Keywords: brachial plexus nerve, innervation, iron, motor dysfunction, muscle atrophy, signal, skeletal muscle, transferrin
Abstract
Previous studies demonstrate an accumulation of transferrin and transferrin receptor 1 (TfR1) in regenerating peripheral nerves. However, the expression and function of transferrin and TfR1 in the denervated skeletal muscle remain poorly understood. In this study, a mouse model of denervation was produced by complete tear of the left brachial plexus nerve. RNA-sequencing revealed that transferrin expression in the denervated skeletal muscle was upregulated, while TfR1 expression was downregulated. We also investigated the function of TfR1 during development and in adult skeletal muscles in mice with inducible deletion or loss of TfR1. The ablation of TfR1 in skeletal muscle in early development caused severe muscular atrophy and early death. In comparison, deletion of TfR1 in adult skeletal muscles did not affect survival or glucose metabolism, but caused skeletal muscle atrophy and motor functional impairment, similar to the muscular atrophy phenotype observed after denervation. These findings suggest that TfR1 plays an important role in muscle development and denervation-induced muscular atrophy. This study was approved by the Institutional Animal Care and Use Committee of Beijing Institute of Basic Medical Sciences, China (approval No. SYXK 2017-C023) on June 1, 2018.
Chinese Library Classification No. R446; R746; Q445
Introduction
Transferrin is necessary for vertebrate cells to bind and transport iron, which is required for cell viability, proliferation and differentiation (Anderson and Frazer, 2017). The transferrin receptor (TfR) is a carrier protein for transferrin, and imports iron by internalizing the transferrin-iron complex through receptor-mediated endocytosis (Qian et al., 2002). TfR1, the predominant TfR, regulates the absorption of iron in multiple organs in vivo (Gammella et al., 2017).
The loss of Tfr1 in embryonic mice leads to severe anemia, resulting in fetal death (Levy et al., 1999). Mice with cardiomyocyte-specific Tfr1 knockout (KO) exhibit severe cardiomyopathy and early death (Xu et al., 2015). Moreover, loss of Tfr1 in skeletal muscles leads to abnormal metabolism throughout the entire body in addition to significant muscular atrophy (Barrientos et al., 2015). Interestingly, previous studies have shown that transferrin is abundant in regenerating peripheral nerves (Markelonis et al., 1982; Mescher et al., 1997), and axotomy results in a strong increase in TfRs in regenerating motor neurons in both amphibians and rats (Graeber et al., 1989; Raivich et al., 1991), suggesting the involvement of TfRs in neural regeneration. However, the expression and function of transferrin and TfR1 in denervated skeletal muscles are still incompletely understood.
In this study, we established a mouse model of brachial plexus avulsion (BPA) injury, and then compared the gene expression patterns in the denervated and contralateral limb muscles by RNA-sequencing. To examine the role of TfR1 in muscle, we generated two lines of muscle-specific TfR1 conditional KO (cKO) mice using conventional and doxycycline (DOX)-inducible muscle-specific human skeletal muscle actin (HSA)-Cre transgenic mice. The inducible HSA-reverse tetracycline-controlled transactivator (rtTA)/TRE-Cre line provides the opportunity to bypass the developmental effect of TfR1 on myogenesis. Using these genetic tools, we investigated the distinct functions of TfR1 in developing and adult skeletal muscles.
Materials and Methods
Animals
The C57BL/6 mice bearing a floxed Tfr1 allele were generously provided by Dr. Fudi Wang of Zhejiang University, China (Chen et al., 2015). These mice were crossed with conventional B6;Cg-Tg (HSA-cre)79Jme/J (also named HSA-Cre) mice (Jackson Laboratory Cat# 006149) (Miniou et al., 1999; Wu et al., 2012a, b, 2015) and inducible B6;C3-Tg(ACTA1-rtTA,tetO-cre)102Monk/J (also named HSA-rtTA/TRE-Cre) mice (Jackson Laboratory Cat# 012433) (Rao and Monks, 2009) to generate embryonic muscle and adult skeletal muscle-specific KO mutant mice, respectively. All of the mutant mice were backcrossed with C57BL/6 mice for more than 10 generations to produce a homogeneous C57BL/6 background. Mice were housed in a room with a 12/12-hour light/dark cycle with ad libitum access to water and rodent chow diet. Experiments with animals were approved by the Institutional Animal Care and Use Committee of the Beijing Institute of Basic Medical Sciences, China (approval No. SYXK 2017-C023) on June 1, 2018. Animals were genotyped by polymerase chain reaction (PCR) using genomic DNA from toe clips (Mizutani et al., 2002). Primers for Tfr1 alleles and Cre are described in Table 1.
Table 1.
Primers used for genotyping and quantitative RT-PCR
| Primer | Sequence | Product size (bp) |
|---|---|---|
| Primers for genotyping | ||
| Tfr1 loxP | F: 5′-CAG TAA CAG AGG AAT CAT TAG-3′ | WT 500 and floxed 650 |
| R: 5′-CTA AAC CGG GTG TAT GAC AAT G-3′ | ||
| HSA-Cre | F: 5′-GCC TGC ATT ACC GGT CGA TGC AAC GA-3′ | 750 |
| R: 5′-GTG GCA GAT GGC GCG GCA ACA CCA TT-3′ | ||
| HSA-rtTA | F: 5′-ACT GAG AGG TGG GAA GCT CA-3′ | 284 |
| R: 5′-GGC GAG TTT ACG GGT TGT TA-3′ | ||
| HSA-rtTA,tetO-Cre | F: 5′-AGG TGT AGA GAA GGC ACT TA-3′ | 412 |
| R: 5′-CTA ATC GCC ATC TTC CAG CA -3′ | ||
| Primers for RT-PCR | ||
| Transferrin | F: 5′-TGG GGG TTG GGT GTA CGA T-3′ | 103 |
| R: 5′-AGC GTA GTA GTA GGT CTG TGG-3′ | ||
| Tfr1 | F: 5′-GTT TCT GCC AGC CCC TTA TTA T-3′ | 152 |
| R: 5′-GCA AGG AAA GGA TAT GCA GCA-3′ | ||
| Tfr2 | F: 5′-TTG GGG TCT ACT TCG GAG AGT-3′ | 179 |
| R: 5′-GAC AGG AGC CTA AGT GCT CAG-3′ | ||
| Myod1 | F: 5′-CCA CTC CGG GAC ATA GAC TTG-3′ | 109 |
| R: 5′-AAA AGC GCA GGT CTG GTG AG-3′ | ||
| Myogenin | F: 5′-GAG ACA TCC CCC TAT TTC TAC CA-3′ | 106 |
| R: 5′-GCT CAG TCC GCT CAT AGC C-3′ | ||
| Mef2c | F: 5′-ATC CCG ATG CAG ACG ATT CAG-3′ | 110 |
| R: 5′-AAC AGC ACA CAA TCT TTG CCT-3′ | ||
| MCK | F: 5′-CTG ACC CCT GAC CTC TAC AAT-3′ | 193 |
| R: 5′-CAT GGC GGT CCT GGA TGAT-3′ | ||
| Foxo1 | F: 5′-CCC AGG CCG GAG TTT AAC C-3′ | 132 |
| R: 5′-GTT GCT CAT AAA GTC GGT GC-3′ | ||
| Actb | F: 5′-GGC TGT ATT CCC CTC CAT CG-3′ | 154 |
| R: 5′-CCA GTT GGT AAC AAT GCC ATG T-3′ |
Actb: Beta-actin; F: forward; Foxo1: forkhead box protein O1; HSA: skeletal muscle actin; MCK: muscle creatine kinase; Mef2C: myocyte enhancer factor 2C; R: reverse; RT-PCR: reverse transcription-polymerase chain reaction; Tfr: transferrin receptor; WT: wild type.
Transgene induction
The tetracycline analog DOX (Cat# D9891; Sigma-Aldrich, St. Louis, MO, USA) was administered to induce HSA-Cre transgene expression specifically in skeletal muscles, but not in brain or any other tissues (Li et al., 2008; Rao and Monks, 2009; Wu et al., 2012a, b, 2015). Briefly, the DOX was dissolved in saline to a final concentration of 10 mg/kg. When the mice were 1 month old, DOX was injected intraperitoneally once per day to induce the expression of the HSA-Cre transgene for 30 consecutive days. At the same time, the control littermate mice were injected with the same volume (200 μL) of saline every day.
Mouse model of BPA injury
The surgical procedure was performed, with modification, as described in a previous report (Hou and Xu, 2018). Briefly, animals were anesthetized with 2% pentobarbital sodium (5.0 mg/100 g, intraperitoneal injection; Cat# 69020194; Sinopharm Chemical Reagent Beijing Co., Ltd., Beijing, China), and then placed in the prone position. The surgery was performed under a M80 Stereo Zoom stereomicroscope (Leica, Buffalo Grove, IL, USA). An incision, 1 cm in length, was made on the neck. Next, hemilaminectomy of the left cervical 4 (C4) to thoracic (T1) segments was performed using microdissection scissors, and the brachial plexus was exposed. Then, the dorsal roots of the BPA were completely avulsed from the spinal cord, followed by avulsion of the ventral roots. The contralateral sham-operated side was not given any intervention and served as the control. The tissue layers and skin were closed with 4.0 silk suture. Animals were returned to their cages after recovery from anesthesia.
RNA-sequencing and data analysis
Total RNA was extracted from ipsilateral BPA injury and contralateral control limb muscles using TRIzol reagent (Cat# 15596018; Invitrogen, Waltham, MA, USA) following the manufacturer’s instructions. RNA quality was determined with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). High-throughput sequencing was performed by BGI Corporation (Beijing, China) using the MGISEQ-500 platform. Data analysis was performed according to Cuffdiff and R as previously described (Trapnell et al., 2012). The identified differentially-expressed genes were uploaded to the Database for Annotation, Visualization and Integrated Discovery for Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses.
Real-time quantitative reverse transcription (RT)-PCR
RT-PCR analysis was performed as described previously (Wu et al., 2012a). Briefly, total RNA was extracted from ipsilateral BPA injury and contralateral control limb muscles using TRIzol reagent (Cat# 15596018; Invitrogen) following the manufacturer’s instructions. A total of 0.5 μg of RNA was used for RT using the Reverse Transcription System (Cat# RR036; TaKaRa, Kusatsu, Japan) according to the manufacturer’s protocol. RT-PCR was conducted in triplicate employing SYBR Green PCR master mix (Cat# CW2601; CWBIO, Beijing, China) with the appropriate forward and reverse primers. For quantitative analysis of gene expression, results were averaged from three replicates in three independent experiments. Values were normalized to Actb levels. Primers used for quantitative RT-PCR analysis are listed in Table 1.
Western blot assay
Experiments were performed as previously described (Wu et al., 2012a; Yang et al., 2019a, b). Briefly, protein lysates were prepared from muscle tissue separated from mouse thighs using radio immunoprecipitation assay lysis buffer (Cat# 89900; Sigma-Aldrich) with inhibitor cocktail (Cat# HY-K0011; MedChemExpress, Princeton, NJ, USA). Quantification was performed using a bicinchoninic acid protein assay kit (Cat# 23227; Sigma-Aldrich). Lysates (10–30 μg, including 5 μL of prestained protein standards) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then transferred to polyvinylidene difluoride membranes (Cat# 10600023; GE Healthcare, Chicago, IL, USA). Membranes were blocked with 5% skim milk in 0.1% Tris-buffered saline/Tween-20 for 1 hour at room temperature, and then incubated in primary antibodies at 4°C overnight. The following antibodies were used: rabbit anti-TfR1 antibody (1:500; Cat# 226290; Biocompare, Salem, MA, USA), mouse anti-myosin (skeletal, fast) antibody (1:1000; Cat# M4276; Sigma-Aldrich), mouse anti-myosin (skeletal, slow) antibody (1:1000; Cat# M8421; Sigma-Aldrich), rabbit anti-MyoD antibody (a muscle transcriptional regulatory factor; 1:1000; Cat# MBS856897; Biocompare, San Diego, CA, USA), goat anti-MyoG antibody (a muscle transcriptional regulatory factor; 1:1000; Cat# SAB2501587; Sigma-Aldrich), mouse anti-glyceraldehyde 3-phosphate dehydrogenase antibody (1:2000; Cat# KM9002T; Sungenebiotech, Tianjin, China). Thereafter, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (1:1000; Cat# ZB-5305; ZSGB-BIO, Beijing, China), horseradish peroxidase-conjugated goat anti-rabbit IgG (1:1000; Cat# ZB-2301; ZSGB-BIO) or horseradish peroxidase-conjugated rabbit anti-goat IgG (1:1000; Cat# ZB-2306; ZSGB-BIO) in 0.1% Tris-buffered saline/Tween-20 for 2 hours at room temperature. Enhanced chemiluminescence was used to detect signals on X-ray film. The signals were quantified using National Institutes of Health (Bethesda, MD, USA) ImageJ software after scanning.
Analysis of muscle histology
Muscle tissue separated from thighs was fixed in 4% paraformaldehyde, and embedded in optimal cutting temperature compound (Cat# 4583; Tissue-Tek, Sakura Finetek, Torrance, CA, USA) and cut into 20-μm frozen sections. The sections were stained with hematoxylin and eosin as in a previous report (Gueneau et al., 2009). Briefly, muscle frozen sections were washed with phosphate buffer (3×), and then stained sequentially with eosin and hematoxylin. The muscle sections were thereafter dehydrated through a graded ethanol series and sealed with neutral balsam mounting medium (Cat#ZLI-9555; ZSGB-BIO). The histological sections were scanned on a Hamamatsu Nanozoomer 6.0 HT (Hamamatsu, Japan) and then analyzed quantitatively using ImageJ software.
Transmission electron microscopy of skeletal muscles was performed as described previously (Wu et al., 2012a, b; Yang et al., 2019b). Briefly, the gastrocnemius muscles from wild-type and KO mice were isolated and fixed in 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer for 1 hour at 25°C and 4°C overnight. The myofibers were isolated and fixed in sodium cacodylate-buffered (pH 7.3) 1% osmium tetroxide for 1 hour at 25°C. After washing with phosphate buffer (3×), 10 minutes each, the samples were dehydrated through a graded ethanol series (30%, 50%, 70%, 80%, 90%, 100%), rinsed with 100% propylene oxide (3×), embedded in plastic resin (EM-bed 812; EM Sciences, Hatfield, PA, USA), and subjected to serial thick-sectioning (2 μm). Some sections were stained with 1% toluidine blue for light microscopic analysis. Adjacent sections were cut into ultra-thin sections, mounted on 200-mesh unsupported copper grids, and stained with uranyl acetate (3% in 50% methanol) and lead citrate (2.6% lead nitrate and 3.5% sodium citrate, pH 12.0). Electron micrographs were taken on a transmission electron microscope (H-7650; Hitachi, Tokyo, Japan) with an AMT 2k CCD camera. Micrographs were digitized, and morphometric analysis was performed using ImageJ software.
Immunofluorescence staining
Immunofluorescence staining of skeletal muscle sections was performed as previously described (Wu et al., 2012b; Yang et al., 2019a). Briefly, the frozen skeletal muscle sections were washed for 10 minutes with 0.5% phosphate-buffered saline/Triton X-100 (PBST) (3×), and then blocked with 5% normal goat serum in PBST for 1 hour at room temperature. The sections were thereafter incubated with primary antibodies in PBST at 4°C overnight. The following antibodies were used: mouse anti-myosin (skeletal, fast) antibody (1:500; Cat# M4276; Sigma-Aldrich) and mouse anti-myosin (skeletal, slow) antibody (1:500; Cat# M8421; Sigma-Aldrich). Then, the sections were washed 10 minutes with PBST (3×), and then incubated with Alexa Fluor 568-conjugated goat anti-mouse IgG (1:500; Cat# 20100; Biotium, Fremont, CA, USA) or Alexa Fluor 488-conjugated goat anti-mouse IgG (1:500; Cat# 20010; Biotium) for 1 hour at room temperature. The sections were thereafter washed with PBST (3×). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Cat# ZLI-9556; ZSGB-BIO) and mounted onto glass slides with anti-fade solution. All images were captured on a Olympus FV-1200 confocal microscope (Olympus, Center Valley, PA, USA) and analyzed with Imaris 9.0.1 software (Abingdon, Oxfordshire, UK).
Muscle fiber area and fiber number
The area and total number of cross-sections of single muscle fibers were quantified as previously described (Bodine and Baar, 2012; Marzuca-Nassr et al., 2016). Briefly, muscle frozen slices with the largest cross-sectional area of each type of muscle was chosen for hematoxylin and eosin staining. Then, the total number of single muscle fibers in the cross-sectional area of each type of muscle was counted, and the cross-sectional area of each type of muscle was measured. Immunofluorescence images were taken on a Olympus FV-1200 confocal microscope. Images of hematoxylin and eosin-stained sections were captured on a Zeiss WF-6R optical microscope (Zeiss, Oberkochen, Germany). The images were analyzed using Imaris 9.0.1 or ImageJ software.
Behavioral analysis
Rotarod test
After the 30-day induction period, the mice were placed on the rod of the accelerating model of a rotarod apparatus (Cat# 47650; Ugo Basile, Gemonio, Italy), which accelerated slowly from 4 to 60 r/min within a 5-minute period, with a cut-off time of 300 seconds, and mice were tested every 8 hours for eight consecutive sessions.
Grip strength test
The mice were allowed to grasp the grip strength meter (TSE systems, Chesterfield, MO, USA) three times per day over 3 days to measure muscle strength after 30 days of induction. The force meter was configured for the recording of the maximal force according to the manual and in the units (lb, kg, N) preferred by the experimenter. The average grip strength for each mouse was recorded and analyzed.
Footprint analysis
The CatWalk quantitative gait analysis system (CatWalk XT 9.0; Noldus Information Technology, Wageningen, The Netherlands) was used to analyze the footprints of mice (Neumann et al., 2009). Briefly, after the 30-day induction period, mice were placed on one side of the 1.3-meter-long glass walkway that emits green light where pressure is placed, and images of the footprints were recorded by a camera as the mice crossed the glass walkway to the other side. The footprint data were analyzed by the CatWalk XT software. Mice were subjected to 3 consecutive runs for each test.
Statistical analysis
Data are presented as the mean ± standard error of mean (SEM), and were analyzed using Student’s t-test or one-way analysis of variance followed by Tukey’s post hoc test. Graphs were generated by GraphPad Prism 8.0 software (GraphPad, San Diego, CA, USA), and P < 0.05 was considered statistically significant.
Results
Peripheral nerve injury leads to elevated levels of transferrin in denervated muscles
To identify the key molecules involved in muscular atrophy and neural regeneration in denervated skeletal muscles, we generated a BPA injury model in male mice. The injured mice showed significantly decreased daily activity and alertness after surgery, as expected after peripheral nerve injury (Gigo-Benato et al., 2010). Three days after surgery, the skeletal muscles of the contralateral and ipsilateral upper limbs were isolated for RNA extraction and deep-sequencing (Figure 1A). We identified the top 20 pathways enriched in the denervated skeletal muscles compared with contralateral controls. Among these, Ribosome, cell cycle, Phagosome and chemokine signaling pathways were significantly activated in the BPA injured muscles (Figure 1B). A total of 1260 genes were upregulated, while 233 were downregulated in BPA injured muscles compared with contralateral muscles (Figure 1C). In Figure 1D, we list 21 upregulated genes in BPA injured muscles. We found that soluble frizzled-related protein 1 and 2 (Sfrp1 and Sfrp2), a soluble modulator of Wnt signaling, was dramatically upregulated over 25 and 28 -fold compared with control, respectively. Cell cycle regulators Ccnb1 and Ccna2 were also significantly upregulated after injury. Muscle transcriptional regulatory factors Myogenin and Myod1 were also significantly increased in BPA denervated muscles, in agreement with previous reports (Ishido et al., 2004; Macpherson et al., 2011), suggesting our BPA injury model is successful and reliable. Interestingly, we found that transferrin was also significantly upregulated in BPA injured muscles. The upregulated expression of transferrin in denervated muscles was confirmed by quantitative RT-PCR (1.62 ± 0.054-fold of control, P < 0.05; Figure 1E). Previous studies show that the expression of TfRs is significantly increased in regenerating motor neurons (Graeber et al., 1989; Raivich et al., 1991). However, here, we found the expression of TfR1, but not TfR2, was significantly decreased in denervated muscles (0.11 ± 0.00-fold of control, P < 0.01; Figure 1E), suggesting a role of TfR1 in peripheral nerve denervation-induced muscular atrophy.
Figure 1.
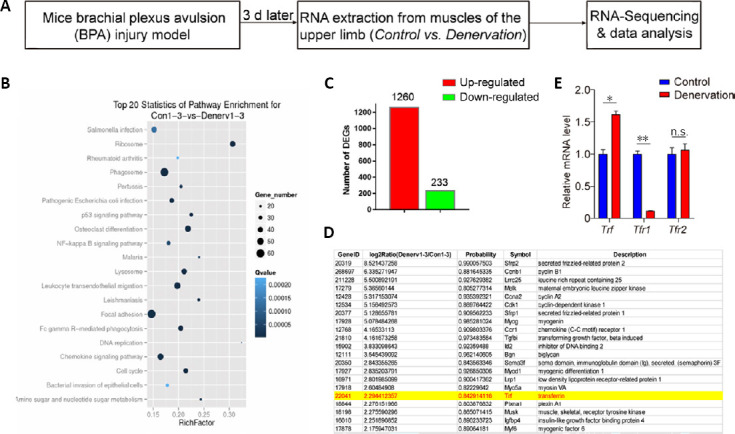
Identification of transferrin as one of the upregulated genes in denervated skeletal muscles.
(A) Schematic of the experimental design for the identification of denervated skeletal muscle-related genes in the brachial plexus avulsion injury mouse model and RNA deep-sequencing. (B) Kyoto Encyclopedia of Genes and Genomes pathway analysis and the top 20 enriched differential signaling pathways between control and denervated skeletal muscles. (C) Quantification of the total number of DEGs between control and denervated skeletal muscles. (D) Representative list of significantly upregulated genes, including transferrin and 20 other genes, in denervated skeletal muscles. (E) Quantitative reverse transcription-polymerase chain reaction analysis indicates significantly increased expression of Trf, but decreased expression of the Tfr1 gene, in denervated skeletal muscles. The expression of the Tfr2 gene was not changed. Data are expressed as the mean ± SEM (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). DEGs: Differentially-expressed genes; n.s.: not significant; Trf: transferrin; Tfr1: transferrin receptor 1; Tfr2: transferrin receptor 2.
Deletion of Tfr1 in developing skeletal muscle causes severe muscular atrophy
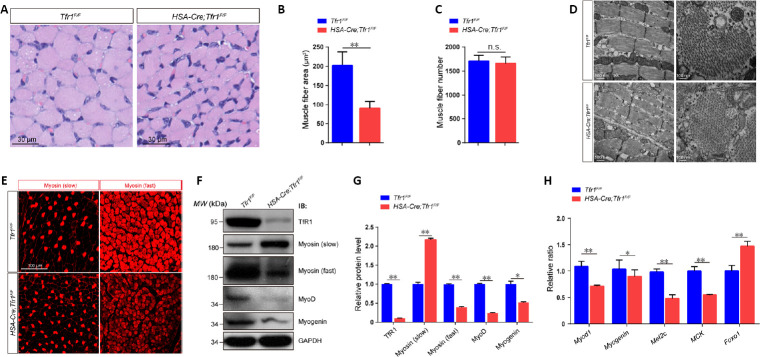
To investigate the function of TfR1 in skeletal muscles, human HSA-Cre transgenic mice, expressing Cre from embryonic day 9 specifically in skeletal muscles, were crossed with Tfr1 loxP mice (flanking Tfr1 exons 3 to 6) to generate HSA-Cre;Tfr1F/F mutant mice (Miniou et al., 1999; Barrientos et al., 2015; Chen et al., 2015) (Figure 2A). These mice were born in a Mendelian ratio. The expression of TfR1 in skeletal muscles was significantly decreased in HSA-Cre;Tfr1F/F cKO mice compared with Tfr1F/F control littermates, and this result was confirmed by western blot and quantitative RT-PCR (P < 0.01; Figure 2B and C). Interestingly, we found that HSA-Cre;Tfr1F/F cKO mice died around P14 with significantly decreased body size and weight (P < 0.01; Figure 2D and E). In addition, the hindlimb skeletal muscle volume in mutant mice was dramatically decreased compared with control littermates (P < 0.01; Figure 2F and G).
Figure 2.
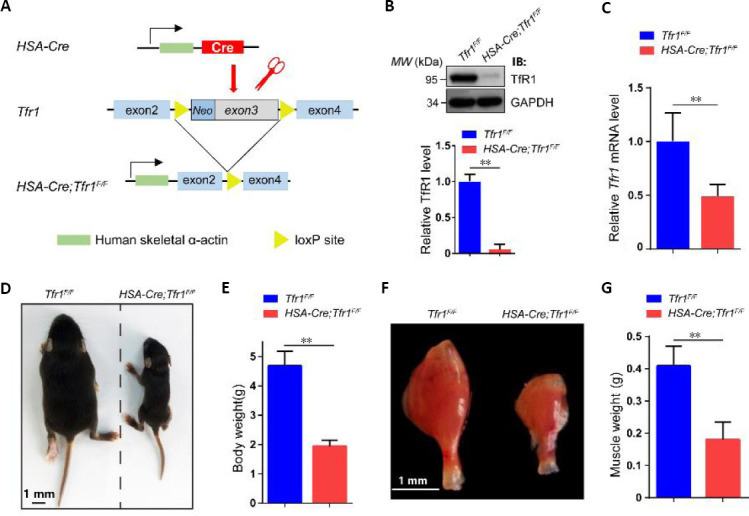
Generation of HSA-Cre;Tfr1F/F cKO mice and characterization of phenotypes.
(A) Schematic of the generation of HSA-Cre;Tfr1F/F cKO mice. The HSA-Cre transgene expressed in skeletal muscle was used to recombine loxP sites flanking Tfr1 exons 3 to generate HSA-Cre;Tfr1F/F cKO mice. (B) Representative western blot showing the expression of TfR1 in skeletal muscles from P7 Tfr1F/F control and HSA-Cre;Tfr1F/F mutants. Quantitative analysis of western blots reveals significantly decreased expression of TfR1 in mutants compared with control littermates. (C) Quantitative reverse transcription-polymerase chain reaction analysis indicates significantly decreased expression of the Tfr1 gene in mutants compared with control littermates. (D) The body size of HSA-Cre;Tfr1F/F mutant mice was significantly reduced compared with the control littermates at P10. (E) Quantification of the body weight shows significant reduction in HSA-Cre;Tfr1F/F mutants compared with control littermates at P10. (F) The size of the gastrocnemius muscle in HSA-Cre;Tfr1F/F mutants was significantly smaller than that in control littermates at P10. Scale bars: 1 mm in D and F. (G) Quantification of the gastrocnemius muscle weight shows significant reduction in HSA-Cre;Tfr1F/F mutants compared with control littermates at P10. Data are expressed as the mean ± SEM (n = 4 in B, C, 10 in E, G). **P < 0.01 (Student’s t-test). cKO: Conditional knockout; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; HSA: skeletal muscle actin; P7 or P10: postnatal day 7 or 10; Tfr1: transferrin receptor 1.
Next, we characterized the histological properties of skeletal muscle in HSA-Cre;Tfr1F/F cKO and control mice. We found the cross-sectional area of muscle fibers from cKO mice was significantly decreased compared with control littermates (P < 0.01; Figure 3A and B), indicative of muscular atrophy in these postnatal mutant mice. Although the total number of muscle fibers in cKO mice was similar to that in control littermates, no central nucleus was detected in these mutant mice (Figure 3A and C). By transmission electron microscopy, the ultrastructure of the muscle appeared intact in cKO mice, including well-aligned single myofibers and Z-lines, as well as nuclei underneath the basal lamina (Figure 3D). The reduced muscle mass in Tfr1 mutant mice suggests impaired myogenesis at the embryonic and early postnatal stages. Moreover, immunofluorescence staining of the anterior thigh muscle revealed that the expression of the fast type myosin was significantly lower in HSA-Cre;Tfr1F/F cKO mice compared with control littermates, and that, in contrast, the expression of slow-type myosin was relatively higher in mutants compared with control littermates (Figure 3E). Western blots from muscle lysates confirmed that the expression of slow-type myosin in HSA-Cre;Tfr1F/F cKO muscles was significantly higher than that in control muscles, while fast type myosin was markedly reduced in mutant muscles (P < 0.05 or P < 0.01; Figure 3F and G), consistent with the immunofluorescence staining results.
Figure 3.
Severe muscular atrophy in HSA-Cre;Tfr1F/F mutant mice.
(A) Hematoxylin-eosin staining of the cross-sections of gastrocnemius muscle in HSA-Cre; Tfr1F/F mutants and control littermates at P10. The outline of a single muscle fiber is indicated by a dashed line, revealing smaller cross-sectional area of muscle fibers in mutants compared with controls. Scale bars: 30 μm. (B, C) Quantitative analysis of the average muscle fiber area shows a significant reduction in HSA-Cre;Tfr1F/F mutants compared with control littermates at P10, but the number of muscle fibers was nearly the same. (D) Ultrastructural analysis of gastrocnemius muscles by electron microscopy. No significant misalignment of Z-lines (arrows) or central nuclei was found in either HSA-Cre;Tfr1F/F mutants or control littermates. Scale bars: 500 nm (left), 100 nm (right). (E) Immunofluorescence staining of gastrocnemius muscle cross-sections labeled with anti-myosin (slow) (arrowheads) or anti-myosin (fast) (asterisks) antibody (red, Alexa Fluor 568) in HSA-Cre;Tfr1F/F mutants and control littermates. Increased expression of slow-type myosin, but reduced expression of fast-type myosin, was found in HSA-Cre;Tfr1F/F mutants compared with control littermates. Scale bar: 100 mm. (F) Representative western blot shows the expression of TfR1, myosin (slow), myosin (fast), MyoD and myogenin in skeletal muscles from HSA-Cre;Tfr1F/F mutants and control littermates at P10. (G) Quantitative analysis of western blots showing significantly decreased expression of TfR1, myosin (fast), MyoD and myogenin, but increased expression of myosin (slow), in HSA-Cre;Tfr1F/F mutants compared with control littermates. (H) Quantitative reverse transcription-polymerase chain reaction analysis indicating significantly downregulated expression of Myod1, Myogenin, Mef2c and MCK genes in HSA-Cre;Tfr1F/F mutant mice. In contrast, Foxo1 was significantly upregulated in HSA-Cre;Tfr1F/F mutants compared with control littermates. Data are expressed as the mean ± SEM (n = 6). *P < 0.05, **P < 0.01 (Student’s t-test). Foxo1: Forkhead box protein O1; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; HSA: skeletal muscle actin; MCK: muscle creatine kinase; Mef2C: myocyte enhancer factor 2C; n.s.: not significant; P10: postnatal day 10; Tfr1: transferrin receptor 1.
MyoD and myogenin, myogenic regulatory factors, were both significantly downregulated in HSA-Cre;Tfr1F/F cKO muscles compared with control muscles (P < 0.05 or P < 0.01; Figure 3F and G). Moreover, significantly altered expression of various mRNAs was detected in HSA-Cre;Tfr1F/F cKO muscles, including myogenic regulatory factors (Myod and Myogenin) and the skeletal muscle fiber identity-related factors Mef2C, muscle creatine kinase and forkhead box protein O1 (Foxo1). The majority of these transcripts were significantly downregulated in cKO muscles, except for Foxo1, a critical regulator of muscle fiber-type specification (Kitamura et al., 2007), which showed relatively higher expression in cKO muscles compared with control muscles (P < 0.05 or P < 0.01; Figure 3H). Interestingly, the increased Foxo1 expression in HSA-Cre;Tfr1F/F cKO muscles caused a reduction in skeletal muscle mass and a decrease in fast-twitch fibers, but an increase in slow-twitch fibers, consistent with previous studies (Kamei et al., 2004; Kitamura et al., 2007). In addition, we observed liver and spleen damage as well as metabolic disorders in HSA-Cre;Tfr1F/F cKO mice (data not shown), as reported previously (Trenor et al., 2000; Barrientos et al., 2015).
Inducible deletion of Tfr1 in adult skeletal muscle results in mild muscular atrophy
Next, to clarify whether Tfr1 is essential for the maintenance of skeletal muscle metabolic homeostasis at the adult stage, HSA-rtTA/TRE-Cre mice expressing Cre recombinase specifically in skeletal muscles, inducible by tetracycline (Rao and Monks, 2009), were bred with Tfr1F/F mice to generate HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice. The tetracycline analog DOX was administered to 1-month-old HSA-rtTA/TRE-Cre;Tfr1F/F mice by intraperitoneal injection once per day for 30 consecutive days to induce the expression of Cre recombinase specifically in skeletal muscles in adulthood to bypass the developmental effects of Tfr1 in skeletal muscles (Figure 4A). The expression of TfR1 was dramatically decreased in hind limb skeletal muscles in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice, including the tibialis anterior, extensor digitorum longus, gastrocnemius and soleus, but not in saline-injected mutant mice, compared with Tfr1F/F control littermates. This suggests that Tfr1 knockout by DOX injection was successful (P < 0.01; Figure 4B and C). In contrast to the significantly decreased body size and reduced skeletal muscle mass in HSA-Cre;Tfr1F/F cKO mice, body size was similar between saline injection control and DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice, and the mice with induced Tfr1 KO survived for 30 days after the DOX injection (data not shown). We also found that almost all of the skeletal muscles from the hind limb displayed a much more intense red color in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice compared with saline-treated controls and Tfr1F/F littermates (Figure 4D). Histological analyses further revealed that the majority of the perimeters of the cross-section of skeletal muscle fibers from the hind limb were relatively smaller in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice, including in the tibialis anterior, extensor digitorum longus and gastrocnemius muscles, but not in the soleus muscles (P < 0.05; Figure 4E and F), suggesting deletion of Tfr1 in adult skeletal muscles may also lead to mild muscular atrophy in developing skeletal muscles (P < 0.05 or P < 0.01; Figures 2F and 3A–E).
Figure 4.
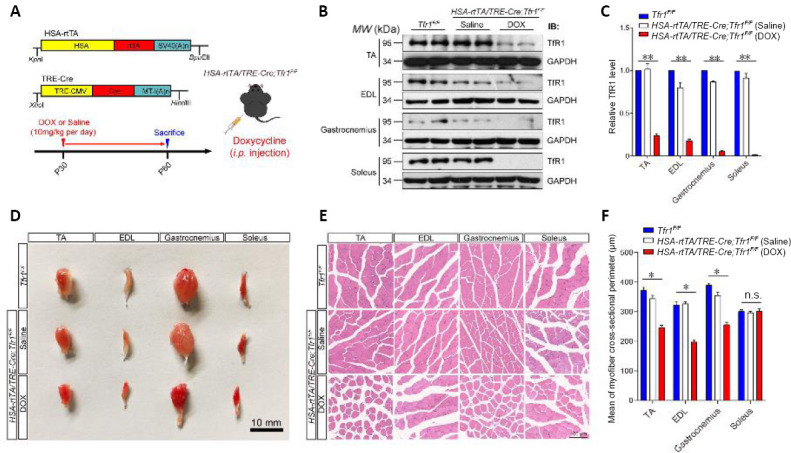
Generation and characterization of the muscle phenotype in HSA-rtTA,tetO-Cre;Tfr1F/F inducible mutant mice.
(A) Schematic of the generation of adult skeletal muscle inducible Tfr1 conditional knockout mice. DOX combines with and activates the rtTA promoter to induce HSA-Cre recombinase to generate the adult skeletal muscle-specific Tfr1 knockout. DOX (10 mg/kg) was injected intraperitoneally once per day to induce HSA-Cre transgene expression for 30 consecutive days. (B) Representative western blots showing the expression of TfR1 in four types of skeletal muscles, including TA, EDL, gastrocnemius and soleus, in the indicated mice at P60. (C) Quantitative analysis of western blots showing significantly decreased expression of TfR1 in all types of skeletal muscles from DOX-treated HSA-rtTA,tetO-Cre;Tfr1F/F mutants compared with saline-treated mutants and Tfr1F/F control littermates. (D) Gross morphology of four types of skeletal muscles, including TA, EDL, gastrocnemius and soleus muscles, dissected from Tfr1F/F control littermates, and saline-treated and DOX-treated HSA-rtTA,tetO-Cre;Tfr1F/F mutants at P60. Significantly reduced muscle volume was found in TA, EDL and gastrocnemius muscles, but not in soleus muscles. Scale bar: 10 mm. (E) Hematoxylin-eosin staining of the cross-sections of four types of skeletal muscles of the indicated genotypes at P60. Scale bar: 100 μm. The cross-sectional area of TA, EDL and gastrocnemius muscles, but not soleus muscles, was markedly reduced in HSA-rtTA,tetO-Cre;Tfr1F/F mutants compared with controls. (F) Quantitative analysis of the average cross-sectional perimeter of single muscle fibers in four types of skeletal muscles of the indicated genotypes at P60. Data are expressed as the mean ± SEM (n = 6). *P < 0.05, **P < 0.01 (one-way analysis of variance followed by Tukey’s post hoc test). DOX: Doxycycline; EDL: extensor digitorum longus; HSA: skeletal muscle actin; i.p.: intraperitoneal injection; n.s.: not significant; P60: postnatal day 60; rtTA: reverse tetracycline-controlled transactivator; TA: tibialis anterior; Tfr1: transferrin receptor 1.
Our immunofluorescence staining results further demonstrated that, as in HSA-Cre;Tfr1F/F cKO mice, the expression of fast type myosin was significantly decreased in almost all hind limb skeletal muscles in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice, including the tibialis anterior, extensor digitorum longus, gastrocnemius and soleus muscles (P < 0.01; Figure 5A and B). Moreover, the average number of myofibers was not affected in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice (Figure 5C), similar to the HSA-Cre;Tfr1F/F cKO mice (Figure 3C). However, in contrast to the increased expression of slow-type myosin in HSA-Cre;Tfr1F/F cKO mice, the expression of slow-type myosin as well as the area of the cross-section of muscle fibers were drastically decreased in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice, including in the tibialis anterior, extensor digitorum longus and gastrocnemius muscles, but not in the soleus muscles (P < 0.01; Figure 5D–F). Taken together, these results demonstrate that the muscular atrophy phenotype induced by deletion of Tfr1 in developing skeletal muscles is much more severe than that produced by the selective deletion of the gene in adult skeletal muscles, suggesting distinct functions of TfR1 in skeletal muscles in the developmental and adult stages.
Figure 5.
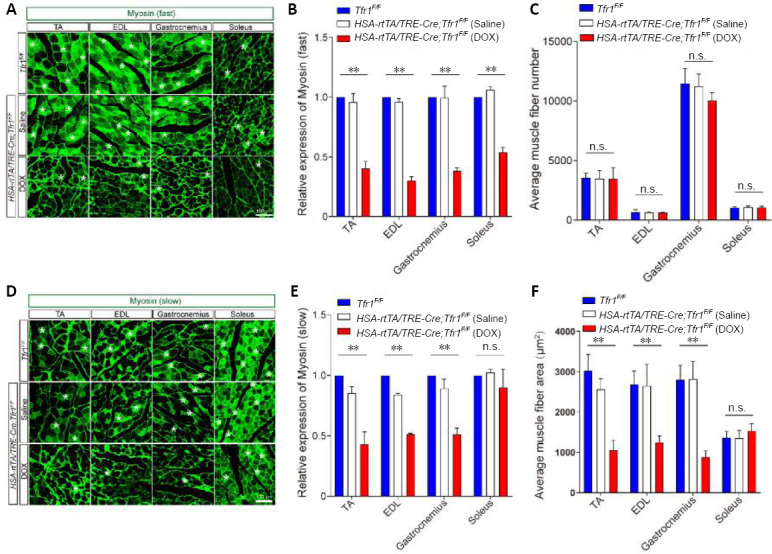
Expression levels of both fast- and slow-type myosin are reduced in HSA-rtTA,tetO-Cre;Tfr1F/F inducible mutant mice.
(A) Immunofluorescence staining of the four different types of skeletal muscle with anti-myosin (fast) (green, Alexa Fluor 488) (asterisk) in Tfr1F/F control littermates, and saline-treated and DOX-treated HSA-rtTA,tetO-Cre;Tfr1F/F mutants at P60. The expression of fast-type myosin was significantly decreased in all four types of muscles in DOX-treated HSA-rtTA,tetO-Cre;Tfr1F/F mutants compared with controls. Scale bar: 100 μm. (B) Quantitative analysis of the expression of myosin (fast) in various skeletal muscles of the indicated genotypes. (C) Quantitative analysis of the number of muscle fibers for each section of the indicated skeletal muscle of the indicated genotype. (D) Immunofluorescence staining of the four types of skeletal muscle with anti-myosin (slow) (green, Alexa Fluor 488) (asterisks) from Tfr1F/F control littermates, and saline-treated and DOX-treated HSA-rtTA,tetO-Cre;Tfr1F/F mutants at P60. The expression of slow-type myosin was significantly decreased in TA, EDL and gastrocnemius muscles, but not in soleus muscles, in DOX-treated HSA-rtTA,tetO-Cre;Tfr1F/F mutants compared with controls. Scale bar: 100 μm. (E) Quantitative analysis of the expression of myosin (slow) in the different skeletal muscles of the indicated genotypes. (F) Quantitative analysis of the average area of each single muscle fiber from the different skeletal muscles of the indicated genotypes at P60. Data are expressed as the mean ± SEM (n = 6). **P < 0.01 (one-way analysis of variance followed by Tukey’s post hoc test). DOX: Doxycycline; EDL: extensor digitorum longus; HSA: skeletal muscle actin; n.s.: not significant; P60: postnatal day 60; rtTA: reverse tetracycline-controlled transactivator; TA: tibialis anterior; Tfr1: transferrin receptor 1.
Loss of Tfr1 in adult skeletal muscle causes deficits in motor behaviors
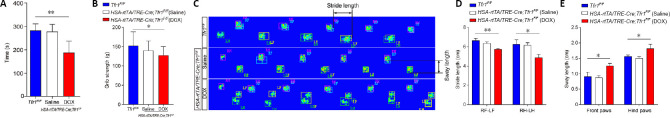
In addition to the histological analyses, we also evaluated motor function in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice, saline-treated controls and Tfr1F/F littermates. Rotarod test demonstrated that the time on the accelerating rod was significantly reduced in DOX-treated mutant mice compared with saline-treated mutants and Tfr1F/F littermates (P < 0.01; Figure 6A). To assess physiological function of skeletal muscles in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice, we performed the conventional forelimb grip strength test, and found that the grip strength was significantly decreased in DOX-treated mutant mice compared with saline-treated mutants and Tfr1F/F control littermates (P < 0.05; Figure 6B).
Figure 6.
Impaired motor function in HSA-rtTA,tetO-Cre;Tfr1F/F inducible mutant mice.
(A) Histogram represents differences in the balance beam assay among the various genotypes at P60. (B) Histogram represents differences in the grip strength test assay among the various genotypes at P60. The grip strength was significantly decreased in DOX-treated mutant mice compared with saline-treated mutants and Tfr1F/F control littermates. (C) Footprints of the indicated control and mutant mice at P60 were evaluated for stride length and sway length. (D, E) Quantitative analysis of footprints reveals differences in stride length (D) and sway length (E) in the indicated genotypes at P60. Data are expressed as the mean ± SEM (n = 10). *P < 0.05, **P < 0.01 (one-way analysis of variance followed by Tukey’s post hoc test). DOX: Doxycycline; HSA: skeletal muscle actin; LF: left front paws; LH: left hind paws; P60: postnatal day 60; RF: right front paws; RH: right hind paws; rtTA: reverse tetracycline-controlled transactivator; Tfr1: transferrin receptor 1.
We used the CatWalk gait analysis system to examine the effects of Tfr1 deletion in adult skeletal muscles (Crowley et al., 2018). We found that the stride length of the girdle paw pairs of both the front and hind paws were significantly decreased in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice compared with saline-treated mutants and Tfr1F/F control littermates (both P < 0.05; Figure 6C and D). In contrast, the sway lengths of both the front and hind paws in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice were significantly increased compared with saline-treated mutants and Tfr1F/F control littermates (both P < 0.05; Figure 6C and E). Collectively, the impaired motor functions in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice suggest that the absence of TfR1 at the adult stage results in physiological disorders of skeletal muscles.
Discussion
In this study, we first identified the significantly increased expression of transferrin, but dramatically downregulated expression of its receptor, TfR1, in denervated skeletal muscles. Thereafter, using different genetic ablation methods, we studied the function of TfR1 in developing and adult skeletal muscles. We found that loss of TfR1 in embryonic skeletal muscles caused significant muscular atrophy and lethal phenotypes at P10 caused by severe metabolic catastrophe, as reported previously (Barrientos et al., 2015). In contrast, selective deletion of Tfr1 in adult skeletal muscles generated much milder muscular atrophy without lethality. Although motor behavioral analyses showed locomotor deficits in mice with deletion of Tfr1 in adult muscle, no metabolic catastrophe was found in these mice, suggesting distinct functions of TfR1 in developing and adult skeletal muscles. Because TfR1 is significantly downregulated in denervated skeletal muscles, TfR1 loss-of-function studies in skeletal muscles indicate the potential function of TfR1 signaling in muscles in the regulation of peripheral nerve denervation-induced muscular atrophy.
From our RNA-sequencing analysis of mice with BPA injury, we found that the expression of transferrin was significantly upregulated in the denervated limbic muscles compared with contralateral muscles, but that its receptor, TfR1, was significantly reduced, suggesting a key role of transferrin/TfR1 signaling in the regulation of neural regeneration or muscular atrophy. Interestingly, it was previously reported that transferrin is also abundant in regenerating nerves (Markelonis et al., 1982; Mescher et al., 1997). This result together with our findings suggest that transferrin might be strongly induced in both nerve terminals and skeletal muscles during neural regeneration. TfR1 is also strongly upregulated in regenerating sciatic nerves (Raivich et al., 1991), suggesting that transferrin might function during neural regeneration in both an autocrine and paracrine manner. It should be noted that TfR1 is ubiquitously expressed, including nerve terminals and skeletal muscles, and interestingly, in our BPA injury mouse model, we found that TfR1 was significantly downregulated in denervated skeletal muscles. Moreover, our genetic studies demonstrate that deletion of Tfr1 specifically in motor neurons has no effect on motor neuron development or neuromuscular junction formation (data not shown). However, deletion of Tfr1 specifically in skeletal muscles produces severe muscular and metabolic deficits (Barrientos et al., 2015), suggesting a critical role of TfR1 in skeletal muscles.
The normal numbers of skeletal muscle fibers in both developing and adult skeletal muscle-specific Tfr1 KO mice indicate that TfR1 is more important for muscle tissue metabolism rather than myogenesis (Barrientos et al., 2015). Compared with the severe muscular atrophy phenotype of HSA-Cre;Tfr1F/F cKO muscles, the muscular atrophy deficits in DOX-treated HSA-rtTA/TRE-Cre;Tfr1F/F mutant mice were much milder, suggesting that TfR1 is distinctly required in skeletal muscle tissue to maintain metabolic homeostasis at the various stages of development. It should be noted that in muscle-specific Tfr1 cKO mice, we noticed that the expression of fast type myosin was significantly reduced, but that, in contrast, the expression of slow-type myosin was increased in the absence of TfR1, suggesting a shift from fast to slow muscle fiber type. The finding also suggests that TfR1 has a more critical role in fast-type muscles. Indeed, we found that the muscular atrophy phenotype is more severe in Tfr1 cKO extensor digitorum longus muscles, which are predominantly composed (87%) of fast-type (type II myosin) fibers (Kalhovde et al., 2005). Conversely, in the soleus muscle, which is comprised mainly (85–87%) of slow-type fibers (type I myosin) (Ng et al., 1998; Kalhovde et al., 2005), we did not find significant muscular atrophy in the Tfr1 cKO, further suggesting that fast type muscle fibers are more sensitive to the loss-of-function of TfR1.
The severe muscular atrophy in the developing skeletal muscle-specific TfR1 KO mice was accompanied by significantly reduced expression of myogenic regulatory factors, including MyoD and myogenin, but increased expression of FoxO1. This result contrasts with the upregulated expression of myogenin observed in denervated skeletal muscles in previous reports (Blagden et al., 2004; Macpherson et al., 2011), indicating that TfR1 has distinct roles in the regulation of muscle development and denervation-induced muscular atrophy. FoxO1 plays a key role in promoting the expression of gluconeogenic enzymes and metabolism (Gross et al., 2008), and it also controls myogenic differentiation and fiber type specification (Kitamura et al., 2007). Here, we found that the expression of FoxO1 was significantly upregulated in Tfr1-deleted skeletal muscles. To our knowledge, this is the first demonstration of a link between TfR1 and FoxO1 signaling in muscular atrophy, although the finding is preliminary.
Taken together, our findings demonstrate that transferrin is significantly upregulated, while TfR1 is dramatically downregulated, in denervated skeletal muscles. We found that TfR1 plays distinct roles in early developmental and adult stages in skeletal muscles. In agreement with a previous report, we found that the deletion of TfR1 in embryonic developing skeletal muscles in mice causes severe muscular atrophy and lethal phenotypes around postnatal day 10 (P10) (Barrientos et al., 2015). In comparison, deletion of Tfr1 in adult skeletal muscle induces muscular atrophy and locomotor deficits, but without lethality. Our key finding that TfR1 is severely downregulated in denervated skeletal muscles suggests that preserving transferrin–TfR1 signaling in these muscles may have potential in the treatment and rehabilitation of peripheral nerve injury-induced muscular atrophy. It should be noted that iron metabolism disorders and transferrin receptor-mediated ferroptosis also play a role in the progression of degenerative diseases of the kidney, heart, liver, and brain (Lu and Black, 2016; Weiland et al., 2019; Feng et al., 2020; Sumbria, 2020). In this study, we did not investigate whether loss-of-function of TfR1 in skeletal muscles leads to the ferroptosis of muscle tissue. Furthermore, we also do not know whether peripheral nerve regeneration could be affected in adult skeletal muscle-specific Tfr1 KO mice. Therefore, further study is needed to elucidate the functions of TfR1 in the regulation of both skeletal muscle ferroptosis and denervated peripheral nerve regeneration.
Additional file:
Additional file 1 (199.5KB, pdf) : Original data of the experiment.
Acknowledgments
We thank Dr. Fudi Wang at Zhejiang University for generously providing the valuable C57BL/6 mice bearing a floxed Tfr1 allele, and thank all members of Hai-Tao Wu’s laboratory for discussion.
Footnotes
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Patel B, Yu J, Song LP; T-Editor: Jia Y
Conflicts of interest: The authors declare that they have no conflicts of interest with the contents of this article.
Financial support: This study was supported by the National Natural Science Foundation of China, Nos. 31770929 (to HTW), 31522029 (to HTW), 81902847 (to HHY), and the Beijing Municipal Science and Technology Commission of China, Nos. Z181100001518001 (to HTW), Z161100000216154 (to HTW). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: This study was approved by the Institutional Animal Care and Use Committee of Beijing Institute of Basic Medical Sciences, China (approval No. SYXK 2017-C023) on June 1, 2018.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Funding: This study was supported by the National Natural Science Foundation of China, Nos. 31770929 (to HTW), 31522029 (to HTW), 81902847 (to HHY), and the Beijing Municipal Science and Technology Commission of China, Nos. Z181100001518001 (to HTW), Z161100000216154 (to HTW).
References
- 1.Anderson GJ, Frazer DM. Current understanding of iron homeostasis. Am J Clin Nutr. 2017;106:1559s–1566s. doi: 10.3945/ajcn.117.155804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrientos T, Laothamatas I, Koves TR, Soderblom EJ, Bryan M, Moseley MA, Muoio DM, Andrews NC. Metabolic catastrophe in mice lacking transferrin receptor in muscle. EBioMedicine. 2015;2:1705–1717. doi: 10.1016/j.ebiom.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blagden CS, Fromm L, Burden SJ. Accelerated response of the myogenin gene to denervation in mutant mice lacking phosphorylation of myogenin at threonine 87. Mol Cell Biol. 2004;24:1983–1989. doi: 10.1128/MCB.24.5.1983-1989.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodine SC, Baar K. Analysis of skeletal muscle hypertrophy in models of increased loading. Methods Mol Biol. 2012;798:213–229. doi: 10.1007/978-1-61779-343-1_12. [DOI] [PubMed] [Google Scholar]
- 5.Chen AC, Donovan A, Ned-Sykes R, Andrews NC. Noncanonical role of transferrin receptor 1 is essential for intestinal homeostasis. Proc Natl Acad Sci U S A. 2015;112:11714–11719. doi: 10.1073/pnas.1511701112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowley ST, Kataoka K, Itaka K. Combined CatWalk Index: an improved method to measure mouse motor function using the automated gait analysis system. BMC Res Notes. 2018;11:263. doi: 10.1186/s13104-018-3374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, Schorpp K, Jin J, Yozwiak CE, Hoffstrom BG, Decker AM, Rajbhandari P, Stokes ME, Bender HG, Csuka JM, Upadhyayula PS, Canoll P, Uchida K, Soni RK, Hadian K, Stockwell BR. Transferrin receptor is a specific ferroptosis marker. Cell Rep. 2020;30:3411–3423e7. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gammella E, Buratti P, Cairo G, Recalcati S. The transferrin receptor: the cellular iron gate. Metallomics. 2017;9:1367–1375. doi: 10.1039/c7mt00143f. [DOI] [PubMed] [Google Scholar]
- 9.Gigo-Benato D, Russo TL, Geuna S, Domingues NR, Salvini TF, Parizotto NA. Electrical stimulation impairs early functional recovery and accentuates skeletal muscle atrophy after sciatic nerve crush injury in rats. Muscle Nerve. 2010;41:685–693. doi: 10.1002/mus.21549. [DOI] [PubMed] [Google Scholar]
- 10.Graeber MB, Raivich G, Kreutzberg GW. Increase of transferrin receptors and iron uptake in regenerating motor neurons. J Neurosci Res. 1989;23:342–345. doi: 10.1002/jnr.490230315. [DOI] [PubMed] [Google Scholar]
- 11.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 12.Gueneau L, Bertrand AT, Jais JP, Salih MA, Stojkovic T, Wehnert M, Hoeltzenbein M, Spuler S, Saitoh S, Verschueren A, Tranchant C, Beuvin M, Lacene E, Romero NB, Heath S, Zelenika D, Voit T, Eymard B, Ben Yaou R, Bonne G. Mutations of the FHL1 gene cause Emery-Dreifuss muscular dystrophy. Am J Hum Genet. 2009;85:338–353. doi: 10.1016/j.ajhg.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou AL, Xu WD. A model of neuropathic pain in brachial plexus avulsion injury and associated spinal glial cell activation. J Pain Res. 2018;11:3171–3179. doi: 10.2147/JPR.S174663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishido M, Kami K, Masuhara M. In vivo expression patterns of MyoD, p21, and Rb proteins in myonuclei and satellite cells of denervated rat skeletal muscle. Am J Physiol Cell Physiol. 2004;287:C484–493. doi: 10.1152/ajpcell.00080.2004. [DOI] [PubMed] [Google Scholar]
- 15.Kalhovde JM, Jerkovic R, Sefland I, Cordonnier C, Calabria E, Schiaffino S, Lømo T. “Fast” and “slow” muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. J Physiol. 2005;562:847–857. doi: 10.1113/jphysiol.2004.073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 17.Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accili D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy JE, Jin O, Fujiwara Y, Kuo F, Andrews NC. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat Genet. 1999;21:396–399. doi: 10.1038/7727. [DOI] [PubMed] [Google Scholar]
- 19.Li XM, Dong XP, Luo SW, Zhang B, Lee DH, Ting AK, Neiswender H, Kim CH, Carpenter-Hyland E, Gao TM, Xiong WC, Mei L. Retrograde regulation of motoneuron differentiation by muscle beta-catenin. Nat Neurosci. 2008;11:262–268. doi: 10.1038/nn2053. [DOI] [PubMed] [Google Scholar]
- 20.Lu Q, Black SM. Iron metabolism, oxidative stress, and neonatal brain injury. Neural Regen Res. 2016;11:725–726. doi: 10.4103/1673-5374.182691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macpherson PC, Wang X, Goldman D. Myogenin regulates denervation-dependent muscle atrophy in mouse soleus muscle. J Cell Biochem. 2011;112:2149–2159. doi: 10.1002/jcb.23136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markelonis GJ, Bradshaw RA, Oh TH, Johnson JL, Bates OJ. Sciatin is a transferrin-like polypeptide. J Neurochem. 1982;39:315–320. doi: 10.1111/j.1471-4159.1982.tb03949.x. [DOI] [PubMed] [Google Scholar]
- 23.Marzuca-Nassr GN, Vitzel KF, De Sousa LG, Murata GM, Crisma AR, Rodrigues Junior CF, Abreu P, Torres RP, Mancini-Filho J, Hirabara SM, Newsholme P, Curi R. Effects of high EPA and high DHA fish oils on changes in signaling associated with protein metabolism induced by hindlimb suspension in rats. Physiol Rep. 2016;4:e12958. doi: 10.14814/phy2.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mescher AL, Connell E, Hsu C, Patel C, Overton B. Transferrin is necessary and sufficient for the neural effect on growth in amphibian limb regeneration blastemas. Dev Growth Differ. 1997;39:677–684. doi: 10.1046/j.1440-169x.1997.t01-5-00003.x. [DOI] [PubMed] [Google Scholar]
- 25.Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27:e27. doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizutani A, Furukawa T, Adachi Y, Ikehara S, Taketani S. A zinc-finger protein, PLAGL2, induces the expression of a proapoptotic protein Nip3, leading to cellular apoptosis. J Biol Chem. 2002;277:15851–15858. doi: 10.1074/jbc.M111431200. [DOI] [PubMed] [Google Scholar]
- 27.Neumann M, Wang Y, Kim S, Hong SM, Jeng L, Bilgen M, Liu J. Assessing gait impairment following experimental traumatic brain injury in mice. J Neurosci Methods. 2009;176:34–44. doi: 10.1016/j.jneumeth.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng JK, Richardson CA, Kippers V, Parnianpour M. Relationship between muscle fiber composition and functional capacity of back muscles in healthy subjects and patients with back pain. J Orthop Sports Phys Ther. 1998;27:389–402. doi: 10.2519/jospt.1998.27.6.389. [DOI] [PubMed] [Google Scholar]
- 29.Qian ZM, Li H, Sun H, Ho K. Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacol Rev. 2002;54:561–587. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 30.Raivich G, Graeber MB, Gehrmann J, Kreutzberg GW. Transferrin receptor expression and iron uptake in the injured and regenerating rat sciatic nerve. Eur J Neurosci. 1991;3:919–927. doi: 10.1111/j.1460-9568.1991.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 31.Rao P, Monks DA. A tetracycline-inducible and skeletal muscle-specific Cre recombinase transgenic mouse. Dev Neurobiol. 2009;69:401–406. doi: 10.1002/dneu.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumbria RK. Targeting the transferrin receptor to develop erythropoietin for Alzheimer’s disease. Neural Regen Res. 2020;15:2251–2252. doi: 10.4103/1673-5374.284994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trenor CC, 3rd, Campagna DR, Sellers VM, Andrews NC, Fleming MD. The molecular defect in hypotransferrinemic mice. Blood. 2000;96:1113–1118. [PubMed] [Google Scholar]
- 35.Weiland A, Wang Y, Wu W, Lan X, Han X, Li Q, Wang J. Ferroptosis and its role in diverse brain diseases. Mol Neurobiol. 2019;56:4880–4893. doi: 10.1007/s12035-018-1403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Lu Y, Barik A, Joseph A, Taketo MM, Xiong WC, Mei L. β-Catenin gain of function in muscles impairs neuromuscular junction formation. Development. 2012a;139:2392–2404. doi: 10.1242/dev.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, Lu Y, Shen C, Patel N, Gan L, Xiong WC, Mei L. Distinct roles of muscle and motoneuron LRP4 in neuromuscular junction formation. Neuron. 2012b;75:94–107. doi: 10.1016/j.neuron.2012.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Barik A, Lu Y, Shen C, Bowman A, Li L, Sathyamurthy A, Lin TW, Xiong WC, Mei L. Slit2 as a β-catenin/Ctnnb1-dependent retrograde signal for presynaptic differentiation. Elife. 2015;4:e07266. doi: 10.7554/eLife.07266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W, Barrientos T, Mao L, Rockman HA, Sauve AA, Andrews NC. Lethal cardiomyopathy in mice lacking transferrin receptor in the heart. Cell Rep. 2015;13:533–545. doi: 10.1016/j.celrep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Zhu Q, Cheng J, Wu Y, Fan M, Zhang J, Wu H. Opposite regulation of Wnt/β-catenin and Shh signaling pathways by Rack1 controls mammalian cerebellar development. Proc Natl Acad Sci U S A. 2019a;116:4661–4670. doi: 10.1073/pnas.1813244116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Yang C, Zhu Q, Wei M, Li Y, Cheng J, Liu F, Wu Y, Zhang J, Zhang C, Wu H. Rack1 controls parallel fiber-purkinje cell synaptogenesis and synaptic transmission. Front Cell Neurosci. 2019b;13:539. doi: 10.3389/fncel.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.