Abstract
In prokaryotes, CRISPR-Cas immune systems recognize and cleave foreign nucleic acids to defend against mobile genetic elements (MGEs). Type III CRISPR-Cas complexes also synthesize cyclic oligoadenylate (cOA) second messengers, which activate CRISPR ancillary proteins involved in antiviral defense. In particular, cOA-stimulated nucleases degrade RNA and DNA nonspecifically, which slows MGE replication but also impedes cell growth, necessitating mechanisms to eliminate cOA in order to facilitate cell recovery. Extant cOA is degraded by a new class of enzyme termed a “ring nuclease,” which cleaves cOA specifically and switches off CRISPR ancillary enzymes. Several ring nuclease families have been characterized to date, including a family used by MGEs to circumvent CRISPR immunity, and encompass diverse protein folds and distinct cOA cleavage mechanisms. In this review we examine cOA signaling, discuss how different ring nucleases regulate the cOA signaling pathway, and reflect on parallels between cyclic nucleotide-based immune systems to reveal new areas for exploration.
Keywords: CARF, CRISPR-Cas, Csm6 ribonuclease, cyclic nucleotides, ring nuclease
TYPE III CRISPR SYSTEMS AND CYCLIC OLIGOADENYLATE SIGNALING
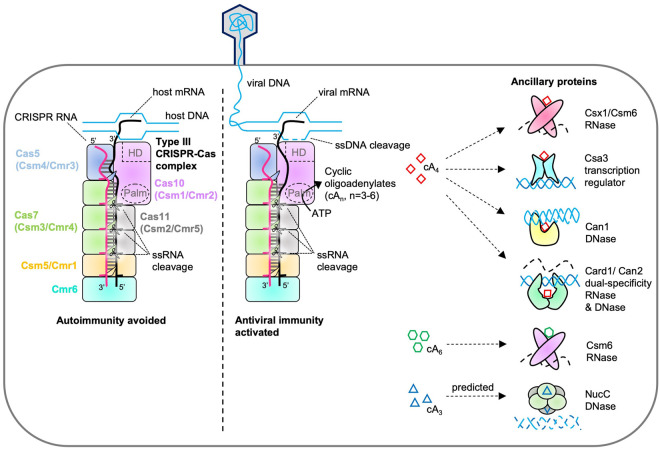
CRISPR-Cas systems capture and store fragments of foreign DNA, transcribe these into CRISPR RNA (crRNA), and carry out homology-directed nucleic acid cleavage to protect prokaryotes from invading mobile genetic elements (MGEs) (Makarova et al. 2020a). Type III CRISPR systems comprise a multisubunit ribonucleoprotein effector complex and ancillary effector proteins, which are recruited for defense by antiviral signaling (Fig. 1). Activation of the immune response relies on the lack of base-pairing between the 8 nt 5′-end of the crRNA and the 3′-end of a target RNA, which is key to distinguishing nonself (Marraffini and Sontheimer 2010; Hale et al. 2012; Zhang et al. 2012; Johnson et al. 2019). When viral mRNA is detected, the large Cas10 subunit is allosterically activated to carry out two enzymatic activities: ssDNA cleavage by an HD (histidine-aspartate) nuclease domain (Elmore et al. 2016; Estrella et al. 2016; Kazlauskiene et al. 2016; Liu et al. 2017; Jia et al. 2019a) and cyclic oligoadenylate (cOA) synthesis by the Palm polymerase domains (Kazlauskiene et al. 2017; Niewoehner et al. 2017; Jia et al. 2019a; You et al. 2019; Sofos et al. 2020). cOA acts as a second messenger and binds to CRISPR-associated Rossmann fold (CARF) domains of CRISPR ancillary proteins, allosterically activating the adjacent effector domains, which are commonly nonspecific nucleases (Fig. 2; Kazlauskiene et al. 2017; Niewoehner et al. 2017; McMahon et al. 2020; Rostøl et al. 2021; Zhu et al. 2021). Importantly, not all Cas10 proteins have a catalytically active HD domain and some lack it altogether (Staals et al. 2013; Grüschow et al. 2019; Makarova et al. 2020a), emphasizing that these systems likely rely on the cOA signaling pathway for antiviral defense.
FIGURE 1.
Cyclic oligoadenylate signaling by type III CRISPR systems. Multisubunit type III CRISPR-Cas complexes (denoted Csm or Cmr) detect foreign RNA and carry out nucleic acid cleavage directly and by recruiting CRISPR ancillary enzymes. RNA bound by the CRISPR-Cas complex, as a result of complementary base-pairing with the crRNA, is degraded by the Cas7 (Csm3/Cmr4) backbone subunits (Benda et al. 2014; Ramia et al. 2014; Staals et al. 2014; Tamulaitis et al. 2014). Bona fide RNA targets contain a 3′-region that is not complementary to the 8 nt 5′-end of the crRNA, which allosterically activates the Cas10 subunit to synthesize cyclic oligoadenylates (cOA) and cleave ssDNA (Kazlauskiene et al. 2017; Niewoehner et al. 2017). Target RNA cleavage by Cas7 subunits switches off both the ssDNase and cOA synthesis activities of Cas10 (Rouillon et al. 2018). cOA can activate Csx1/Csm6 ribonucleases that cleave RNA nonspecifically, DNases such as NucC (Lau et al. 2020) and the CRISPR ancillary nuclease 1 (Can1) (McMahon et al. 2020), and the related dual-specificity cOA-activated RNase and DNase (Card1)/Can2 (Rostøl et al. 2021; Zhu et al. 2021), which help eliminate invading mobile genetic elements (MGE). cOA can also stimulate the transcription regulator Csa3, which alters CRISPR loci and cas gene expression to promote MGE elimination (Lawrence et al. 2020).
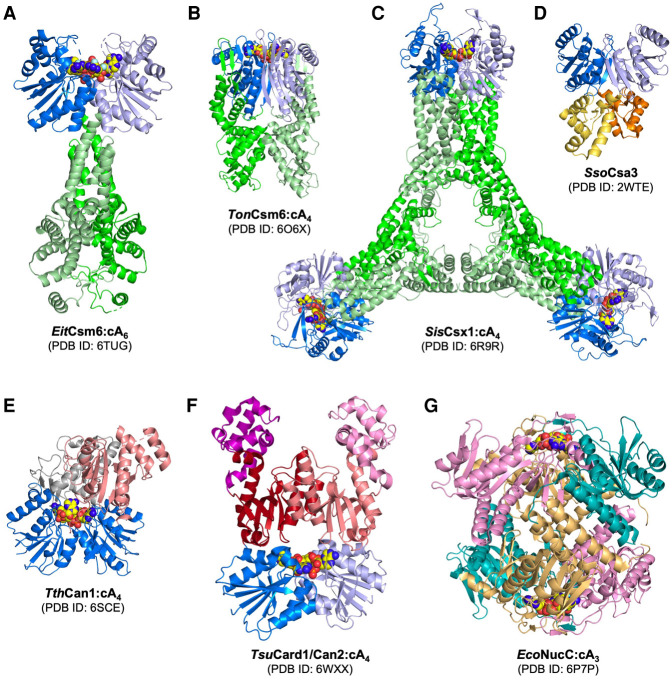
FIGURE 2.
Structures of type III CRISPR ancillary proteins. (A) E. italicus (Eit) Csm6 dimer in complex with its cyclic hexa-adenylate (cA6) activator (shown in sphere form). EitCsm6 is a nonspecific ribonuclease containing CARF (dark and light blue) and HEPN (dark and light green) domains (Garcia-Doval et al. 2020). (B) T. onnurineus (Ton) Csm6 dimer in complex with its cyclic tetra-adenylate (cA4) activator. TonCsm6 is a nonspecific ribonuclease consisting of CARF and HEPN domains (Jia et al. 2019c). (C) S. islandicus (Sis) Csx1 hexamer in complex with its cA4 activator. SisCsx1 dimers form a hexamer upon cA4 binding and RNA is cleaved at three distinct active sites within the interior of the hexamer (Molina et al. 2019). (D) S. solfataricus (Sso) Csa3 dimer. SsoCsa3 is a cA4 stimulated transcription regulator consisting of CARF and a helix-turn-helix DNA binding domain (yellow and orange) (Lintner et al. 2011). (E) T. thermophilus (Tth) CRISPR ancillary nuclease 1 (Can1) monomer in complex with its cA4 activator. TthCan1 nicks super-coiled DNA and is comprised of two CARF domains and a PD-D/ExK family nuclease domain (salmon colored) (McMahon et al. 2020). (F) T. succinifaciens cyclic oligoadenylate activated RNase and DNase 1 (Card1)/Can2 dimer in complex with its cA4 activator. Card1/Can2 is related to Can1 and is a dual-specificity nuclease with CARF and PD-D/ExK nuclease domains (red and salmon) (Rostøl et al. 2021; Zhu et al. 2021). (G) E. coli (Eco) NucC hexamer in complex with its cyclic tri-adenylate (cA3) activator. EcoNucC trimers assemble into a hexamer upon cA3 binding and degrades dsDNA (Lau et al. 2020). NucC is related to restriction endonucleases and binds cA3 at a protein domain unrelated to the CARF family.
Cas7 subunits comprise the backbone of the type III effector complex and cleave bound RNA, regardless of host or viral genomic origin. This is predicted to allow for multiple-turnover detection and continued surveillance by the effector complex (Rouillon et al. 2018). A conserved aspartate residue in Cas7 is responsible for RNA cleavage (Benda et al. 2014; Ramia et al. 2014; Staals et al. 2014; Tamulaitis et al. 2014), and whereas RNA cleavage by the effector complex is not essential for resistance to phage infection in vivo (Samai et al. 2015), it switches off the ssDNase and cOA synthesis activities of Cas10 that are activated by bona fide targets (Kazlauskiene et al. 2017; Niewoehner et al. 2017; Rouillon et al. 2018). Therefore, it is likely that RNA cleavage by the effector complex serves to exert control over the potent HD nuclease and cOA signaling activities, rather than as a primary mode of defense (Rouillon et al. 2018). Nevertheless, if extant cOA is not eliminated from cells it can sustain the immune response (Garcia-Doval et al. 2020) and has the potential to compromise cell survival (Koonin and Makarova 2018).
Cas10 can synthesize cOA of different sizes, containing between three and six 3′–5′ linked AMP units, although in vitro most type III systems generate a single cOA species in greater abundance (Kazlauskiene et al. 2017; Niewoehner et al. 2017; Rouillon et al. 2018; Grüschow et al. 2019; Smalakyte et al. 2020). The mechanism for cOA synthesis has been elucidated using structural biology and involves a nucleophilic attack by the 3′-hydroxyl of one ATP molecule, held in one Palm polymerase pocket of Cas10, on the α-phosphate of another ATP molecule held in the other Palm polymerase pocket (Jia et al. 2019b). Once the first pppApA intermediate is formed, it is then subjected to further rounds of AMP incorporation in the same manner (Kazlauskiene et al. 2017; Niewoehner et al. 2017; Jia et al. 2019b). Finally, intermediates are cyclised by an intramolecular nucleophilic attack by the 3′-hydroxyl on the α-position of its 5′-triphosphate (Jia et al. 2019b). The factors governing the type(s) and abundance of cOA molecules made, which may be determined by the protein architecture of Cas10 homologs and/or cellular ATP levels, remain to be fully explored.
Most type III systems characterized to date use cyclic tetra-adenylate (cA4) as the second messenger (Kazlauskiene et al. 2017; Han et al. 2018; Rouillon et al. 2018; Jia et al. 2019b; Foster et al. 2020; Steens et al. 2021), but several use cyclic hexa-adenylate (cA6) instead (Niewoehner et al. 2017; Nasef et al. 2019; Sridhara et al. 2021). Within these systems, the CARF domains of cognate CARF family effector(s) specifically recognize one of these signals (Rouillon et al. 2018; Molina et al. 2019; Garcia-Doval et al. 2020; McMahon et al. 2020). Csx1/Csm6 ribonucleases are the most common CARF family proteins associated with type III CRISPR systems (Makarova et al. 2020a). When activated by cA4 or cA6, as determined by the CARF domain architecture, Csx1/Csm6 cleaves RNA using its higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain (Kazlauskiene et al. 2017; Niewoehner et al. 2017). The nonspecific RNase activity of Csm6 has been shown to result in cell growth arrest, and cell growth is restored when infection is cleared from cells (Rostøl and Marraffini 2019). cA4 has also been shown to bind to the CARF family transcription regulator Csa3 (Lawrence et al. 2020), which up-regulates CRISPR loci and cas gene expression (Liu et al. 2015; Lawrence et al. 2020; Ye et al. 2020b), presumably acting as a positive-feedback mechanism linked to MGE detection. Several cOA-activated DNA nucleases also feature in CRISPR immunity. CRISPR ancillary nuclease 1 (Can1) has been shown to nick super-coiled DNA when activated by cA4 and is anticipated to slow phage replication by collapsing replication forks (McMahon et al. 2020). The related Can2 enzyme, also termed cOA-activated RNase and DNase 1 (Card1), was demonstrated to be a dual-specificity nuclease capable of both RNA and DNA cleavage (Rostøl et al. 2021; Zhu et al. 2021). While one study showed that select Card1/Can2 enzymes cleaved ssRNA and nicked super-coiled DNA to provide antiphage immunity (Zhu et al. 2021), another identified a Card1/Can2 that cleaved ssRNA and exclusively ssDNA and provided antiphage immunity (Rostøl et al. 2021). Interestingly, the authors attributed the accompanying cell growth arrest phenotype to the ssDNase activity, which is presumed to cause DNA lesions in the host chromosome, rather than the RNase activity of Card1/Can2 (Rostøl et al. 2021). Another DNase, NucC, is a cA3-activated enzyme associated with both type III CRISPR systems and cyclic oligonucleotide-based antiphage signaling systems (CBASS) (Lau et al. 2020; Ye et al. 2020a). As a CBASS component, NucC has been shown to degrade dsDNA nonspecifically and trigger cell death before phage replication is complete (Lau et al. 2020; Ye et al. 2020a). NucC is expected to act similarly during type III CRISPR immunity, and its activation may represent an instance when adaptive immunity leads to abortive infection.
REGULATION OF CYCLIC OLIGOADENYLATE SIGNALING
Nonspecific degradation of DNA and RNA by cOA-activated nucleases imbue type III CRISPR systems with an intrinsic potential for self-destruction. While collateral cleavage of host nucleic acids may instigate cell dormancy to slow viral propagation, as has been uncovered for the in trans nonspecific RNase activity of a type VI (Cas13) CRISPR system (Meeske et al. 2019), it will compromise cell survival if not carefully regulated. To overcome this problem, type III CRISPR systems harbor two regulatory mechanisms. First, cOA synthesis is switched off by target RNA cleavage (Kazlauskiene et al. 2017; Han et al. 2018; Rouillon et al. 2018; Grüschow et al. 2019; Nasef et al. 2019), and second, extant cOA is eliminated within cells to switch off CRISPR ancillary enzymes (Athukoralage et al. 2018).
Switching off cOA synthesis is not enough
Although target RNA cleavage by the type III effector complex switches off cOA synthesis (Kazlauskiene et al. 2017; Han et al. 2018; Rouillon et al. 2018; Grüschow et al. 2019; Nasef et al. 2019), extant cOA is predicted to sustain the immune response (Athukoralage et al. 2020a). In vitro, the Sulfolobus solfataricus type III-D CRISPR system has been shown to synthesize approximately 1000 molecules of cA4 per viral mRNA (Rouillon et al. 2018, 2019; Athukoralage et al. 2020a), which equates to ∼6 µM cA4 in a S. solfataricus cell, in significant excess of what is required to activate the Csx1 RNase (Athukoralage et al. 2020a). While cOA synthesis is ended by viral mRNA cleavage, the micromolar quantities of cOA already made must be eliminated if the immune response is to be switched off in a timely manner. The need to control the immune response and mitigate collateral damage has likely led to the evolution of cOA degradative enzymes, which have been termed “ring nucleases” (Athukoralage et al. 2018).
Ring nucleases
CRISPR ring nuclease 1
cOA degradative enzymes were first identified by activity-guided fractionation of S. solfataricus cellular lysates and identification of enriched proteins by mass spectrometry (Athukoralage et al. 2018). The identified protein Sso2081, and the related protein Sso1393, were shown to act as standalone enzymes that degraded cA4 specifically and were named CRISPR ring nucleases 1 (Crn1) (Fig. 3A; Athukoralage et al. 2018). Crn1 enzymes are CARF domain only proteins that have evolved to catalyze cOA cleavage and do so by cleaving the symmetrical cA4 molecule as a protein dimer (Athukoralage et al. 2018). The two S. solfataricus Crn1 enzymes exhibited a 10-fold difference in the rate of cA4 cleavage, which result in variable capacities for Csx1 deactivation, and may therefore be leveraged temporally or synergistically for cA4 elimination (Athukoralage et al. 2018). Protein sequence analyses and structural modeling suggest that Sso1393, Sso2081, and the uncharacterized Sso1397, which is predicted to be a ring nuclease, are orthologs and likely originated from gene duplication events (Athukoralage et al. 2018; Makarova et al. 2020b). Crn1 enzymes are exclusive to the crenarchaea, and Sulfolobales typically encode several Crn1 orthologs alongside Csx1/Csm6 (for review, see Zink et al. 2020). For example, S. islandicus REY15A encodes two Crn1 enzymes that degrade cA4 made by its type III-B CRISPR system, and like Crn1 enzymes from S. solfataricus, one ring nuclease appears to degrade cA4 at a higher rate compared to the other (Molina et al. 2019). As Crn1 enzymes degrade cOA slowly (Athukoralage et al. 2018), the presence of multiple Crn1 enzymes may reflect the regulatory needs of a given type III CRISPR system, particularly where large quantities of cOA are made even at low levels of infection as per the S. solfataricus type III-D CRISPR system (Athukoralage et al. 2020a).
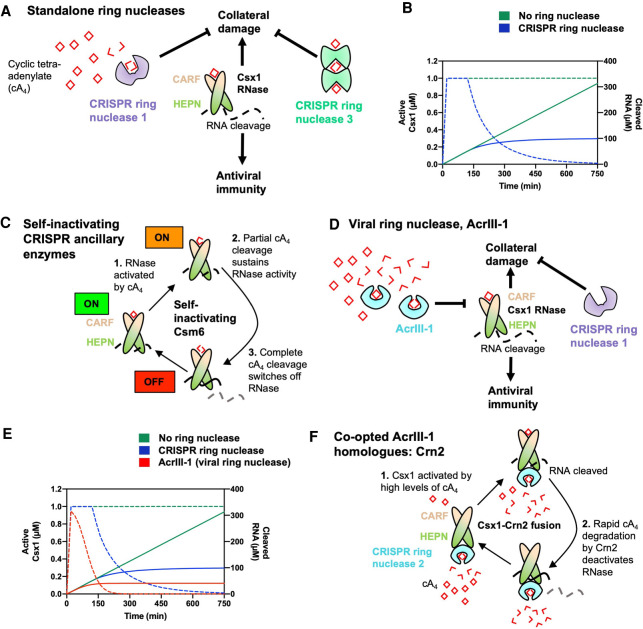
FIGURE 3.
Ring nucleases regulate the type III CRISPR immune response. (A) Cyclic tetra-adenylate (cA4)-activated Csx1 ribonucleases cleave RNA nonspecifically, which provides antiviral immunity but also causes collateral damage to cells. cA4 binds the CRISPR-associated Rossmann fold (CARF) domain of Csx1 and allosterically activates its higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain, which cleaves RNA. CRISPR ring nucleases 1 and 3 eliminate extant cA4 and deactivate Csx1, mitigating sustained collateral damage to cells. (B) Graph depicting kinetic modeling of the type III CRISPR response in a cell where 60 µM of cA4 is generated upon infection, representative of a medium-level infection. In the absence of ring nucleases, Csx1 remains in an active state (dotted green line, corresponding to left-hand side y-axis) and RNA cleavage (solid green line, corresponding to right-hand side y-axis) continues unimpeded. Csx1 is slowly deactivated when CRISPR ring nuclease 1 is present (dotted blue line) and thus RNA cleavage is limited (solid blue line). Simulations were carried out using KinTek Global Kinetic Explorer software (Johnson et al. 2009), using a previously published model of the S. solfataricus type III CRISPR defense pathway (Athukoralage et al. 2020a), and data were plotted using GraphPad Prism. (C) Some Csm6 enzymes act as bifunctional ribonucleases and ring nucleases. These enzymes cleave cA4 at the CARF domain upon cA4 binding and activating the HEPN RNase. Some Csm6 enzymes also cleave cOA at the HEPN domain. (D) Prokaryotic viruses encode an anti-CRISPR viral ring nuclease (AcrIII-1). AcrIII-1 rapidly degrades cA4 and attenuates RNA cleavage by swiftly deactivating Csx1. (E) Graph depicting the effect of having no ring nuclease (green), a host cell CRISPR ring nuclease 1 (blue), and both CRISPR ring nuclease 1 and AcrIII-1 on the active form of Csx1 (dotted lines, left-hand side y-axis) and consequent RNA cleavage (solid lines, right-hand side y-axis). When AcrIII-1 is present, Csx1 is deactivated much more quickly. (F) In some bacteria, AcrIII-1 homologs are found associated with type III CRISPR systems and these proteins have been named CRISPR ring nuclease 2 (Crn2). In Marinitoga piezophile, Csx1 is fused to Crn2, which limits Csx1 activity by rapidly and constitutively degrading cA4. The Crn2 domain only permits Csx1 activation when a high cA4 threshold, determined by the balance between cA4 affinity of the CARF domain of Csx1 and the high cA4 affinity and rate of degradation by Crn2, is reached.
While cyclic nucleotide phosphodiesterases typically coordinate metal ions to catalyze cyclic nucleotide cleavage (for review, see Conti and Beavo 2007), Crn1 is metal-independent and yields cA4 cleavage products containing 2′,3′-cyclic phosphates (Athukoralage et al. 2018). Crn1 enzymes catalyze cA4 cleavage by positioning and/or activating the 2′-hydroxyl group of a ribose for an in-line nucleophilic attack on the adjacent scissile phosphodiester bond (Athukoralage et al. 2018). The first nucleophilic attack generates a linear tetra-adenylate containing a 2′,3′-cyclic phosphate (A4 > P), which is then converted to two molecules of di-adenylate containing a 2′,3′-cyclic phosphate (A2 > P) by the second nucleophilic attack on the other side of the molecule (Athukoralage et al. 2018). The final A2 > P products do not stimulate CARF family CRISPR ancillary effectors such as Csx1 (Athukoralage et al. 2018; Molina et al. 2019). Consequently, ring nucleases are poised to act as crucial regulators of the type III CRISPR immune response. Indeed, computational modeling demonstrates that Crn1 curtails RNA cleavage by decreasing the active form of Csx1 over time, and without Crn1 cell death is the most probable outcome (Fig. 3B; Athukoralage et al. 2020a).
Self-inactivating CRISPR ancillary ribonucleases
In the absence of standalone ring nucleases some Csm6 enzymes have adapted to intrinsically degrade their cOA activators. Several Csm6 enzymes have a CARF domain that acts as a dual cOA sensor and ring nuclease and this has been proposed to facilitate timed deactivation of the RNase component (Athukoralage et al. 2019; Jia et al. 2019c; Garcia-Doval et al. 2020; Smalakyte et al. 2020). In this model, cOA is cleaved at the CARF domain as it allosterically stimulates the HEPN RNase domain (Fig. 3C). First, cOA is cleaved to generate an A4 > P intermediate, which is known to support HEPN RNase activation (Niewoehner et al. 2017; Rouillon et al. 2018, 2019), whereas the second cleavage event generates two A2 > P products and deactivates the enzyme, similar to the catalytic mechanism of Crn1 (Athukoralage et al. 2018). Notably, no standalone ring nucleases have yet been identified that degrade cA6, although several Csm6 enzymes possess cA6 degradative CARF domains (Garcia-Doval et al. 2020; Smalakyte et al. 2020). Site-directed mutagenesis to abolish cOA cleavage at one such CARF domain was found to result in sustained RNA cleavage by Csm6, which led to a strong growth inhibition phenotype, highlighting the crucial role of ring nuclease activity in cell recovery (Garcia-Doval et al. 2020).
Some Csm6/Csx1 enzymes also degrade cA4 and cA6 by their HEPN RNase domain (Jia et al. 2019c; Foster et al. 2020; Smalakyte et al. 2020), although this may be a side reaction. For S. thermophilus Csm6, the HEPN domain has been shown to exhibit higher affinity for single-stranded RNA substrates compared to cA6, and the CARF domain was found to have higher affinity for cA6 compared to the HEPN domain (Smalakyte et al. 2020). These observations suggest that CARF ring nuclease activity will be particularly important at low cA6 levels, as the lower cA6 binding affinity of the HEPN domain may prevent it from eliminating cOA sufficiently to preclude Csm6 activation. Nevertheless, the S. thermophilus Csm6 HEPN domain was highly efficient at cA6 degradation compared to its CARF domain (Smalakyte et al. 2020), and lesser conformational restraints associated with cA6 compared to other cOA species may enable its rapid turnover at the HEPN active site. While single-stranded RNA substrates and cOA appear to be cleaved similarly by the HEPN domain (Jia et al. 2019c), it is worth noting that only Csx1/Csm6 that lack cleavage preferences (Kazlauskiene et al. 2017; Athukoralage et al. 2019), or cleave selectively after adenosines (Sheppard et al. 2016; Jia et al. 2019c), are likely to support cOA cleavage at the HEPN active site.
Viral ring nuclease AcrIII-1
Type III CRISPR immunity is capable of driving viruses to extinction (Pyenson et al. 2017). cOA-activated effector proteins that inhibit viral replication are therefore anticipated to drive viruses to evolve mechanisms to inhibit cOA signaling. Indeed, diverse archaeal viruses, proviruses, bacteriophages, prophages, and plasmids encode a highly efficient cA4 degradative enzyme (Athukoralage et al. 2020b). The DUF1874-family viral ring nuclease was shown to degrade cA4 at a rate ∼50-fold greater than cellular Crn1 enzymes, enabling MGEs to circumvent type III CRISPR immunity (Fig. 3D; Athukoralage et al. 2020b). As an anti-CRISPR protein, the viral ring nuclease was named (AcrIII-1), in keeping with established anti-CRISPR nomenclature (Bondy-Denomy et al. 2018) but incorporating a hyphen to denote that it is not type III subtype-specific. In agreement with biochemistry and microbiology studies, kinetic modeling has been used to show that AcrIII-1 is able to quickly decrease the level of activated Csx1 in cells by rapidly degrading cA4, which underlies its potent anti-CRISPR function (Fig. 3E; Athukoralage et al. 2020a).
AcrIII-1 is a metal-independent ring nuclease that forms A2 > P products similar to Crn1 enzymes (Athukoralage et al. 2020b). Both AcrIII-1 and Crn1 bind cA4 with high affinity (Athukoralage et al. 2020a), therefore its greater catalytic efficiency is ascribed to conserved active site residues that better position the 2′-hydroxyl of the ribose for an in-line nucleophilic attack, stabilize the transition state, and protonate the leaving group (Athukoralage et al. 2020b). In particular, a highly conserved active site histidine residue is critical for cA4 cleavage and is postulated to act as the general acid involved in protonating the oxyanion leaving group (Athukoralage et al. 2020b).
Co-option of viral ring nucleases by bacteria: CRISPR ring nuclease 2
Intriguingly, AcrIII-1 is found in association with type III CRISPR systems in several bacteria, independent of MGEs (Athukoralage et al. 2020b). Due to the wide distribution of AcrIII-1 among MGEs, it is plausible that bacteria acquired AcrIII-1 by horizontal gene transfer from MGEs and harnessed the enzyme to regulate cOA signaling. AcrIII-1 associated with type III CRISPR systems have been termed CRISPR ring nuclease 2 (Crn2) (Athukoralage et al. 2020b).
In Marinitoga piezophila (Mpi), a Csx1 protein is fused to Crn2, directly implicating this Crn2 in Csx1 regulation (Samolygo et al. 2020). The Crn2 domain was shown to exhibit constitutive cA4 degradation at a rate comparable to AcrIII-1 enzymes (Samolygo et al. 2020). However, cA4 degradation by the Crn2 domain limited but did not abolish RNA cleavage by Csx1, consistent with MpiCsx1–Crn2 acting as a self-limiting ribonuclease (Samolygo et al. 2020). High micromolar levels of cA4 were required to detect MpiCsx1–Crn2 RNase activity, which is attributed to the higher cA4 affinity and potent ring nuclease activity of the Crn2 domain (Samolygo et al. 2020). The Crn2 domain likely prevents spurious activation of Csx1, permitting RNA degradation only once a set cA4 threshold—perhaps a molecular signature of a bona fide infection—has been reached (Fig. 3F). Interestingly, the majority of crn2 genes encode standalone enzymes (Athukoralage et al. 2020b), and in these cases Crn2 expression may be temporally regulated so as not to compromise the antiviral response by rapid elimination of cOA.
CRISPR ring nuclease 3
In prokaryotic phyla where crn1 genes are absent, csx3 is commonly found associated with type III CRISPR systems, and csx1 or csm6 are its most common adjacent genes (Shah et al. 2019; Makarova et al. 2020b). Early studies on Archaeoglobus fulgidus (Afu) Csx3 identified the protein as a manganese dependent RNA exoribonuclease that specifically cleaved 3′ poly(A) tails (Yan et al. 2015), although its precise role in antiviral immunity remained unclear. Recently, the enzymatic activities of AfuCsx3 were reassessed and the protein was demonstrated to be a cA4-specific ring nuclease (Athukoralage et al. 2020c; Brown et al. 2020). Csx3 has been shown to regulate Csx1-mediated immunity in vivo and was therefore renamed CRISPR ring nuclease 3 (Crn3) (Athukoralage et al. 2020c). Surprisingly, Crn3 degraded cA4 at a rate comparable to AcrIII-1 enzymes in vitro; however, in vivo studies showed that Crn3 provided only partial protection from Csx1 toxicity, whereas AcrIII-1 provided near complete protection by fully deactivating the pathway (Athukoralage et al. 2020c).
Based on structural topology, Crn3 was initially postulated to be a divergent member of the CARF family (Topuzlu and Lawrence 2016). However, recent structural analysis indicates that Crn3 is more closely related to sulfate transporter and anti-sigma factor antagonist (STAS) domains (Makarova et al. 2020b). X-ray structures revealed that Crn3 forms head to tail filaments containing cA4 bound between adjacent dimers (Fig. 3A; Athukoralage et al. 2020c). In contrast to the metal-independent Crn1 family, Crn3 relies on manganese ions to cleave cA4 into A2 > P and A2P (di-adenylate containing 5′-hydroxyl and 3′-phosphate moieties) products. One face of Crn3 harbors three highly conserved histidine residues, which may coordinate up to three manganese ions required for catalysis, whereas the other face harbors a conserved aspartate that is also catalytically important. Dimer-dimer stacking leads to the union of these two faces, otherwise over 20 Å apart, and forms the composite active site (Athukoralage et al. 2020c). While the cOA cleavage mechanism is not fully understood, kinetic studies show cooperativity in the Crn3 reaction cycle, which suggests that the rate of cA4 elimination may be controlled by altering Crn3 levels in cells (Athukoralage et al. 2020c).
Intriguingly, cyanobacteria encode a protein family containing an amino-terminal Crn3 domain and a carboxy-terminal AAA+ (ATPases associated with diverse cellular activities) ATPase domain (Shah et al. 2019). AAA+ ATPase domains hydrolyze ATP to drive a range of cellular processes including protein unfolding and degradation and DNA repair, replication, and recombination (for review, see Ogura and Wilkinson 2001). Hence the coupling of Crn3 and an AAA+ ATPase may indicate that cOA degradation is linked to other cellular pathways. Additionally, recent work has uncovered a gene containing both Csx1 and Crn3 domains in Thermodesulfobium narugense (Makarova et al. 2020b), directly implicating Crn3 in Csx1 regulation in bacteria.
Predicted ring nucleases
Recent analysis of the CARF superfamily has led to identification of several genes which are expected to comprise new ring nuclease families (Makarova et al. 2020b). Namely, Csx16, Csx20, and Unk_01 have been identified to contain polar resides (histidine, arginine, and aspartate or glutamate) compatible with ring nuclease function (Makarova et al. 2020b). Csx16 and Csx20 share sequence identity, and Csx16 has been noted as partially similar to the DUF1874 protein family which comprises both AcrIII-1 and Crn2, suggesting homology with viral ring nucleases (Makarova et al. 2020b). Csx16, Csx20, and Unk_01 have also been detected fused to Csx1, and based on the characterized MpiCsx1–Crn2 protein, such fusions are good indicators of regulatory function. Csx14 is further predicted to constitute a new ring nuclease family based on conservation of serine or threonine and lysine residues which are crucial for cA4 cleavage in Crn1 enzymes (Makarova et al. 2020b). On the other hand, additional cOA sensing and/or degradative enzymes unrelated to the CARF and DUF1874 protein families should not be ruled out. Indeed, a membrane-associated DHH-DHHA1 (MAD) family nuclease has recently been uncovered that nonspecifically degrades cA4 (Zhao et al. 2020). In several cases, CorA, which are associated with type III CRISPR systems and anticipated to be cOA-activated membrane channels, are found adjacent or fused to DHH family nucleases (Shah et al. 2019), and it is possible that the DHH family nuclease may cleave cOA to regulate the CorA component.
PARALLELS BETWEEN CYCLIC NUCLEOTIDE-BASED IMMUNE SYSTEMS IN PROKARYOTES
Similar to type III CRISPR systems, CBASS systems generate cyclic nucleotide second messengers in response to stimuli from phage infection (Fig. 4; for review, see Millman et al. 2020). Antiviral signaling by CBASS comprises a greater repertoire of signals, which include cyclic nucleotides containing noncanonical linkages (Lowey et al. 2020). CBASS signals identified to date include cyclic GMP-AMP (c-GAMP), cyclic di-GMP, cyclic UMP–UMP, cyclic UMP–AMP, cyclic AMP–AMP–GMP, cA3, and cA4 (Cohen et al. 2019; Whiteley et al. 2019; Morehouse et al. 2020; Ye et al. 2020a). These activate NucC and Cap4 DNases (Lau et al. 2020; Lowey et al. 2020), as well as phospholipases (Cohen et al. 2019) and NADases (Morehouse et al. 2020) that are involved in precipitating cell death as mechanisms of antiphage immunity. In the Cap4 enzyme, cA3 is recognized by a SMODS associated and fused to various effectors sensor domain (SAVED), which structural study indicates is evolutionarily linked to the CARF family (Lowey et al. 2020), and SAVED domain containing effectors are also associated with several type III CRISPR systems (Shmakov et al. 2018). Many of the characterized CBASS-associated effectors are cytotoxic (Severin et al. 2018; Cohen et al. 2019; Lau et al. 2020), revealing another similarity between type III CRISPR and CBASS immune responses. Indeed, when type III CRISPR-Cas and CBASS co-occur, cOA synthesized by one or the other system might cross-activate ancillary effectors associated with either system. Importantly, it remains to be elucidated whether CBASS systems function exclusively via abortive infection, or if as of yet unidentified ring nucleases or regulatory mechanisms play a role in deactivating these systems.
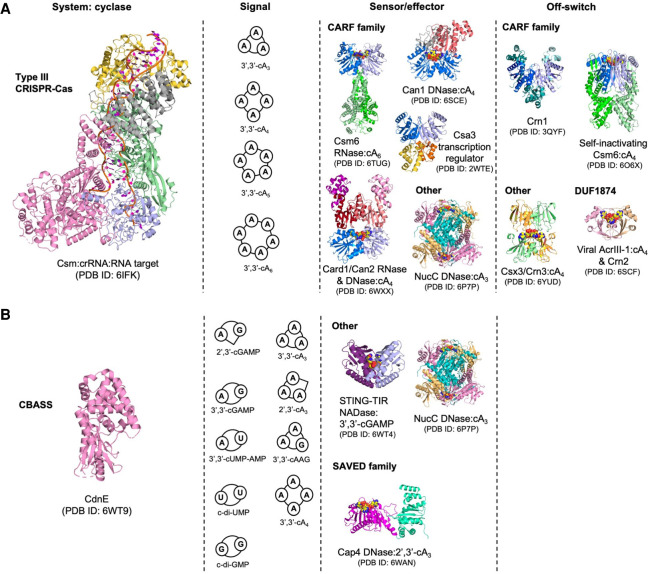
FIGURE 4.
Comparisons of cyclic nucleotide-based immune systems in prokaryotes. (A) Protein Data Bank (PDB) identifiers are shown alongside all structures. Type III CRISPR immunity comprises four key components; a detection and signaling platform, cyclic nucleotide signals, ancillary signal sensors fused to effectors, and mechanisms to eliminate the signal. The type III CRISPR complex detects viral mRNA, and the Cas10 nucleotidyl cyclase generates cyclic oligoadenylates (cOA; cAn, n = 3–6) containing 3′–5′ phosphodiester linkages. cOA allosterically stimulates downstream CRISPR ancillary effector proteins, typically by binding to a CRISPR-associated Rossmann fold (CARF) domain (colored in marine and light blue in protein dimers). Finally, ring nucleases eliminate cOA and deactivate CRISPR ancillary effectors, controlling the immune response. Some ring nucleases are CARF family proteins, whereas others have unique cOA sensing domains. Viruses encode variant ring nucleases (AcrIII-1), which rapidly degrade cOA and suppress the type III CRISPR immune response. In some bacteria, AcrIII-1 is found associated with type III CRISPR systems and has been termed Crn2 because it appears to be harnessed by bacteria to regulate CRISPR immunity. (B) The cyclic oligonucleotide-based antiphage signaling system (CBASS) resembles the type III CRISPR immune system in some respects. The cyclase (CdnE) is activated by unknown stimuli during phage infection, and different CdnE proteins synthesize different cyclic nucleotide molecules which activate downstream effector proteins. CBASS can synthesize cyclic nucleotides containing both 3′–5′ and 2′–5′ phosphodiester linkages, giving rise to an enormous repertoire of possible cyclic nucleotide signals. These signals are detected by distinct protein domains, including the SMODS associated and fused to various effectors sensor domain (SAVED), which is evolutionarily linked to the CARF domain. Although ring nucleases may degrade cA4, no prokaryotic enzymes have been identified that degrade any of the other cyclic nucleotide signals generated by CBASS. Some CBASS systems may function exclusively via abortive infection, whereas others may have novel regulatory mechanisms.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
cOA signaling by type III CRISPR complexes to activate downstream effectors and eliminate foreign intruders represents a highly potent antiviral defense strategy. Ring nucleases are a crucial component in this pathway and deactivate CRISPR ancillary enzymes by degrading cOA. CARF family ring nucleases exhibit significant sequence variation, highlighting convergent evolution on a crucial catalytic function. From a microbiological standpoint, ring nucleases are undoubtedly important, this is emphasized by MGEs that use ring nucleases as anti-immune weaponry. By eliminating extant cOA, ring nucleases have the potential to regulate diverse cOA-activated effectors, including putative proteases, RelE RNases predicted to cleave mRNA at ribosomes, and WYL-domain containing transcription regulators that may promote antiviral immunity and/or cell recovery (Makarova et al. 2014; Shmakov et al. 2018; Shah et al. 2019). While rapid progress has been made in understanding cOA signaling, many cOA-activated enzymes await biochemical study, and some may be particularly useful for biotechnological applications. For example, Csm6 has been used to cleave reporter molecules as a second amplification step in several diagnostic assays (Gootenberg et al. 2017, 2018), including in recent studies using type III CRISPR-Cas systems and cOA signaling as viral RNA diagnostic platforms (Santiago-Frangos et al. 2020; Steens et al. 2021). Likewise, newly predicted ring nucleases based on CARF superfamily analysis may harbor new catalytic mechanisms of broad interest to enzymology.
Importantly, the discovery of cOA elimination by ring nucleases should not exclude consideration of other fates for cOA, such as the possibility that cOA is used to warn neighboring cells of infection. cOA may be disseminated within bacterial aggregates such as biofilms by the action of efflux and influx transporters or accompanying cell lysis. For example, L. monocytogenes is known to secrete the second messenger c-di-AMP using a multidrug efflux pump (Woodward et al. 2010). In mammals, 2′,3′-cGAMP synthesized by cGAS (cyclic GMP-AMP synthase) is disseminated between cells via gap junctions and confer immunity to bystander cells (Ablasser et al. 2013). cGAS detects cytosolic DNA and synthesizes 2′,3′-cGAMP, which stimulates STING (stimulator of interferon genes) signalosome assembly and downstream stimulation of transcription regulators critical to the antiviral interferon response (Ishikawa et al. 2009; Sun et al. 2013). 2′,3′-cGAMP has also been found packaged into virions, whereby its transfer into newly infected cells triggers STING-dependent innate immunity (Bridgeman et al. 2015; Gentili et al. 2015). As cOA levels reach high µM concentrations during infection (Smalakyte et al. 2020), cOA may similarly accumulate in new virions and serve to accelerate immune activation in newly infected cells. Accelerated immune activation would be highly advantageous because type III CRISPR immunity is transcription-dependent and effective during middle to late viral gene expression (Deng et al. 2013; Goldberg et al. 2014; Tamulaitis et al. 2014; Samai et al. 2015).
Mobile genetic elements are known to encode multiple anti-CRISPR proteins to inhibit the same CRISPR-Cas system (Bondy-Denomy et al. 2013; Pawluk et al. 2014). In addition to AcrIII-1, Sulfolobus islandicus rod-shaped virus 2 (SIRV2) encodes AcrIIIB1, a small protein that binds the S. islandicus LAL14/1 type III-B Cmr complex and blocks cOA synthesis (Bhoobalan-Chitty et al. 2019). However, AcrIIIB1 is subtype-specific, whereas AcrIII-1 is effective against any type III subtype that uses cA4 as a second messenger (Athukoralage et al. 2020b). Consequently, AcrIII-1 may drive type III CRISPR systems to select alternative cOA activators to precipitate the immune response. As AcrIII-1 is cA4-specific (Athukoralage et al. 2020b), viral enzymes that degrade cA3 and cA6 are highly anticipated. In turn, cyclic nucleotide-based immune systems may protect signals by synthesizing cyclic nucleotides containing noncanonical linkages, as seen for CBASS systems (Lowey et al. 2020), that may be resistant to cleavage by virus encoded nucleases. Thus, the continued study of cyclic nucleotide-based immune systems, particularly under viral selection, should provide valuable insights into the dynamics of virus-host coevolution, and uncover further troves of natural products and novel enzymes.
Interestingly, similar to MGEs targeting prokaryotes, eukaryotic viruses use second messenger degradative enzymes, indicating that it is a widely effective immune evasion strategy. Poxviridae that infect eukaryotes produce a 2′,3′-cGAMP phosphodiesterase, termed poxvirus immune nuclease or poxin, which interferes with cGAS-STING immunity (Eaglesham et al. 2019). Furthermore, mirroring bacterial co-option of AcrIII-1 homologs, Lepidoptera (moths and butterflies) encode poxin homologs that degrade 2′,3′-cGAMP (Eaglesham et al. 2019). Phosphodiesterases that degrade 2′,3′-cGAMP have not yet been identified in a eukaryotic cell cytoplasm, therefore akin to acquisition of AcrIII-1 by bacteria, Lepidopterans may have acquired poxins from viruses in order to regulate cGAS-STING immunity. Considering current knowledge of cyclic nucleotide-based virus-host conflicts, eukaryotic viruses are the best studied and are known to harbor diverse mechanisms that block and counteract antiviral signaling. For example, eukaryotic viruses are known to shield DNA to prevent detection by cGAS (Lahaye et al. 2013; Sun et al. 2015), degrade cGAS using proteases (Aguirre et al. 2017), prevent cyclic nucleotide synthesis by disabling DNA sensing or promoting DNA dissociation (Wu et al. 2015; Biolatti et al. 2018; Huang et al. 2018a,b), and inhibit various stages of downstream signaling by proteolytic degradation of STING (Aguirre et al. 2012; Ding et al. 2018). It is likely that prokaryotic viruses adopt similar strategies to inhibit different stages of the type III CRISPR pathway, and continued study of virus-host interactions promises to yield exciting discoveries at every turn.
ACKNOWLEDGMENTS
We thank the RNA society for an invitation to submit this work as part of a 2021 RNA Society/Sacringe Graduate Student Award. The work in the authors’ laboratory described in this article was supported by research grants from the Biotechnology and Biological Sciences Research Council (REF: BB/S000313/1 and BB/T004789/1).
Author contributions: J.S.A. and M.F.W. conceptualized and prepared the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078739.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503: 530–534. 10.1038/nature12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, et al. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8: e1002934. 10.1371/journal.ppat.1002934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, et al. 2017. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2: 17037. 10.1038/nmicrobiol.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, Rouillon C, Graham S, Grüschow S, White MF. 2018. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562: 277–280. 10.1038/s41586-018-0557-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, Graham S, Grüschow S, Rouillon C, White MF. 2019. A type III CRISPR ancillary ribonuclease degrades its cyclic oligoadenylate activator. J Mol Biol 431: 2894–2899. 10.1016/j.jmb.2019.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, Graham S, Rouillon C, Grüschow S, Czekster CM, White MF. 2020a. The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. Elife 9: e55852. 10.7554/eLife.55852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, McMahon SA, Zhang C, Grüschow S, Graham S, Krupovic M, Whitaker RJ, Gloster TM, White MF. 2020b. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577: 572–575. 10.1038/s41586-019-1909-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, McQuarrie S, Grüschow S, Graham S, Gloster TM, White MF. 2020c. Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage. Elife 9: e57627. 10.7554/eLife.57627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgärtner M, Bonneau F, Mann M, Conti E. 2014. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell 56: 43–54. 10.1016/j.molcel.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Bhoobalan-Chitty Y, Johansen TB, Di Cianni N, Peng X. 2019. Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell 179: 448–458.e11. 10.1016/j.cell.2019.09.003 [DOI] [PubMed] [Google Scholar]
- Biolatti M, Dell'Oste V, Pautasso S, Gugliesi F, von Einem J, Krapp C, Jakobsen MR, Borgogna C, Gariglio M, De Andrea M, et al. 2018. Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J Virol 92: e01774-17. 10.1128/JVI.01774-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493: 429–432. 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Davidson AR, Doudna JA, Fineran PC, Maxwell KL, Moineau S, Peng X, Sontheimer EJ, Wiedenheft B. 2018. A unified resource for tracking anti-CRISPR names. Cris J 1: 304–305. 10.1089/crispr.2018.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, Rehwinkel J. 2015. Viruses transfer the antiviral second messenger cGAMP between cells. Science 349: 1228–1232. 10.1126/science.aab3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Gauvin CC, Charbonneau AA, Burman N, Lawrence CM. 2020. Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type-III CRISPR-Cas that degrades the second messenger cA4. J Biol Chem 295: 14963–14972. 10.1074/jbc.RA120.014099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, Doron S, Amitai G, Sorek R. 2019. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574: 691–695. 10.1038/s41586-019-1605-5 [DOI] [PubMed] [Google Scholar]
- Conti M, Beavo J. 2007. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem 76: 481–511. 10.1146/annurev.biochem.76.060305.150444 [DOI] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, Peng X, She Q. 2013. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87: 1088–1099. 10.1111/mmi.12152 [DOI] [PubMed] [Google Scholar]
- Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, Ploss A. 2018. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc Natl Acad Sci 115: E6310–E6318. 10.1073/pnas.1803406115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglesham JB, Pan Y, Kupper TS, Kranzusch PJ. 2019. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 566: 259–263. 10.1038/s41586-019-0928-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP. 2016. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev 30: 447–459. 10.1101/gad.272153.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella MA, Kuo F-T, Bailey S. 2016. RNA-activated DNA cleavage by the Type III-B CRISPR–Cas effector complex. Genes Dev 30: 460–470. 10.1101/gad.273722.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K, Grüschow S, Bailey S, White MF, Terns MP. 2020. Regulation of the RNA and DNA nuclease activities required for Pyrococcus furiosus Type III-B CRISPR-Cas immunity. Nucleic Acids Res 48: 4418–4434. 10.1093/nar/gkaa176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Doval C, Schwede F, Berk C, Rostøl JT, Niewoehner O, Tejero O, Hall J, Marraffini LA, Jinek M. 2020. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat Commun 11: 1596. 10.1038/s41467-020-15334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G, et al. 2015. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science 349: 1232–1236. 10.1126/science.aab3628 [DOI] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, Marraffini LA. 2014. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514: 633–637. 10.1038/nature13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356: 438–442. 10.1126/science.aam9321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. 2018. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360: 439–444. 10.1126/science.aaq0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüschow S, Athukoralage JS, Graham S, Hoogeboom T, White MF. 2019. Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence. Nucleic Acids Res 47: 9259–9270. 10.1093/nar/gkz676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CVC, Graveley BR, Terns RM, et al. 2012. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell 45: 292–302. 10.1016/j.molcel.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Stella S, Zhang Y, Guo T, Sulek K, Peng-Lundgren L, Montoya G, She Q. 2018. A Type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res 46: 10319–10330. 10.1093/nar/gky844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, You H, Su C, Li Y, Chen S, Zheng C. 2018a. Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J Virol 92: e00841-18. 10.1128/JVI.00841-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z-F, Zou H-M, Liao B-W, Zhang H-Y, Yang Y, Fu Y-Z, Wang S-Y, Luo M-H, Wang Y-Y. 2018b. Human cytomegalovirus protein UL31 inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microbe 24: 69–80.e4. 10.1016/j.chom.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792. 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Mo CY, Wang C, Eng ET, Marraffini LA, Patel DJ. 2019a. Type III-A CRISPR-Cas Csm complexes: assembly, periodic RNA cleavage, DNase activity regulation, and autoimmunity. Mol Cell 73: 264–277.e5. 10.1016/j.molcel.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Jones R, Sukenick G, Patel DJ. 2019b. Second messenger cA4 formation within the composite Csm1 palm pocket of type III-A CRISPR-Cas Csm complex and its release path. Mol Cell 75: 933–943.e6. 10.1016/j.molcel.2019.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Jones R, Yang G, Ouerfelli O, Patel DJ. 2019c. CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol Cell 75: 944–956.e6. 10.1016/j.molcel.2019.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Simpson ZB, Blom T. 2009. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem 387: 20–29. 10.1016/j.ab.2008.12.024 [DOI] [PubMed] [Google Scholar]
- Johnson K, Learn BA, Estrella MA, Bailey S. 2019. Target sequence requirements of a type III-B CRISPR-Cas immune system. J Biol Chem 294: 10290–10299. 10.1074/jbc.RA119.008728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas Č, Siksnys V. 2016. Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell 62: 295–306. 10.1016/j.molcel.2016.03.024 [DOI] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, Siksnys V. 2017. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357: 605–609. 10.1126/science.aao0100 [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. 2018. Discovery of oligonucleotide signaling mediated by CRISPR-associated polymerases solves two puzzles but leaves an enigma. ACS Chem Biol 13: 309–312. 10.1021/acschembio.7b00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, El Marjou A, Lacabaratz C, Lelièvre J-D, Manel N. 2013. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39: 1132–1142. 10.1016/j.immuni.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Lau RK, Ye Q, Birkholz EA, Berg KR, Patel L, Mathews IT, Watrous JD, Ego K, Whiteley AT, Lowey B, et al. 2020. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol Cell 77: 723–733.e6. 10.1016/j.molcel.2019.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CM, Charbonneau A, Gauvin C. 2020. Cyclic tetra-adenylate (cA4) activates CRISPR associated transcription factor Csa3, providing feedback activation of protospacer acquisition and crRNA expression. FASEB J 34: 1–1. 10.1096/fsb2.21134 [DOI] [Google Scholar]
- Lintner NG, Frankel KA, Tsutakawa SE, Alsbury DL, Copié V, Young MJ, Tainer JA, Lawrence CM. 2011. The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J Mol Biol 405: 939–955. 10.1016/j.jmb.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li Y, Wang X, Ye Q, Li H, Liang Y, She Q, Peng N. 2015. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res 43: 1044–1055. 10.1093/nar/gku1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Iavarone AT, Doudna JA. 2017. RNA and DNA targeting by a reconstituted Thermus thermophilus type III-A CRISPR-Cas system. PLoS One 12: e0170552. 10.1371/journal.pone.0170552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey B, Whiteley AT, Keszei AFA, Morehouse BR, Mathews IT, Antine SP, Cabrera VJ, Kashin D, Niemann P, Jain M, et al. 2020. CBASS immunity uses CARF-related effectors to sense 3′–5′- and 2′–5′-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell 182: 38–49.e17. 10.1016/j.cell.2020.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Grishin N V, Koonin EV, Aravind L. 2014. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet 5: 102. 10.3389/fgene.2014.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, et al. 2020a. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18: 67–83. 10.1038/s41579-019-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Timinskas A, Wolf YI, Gussow AB, Siksnys V, Venclovas Č, Koonin EV. 2020b. Evolutionary and functional classification of the CARF domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic Acids Res 48: 8828–8847. 10.1093/nar/gkaa635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463: 568–571. 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SA, Zhu W, Graham S, Rambo R, White MF, Gloster TM. 2020. Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate. Nat Commun 11: 500. 10.1038/s41467-019-14222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Nakandakari-Higa S, Marraffini LA. 2019. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570: 241–245. 10.1038/s41586-019-1257-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman A, Melamed S, Amitai G, Sorek R. 2020. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat Microbiol 5: 1608–1615. 10.1038/s41564-020-0777-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina R, Stella S, Feng M, Sofos N, Jauniskis V, Pozdnyakova I, López-Méndez B, She Q, Montoya G. 2019. Structure of Csx1-cOA4 complex reveals the basis of RNA decay in type III-B CRISPR-Cas. Nat Commun 10: 4302. 10.1038/s41467-019-12244-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse BR, Govande AA, Millman A, Keszei AFA, Lowey B, Ofir G, Shao S, Sorek R, Kranzusch PJ. 2020. STING cyclic dinucleotide sensing originated in bacteria. Nature 586: 429–433. 10.1038/s41586-020-2719-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasef M, Muffly MC, Beckman AB, Rowe SJ, Walker FC, Hatoum-Aslan A, Dunkle JA. 2019. Regulation of cyclic oligoadenylate synthesis by the Staphylococcus epidermidis Cas10-Csm complex. RNA 25: 948–962. 10.1261/rna.070417.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostøl JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, Jinek M. 2017. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548: 543–548. 10.1038/nature23467 [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. 2001. AAA+ superfamily ATPases: common structure-diverse function. Genes Cells 6: 575–597. 10.1046/j.1365-2443.2001.00447.x [DOI] [PubMed] [Google Scholar]
- Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL, Davidson AR. 2014. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 5: e00896. 10.1128/mBio.00896-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyenson NC, Gayvert K, Varble A, Elemento O, Marraffini LA. 2017. Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 22: 343–353.e3. 10.1016/j.chom.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramia NF, Spilman M, Tang L, Shao Y, Elmore J, Hale C, Cocozaki A, Bhattacharya N, Terns RM, Terns MP, et al. 2014. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep 9: 1610–1617. 10.1016/j.celrep.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostøl JT, Marraffini LA. 2019. Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR–Cas immunity. Nat Microbiol 4: 656–662. 10.1038/s41564-018-0353-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostøl JT, Xie W, Kuryavyi V, Maguin P, Kao K, Froom R, Patel DJ, Marraffini LA. 2021. The Card1 nuclease provides defence during type III CRISPR immunity. Nature 590: 624–629. 10.1038/s41586-021-03206-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Athukoralage JS, Graham S, Grüschow S, White MF. 2018. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. Elife 7: e36734. 10.7554/eLife.36734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Athukoralage JS, Graham S, Grüschow S, White MF. 2019. Investigation of the cyclic oligoadenylate signaling pathway of type III CRISPR systems. Methods Enzymol 616: 191–218. 10.1016/bs.mie.2018.10.020 [DOI] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. 2015. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell 161: 1164–1174. 10.1016/j.cell.2015.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samolygo A, Athukoralage JS, Graham S, White MF. 2020. Fuse to defuse: a self-limiting ribonuclease-ring nuclease fusion for type III CRISPR defence. Nucleic Acids Res 48: 6149–6156. 10.1093/nar/gkaa298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Frangos A, Hall LN, Nemudraia A, Nemudryi A, Krishna P, Wiegand T, Wilkinson RA, Snyder DT, Hedges JF, Jutila MA, et al. 2020. Intrinsic signal amplification by type-III CRISPR-Cas systems provides a sequence-specific viral diagnostic. medRxiv. 10.1101/2020.10.14.20212670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin GB, Ramliden MS, Hawver LA, Wang K, Pell ME, Kieninger A-K, Khataokar A, O'Hara BJ, Behrmann LV, Neiditch MB, et al. 2018. Direct activation of a phospholipase by cyclic GMP-AMP in El Tor Vibrio cholerae. Proc Natl Acad Sci 115: E6048–E6055. 10.1073/pnas.1801233115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Alkhnbashi OS, Behler J, Han W, She Q, Hess WR, Garrett RA, Backofen R. 2019. Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-Cas gene cassettes reveals 39 new cas gene families. RNA Biol 16: 530–542. 10.1080/15476286.2018.1483685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard NF, Glover CVC, Terns RM, Terns MP. 2016. The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 22: 216–224. 10.1261/rna.039842.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov SA, Makarova KS, Wolf YI, Severinov K V, Koonin EV. 2018. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci 115: E5307–E5316. 10.1073/pnas.1803440115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalakyte D, Kazlauskiene M, Havelund J F, Rukše˙naite˙ A, Rimaite A, Tamulaitiene G, Færgeman NJ, Tamulaitis G, Siksnys V. 2020. Type III-A CRISPR-associated protein Csm6 degrades cyclic hexa-adenylate activator using both CARF and HEPN domains. Nucleic Acids Res 48: 9204–9217. 10.1093/nar/gkaa634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofos N, Feng M, Stella S, Pape T, Fuglsang A, Lin J, Huang Q, Li Y, She Q, Montoya G. 2020. Structures of the Cmr-β complex reveal the regulation of the immunity mechanism of type III-B CRISPR-Cas. Mol Cell 79: 741–757.e7. 10.1016/j.molcel.2020.07.008 [DOI] [PubMed] [Google Scholar]
- Sridhara S, Rai J, Whyms C, Woodside W, Terns MP, Li H. 2021. Structure and function of an in vivo assembled type III-A CRISPR-Cas complex reveal critical roles of dynamics in activity control. bioRxiv. 10.1101/2021.01.27.428455 [DOI] [Google Scholar]
- Staals RHJ, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. 2013. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 52: 135–145. 10.1016/j.molcel.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RHJ, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, et al. 2014. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell 56: 518–530. 10.1016/j.molcel.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steens JA, Zhu Y, Taylor DW, Bravo JPK, Prinsen SHP, Schoen CD, Keijser BJF, Ossendrijver M, Hofstra LM, Brouns SJJ, et al. 2021. SCOPE: flexible targeting and stringent CARF activation enables type III CRISPR-Cas diagnostics. bioRxiv. 10.1101/2021.02.01.429135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, et al. 2015. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol 194: 1819–1831. 10.4049/jimmunol.1402495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas Č, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. 2014. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell 56: 506–517. 10.1016/j.molcel.2014.09.027 [DOI] [PubMed] [Google Scholar]
- Topuzlu E, Lawrence CM. 2016. Recognition of a pseudo-symmetric RNA tetranucleotide by Csx3, a new member of the CRISPR associated Rossmann fold superfamily. RNA Biol 13: 254–257. 10.1080/15476286.2015.1130209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee ASY, Mekalanos JJ, et al. 2019. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567: 194–199. 10.1038/s41586-019-0953-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328: 1703–1705. 10.1126/science.1189801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, et al. 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18: 333–344. 10.1016/j.chom.2015.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Guo W, Yuan YA. 2015. Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease. RNA Biol 12: 749–760. 10.1080/15476286.2015.1051300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Lau RK, Mathews IT, Birkholz EA, Watrous JD, Azimi CS, Pogliano J, Jain M, Corbett KD. 2020a. HORMA domain proteins and a trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol Cell 77: 709–722.e7. 10.1016/j.molcel.2019.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Zhao X, Liu J, Zeng Z, Zhang Z, Liu T, Li Y, Han W, Peng N. 2020b. CRISPR-associated factor Csa3b regulates CRISPR adaptation and Cmr-mediated RNA interference in Sulfolobus islandicus. Front Microbiol 11: 2038. 10.3389/fmicb.2020.02038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Ma J, Wang J, Artamonova D, Wang M, Liu L, Xiang H, Severinov K, Zhang X, Wang Y. 2019. Structure studies of the CRISPR-Csm complex reveal mechanism of co-transcriptional interference. Cell 176: 239–253.e16. 10.1016/j.cell.2018.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. 2012. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 45: 303–313. 10.1016/j.molcel.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Yang Y, Zheng F, Zeng Z, Feng W, Jin X, Wang J, Yang K, Liang YX, She Q, et al. 2020. A membrane-associated DHH-DHHA1 nuclease degrades type III CRISPR second messenger. Cell Rep 32: 108133. 10.1016/j.celrep.2020.108133 [DOI] [PubMed] [Google Scholar]
- Zhu W, McQuarrie S, Grüschow S, McMahon SA, Graham S, Gloster TM, White MF. 2021. The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence. Nucleic Acids Res 49: 2777–2789. 10.1093/nar/gkab073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink IA, Wimmer E, Schleper C. 2020. Heavily armed ancestors: CRISPR immunity and applications in archaea with a comparative analysis of CRISPR types in sulfolobales. Biomolecules 10: 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]