Abstract
Measuring the methylation status of cell-free DNA (cfDNA) in plasma holds great potential for the early, noninvasive detection of cancer. Two recent papers published in Nature Medicine showcase the successful application of cfDNA methylation- based cancer detection to two highly challenging scenarios.
Liquid biopsy holds great promise as a method of noninvasive cancer detection, especially through the analysis of cell-free DNA (cfDNA) fragments. In patients with cancer, cfDNA often includes tumour DNA released into the bloodstream through apoptosis, necrosis and/or active secretion. However, sensitively detecting the usually very limited amounts of tumour DNA in the blood of patients with early stage cancers remains an ongoing challenge. Recently, in two studies with results published in Nature Medicine, investigators used cfDNA methylome analysis to detect and classify intracranial tumours from plasma cfDNA1, and renal cell carcinomas (RCCs) from plasma and urinary cfDNA2. Tumours in both locations are often difficult to detect: in patients with intracranial tumours, the blood–brain barrier restricts the release of tumour DNA into blood3, and, among all major extracranial tumour types, RCCs shed the least cfDNA into blood4.
Noninvasive, early detection is enormously important for patients with intracranial tumours or RCCs. Intracranial tumours include a diverse group of conditions, and standard-of-care MRI often fails to reliably differentiate benign from malignant tumours, or primary intracranial tumours from metastatic lesions of an extracranial origin. Currently, the accurate diagnosis of brain lesions found on imaging requires invasive and risky tissue biopsy sampling or excision. Any noninvasive strategy that provides reliable diagnostic information would vastly improve neurosurgical planning for some patients and enable others to avoid surgical procedures altogether. In RCC, no clinically validated, noninvasive biomarker currently exists. Furthermore, RCC is the most lethal urological malignancy, killing 50% of patients that develop the disease5. Incidental renal masses are seen in up to one in four patients in abdominal imaging studies6, and accurate diagnosis requires invasive histopathological examinations. Given the ease of routine clinical sampling, urine is an attractive source of cfDNA for the noninvasive detection of RCCs.
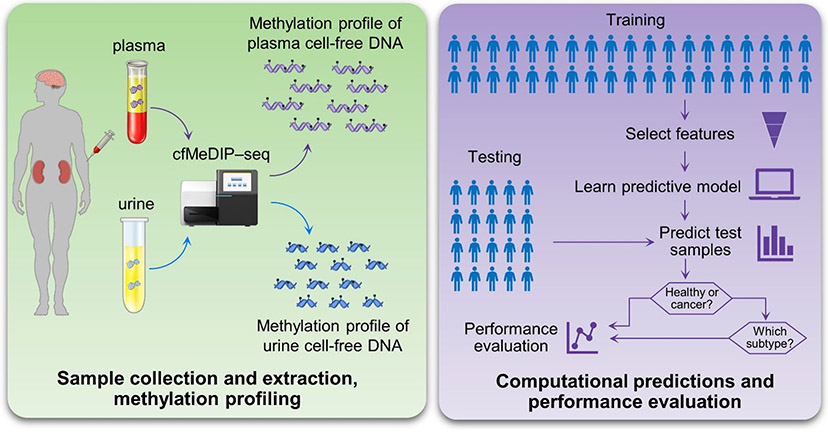
Nassiri et al.1 and Nuzzo et al.2 not only overcome numerous technical challenges to sensitively detect intracranial tumours and RCCs, respectively, but also demonstrate feasibility in reliably discriminating between distinct subtypes (Fig. 1). Analysis of DNA methylation provides a higher level of sensitivity than mutation-based cancer detection because aberrant methylation is both more prevalent and more pervasive. DNA methylation is also tissue specific, therefore analysis of cfDNA methylation can reveal the tissue of origin of a tumour7. This information can be challenging to obtain using MRI alone. In both papers, investigators use the cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) method, which is a sensitive, low-input, cost-efficient and bisulfite-free technology for the profiling of cfDNA methylomes7. This approach enriches methylated DNA fragments in cfDNA by immunoprecipitation, followed by high-throughput sequencing. cfMeDIP-seq has been previously demonstrated to provide a high level of sensitivity for both the detection and classification of several tumour types, including both localized and metastatic cancers8.
Fig. 1 ∣. Applications of cfMeDIP-seq for the detection of intracranial tumours and renal cell carcinomas.
Cell-free DNA (cfDNA) from plasma or urine is extracted and subjected to methylation profiling using the cell-free methylated DNA immunoprecipitation and high- throughput sequencing (cfMeDIP-seq) method. The training samples are used to select features and develop predictive models that reliably discriminate between patients with cancer and those without, as well as between various cancer subtypes. The held-out testing samples are used to validate the classification performance. If a patient is classified as having cancer, additional classifications can be performed to determine the subtype.
To demonstrate the accuracy of cfMeDIP-seq for the detection of intracranial tumours, Nassiri et al.1 obtained 447 cfDNA samples across eight tumour types and from individuals without cancer. The samples were split into training and test sets, and random forest classifiers were trained using the top 300 differentially methylated regions (DMRs) for gliomas versus each other class, and model performance was then tested on the held-out test sets (Fig. 1). The final models were highly precise and sensitive for classifying gliomas among patients with this cancer and in those without (mean area under the receiver operating characteristic (AUROC) curve = 0.99). This team collected an additional 161 samples from patients with solitary extra-axial tumours and intra-axial tumours, to demonstrate the feasibility of cfMeDIP-seq in reliably discriminating between common extra-axial tumours (AUROC of meningioma versus others = 0.89; AUROC of haemangiopericytoma versus others = 0.95), as well as intra-axial tumours (AUROC of low-grade indolent glial–neuronal tumours versus others = 0.93; AUROC of IDH-mutant glioma versus others = 0.82; and AUROC of IDH wild-type glioma versus others = 0.71).
Nuzzo et al.2 used cfMeDIP-seq to generate data from both plasma and urine samples obtained from a cohort of 120 patients with RCC and 28 individuals without cancer (Fig. 1). Similar to the study conducted by Nassiri et al.1 they selected 300 DMRs as discriminative features, then trained a machine learning classifier (GLMnet) on the dataset. These investigators also report an AUROC of 0.99. Notably, 33.3% of plasma samples and 66.7% of urine samples came from patients with stage I or stage II disease, according to the TNM Classification of Malignant Tumors. To evaluate classification performance between different genitourinary tumours, the team performed the same analysis comparing samples from patients with stage IV RCC versus stage IV urothelial carcinoma, resulting in a mean AUROC of 0.98. To investigate the performance of cfMeDIP–seq of urine samples, they conducted the same analyses on urinary cfDNA from patients with RCC and those without. The mean AUROC across 100 train–test partitions was 0.86. Note that the protocol used to generate these data was optimized for the analysis of plasma, therefore the performance of urine-based classifications could potentially be further improved by tailoring the experimental and computational approaches.
These two studies highlight the superb capabilities of liquid biopsy plus methylation analysis as a resource for accurate and noninvasive cancer detection and subtyping. These data also validate the performance of the cfMeDIP–seq platform, which can sensitively detect tumour epigenetic signatures in body fluids even in the highly challenging scenarios described in these reports. This methylome-based approach is noninvasive, cost-saving, efficient (turn-around time of a week) and applicable to various sources of cfDNA (including blood and urine). If validated in larger cohort studies, such a liquid biopsy test could be a game-changer for the diagnosis and management of multiple types of cancer. Furthermore, given that many diseases of different organs can cause epigenetic changes that are potentially detectable in cfDNA, we anticipate that noninvasive, cfDNA methylome-based diagnostics will have an increasingly important role in disease screening and health monitoring in future.
Acknowledgements
The authors’ work is supported by the NIH grants U01CA230705, R01CA246329, and U01CA237711.
Footnotes
Competing interests
W.L. and X.J.Z. are co-founders and shareholders of EarlyDiagnostics Inc.
References
- 1.Nassiri F et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med 26, 1044–1047 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuzzo PV et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med 26, 1041–1043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettegowda C et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med 6, 224ra24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zill OA et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res 24, 3528–3538 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Reynard J, Brewster S & Biers S Oxford Handbook of Urology. (Oxford University Press, 2013). [Google Scholar]
- 6.Gill IS, Aron M, Gervais DA & Jewett MAS Small renal mass. N. Engl. J. Med 362, 624–634 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Kang S et al. CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol. 18, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen SY, Burgener JM, Bratman SV & De Carvalho DD Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat. Protoc 14, 2749–2780 (2019). [DOI] [PubMed] [Google Scholar]