ABSTRACT
At a hospital system (H1) in Ontario, Canada, we investigated whether whole-genome sequencing (WGS) altered initial epidemiological interpretation of carbapenemase-producing Enterobacterales (CPE) transmission. We included patients with CPE colonization/infection identified by population-based surveillance from October 2007 to August 2018 who received health care at H1 in the year before/after CPE detection. H1 reported epidemiological transmission clusters. We combined single nucleotide variant (SNV) analysis, plasmid characterization, and epidemiological data. Eighty-five patients were included. H1 identified 7 epidemiological transmission clusters, namely, A to G, involving 24/85 (28%) patients. SNV analysis confirmed transmission clusters C, D, and G and identified two additional cases belonging to cluster A. One was a travel-related case that was the likely index case (0 to 6 SNVs from other isolates); this case stayed on the same unit as the initially presumed index case 4 months prior to detection of the initially presumed index case on another unit. The second additional case occupied a room previously occupied by 5 cluster A cases. Plasmid sequence analysis excluded a case from cluster A and identified clusters E and F as possibly two parts of a single cluster. SNV analysis also identified a case without direct epidemiologic links that was 18 to 21 SNVs away from cluster B, suggesting possible undetected interhospital transmission. SNV and plasmid sequence analysis identified cases belonging to transmission clusters that conventional epidemiology missed and excluded other cases. Implementation of routine WGS to complement epidemiological transmission investigations has the potential to improve prevention and control of CPE in hospitals.
KEYWORDS: carbapenem-resistant Enterobacteriaceae, beta-lactamases, disease transmission, genomic epidemiology, public health
INTRODUCTION
Carbapenemase-producing Enterobacterales (CPE) are among the most concerning antibiotic-resistant organisms. They are resistant to all beta-lactams and many other antibiotics (1). Invasive CPE infections have case fatality rates of up to 70% (2–5).
CPE are not yet widespread in Canada, with 0.03 CPE infections per 10,000 patient-days at sentinel sites across the country from 2012 to 2017 (6). However, the incidence of CPE in Toronto and Peel, south-central Ontario, has approximately doubled every 2 years since its emergence in 2007 to 1.3 clinical isolates per 100,000 people in 2017 (7). Notably, over one-third of all CPE cases in south-central Ontario are associated with local health care and not health care or travel abroad (8). Furthermore, the incidence of CPE associated with health care in south-central Ontario has approximately doubled since 2007 (9). These data highlight an urgent need to investigate CPE transmission dynamics locally.
We combined population-based surveillance data in south-central Ontario with isolate whole-genome sequencing (WGS) to explore CPE transmission among patients who received health care at a large hospital system in Toronto over a 10-year period. Specifically, we aimed to determine whether WGS altered the interpretation of CPE transmission that had been made using conventional epidemiology at the time of initial investigation.
RESULTS
This study included 85 patients with CPE colonization/infection. Table 1 shows baseline characteristics. Fifty-one (60%) patients had CPE first detected at hospital system (H1), 14 (16%) had CPE first detected at another Toronto Invasive Bacterial Diseases Network (TIBDN) hospital with health care at H1 in the year before detection, and 20 (24%) had CPE first detected at another TIBDN hospital with health care at H1 in the year after detection (Table 1). Forty-five (53%) patients had a surveillance (rectal) isolate and the remaining 40 (47%) had a clinical isolate (Table 1).
TABLE 1.
Baseline characteristics of patientsa with carbapenemase-producing Enterobacterales colonization/infection that received health care at H1 from 2007 to 2018
| Study characteristicb | Valuec |
|---|---|
| Characteristics of participants | |
| Age in yrs (median [IQR]) | 65 (51–76) |
| Female sex | 34 (40) |
| Nursing home resident | 4 (5) |
| Hospitalization in prior yrd | |
| Abroad only | 22 (26) |
| Canada only | 38 (45) |
| Abroad and Canada | 12 (14) |
| None | 12 (14) |
| Travel in prior yrd | |
| Indian subcontinent | 22 (26) |
| Other countries (except United States/northern Europe) | 13 (15) |
| None (or United States/northern Europe) | 49 (58) |
| Characteristics of participants’ first detected CPE | |
| Hospital CPE first detected | |
| H1 | 51 (60) |
| Another TIBDN hospital with receipt of healthcare at H1 in the yr after | 20 (24) |
| Another TIBDN hospital with receipt of healthcare at H1 in the yr before | 14 (16) |
| Sample type | |
| Rectal | 45 (53) |
| Urine | 24 (28) |
| Blood | 6 (7) |
| Wound | 8 (9) |
| Tissue | 2 (2) |
| Species | |
| Klebsiella pneumoniae | 39 (46) |
| Escherichia coli | 23 (27) |
| Enterobacter cloacae complex | 13 (15) |
| Othere | 10 (12) |
| Carbapenemase produced | |
| NDM | 32 (38) |
| KPCf | 27 (32) |
| OXA | 16 (19) |
| VIM | 6 (7) |
| NDM/OXA | 2 (2) |
| SME | 2 (2) |
n = 85.
IQR, interquartile range; CPE, carbapenemase-producing Enterobacterales; H1, hospital system 1; TIBDN, Toronto Invasive Bacterial Diseases Network.
Values are n (%) except where otherwise noted.
Missing travel and hospitalization data for one patient.
Other species included Citrobacter freundii (n = 3), Serratia marcescens (2), Providencia rettgeri (2), Mixta calida (2), Klebsiella oxytoca (1).
One was a KPC/VIM coproducer, and one was a KPC/NDM coproducer.
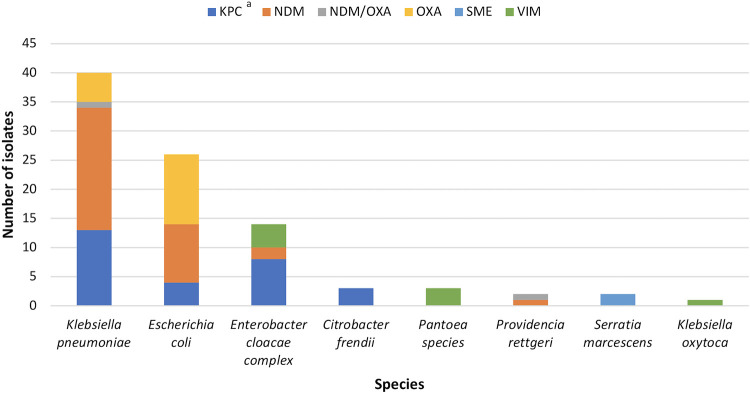
Seventy-nine (93%) patients had one carbapenemase/species and 6 (7%) patients had 2, for 91 CPE isolates total. Most species were Klebsiella pneumoniae (n = 40, 44%) and Escherichia coli (26, 29%), and most carbapenemases were NDM (34, 37%), KPC (28, 31%), or OXA-48-like (17, 19%) (Fig. 1).
FIG 1.
Carbapenemase/species of 91 carbapenemase-producing Enterobacterales (CPE) isolates belonging to patients who received health care at H1 from 2007 to 2018. a, One was a KPC/VIM coproducer, and one was a KPC/NDM coproducer.
Of 91 CPE isolates, 87 (96%) underwent WGS (4 could not be retrieved). Multilocus sequence typing (MLST) revealed a diverse isolate collection within each species. Only three sequence types (STs) were represented in more than 2 isolates; more than one-half of K. pneumoniae isolates belonged to ST147 (n =15, 39%) or ST258 (10, 32%), and almost one-quarter of E. coli isolates belonged to ST38 (6, 23%) (see Table S1 in the supplemental material).
Interpretation of transmission using conventional epidemiology.
The Infection Prevention and Control department at H1 (H1-IPAC) reported its interpretation of CPE transmission using conventional epidemiology at the time of initial investigation for the 51 patients whose CPE was first detected at H1 and, if known, for the 20 patients who received health care at H1 in the year after first CPE detection at another TIBDN hospital (Table 2). H1-IPAC reported that most patients acquired CPE via health care abroad (n = 24, 43%) or health care at H1 (27, 47%) (Table 2). Of the 20 patients who received health care at H1 in the year after CPE detection at another hospital, only 6 (30%) were known to H1-IPAC (Table 2); of the 14 patients not known to H1-IPAC, 11 (55%) had only outpatient visits at H1 and 3 (15%) were admitted without admission screening (see Table S2 in the supplemental material).
TABLE 2.
H1-IPAC’s interpretation of carbapenemase-producing Enterobacterales acquisition source among patients with CPE infection/colonization that received health care at H1 from 2007 to 2018a
| Hospital CPE first detected (n = 85) | No. (%) of cases known to H1-IPAC | Interpretation of CPE acquisition source as determined by H1-IPAC at initial investigation (n [%]) |
||||
|---|---|---|---|---|---|---|
| Healthcare abroad | Travel to the Indian subcontinent without healthcare | Healthcare in Canada (not at H1) | Healthcare at H1 | Unknown | ||
| H1 (n = 51) | 51 (100) | 20 (39) | 2 (4) | 0 | 26 (51) | 3 (6) |
| Another TIBDN hospital with receipt of healthcare at H1 in the yr after (n = 20) | 6 (30) | 4 (46) | 0 | 1 (17) | 1 (17) | 0 |
| Another TIBDN hospital with receipt of healthcare at H1 in the yr before (n = 14)b | 0 | NA | NA | NA | NA | NA |
CPE, carbapenemase-producing Enterobacterales; H1-IPAC, Infection Prevention and Control department at H1; TIBDN, Toronto Invasive Bacterial Diseases Network; NA, not applicable.
Two also received health care at H1 in the year after CPE detection at another hospital.
H1-IPAC identified 7 epidemiological clusters, namely, A to G, involving 24 (28%) of the 85 total patients (Table 3, with additional details in Table S3 in the supplemental material).
TABLE 3.
Characteristics of CPE transmission clusters, as determined using conventional epidemiology at the time of initial investigation at H1 as well as WGS, 2007 to 2018a
| Cluster | Yr | No. of cases in epidemiological cluster | No. of cases WGS added to cluster | No. of cases WGS removed from cluster | Carbapenemase gene | Plasmid replicon typeb | Species and ST | SNV differences between isolatesb | Affected units | Time between detection of first and last case |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 2011 | 9c | 2 | 1 | blaNDM-1 | IncFIB/IncR | K. pneumoniae ST147 | 0–31d | ICU A, ICU B, Unit A | 39 mo |
| B | 2010 | 4 | 1 | 0 | blaKPC-2 | IncFIB/IncFII | K. pneumoniae ST258 | 1–21e | ICU C | 6 mo |
| C | 2016 | 3 | 0 | 0 | blaKPC-2 | NA | K. pneumoniae ST258 | 1–2 | ICU C | 3 mo |
| D | 2015 | 2 | 0 | 0 | blaKPC-3 | NA | K. pneumoniae ST258 | 4 | Unit M | 4 mo |
| E | 2017 | 2 | 0 | 0 | blaVIM-1 | IncR, IncR/HI2/HI2A hybrid | ECC ST148, KO | NA | ICU C | 2 mo |
| F | 2018 | 2 | 0 | 0 | blaVIM-1 | IncR, IncR/HI2/HI2A hybrid | M. calida | 10 | ICU C | 4 days |
| G | 2016 | 2 | 0 | 0 | blaKPC-3 | NA | ECC ST97 | 3 | Unit B | 1 mo |
| H | 2017 | 2f | 0 | 0 | blaOXA-48 | NA | E. coli ST38 | 10 | NA | 2 mo |
CPE, carbapenemase-producing Enterobacterales; WGS, whole-genome sequencing; ST, sequence type; SNV, single nucleotide variant; ECC, E. cloacae complex; NA, not applicable.
For clusters where plasmid analysis or SNV analysis were not conducted, NA is marked.
An isolate of 1 of these 9 cases (A7) could not be retrieved.
There were 0–6 SNV differences between A1, A2, A3, A4, A5, A6, A10, and A11. A9 was 27–31 SNVs away from the rest of the cluster. A8 was excluded from the cluster by ST and replicon type (ST340, blaNDM-1 on an IncA/C-type plasmid).
There were 1–3 SNVs between B1, B2, B3, and B4. B5 was 18–21 SNVs away from the rest of the cluster.
Cluster was initially identified by SNV analysis and not by conventional epidemiology. No direct epidemiological link between cases could be identified.
Interpretation of transmission using conventional epidemiology and WGS.
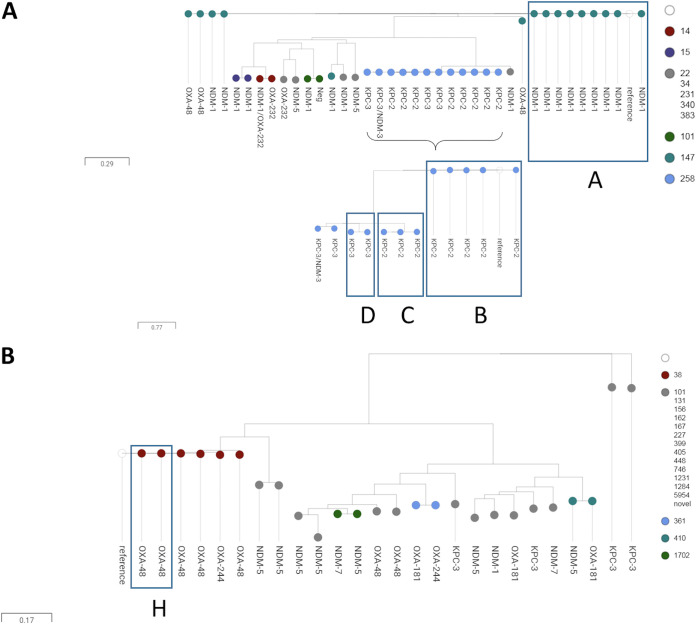
Single nucleotide variant (SNV) analysis was conducted on all isolates by species, with K. pneumoniae and E. coli isolate collections being the largest (Figure 2a and b). This analysis clustered isolates by their STs. SNV analysis was rerun by ST and carbapenemase variant, using internal references to each ST (data not shown). SNV analysis confirmed epidemiological clusters C, D, and G with 100% agreement (Table 3, Table S3). However, SNV data combined with molecular characterization of isolates revealed additional cases potentially related to epidemiological clusters A, B, E, and F, which were further investigated. SNV data also identified cluster H, including two cases that H1-IPAC had not identified as an epidemiological cluster.
FIG 2.
Single nucleotide variant tree of all carbapenemase-producing Klebsiella pneumoniae (n = 38) (A) and Escherichia coli (n = 26) (B) isolates belonging to patients who received health care at H1 from 2007 to 2018. Transmission clusters are boxed and labeled.
Cluster A.
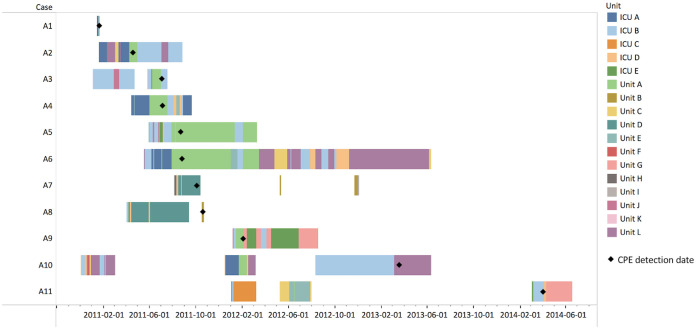
SNV analysis revealed two cases (A1 and A11) related to epidemiological cluster A (Table 3, Table S3). A1, whose CPE was associated with health care in the Indian subcontinent, was the likely index case (differing by as few as 0 SNVs from epidemiological cluster A). On further investigation, A1 could be linked to A2 (H1-IPAC’s epidemiologically presumed index case); 3 days after A1 was discharged from intensive care unit A (ICU A), A2 was admitted to a room two rooms away from A1 in ICU A. This occurred 4 months before A2’s CPE was first detected in a clinical isolate on unit A (Fig. 3). A11 (differing by as few as 0 SNVs from epidemiological cluster A) was first detected 13 months after the last epidemiologically defined cluster A case; A11 occupied a room in ICU B that had been occupied by 5 other epidemiological cluster A cases.
FIG 3.
Units that cluster A cases stayed on during their admissions to H1. Cases that stayed on unit A all stayed in unit A’s satellite room for ICU B.
SNV analysis flagged two cases (A8 and A9) in epidemiological cluster A requiring further investigation (Table 3, Table S3). A8 was a different sequence type (ST340) than other cluster isolates (ST147), and A9 was an outlier (27 to 31 SNVs away from other ST147 cluster isolates). To resolve the blaNDM-1-containing plasmids and determine potential transfer events, long-read sequencing was performed on A8, A9, and the index case A1 (for comparison). A9 harbored blaNDM-1 on a plasmid sharing 99.9% identity with A1’s 217-kb IncFIB/IncR-type plasmid (Fig. S1). Conversely, A8 harbored blaNDM-1 on an IncA/C-type plasmid. No other cluster A isolates harbored an IncA/C replicon, excluding A8 from cluster A.
Cluster B.
SNV analysis revealed a case (B5) whose blaKPC-2-containing K. pneumoniae ST258 isolate was 18 to 21 SNVs from epidemiological cluster B isolates (Table 3, Table S3). B5 had no direct epidemiological links to cluster B; B5 was identified a year before cluster B at another TIBDN hospital that shares patients with H1 and was admitted to H1 after all cluster B cases were detected. Given the number of SNV differences between B5 and cluster B isolates, long-read sequencing was performed to resolve and compare the blaKPC-2-containing plasmids of B5 and B1 (H1-IPAC’s epidemiologically presumed cluster B index case, whose CPE bacteremia was acquired at H1). Both isolates harbored a 114-kb blaKPC-2-containing IncFIB-type plasmid sharing 99.9% identity, suggesting that B5 was possibly related to epidemiological cluster B (see Fig. S2 in the supplemental material).
Clusters E and F.
Two cases (E1 and E2) with blaVIM-1-containing isolates of different species belonged to epidemiological cluster E (Table 3, Table S3). E2 was admitted to ICU C 3 weeks after E1 was discharged. Long-read sequencing was used to resolve the blaVIM-1-containing plasmids. The first identified case (E1) had a 32-kb blaVIM-1-containing IncR-type plasmid. The second case (E2) had a 497-kb blaVIM-1-containing IncR/HI2/HI2A-type hybrid plasmid. All (100%) of the IncR plasmid in E1 was present in E2’s HI2/HI2A plasmid, likely through homologous recombination (see Fig. S3 in the supplemental material). Six months after E2 was detected, epidemiological cluster F was identified. This cluster included two cases (F1 and F2) whose ICU C stays overlapped, with blaVIM-1-containing Mixta calida isolates differing by 10 SNVs. Clusters E and F were thought to be separate, given their weak spatial link (same unit but different rooms) and absence of a temporal link (3 months between discharge of cluster E cases and admission of F1).
The rare isolation of blaVIM-1 at H1 prompted further molecular analysis of all 4 isolates. Using a mapping-based approach and cluster E’s closed blaVIM-1-containing plasmids as a reference, we searched for the presence of the IncR and IncR/HI2/IncHI2A plasmids in cluster F. The IncR/HI2/HI2A hybrid plasmid was observed in both F1 and F2 at 99.9% and 80.9%, respectively. When we mapped the IncR plasmid alone, it was observed at over 99% identity in both cluster F isolates. It is possible that clusters E and F are related by plasmid transfer and that the mobilization of blaVIM-1 was driven by the movement of the IncR plasmid.
Cluster H.
Cluster H was identified by SNV analysis of E. coli isolates (Figure 2b, Table 3, and Table S3). Further analysis of the ST38 cluster revealed a pair of blaOXA-48-containing isolates differing by 10 SNVs; the OXA-48 region was integrated into the same chromosomal location in both isolates (data not shown). No direct epidemiological link between cases could be identified. The first identified case was detected on admission to H1, which occurred the day after return from travel to Egypt with health care. Two months later, the second identified case was detected at an outpatient visit at H1 within weeks of travel to the Indian subcontinent without health care.
DISCUSSION
Among CPE cases detected at or exposed to a large hospital system in Canada over 10 years, we examined whether WGS altered the initial epidemiological interpretation of transmission. Overall, there was diversity in CPE carbapenemases and STs. A minority of cases received health care at H1 after first detection at another local hospital, without being known to H1-IPAC. Over one-quarter of cases belonged to a cluster supported by short- and long-read WGS. Among 7 epidemiological clusters, WGS altered the initial epidemiologic interpretation of transmission in one (A), identified that two others (E and F) were possibly part of a single cluster, and linked one (B) to a case identified at another hospital. SNV analysis also identified a cluster (H) including two cases that H1-IPAC had not initially identified as epidemiologically linked.
Two-thirds of all CPE identified in south-central Ontario are associated with health care or travel abroad (8), explaining our diversity in carbapenemases/STs. As expected, the most common K. pneumoniae isolates were major clones (ST147 and ST258) (10–12).
Fourteen of the 20 cases that received health care at H1 in the year after first CPE detection at another hospital were not known to H1-IPAC. Although it is possible that these cases were no longer colonized by the time they received health care at H1, CPE colonization is generally prolonged (13). We did not detect transmission from these cases at H1, possibly because most of these cases were only outpatients (transmission in outpatient clinics is possible [14, 15] but less common), but also perhaps because contacts of the three admitted cases were not screened. Nonetheless, this observation highlights the potential for exposure when CPE reporting and transmission investigations are not centralized and when screening is limited to potential exposures outside the region. In our region in 2020, fewer than one-fifth of hospitals screened patients with prior local health care on admission (16), which hampers the “search and contain” strategy deemed important in CPE control.
SNV analysis supported transmission in clusters A, B, C, D, F, and G, and plasmid long-read sequence analysis supported transmission in clusters A, B, and E. Notably, plasmid analysis also suggested that clusters E and F were possibly linked. Although there was no temporal overlap between these clusters in ICU C, an undetected environmental reservoir or intermediate undetected cases may explain this link. If WGS had been available, the initial IPAC investigation would have included both environmental testing and additional patient screening. This also applies to cluster A, where SNV analysis identified the most likely index case in ICU A (a unit not initially thought to be part of the outbreak). This index case was likely linked to the initially presumed index case via intermediary transmission in ICU A, which was not obvious until WGS was performed. Overall, WGS also emphasized the covert dissemination of CPE at our center and others (17). In our setting, the availability of routine WGS might have reduced the size of some clusters, and the insight into undetected transmission might have been used to modify and improve investigations into transmission.
Our transmission analyses also emphasized that clusters could include both clonal and plasmid-mediated transmission, necessitating long-read sequencing in addition to short-read sequencing (18). Use of plasmid analysis was important in clusters E and F and also in cluster A, where it excluded one case and confirmed inclusion of another. Plasmid analysis was also useful in further assessing the individual case that SNV analysis linked to epidemiological cluster B; the carbapenemase gene-containing plasmids of the individual case and first cluster B case were highly related, further supporting the possibility of undetected transmission at H1 or elsewhere. The CPE bacteremia that epidemiological cluster B’s index case sporadically acquired at H1 may have been prevented had a more aggressive case finding been in place.
While WGS offered insight into transmission, it did not add information about the 3 cases known to H1-IPAC with an unknown source, nor did it clarify transmission chains for all cases thought to be acquired at H1. This may have been because of incomplete detection of colonized cases or due to plasmid-mediated transmission that we did not investigate.
There are limitations to this study. First, our transmission analyses are limited by undetected transmission, which undoubtedly occurs given gaps in our patient screening programs (16). Second, there is no SNV threshold that confirms CPE transmission, which makes interpretation of results challenging. In this study, there were up to 31 SNV differences between isolates of epidemiologically linked cases. At the same time, there were 8 SNV differences between our 2 travel-associated cases with OXA-48-producing E. coli ST38 isolates. Factors that influence SNV differences between isolates include but are not limited to species, reference strain, and SNV calling parameters (19). It is therefore critical to interpret genomic data with epidemiological data in transmission analyses.
In this study, WGS confirmed initial epidemiological interpretation of transmission in most clusters and altered the initial interpretation of transmission in some clusters. In our settings and most others, conventional epidemiology is used to investigate CPE transmission in individual hospitals. However, we and others (17, 20–22) demonstrate that WGS can offer insights into transmission that may alter hospital IPAC practices and more rapidly halt transmission chains. Evidence is also emerging that use of WGS in hospital outbreaks is cost saving (23). Beyond individual hospitals, a centralized genomic and epidemiological surveillance and response program for CPE has been successfully implemented in a low-prevalence setting (22). This program demonstrated a greater number of cases identified through IPAC interventions, rapid identification of small transmission clusters facilitating early response, and a decrease in KPC-2 infections (22). Collaboration and investment are required to ensure that this valuable technology can be incorporated into clinical and public health microbiology laboratories in low-prevalence settings.
MATERIALS AND METHODS
Setting and population.
The Toronto Invasive Bacterial Diseases Network (TIBDN) has performed population-based surveillance for CPE in metropolitan Toronto and the regional municipality of Peel, south-central Ontario, Canada, since its emergence in October 2007 (8). The microbiology laboratories serving all 33 hospitals that provide care to residents of the population area, as well as the two largest microbiology laboratories serving the community (who provide >80% of community-based microbiology testing), participate in this surveillance. As part of surveillance, all inpatients and outpatients identified as CPE-colonized/infected are asked about health care visits in the year prior to identification and are followed to identify health care visits in the year after identification.
H1 is a 1,055-bed hospital system in Toronto, ON, that is a member of TIBDN. This analysis included all patients with CPE colonization/infection identified via TIBDN surveillance from October 2007 to August 2018, if their CPE was first detected at H1 or first detected at another TIBDN hospital with receipt of health care (admission or outpatient visit) at H1 in the year before or after first detection.
During the study period, the microbiology laboratories serving all TIBDN hospitals (including H1) identified carbapenemase production in clinical isolates of Enterobacterales with reduced susceptibility to carbapenems using provincial recommendations (8). All clinical isolates with an ertapenem MIC of >1 mg/liter or a meropenem disc diffusion diameter of <25 mm were screened for carbapenemase production using the modified Hodge test prior to 2010 and other screening methods approved by the provincial laboratory accreditation body (e.g., KPC + MBL confirm identification [ID] kit; Rosco Diagnostica, Taastrup, Denmark) or by direct in-house PCR after that time. All isolates with a positive screen in all years were tested by PCR for the presence of blaKPC, blaOXA-48-like, blaVIM, blaNDM, blaIMP, and blaSME genes at a public health reference laboratory. All hospitals screened patients on admission for CPE rectal colonization if previously known to be CPE colonized/infected or recently hospitalized abroad. Two of the 33 hospitals also screened patients recently hospitalized in Canada on admission.
CPE surveillance was approved by the research ethics boards of all TIBDN hospitals. This study was approved by the research ethics boards of Sinai Health System and Sunnybrook Health Sciences Centre (Toronto, Canada).
Data collection.
For all included patients, study staff collected demographic, clinical, and CPE acquisition risk factor data by chart review and patient interview (8). H1 provided room location data. For each patient whose CPE was known to the Infection Prevention and Control department at H1 (H1-IPAC), we requested CPE acquisition source as determined using conventional epidemiology at the time of initial investigation (health care abroad, travel to the Indian subcontinent without health care, health care at H1, health care at another facility in Canada, or other) (supplemental materials). We also requested a line list for each epidemiological transmission cluster (supplemental materials). H1-IPAC deemed cases to be epidemiologically linked if their CPE produced the same carbapenemase and if there was overlap in time and/or space and/or exposure to procedures.
Whole-genome sequencing (WGS) and transmission analysis.
The first CPE isolate of each included patient underwent short-read Illumina (San Diego, USA) sequencing at the National Microbiology Laboratory (Winnipeg, Canada). For patients with more than one CPE carbapenemase/species, the first isolate of each new carbapenemase/species underwent Illumina sequencing. A subset of isolates also underwent long-read MinION sequencing (Nanopore Technologies, Oxford, UK) (supplemental methods).
De novo assembly of short reads was performed using Shovill v1.0 for all isolates. Assembled sequence data were analyzed for resistance genes (ResFinder), plasmid genes (PlasmidFinder), and multilocus sequence typing (MLST) (starAMR).
Single nucleotide variant (SNV) analysis was performed on all isolates of the same species using the SNVPhyl pipeline (24) to identify genomic clusters. SNV analysis was then rerun by ST and carbapenemase variant, using an internal reference to each ST to achieve the largest core genome for SNV comparison. These genomic data were combined with epidemiological data to confirm epidemiological clusters (defined as transmission clusters identified using conventional epidemiology at the time of initial investigation at H1) and to identify additional cases potentially belonging to these epidemiological clusters. For epidemiologically linked cases whose isolates shared carbapenemase variants but were of different species (or were of the same species with ≥10 SNVs between isolates) (25), MinION sequencing was performed to resolve plasmids and assess for plasmid-mediated transmission. Plasmids were compared using GView alignment, Kmer mapping, and BLASTn (supplemental methods). For the purpose of this study, plasmid pairs that shared less than 80% coverage and 80% nucleotide identity coupled with consistent epidemiological data were considered unrelated.
Statistical analysis.
Descriptive statistics were used to describe patients and their CPE. Analyses were performed using SAS 9.4M6 (Cary, USA).
Data availability.
Illumina sequences have been deposited on NCBI under BioProject PRJNA725227. Accession numbers are listed in Table S3.
ACKNOWLEDGMENTS
We thank TIBDN microbiology laboratory technologists, infection control practitioners, and public health professionals. We also thank National Microbiology Laboratory employees Ken Fakharuddin for technical services and M. Graham and the DNA Core staff for isolate short-read sequencing. We thank Marion Elligson and Nick Daneman for providing room location information at H1.
A.J.J. was supported by the Vanier Canada Graduate Scholarship at the time of this work. This work was supported by the Canadian Institutes of Health Research (grant number 313039).
We declare that we have no conflicts of interest.
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol 30:972–976. 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 3.Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H. 2010. An outbreak of infection due to beta‐lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis 50:364–373. 10.1086/649865. [DOI] [PubMed] [Google Scholar]
- 4.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedeschi S, Cauda R, Bassetti M. 2012. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis 55:943–950. 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 5.Kontopoulou K, Protonotariou E, Vasilakos K, Kriti M, Koteli A, Antoniadou E, Sofianou D. 2010. Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 beta-lactamase resistant to colistin. J Hosp Infect 76:70–73. 10.1016/j.jhin.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Public Health Agency of Canada. 2018. Canadian Resistance Surveillance System Update 2018. https://www.canada.ca/content/dam/phac-aspc/documents/services/publications/drugs-health-products/canadian-antimicrobial-resistance-surveillance-system-2018-report-executive-summary/pub1-eng.pdf. Accessed 10 January 2021.
- 7.Zhong Z, Faheem A, Farooqi L, Armstrong I, Borgundvaag E, Coleman B, Green K, Jayasinghe K, Johnstone J, Katz K, Kohler P, Li A, Melano R, Muller M, Nayani S, Patel S, Paterson A, Poutanen S, Rebbapragada A, Richardson D, Sarabia A, Shafinaz S, Simor AE, Willey B, Wisely L, McGeer A, Toronton Invasive Bacterial Diseases Network. 2018. Emergence of carbapenemase producing Enterobacteriaceae in South Central Ontario, Canada. Open Forum Infect Dis 5:S365–S365. 10.1093/ofid/ofy210.1038. [DOI] [Google Scholar]
- 8.Kohler PP, Melano RG, Patel SN, Shafinaz S, Faheem A, Coleman BL, Green K, Armstrong I, Almohri H, Borgia S, Borgundvaag E, Johnstone J, Katz K, Lam F, Muller MP, Powis J, Poutanen SM, Richardson D, Rebbapragada A, Sarabia A, Simor A, McGeer A, Toronto Invasive Bacterial Diseases Network (TIBDN). 2018. Emergence of carbapenemase-producing Enterobacteriaceae, South-Central Ontario. Canada Emerg Infect Dis 24:1674–1682. 10.3201/eid2409.180164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamal A, Coleman B, Johnstone J, Katz K, Muller MP, Patel S, Melano R, Rebbapragada A, Richardson D, Sarabia A, Mubareka S, Poutanen S, Zhong Z, Kohler P, McGeer A. 2019. Healthcare-acquired (HA) carbapenemase-producing Enterobacteriales (CPE) in Southern Ontario, Canada: to whom are we transmitting CPE? Open Forum Infect Dis 6:S247–S248. 10.1093/ofid/ofz360.581. [DOI] [Google Scholar]
- 10.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, EuSCAPE Working Group; ESGEM Study Group, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H. 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitout JDD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. 2016. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 7:895. 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-Yoseph H, Hussein K, Braun E, Paul M. 2016. Natural history and decolonization strategies for ESBL/carbapenem-resistant Enterobacteriaceae carriage: systematic review and meta-analysis. J Antimicrob Chemoter 71:2729–2739. 10.1093/jac/dkw221. [DOI] [PubMed] [Google Scholar]
- 14.Klein S, Boutin S, Spath I, Kimmich C, Brandt J, Müller-Tidow C, Heeg K, Nurjadi D. 2020. Acquisition and transmission of carbapenemase producing (blaKPC-2) Enterobacter cloacae in a highly frequented outpatient clinic. Clin Infect Dis 72:e158–e161. 10.1093/cid/ciaa1734. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs A, Argudin MA, De Mendonca R, Deplano A, Roisin S, Dodémont M, Coussement J, Filippin L, Dombrecht J, De Bruyne K, Huang TD, Supply P, Byl B, Glupczynski Y, Denis O. 2019. An outpatient clinic as a potential site of transmission for an outbreak of New Delhi metallo-beta-lactamase-producing Klebsiella pneumoniae sequence type 716: a study using whole-genome sequencing. Clin Infect Dis 68:993–1000. 10.1093/cid/ciy581. [DOI] [PubMed] [Google Scholar]
- 16.Jamal AJ, Garcia-Jeldes F, Baqi M, Borgia S, Johnstone J, Katz K, Kohler P, Muller MP, McGeer AJ, CPE Investigators of the Toronto Invasive Bacterial Diseases Network. 2019. Infection prevention and control practices related to carbapenemase-producing Enterobacteriaceae (CPE) in acute-care hospitals in Ontario, Canada. Infect Control Hosp Epidemiol 40:1006–1012. 10.1017/ice.2019.173. [DOI] [PubMed] [Google Scholar]
- 17.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA, NISC Comparative Sequencing Program Group. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin J, Phan HTT, Findlay J, Stoesser N, Pankhurst L, Navickaite I, De Maio N, Eyre DW, Toogood G, Orsi NM, Kirby A, Young N, Turton JF, Hill RLR, Hopkins KL, Woodford N, Peto TEA, Walker AS, Crook DW, Wilcox MH. 2017. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J Antimicrob Chemother 72:3025–3034. 10.1093/jac/dkx264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry NL, Lane CR, Kwong JC, Schultz M, Sait M, Stevens K, Ballard S, Gonçalves da Silva A, Seemann T, Gorrie CL, Stinear TP, Williamson DA, Brett J, van Diemen A, Easton M, Howden BP. 2019. Genomics for molecular epidemiology and detecting transmission of carbapenemase-producing Enterobacterales in Victoria, Australia, 2012 to 2016. J Clin Microbiol 57:e00573-19. 10.1128/JCM.00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellington MJ, Davies F, Jauneikaite E, Hopkins KL, Turton JF, Adams G, Pavlu J, Innes AJ, Eades C, Brannigan ET, Findlay J, White L, Bolt F, Kadhani T, Chow Y, Patel B, Mookerjee S, Otter JA, Sriskandan S, Woodford N, Holmes A. 2020. A multi-species cluster of GES-5 carbapenemase producing Enterobacterales linked by a geographically disseminated plasmid. Clin Infect Dis 71:2553–2560. 10.1093/cid/ciz1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The HC, Karkey A, Pham Thanh D, Boinett CJ, Cain AK, Ellington M, Baker KS, Dongol S, Thompson C, Harris SR, Jombart T, Le Thi Phuong T, Tran Do Hoang N, Ha Thanh T, Shretha S, Joshi S, Basnyat B, Thwaites G, Thomson NR, Rabaa MA, Baker S. 2015. A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol Med 7:227–239. 10.15252/emmm.201404767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane C, Brett J, Schultz M, Gorrie CL, Stevens K, Cameron DRM, St George S, van Diemen A, Easton M, Stuart RL, Sait M, Peleg AY, Stewardson AJ, Cheng AC, Spelman DW, Waters MJ, Ballard SA, Sherry NL, Williamson DA, Romanes F, Sutton B, Kwong JC, Seemann T, Goncalves da Silva A, Stephens N, Howden BP. 2020. Search and contain: impact of an integrated genomic and epidemiological surveillance and response program for control of carbapenemase-producing Enterobacterales. Clin Infect Dis ciaa972. 10.1093/cid/ciaa972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee XJ, Elliott TM, Harris PNA, Douglas J, Henderson B, Watson C, Paterson DL, Schofield DS, Graves N, Gordon L. 2020. Clinical and economic outcomes of genome sequencing availability on containing a hospital outbreak of resistant Escherichia coli in Australia. Value Health 23:994–1002. 10.1016/j.jval.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Petkau A, Mabon P, Sieffert C, Knox NC, Cabral J, Iskander M, Iskander M, Weedmark K, Zaheer R, Katz LS, Nadon C, Reimer A, Taboada E, Beiko RG, Hsiao W, Brinkman F, Graham M, Van Domselaar G. 2017. SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb Genom 3:e000116. 10.1099/mgen.0.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamal AJ, Faheem A, Farooqi L, Zhong XZ, Armstrong I, Boyd DA, Borgundvaag E, Coleman BL, Green K, Jayasinghe K, Johnstone J, Katz K, Kohler P, Li AX, Mataseje L, Melano R, Muller MP, Mulvey MR, Nayani S, Patel SN, Paterson A, Poutanen S, Rebbapragada A, Richardson D, Sarabia A, Shafinaz S, Simor AE, Willey BM, Wisely L, McGeer AJ. 2020. Household transmission of carbapenemase-producing Enterobacterales in Ontario, Canada. Clin Infect Dis ciaa1295. 10.1093/cid/ciaa1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Illumina sequences have been deposited on NCBI under BioProject PRJNA725227. Accession numbers are listed in Table S3.