Abstract
Mammalian C-to-U RNA editing was described more than 30 yr ago as a single nucleotide modification in small intestinal Apob RNA, later shown to be mediated by the RNA-specific cytidine deaminase APOBEC1. Reports of other examples of C-to-U RNA editing, coupled with the advent of genome-wide transcriptome sequencing, identified an expanded range of APOBEC1 targets. Here we analyze the cis-acting regulatory components of verified murine C-to-U RNA editing targets, including nearest neighbor as well as flanking sequence requirements and folding predictions. RNA secondary structure of the editing cassette was associated with editing frequency and exhibited minimal free energy values comparable to small nuclear RNAs. We summarize findings demonstrating the relative importance of trans-acting factors (A1CF, RBM47) acting in concert with APOBEC1. Cofactor dominance was associated with editing frequency, with RNAs targeted by both RBM47 and A1CF edited at a lower frequency than RBM47-dominant targets. Using this information, we developed a multivariable linear regression model to predict APOBEC1 dependent C-to-U RNA editing efficiency, incorporating factors independently associated with editing frequencies based on 103 Sanger-confirmed editing sites, which accounted for 84% of the observed variance. This model also predicted a composite score for available human C-to-U RNA targets, which again correlated with editing frequency.
Keywords: RNA folding, A1CF, RBM47
INTRODUCTION
Mammalian C-to-U RNA editing was identified as the molecular basis for human intestinal APOB48 production more than three decades ago (Chen et al. 1987; Hospattankar et al. 1987; Powell et al. 1987). A site-specific enzymatic deamination of C6666 to U of Apob mRNA was originally considered the sole example of mammalian C-to-U RNA editing, occurring at a single nucleotide in a 14 kilobase transcript and mediated by an RNA specific cytidine deaminase (APOBEC1) (Teng et al. 1993). Earlier studies identified RNA motifs (Davies et al. 1989) contained within a 26-nt segment flanking the Apob mRNA edited cytidine base or within 55 nt using S100 extracts from rat hepatoma cells (Bostrom et al. 1989; Driscoll et al. 1989). Those, and other studies, established that Apob RNA editing reflects both the tissue/cell of origin as well as RNA elements remote and adjacent to the edited base (Bostrom et al. 1989; Davies et al. 1989). A granular examination of the regions flanking the edited base in Apob RNA demonstrated a critical 3′ sequence 6671–6681, downstream from C6666, in which mutations reduced or abolished editing activity (Shah et al. 1991). This 3′ site, termed a “mooring sequence” was associated with a 27s- “editosome” complex (Smith et al. 1991), which was both necessary and sufficient for site-specific Apob RNA editing and editosome assembly (Backus and Smith 1991). Other cis-acting elements include a 5-nt spacer region between the edited cytidine and the mooring sequence, and also sequences 5′ of the editing site that regulate editing efficiency (Backus and Smith 1992; Backus et al. 1994) along with AU-rich regions both 5′ and 3′ of the edited cytidine that together function in concert with the mooring sequence (Hersberger and Innerarity 1998).
Computational methods using homology matching surrounding the mooring sequence led to the identification of C-to-U RNA editing in human neurofibromatosis type 1 mRNA by endogenously expressed APOBEC1 and its auxiliary factors (Skuse et al. 1996). With the advent of massively parallel RNA sequencing technology, we now appreciate that APOBEC1 mediated RNA editing targets hundreds of sites (Rosenberg et al. 2011; Blanc et al. 2014) mostly within 3′ untranslated regions of mRNA transcripts. These computational methods and RNA sequencing also led to identification of the RNA editing targets for other members of the AID/APOBEC family (Zaranek et al. 2010; Roth et al. 2018). This expanded range of targets of C-to-U RNA editing prompted us to reexamine key functional attributes in the regulatory motifs and cis-acting elements that impact editing frequency, focusing primarily on data emerging from in vivo studies of mouse tissue-specific C-to-U RNA editing.
Advances in our understanding of physiological Apob RNA editing were propelled with the identification of components of the Apob RNA editosome (Sowden et al. 1996). APOBEC1, the catalytic deaminase (Teng et al. 1993) is necessary for physiological C-to-U RNA editing in vivo (Hirano et al. 1996) and in vitro (Giannoni et al. 1994) along with other cofactors likely acting in concert with RNA elements (Sowden et al. 1998). Using the mooring sequence of Apob RNA as bait, two groups identified APOBEC1 complementation factor (A1CF), an RNA-binding protein sufficient in vitro to support efficient editing in the presence of APOBEC1 and Apob mRNA (Lellek et al. 2000; Mehta et al. 2000). Those findings reinforced the importance of both the mooring sequence and an RNA binding component of the editosome in promoting Apob RNA editing. However, while A1CF and APOBEC1 are sufficient to support in vitro Apob RNA editing, neither heterozygous (Blanc et al. 2005) or homozygous genetic deletion of A1cf impaired Apob RNA editing in vivo in mouse tissues (Snyder et al. 2017), suggesting that an alternate complementation factor was likely involved. Other work identified a homologous RNA binding protein, RBM47, that functioned to promote Apob RNA editing both in vivo and in vitro (Fossat et al. 2014), and more recent studies utilizing conditional, tissue-specific deletion of A1cf and Rbm47 indicate that both factors play distinctive roles in APOBEC1-mediated C-to-U RNA editing, including Apob as well as a range of other APOBEC1 targets (Blanc et al. 2019).
These findings together establish important regulatory roles for both cis-acting elements and trans-acting factors in C-to-U mRNA editing. However, the majority of studies delineating cis-acting elements reflect earlier in vitro experiments, many in artificial cell systems using fragments of Apob mRNA, and relatively little is known regarding the role of cis-acting elements in tissue-specific C-to-U RNA editing of other transcripts, particularly in a physiological setting in vivo. Here we use statistical modeling to investigate the independent roles of candidate regulatory factors in mouse C-to-U mRNA editing using data from in vivo studies from over 170 editing sites in 119 transcripts (Meier et al. 2005; Rosenberg et al. 2011; Gu et al. 2012; Blanc et al. 2014, 2019; Rayon-Estrada et al. 2017; Snyder et al. 2017; Kanata et al. 2019). We also examined these regulatory factors in known human mRNA targets (Chen et al. 1987; Powell et al. 1987; Skuse et al. 1996; Mukhopadhyay et al. 2002; Grohmann et al. 2010; Schaefermeier and Heinze 2017).
RESULTS
Descriptive data
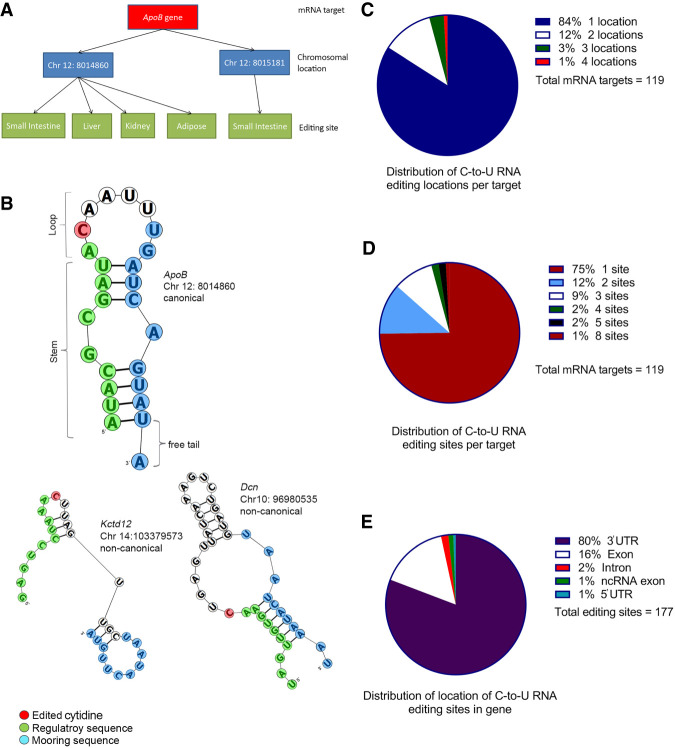
Primary records (N = 528) from our review literature were screened for relevance and in vivo studies reporting editing frequencies of individual or transcriptome-wide APOBEC1-dependent C-to-U mRNA targets selected, using a threshold of 10% editing frequency. For analyses based on RNA sequence information, only targets with available sequence information or chromosomal location for the edited cytidine were included. Exclusion criteria included: studies that reported C-to-U mRNA editing frequencies of target genes in other species, studies reporting editing frequencies of target genes in animal models overexpressing APOBEC1, in vitro studies, and conference abstracts. For each RNA target, chromosomal and strand location of the edited cytidine, tissue site, and secondary structure were determined among other characteristics (Fig. 1A,B). 177 C-to-U RNA editing sites were identified based on eight studies that met inclusion and exclusion criteria (Meier et al. 2005; Rosenberg et al. 2011; Gu et al. 2012; Blanc et al. 2014, 2019; Rayon-Estrada et al. 2017; Snyder et al. 2017; Kanata et al. 2019), representing 119 distinct RNA editing targets. 84% (100/119) of RNA targets were edited at one chromosomal location (Fig. 1C) and 75% (89/119) of mRNA targets were edited at both a single chromosomal location and also within a single tissue (Fig. 1D). The majority of editing sites occur in the 3′ untranslated region (142/177; 80%) as previously noted (Rosenberg et al. 2011; Blanc et al. 2014), with exonic editing sites the next most abundant subgroup (28/177; 16%, Fig. 1E). 103/177 editing sites were confirmed by Sanger sequencing, with a mean editing frequency of 37 ± 22%.
FIGURE 1.
Characteristics of murine APOBEC1-mediated C-to-U mRNA editing sites. (A) Schematic presentation of mRNA target, chromosomal editing location, and editing sites considered. Each mRNA target could be edited at one or more chromosomal location(s) (blue boxes). Each editing location could be edited in one or more tissues, giving rise to one or more editing site(s) per location (green boxes). Editing site(s) of each mRNA target are the sum of editing sites from all editing locations reported for that target. (B) Examples of canonical (Apob chr12: 8014860, top) and two types of noncanonical (Kctd12 chr14: 103379573 and Dcn chr10: 96980535) secondary structures. (C) Distribution of number of chromosomal editing location(s), or targeted cytidine(s), per mRNA target. (D) Distribution of number of total editing sites per mRNA target considering all chromosomal location(s) edited at different tissue(s). (E) Distribution of location of editing sites within gene structure.
Factors influencing editing frequency
Regulatory-spacer-mooring cassette
Using the mooring sequence model (Backus and Smith 1992), three cis-acting elements were considered for each RNA editing site: These elements included (i) a 10-nt segment immediately upstream of the edited cytidine “regulatory sequence”; (ii) a 10-nt segment downstream from the edited cytidine with complete or partial consensus with the canonical “mooring sequence” of Apob mRNA; (iii) the sequence between the edited cytidine and the 5′ end of the mooring sequence, referred to as “spacer.” We used an unbiased approach to identify potential mooring sequences by taking the nearest segment to the edited cytidine with the lowest number of mismatch(es) compared to the canonical mooring sequence of ApoB RNA. For each of the three segments, we investigated the number of mismatches compared to the corresponding segment of the Apob gene (Blanc et al. 2014) as well as length of spacer, the abundance of A and U nucleotides (AU content) and the G to C abundance ratio (G/C fraction, Arbab et al. 2020).
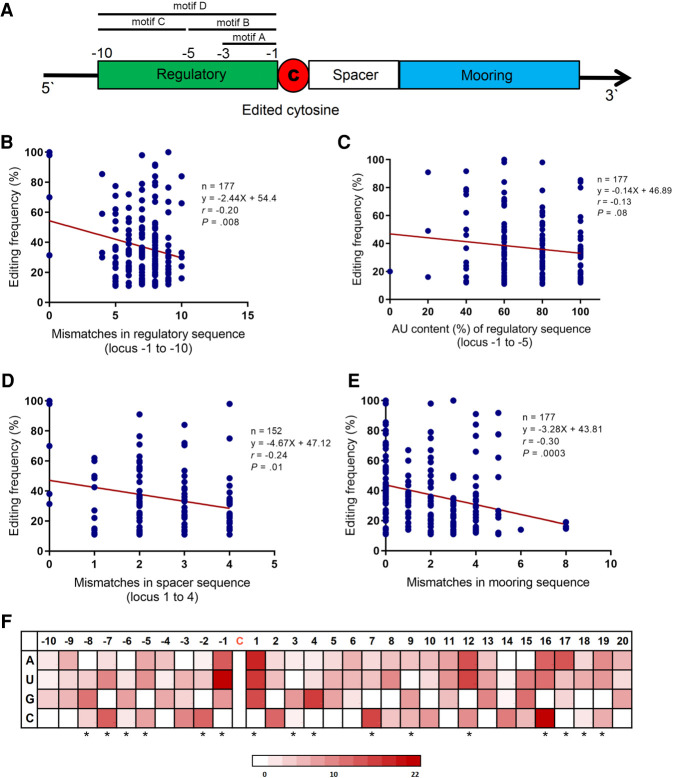
Although no significant associations were found between editing frequency and mismatches in regulatory motif A or motif B (Supplemental Fig. 1), we found that mismatches in motif C and D negatively impacted editing frequency (r = −0.24, P = 0.001) (motif D r = −0.20, P = 0.008, Fig. 2B). AU content of motif B showed a (nonsignificant) trend toward negative association with editing frequency (r = −0.13, P = 0.08 Fig. 2C), but AU content of motifs A, C, and D did not impact editing frequency (Supplemental Fig. 1). The abundance of G (r = 0.17, P = 0.02) and G/C fraction (r = 0.14, P = 0.04) in motif C showed significant associations with editing frequency and the abundance of C in motif B (r = 0.13, P = 0.08) exhibited a (nonsignificant) trend toward positive association. The spacer sequence averaged 5 ± 4 nt with a (nonsignificant) trend toward negative association between length and editing frequency (r = −0.14, P = 0.09). The spacer AU content was not significantly associated with editing frequency (Supplemental Fig. 2). However, G abundance (r = −0.23, P = 0.01) and G/C fraction (r = −0.20, P = 0.03) of spacer showed significant associations with editing frequency in Sanger-confirmed targets. The mismatches in the first 4 nt of the spacer exerted a significant negative impact on editing frequency (r = −0.24, P = 0.01) (Fig. 2D). Similarly, the mismatches in the mooring sequence showed a significant negative association with editing frequency (r = −0.30, P = 0.0003, Fig. 2E). The base content of individual nucleotides surrounding the edited cytidine showed significant associations with editing frequency, which was more emphasized in nucleotides closer to the edited cytidine (Fig. 2F; Supplemental Table 1). Furthermore, overall AU content of downstream sequence +16 to +20 had positive impact on editing frequency (r = 0.17, P = 0.02) (Supplemental Fig. 2). The positive influence of AU content was stronger in the subgroup of editing sites with 2 or more bases in the sequence +16 to +20 as part of the dsRNA region (r = 0.21, P = 0.02). However, G abundance in downstream 20 nt (r = −0.24, P = 0.01) showed significant negative association with editing frequency in Sanger-confirmed targets.
FIGURE 2.
Characteristics of regulatory-spacer-mooring cassette and base content of individual nucleotides flanking edited cytidine in association with editing frequency. (A) Schematic illustration of regulatory-spacer-mooring cassette. Four motifs were defined for regulatory sequence: motif A for nucleotides −1 to −3; motif B for nucleotides −1 to −5; motif C for nucleotides −6 to −10; motif D representative of the whole sequence. (B) Association of the mismatches in motif D of regulatory sequence with editing frequency. (C) Association between the AU content (%) of regulatory sequence (motif B) and editing frequency. (D) Association of the mismatches in spacer (nucleotides +1 to +4 downstream from the edited cytidine) with editing frequency. (E) Association of the mismatches in mooring sequence with editing frequency. (F) Heatmap plot illustrating the association between base content of 30 nt flanking the edited cytidine with editing frequency. Red color density in each cell represents the β coefficient value of corresponding base in the multivariable linear regression model fit including that nucleotide. The asterisks refer to the nucleotides that were retained in the final model. Mismatches in regulatory, spacer, and mooring sequences were determined in comparison to the corresponding sequences in Apob mRNA (as reference). (r) Pearson correlation coefficient.
Secondary structure
We generated a predicted secondary structure for 172 editing sites, with four subgroups based on overall structure and location of the edited cytidine: loop (Cloop), stem (Cstem), tail (Ctail), and noncanonical structure (NC). The majority of editing sites were in the Cloop subgroup (59%), followed by Cstem (20%), Ctail (13%), and NC (8%) subgroups (Fig. 3A). Editing sites in the Ctail subgroup exhibited lower editing frequencies compared to editing sites in Cloop (29 ± 12 vs. 41 ± 23%, P = 0.02) or Cstem (37 ± 21%, P = 0.04) subgroups. No significant differences were detected in other comparisons (Fig. 3B). The edited cytidine was located in loop, stem, and tail of the secondary structure in 110 (64%), 38 (22%), and 24 (14%) of the edited RNAs, respectively. Editing sites with the edited cytidine within the loop exhibited significantly higher editing frequency compared to those with the edited cytidine in the tail (40 ± 24% vs. 28 ± 12%, P = 0.04). Other subgroups exhibited comparable editing frequencies (Supplemental Fig. 3). The majority (78%) of editing sites contained a mooring sequence located in the main stem–loop structure (Fig. 3C), with the remainder located in the tail or secondary loop. Average editing efficiency was significantly higher in targets where the mooring sequence was located in the main stem–loop (Fig. 3D). We also calculated the proportion of total nucleotides that constitute the main stem–loop in the secondary structure. The average ratio was 0.62 ± 0.18 ranging from 0.28 to 1 (Supplemental Table 2) with higher ratios associated with higher editing frequency of the corresponding editing site (r = 0.20, P = 0.007) (Fig. 3E). Finally, we considered the orientation of free tails in the secondary structure in terms of length and symmetry. Symmetric free tails were observed in 59% of editing sites (Supplemental Fig. 3). The length of 5′ free tail showed negative association with editing frequency (r = −0.14, P = 0.04, Fig. 3F) while no significant associations were detected between either the length of 3′ tail or symmetry of tails and editing frequency (Supplemental Fig. 3).
FIGURE 3.
Secondary structure-related features in association with editing frequency. (A) Distribution of different types of overall secondary structure in editing sites. Cloop, Cstem, and Ctail are three subtypes of canonical secondary structure based on the location of the edited cytidine. (B) Association between type of secondary structure and editing frequency. (C) Distribution of the mooring sequence location in editing sites. “Other” refers to mooring sequences located in tail or stem/loop and not part of the main stem–loop structure. (D) Association of mooring sequence location with editing frequency. (E) Association between ratio of main stem–loop bases to total bases count and editing frequency. (F) Association of the 5′ free tail length with editing frequency. (*) P < 0.05; (**) P < 0.001. (r) Pearson correlation coefficient.
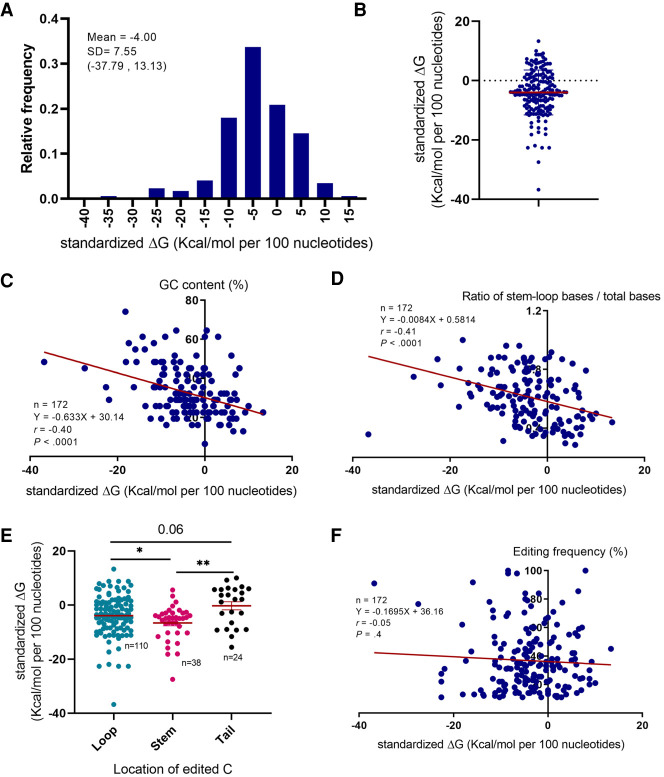
We used Mfold and the RNA-structure software to calculate the minimum Gibbs free energy (MFE also referred to as ΔG) for the secondary structure of the mRNA editing cassettes. Because mRNA strand length significantly influences ΔG (Seffens and Digby 1999; Trotta 2014), we generated standardized ΔG (S-ΔG) by dividing the ΔG by the length of the corresponding mRNA strand in order to normalize ΔGs regardless of the mRNA strand length. We found that the majority of the mRNA editing targets exhibited negative predicted S-ΔGs with a mean value of −4 Kcal/mol per 100 nt (ranging from −38 to13) (Fig. 4A,B). The S-ΔGs exhibit significant inverse correlation with the GC content of the mRNA strands (r = −0.40, P < 0.0001 Fig. 4C). Furthermore, mRNA strands with higher stem–loop base/total base ratios exhibited more negative S-ΔGs (r = −0.41, P < 0.0001 Fig. 4D). Similarly, mRNA strands with their edited cytosine located in the stem–loop part of the secondary structure exhibited more negative S-ΔGs (Fig. 4E). These observations are in line with earlier work showing that mRNA strands with higher GC content and those with base-pair stacking in their secondary structure exhibit more negative ΔGs (Seffens and Digby 1999; Trotta 2014). On the other hand, the S-ΔG did not exhibit a significant association with the editing frequencies of mRNA targets (r = −0.05, P = 0.4 Fig. 4F). The association of S-ΔG with both editing-promoting (stem–loop base/total ratio) and -inhibitory (GC content) factors may explain the overall null association of the S-ΔG with editing frequency. Earlier work suggested MFE density (MFEden) as an alternative strategy to adjust the ΔG (i.e., MFE) for the length of mRNA strand (Trotta 2014). Accordingly, we calculated MFEden and found that the majority of the mRNA editing targets exhibit negative MFEden (Supplemental Fig. 4A,B). The association of MFEden with other characteristics of mRNA editing targets also followed the same pattern as S-ΔG (Supplemental Fig. 4C–E). As noted above, we found no significant association between MFEden and the editing efficiency of mRNA targets (Supplemental Fig. 4F).
FIGURE 4.
Secondary structure standardized minimum free energy. (A,B) Distribution of the RNA editing sites based on the standardized minimal free Gibbs energy of the editing cassette secondary structure (S-ΔG). (C) Association of S-ΔG with GC content of the editing cassette. (D) Association between S-ΔG and ratio of main stem–loop bases to total bases count. (E) Association of the edited cytosine location with S-ΔG. (F) Association of S-ΔG with editing frequency.
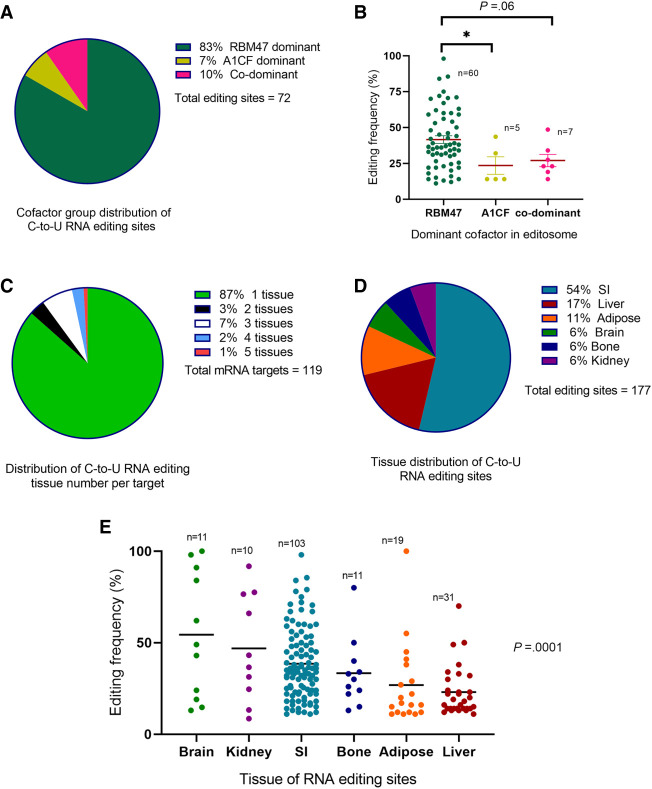
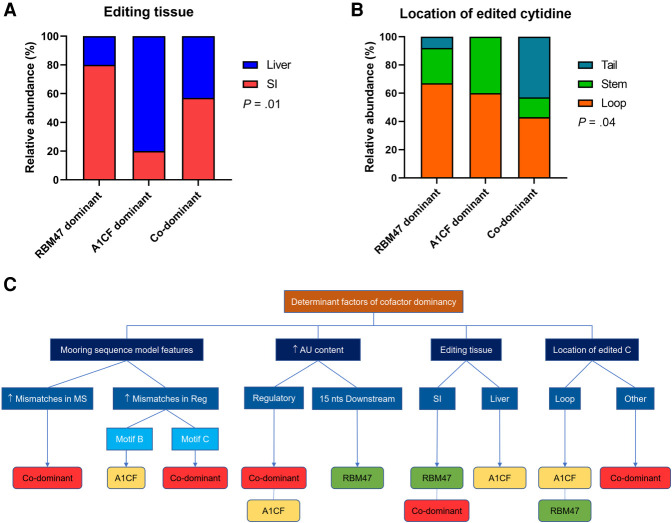
Trans-acting factors and tissue specificity: Data for relative dominance of cofactors in APOBEC1- dependent RNA editing were available for 72 editing sites for targets in small intestine or liver (Blanc et al. 2019). Cofactor dominancy was determined based on the relative contribution of each cofactor to editing frequency. In each editing site, editing frequencies in mouse tissues deficient in A1cf or Rbm47 were compared to that of wild-type mice. The relative contribution of each cofactor was calculated by subtracting the editing frequency for each target in A1cf or Rbm47 knockout tissue from the total editing frequency in wild-type control. Editing sites with <20% difference between contributions of RBM47 and A1CF were considered codominant. Sites with ≥20% difference were considered either RBM47- or A1CF-dominant, depending on the cofactor with higher contribution (Blanc et al. 2019). RBM47 was identified as the dominant factor in 60/72 (83%) sites; A1CF was the dominant factor in 5/72 (7%) editing sites with the remaining sites (7/72; 10%), exhibiting equal dominancy (Fig. 5A). The average editing frequencies at editing sites revealed differences across the groups with 41 ± 20% in RBM47-dominant targets, 23 ± 14% in A1CF-dominant, and 27 ± 11% in the codominant group (P = 0.03) (Fig. 5B). The majority of RNA editing targets were edited in one tissue (103/119; 86% Fig. 5C), while the maximum number of tissues in which an editing target is edited (at the same site) is 5 (Cd36). The small intestine harbors the highest number of verified editing sites (95/177; 54%), followed by liver (31/177; 17%), and adipose tissue (19/177; 11% Fig. 5D). Editing sites found in more than one tissue were counted separately in examining the tissue distribution. Sites edited in brain tissue showed the highest average editing frequency (54 ± 35%, n = 11), followed by bone marrow myeloid cells (50 ± 22%, n = 4), and kidney (47 ± 29%, n = 10, P = 0.0001) (Fig. 5E).
FIGURE 5.
Dominance and tissue-specific cofactor patterns among editing sites. (A) Distribution of dominant cofactor in editosomes of editing sites. (B) Association of dominant cofactor with editing frequency. (C) Distribution of number of editing tissue(s) per mRNA target. (D) Tissue distribution of editing sites. (E) Average editing frequency of editing sites edited at different tissues. SI, small intestine.
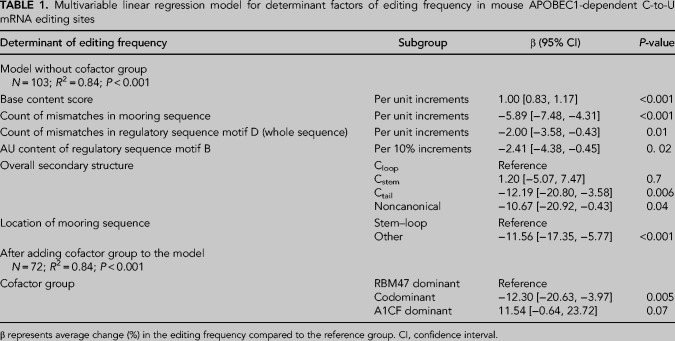
We then developed a multivariable linear regression model to predict APOBEC1 dependent C-to-U RNA editing efficiency, incorporating factors independently associated with editing frequencies (Table 1). This model, based on 103 Sanger-confirmed editing sites with available data for all of the parameters mentioned, accounted for 84% of variance in editing frequency of editing sites included (R2 = 0.84, P < 0.001, Table 1). The final multivariable model revealed several factors independently associated with editing frequency. These included the number of mismatches in mooring sequence; regulatory sequence motif D; AU content of regulatory sequence motif B; overall secondary structure for group Ctail versus group Cloop; location of mooring sequence in secondary structure; “base content score” parameter that represents base content of the sequences flanking edited cytidine (Table 1). Removing “base content score” from the model reduced the power from R2 = 0.84 to R2 = 0.59. Next, we added a cofactor dominance variable and fit the model using the 72 editing sites with available data for cofactor dominance. Along with other factors mentioned above, cofactor dominance showed significant association with editing frequency (Table 1) with RNAs targeted by both RBM47 and A1CF observed to be edited at a lower frequency than RBM47-dominant targets.
TABLE 1.
Multivariable linear regression model for determinant factors of editing frequency in mouse APOBEC1-dependent C-to-U mRNA editing sites
Factors associated with cofactor dominance (Fig. 6; Supplemental Table 3; Supplemental Fig. 5), included tissue-specificity, with higher frequency of RBM47-dominant sites in small intestine compared to liver (91 vs. 63%, P = 0.008) and A1CF-dominant and codominant editing sites more prevalent in liver. The number of mooring sequence mismatches also varied among three subgroups: 1.1 ± 1.3 in RBM47-dominant subgroup; 2.0 ± 2.5 in A1CF-dominant subgroup; and 2.9 ± 0.4 in codominant subgroup (P = 0.004). This was also the case regarding mismatches in the spacer: 2.4 ± 1.2 in RBM47-dominant subgroup; 2.7 ± 1.5 in A1CF-dominant subgroup; 3.8 ± 0.4 in codominant subgroup (P = 0.02). AU content (%) of downstream sequence +6 to +10 was higher in RBM47-dominant subgroup (P = 0.01). Finally, the location of the edited cytidine in secondary structure of mRNA strand was different across three subgroups (P = 0.04, Fig. 6). We used pairwise multinomial logistic regression to determine factors independently associated with cofactor dominance (Fig. 6C; Supplemental Table 4). Ctail editing sites, those with more mismatches in mooring and regulatory motif C, lower AU content in downstream sequence, and higher AU content in regulatory motif D were more likely codominant. Editing sites from small intestine and those with higher AU content of downstream sequence were more likely RBM47-dominant. Editing sites from liver and those with higher mismatches in regulatory motif B were more likely A1CF-dominant (Fig. 6C).
FIGURE 6.
Cofactor pattern and tissue-specific role in murine C-to-U mRNA editing sites. (A) Distribution of editing tissue across subgroups of editing sites with different dominant cofactor patterns. (B) Location of edited cytidine in secondary structure of editing sites with different dominant cofactor patterns. (C) Schematic presentation of factors that correlate with dominant cofactor pattern in editing sites. This graph is based on the findings derived from pairwise multinomial logistic regression models.
Comparison of base content of sequences flanking edited and mutated cytidines
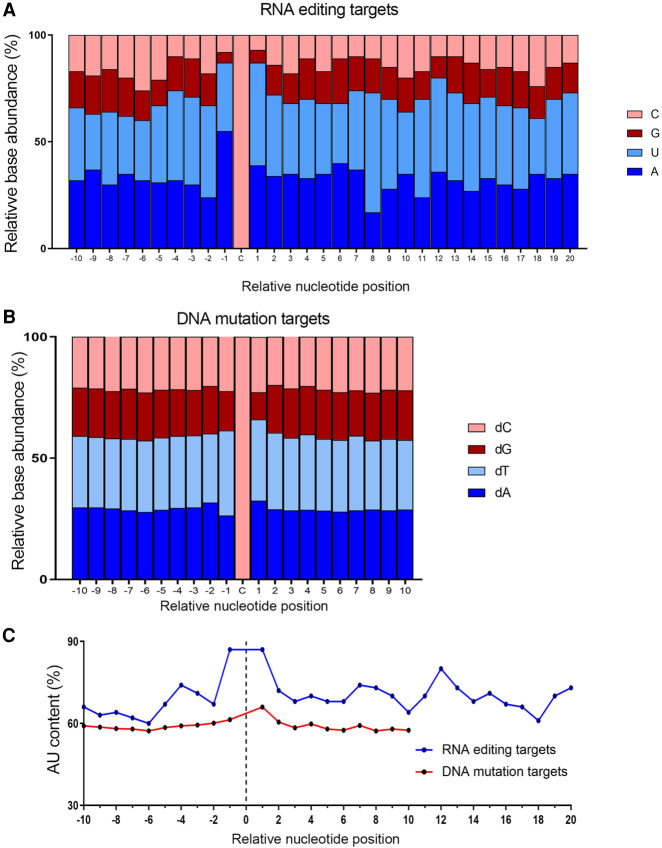
AU content was enriched (∼87%) in nucleotides both immediately upstream and downstream from the edited cytidine across mouse RNA editing targets (Fig. 7A,C). The average AU content across the region 10 nt upstream to 20 nt downstream from the edited cytidine was ∼70% (60%–87%). Because APOBEC1 has been shown to be a DNA mutator (Harris et al. 2003; Wolfe et al. 2019, 2020), we also determined the AU content of the mutated deoxycytidine (C to X) region flanking over 6000 human DNA targets (Nik-Zainal et al. 2012) to be ∼66% at a site 1 nt downstream from the edited base (Fig. 7B,C). The average AU content in the sequence 10 nt upstream and 10 nt downstream from mutated deoxycytidines is 59% (57%–66.0%). These DNA mutation targets reside in both coding and noncoding regions, with no relationship to the RNA editing sites discussed. The average AU content was 90% and 80% in nucleotides immediately upstream and downstream, respectively, of the targeted deoxycytidine in a subgroup of over 700 DNA editing events of the C to T type (Nik-Zainal et al. 2012), which is closer to the distribution found in C to U RNA editing targets. These features suggest that AU enrichment in nucleotides immediately flanking the modified cytosine is an important component to editing function of APOBEC1 on both RNA and DNA targets, especially for the C/dC to U/dT change.
FIGURE 7.
Base content of sequences flanking modified cytidine in RNA editing and DNA mutation targets. (A) Base content of 10 nt upstream and 20 nt downstream from edited cytidine in mouse APOBEC1-mediated C-to-U mRNA editing targets. (B) Base content of 10 nt upstream and 10 nt downstream from mutated cytidine in proposed human APOBEC-mediated DNA mutation targets in patients with breast cancer. (C) Comparison of AU base content (%) of nucleotides flanking modified cytidine in RNA editing targets and DNA mutation targets in mouse and human breast cancer patients, respectively.
Human mRNA targets
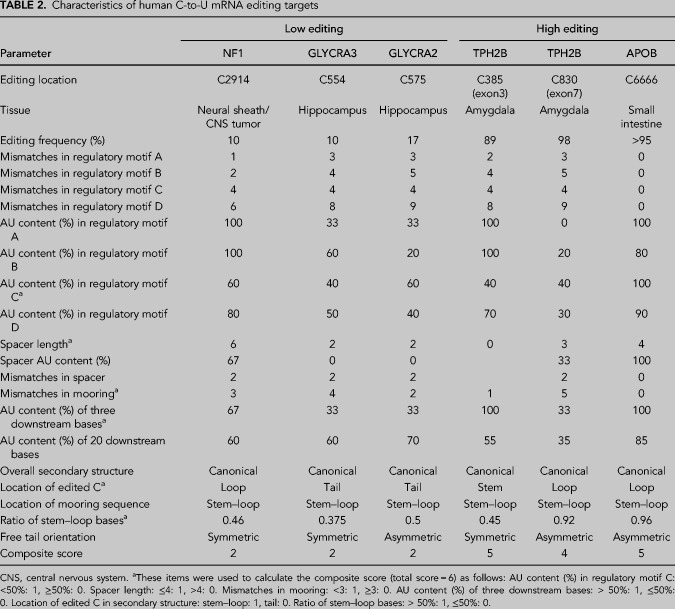
Finally, we turned to an analysis of human C-to-U RNA editing targets for which this same panel of parameters was available (Table 2). Aside from APOB RNA, which is known to be edited in the small intestine (Chen et al. 1987; Powell et al. 1987), other targets have been identified in central or peripheral nervous tissue (Skuse et al. 1996; Mukhopadhyay et al. 2002; Meier et al. 2005; Schaefermeier and Heinze 2017). The human targets were categorized into low editing (NF1, GLYRα2, GLYRα3) and high editing (APOB, TPH2B exon3, TPH2B exon7) subgroups using 20% as cut-off. A composite score (maximum = 6) was generated based on six parameters introduced in the mouse model with notable variance between the two subgroups including mismatches in mooring sequence, spacer length, location of the edited cytidine, and relative abundance of stem–loop bases (Table 2). High editing targets exhibited a significantly higher composite score (4.7 vs. 2, P = 0.001) compared to low editing targets and the composite score significantly correlated with editing frequency in individual targets (r = 0.95, P = 0.005). The canonical editing target APOB (Chen et al. 1987; Powell et al. 1987) achieved a score of 5 (out of 6), reflecting the observation that one of the six parameters (AU% of regulatory motifs) in human APOB is nonpreferential compared to the editing-promoting features identified in the mouse multivariable model.
TABLE 2.
Characteristics of human C-to-U mRNA editing targets
DISCUSSION
The current study reflects our analysis of 177 C-to-U RNA editing sites from 119 target mRNAs, with the majority residing within the 3′ untranslated region. Our multivariable model identified several key factors influencing editing frequency, including host tissue, base content of nucleotides surrounding the edited cytidine, number of mismatches in regulatory and mooring sequences, AU content of the regulatory sequence, overall secondary structure, location of the mooring sequence, and cofactor dominance. These factors, each exerting independent effects, together accounted for 84% of the variance in editing frequency. Our findings also showed that mismatches in the mooring and regulatory sequences, AU content of regulatory and downstream sequences, host tissue and secondary structure of target mRNA were associated with the pattern of cofactor dominance. Several aspects of these primary conclusions merit further discussion.
Previous studies investigating the key factors that regulate C-to-U mRNA editing were confined to in vitro studies and predicated on a single mRNA target (Apob) (Backus and Smith 1991, 1992; Shah et al. 1991; Smith et al. 1991; Hersberger and Innerarity 1998). With the expanded range of verified C-to-U RNA editing targets now available for interrogation, we revisited the original assumptions to understand more globally the determinants of C-to-U mRNA editing efficiency. In undertaking this analysis, we were reminded that the requirements for C-to-U mRNA editing in vitro often appear more stringent than in vivo (Backus and Smith 1991; Shah et al. 1991), which further emphasizes the importance of our findings. In addition, our approach included both cis-acting sequence- and folding-related predictions along with the role of trans-acting factors and took advantage of statistical modeling to adjust for confounding or modifier effects between these factors to identify their role in editing frequency.
We began with the assumptions established for Apob RNA editing which identified a 26-nt segment encompassing the edited base, spacer, mooring sequence, and part of regulatory sequence as the minimal sequence competent for physiological editing in vitro and in vivo (Davies et al. 1989; Shah et al. 1991; Backus and Smith 1992). Those studies identified an 11-nt mooring sequence as essential and sufficient for editosome assembly and site-specific C-to-U editing (Backus and Smith 1991, 1992; Shah et al. 1991) and established optimal positioning of the mooring sequence relative to the edited base in Apob RNA (Backus and Smith 1992). The current work supports the key conclusions of this original mooring sequence model as applied to the entire range of C-to-U RNA editing targets. We observed that mismatches in either the mooring or regulatory sequences were independent factors governing editing frequency. In contrast, while mismatches in the spacer sequence also showed negative association with editing frequency, the impact of spacer mismatches was not retained in the final model, nor was the length of the spacer associated with editing frequency. Furthermore, we found mismatches in the regulatory sequence motif C to be more important than mismatches in motif B. These inconsistencies might conceivably reflect the context in which an RNA segment is studied (Backus and Smith 1992). For example, our analysis reflects physiological conditions in which naturally occurring mRNA targets are edited, while the aforementioned study used in vitro data based on varying lengths of Apob mRNA embedded within different mRNA contexts (Apoe RNA) (Backus and Smith 1992).
Earlier work revealed that not all mooring sequences support RNA editing (Skuse et al. 1996). Therefore, in addition to the components of the mooring sequence model, we examined variations in the base content in different segments/motifs as well as among individual nucleotides surrounding the edited cytidine. As expected, we found that sequences flanking the edited cytidine exhibited high AU content. We further observed a similarly high AU content in the flanking sequences of a range of proposed APOBEC-mediated DNA mutation targets in human cancer tissues and cell lines (Alexandrov et al. 2013; Petljak et al. 2019), especially in targets with dC/dT change (Nik-Zainal et al. 2012). This observation implies that APOBEC-mediated DNA and RNA editing frequency may each be functionally modified by AU enrichment in the flanking sequences surrounding modifiable bases. The base content in individual nucleotides surrounding the edited cytidine also exerted significant impact on editing frequency, particularly in a 10-nt segment spanning the edited cytidine (Supplemental Table 1), accounting for 25% of the variance in editing frequency independent of the mooring sequence model. Our findings regarding individual nucleotides surrounding the edited cytidine are consistent with findings for both DNA and RNA editing targets, particularly in the setting of cancers (Backus and Smith 1992; Conticello 2012; Roberts et al. 2013; Saraconi et al. 2014; Gao et al. 2018; Arbab et al. 2020). Recent work examining the sequence-editing relationship of a large in vitro library of DNA targets edited by a different synthetic cytidine base editor (CBE)s (Arbab et al. 2020) showed that the base content of a 6-nt window spanning the edited cytidine explained 23%–57% of the editing variance, in particular 1 or 2 nt immediately 5′ of the edited nucleotide. That study also demonstrated that occurrence of T and C nucleotides at the position −1 increased, while a G nucleotide at that position decreased editing frequency (Arbab et al. 2020). However, in contrast to our findings, the presence of A at position −1 had either a negative or null effect on DNA editing activity (Arbab et al. 2020). This latter finding is consistent with the lower AU content observed in nucleotides adjacent to the edited cytidine in APOBEC1 DNA targets compared to the AU content in RNA targets. Our findings assign a greater importance of adjacent nucleotides in RNA editing frequency, similar to earlier reports that the five bases immediately 5′ of the edited cytidine in Apob mRNA exert a greater impact on editing activity compared to nucleotides further upstream of this segment (Backus and Smith 1991, 1992; Shah et al. 1991).
G/C fraction of a 6-nt window spanning the edited cytidine in DNA targets is associated with editing activity of the synthetic CBEs (Arbab et al. 2020). Similarly, we found that the G/C content of the region 20 nt downstream exerts a negative impact, while the AU content of the region 15–20 nt downstream exerts a positive impact on C-to-U RNA editing. Using NMR-based structural analysis, Maris et al proposed that A1CF melts the structured double-stranded stem of the RNA substrate and exposes the unfolded RNA to the APOBEC-1 catalytic site (Maris et al. 2005). Strong G:C hydrogen bonds are proposed to impair the ability of A1CF to unfold the RNA and might explain the negative association we observed between downstream sequence G content and RNA editing efficiency.
The conserved 26-nt sequence around the edited C forms a stem–loop secondary structure, where the editing site is in an octa-loop (Richardson et al. 1998) as predicted for the 55-nt sequence of Apob mRNA (Shah et al. 1991) and confirmed by others (Navaratnam et al. 1993; Hersberger et al. 1999) Mutations resulting in loss of base-pairing in peripheral parts of the stem did not impact the editing frequency (Shah et al. 1991). Editing sites with the cytidine located in central parts (e.g., loop) exhibited higher editing frequencies than those with the edited cytidine located in peripheral parts (e.g., tail) and it is worth noting that the computer-based stem–loop structure was independently confirmed by NMR studies of a 31-nt human APOB mRNA (Maris et al. 2005). Those studies demonstrated that the location of the mooring sequence in the APOB mRNA secondary structure plays a critical role in the RNA recognition by A1CF (Maris et al. 2005). In line with those findings, the current findings emphasize that the location of the mooring sequence in secondary structure of the target mRNA exerts significant independent impact on editing frequency. These predictions were confirmed in crystal structure studies of the carboxyl-terminal domain of APOBEC-1 and its interaction with cofactors and substrate RNA (Wolfe et al. 2020). Our conclusions regarding murine C-to-U editing frequency, such as mooring sequence, base content, and secondary structure appear consistent with a similar regulatory role among the smaller number of verified human targets. That being said, further study and expanded understanding of the range of C-to-U editing targets in human tissues will be needed as recently suggested (Destefanis et al. 2020), analogous to that for A-to-I editing (Bahn et al. 2012; Bazak et al. 2014).
There is continued uncertainty regarding the role of secondary structure for APOBEC1-dependent C-to-U mRNA editing, with findings showing the sequence flanking the Apob editing site contains no predictable stable secondary structure (Smith 1998). Some of this controversy reflects limitations of these earlier in vitro experiments using a single artificial RNA target in cell-free systems. We found that the regulatory-spacer-mooring cassette of the majority of mRNA editing targets exhibit negative S-ΔG and MFEden. Given an average GC content of 30% and an MFEden range of −46 to 21 Kcal/mol, these RNA editing targets exhibit similarity to snRNAs such as H/ACA box RNAs (Trotta 2014), which contain an evolutionarily conserved secondary structure (Ganot et al. 1997), similar to the stem–loop or hairpin-like canonical secondary structure observed in the Apob mRNA editing cassette. RNA stem–loops provide necessary structure for the recognition of RNA by various functional proteins, such as R17 phage coat, the iron-responsive binding protein, and several splicing proteins (Navaratnam et al. 1993). H/ACA box RNAs also interact with RNA binding proteins (Ganot et al. 1997), implying shared features of a conserved stem–loop RNA structure in post-transcriptional events including RNA editing.
We recognize that other factors likely contribute to the variance in RNA editing frequency not covered by our model. We did not consider the role of naturally occurring Apobec1 variants which may be relevant since mutant alleles were shown to modify the editing activity of related hybrid DNA cytosine base editors (Arbab et al. 2020). Furthermore, genetic variants of APOBEC1 in humans were associated with altered frequency of GlyR editing (Kankowski et al. 2017). In addition, we did not consider variants in A1CF alternative splicing, which have been implicated in regulating Apob RNA editing efficiency (Sowden et al. 2004). Other factors not included in our approach included tertiary structure of the mRNA target and other regulatory cofactors. Another limitation in the tissue-specific designation used to categorize editing frequency is that cell specific features of RNA binding protein distribution and editing frequency may have been overlooked (Brannan et al. 2021). This is a relevant concern because small intestinal and liver preparations are a heterodisperse blend of cell types (MacParland et al. 2018; Elmentaite et al. 2020) and tumor tissues are highly heterogeneous in cellular composition (Barker et al. 2009). The current findings provide a platform for future approaches to resolve these questions.
MATERIALS AND METHODS
Search strategy
A comprehensive literature review from 1987 (when Apob RNA editing was first reported, Chen et al. 1987; Powell et al. 1987) to November 2020, using studies published in English reported C-to-U mRNA editing frequencies of individual or transcriptome-wide target genes. Databases searched included Medline, Scopus, Web of Science, Google Scholar, and ProQuest (for thesis). The references of full texts retrieved were also scrutinized for additional papers not indexed in the initial search.
Human targets
We included studies reporting human C-to-U mRNA targets (Chen et al. 1987; Powell et al. 1987; Skuse et al. 1996; Mukhopadhyay et al. 2002; Grohmann et al. 2010; Schaefermeier and Heinze 2017). We also included work describing APOBEC1-mediated mutagenesis in human breast cancer (Nik-Zainal et al. 2012).
Data extraction
Two reviewers (SS and VB) conducted the extraction process independently and discrepancies were addressed upon consensus and input from a third reviewer (NOD). The parameters were categorized as follows:
General parameters
Gene name (RNA target), chromosomal and strand location of the edited cytidine, tissue site, editing frequency were determined by RNA-seq or Sanger sequencing as illustrated for Apob (Fig. 1A). Editing frequency was highly correlated by both approaches (r = 0.8, P < 0.0001), and where both methodologies were available we used RNA-seq. We also defined relative dominance of editing cofactors (A1CF-dominant, RBM47-dominant, or codominant), relative mRNA expression (edited gene vs. unedited gene) by RNA-seq or quantitative RT-PCR, and abundance of corresponding protein (edited gene vs. unedited gene) by western blotting or proteomic comparison.
Sequence-related parameters
A sequence spanning 10 nt upstream and 30 nt downstream from the edited cytidine was extracted for each C-to-U mRNA editing site. These sequences were extracted either directly from the full-text or using the online UCSC Genome Browser on Mouse (NCBI37/mm9) and Human (Grch38/hg38) (https://genome.ucsc.edu/cgi-bin/hgGateway). We calculated relative abundance of A, G, C, and U individually across a region 10 nt upstream and 20 nt downstream from the edited cytidine across all editing sites. For comparison, we examined the base content of a sequence spanning 10 nt upstream and downstream from mutated deoxycytidine for over 6000 proposed C to X (T, A, and G) DNA mutation targets of the APOBEC family in human breast cancer (Nik-Zainal et al. 2012) along with relative deoxynucleotide distribution in proximity to the edited site.
Secondary structure parameters
We used RNA-structure (Reuter and Mathews 2010) and Mfold (Zuker 2003) to determine the secondary structure of an RNA cassette consisting of regulatory sequence, edited cytidine, spacer, and mooring sequence. Secondary structures similar to that of the cassette for Apob chr12: 8014860 consisting of one loop and stem (with or without unassigned nucleotides with ≤4 unpaired bases inside the stem) as the main stem–loop with or without free tail(s) in one or both ends of the stem were considered as canonical. Two other types of secondary structure were considered as noncanonical structures (Fig. 1B), with ≥2 loops located either at ends of the stem or inside the stem. Loops inside the stem were circular open structures with ≥5 unpaired bases. Editing sites with canonical structure were further categorized into three subgroups based on location of the edited cytidine: specifically (Cloop), stem (Cstem), or tail (Ctail). In addition to overall secondary structure, we considered location of the edited cytidine, location of mooring sequence, symmetry of the free tails, and proportion of the nucleotides in the target cassette that constitute the main stem–loop. This proportion is 1.0 in the case of Apob chr12: 8014860 where all the bases are part of the main stem–loop structure. Symmetry was defined based on existence of free tails in both ends of the RNA strand.
We used Mfold and the RNA-structure software to calculate the minimum Gibbs free energy (MFE also referred to as ΔG) for the secondary structure of the mRNA editing cassettes. Because mRNA strand length significantly influences ΔG (Seffens and Digby 1999; Trotta 2014), we generated standardized ΔG (S-ΔG) by dividing the ΔG by the length of the corresponding mRNA strand in order to normalize ΔGs regardless of the mRNA strand length. Earlier work suggested MFE density (MFEden) as an alternative strategy to adjust the ΔG (i.e., MFE) for the length of mRNA strand and found that MFEden outperforms S-ΔG over a wide range of mRNA strand length (Trotta 2014). The other advantage for MFEden is that we could use it to compare MFE of the RNA editing sites with the MFEs of other groups of published RNA (Trotta 2014). Accordingly, we also calculated MFEden for the mRNA editing sites.
Statistical methodology
Continuous variables are reported as means ± SD with relative proportions for binary and categorical variables. T-test and ANOVA tests were used to compare continuous parameters of interest between two or more than two groups, respectively. χ2 testing was used to compare binary or categorical variables among different groups. Pearson r testing was used to investigate correlation of two continuous variables. Overall, P-values <0.05 were considered significant, and P-values ≥0.05 but smaller than 0.1 were considered nonsignificant despite a trend toward significance.
We used linear regression analyses to develop the final model of independent factors that correlate with editing frequency. We used the Hosmer and Lemeshow approach for model building (Hosmer et al. 2013) to fit the multivariable regression model. In brief, we first used bivariate and/or simple regression analyses with P-value of 0.2 as the cut-off point to screen the variables and detect primary candidates for the multivariable model. Subsequently, we fitted the primary multivariable model using candidate variables from the screening phase. A backward elimination method was used to reach the final multivariable model. Parameters with P-values <0.05 or those that added to the model fitness were retained. Next, the eliminated parameters were added back individually to the final model to determine their impact. Plausible interaction terms between final determinants were also checked. The final model was screened for collinearity. We used the same approach to develop a multinomial logistic regression model to identify factors that were independently associated with cofactor dominance in RNA editing sites. Squared R and pseudo squared R were used to estimate the proportion of variance in responder parameter that could be explained by multivariable linear regression and multinomial logistic regression models, respectively. The same screening and retaining methods were used to investigate association of base content in a sequence 10 nt upstream and 20 nt downstream from the edited cytidine, with editing frequency. However, after determining the nucleotides that were retained in final regression model, a proxy parameter named “base content score” was calculated for each editing site based on the β coefficient values retrieved for individual nucleotides in the model. This parameter was used in the final model as representative variable for base content of the aforementioned sequence in each editing site.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants DK-119437, DK-112378, HL-151328, and Washington University Digestive Diseases Research Core Center P30 DK-52574 (to N.O.D.)
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078678.121.
REFERENCES
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbab M, Shen MW, Mok B, Wilson C, Matuszek Z, Cassa CA, Liu DR. 2020. Determinants of base editing outcomes from target library analysis and machine learning. Cell 182: 463–480.e430. 10.1016/j.cell.2020.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus JW, Smith HC. 1991. Apolipoprotein B mRNA sequences 3′ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res 19: 6781–6786. 10.1093/nar/19.24.6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus JW, Smith HC. 1992. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res 20: 6007–6014. 10.1093/nar/20.22.6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus JW, Schock D, Smith HC. 1994. Only cytidines 5′ of the apolipoprotein B mRNA mooring sequence are edited. Biochim Biophys Acta 1219: 1–14. 10.1016/0167-4781(94)90240-2 [DOI] [PubMed] [Google Scholar]
- Bahn JH, Lee JH, Li G, Greer C, Peng G, Xiao X. 2012. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res 22: 142–150. 10.1101/gr.124107.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611. 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. 10.1101/gr.164749.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Henderson JO, Newberry EP, Kennedy S, Luo J, Davidson NO. 2005. Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol Cell Biol 25: 7260–7269. 10.1128/MCB.25.16.7260-7269.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Park E, Schaefer S, Miller M, Lin Y, Kennedy S, Billing AM, Ben Hamidane H, Graumann J, Mortazavi A, et al. 2014. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol 15: R79. 10.1186/gb-2014-15-6-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Xie Y, Kennedy S, Riordan JD, Rubin DC, Madison BB, Mills JC, Nadeau JH, Davidson NO. 2019. Apobec1 complementation factor (A1CF) and RBM47 interact in tissue-specific regulation of C to U RNA editing in mouse intestine and liver. RNA 25: 70–81. 10.1261/rna.068395.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom K, Lauer SJ, Poksay KS, Garcia Z, Taylor JM, Innerarity TL. 1989. Apolipoprotein B48 RNA editing in chimeric apolipoprotein EB mRNA. J Biol Chem 264: 15701–15708. 10.1016/S0021-9258(19)84889-5 [DOI] [PubMed] [Google Scholar]
- Brannan KW, Chaim IA, Marina RJ, Yee BA, Kofman ER, Lorenz DA, Jagannatha P, Dong KD, Madrigal AA, Underwood JG, et al. 2021. Robust single-cell discovery of RNA targets of RNA-binding proteins and ribosomes. Nat Methods 18: 507–519. 10.1038/s41592-021-01128-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, Silberman SR, Cai SJ, Deslypere JP, Rosseneu M, et al. 1987. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science 238: 363–366. 10.1126/science.3659919 [DOI] [PubMed] [Google Scholar]
- Conticello SG. 2012. Creative deaminases, self-inflicted damage, and genome evolution. Ann N Y Acad Sci 1267: 79–85. 10.1111/j.1749-6632.2012.06614.x [DOI] [PubMed] [Google Scholar]
- Davies MS, Wallis SC, Driscoll DM, Wynne JK, Williams GW, Powell LM, Scott J. 1989. Sequence requirements for apolipoprotein B RNA editing in transfected rat hepatoma cells. J Biol Chem 264: 13395–13398. 10.1016/S0021-9258(18)80008-4 [DOI] [PubMed] [Google Scholar]
- Destefanis E, Avsar G, Groza P, Romitelli A, Torrini S, Pir P, Conticello SG, Aguilo F, Dassi E. 2020. A mark of disease: how mRNA modifications shape genetic and acquired pathologies. RNA 27: 367–389. 10.1261/rna.077271.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Wynne JK, Wallis SC, Scott J. 1989. An in vitro system for the editing of apolipoprotein B mRNA. Cell 58: 519–525. 10.1016/0092-8674(89)90432-7 [DOI] [PubMed] [Google Scholar]
- Elmentaite R, Ross ADB, Roberts K, James KR, Ortmann D, Gomes T, Nayak K, Tuck L, Pritchard S, Bayraktar OA, et al. 2020. Single-cell sequencing of developing human gut reveals transcriptional links to childhood Crohn's disease. Dev Cell 55: 771–783.e5. 10.1016/j.devcel.2020.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N, Tourle K, Radziewic T, Barratt K, Liebhold D, Studdert JB, Power M, Jones V, Loebel DA, Tam PP. 2014. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep 15: 903–910. 10.15252/embr.201438450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956. 10.1101/gad.11.7.941 [DOI] [PubMed] [Google Scholar]
- Gao J, Choudhry H, Cao W. 2018. Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like family genes activation and regulation during tumorigenesis. Cancer Sci 109: 2375–2382. 10.1111/cas.13658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoni F, Bonen DK, Funahashi T, Hadjiagapiou C, Burant CF, Davidson NO. 1994. Complementation of apolipoprotein B mRNA editing by human liver accompanied by secretion of apolipoprotein B48. J Biol Chem 269: 5932–5936. 10.1016/S0021-9258(17)37551-8 [DOI] [PubMed] [Google Scholar]
- Grohmann M, Hammer P, Walther M, Paulmann N, Buttner A, Eisenmenger W, Baghai TC, Schule C, Rupprecht R, Bader M, et al. 2010. Alternative splicing and extensive RNA editing of human TPH2 transcripts. PLoS One 5: e8956. 10.1371/journal.pone.0008956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Buaas FW, Simons AK, Ackert-Bicknell CL, Braun RE, Hibbs MA. 2012. Canonical A-to-I and C-to-U RNA editing is enriched at 3′UTRs and microRNA target sites in multiple mouse tissues. PLoS One 7: e33720. 10.1371/journal.pone.0033720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113: 803–809. 10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Hersberger M, Innerarity TL. 1998. Two efficiency elements flanking the editing site of cytidine 6666 in the apolipoprotein B mRNA support mooring-dependent editing. J Biol Chem 273: 9435–9442. 10.1074/jbc.273.16.9435 [DOI] [PubMed] [Google Scholar]
- Hersberger M, Patarroyo-White S, Arnold KS, Innerarity TL. 1999. Phylogenetic analysis of the apolipoprotein B mRNA-editing region. Evidence for a secondary structure between the mooring sequence and the 3′ efficiency element. J Biol Chem 274: 34590–34597. 10.1074/jbc.274.49.34590 [DOI] [PubMed] [Google Scholar]
- Hirano K, Young SG, Farese RV Jr, Ng J, Sande E, Warburton C, Powell-Braxton LM, Davidson NO. 1996. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J Biol Chem 271: 9887–9890. 10.1074/jbc.271.17.9887 [DOI] [PubMed] [Google Scholar]
- Hosmer DW Jr, Lemeshow S, Sturdivant RX. 2013. Applied logistic regression. John Wiley & Sons. [Google Scholar]
- Hospattankar AV, Higuchi K, Law SW, Meglin N, Brewer HB Jr. 1987. Identification of a novel in-frame translational stop codon in human intestine apoB mRNA. Biochem Biophys Res Commun 148: 279–285. 10.1016/0006-291X(87)91107-7 [DOI] [PubMed] [Google Scholar]
- Kanata E, Llorens F, Dafou D, Dimitriadis A, Thune K, Xanthopoulos K, Bekas N, Espinosa JC, Schmitz M, Marin-Moreno A, et al. 2019. RNA editing alterations define manifestation of prion diseases. Proc Natl Acad Sci 116: 19727–19735. 10.1073/pnas.1803521116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankowski S, Forstera B, Winkelmann A, Knauff P, Wanker EE, You XA, Semtner M, Hetsch F, Meier JC. 2017. A novel RNA editing sensor tool and a specific agonist determine neuronal protein expression of RNA-edited glycine receptors and identify a genomic APOBEC1 dimorphism as a new genetic risk factor of epilepsy. Front Mol Neurosci 10: 439. 10.3389/fnmol.2017.00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellek H, Kirsten R, Diehl I, Apostel F, Buck F, Greeve J. 2000. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem 275: 19848–19856. 10.1074/jbc.M001786200 [DOI] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, et al. 2018. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 9: 4383. 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Masse J, Chester A, Navaratnam N, Allain FH. 2005. NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor. RNA 11: 173–186. 10.1261/rna.7190705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Kinter MT, Sherman NE, Driscoll DM. 2000. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol 20: 1846–1854. 10.1128/MCB.20.5.1846-1854.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JC, Henneberger C, Melnick I, Racca C, Harvey RJ, Heinemann U, Schmieden V, Grantyn R. 2005. RNA editing produces glycine receptor 3P185L, resulting in high agonist potency. Nat Neurosci 8: 736–744. 10.1038/nn1467 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Anant S, Lee RM, Kennedy S, Viskochil D, Davidson NO. 2002. C→U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am J Hum Genet 70: 38–50. 10.1086/337952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnam N, Shah R, Patel D, Fay V, Scott J. 1993. Apolipoprotein B mRNA editing is associated with UV crosslinking of proteins to the editing site. Proc Natl Acad Sci 90: 222–226. 10.1073/pnas.90.1.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149: 979–993. 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petljak M, Alexandrov LB, Brammeld JS, Price S, Wedge DC, Grossmann S, Dawson KJ, Ju YS, Iorio F, Tubio JMC, et al. 2019. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176: 1282–1294.e1220. 10.1016/j.cell.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. 1987. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell 50: 831–840. 10.1016/0092-8674(87)90510-1 [DOI] [PubMed] [Google Scholar]
- Rayon-Estrada V, Harjanto D, Hamilton CE, Berchiche YA, Gantman EC, Sakmar TP, Bulloch K, Gagnidze K, Harroch S, McEwen BS, et al. 2017. Epitranscriptomic profiling across cell types reveals associations between APOBEC1-mediated RNA editing, gene expression outcomes, and cellular function. Proc Natl Acad Sci 114: 13296–13301. 10.1073/pnas.1714227114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson N, Navaratnam N, Scott J. 1998. Secondary structure for the apolipoprotein B mRNA editing site. Au-binding proteins interact with a stem loop. J Biol Chem 273: 31707–31717. 10.1074/jbc.273.48.31707 [DOI] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976. 10.1038/ng.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. 2011. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol 18: 230–236. 10.1038/nsmb.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SH, Danan-Gotthold M, Ben-Izhak M, Rechavi G, Cohen CJ, Louzoun Y, Levanon EY. 2018. Increased RNA editing may provide a source for autoantigens in systemic lupus erythematosus. Cell Rep 23: 50–57. 10.1016/j.celrep.2018.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraconi G, Severi F, Sala C, Mattiuz G, Conticello SG. 2014. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biol 15: 417. 10.1186/s13059-014-0417-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefermeier P, Heinze S. 2017. Hippocampal characteristics and invariant sequence elements distribution of GLRA2 and GLRA3 C-to-U editing. Mol Syndromol 8: 85–92. 10.1159/000453300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seffens W, Digby D. 1999. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res 27: 1578–1584. 10.1093/nar/27.7.1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RR, Knott TJ, Legros JE, Navaratnam N, Greeve JC, Scott J. 1991. Sequence requirements for the editing of apolipoprotein B mRNA. J Biol Chem 266: 16301–16304. 10.1016/S0021-9258(18)55296-0 [DOI] [PubMed] [Google Scholar]
- Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC. 1996. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res 24: 478–485. 10.1093/nar/24.3.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HC. 1998. Analysis of protein complexes assembled on apolipoprotein B mRNA for mooring sequence-dependent RNA editing. Methods 15: 27–39. 10.1006/meth.1998.0603 [DOI] [PubMed] [Google Scholar]
- Smith HC, Kuo SR, Backus JW, Harris SG, Sparks CE, Sparks JD. 1991. In vitro apolipoprotein B mRNA editing: identification of a 27S editing complex. Proc Natl Acad Sci 88: 1489–1493. 10.1073/pnas.88.4.1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EM, McCarty C, Mehalow A, Svenson KL, Murray SA, Korstanje R, Braun RE. 2017. APOBEC1 complementation factor (A1CF) is dispensable for C-to-U RNA editing in vivo. RNA 23: 457–465. 10.1261/rna.058818.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowden M, Hamm JK, Spinelli S, Smith HC. 1996. Determinants involved in regulating the proportion of edited apolipoprotein B RNAs. RNA 2: 274–288. [PMC free article] [PubMed] [Google Scholar]
- Sowden MP, Eagleton MJ, Smith HC. 1998. Apolipoprotein B RNA sequence 3′ of the mooring sequence and cellular sources of auxiliary factors determine the location and extent of promiscuous editing. Nucleic Acids Res 26: 1644–1652. 10.1093/nar/26.7.1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowden MP, Lehmann DM, Lin X, Smith CO, Smith HC. 2004. Identification of novel alternative splice variants of APOBEC-1 complementation factor with different capacities to support apolipoprotein B mRNA editing. J Biol Chem 279: 197–206. 10.1074/jbc.M307920200 [DOI] [PubMed] [Google Scholar]
- Teng B, Burant CF, Davidson NO. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260: 1816–1819. 10.1126/science.8511591 [DOI] [PubMed] [Google Scholar]
- Trotta E. 2014. On the normalization of the minimum free energy of RNAs by sequence length. PLoS One 9: e113380. 10.1371/journal.pone.0113380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AD, Arnold DB, Chen XS. 2019. Comparison of RNA editing activity of APOBEC1-A1CF and APOBEC1-RBM47 complexes reconstituted in HEK293T cells. J Mol Biol 431: 1506–1517. 10.1016/j.jmb.2019.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AD, Li S, Goedderz C, Chen XS. 2020. The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity. NAR Cancer 2: zcaa027. 10.1093/narcan/zcaa027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaranek AW, Levanon EY, Zecharia T, Clegg T, Church GM. 2010. A survey of genomic traces reveals a common sequencing error, RNA editing, and DNA editing. PLoS Genet 6: e1000954. 10.1371/journal.pgen.1000954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.