ABSTRACT
A total of 1,281 specimens from 1,024 patients were screened. Phylogenetic analysis classified 44 of these isolates as Klebsiella quasipneumoniae subsp. similipneumoniae (44/1,281 [3.4%]) and the remaining three as K. quasipneumoniae subsp. quasipneumoniae. The most common specimen source was urine (21/47 [44.7%]) followed by blood (14/47 [29.8%]). K. quasipneumoniae isolates were nonclonal. Carbapenemase-encoding genes (blaNDM and blaOXA-181) were detected in only two isolates (2/47 [4.3%]). K. quasipneumoniae appears to cause a spectrum of infections similar to those of K. pneumoniae, although higher rates of susceptibility to many commonly tested antimicrobials and low prevalence of virulence genes were demonstrated.
KEYWORDS: antimicrobial resistance, clinical microbiology, Enterobacteriaceae, whole genome
INTRODUCTION
Klebsiella pneumoniae is a pathogenic Gram-negative organism capable of causing serious infections in humans and can acquire significant antibiotic resistance. The spectrum of infection is wide, including urinary tract infections and respiratory tract infections. Invasive infections such as liver abscess, endophthalmitis, and meningitis have been attributed to hypervirulent Klebsiella pneumoniae (1, 2). Hypervirulence has been associated with a mucoid phenotype in strains with particular capsular serotypes (K1, K2, and K5) and virulence genes, such as siderophore systems yersiniabactin, aerobactin, and salmochelin (ybt, iuc, and iro), genotoxin colibactin (clb), and the regulators of mucoid phenotype, rmpA/rmpA2 (3).
Genomic analysis has distinguished Klebsiella pneumoniae into at least seven phylogroups: K. pneumoniae sensu stricto (Kp1), K. quasipneumoniae subsp. quasipneumoniae (Kp2), K. variicola (Kp3), K. quasipneumoniae subsp. similipneumoniae (Kp4), K. variicola subsp. tropicalensis (Kp5), K. quasivariicola (Kp6), and K. africanensis (Kp7) (4–8).
There is increasing recognition that both K. quasipneumoniae subspecies cause infections in humans (5, 9–11). Of particular concern were reports of K. quasipneumoniae subsp. similipneumoniae identified as the causative microorganism of neonate septicemia in China (12) and also the cause of a neonatal bacteremia outbreak at a tertiary hospital in Nigeria (13). The K. quasipneumoniae subspecies have also been observed to acquire plasmid-mediated carbapenem resistance (14–16), which may complicate treatment regimens. The true prevalence of K. quasipneumoniae remains unknown, as it is indistinguishable from K. pneumoniae (Kp1) in clinical laboratories which utilize matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) for routine microbial identification (17).
In this study, we investigated the distribution of K. quasipneumoniae within the K. pneumoniae complex and performed whole-genome sequencing to describe their genomic characteristics with a focus on acquired antimicrobial resistance and virulence genes.
Blood culture isolates in 2019 and all isolates between February and August 2020 identified as K. pneumoniae by MALDI-TOF (Bruker MALDI Biotyper; Bruker, Billerica, MA, USA) were included. Further identification of these isolates was performed using PCR for detection of OKP β-lactamase (blaOKP), a known chromosomal species-specific marker for K. quasipneumoniae (16). The primers okpBF, 5′-GCCAGCCCTCAGCCGCTTGAG-3′, and okpBR, 5′-ATAGATCACCACGATACGCTCCGC-3′, were used for PCR amplification, producing 714-bp amplicons, which then underwent Sanger sequencing. Routine susceptibility testing was performed using Vitek 2 (bioMérieux, Marcy-l’Étoile, France), and antimicrobial susceptibility was interpreted based on European Committee on Antimicrobial Susceptibility Testing (EUCAST) susceptibility break points (version 11.0).
Illumina NovaSeq 6000 sequencing (Illumina Inc., San Diego, CA, USA) was used to generate and assemble 150-bp paired-end reads. An average sequencing depth of 250× was achieved for the genomes. Raw reads were trimmed using Trimmomatic v. 0.38 (18) and then assembled with SPAdes version 3.14.0 (19). Genome annotation was carried out using Prokka (20). Kleborate (21) was used to identify the multilocus sequence type, species identity, K. pneumoniae integrating conjugative element (ICEKp)-associated and plasmid-associated virulence loci and antimicrobial resistance. Kleborate also assigns a virulence score based on the presence of ybt, clb, and iuc as follows: 0, none present; 1, ybt only; 2, clb without iuc (regardless of ybt; however, ybt is almost always present when clb is); 3, iuc only; 4, iuc and ybt without clb; and 5, all three genes present. Abricate using the database PlasmidFinder 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) was used to determine the presence of plasmid replicons. Single-nucleotide polymorphisms (SNPs) were identified using Snippy (https://github.com/tseemann/snippy). A total of 71,339 SNPs were included for phylogenetic analysis with 1,000 bootstrap replicates using FastTree (22). Tree editing and annotation were performed using interactive Tree Of Life (iTOL) (23). Average nucleotide identity (ANI) values were calculated using the Pyani package (https://github.com/widdowquinn/pyani). The following reference genomes were used for ANI comparisons: K. quasipneumoniae subsp. similipneumoniae strain ATCC 700603 (GenBank assembly accession GCA_003181175.1), K. quasipneumoniae subsp. quasipneumoniae MGH96 (GenBank assembly accession GCA_001033665.1), and K. pneumoniae ATCC 35657 strain KPN1482 (GenBank CP015134.1).
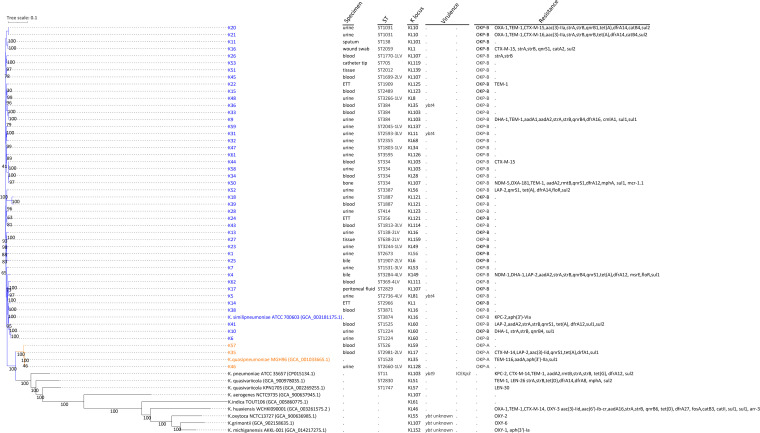
A total of 1,281 isolates from 1,024 patients were included. The specimen sources were diverse and included respiratory samples (bronchoalveolar lavage fluid and sputum), urine, rectal swabs, wound swabs, tissue, bone, pus, bile, and pleural and peritoneal fluid. Of the 1,281 isolates, 47 were blaOKP positive. blaOKP-A is associated with K. quasipneumoniae subsp. quasipneumoniae and blaOKP-B with K. quasipneumoniae subsp. similipneumoniae (Fig. 1). All 47 isolates were whole-genome sequenced, and both phylogenetic analysis and Kleborate classified 44 of these isolates as K. quasipneumoniae subsp. similipneumoniae (44/1,281 [3.43%]) and the remaining three as K. quasipneumoniae subsp. quasipneumoniae (3/1,281 [0.23%]) (Fig. 1). The species identification was also supported by ANI values of >99% compared to their species reference genome (data not shown). The most common specimen source was urine (21/47 [44.7%]) followed by blood (14/47 [29.8%]) (Fig. 1). Our results are consistent with previous studies in which K. quasipneumoniae typically forms a smaller proportion of the K. pneumoniae complex, with K. pneumoniae sensu stricto being the dominant species (4, 24). Likewise, Long et al. (16) suggested that approximately 2% of human infections attributed to K. pneumoniae were actually caused by K. quasipneumoniae.
FIG 1.
Core SNP phylogenetic tree of 47 Klebsiella quasipneumoniae. The metadata include specimen source, sequence type (ST), K-loci, virulence factors, presence of blaOKP, and other acquired resistance determinants. LV, locus variant; ETT, endotracheal aspirate; ybt, yersiniabactin; ICEKp, Klebsiella integrative conjugative element. The blue branch labels belong to K. quasipneumoniae subsp. similipneumoniae, and the orange labels belong to K. quasipneumoniae subsp. quasipneumoniae. The bootstrap values are indicated on the nodes, where black asterisks represent a bootstrap value of 100.
The K. quasipneumoniae isolates were nonclonal, with 39 diverse sequence types (STs) (Fig. 1). The most commonly identified ST was ST334 (4/47 [8.5%]). K. quasipneumoniae ST334 has been reported as a potential emerging outbreak-associated antimicrobial-resistant clone (25). In a Cambodian neonatal unit, ST334 was the most prevalent K. quasipneumoniae subsp. similipneumoniae sequence type, representing 30.5% of all isolates analyzed (26). Additionally, in a Pakistani hospital, the major sequence type observed was ST334, making up 29.2% of all K. quasipneumoniae hospital isolates. In our cohort, none of our K. quasipneumoniae isolates came from neonates. Of the ST334 isolates, one particular isolate was phenotypically multidrug resistant and was also the only isolate bearing dual carbapenemases (NDM-5 and OXA-181) and plasmid-mediated mcr-1.1 (Fig. 1). ST2727 K. quasipneumoniae subsp. similipneumoniae has been reported in an outbreak event in a neonatal intensive care unit (NICU) (12). However, none of the isolates in our study belonged to this sequence type. Typically, the number of SNP differences for isolates with the same sequence type was observed to be <500, while nonrelated sequence types had SNP differences of >1,100 (see Fig. S1 in the supplemental material).
Acquired antimicrobial resistance genes were observed in 13 isolates (13/47 [27.6%]). Carbapenemase-encoding genes (blaNDM and blaOXA-181) were detected in two isolates (2/47 [4.3%]). Other observed resistance genes included those encoding extended-spectrum β-lactamases (blaCTX-M), oxacillinases (blaOXA-1), penicillinases (blaTEM), aminoglycoside-modifying enzymes (aacA, aadA, strA, and strB), fluoroquinolone resistance proteins (qnrB4 and qnrS), dihydrofolate reductase (dfr), and dihydropteroate synthase (sul1 and sul2) conferring trimethoprim-sulfamethoxazole resistance as well as plasmid-mediated colistin resistance (mcr-1.1) (Fig. 1).
We reviewed the phenotypic drug susceptibility data for K. quasipneumoniae; in addition, antibiograms from K. pneumoniae identified during the study period were used as a comparator. Overall, there was a trend toward higher rates of susceptibility to the tested antibiotics (with the exceptions of those for meropenem and amikacin, which were similar) for K. quasipneumoniae than for K. pneumoniae (Table 1).
TABLE 1.
Susceptibility rates of K. quasipneumoniae and K. pneumoniae isolated during the 6-month study period from February to August 2020
| Species | Antibiotic susceptibility (% [no. of susceptible isolates/total no. of isolates]) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Amoxicillin/clavulanic acid | Ceftriaxone | Piperacillin-tazobactam | Meropenem | Gentamicin | Amikacin | Ciprofloxacin | Sulfamethoxazole-trimethoprim | |
| K. quasipneumoniae | 80.9 (38/47) | 72. (34/47) | 87.2 (41/47) | 97.9 (46/47) | 91.5 (43/47) | 97.9 (46/47) | 80.9 (38/47) | 83.0 (39/47) |
| K. pneumoniae | 70.8 (862/1,218) | 69.6 (848/1,218) | 75.8 (920/1,214) | 98.1 (1,194/1,218) | 87.4 (1,065/1,218) | 97.8 (1,191/1,218) | 64.4 (784/1,218) | 71.2 (867/1,218) |
Our observations paralleled the study by Lam et al. (27), in which no ICEKp was detected, and ybt4 (plasmid encoded and not mobilized by ICEKp) was the only virulence factor detected in three K. quasipneumoniae subsp. similipneumoniae isolates (Fig. 1). It has been noted that ICEKp, an integrative conjugative element which mobilizes the ybt locus, was rare or absent from K. pneumoniae’s closest relatives, K. variicola and K. quasipneumoniae (27). iuc, clb, iro, and rmpA/rmpA2 genes were not detected in any isolate.
Of the three ybt+ isolates, one was from blood and the other two were from urine. The overall virulence score as predicted by the Kleborate tool (21) was 0 (no virulence factors detected) or 1 (only ybt detected) (21). In contrast, it was common to observe scores of >2 in the more pathogenic K. pneumoniae isolates (21). The ybt+ isolates also did not have either K1, K2, or K5 capsular serotypes. In addition, a capsule synthesis locus (K-locus) was detected for every isolate (Fig. 1). Thirty-nine distinct K-loci were identified, reflecting the immense diversity of the capsular serotype. The K-loci did not appear to associate with phylogenetic clusters (Fig. 1). Two of our isolates had the K1 serotype and were recovered from a wound swab and an endotracheal tube specimen (Fig. 1), which are not sample sites typically associated with hypervirulent K. pneumoniae infections. Genomes from the Klebsiella liver abscess syndrome study (28) were also investigated to identify potential K. quasipneumoniae isolates that were misidentified as K. pneumoniae. We performed the same bioinformatic analysis described in that study on the 70 genomes (NCBI BioProject PRJNA351910) and found that one of the genomes was blaOKP-B-positive K. quasipneumoniae subsp. similipneumoniae. Interestingly, this clinical Klebsiella liver abscess isolate (TTSH04) harbored a novel multilocus sequence type (MLST; ST2037) and capsule serotype (28) and had a virulence score of 0. K. quasipneumoniae isolates with the K1 capsule as well as the complement of virulence factors (ybt, iro, and rmpA) have also been reported as the causative agent of liver abscess (29, 30).
In our study, the proportion of K. quasipneumoniae isolates identified within K. pneumoniae was small (3.67%). Consistent with other reports, K. quasipneumoniae isolates bearing virulence genes were the minority (three ybt+ isolates) (31), which may indicate that the virulence potential of K. quasipneumoniae is limited. Despite this, the sample source distribution of K. quasipneumoniae isolates suggests that the spectrum of infections is similar to that of K. pneumoniae sensu stricto and cannot be dismissed as nonpathogenic. Also, K. quasipneumoniae has been sporadically reported in the literature as the causative agent of liver abscesses exhibiting a hypervirulent phenotype. K. quasipneumoniae may still represent significant pathogen potential despite higher overall susceptibility rates than for K. pneumoniae isolates.
Data availability.
The raw reads of the isolates sequenced in this study have been deposited under BioProject number PRJNA704495.
ACKNOWLEDGMENT
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Catalán-Nájera JC, Garza-Ramos U, Barrios H, Catal JC. 2017. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 8:1111–1113. 10.1080/21505594.2017.1317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chew KL, Lin RTP, Teo JWP. 2017. Klebsiella pneumoniae in Singapore: hypervirulent infections and the carbapenemase threat. Front Cell Infect Microbiol 7:515. 10.3389/fcimb.2017.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NTK, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brisse S, Verhoef J. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol Microbiol 51:915–924. 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 5.Brisse S, Passet V, Grimont PAD. 2014. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol 64:3146–3152. 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 6.Rosenblueth M, Martínez L, Silva J, Martínez-Romero E. 2004. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol 27:27–35. 10.1078/0723-2020-00261. [DOI] [PubMed] [Google Scholar]
- 7.Long SW, Linson SE, Ojeda Saavedra M, Cantu C, Davis JJ, Brettin T, Olsen RJ. 2017. Whole-genome sequencing of a human clinical isolate of the novel species Klebsiella quasivariicola sp. nov. Genome Announc 5:e01057-17. 10.1128/genomeA.01057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues C, Passet V, Rakotondrasoa A, Diallo TA, Criscuolo A, Brisse S. 2019. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res Microbiol 170:165–170. 10.1016/j.resmic.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Brisse S, van Himbergen T, Kusters K, Verhoef J. 2004. Development of a rapid identification method for Klebsiella pneumoniae phylogenetic groups and analysis of 420 clinical isolates. Clin Microbiol Infect 10:942–945. 10.1111/j.1469-0691.2004.00973.x. [DOI] [PubMed] [Google Scholar]
- 10.Maatallah M, Vading M, Kabir MH, Bakhrouf A, Kalin M, Nauclér P, Brisse S, Giske CG. 2014. Klebsiella variicola is a frequent cause of bloodstream infection in the Stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS One 9:e113539. 10.1371/journal.pone.0113539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai K, Ishibashi N, Kodana M, Tarumoto N, Sakai J, Kawamura T, Takeuchi S, Taji Y, Ebihara Y, Ikebuchi K, Murakami T, Maeda T, Mitsutake K, Maesaki S. 2019. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014–2017. BMC Infect Dis 19:946. 10.1186/s12879-019-4498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perlaza-Jiménez L, Wu Q, Torres VVL, Zhang X, Li J, Rocker A, Lithgow T, Zhou T, Vijaykrishna D. 2020. Forensic genomics of a novel Klebsiella quasipneumoniae type from a neonatal intensive care unit in China reveals patterns of genetic diversity, evolution and epidemiology. Microb Genom 6:mgen000433. 10.1099/mgen.0.000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okomo U, Senghore M, Darboe S, Bojang E, Zaman SMA, Hossain MJ, Nwakanma D, Le Doare K, Holt KE, Hos NJ, Lawn JE, Bentley SD, Kampmann B. 2020. Investigation of sequential outbreaks of Burkholderia cepacia and multidrug-resistant extended spectrum β-lactamase producing Klebsiella species in a West African tertiary hospital neonatal unit: a retrospective genomic analysis. Lancet Microbe 1:e119–e129. 10.1016/S2666-5247(20)30061-6. [DOI] [PubMed] [Google Scholar]
- 14.Nicolas MF, Ramos PIP, Marques de Carvalho F, Camargo DRA, de Fatima Morais Alves C, Loss de Morais G, Almeida LGP, Souza RC, Ciapina LP, Vicente ACP, Coimbra RS, Ribeiro de Vasconcelos AT. 2018. Comparative genomic analysis of a clinical isolate of Klebsiella quasipneumoniae subsp. similipneumoniae, a KPC-2 and OKP-B-6 beta-lactamases producer harboring two drug-resistance plasmids from southeast Brazil. Front Microbiol 9:220. 10.3389/fmicb.2018.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathers AJ, Crook D, Vaughan A, Barry KE, Vegesana K, Stoesser N, Parikh HI, Sebra R, Kotay S, Walker AS, Sheppard AE. 2019. Klebsiella quasipneumoniae provides a window into carbapenemase gene transfer, plasmid rearrangements, and patient interactions with the hospital environment. Antimicrob Agents Chemother 63:e02513-18. 10.1128/AAC.02513-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long SW, Linson SE, Saavedra MO, Cantu C, Davis JJ, Brettin T, Olsen RJ. 2017. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mBio 2:e00290-17. 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues C, Passet V, Rakotondrasoa A, Brisse S. 2018. Identification of Klebsiella pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and related phylogroups by MALDI-TOF mass spectrometry. Front Microbiol 9:3000. 10.3389/fmicb.2018.03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 21.Lam MMC, Wick RR, Wyres KL, Holt KE. 14 December 2020. Genomic surveillance framework and global population structure for Klebsiella pneumoniae. bioRxiv 10.1101/2020.12.14.422303. [DOI]
- 22.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letunic I, Bork P. 2016. Interactive Tree Of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Octavia S, Kalisvar M, Venkatachalam I, Ng OT, Xu W, Sridatta PSR, Ong YF, De Wang L, Chua A, Cheng B, Lin RTP, Teo JWP. 2019. Klebsiella pneumoniae and Klebsiella quasipneumoniae define the population structure of blaKPC-2Klebsiella: a 5 year retrospective genomic study in Singapore. J Antimicrob Chemother 74:3205–3210. 10.1093/jac/dkz332. [DOI] [PubMed] [Google Scholar]
- 25.Wyres KL, Holt KE. 2016. Klebsiella pneumoniae Population genomics and antimicrobial-resistant clones. Trends Microbiol 24:944–956. 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Crellen T, Turner P, Pol S, Baker S, Thi Nguyen TN, Stoesser N, Day NP, Turner C, Cooper BS. 2019. Transmission dynamics and control of multidrug-resistant Klebsiella pneumoniae in neonates in a developing country. Elife 8:e50468. 10.7554/eLife.50468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE. 2018. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 4:e000196. 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee IR, Molton JS, Wyres KL, Gorrie C, Wong J, Hoh CH, Teo J, Kalimuddin S, Lye DC, Archuleta S, Holt KE, Gan Y-H. 2016. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep 6:29316. 10.1038/srep29316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. 2018. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol 18:6. 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breurec S, Melot B, Hoen B, Passet V, Schepers K, Bastian S, Brisse S. 2016. Liver abscess caused by infection with community-acquired Klebsiella quasipneumoniae subsp. quasipneumoniae. Emerg Infect Dis 22:529–531. 10.3201/eid2203.151466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam MMC, Wyres KL, Duchene S, Wick RR, Judd LM, Gan Y-H, Hoh C-H, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. 2018. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 9:2703. 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC00412-21_Supp_S1_seq4.pdf, PDF file, 0.7 MB (687.1KB, pdf)
Data Availability Statement
The raw reads of the isolates sequenced in this study have been deposited under BioProject number PRJNA704495.