Supplemental Digital Content is available in the text.
Keywords: blood cells, coronary artery disease, inflammation, ligands, macrophages
Abstract
Background:
Randomized clinical trials indicate that the immune response plays a significant role in coronary artery disease (CAD), a disorder impacting the lifespan potential. However, the identification of targets critical to the immune response in atheroma is still hampered by a lack of solid inference.
Methods:
Herein, we implemented a system genetics approach to identify causally associated immune targets implicated in atheroma. We leveraged genome-wide association studies to perform mapping and Mendelian randomization to assess causal associations between gene expression in blood cells with CAD and the lifespan. Expressed genes (eGenes) were prioritized in network and in single-cell expression derived from plaque immune cells.
Results:
Among 840 CAD-associated blood eGenes, 37 were predicted causally associated with CAD and 6 were also associated with the parental lifespan in Mendelian randomization. In multivariable Mendelian randomization, the impact of eGenes on the lifespan potential was mediated by the CAD risk. Predicted causal eGenes were central in network. FLT1 and CCR5 were identified as targets of approved drugs, whereas 22 eGenes were deemed tractable for the development of small molecules and antibodies. Analyses of plaque immune single-cell expression identified predicted causal eGenes enriched in macrophages (GPX1, C4orf3) and involved in ligand-receptor interactions (CCR5).
Conclusions:
We identified 37 blood eGenes predicted causally associated with CAD. The predicted expression for 6 eGenes impacted the lifespan potential through the risk of CAD. Prioritization based on network, annotations, and single-cell expression identified targets deemed tractable for the development of drugs and for drug repurposing.
Coronary artery disease (CAD) is a leading cause of morbidity and mortality worldwide1 and thus impacts the lifespan potential. There is evidence that the immune response is involved in the development of plaque and clinical events.2 Randomized clinical trials including CANTOS (Canakinumab Anti-inflammatory Thrombosis Outcomes Study) and COLCOT (Colchicine Cardiovascular Outcomes Trial), which administered an anti-IL1B (interleukin-1β) antibody and colchicine respectively, showed modest but significant reduction of cardiovascular events in at-risk patients.3 Post hoc analysis of CANTOS suggested that anti-IL1B therapy may provide a benefit through the reduction of circulating IL6 (interleukin 6).4 These data are supported by Mendelian randomization (MR) analysis showing that IL6 signaling is associated with different cardiovascular outcomes.5 Colchicine, an antimitotic agent used to treat gout, has anti-inflammatory properties including among other proposed mechanisms the blockade of the NLRP3 inflammasome.6 On the contrary, the CIRT trial (Cardiovascular Inflammation Reduction Trial), which tested low-dose methotrexate, an adenosine-dependent immunomodulatory agent used to treat rheumatoid arthritis, did not reduce major adverse cardiovascular events.7 Hence, these randomized clinical trials provided some evidence that the immune response is causally associated with CAD and cardiovascular events.2 However, targeting the immune response in CAD is compounded by several factors such as the risk of serious infections as observed in CANTOS and target selection. The failure of methotrexate to reduce cardiovascular events illustrates that the design of trials should rely on validated disease-related targets.
Genome-wide association studies (GWAS) have identified thousands of risk variants that are associated with different traits/disorders. For CAD, 159 risk loci have been associated at a genome-wide level.8 Growing evidence suggests that genetically supported targets have a higher chance of success in randomized clinical trials.9 System genetics and MR provide powerful agnostic inference and integrative tools to identify underpinning molecular processes involved in complex trait disorders.10 System genetics is a holistic approach, which integrates multidimensional genomic data (eg, genome-wide gene expression, chromatin contacts in 3D genome, single-cell expression) and uses the graph theory to interrogate biologic and disease-perturbed networks.10 In network, proteins (nodes) that interact together (edges) provide information about the function of the system and allow the identification of nodes acting as hub (elevated number of edges) and or bottleneck (shortest path in the network), 2 metrics linked to prominence in the network and enriched in drug targets.11 Gene variants related to the expression of genes explains a significant proportion of the heritability for disorders.12 Gene variants associated with quantitative traits such as gene expression (expression quantitative trait loci [eQTL]) can be leveraged for causal inference using MR technique. In MR, inherited alleles driving the expression of genes (eGenes; exposure) are used to infer causality for the risk of disorders (outcome) while limiting the impact of confounders.13 MR is a powerful approach but may be subject to an inflation of type 1 error rate if it is not appropriately controlled for instrumental variables (IVs; gene variants) with horizontal pleiotropy (ie, IVs related to the outcome through an alternative pathway). Statistical detection of horizontal pleiotropy by the Cochran Q test provides robust causal inference. Also, MR with the weighted median allow causal inference with up to 50% of the IVs that are invalid and is thus another strategy to assess causal inference while minimizing the risk of horizontal pleiotropy.14 Single-cell expression is new among the armamentarium of system genetics and provides inference in a cell- and disease-relevant context.15 Herein, we implemented a multimodal approach including eQTL from blood cells, MR, network prioritization, and the modeling of single-cell gene expression derived from plaques to identify CAD-related immune molecules. Overall, in MR, 37 blood eGenes were associated with CAD and included druggable genes as well as targets of approved drugs with a strong potential for drug repurposing. We also identified 6 blood eGenes predicted to impact the lifespan potential through their effect on the CAD risk.
Methods
All materials and methods are available in the Data Supplement.
The authors declare that all supporting data are available within the article and in the Data Supplement. We performed analyses based on publicly available data. As all analyses were based on publicly available data, no ethical approval was required.
Results
Mapping and Identification of eGenes Associated With CAD
Figure 1A illustrates our pipeline of analyses. We leveraged blood eQTLs derived from 31 684 samples16 to map eGenes associated with CAD by using a GWAS8 totaling 547 261 individuals. In 159 CAD risk loci, we identified the individual significant single nucleotide polymorphism (SNPs; PGWAS<5×10−8, r2<0.6) and variants in linkage disequilibrium (methods) associated with blood eQTLs. In total, 79 398 SNP-gene pairs (false discovery rate <0.05) tagging 840 eGenes were mapped (Table I in the Data Supplement). In ARCHS4, blood eGenes were enriched in macrophages (Padjusted=0.04; Table II in the Data Supplement), which is consistent with the important role of these cells in atheroma. Gene regulation is linked to chromatin interactions between distant regulatory regions and gene promoters. To this effect, disease-associated gene variants residing in the noncoding genome may regulate gene expression, often located at distance, by chromatin folding. Investigations have shown that eQTL and GWAS variants were enriched in chromatin loops.17,18 We wondered whether eGenes were also mapped by distant promoter interacting regions (PIRs) in macrophages. To that end, we analyzed a data set of promoter capture Hi-C obtained in human primary macrophages.19 This analysis revealed an enrichment of CAD gene variants (individual significant SNPs) in PIRs identified in promoter capture Hi-C from macrophages (P<0.01). In total, 181 individual significant SNPs associated with CAD were mapped to 142 significant PIRs (Tables III and IV in the Data Supplement). These CAD-associated PIRs interacted with 291 gene promoters which are listed in Table V in the Data Supplement. The mean distance between CAD-associated PIRs and gene promoters was 354 kb, and the average number of promoters targeted by a PIR was 2.05 (Table V in the Data Supplement). There were 48 PIRs targeting 1 gene promoter, whereas 27 PIRs targeted 2 gene promoters, and 67 PIRs targeted >2 gene promoters. Among the 840 CAD-associated eGenes, we found 94 promoters of these eGenes (Table VI and Figure IA in the Data Supplement) interacting with at least 1 CAD-associated PIR in macrophages (fold enrichment, 7.47; P<2.20×10−16, hypergeometric test). Figure 1B and 1C show 2 illustrative cases for 16q23.1 and 10p13 chromosomal loci where BCAR1 and CAMK1D were mapped by eQTL and chromatin contact in macrophages, whereas Figure IB and IC in the Data Supplement present data for TSPAN14 and RAB5C.
Figure 1.
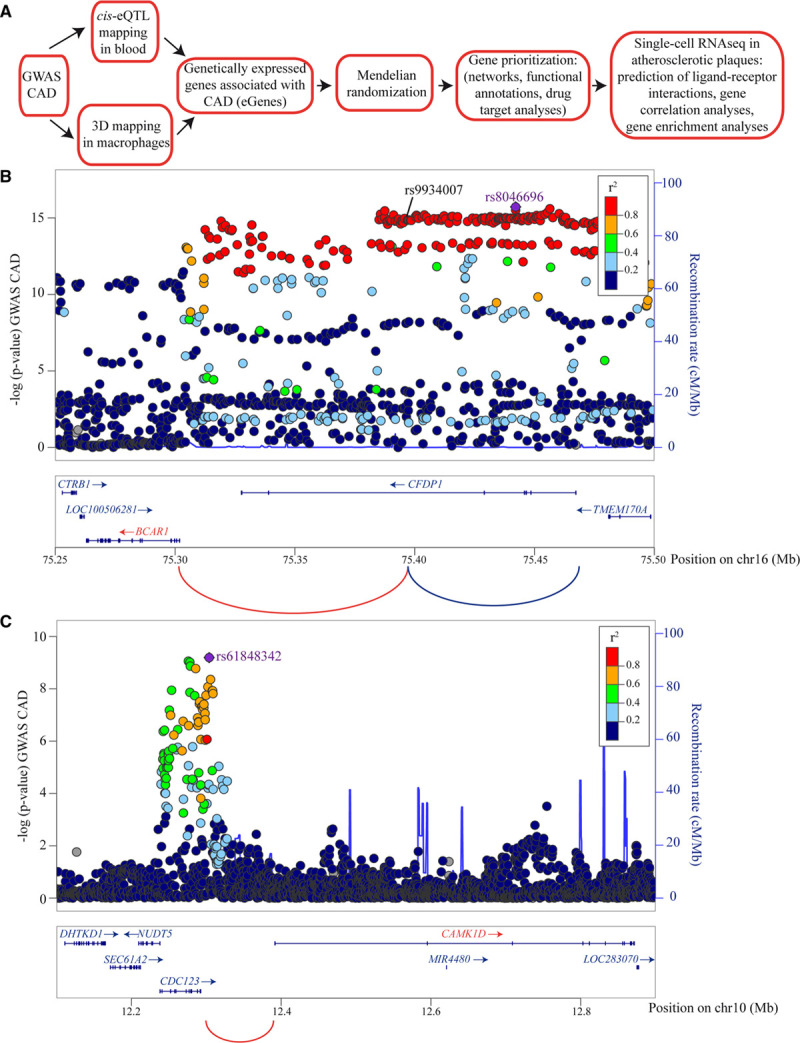
Mapping the coronary artery disease (CAD)-associated genes in blood (eGenes). A, Representative scheme showing the pipeline of analyses. B, Example of 3-dimensional (3D)-mapped BCAR1 gene where rs9934007, an intronic variant in CFDP1, intersected with a promoter interacting region (PIR) that interacted with CFDP1 and BCAR1 gene promoters in macrophages. C, Example of 3D-mapped CAMK1D gene where rs61848342, an intergenic variant, intersected with a PIR that interacted with CAMK1D gene promoter in macrophages. Arcs are significant 3D genomic interactions. Genes and arcs colored in red are 3D-mapped genes and also predicted as causally associated with CAD in Mendelian randomization (MR). Genes not predicted as causally associated with CAD are in blue. eQTL indicates expression quantitative trait loci; and GWAS, genome-wide association studies.
MR of Blood cis-eGenes
MR analysis was performed to infer the causal relationships between the blood cis-eGenes and CAD. Enough IVs (minimum of 3 variants) were available to perform 795 MR analyses with a mean of 27 IVs per gene (Table VII in the Data Supplement). By using inverse variance weighted analysis and a correction for multiple testing (Bonferroni correction, Pcausal <6.29×10−5, 0.05/795), we identified 97 eGenes associated with CAD (Table VII in the Data Supplement). Among the 97 significant eGenes in inverse variance weighted MR, 37 did not show heterogeneity according to the Cochran Q test (P>0.05) and were thus considered as predicted causally associated with CAD (DHX36, HOXC4, POC1B, TSPAN14, CDC123, C4orf3, SREBF1, BCAR1, N4BP2L2, CCDC167, RAB5C, GID4, SF3A3, NUDT5, EIF2B2, DAGLB, UHRF1BP1, FHL3, EFCAB5, COL17A1, KAT2A, CKM, DDX59, HBP1, FLT1, CLUH, CAMK1D, GPX1, BLMH, RPS6KL1, CDC25A, ZHX3, ARID4A, CCR5, TRIP4, CCDC181, MED9; Figure 2A; Table VIII in the Data Supplement). Predicted causal eGenes BCAR1, CAMK1D, TSPAN14, and RAB5C were also identified by chromatin contact mapping of individual significant SNPs in macrophages (Table VI in the Data Supplement). Figure 2B shows that the 37 eGenes associated with CAD in MR were located in 28 CAD risk loci. Among the 37 predicted causal eGenes, 24 genes were not previously mapped in the CAD GWAS meta-analysis8 (Table IX in the Data Supplement). In sensitivity analysis, the 37 blood eGenes were also significant in weighted median MR, which provides an assessment that is more robust to IVs with potential pleiotropy (Table VIII in the Data Supplement).
Figure 2.
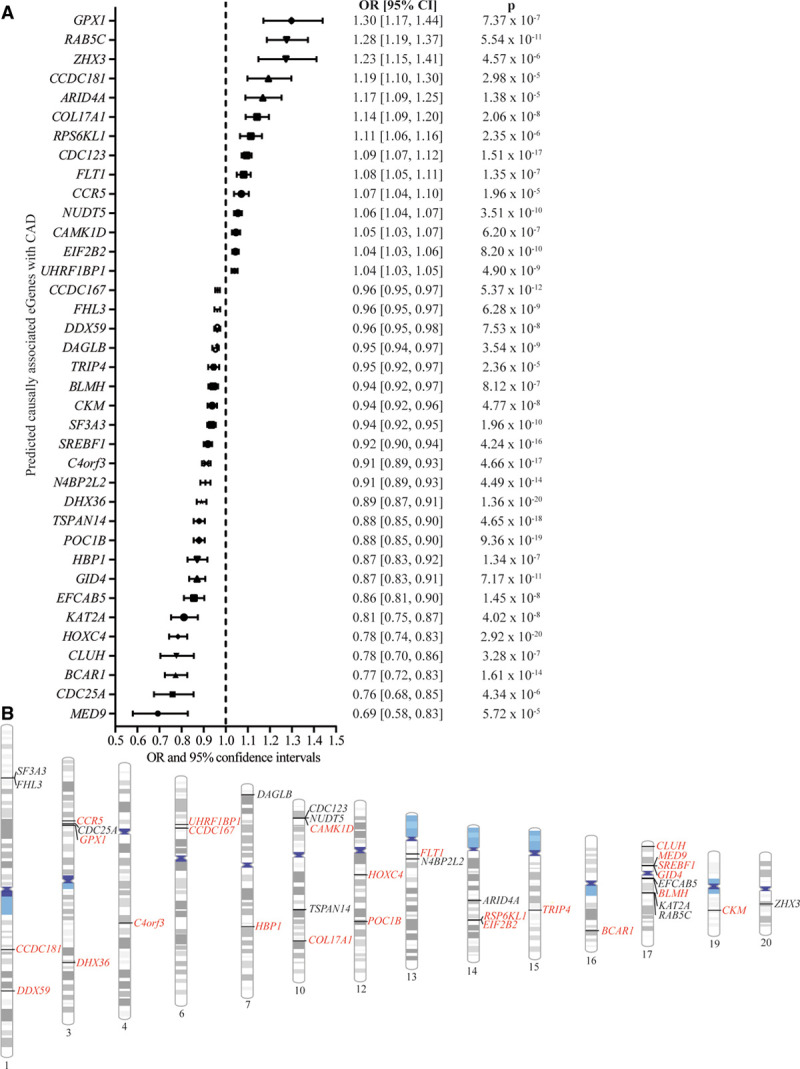
Coronary artery disease (CAD)-predicted causally associated eGenes in blood and their respective loci. A, Forest plot showing the odds ratio (OR; with 95% CI) and the inverse variance weighted Mendelian randomization P value for each CAD predicted causally associated eGenes. B, Chromosome ideogram representing the loci for each CAD predicted causally associated eGenes. Genes in black were mapped by van der Harst et al,8 and the genes in red are the novel CAD predicted causally associated eGenes.
Network Analysis and Prioritization of CAD Predicted Causally Associated eGenes
Networks provide inference about the function of complex systems. We extracted a gene-set network based on gene ontology by using the 37 predicted causally associated eGenes to capture functional modules by which these genes operate in CAD. Several gene ontology such as cell migration (CCR5, FLT1, CAMK1D, BCAR1, and GPX1), regulation of endocytosis (CAMK1D and RAB5C), insulin receptor signaling (BCAR1 and SREBF1), and triglyceride metabolic process (GPX1 and SREBF1) were highly connected and relevant to CAD (Figure 3A; Table X in the Data Supplement). We next wondered whether predicted causally associated eGenes and their derived proteins were enriched in a protein-protein interaction network. From the predicted causally associated eGenes, we extracted a blood protein-protein interaction network by using DifferentialNet20 data. Blood eGenes identified in MR generated a network with 376 nodes (protein coding genes) and 400 edges (connections by physical interactions; Figure 3B). By using the 16 532 proteins in DifferentialNet20 data as a background, we found that the network was enriched for developmental biology (fold enrichment, 4.08, P<2.20×10−16; hypergeometric test), transcriptional regulation of white adipocyte differentiation (fold enrichment, 12.80, P<2.20×10−16; hypergeometric test), and in signaling by VEGF (vascular endothelial growth factor; fold enrichment, 5.49, P=2.88×10−16; hypergeometric test) in Reactome (Figure 3C). In graph theory, nodes with indices of centrality are acting as hub (degree) or bottleneck (betweenness), 2 metrics associated with prominence in networks and enriched in pharmacological targets.11 Among the first 32 genes with an elevated betweenness (>99th percentile), there were 19 cis-regulated eGenes predicted as causally associated with CAD (fold enrichment, 6.03, P=5.21×10−7; hypergeometric test; FHL3, KAT2A, BCAR1, CDC25A, SREBF1, RAB5C, MED9, TRIP4, CCR5, FLT1, DHX36, SF3A3, BLMH, HOXC4, COL17A1, CAMK1D, EIF2B2, NUDT5, N4BP2L2; Figure 3B; Table XI in the Data Supplement). Network provides a robust method for gene prioritization. We were curious if a prioritization based on functional annotation would provide similar results. We implemented ToppGene,21 an algorithm based on functional annotation to prioritize genes against a training data set (DISEASES database for CAD, DOID 3393). Among the 37 predicted causally associated blood eGenes, FLT1, CCR5, SREBF1, HOXC4, COL17A1, BCAR1, and CAMK1D were among the top-10 genes prioritized by ToppGene21 (Table XII in the Data Supplement). These eGenes were also acting as bottleneck (elevated betweenness) in the protein-protein interaction network (Figure 3B; Table XI in the Data Supplement).
Figure 3.
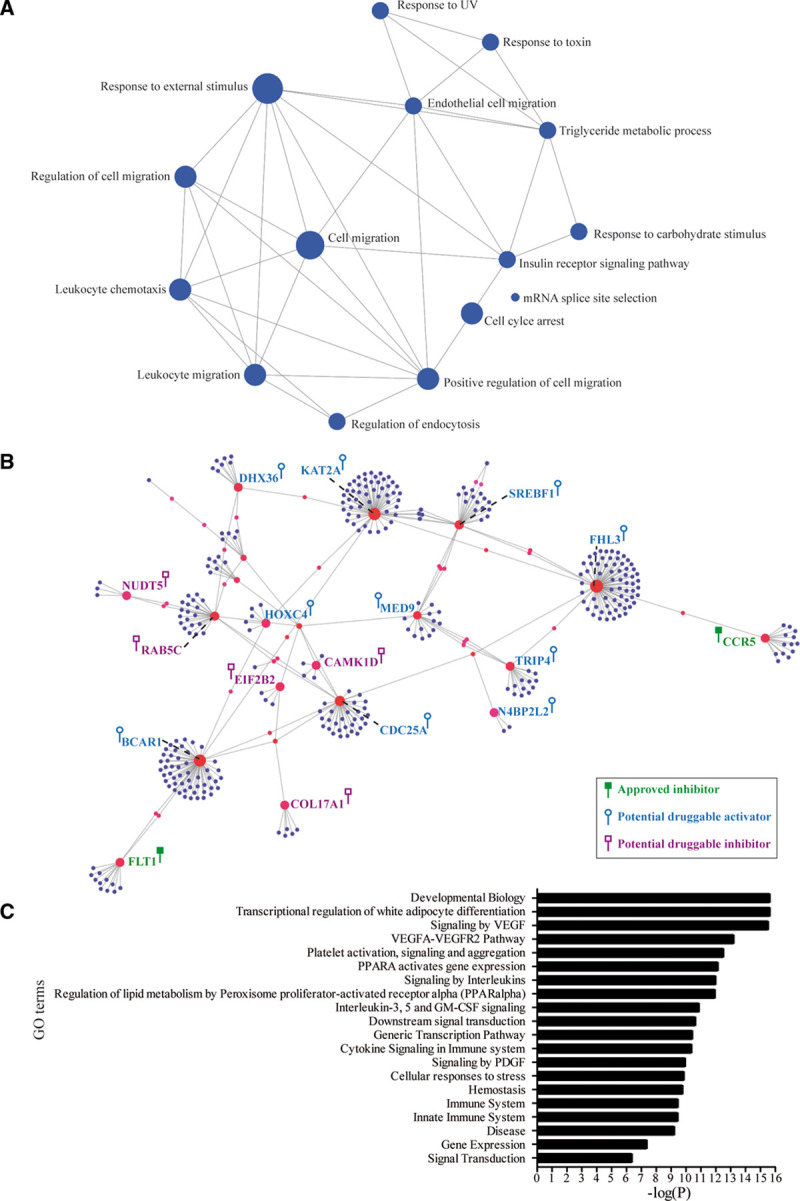
Networks for the 37 predicted causal eGenes. A, Gene ontology (GO) enrichment network of the 37 predicted causal eGenes. B, Protein-protein interaction (PPI) network of the CAD predicted causally associated eGenes in whole blood. The genes highlighted in green are targeted by approved inhibitors; genes in blue are potentially druggable by activators; and genes in purple are potentially druggable by inhibitors. C, Bar graph showing the enrichment in Reactome for genes of the PPI network (hypergeometric tests). VEGF indicates vascular endothelial growth factor.
Druggable Genome
The enrichment of CAD predicted causally associated eGenes in nodes with an elevated central betweenness suggests that these genes orchestrate the communication between the nodes and are thus potential targets of interest. We thus investigated whether eGenes could be targeted by pharmaceutical compounds. In the Open Targets database,22 there were 47 compounds in different phase of development or approved, targeting predicted causally associated eGenes FLT1 and CCR5 (Tables XIII and XIV in the Data Supplement). Among the 37 predicted causal eGenes, 22 of them were deemed tractable in the Open Targets database22 (Table XV in the Data Supplement). Genes with a specific expression have been highlighted to be enriched as drug targets.23 Of interest, by using genotype-tissue expression data and τ, a metric evaluating tissue-specific expression (0 is broadly expressed, and 1 is specific), we found that among the predicted causal eGenes, CCR5 was the second most tissue-specific gene (τ=0.97) with highest expression in the blood, spleen, and lung (Table XVI in the Data Supplement). In genotype-tissue expression, CAD variants were not mapped to the expression of CCR5 in the spleen or lungs (false discovery rate >0.05). In MR, the expression of FLT1 and CCR5 in the blood was positively associated with the risk of CAD. Hence, inhibitors such as Pazopanib and Maraviroc, which are inhibitors of FLT1 (Fms related receptor tyrosine kinase 1) and CCR5 (C-C chemokine receptor type 5), respectively, could represent drugs for repurposing. Pazopanib and Maraviroc are approved drugs for cancer24 and for the infection with the HIV,25 respectively. Figure 3B illustrates druggable genes (genes with approved drugs and also genes deemed tractable) in the network along with their requirement for the development of an agonist or an antagonist based on the direction of causal inference in MR. For instance, CAMK1D, a gene mapped in eQTL and 3D analyses, with a high betweenness in the network and prioritized by ToppGene,21 is positively associated with the CAD risk and could be targeted by inhibitors (gene deemed tractable by the Open Targets database).
Impact on the Lifespan
The lifespan potential is partially determined by gene variants and the expression of genes in the blood.15 CAD is a disorder impacting the lifespan potential. It is presently unknown whether CAD-associated blood eGenes could also impact the lifespan. To test this hypothesis, we leveraged summary statistics of 1 012 240 parental lifespans26 to infer the potential causal relationships between blood CAD predicted causally associated eGenes and the lifespan potential (methods). After correction for multiple testing (Bonferroni threshold, P<1.35×10−3; 0.05/37), we found that 6 predicted causally associated eGenes with CAD were also associated with the lifespan in MR (ARID4A, CCDC167, CCR5, DAGLB, RPS6KL1, and UHRF1BP1) and were without heterogeneity on the Cochran Q test (Figure 4A; Table XVII in the Data Supplement). The strongest association for the lifespan was for the blood expression of CCDC167 (Pcausal=3.38×10−14; Table XVII in the Data Supplement). In MR, the directional effects of the 6 blood predicted causal eGenes were concordant between the risk of CAD and the lifespan (eg, blood eGenes positively associated with the risk of CAD were associated with a decreased lifespan). For instance, Figure 4B and 4C show that predicted expression of CCR5 in blood cells was associated with an increased risk of CAD (per 1 SD odds ratio [OR], 1.07 [95% CI, 1.04–1.10], Pcausal=1.96×10−5) and a decreased lifespan (per 1 SD, −0.31 year [95% CI, −0.49 to −0.13], Pcausal=8.63×10−4; methods). We next hypothesized that these 6 predicted causal eGenes could mediate their effect on the lifespan through a modulation of the CAD risk. To verify this hypothesis, we performed a mediation analysis by conducting multivariable MR (MMR). In MMR, the associations with the lifespan were lost for the 6 blood eGenes (ARID4A, CCDC167, CCR5, DAGLB, RPS6KL1, and UHRF1BP1) after correction for the associations with CAD (Table XVIII in the Data Supplement). Thus, these data underlined that predicted causal eGenes impact the lifespan potential through a significant modulation of the CAD risk.
Figure 4.
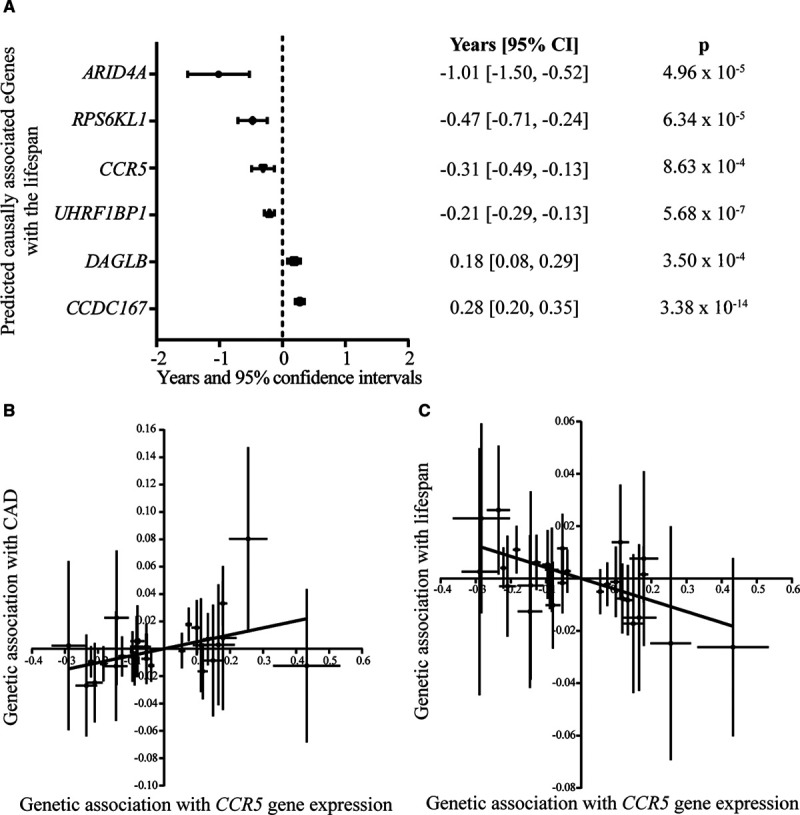
Identification of lifespan predicted causally associated eGenes. A, Forest plot showing the years gained or lost for 6 lifespan predicted causally associated eGenes with their corresponding inverse variance weighted Mendelian randomization P values. B, Graph showing the genetic association of CCR5 expression in blood with the risk of coronary artery disease (CAD). C, Graph showing the genetic association of CCR5 expression in blood with the lifespan potential; each point represents an instrumental variable (gene variant).
Analysis of Single-Cell Expression in Plaques
Gene expression from bulk tissue does not capture cell- and context-specific information. Single-cell expression provides mapping of cell types in different states in a disease-relevant context to evaluate gene program and cell interaction pathways. We interrogated single-cell expression (CITE-seq; n=1186 cells) from plaque-derived immune cells.27 Specifically, we were curious if predicted causally CAD-associated eGenes were enriched in ligand-receptor interactions28 and in cell clusters of plaque. We identified in plaque immune single-cell, clusters of macrophage (CD14, CD68), CD4 T-cell (CD3D), CD8 T-cell (CD8A), natural killer cell (KLRD1), and plasma cell (IGHG2; Figure 5A; Figure IIA and IIB in the Data Supplement). GPX1 was among the top enriched genes in the macrophage cluster (Padjusted=8.19×10−119; Figure 5B; Table XIX in the Data Supplement). The expression of GPX1 was positively correlated with markers of the macrophage lineage (CD14, CD68, CD163) and inflammation (CTSB, FCN1, S100A9; Table XX in the Data Supplement). GPX1 encodes for a glutathione peroxidase for which a common nonsynonymous coding variant rs1050450 (MAF in CEU, 0.28; Figure III in the Data Supplement) affects the enzymatic function. Gene variant A-rs1050450 changes a proline for a leucine at position 200 of GPX1 (glutathione peroxidase 1; p.Pro200Leu). In the CAD GWAS, carriers of the allele A, which decreases the activity of GPX1 by 40%,29 have a reduced risk of CAD (OR, 0.97; PGWAS=2.06×10−6).8 Together, the enrichment of GPX1 in macrophage along with markers of inflammation is consistent with the genetic association data for rs1050450 and MR, which showed a positive association for the expression of GPX1 in the blood with the CAD risk (per 1 SD OR, 1.30 [95% CI, 1.17–1.44], Pcausal=7.37×10−7). Also, C4orf3 is a CAD predicted causally associated eGene (per 1 SD OR, 0.91 [95% CI, 0.89–0.93], Pcausal=4.66×10−17) significantly enriched in the macrophage cluster (Padjusted=1.09×10−4; Table XIX in the Data Supplement). C4orf3 encodes for an uncharacterized protein. In The Bioplex Interactome (Bioplex 3.0),30 C4orf3 protein interaction partners are enriched in gene ontology for endosomal sorting complexes required for transport III complex disassembly (Padjusted=6.51×10−5) and in multivesicular body assembly (Padjusted=5.05×10−4; Table XXI in the Data Supplement). The endosomal sorting complexes required for transport III complex participates into the biogenesis of the multivesicular body, which allows the destruction of misfolded proteins.
Figure 5.
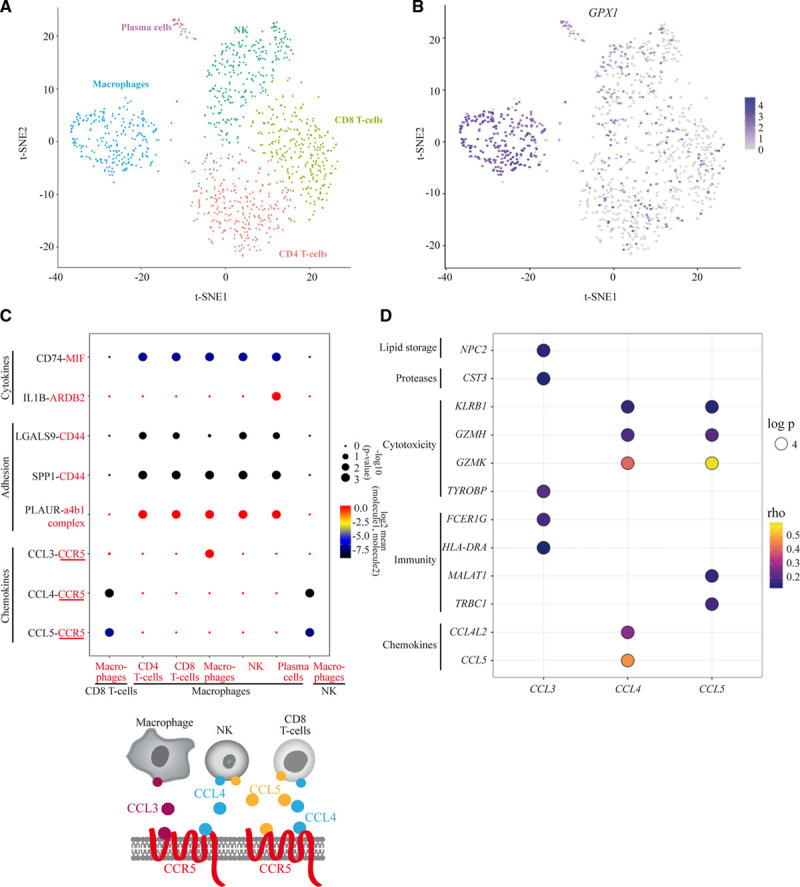
Analyses of single-cell RNA-seq data from immune cells of human atherosclerotic plaques. A, tSNE plot for immune cells from atherosclerotic plaques showing the presence of macrophages, T-CD4, T-CD8, natural killer (NK), and plasma cells. B, tSNE plot showing the enrichment of GPX1 in the cluster of macrophages. C, Ligand-receptor interactions between immune cells using CellPhoneDB.28 Ligands are in black and receptors in red. D, Balloon plot representing the correlations between CCL3, CCL4, CCL5 and genes coding for chemokines, immune, cytotoxic, proteases, and lipid storage proteins. The color of the circle indicates the ρ correlation score and the size corresponds to the log Padjusted. CCL indicates C-C motif chemokine ligand; and IL1B, interleukin 1β.
Ligands and receptors represent a class of molecules accessible for the development of therapeutic agents. Among the 37 CAD predicted causally associated eGenes, CCR5, FLT1, and COL17A1 encode for molecules involved in ligand-receptor interactions as listed by Ramilowski et al.31 In plaque single-cell expression,27 we examined the predicted ligand-receptor interactions derived from a repository of ligand-receptor heteromeric complexes28 (methods). Figure 5C shows predicted cell-cell interactions in plaque based on ligand-receptor gene expression level with different functions such as chemoattraction, immune response, and adhesion. Several interactions based on the expression of CCR5 were predicted. Macrophage-macrophage interactions involving CCL (C-C motif chemokine ligand) 3 and CCR5 were predicted. In addition, CD8+ and natural killer cells that expressed CCL4 and CCL5 were predicted to interact with CCR5 expressed by macrophages (Figure 5C). Interactions also included several molecules known to be involved in atheroma such as IL1B, MIF, CD44, and PLAUR (Figure 5C). In plaque-derived immune cells, no significant interaction was predicted based on the expression level of FLT1 or COL17A1. We sorted the top positively correlated genes with CCL3, CCL4, and CCL5 in plaque immune single-cell expression data. These chemokines were correlated with genes involved in lipid storage (NPC2), cytotoxicity (GZMH, GZMK, TYROB, and KLRB1), and immune response (FCER1G, HLA-DRA, TRBC1, and MALAT1; Figure 5D). Taken together, data derived from plaque single-cell expression provides a disease- and cell-context relevant information about the potential function of genes, which warrants further investigation in follow-up studies (eg, C4orf3 and GPX1).
Discussion
By using a system genetics approach including causal inference, we provide robust evidence based on MR that gene expression in blood cells is associated with CAD. A subset of predicted causally associated CAD-eGenes were also linked to the lifespan. Among the 37 blood eGenes predicted causally associated with CAD, 22 are potentially druggable, whereas 2 (FLT1 and CCR5) are targets of approved drugs. Among the druggable candidate genes, several are acting as hub in a network and were also prioritized by ToppGene21 (SREBF1, HOXC4, COL17A1, BCAR1, and CAMK1D) and could be prioritized in follow-up investigations. Analysis of plaque immune single-cell expression data provides evidence that some predicted CAD causal blood eGenes (GPX1 and C4orf3) are enriched in macrophages, whereas several ligands are predicted to interact with CCR5 expressed by macrophages. Together, these data pinpoint CAD predicted-causally associated eGenes with important regulatory functions and provide strong inference supporting drug repurposing or for follow-up studies and the development of novel molecules to modulate the immune response in atheroma.
Studies conducted in animal models and humans strongly support a role for the immune response in CAD.2 However, the immune system relies on a highly connected network with a vast array of response. Hence, it is crucial to identify disease-specific genes with prominent functions, which could also represent suitable drug targets to reduce events. We identified that CAD-associated eGenes were enriched in macrophages, which is consistent with the important role of these cells in the pathophysiology of atheroma.2 Among the 37 predicted causally associated eGenes, several were highly connected in a blood protein-protein network. These data thus highlighted blood cell eGenes with potential regulatory function in atheroma. In this regard, SREBF1, a highly connected gene, which encodes SREBP-1, is a basic helix-loop-helix-leucine zipper transcription factor that promotes in macrophages reprogramming and the resolution of inflammation.32 These data are consistent with the directional effect in MR for the expression of SREBF1 and the risk of CAD (per 1 SD OR, 0.92 [95% CI, 0.90–0.94], Pcausal=4.24×10−16). Among the other highly connected eGenes, CAMK1D encodes for a calcium calmodulin-dependent protein kinase involved in the activation of neutrophils.33 CAMK1D is predicted tractable for the development of small inhibitory molecules and our data in MR indicate a positive association with the risk of CAD (per 1 SD OR, 1.05 [95% CI, 1.03–1.07], Pcausal=6.20×10−7). Emerging data indicates that homeobox genes play complex roles in the development of atheroma.34 Our results indicate that the expression of HOXC4, a densely connected gene, is negatively associated with the risk of atheroma (per 1 SD OR, 0.78 [95% CI, 0.74–0.83], Pcausal=2.92×10−20). Of interest, several homeobox genes including HOXC4 were shown to be downregulated in atherosclerotic carotid arteries.35 Hence, further studies on interconnected eGenes could provide novel mechanistic data with regard to molecular processes related to atheroma.
Analysis of plaque-derived single-cell data provided evidence that GPX1 and C4orf3 were highly enriched in macrophages. GPX1 encodes for a glutathione peroxidase that decreases the level of H2O2, a reactive oxygen species.36 In MR, the blood expression of GPX1 was positively associated with the risk of CAD (per 1 SD OR, 1.30 [95% CI, 1.17–1.44], Pcausal=7.37×10−7). In the same line, carriers of A-rs1050450 (p.Pro200Leu), which decreases the activity of GPX1 by 40%,29 had a lower risk of CAD. These data may seem counterintuitive as reactive oxygen species have been incriminated to play a detrimental role in several diseases including CAD.37 However, the impact of GPX1 is likely cell- and context-specific as overexpression of GPX1 in mice worsen insulin sensitivity.36 In this regard, hydroperoxides affect the function of different kinases involved in insulin signaling.36 We underlined that C4orf3, a protein with an unknown function, was enriched in plaque-infiltrating macrophages. In MR, the expression of C4orf3 in the blood was negatively associated with the risk of CAD (per 1 SD OR, 0.91 [95% CI, 0.89–0.93], Pcausal=4.66×10−17). Though follow-up studies are required, interrogation of the C4orf3 protein interactome in Bioplex 3.030 suggests that C4orf3 may play a role in the destruction of misfolded proteins.
Studies have highlighted that nodes with an elevated central betweenness (shortest path) are enriched in drug targets.11 Considering the cost for the development of small molecules and the high attrition rate of drugs under development, drug repurposing is an alternative and attractive approach. We identified 2 CAD predicted causally associated eGenes that are targets of approved drugs. FLT1 is targeted by several kinase inhibitors with indications in cancer.24 However, several kinase inhibitors have important side-effects, which may limit their use for chronic administration.38 On the contrary, drugs such as Maraviroc, which targets CCR5, are approved for chronic administration in the treatment of HIV-AIDS.25 Genes with an elevated tissue-specific expression are ideally suited for the development of inhibitors as it may limit drug-related side effects.23 Among CAD-associated eGenes, CCR5 was among the top tissue-selective genes having a significant expression in the blood. To this effect, Maraviroc, which targets CCR5, is generally well tolerated with reported side effects such as upper respiratory tract infection and rash.39 In the present work, we found that CCR5 and interacting chemokines (CCL3, CCL4, and CCL5) were highly expressed in plaque immune cells. Data derived from plaque single-cell expression showed that the expression of chemokines were correlated with several genes involved in atherosclerosis such as NPC2, FCER1G, and MALAT1.40 In mice, the deletion of CCR5 lowers the formation of plaques.41 These data are consistent with the present investigation in which we found in MR a positive association between the expression of CCR5 and the risk of CAD (per 1 SD OR, 1.07 [95% CI, 1.04–1.10], Pcausal=1.96×10−5). Taken together, these data strongly militate for a causal role of CCR5 in CAD.
We hypothesized that gene variants affecting the expression of CAD-associated genes may also impact the lifespan. By analyzing 1 012 240 parental lifespan,26 we found that 6 CAD-associated eGenes (ARID4A, CCDC167, CCR5, DAGLB, RPS6KL1, and UHRF1BP1) were also associated with the lifespan potential. The effects were concordant between CAD risk and the lifespan (eg, eGenes positively associated with the risk of CAD decreased the lifespan potential). For instance, the blood expression of ARID4A was negatively associated with the lifespan (per 1SD, −1.01 year [95% CI, −1.50 to −0.52], Pcausal=4.96×10−5) and was positively associated with the CAD risk (per 1 SD OR, 1.17 [95% CI, 1.09–1.25], Pcausal=1.38×10−5). ARID4A is involved in chromatin remodeling and cancer.42 Its role in atheroma remains to be investigated. In mediation analysis by using MMR, the impact of the 6 blood eGenes (ARID4A, CCDC167, CCR5, DAGLB, RPS6KL1, and UHRF1BP1) on the lifespan potential was determined by the risk of CAD. To our knowledge, this is the first MR study to provide evidence that the expression of genes impacting the risk of CAD also affect the lifespan potential.
Causal inference, though a powerful approach, has some limitations such as the risk of horizontal pleiotropy (ie, genes variants affecting the outcome through an alternative pathway), which is, however, minimized by different sensitivity tools used in the present work. Moreover, MR along with a genetics system approach provide multi-level data, which when integrated allow a robust assessment of causality.43
The present findings underlined by using MR that genotype-based expression of genes in the blood is associated with the risk of CAD. In total, by using robust causal inference, we identified 37 blood eGenes involved in the immune response that are associated with the risk of CAD. We provided a map of eGenes involved the development of atheroma, and we highlighted genes with a druggable potential for follow-up studies. A multi-pronged analysis scheme pinpointed CCR5 as a CAD target, which may also favorably impact the lifespan, possibly in at-risk population, for drug repurposing, and further investigations in randomized clinical trials.
Acknowledgments
V. Bon-Baret, A. Chignon, and Dr Mathieu designed the study. V. Bon-Baret and A. Chignon performed mapping and MR analyses. V. Bon-Baret performed enrichment and network analyses. V. Bon-Baret and A. Chignon analyzed single-cell expression data. Z. Li performed tissue-specific enrichment analyses. Dr Mathieu and V. Bon-Baret drafted the article. All the authors critically revised the article and provided important scientific inputs.
Sources of Funding
This study was supported by the Canadian Institutes of Health Research grants to Dr Mathieu (FRN148778 and FRN159697) and the Quebec Heart and Lung Institute Fund. Dr Bossé holds a Canada Research Chair in Genomics of Heart and Lung Diseases. Drs Thériault and Arsenault hold a junior scholarship from Fonds de Recherche du Québec-Santé (FRQS). Dr Mathieu holds a FRQS Research Chair.
Disclosures
None.
Supplemental Materials
Expanded Methods
Online Tables I–XXI
Online Figures I–III
Nonstandard Abbreviations and Acronyms
- CAD
- coronary artery disease
- CCL
- C-C motif chemokine ligand
- CCR5
- C-C chemokine receptor type 5
- eGenes
- expressed genes
- eQTL
- expression quantitative trait loci
- FLT1
- Fms related receptor tyrosine kinase 1
- GPX1
- glutathione peroxidase 1
- GWAS
- genome-wide association studies
- IL1B
- interleukin 1β
- IL6
- interleukin 6
- IV
- instrumental variable
- MMR
- multivariable Mendelian randomization
- MR
- Mendelian randomization
- OR
- odds ratio
- pcHi-C
- promoter capture Hi-C
- PIR
- promoter interacting region
- SNP
- single nucleotide polymorphism
- VEGF
- vascular endothelial growth factor
For Sources of Funding and Disclosures, see page 180.
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.120.003196.
Contributor Information
Valentin Bon-Baret, Email: valentin.bon.1@ulaval.ca.
Arnaud Chignon, Email: arnaud.chignon@criucpq.ulaval.ca.
Marie-Chloé Boulanger, Email: marie-chloe.boulanger@criucpq.ulaval.ca.
Zhonglin Li, Email: Zhonglin.LI@criucpq.ulaval.ca.
Deborah Argaud, Email: deborah.argaud@criucpq.ulaval.ca.
Benoit J. Arsenault, Email: benoit.arsenault@criucpq.ulaval.ca.
Sébastien Thériault, Email: sebastien.theriault@criucpq.ulaval.ca.
Yohan Bossé, Email: yohan.bosse@criucpq.ulaval.ca.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després J-P, Fullerton HJ, et al. Heart disease and stroke statistics—2016 update: a report from the American Heart Association. Circulation. 2016; 133:e38–e60. doi: 10.1161/CIR.0000000000000350 [DOI] [PubMed] [Google Scholar]
- 2.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005; 352:1685–1695. doi: 10.1056/NEJMra043430 [DOI] [PubMed] [Google Scholar]
- 3.Ridker PM. From CANTOS to CIRT to COLCOT to clinic: will all atherosclerosis patients soon be treated with combination lipid-lowering and inflammation-inhibiting agents? Circulation. 2020; 141:787–789. doi: 10.1161/CIRCULATIONAHA.119.045256 [DOI] [PubMed] [Google Scholar]
- 4.Ridker PM, Libby P, MacFadyen JG, Thuren T, Ballantyne C, Fonseca F, Koenig W, Shimokawa H, Everett BM, Glynn RJ. Modulation of the interleukin-6 signalling pathway and incidence rates of atherosclerotic events and all-cause mortality: analyses from the Canakinumab Anti-Inflammatory Thrombosis Outcomes Study (CANTOS). Eur Heart J. 2018; 39:3499–3507. doi: 10.1093/eurheartj/ehy310 [DOI] [PubMed] [Google Scholar]
- 5.Rosa M, Chignon A, Li Z, Boulanger M-C, Arsenault BJ, Bossé Y, Thériault S, Mathieu P. A Mendelian randomization study of IL6 signaling in cardiovascular diseases, immune-related disorders and longevity. NPJ Genom Med. 2019; 4:23. doi: 10.1038/s41525-019-0097-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez GJ, Celermajer DS, Patel S. The NLRP3 inflammasome and the emerging role of colchicine to inhibit atherosclerosis-associated inflammation. Atherosclerosis. 2018; 269:262–271. doi: 10.1016/j.atherosclerosis.2017.12.027 [DOI] [PubMed] [Google Scholar]
- 7.Ridker PM, Everett BM, Pradhan A, MacFadyen JG, Solomon DH, Zaharris E, Mam V, Hasan A, Rosenberg Y, Iturriaga E, et al. ; CIRT Investigators. Low-dose methotrexate for the prevention of atherosclerotic events. N Engl J Med. 2019; 380:752–762. doi: 10.1056/NEJMoa1809798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018; 122:433–443. doi: 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015; 47:856–860. doi: 10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- 10.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014; 15:34–48. doi: 10.1038/nrg3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrold JM, Ramanathan M, Mager DE. Network-based approaches in drug discovery and early development. Clin Pharmacol Ther. 2013; 94:651–658. doi: 10.1038/clpt.2013.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh PR, Lareau C, Shoresh N, et al. ; Brainstorm Consortium. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018; 50:621–629. doi: 10.1038/s41588-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003; 32:1–22. doi: 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- 14.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016; 40:304–314. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chignon A, Bon-Baret V, Boulanger MC, Li Z, Argaud D, Bossé Y, Thériault S, Arsenault BJ, Mathieu P. Single-cell expression and Mendelian randomization analyses identify blood genes associated with lifespan and chronic diseases. Commun Biol. 2020; 3:206. doi: 10.1038/s42003-020-0937-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Võsa U, Claringbould A, Westra H-J, Bonder MJ, Deelen P, Zeng B, Kirsten H, Saha A, Kreuzhuber R, Kasela S, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. 2018. BioRxiv. doi: 10.1101/447367
- 17.Pan DZ, Garske KM, Alvarez M, Bhagat YV, Boocock J, Nikkola E, Miao Z, Raulerson CK, Cantor RM, Civelek M, et al. Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS. Nat Commun. 2018; 9:1512. doi: 10.1038/s41467-018-03554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet. 2017; 49:1602–1612. doi: 10.1038/ng.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, et al. ; BLUEPRINT Consortium. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016; 167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basha O, Shpringer R, Argov CM, Yeger-Lotem E. The DifferentialNet database of differential protein-protein interactions in human tissues. Nucleic Acids Res. 2018; 46(D1):D522–D526. doi: 10.1093/nar/gkx981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009; 37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho-Silva D, Pierleoni A, Pignatelli M, Ong C, Fumis L, Karamanis N, Carmona M, Faulconbridge A, Hercules A, McAuley E, et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 2019; 47(D1):D1056–D1065. doi: 10.1093/nar/gky1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryaboshapkina M, Hammar M. Tissue-specific genes as an underutilized resource in drug discovery. Sci Rep. 2019; 9:7233. doi: 10.1038/s41598-019-43829-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motzer RJ, Hutson TE, Cella D, Reeves J, Hawkins R, Guo J, Nathan P, Staehler M, de Souza P, Merchan JR, et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med. 2013; 369:722–731. doi: 10.1056/NEJMoa1303989 [DOI] [PubMed] [Google Scholar]
- 25.Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, Nadler J, Clotet B, Karlsson A, Wohlfeiler M, et al. ; MOTIVATE Study Teams. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med. 2008; 359:1429–1441. doi: 10.1056/NEJMoa0803152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timmers PR, Mounier N, Lall K, Fischer K, Ning Z, Feng X, Bretherick AD, Clark DW, Shen X, Esko T, et al. ; eQTLGen Consortium. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. eLife. 2019; 8:e39856. doi: 10.7554/eLife.39856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, et al. Single-cell immune landscape of human atherosclerotic plaques. Nat Med. 2019; 25:1576–1588. doi: 10.1038/s41591-019-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020; 15:1484–1506. doi: 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- 29.Hamanishi T, Furuta H, Kato H, Doi A, Tamai M, Shimomura H, Sakagashira S, Nishi M, Sasaki H, Sanke T, et al. Functional variants in the glutathione peroxidase-1 (GPx-1) gene are associated with increased intima-media thickness of carotid arteries and risk of macrovascular diseases in japanese type 2 diabetic patients. Diabetes. 2004; 53:2455–2460. doi: 10.2337/diabetes.53.9.2455 [DOI] [PubMed] [Google Scholar]
- 30.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015; 162:425–440. doi: 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 2015; 6:7866. doi: 10.1038/ncomms8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oishi Y, Spann NJ, Link VM, Muse ED, Strid T, Edillor C, Kolar MJ, Matsuzaka T, Hayakawa S, Tao J, et al. SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 2017; 25:412–427. doi: 10.1016/j.cmet.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verploegen S, Ulfman L, van Deutekom HW, van Aalst C, Honing H, Lammers JW, Koenderman L, Coffer PJ. Characterization of the role of CaMKI-like kinase (CKLiK) in human granulocyte function. Blood. 2005; 106:1076–1083. doi: 10.1182/blood-2004-09-3755 [DOI] [PubMed] [Google Scholar]
- 34.Souilhol C, Gauci I, Feng S, Tardajos Ayllon B, Mahmoud M, Canham L, Fragiadaki M, Serbanovic-Canic J, Ridger V, Evans PC. Homeobox B9 integrates bone morphogenic protein 4 with inflammation at atheroprone sites. Cardiovasc Res. 2020; 116:1300–1310. doi: 10.1093/cvr/cvz235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steenman M, Espitia O, Maurel B, Guyomarch B, Heymann MF, Pistorius MA, Ory B, Heymann D, Houlgatte R, Gouëffic Y, et al. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci Rep. 2018; 8:3940. doi: 10.1038/s41598-018-22292-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClung JP, Roneker CA, Mu W, Lisk DJ, Langlais P, Liu F, Lei XG. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc Natl Acad Sci U S A. 2004; 101:8852–8857. doi: 10.1073/pnas.0308096101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moris D, Spartalis M, Spartalis E, Karachaliou GS, Karaolanis GI, Tsourouflis G, Tsilimigras DI, Tzatzaki E, Theocharis S. The role of reactive oxygen species in the pathophysiology of cardiovascular diseases and the clinical significance of myocardial redox. Ann Transl Med. 2017; 5:326. doi: 10.21037/atm.2017.06.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caldemeyer L, Dugan M, Edwards J, Akard L. Long-term side effects of tyrosine kinase inhibitors in chronic myeloid leukemia. Curr Hematol Malig Rep. 2016; 11:71–79. doi: 10.1007/s11899-016-0309-2 [DOI] [PubMed] [Google Scholar]
- 39.Borrás-Blasco J, Navarro-Ruiz A, Borrás C, Casterá E. Adverse cutaneous reactions associated with the newest antiretroviral drugs in patients with human immunodeficiency virus infection. J Antimicrob Chemother. 2008; 62:879–888. doi: 10.1093/jac/dkn292 [DOI] [PubMed] [Google Scholar]
- 40.Cremer S, Michalik KM, Fischer A, Pfisterer L, Jaé N, Winter C, Boon RA, Muhly-Reinholz M, John D, Uchida S, et al. Hematopoietic deficiency of the long noncoding RNA MALAT1 promotes atherosclerosis and plaque inflammation. Circulation. 2019; 139:1320–1334. doi: 10.1161/CIRCULATIONAHA.117.029015 [DOI] [PubMed] [Google Scholar]
- 41.Braunersreuther V, Zernecke A, Arnaud C, Liehn EA, Steffens S, Shagdarsuren E, Bidzhekov K, Burger F, Pelli G, Luckow B, et al. Ccr5 but not Ccr1 deficiency reduces development of diet-induced atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 2007; 27:373–379. doi: 10.1161/01.ATV.0000253886.44609.ae [DOI] [PubMed] [Google Scholar]
- 42.Wu MY, Eldin KW, Beaud AL. Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst. 2008; 100:1247–1259. doi: 10.1093/jnci/djn253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005; 37:710–717. doi: 10.1038/ng1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017; 8:1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44(W1):W90–W97. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trynka G, Westra H-J, Slowikowski K, Hu X, Xu H, Stranger BE, Klein RJ, Han B, Raychaudhuri S. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am J Hum Genet. 2015; 97:139–152. doi: 10.1016/j.ajhg.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017; 46:1734–1739. doi: 10.1093/ije/dyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016; 48:481–487. doi: 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 49.Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019; 47(W1):W234–W241. doi: 10.1093/nar/gkz240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kryuchkova-Mostacci N, Robinson-Rechavi M. A benchmark of gene expression tissue-specificity metrics. Brief Bioinform. 2017; 18:205–214. doi: 10.1093/bib/bbw008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018; 36:411–420. doi: 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]


