but little effect on mRNA stability. Importantly, the effect of codon usage on the mRNA levels of reporter genes is largely due to its influence on transcription and on transcription-factor binding at promoter regions and is independent of translation. Furthermore, codon usage affects chromatin structures around ORFs. For example, the heterochromatic H3K9me3 modification is partly responsible for the suppression of transcription by rare codons in some genes. The importance of codon usage for transcription and chromatin structures has also been demonstrated in a study of the human oncogene KRAS. Increasing codon optimality results in induction of transcription, recruitment of histone acetyltransferases, and increases in histone modifications associated with gene activation (18). Consistent with a broad impact of codon usage on transcription, codon usage was found to be positively correlated with the predicted mRNA synthesis rate genome wide in both S. cerevisiae and Schizosaccharomyces pombe (176). Together, these studies demonstrate that the role of codon usage in transcription is conserved in eukaryotic organisms.
Unlike the translation-dependent codon usage effects, such as those on mRNA decay, the translation-independent transcriptional effect is counterintuitive, especially in eukaryotes, where transcription and translation are decoupled. The observed translation-independent transcriptional effects suggest that gene codon usage is due to the coevolution of coding region sequences with both the transcription and translation machineries. Selection for optimal amounts of each protein led to gene codon usage optimized for transcription, mRNA stability, or both. As a result, codon usage is adapted to both the translation and transcription processes. Codon usage information is read by the transcriptional machinery in the form of DNA elements, which suppress or activate transcription. For example, GC content or CpG islands are known to affect nucleosome occupancy over genes (177), which may influence chromatin structures that are important for transcription. Although most known transcriptional regulatory mechanisms affect promoter regions, the mechanism mediating the transcriptional effect of codon usage is largely unknown. Our current knowledge does, however, highlight the importance of the gene coding sequence for determining transcription levels.
Other Translation-Independent Cotranscriptional and Posttranscriptional Processes
The evolution of codon usage is also known to be under selective pressure due to features independent of translation, such as the Shine-Dalgarno sequence in E. coli (27) and splicing junctions in eukaryotes (178). Splice site and exonic splicing enhancer and silencer sequence requirements may also influence the selection of synonymous codons (178, 179). In addition, we showed that codon usage bias promotes gene transcription in both Neurospora and mammalian cells by suppressing premature transcription termination. Clusters of rare codons can lead to the formation of noncanonical poly(A) signals within ORFs and premature termination (180).
More recently, the interplay between codon usage and splicing has been proposed to regulate gene expression levels in human cells by regulating mRNA localization. Splicing can specifically increase the expression level of genes with low GC content (enriched in rare codons), and genes with high GC content (enriched in common codons) can promote the cytoplasmic localization of reporter mRNAs (181). In addition, P-body transcriptome analyses suggest that GC content can influence mRNA storage in P-bodies and mRNA decay in human cells. Most mRNAs in P-bodies are AU rich (i.e., rare-codon rich) with a low protein yield (182). Although the mechanisms underlying these phenomena are unclear, they indicate that codon usage can have multiple roles affecting the cotranscriptional and posttranscriptional control of gene expression (Figure 3b).
In E. coli, expression screening of hundreds of synonymous GFP gene variants revealed that some variants were toxic to cells independently from translation, suggesting that synonymous codon changes in bacteria can produce toxic mRNA molecules (183). Mutants that can reduce the mRNA expression levels of these variants lead to suppression of their toxicity. The mechanism for this mRNA toxicity is not known.
SUMMARY AND PERSPECTIVES
Although the phenomenon of codon usage bias has been known for more than four decades, its biological importance has been experimentally demonstrated primarily in the last decade. A large body of biochemical, genetic, biophysical, and bioinformatic evidence demonstrates that codon usage affects multiple gene regulatory processes, including translation, cotranslational protein folding, transcription, and posttranscriptional regulatory processes. In addition, codon usage influences gene expression and protein function in organisms from prokaryotes to humans. These findings support the proposal that codon usage is a previously underappreciated layer of genetic information that is fundamental to the regulation of gene expression.
Despite recent advances, there are many unanswered questions regarding the diverse functions of codon usage and the mechanisms responsible for these effects. For example, the contribution of codon usage to tissue- and cell type–specific gene regulation and its mechanism of action have yet to be characterized in vivo. It is not clear how the translation elongation rate is influenced by the effects of codon usage on translation initiation. With regard to protein folding, it is not known how codon usage–mediated effects on elongation speed are adapted to different cotranslational protein folding processes at the individual codon level. Although there are now a few clear genetic examples of the importance of codon usage in protein folding and gene expression, more genetic evidence is needed to demonstrate the broad physiological impact of these mechanism. Many human diseases are associated with silent SNPs, but whether codon usage changes contribute to human diseases caused by these SNPs is still not clear. Although a minute change in protein function and expression can have a major effect on organism fitness during evolution, demonstration of physiological relevance in a laboratory is still limited by experimental sensitivity. The translation-dependent transcriptional effect of codon usage was unexpected, and the mechanism involved is almost completely unknown. Future studies of this conserved phenomenon will likely reveal an important gene regulatory mechanism.
ACKNOWLEDGMENTS
This work is supported by grants from the National Institutes of Health (R35GM118118) and the Welch Foundation (I-1560) to Y.L.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ikemura T 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol 2:13–34 [DOI] [PubMed] [Google Scholar]
- 2.Sharp PM, Tuohy TM, Mosurski KR. 1986. Codon usage in yeast: Cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14:5125–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet 12:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershberg R, Petrov DA. 2008. Selection on codon bias. Annu. Rev. Genet 42:287–99 [DOI] [PubMed] [Google Scholar]
- 5.Zhou M, Guo J, Cha J, Chae M, Chen S, et al. 2013. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495:111–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaney JL, Clark PL. 2015. Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys 44:143–66 [DOI] [PubMed] [Google Scholar]
- 7.Hanson G, Coller J. 2018. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol 19:20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quax TE, Claassens NJ, Soll D, van der Oost J. 2015. Codon bias as a means to fine-tune gene expression. Mol. Cell 59:149–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duret L 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16:287–89 [DOI] [PubMed] [Google Scholar]
- 10.Sabi R, Tuller T. 2014. Modelling the efficiency of codon-tRNA interactions based on codon usage bias. DNA Res. 21:511–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spencer PS, Siller E, Anderson JF, Barral JM. 2012. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol 422:328–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou M, Wang T, Fu J, Xiao G, Liu Y. 2015. Nonoptimal codon usage influences protein structure in intrinsically disordered regions. Mol. Microbiol 97:974–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Dang Y, Zhou M, Li L, Yu CH, et al. 2016. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. PNAS 113:E6117–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duret L, Mouchiroud D. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. PNAS 96:4482–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. 2013. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495:116–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hense W, Anderson N, Hutter S, Stephan W, Parsch J, Carlini DB. 2010. Experimentally increased codon bias in the Drosophila Adh gene leads to an increase in larval, but not adult, alcohol dehydrogenase activity. Genetics 184:547–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampson BL, Pershing NL, Prinz JA, Lacsina JR, Marzluff WF, et al. 2013. Rare codons regulate KRas oncogenesis. Curr. Biol 23:70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu J, Dang Y, Counter C, Liu Y. 2018. Codon usage regulates human KRAS expression at both transcriptional and translational levels. J. Biol. Chem 293:17929–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorensen MA, Kurland CG, Pedersen S. 1989. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol 207:365–77 [DOI] [PubMed] [Google Scholar]
- 20.Pedersen S 1984. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 3:2895–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM. 2010. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J. Mol. Biol 396:1310–18 [DOI] [PubMed] [Google Scholar]
- 22.Bonekamp F, Dalboge H, Christensen T, Jensen KF. 1989. Translation rates of individual codons are not correlated with tRNA abundances or with frequencies of utilization in Escherichia coli. J. Bacteriol 171:5812–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chevance FF, Le Guyon S, Hughes KT. 2014. The effects of codon context on in vivo translation speed. PLOS Genet. 10:e1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varenne S, Buc J, Lloubes R, Lazdunski C. 1984. Translation is a non-uniform process: effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol 180:549–76 [DOI] [PubMed] [Google Scholar]
- 25.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147:789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li GW, Oh E, Weissman JS. 2012. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484:538–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. 2012. Balanced codon usage optimizes eukaryotic translational efficiency. PLOS Genet. 8:e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dana A, Tuller T. 2014. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res. 42:9171–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artieri CG, Fraser HB. 2014. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 24:2011–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, et al. 2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 59:744–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao F, Yu CH, Liu Y. 2017. Codon usage regulates protein structure and function by affecting translation elongation speed in Drosophila cells. Nucleic Acids Res. 45:8484–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Q, Yu CH, Zhao F, Dang Y, Wu C, et al. 2019. eRF1 mediates codon usage effects on mRNA translation efficiency through premature termination at rare codons. Nucleic Acids Res. 47:9243–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang G, Hubalewska M, Ignatova Z. 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol 16:274–80 [DOI] [PubMed] [Google Scholar]
- 35.Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP. 2016. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 14:1787–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. 2015. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLOS Genet. 11:e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CC, Zinshteyn B, Wehner KA, Green R. 2019. High-resolution ribosome profiling defines discrete ribosome elongation states and translational regulation during cellular stress. Mol. Cell 73:959–70.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan X, Hoek TA, Vale RD, Tanenbaum ME. 2016. Dynamics of translation of single mRNA molecules in vivo. Cell 165:976–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeacock L, Faria J, Horn D. 2018. Codon usage bias controls mRNA and protein abundance in trypanosomatids. eLife 7:e32496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kudla G, Murray AW, Tollervey D, Plotkin JB. 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol 25:117–24 [DOI] [PubMed] [Google Scholar]
- 43.Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, et al. 2010. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141:344–54 [DOI] [PubMed] [Google Scholar]
- 44.Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, et al. 2014. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol 10:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dittmar KA, Goodenbour JM, Pan T. 2006. Tissue-specific differences in human transfer RNA expression. PLOS Genet. 2:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. 2016. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165:1416–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, et al. 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun 3:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chionh YH, McBee M, Babu IR, Hia F, Lin W, et al. 2016. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun 7:13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goffena J, Lefcort F, Zhang Y, Lehrmann E, Chaverra M, et al. 2018. Elongator and codon bias regulate protein levels in mammalian peripheral neurons. Nat. Commun 9:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyu X, Yang Q, Li L, Dang Y, Zhou Z, et al. 2020. Adaptation of codon usage to tRNA I34 modification controls translation kinetics and proteome landscape. PLOS Genet. 16:e1008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letzring DP, Wolf AS, Brule CE, Grayhack EJ. 2013. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA 19:1208–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wada M, Ito K. 2018. Misdecoding of rare CGA codon by translation termination factors, eRF1/eRF3, suggests novel class of ribosome rescue pathway in S. cerevisiae. FEBS J. 286:788–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curran JF. 1995. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res. 23:683–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramaniam AR, Zid BM, O’Shea EK. 2014. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159:1200–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin H, Bjornsson A, Isaksson LA. 2002. Cis control of gene expression in E. coli by ribosome queuing at an inefficient translational stop signal. EMBO J. 21:4357–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diament A, Feldman A, Schochet E, Kupiec M, Arava Y, Tuller T. 2018. The extent of ribosome queuing in budding yeast. PLOS Comput. Biol 14:e1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T. 2014. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33:21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrin MA, Subramaniam AR. 2017. Kinetic modeling predicts a stimulatory role for ribosome collisions at elongation stall sites in bacteria. eLife 6:e23629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, et al. 2014. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345:455–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. 2013. Rate-limiting steps in yeast protein translation. Cell 153:1589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhattacharyya S, Jacobs WM, Adkar BV, Yan J, Zhang W, Shakhnovich EI. 2018. Accessibility of the Shine-Dalgarno sequence dictates N-terminal codon bias in E. coli. Mol. Cell 70:894–905.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yarus M, Folley LS. 1985. Sense codons are found in specific contexts. J. Mol. Biol 182:529–40 [DOI] [PubMed] [Google Scholar]
- 63.Gutman GA, Hatfield GW. 1989. Nonrandom utilization of codon pairs in Escherichia coli. PNAS 86:3699–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexaki A, Kames J, Holcomb DD, Athey J, Santana-Quintero LV, et al. 2019. Codon and codon-pair usage tables (CoCoPUTs): facilitating genetic variation analyses and recombinant gene design. J. Mol. Biol 431:2434–41 [DOI] [PubMed] [Google Scholar]
- 65.Pouyet F, Mouchiroud D, Duret L, Semon M. 2017. Recombination, meiotic expression and human codon usage. eLife 6:e27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunec D, Osterrieder N. 2016. Codon pair bias is a direct consequence of dinucleotide bias. Cell Rep. 14:55–67 [DOI] [PubMed] [Google Scholar]
- 67.Irwin B, Heck JD, Hatfield GW. 1995. Codon pair utilization biases influence translational elongation step times. J. Biol. Chem 270:22801–6 [DOI] [PubMed] [Google Scholar]
- 68.Buchan JR, Aucott LS, Stansfield I. 2006. tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res. 34:1015–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. 2016. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166:679–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curran JF, Poole ES, Tate WP, Gross BL. 1995. Selection of aminoacyl-tRNAs at sense codons: The size of the tRNA variable loop determines whether the immediate 3′ nucleotide to the codon has a context effect. Nucleic Acids Res. 23:4104–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chin JX, Chung BK, Lee DY. 2014. Codon Optimization OnLine (COOL): a web-based multi-objective optimization platform for synthetic gene design. Bioinformatics 30:2210–12 [DOI] [PubMed] [Google Scholar]
- 72.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li P, Ke X, Wang T, Tan Z, Luo D, et al. 2018. Zika virus attenuation by codon pair deoptimization induces sterilizing immunity in mouse models. J. Virol 92:e00701–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Atkinson NJ, Witteveldt J, Evans DJ, Simmonds P. 2014. The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication. Nucleic Acids Res. 42:4527–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akashi H 1994. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136:927–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodnina MV, Fischer N, Maracci C, Stark H. 2017. Ribosome dynamics during decoding. Philos. Trans. R. Soc. B 372:20160182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Precup J, Parker J. 1987. Missense misreading of asparagine codons as a function of codon identity and context. J. Biol. Chem 262:11351–55 [PubMed] [Google Scholar]
- 78.Stansfield I, Jones KM, Herbert P, Lewendon A, Shaw WV, Tuite MF. 1998. Missense translation errors in Saccharomyces cerevisiae. J. Mol. Biol 282:13–24 [DOI] [PubMed] [Google Scholar]
- 79.Ribas de Pouplana L, Santos MA, Zhu JH, Farabaugh PJ, Javid B. 2014. Protein mistranslation: friend or foe? Trends Biochem. Sci 39:355–62 [DOI] [PubMed] [Google Scholar]
- 80.Stoletzki N, Eyre-Walker A. 2007. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol 24:374–81 [DOI] [PubMed] [Google Scholar]
- 81.Gilchrist MA, Shah P, Zaretzki R. 2009. Measuring and detecting molecular adaptation in codon usage against nonsense errors during protein translation. Genetics 183:1493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Drummond DA, Wilke CO. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134:341–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kramer EB, Farabaugh PJ. 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Punde N, Kooken J, Leary D, Legler PM, Angov E. 2019. Codon harmonization reduces amino acid misincorporation in bacterially expressed P. falciparum proteins and improves their immunogenicity. AMB Express 9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wohlgemuth I, Pohl C, Rodnina MV. 2010. Optimization of speed and accuracy of decoding in translation. EMBO J. 29:3701–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johansson M, Zhang J, Ehrenberg M. 2012. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. PNAS 109:131–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mordret E, Dahan O, Asraf O, Rak R, Yehonadav A, et al. 2019. Systematic detection of amino acid substitutions in proteomes reveals mechanistic basis of ribosome errors and selection for translation fidelity. Mol. Cell 75:427–41.e5 [DOI] [PubMed] [Google Scholar]
- 88.Huang Y, Koonin EV, Lipman DJ, Przytycka TM. 2009. Selection for minimization of translational frameshifting errors as a factor in the evolution of codon usage. Nucleic Acids Res. 37:6799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korniy N, Goyal A, Hoffmann M, Samatova E, Peske F, et al. 2019. Modulation of HIV-1 Gag/Gag-Pol frameshifting by tRNA abundance. Nucleic Acids Res. 47:5210–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Caliskan N, Peske F, Rodnina MV. 2015. Changed in translation: mRNA recoding by −1 programmed ribosomal frameshifting. Trends Biochem. Sci 40:265–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R. 2015. Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife 4:e05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ikemura T 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol 151:389–409 [DOI] [PubMed] [Google Scholar]
- 93.Moriyama EN, Powell JR. 1997. Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol 45:514–23 [DOI] [PubMed] [Google Scholar]
- 94.dos Reis M, Savva R, Wernisch L. 2004. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 32:5036–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pechmann S, Frydman J. 2013. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol 20:237–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bulmer M 1987. Coevolution of codon usage and transfer RNA abundance. Nature 325:728–30 [DOI] [PubMed] [Google Scholar]
- 97.Kirchner S, Cai Z, Rauscher R, Kastelic N, Anding M, et al. 2017. Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLOS Biol. 15:e2000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Letzring DP, Dean KM, Grayhack EJ. 2010. Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16:2516–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frumkin I, Lajoie MJ, Gregg CJ, Hornung G, Church GM, Pilpel Y. 2018. Codon usage of highly expressed genes affects proteome-wide translation efficiency. PNAS 115:E4940–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dong H, Nilsson L, Kurland CG. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol 260:649–63 [DOI] [PubMed] [Google Scholar]
- 101.Berg OG, Kurland CG. 1997. Growth rate-optimised tRNA abundance and codon usage. J. Mol. Biol 270:544–50 [DOI] [PubMed] [Google Scholar]
- 102.Avcilar-Kucukgoze I, Bartholomaus A, Cordero Varela JA, Kaml RF, Neubauer P, et al. 2016. Discharging tRNAs: a tug of war between translation and detoxification in Escherichia coli. Nucleic Acids Res. 44:8324–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wohlgemuth SE, Gorochowski TE, Roubos JA. 2013. Translational sensitivity of the Escherichia coli genome to fluctuating tRNA availability. Nucleic Acids Res. 41:8021–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pang YL, Abo R, Levine SS, Dedon PC. 2014. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 42:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Torrent M, Chalancon G, de Groot NS, Wuster A, Madan Babu M. 2018. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal 11:eaat6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sagi D, Rak R, Gingold H, Adir I, Maayan G, et al. 2016. Tissue- and time-specific expression of otherwise identical tRNA genes. PLOS Genet. 12:e1006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, et al. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158:1281–92 [DOI] [PubMed] [Google Scholar]
- 108.Hernandez-Alias X, Benisty H, Schaefer MH, Serrano L. 2020. Translational efficiency across healthy and tumor tissues is proliferation-related. Mol. Syst. Biol 16:e9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rudolph KL, Schmitt BM, Villar D, White RJ, Marioni JC, et al. 2016. Codon-driven translational efficiency is stable across diverse mammalian cell states. PLOS Genet. 12:e1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saikia M, Wang X, Mao Y, Wan J, Pan T, Qian SB. 2016. Codon optimality controls differential mRNA translation during amino acid starvation. RNA 22:1719–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA. 2014. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLOS Biol. 12:e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. 2015. Trm9-catalyzed tRNA modifications regulate global protein expression by codon-biased translation. PLOS Genet. 11:e1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chan C, Pham P, Dedon PC, Begley TJ. 2018. Lifestyle modifications: coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 19:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421:290–94 [DOI] [PubMed] [Google Scholar]
- 115.Yang J, Smith DK, Ni H, Wu K, Huang D, et al. 2020. SOX4-mediated repression of specific tRNAs inhibits proliferation of human glioblastoma cells. PNAS 117:5782–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. 2005. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 6:151–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elf J, Nilsson D, Tenson T, Ehrenberg M. 2003. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300:1718–22 [DOI] [PubMed] [Google Scholar]
- 118.Subramaniam AR, Pan T, Cluzel P. 2013. Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. PNAS 110:2419–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Darnell AM, Subramaniam AR, O’Shea EK. 2018. Translational control through differential ribosome pausing during amino acid limitation in mammalian cells. Mol. Cell 71:229–43.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kothe U, Rodnina MV. 2007. Codon reading by tRNAAla with modified uridine in the wobble position. Mol. Cell 25:167–74 [DOI] [PubMed] [Google Scholar]
- 121.Ran W, Higgs PG. 2010. The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol. Biol. Evol 27:2129–40 [DOI] [PubMed] [Google Scholar]
- 122.Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. 2012. A role for tRNA modifications in genome structure and codon usage. Cell 149:202–13 [DOI] [PubMed] [Google Scholar]
- 123.Bornelov S, Selmi T, Flad S, Dietmann S, Frye M. 2019. Codon usage optimization in pluripotent embryonic stem cells. Genome Biol. 20:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nedialkova DD, Leidel SA. 2015. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161:1606–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zinshteyn B, Gilbert WV. 2013. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLOS Genet. 9:e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thommen M, Holtkamp W, Rodnina MV. 2017. Co-translational protein folding: progress and methods. Curr. Opin. Struct. Biol 42:83–89 [DOI] [PubMed] [Google Scholar]
- 127.Komar AA. 2018. Unraveling co-translational protein folding: concepts and methods. Methods 137:71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Holtkamp W, Kokic G, Jager M, Mittelstaet J, Komar AA, Rodnina MV. 2015. Cotranslational protein folding on the ribosome monitored in real time. Science 350:1104–7 [DOI] [PubMed] [Google Scholar]
- 129.Shiber A, Doring K, Friedrich U, Klann K, Merker D, et al. 2018. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561:268–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Purvis IJ, Bettany AJ, Santiago TC, Coggins JR, Duncan K, et al. 1987. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol 193:413–17 [DOI] [PubMed] [Google Scholar]
- 131.O’Brien EP, Vendruscolo M, Dobson CM. 2012. Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun 3:868. [DOI] [PubMed] [Google Scholar]
- 132.Sharma AK, O’Brien EP. 2018. Non-equilibrium coupling of protein structure and function to translation-elongation kinetics. Curr. Opin. Struct. Biol 49:94–103 [DOI] [PubMed] [Google Scholar]
- 133.Komar AA, Lesnik T, Reiss C. 1999. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 462:387–91 [DOI] [PubMed] [Google Scholar]
- 134.Cortazzo P, Cervenansky C, Marin M, Reiss C, Ehrlich R, Deana A. 2002. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun 293:537–41 [DOI] [PubMed] [Google Scholar]
- 135.Sander IM, Chaney JL, Clark PL. 2014. Expanding Anfinsen’s principle: contributions of synonymous codon selection to rational protein design. J. Am. Chem. Soc 136:858–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, et al. 2016. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell 61:341–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Walsh IM, Bowman MA, Soto Santarriaga IF, Rodriguez A, Clark PL. 2020. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. PNAS 117:3528–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, et al. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525–28 [DOI] [PubMed] [Google Scholar]
- 139.Fu J, Murphy KA, Zhou M, Li YH, Lam VH, et al. 2016. Codon usage affects the structure and function of the Drosophila circadian clock protein PERIOD. Genes Dev. 30:1761–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, et al. 2015. Translational tuning optimizes nascent protein folding in cells. Science 348:444–48 [DOI] [PubMed] [Google Scholar]
- 141.Wan Makhtar WR, Browne G, Karountzos A, Stevens C, Alghamdi Y, et al. 2017. Short stretches of rare codons regulate translation of the transcription factor ZEB2 in cancer cells. Oncogene 36:6640–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alexaki A, Hettiarachchi GK, Athey JC, Katneni UK, Simhadri V, et al. 2019. Effects of codon optimization on coagulation factor IX translation and structure: implications for protein and gene therapies. Sci. Rep 9:15449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hunt R, Hettiarachchi G, Katneni U, Hernandez N, Holcomb D, et al. 2019. A single synonymous variant (c.354G>A [p.P118P]) in ADAMTS13 confers enhanced specific activity. Int. J. Mol. Sci 20:5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Thanaraj TA, Argos P. 1996. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 5:1973–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oresic M, Shalloway D. 1998. Specific correlations between relative synonymous codon usage and protein secondary structure. J. Mol. Biol 281:31–48 [DOI] [PubMed] [Google Scholar]
- 146.Gu W, Zhou T, Ma J, Sun X, Lu Z. 2004. The relationship between synonymous codon usage and protein structure in Escherichia coli and Homo sapiens. Biosystems 73:89–97 [DOI] [PubMed] [Google Scholar]
- 147.Saunders R, Deane CM. 2010. Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res. 38:6719–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chaney JL, Steele A, Carmichael R, Rodriguez A, Specht AT, et al. 2017. Widespread position-specific conservation of synonymous rare codons within coding sequences. PLOS Comput. Biol 13:e1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang G, Ignatova Z. 2009. Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLOS ONE 4:e5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhou T, Weems M, Wilke CO. 2009. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol 26:1571–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jacobs WM, Shakhnovich EI. 2017. Evidence of evolutionary selection for cotranslational folding. PNAS 114:11434–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang F, Saha S, Shabalina SA, Kashina A. 2010. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329:1534–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Clarke TF 4th, Clark PL. 2010. Increased incidence of rare codon clusters at 5′ and 3′ gene termini: implications for function. BMC Genom. 11:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pechmann S, Chartron JW, Frydman J. 2014. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol 21:1100–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sanguinetti M, Iriarte A, Amillis S, Marin M, Musto H, Ramon A. 2019. A pair of non-optimal codons are necessary for the correct biosynthesis of the Aspergillus nidulans urea transporter, UreA. R. Soc. Open Sci 6:190773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hoekema A, Kastelein RA, Vasser M, de Boer HA. 1987. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol. Cell. Biol 7:2914–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Koda A, Bogaki T, Minetoki T, Hirotsune M. 2005. High expression of a synthetic gene encoding potato α-glucan phosphorylase in Aspergillus niger. J. Biosci. Bioeng 100:531–37 [DOI] [PubMed] [Google Scholar]
- 158.Coghlan A, Wolfe KH. 2000. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast 16:1131–45 [DOI] [PubMed] [Google Scholar]
- 159.dos Reis M, Wernisch L, Savva R. 2003. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res. 31:6976–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stimac E, Groppi VE, Coffino P. 1983. Increased histone mRNA levels during inhibition of protein synthesis. Biochem. Biophys. Res. Commun 114:131–37 [DOI] [PubMed] [Google Scholar]
- 161.Roy B, Jacobson A. 2013. The intimate relationships of mRNA decay and translation. Trends Genet. 29:691–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160:1111–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boel G, Letso R, Neely H, Price WN, Wong KH, et al. 2016. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 529:358–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, et al. 2016. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 35:2087–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mishima Y, Tomari Y. 2016. Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol. Cell 61:874–85 [DOI] [PubMed] [Google Scholar]
- 166.Narula A, Ellis J, Taliaferro JM, Rissland OS. 2019. Coding regions affect mRNA stability in human cells. RNA 25:1751–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hia F, Yang SF, Shichino Y, Yoshinaga M, Murakawa Y, et al. 2019. Codon bias confers stability to human mRNAs. EMBO Rep. 20:e48220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, et al. 2019. Translation affects mRNA stability in a codon-dependent manner in human cells. eLife 8:e45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Burow DA, Martin S, Quail JF, Alhusaini N, Coller J, Cleary MD. 2018. Attenuated codon optimality contributes to neural-specific mRNA decay in Drosophila. Cell Rep. 24:1704–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Carlini DB. 2005. Context-dependent codon bias and messenger RNA longevity in the yeast transcriptome. Mol. Biol. Evol 22:1403–11 [DOI] [PubMed] [Google Scholar]
- 171.Radhakrishnan A, Chen Y-H, Martin S, Alhusaini N, Green R, Coller J. 2016. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167:122–32.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Webster MW, Chen YH, Stowell JAW, Alhusaini N, Sweet T, et al. 2018. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Mol. Cell 70:1089–100.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Buschauer R, Matsuo Y, Sugiyama T, Chen YH, Alhusaini N, et al. 2020. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 368:eaay6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. 2006. High guanine and cytosine content increases mRNA levels in mammalian cells. PLOS Biol. 4:e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Newman ZR, Young JM, Ingolia NT, Barton GM. 2016. Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9. PNAS 113:E1362–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Harigaya Y, Parker R. 2016. Analysis of the association between codon optimality and mRNA stability in Schizosaccharomyces pombe. BMC Genom. 17:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tillo D, Hughes TR. 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinform. 10:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chamary JV, Parmley JL, Hurst LD. 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet 7:98–108 [DOI] [PubMed] [Google Scholar]
- 179.Cartegni L, Chew SL, Krainer AR. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet 3:285–98 [DOI] [PubMed] [Google Scholar]
- 180.Zhou Z, Dang Y, Zhou M, Yuan H, Liu Y. 2018. Codon usage biases co-evolve with transcription termination machinery to suppress premature cleavage and polyadenylation. eLife 7:e33569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mordstein C, Savisaar R, Young RS, Bazile J, Talmane L, et al. 2020. Codon usage and splicing jointly influence mRNA localization. Cell Syst. 10:351–62.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Courel M, Clement Y, Bossevain C, Foretek D, Vidal Cruchez O, et al. 2019. GC content shapes mRNA storage and decay in human cells. eLife 8:e49708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mittal P, Brindle J, Stephen J, Plotkin JB, Kudla G. 2018. Codon usage influences fitness through RNA toxicity. PNAS 115:8639–44 [DOI] [PMC free article] [PubMed] [Google Scholar]


