ABSTRACT
In the present report, we describe two small molecules with broad-spectrum antiviral activity. These drugs block the formation of the nodosome. The studies were prompted by the observation that infection of human fetal brain cells with Zika virus (ZIKV) induces the expression of nucleotide-binding oligomerization domain-containing protein 2 (NOD2), a host factor that was found to promote ZIKV replication and spread. A drug that targets NOD2 was shown to have potent broad-spectrum antiviral activity against other flaviviruses, alphaviruses, enteroviruses, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19). Another drug that inhibits receptor-interacting serine/threonine protein kinase 2 (RIPK2), which functions downstream of NOD2, also decreased the replication of these pathogenic RNA viruses. The antiviral effect of this drug was particularly potent against enteroviruses. The broad-spectrum action of nodosome-targeting drugs is mediated in part by the enhancement of the interferon response. Together, these results suggest that further preclinical investigation of nodosome inhibitors as potential broad-spectrum antivirals is warranted.
KEYWORDS: antiviral, broad spectrum, NOD2, RIPK2, arbovirus, coxsackievirus, SARS-CoV-2, COVID-19, nodosome, interferon
INTRODUCTION
Reemerging and emerging RNA viruses represent a major threat to global public health. Vaccines and antiviral drugs, which usually target a single virus species, are critical measures to prevent and control the spread of these pathogens. However, prophylactic or therapeutic drugs are not available for many of the most important RNA viruses in circulation today (1). While highly effective direct-acting antiviral drugs have been developed for a number of important human pathogens such as HIV-1, herpesvirus family members, and hepatitis C virus (2), these drugs tend to be highly specific and are of limited use for treating other viral infections. In contrast, broad-spectrum antivirals would be expected to inhibit the replication of multiple viruses, including emerging and reemerging RNA viruses. Although several broad-spectrum antiviral compounds are in preclinical studies or clinical trials, to date, no drug in this class has been licensed (3). Because hundreds of cellular factors are required for productive viral infection, targeting common host factors that are utilized by multiple viruses may be a viable approach for developing broad-spectrum antivirals (4).
Here, we report the identification and characterization of a novel class of small molecules with broad-spectrum antiviral activity. These drugs selectively block the intracellular pattern recognition receptor NOD2 (nucleotide-binding oligomerization domain-containing protein 2) and a critical mediator of NOD2 signaling, RIPK2 (receptor-interacting serine/threonine protein kinase 2). NOD2 recognizes the peptidoglycan muramyl dipeptide (MDP) that is found in bacterial cell walls, but it can also bind to viral RNA. In doing so, NOD2 induces the formation of the nodosome and stimulates host defense against infections (5). Although NOD2 is important for the innate immune response against HIV-1 (6), cytomegalovirus (7), and respiratory syncytial virus (8), it has also been reported as a major pathogenic mediator of coxsackievirus B3-induced myocarditis (9).
Previously, we reported that Zika virus (ZIKV) infection of human fetal brain cells upregulates the expression of NOD2 (10), and here, we show that the expression of this host protein promotes ZIKV replication. Using multiple human primary cell types and cell lines, we found that the NOD2-blocking drug GSK717 inhibits the replication of flaviviruses, alphaviruses, enteroviruses, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19). The RIPK2-blocking agent GSK583 also potently inhibits these pathogenic RNA viruses but was particularly effective against enteroviruses. The broad-spectrum activity of these drugs is mediated in part by an enhancement of the innate immune response. Together, the data from our in vitro and ex vivo experiments suggest that nodosome inhibitors should be further investigated as broad-spectrum antivirals in preclinical studies.
RESULTS
ZIKV infection induces the inflammasome in primary human fetal brain cells.
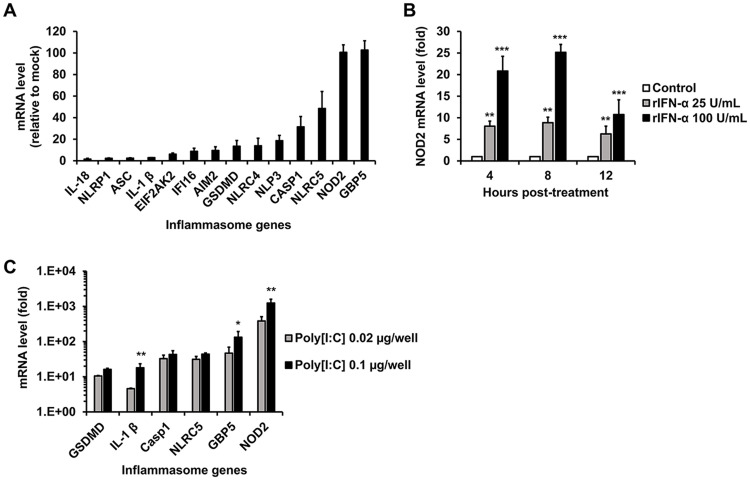
Previously, we reported that human fetal astrocytes (HFAs) are likely the principal reservoirs for ZIKV infection and persistence in the human fetal brain (11). In a subsequent study (10), transcriptome sequencing (RNAseq) analyses revealed that ZIKV infection of these cells upregulates multiple inflammasome genes, including GSDMD, IL-1β, Casp1, NLRC5, GBP5, and NOD2. In light of the recently identified links between the inflammasome and ZIKV neuropathogenesis (12, 13), we asked whether the activity of this multiprotein complex affected virus replication. Since our previous analysis (10) was performed using a strain of ZIKV not associated with microcephaly, we first confirmed that infection of HFAs with the pandemic ZIKV strain PRVABC-59 induced the expression of multiple inflammasome genes. Indeed, NOD2 and GBP5 were upregulated more than 100-fold by ZIKV infection, whereas other genes in this pathway were induced less than 50-fold (Fig. 1A). The expression of inflammasome genes could also be induced by treatment of HFAs with human recombinant interferon alpha (IFN-α) but less so than with the double-strand RNA mimic poly(I·C) (Fig. 1B and C; see also Fig. S1A to D in the supplemental material). This may indicate that detection of viral RNA per se triggers inflammasome induction in HFAs.
FIG 1.
Inflammasome gene expression in HFAs is induced by ZIKV, IFN-α, and poly(I·C). (A) Relative inflammasome gene expression in HFAs infected with ZIKV PRVABC-59 (MOI = 0.3) was determined by qRT-PCR at 48 h postinfection. (B) HFAs were treated with human recombinant IFN-α (rIFN-α) for 4, 8, and 12 h, after which relative NOD2 expression was determined. (C) HFAs were transfected with poly(I·C) for 12 h, after which relative inflammasome gene expression was determined. Error bars represent standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student’s t test).
NOD2 expression promotes ZIKV multiplication by suppression of the innate immune response in HFAs.
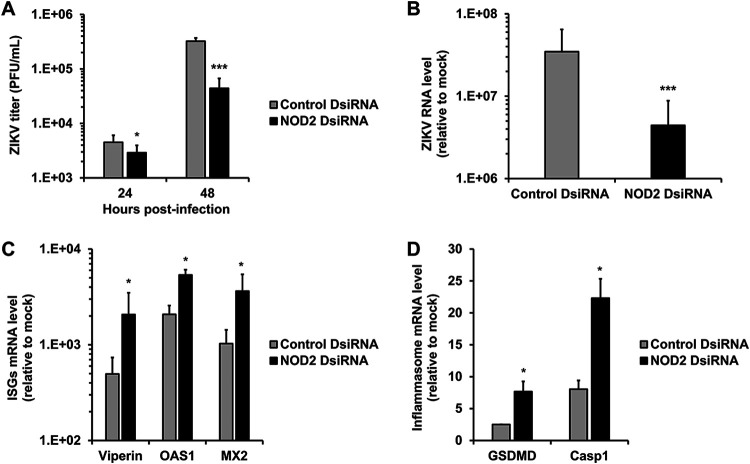
As NOD2 was one of the most upregulated inflammasome genes, we examined how reduced NOD2 expression in HFAs affected the replication of the PRVABC-59 strain of ZIKV. Compared to cells transfected with a nontargeting small interfering RNA (siRNA), the replication of ZIKV in HFAs transfected with NOD2-specific siRNAs was significantly reduced (Fig. 2A and B).
FIG 2.
NOD2 silencing suppresses ZIKV multiplication and enhances the expression of interferon-stimulated and inflammasome genes. HFAs were transfected with NOD2-specific or nonsilencing siRNAs for 24 h and then infected with ZIKV (MOI = 0.05). (A) Cell culture medium or total cellular RNA was harvested after 24 and 48 h for plaque assays or qRT-PCR at 48 h postinfection. (B to D) Relative levels of viral genome (B); the interferon-stimulated genes Viperin, 2′-5′-oligoadenylate synthetase 1 (OAS1), and myxovirus resistance protein 2 (MX2) (C); as well as the inflammasome genes gasdermin D (GSDMD) and caspase 1 (Casp1) (D). Values are expressed as the means from three independent experiments. Error bars represent standard errors of the means. *, P < 0.05; ***, P < 0.001 (by Student’s t test).
Recently, we reported that ZIKV-induced expression of fibroblast growth factor 2 in fetal brain increases viral replication by inhibiting the interferon response (10). As NOD2 is an intracellular pattern recognition receptor that recognizes bacterial MDP as well as viral RNA (5), we questioned whether the effect of NOD2 expression on viral replication was related to its actions on the innate immune response. To address this, the relative expression levels of interferon-stimulated genes (ISGs) were determined in ZIKV-infected HFAs after NOD2 silencing. NOD2 knockdown was associated with a significant upregulation of several important ISGs, including Viperin, OAS1, and MX2 (Fig. 2C), as well as the prototypical inflammasome genes GSDMD and Casp1 (Fig. 2D). Of note, NOD2 silencing reduced NOD2 mRNA expression and was not cytotoxic to HFAs (Fig. S1E and F).
ZIKV replication and spread are inhibited by blocking NOD2 function in fetal brain.
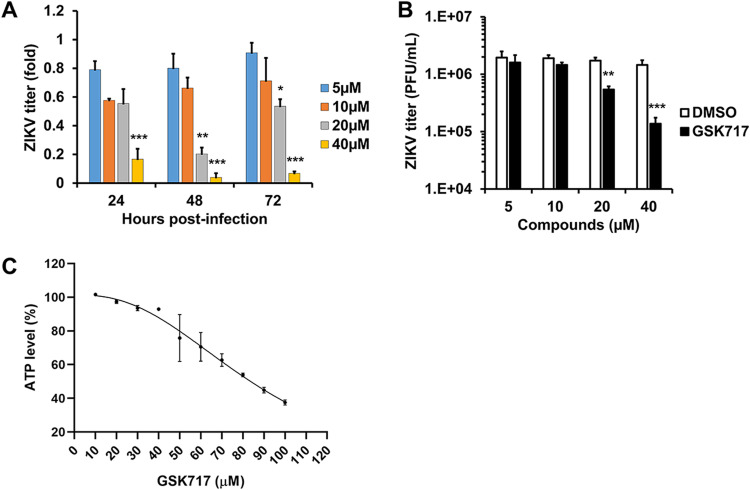
A number of specific NOD2 inhibitors have been developed to treat inflammatory diseases (14). To determine if these drugs have antiviral activity, HFAs infected with ZIKV were treated with or without subcytotoxic concentrations of the anti-NOD2 compound GSK717 for up to 72 h. GSK717 reduced viral titers in a concentration-dependent manner (50% inhibitory concentration [IC50], 14.8 to 17.9 μM), regardless of whether cells were infected at a low or a high multiplicity of infection (MOI) (Fig. 3A to C and Tables 1 and 2). Similar results were observed in human primary embryonic pulmonary fibroblasts (HEL-18), thus demonstrating that the antiviral effect of GSK717 is not limited to fetal brain tissue (Fig. S2A to D).
FIG 3.
The anti-NOD2 drug GSK717 inhibits ZIKV replication. ZIKV-infected HFAs (MOI = 0.05 to 5) were treated with DMSO or the NOD2-blocking agent GSK717, after which viral titers were determined daily up to 72 h postinfection. (A and B) ZIKV titers as relative fold changes (MOI = 0.05) at 72 h (A) and as PFU per milliliter at 48 h postinfection (MOI = 5) (B). (C) Cellular ATP levels were determined in uninfected HFAs treated with GSK717 or DMSO for 72 h. Values are expressed as the means from three independent experiments. Error bars represent standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student’s t test).
TABLE 1.
IC50, CC50, and SI values for GSK717 and GSK583 at low MOIs (0.05 to 0.1)a
| Virusb | GSK717 |
GSK583 |
||||
|---|---|---|---|---|---|---|
| IC50 (μM) | CC50 (μM) | SI | IC50 (μM) | CC50 (μM) | SI | |
| ZIKV | 14.84 | 82.84 | 5.58 | 22.03 | 104.76 | 4.76 |
| DENV | 20.26 | 63.59 | 3.14 | 18.90 | 104.76 | 5.54 |
| MAYV | 12.69 | 63.59 | 5.01 | 19.85 | 104.76 | 5.28 |
| CVB5 | 9.57 | 81.95 | 8.56 | 0.11 | 104.76 | 952.36 |
| SARS-CoV-2 | 10.22 | 75.30 | 7.37 | 11.70 | 61.29 | 5.24 |
CC50, 50% cytotoxic concentration.
Results are from infected A549 (Zika virus [ZIKV], dengue virus 2 [DENV], coxsackievirus B5 [CVB5], or Mayaro virus [MAYV]) and ACE2-SK-N-SH (severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]) cells. Values for GSK717 were also determined using ZIKV-infected human fetal astrocytes. While GSK583 data were generated at 24 h, GSK717 values were generated at 24 h (CVB5) and 48 h (ZIKV, DENV-2, MAYV, and SARS-CoV-2).
TABLE 2.
IC50, CC50, and SI values for GSK717 and GSK583 at a high MOI (5)
| Virusa | GSK717 |
GSK583 |
||||
|---|---|---|---|---|---|---|
| IC50 (μM) | CC50 (μM) | SI | IC50 (μM) | CC50 (μM) | SI | |
| ZIKV | 17.87 | 82.84 | 4.64 | 24.24 | 104.76 | 4.32 |
| DENV | 19.39 | 63.59 | 3.28 | 22.72 | 104.76 | 4.61 |
| MAYV | 21.74 | 63.59 | 2.93 | 20.07 | 104.76 | 5.22 |
| SARS-CoV-2 | 14.38 | 75.30 | 5.24 | 17.95 | 61.29 | 3.41 |
Results are from infected A549 (Zika virus [ZIKV], dengue virus 2 [DENV], or Mayaro virus [MAYV]) and ACE2-SK-N-SH (severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]) cells. Values for GSK717 were also determined using ZIKV-infected human fetal astrocytes. For GSK583 and GSK717, the data were generated at 24 and 48 h, respectively.
A NOD2 inhibitor blocks ZIKV infection and spread in multiple human cell lines.
We next examined whether GSK717 also inhibits ZIKV replication in human nonprenatal cell types, including cell lines usually used for antiflavivirus drug screening, such as A549 (pulmonary) and Huh7 (hepatoma) cells (15), as well as the astrocytoma cell line U251. GSK717 significantly reduced ZIKV titers in these human cell lines in a dose-dependent manner regardless of whether a low (0.05) or a high (5) MOI was used for infection (data not shown).
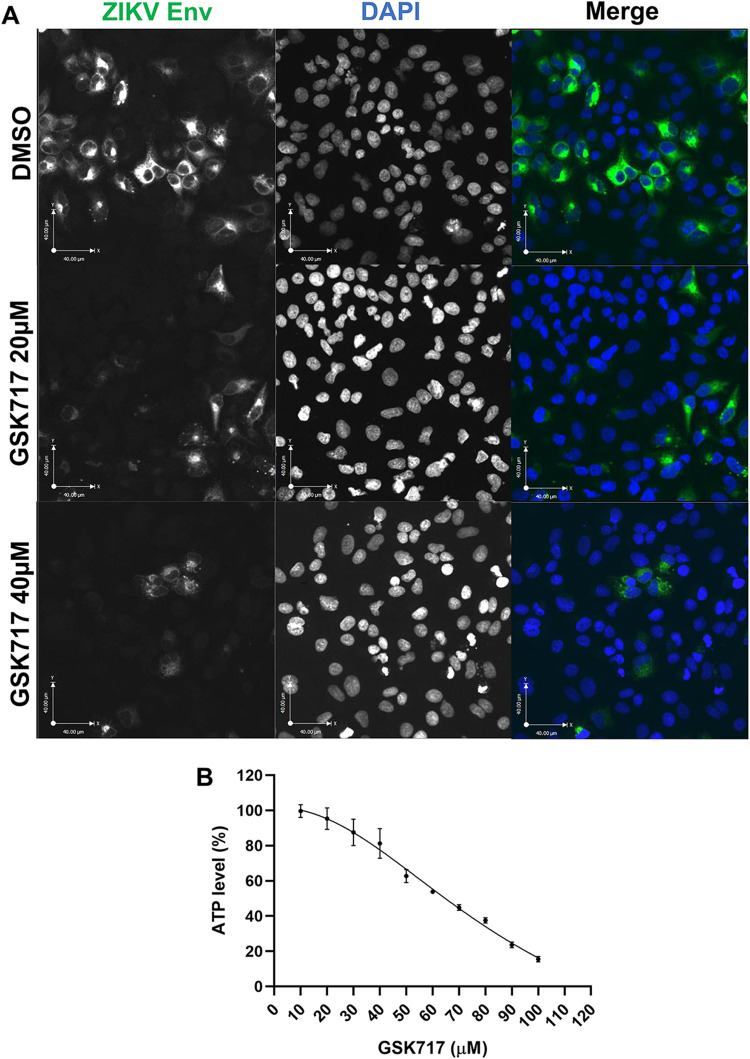
Consistent with its ability to reduce viral titers, treatment with GSK717 reduced the infection and spread of viruses in A549 cultures (Fig. 4A). Interestingly, GSK717 inhibited ZIKV replication even when added at 12 or 24 h postinfection (Fig. S2E). None of the GSK717 concentrations used in the cell-based assays were cytotoxic at the examined time points (Fig. 4B).
FIG 4.
The anti-NOD2 drug GSK717 blocks the spread of ZIKV infection. (A) Representative confocal imaging (magnification, ×20) showing the antiviral effect of GSK717 at 20 μM and 40 μM. A549 cells were infected with ZIKV (MOI = 1) followed by treatment with DMSO or GSK717 at 20 or 40 μM for 48 h before processing for indirect immunofluorescence. ZIKV-infected cells were identified using a mouse monoclonal antibody (4G2) to the envelope protein and Alexa Fluor 488 donkey anti-mouse to detect the primary antibody. Nuclei were stained with DAPI. Images were acquired using a spinning-disk confocal microscope equipped with Volocity 6.2.1 software. (B) Cellular ATP levels in A549 cells after 48 h of GSK717 or DMSO treatment. Values are expressed as the means from three independent experiments. Error bars represent standard errors of the means.
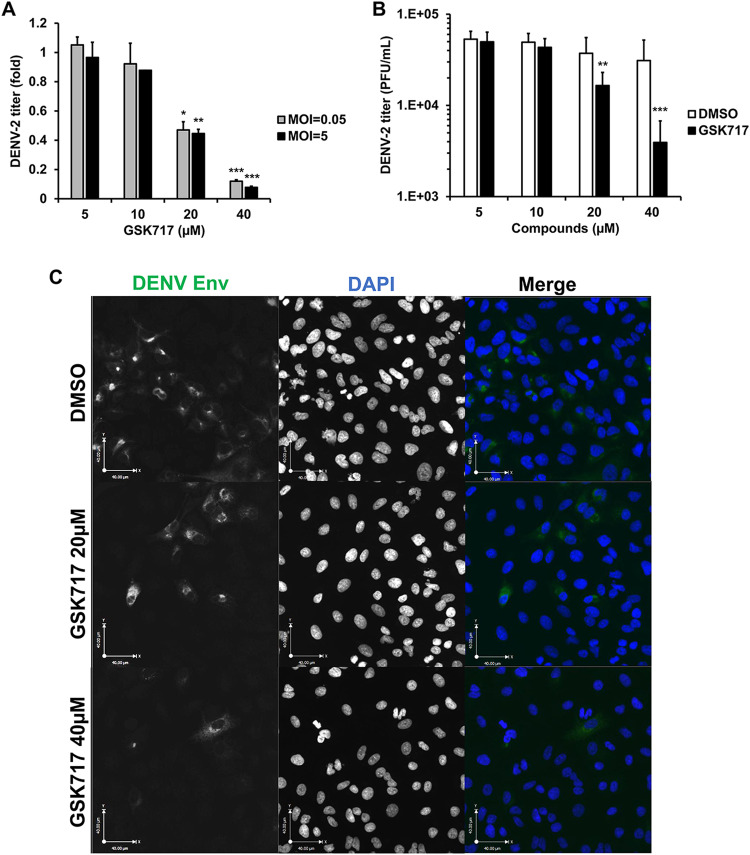
DENV replication is inhibited by the anti-NOD2 drug GSK717.
To determine if GSK717 could inhibit the replication of other flaviviruses, we next focused on dengue virus (DENV), the most important arbovirus in terms of morbidity and mortality and the causative agent of dengue hemorrhagic fever/dengue shock syndrome (16). A549 cells infected with DENV-2 strain 16681 (17) were treated with or without increasing concentrations of GSK717. The data in Fig. 5A and B show that GSK717 reduced DENV titers by >90% when used at 20 to 40 μM (Tables 1 and 2). Blocking NOD2 function with GSK717 also dramatically reduced the number of viral antigen-positive A549 cells after 48 h of infection (Fig. 5C and Fig. S3A).
FIG 5.
The anti-NOD2 drug GSK717 inhibits DENV replication. A549 cells were infected with DENV-2 (MOI = 0.05 to 5) and treated with GSK717 or DMSO for 48 h, after which cell culture media were harvested for plaque assays. (A and B) Viral titers as relative fold changes at an MOI of 0.05 or 5 (A) and as PFU per milliliter at an MOI of 0.05 (B). (C) Representative confocal imaging (magnification, ×20) of DENV-2-infected cells treated with GSK717. A549 cells were infected with DENV-2 (MOI = 1) followed by treatment with DMSO or GSK717 at 20 or 40 μM for 48 h before fixation and immunostaining. DENV-infected cells were detected using mouse monoclonal antibody (4G2) to the envelope protein and Alexa Fluor 488 donkey anti-mouse. Nuclei were stained with DAPI. Images were acquired using a spinning-disk confocal microscope equipped with Volocity 6.2.1 software. Values are expressed as the means from three independent experiments. Error bars represent standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student’s t test).
NOD2 function is important for replication of alphaviruses, enteroviruses, and coronaviruses.
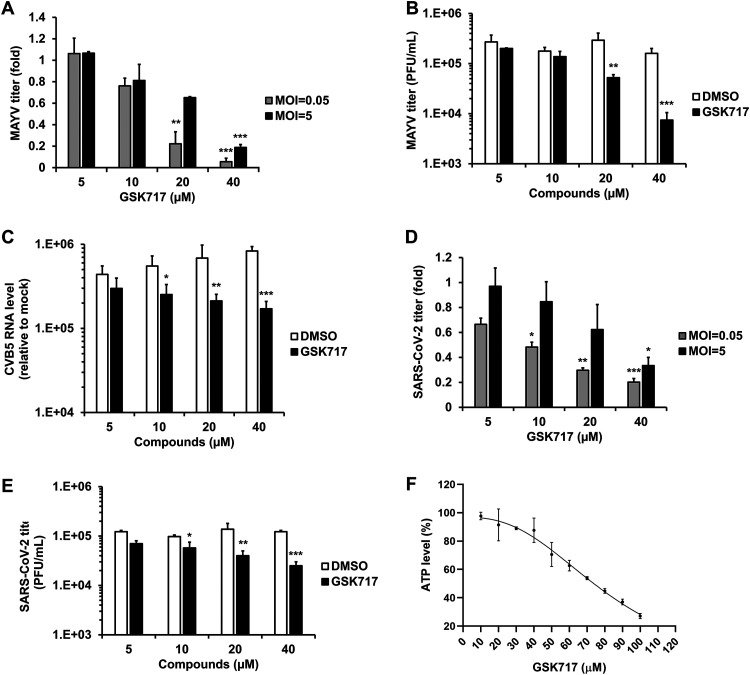
Nodosome formation can be induced following infection by multiple types of RNA viruses (18). As such, we next determined whether GSK717 could inhibit the replication of the mosquito-transmitted alphavirus Mayaro virus (MAYV). The recent MAYV outbreak strain TRVL 15537 was used for these experiments. Supernatants from MAYV-infected A549 cell cultures treated with or without GSK717 were collected for viral titer determinations at 48 h postinfection, after which plaque assays were performed. Similar to what was observed in flavivirus-infected cells, we found that GSK717 reduced MAYV titers by as much as 95% (Fig. 6A and B and Tables 1 and 2).
FIG 6.
The anti-NOD2 drug GSK717 inhibits the replication of MAYV, CVB5, and SARS-CoV-2. A549 cells infected with a low MOI (0.05) and a high MOI (5) of MAYV were treated with GSK717 or DMSO for 48 h, followed by collection of supernatants for plaque assays. (A and B) Relative (A) and absolute (B) viral titers at both MOIs and the MOI of 0.05, respectively. (C) A549 cells were infected with CVB5 (MOI = 0.1) followed by treatment with GSK717 or DMSO as a control for 24 h before total cellular RNA was collected for viral genome quantification by qRT-PCR. Viral RNA levels relative to mock are shown. ACE2-SK-N-SH cells were infected with SARS-CoV-2 (MOI = 0.05 to 5) and treated with GSK717 or DMSO for 48 h, after which culture supernatants were harvested for plaque assays. (D and E) Viral titers as relative fold changes (D) and as PFU per milliliter (E) at both MOIs and the MOI of 0.05, respectively. (F) Cellular ATP levels in ACE2-SK-N-SH cells after 48 h of GSK717 or DMSO treatment. Values are expressed as the means from three independent experiments. Error bars represent standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student’s t test).
In light of the reported pathogenic role for NOD2 in coxsackievirus B3 infections (9), we assessed how NOD2 inhibition by GSK717 affected the replication of the prototype Faulkner strain of coxsackievirus B5 (CVB5) in A549 cells. GSK717 had a modest but significant concentration-dependent effect on CVB5 replication (Fig. 6C). Moreover, compared to its effect on other viruses tested in this study, GSK717 had the lowest IC50 and the highest selectivity index (SI) values against CVB5 (Table 1).
Because SARS-CoV-2 infection reportedly activates the inflammasome in vitro (19) and in patients (20, 21), we tested how NOD2 inhibition affected the replication of this pandemic coronavirus. A human neuroblastoma cell line (SK-N-SH) stably expressing angiotensin-converting enzyme 2 (ACE2) was infected with SARS-CoV-2 in the presence or absence of GSK717. At 48 h postinfection, culture supernatants were collected for viral titer determinations by plaque assays using Vero-E6 cells. The data in Fig. 6D to F show that pharmacological inhibition of NOD2 had an inhibitory effect on SARS-CoV-2 titers similar to that observed with arboviruses (Tables 1 and 2).
Finally, since inhibition of NOD2 reduced the replication of multiple RNA viruses, we assessed whether, similar to what was observed during ZIKV infection, the upregulation of NOD2 was a common occurrence during infection with these viruses. The data in Fig. S3B and C show that NOD2 RNA levels were upregulated in cells infected with DENV-2, SARS-CoV-2, and CVB5.
Inhibition of downstream RIPK2 suppresses replication of arboviruses, coxsackievirus B5, and SARS-CoV-2.
RIPK2 is a critical mediator of NOD2 signaling. The binding of MDP to NOD2 leads to the self-oligomerization of NOD2 molecules, followed by homotypic interactions between the C-terminal caspase activation and recruitment domains of NOD2 and RIPK2. This results in the activation of transcription factors that drive the expression of multiple proinflammatory cytokines, chemokines, and antibacterial proteins (22).
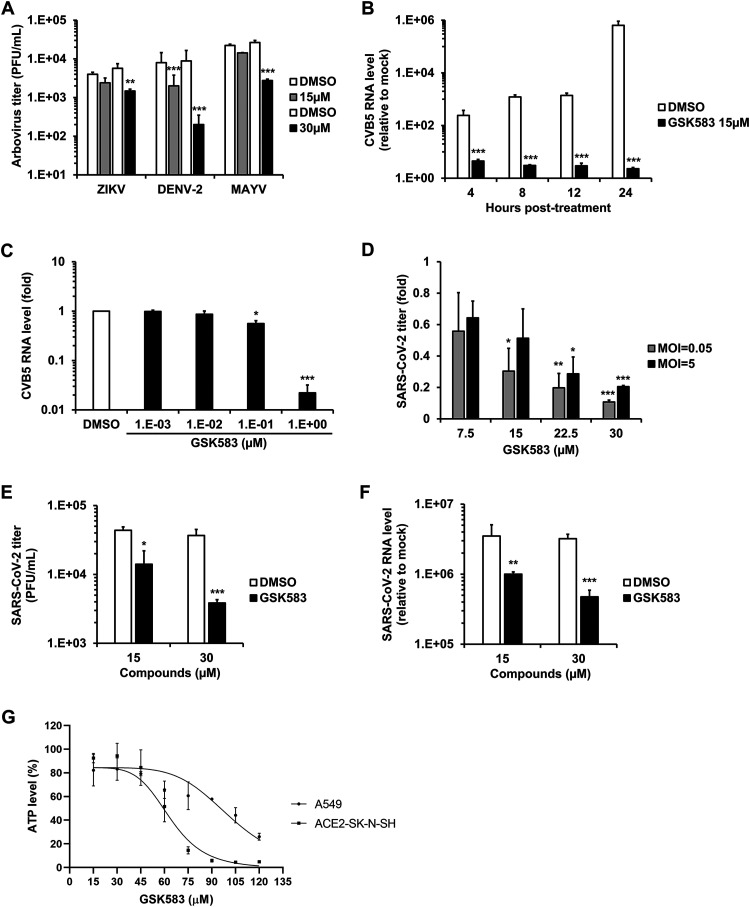
Although RIPK2 mRNA was not upregulated in ZIKV-infected HFAs (10), we questioned whether pharmacological inhibition of this protein would also reduce the replication of arboviruses such as ZIKV, DENV-2, and MAYV. Arbovirus-infected A549 cells were treated with or without GSK583, a highly potent and selective inhibitor of RIPK2 (23). A significant reduction in viral titers was observed in GSK583-treated cells infected with ZIKV, DENV-2, or MAYV at 12 and 24 h postinfection (Fig. 7A, Tables 1 and 2, and Fig. S4A to E). Similarly, indirect immunofluorescence microscopy analyses confirmed that inhibition of RIPK2 reduced the number of viral antigen-positive cells in ZIKV-, DENV-2-, and MAYV-infected A549 cultures (Fig. S5A to D).
FIG 7.
The anti-RIPK2 drug GSK583 has broad-spectrum antiviral activity. (A) A549 cells infected separately with three different arboviruses (MOI = 0.1) were treated with the RIPK2 inhibitor GSK583 or DMSO alone for 24 h, followed by supernatant collection for plaque assays. Arbovirus titers as PFU per milliliter are presented. (B) A549 cells infected with CVB5 (MOI = 0.1) were treated with GSK583 (15 μM) or DMSO alone, and total cellular RNA was then collected at 4, 8, 12, and 24 h postinfection, followed by qRT-PCR to quantify relative levels of viral genomic RNA. (C) A549 cells infected with CVB5 (MOI = 0.1) were treated with increasing concentrations of GSK583 or DMSO. At 24 h postinfection, total cellular RNA was collected and then subjected to qRT-PCR to quantify the relative levels of viral genome. ACE2-SK-N-SH cells infected with SARS-CoV-2 at a low MOI (0.05) and a high MOI (5) were treated with GSK583 or DMSO for 24 h, followed by supernatant and total cellular RNA collection. (D and E) Viral titers at both MOIs as relative fold changes (D) and at the MOI of 0.05 as PFU per milliliter (E). (F) Viral RNA levels relative to the mock level at the MOI of 0.05. (G) Cellular ATP levels were measured in uninfected A549 and ACE2-SK-N-SH cells after 48 h of treatment with GSK583 or DMSO alone. Values are expressed as the means from three independent experiments. Error bars represent standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student’s t test).
As well as inhibiting the replication of arboviruses, GSK583 was even more effective in blocking the replication of the picornavirus CVB5. Levels of CVB5 genomic RNA were reduced more than 250,000-fold by GSK583 treatment compared to dimethyl sulfoxide (DMSO)-treated cells (Fig. 7B). The IC50 for GSK583 against CVB5 (0.11 μM) was lower than the values obtained for all other viruses tested in this study (Fig. 7C and Table 1).
Finally, we tested the effect of GSK583 on the replication of SARS-CoV-2 in ACE2-SK-N-SK cells. At 12 and 24 h postinfection, culture supernatants and cell lysates were collected for viral titer determinations by plaque assays and viral RNA quantification using reverse transcription-quantitative PCR (qRT-PCR). A concentration-dependent reduction in viral multiplication was observed in GSK583-treated cells (Fig. 7D to F and Fig. S6A). A time-of-addition assay (drug treatment at 0 and 24 h postinfection) demonstrated that the RIPK2 inhibitor was able to reduce virus replication even when the drug was added well after viral infection had occurred (Fig. S6B). Quantitation of infection by indirect immunofluorescence showed that GSK583 treatment reduced the number of infected cells in a monolayer culture (Fig. S6C and D). GSK583 was not cytotoxic at a concentration of 30 μM or lower in A549 or ACE2-SK-N-SH cells (Fig. 7G).
Antinodosome drugs boost the innate immune response.
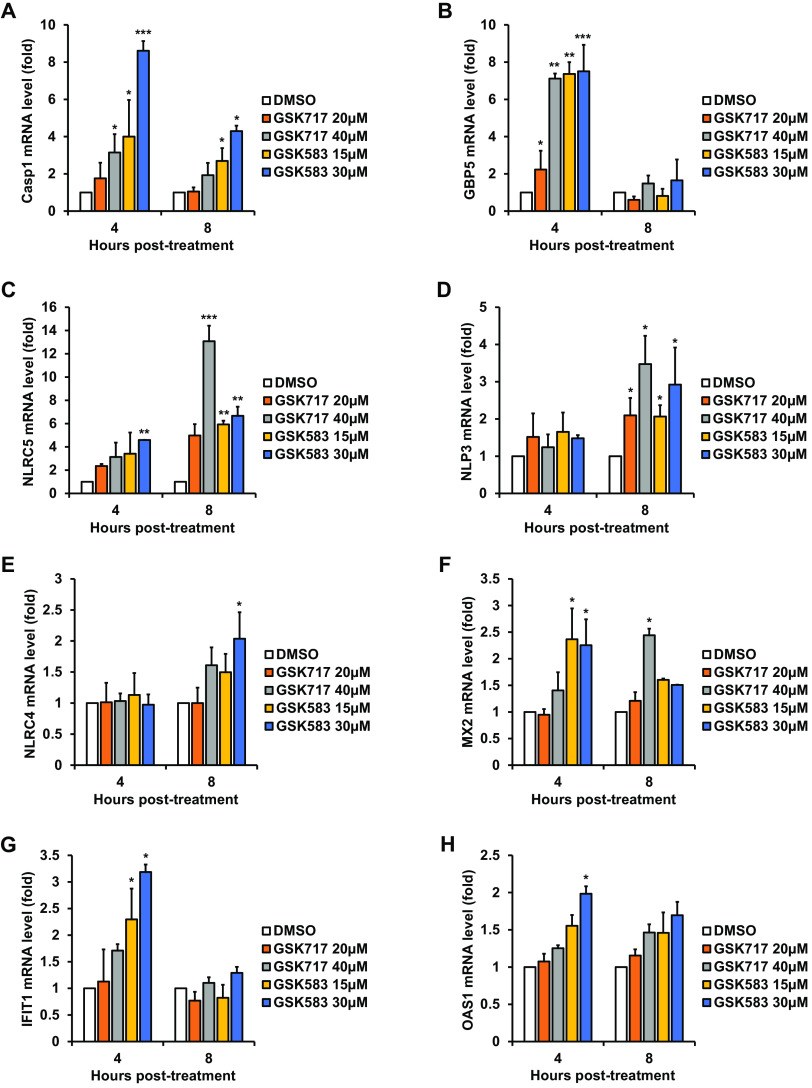
Given that nodosome-targeting drugs exhibit antiviral activity against a variety of RNA viruses, we questioned whether these drugs act by enhancing the innate immune response. To investigate this possible scenario, A549 cells were pretreated with GSK717 or GSK583, after which human recombinant IFN-α was added, or the cells were transfected with poly(I·C). Total RNA was harvested, and levels of ISGs were quantified by qRT-PCR. A number of pivotal ISGs, including inflammasome genes, were variably upregulated at different time points, showing that these antinodosome drugs enhance the innate immune response (Fig. 8A to H and Fig. S7A to H).
FIG 8.
Antinodosome drugs enhance the IFN response. A549 cells were treated with GSK583, GSK717, or DMSO alone for 16 h, followed by the addition of human recombinant IFN-α (100 U/ml). Total cellular RNA was harvested for qRT-PCR at 4 and 8 h posttreatment. Relative expression levels of transcripts for eight ISGs (Casp1, GBP5, NLRC5, NLRP3, NLRC4, MX2, IFIT1, and OAS1) in drug- and DMSO-treated cells are shown. Values are expressed as the means from three independent experiments. Error bars represent standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student’s t test).
DISCUSSION
ZIKV cocirculates in some of the same regions of endemicity as other arboviruses, including chikungunya virus, DENV, and MAYV. Symptoms of acute infection caused by these arboviruses, such as fever, rash, joint pain, and ocular manifestations, are common, which complicates the clinical diagnosis of mono- and coinfections (16, 24, 25). Differential serological diagnosis is further hindered by flavivirus antigen cross-reactivity (16, 25). Given the issues with clinical/laboratory diagnosis and the lack of effective vaccines against most arboviruses, the development of broad-spectrum antivirals against these pathogens should be a high priority.
Nonpolio enterovirus infections in humans can result in mild or self-limited conditions (common cold; herpangina; hand, foot, and mouth disease; pleurodynia; acute hemorrhagic conjunctivitis; and aseptic meningitis) or severe/life-threatening conditions (myocarditis, encephalitis, neonatal disseminated sepsis, and acute flaccid paralysis) (26). There is a clear need for antiviral drugs to treat enteroviral infections, particularly in children, but currently licensed drugs are not available (27).
The ongoing pandemic caused by SARS-CoV-2 poses a different set of challenges. Despite concerted efforts to repurpose and find new antiviral drugs (28, 29), so far, only remdesivir has shown modest clinical efficacy (30, 31), while anti-SARS-CoV-2 monoclonal antibodies appear to be useful in the acute stages of COVID-19 (32, 33). From more than 200 SARS-CoV-2 vaccine candidates in accelerated development at preclinical and clinical stages (34), a limited number of vaccines have been licensed for use, but their efficacies against emerging coronavirus variants are still being evaluated (35, 36).
In this study, we characterized the broad-spectrum antiviral activities of the nodosome inhibitors GSK717 and GSK583. These small molecules display robust antiviral action against multiple RNA viruses and may hold promise as panflavivirus inhibitors. First, we showed that NOD2 expression promotes ZIKV multiplication in HFAs, which are the main target of this flavivirus in the fetal brain (10, 11). Next, we demonstrated that the NOD2 inhibitor GSK717 blocks infection by and spread of ZIKV in human fetal brain and cell lines. NOD2 inhibition also reduced the replication of the related virus DENV, the alphavirus MAYV, the prototype CVB5 strain, and the pandemic coronavirus SARS-CoV-2. Blocking the NOD2 downstream signaling kinase RIPK2 with GSK583 significantly inhibited the replication of these viral pathogens, in particular the enterovirus CVB5.
Gefitinib is an FDA-approved drug for the treatment of lung, breast, and other cancers. It works by reducing the activity of the epidermal growth factor receptor (EGFR) tyrosine kinase domains. Of note, this drug also inhibits the tyrosine kinase activity of RIPK2 (37) and has been shown to inhibit the replication of DENV and the release of proinflammatory cytokines from infected human primary monocytes (38). The authors of those studies suggested a role for EGFR/RIPK2 in DENV pathogenesis and that gefitinib may be beneficial in the treatment of dengue patients. Similarly, the work here, which demonstrated the antiviral activity of NOD2 and RIPK2 inhibitors using primary cells and cell lines, supports the potential clinical use of these compounds in infections by arboviruses and enteroviruses as well as coronavirus infections at early and/or advanced stages.
As GSK717 and GSK583 were developed primarily for immune-mediated inflammatory conditions, their anti-inflammatory effects may have the added benefit of reducing the hyperinflammatory state associated with flavi-, alpha-, entero-, and coronavirus diseases (16, 24–26, 39). Finally, our findings raise potential concerns regarding adjuvants in viral vaccines that augment NOD2 as an immune strategy (40, 41) since this immune signaling protein is not a restriction factor but rather an enhancement factor for multiple pathogenic RNA viruses.
The current study illustrates how the identification of a drug target through transcriptomic analyses of virus-infected cells can lead to novel broad-acting host-directed antiviral strategies with a high barrier of resistance. Increased NOD2 expression may be a novel mechanism of immune evasion that viruses use to evade the innate immune response. Conversely, drugs that block nodosome formation appear to have broad-spectrum antiviral activity by enhancing the interferon response. Collectively, our results warrant consideration of these and related compounds as broad-spectrum antiviral drug candidates for further preclinical development.
MATERIALS AND METHODS
Ethical approval.
Human fetal brain tissues were obtained from 15- to 19-week aborted fetuses with written consent from the donor parents and prior approval under protocol 1420 (University of Alberta Human Research Ethics Board).
Cells and viruses.
ZIKV (PRVABC-59), dengue virus (DENV-2) (16681), and Mayaro virus (MAYV) (TRVL 15537) were propagated in Aedes albopictus C6/36 cells grown in minimum essential medium (MEM; Thermo Fisher Scientific, Waltham, MA). SARS-CoV-2 (SARS-CoV-2/CANADA/VIDO 01/2020) was propagated in Vero-E6 cells grown in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific). CBV5 (Faulkner strain) was propagated in MA104 cells (ATCC, Manassas, VA) in DMEM. A549, Huh7, U251, Vero (ATCC), and ACE2-hyperexpressing SK-N-SH cells were maintained in DMEM, while HEL-18 human primary embryonic pulmonary fibroblasts were maintained in RPMI 1640 medium (Thermo Fisher Scientific). HFAs were prepared from multiple donations (n = 5), as described previously (42). For infection, cells were incubated with virus (MOI of 0.05 to 5) for 1 to 2 h at 37°C using fresh medium supplemented with fetal bovine serum (Thermo Fisher Scientific). Culture of cells, construction of ACE2-SK-N-SH cells, and viral infections are described in more detail in the supplemental material.
qRT-PCR.
RNA from cells was extracted using NucleoSpin RNA kits (Macherey-Nagel GmbH & Co., Düren, Germany). Real-time qRT-PCR was performed in a CFX96 Touch real-time PCR detection system instrument (Bio-Rad, Hercules, CA) using ImProm-II reverse transcriptase (Promega, Madison, WI). For more details about the protocols and primers used in this work, see the supplemental material.
Poly(I·C) transfection.
HFAs grown in 96-well plates (Greiner, Kremsmünster, Austria) were transfected with poly(I·C) (Sigma-Aldrich, St. Louis, MO) at a concentration of 0.02 or 0.1 μg/well using TransIT (0.3 μl/well; Mirus Bio LLC, Madison, WI). At 12 h posttransfection, total RNA was extracted, and transcript levels of IFN-stimulated genes (ISGs) were quantified by qRT-PCR.
Human recombinant IFN-α assay.
HFAs in 96-well plates (Greiner) were treated with or without 25 to 100 U/ml of human recombinant IFN-α (Sigma-Aldrich) for 4 to 12 h, after which total RNA was isolated and subjected to qRT-PCR in order to measure the expression of ISGs.
Viral titer assay.
Titers were determined in Vero ATCC CCL-81 and Vero-E6 cells for arboviruses (flaviviruses and alphaviruses) and coronaviruses, respectively. The supplemental material provides a more detailed description of the assay.
NOD2 silencing.
Cells were seeded in 96-well plates (Greiner) overnight before transfection with 20 nM NOD2 Dicer-substrate short interfering RNA (DsiRNA) hs.Ri.NOD2.13.2 from Integrated DNA Technologies (IDT) (Coralville, IA) using 0.3 μg/well RNAiMax (Invitrogen, Waltham, MA). The nontargeting IDT control DsiRNA was used as a negative control for transfection. Twenty-four hours later, cells were infected with ZIKV at an MOI of 0.05. At 24 and 48 h postinfection, culture supernatants were collected for plaque assays. Total RNA isolated from cells at 48 h postinfection was subjected to qRT-PCR to determine the levels of viral genomic RNA and ISGs.
Measurement of cell viability.
Cell viability assays in response to drug or DMSO treatment were performed using a CellTiter-Glo luminescent cell viability kit (Promega) in cells grown in 96-well plates (Greiner) as described in the supplemental material.
In vitro and ex vivo drug assays.
After drug or DMSO treatment, viral replication and titers were determined by qRT-PCR on total RNA extracted from cells and plaque assays of culture supernatants, respectively, at 12 to 72 h postinfection. Cells seeded into 96-well plates (Greiner) were infected with ZIKV, DENV-2, MAYV, CVB5, or SARS-CoV-2 (MOI = 0.05 to 5), followed by treatment with 5, 10, 20, and 40 μM GSK717 (Sigma-Aldrich) or DMSO. GSK717 selectively inhibits the activation of NOD2 by blocking interaction with NOD2 agonists (14).
A549 or ACE2-SK-N-SH cells on coverslips in 12-well plates (Greiner) were infected (MOI of 1.0) with arboviruses (ZIKV, DENV-2, or MAYV) or SARS-CoV-2, respectively, and then processed for indirect immunofluorescence. Arbovirus time-of-addition assays were conducted in A549 cells, while SARS-CoV-2 time-of-addition assays were performed in ACE2-SK-N-SH cells, at an MOI of 0.1. Viral genome quantification and viral titer determination were performed.
A549 cells infected with arboviruses or CVB5 or ACE2-SK-N-SH cells infected with SARS-CoV-2 (MOI = 0.05 to 5) were treated with the RIPK2 inhibitor GSK583 (23) (Sigma-Aldrich) for 24 h. Cell supernatants and total cellular RNA were collected for determining viral titers and viral RNA levels, respectively.
To assess how GSK583 and GSK717 affected ISG induction, drug or DMSO alone was added to A549 cells for 16 h, after which they were treated with human recombinant IFN-α (100 U/ml) or transfected with poly(I·C) (0.2 μg/well) for 4 or 8 h. Total RNA extracted from the cells as described above was subjected to qRT-PCR using ISG-specific primers. See the supplemental material for additional information about the drug assays.
Immunofluorescence staining and cell imaging.
Infected cells grown on coverslips were fixed with 4% paraformaldehyde; permeabilized/blocked with a Triton X-100 (0.2%)–bovine serum albumin (BSA) (3%) solution; incubated with mouse anti-flavivirus group antigen 4G2 (Millipore, Burlington, MA), rabbit anti-alphavirus capsid (kindly provided by Andres Merits at the University of Tartu), or mouse anti-spike SARS-CoV/SARS-CoV-2 (GenTex, Irvine, CA) at room temperature for 1.5 h; washed; and then incubated with Alexa Fluor secondary antibodies against mouse or rabbit and 4′,6-diamidino-2-phenylindole (DAPI) for 1 h at room temperature. Antibodies were diluted in blocking buffer. Phosphate-buffered saline (PBS) containing 0.3% BSA was used for wash steps. Samples were examined using an Olympus (Tokyo, Japan) 1x81 spinning-disk confocal microscope or a Cytation 5 cell imaging multimode reader instrument (BioTek, Winooski, VT). Images were analyzed using Volocity or Gen5 software. More experimental details are provided in the supplemental material.
Statistical analyses.
Paired Student’s t test was used for pairwise statistical comparisons. The standard errors of the means are shown in all bar graphs. GraphPad Prism software 6.0 (GraphPad Software Inc., La Jolla, CA) was used for all statistical analyses.
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institutes of Health Research (grants PJT-148699, OV3-172302, and PJT-162417) and the Li Ka Shing Institute of Virology to T.C.H. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Anil Kumar, Joaquin Lopez Orozco, Adriana Airo, Zaikun Xu, Cheung Pang Wong, and Ray Ishida in the Hobman lab for helpful discussions and support. Confocal imaging was performed at the Cell Imaging Centre core in the Faculty of Medicine and Dentistry of the University of Alberta. We thank Darryl Falzarano (Vaccine and Infectious Disease Organization-International Vaccine Centre, University of Saskatchewan, Canada) for providing the Canadian SARS-CoV-2 strain for this work, David Safronetz (Public Health Agency of Canada, Canada) for providing the ZIKV strain, Bart Bartenschlager (Heidelberg University Hospital, Germany) for donating the DENV-2 clone, Xiao-Li Pang (Department of Laboratory Medicine and Pathology, University of Alberta, Canada) for providing the Faulkner strain of CVB5, Eva Gönczöl (The Wistar Institute, USA) for providing the HEL-18 cells, and Andres Merits (University of Tartu, Estonia) for kindly donating the anti-alphavirus capsid antibody. Furthermore, we thank Eileen Reklow, Yuanyuan Qiu, and Nikki Atanasova for excellent technical support.
We declare no competing financial interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Brechot C, Bryant J, Endtz H, Garry RF, Griffin DE, Lewin SR, Mercer N, Osterhaus A, Picot V, Vahlne A, Verjans GMGM, Weaver S. 2019. 2018 international meeting of the Global Virus Network. Antiviral Res 163:140–148. 10.1016/j.antiviral.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Clercq E, Li G. 2016. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29:695–747. 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ianevski A, Andersen PI, Merits A, Bjørås M, Kainov D. 2019. Expanding the activity spectrum of antiviral agents. Drug Discov Today 24:1224–1228. 10.1016/j.drudis.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Ianevski A, Zusinaite E, Kuivanen S, Strand M, Lysvand H, Teppor M, Kakkola L, Paavilainen H, Laajala M, Kallio-Kokko H, Valkonen M, Kantele A, Telling K, Lutsar I, Letjuka P, Metelitsa N, Oksenych V, Bjørås M, Nordbø SA, Dumpis U, Vitkauskiene A, Öhrmalm C, Bondeson K, Bergqvist A, Aittokallio T, Cox RJ, Evander M, Hukkanen V, Marjomaki V, Julkunen I, Vapalahti O, Tenson T, Merits A, Kainov D. 2018. Novel activities of safe-in-human broad-spectrum antiviral agents. Antiviral Res 154:174–182. 10.1016/j.antiviral.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, Girardin SE. 2019. NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys 670:69–81. 10.1016/j.abb.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Cardinaud S, Urrutia A, Rouers A, Coulon PG, Kervevan J, Richetta C, Bet A, Maze EA, Larsen M, Iglesias MC, Appay V, Graff-Dubois S, Moris A. 2017. Triggering of TLR-3, -4, NOD2, and DC-SIGN reduces viral replication and increases T-cell activation capacity of HIV-infected human dendritic cells. Eur J Immunol 47:818–829. 10.1002/eji.201646603. [DOI] [PubMed] [Google Scholar]
- 7.Kapoor A, Forman M, Arav-Boger R. 2014. Activation of nucleotide oligomerization domain 2 (NOD2) by human cytomegalovirus initiates innate immune responses and restricts virus replication. PLoS One 9:e92704. 10.1371/journal.pone.0092704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vissers M, Remijn T, Oosting M, de Jong DJ, Diavatopoulos DA, Hermans PW, Ferwerda G. 2012. Respiratory syncytial virus infection augments NOD2 signaling in an IFN-β-dependent manner in human primary cells. Eur J Immunol 42:2727–2735. 10.1002/eji.201242396. [DOI] [PubMed] [Google Scholar]
- 9.Tschöpe C, Müller I, Xia Y, Savvatis K, Pappritz K, Pinkert S, Lassner D, Heimesaat MM, Spillmann F, Miteva K, Bereswill S, Schultheiss H-P, Fechner H, Pieske B, Kühl U, Van Linthout S. 2017. NOD2 (nucleotide-binding oligomerization domain 2) is a major pathogenic mediator of coxsackievirus B3-induced myocarditis. Circ Heart Fail 10:e003870. 10.1161/CIRCHEARTFAILURE.117.003870. [DOI] [PubMed] [Google Scholar]
- 10.Limonta D, Jovel J, Kumar A, Lu J, Hou S, Airo AM, Lopez-Orozco J, Wong CP, Saito L, Branton W, Ka-Shu Wong G, Mason A, Power C, Hobman TC. 2019. Fibroblast growth factor 2 enhances Zika virus infection in human fetal brain. J Infect Dis 220:1377–1387. 10.1093/infdis/jiz073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limonta D, Jovel J, Kumar A, Airo AM, Hou S, Saito L, Branton W, Ka-Shu Wong G, Mason A, Power C, Hobman TC. 2018. Human fetal astrocytes infected with Zika virus exhibit delayed apoptosis and resistance to interferon: implications for persistence. Viruses 10:646. 10.3390/v10110646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Sousa JR, Azevedo RDSDS, Martins Filho AJ, de Araujo MTF, Cruz EDRM, Vasconcelos BCB, Cruz ACR, de Oliveira CS, Martins LC, Vasconcelos BHB, Casseb LMN, Chiang JO, Quaresma JAS, Vasconcelos PFDC. 2018. In situ inflammasome activation results in severe damage to the central nervous system in fatal Zika virus microcephaly cases. Cytokine 111:255–264. 10.1016/j.cyto.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Tricarico PM, Caracciolo I, Crovella S, D’Agaro P. 2017. Zika virus induces inflammasome activation in the glial cell line U87-MG. Biochem Biophys Res Commun 492:597–602. 10.1016/j.bbrc.2017.01.158. [DOI] [PubMed] [Google Scholar]
- 14.Rickard DJ, Sehon CA, Kasparcova V, Kallal LA, Zeng X, Montoute MN, Chordia T, Poore DD, Li H, Wu Z, Eidam PM, Haile PA, Yu J, Emery JG, Marquis RW, Gough PJ, Bertin J. 2013. Identification of benzimidazole diamides as selective inhibitors of the nucleotide-binding oligomerization domain 2 (NOD2) signaling pathway. PLoS One 8:e69619. 10.1371/journal.pone.0069619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mumtaz N, Jimmerson LC, Bushman LR, Kiser JJ, Aron G, Reusken CBEM, Koopmans MPG, van Kampen JJA. 2017. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antiviral Res 146:161–163. 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Katzelnick LC, Coloma J, Harris E. 2017. Dengue: knowledge gaps, unmet needs, and research priorities. Lancet Infect Dis 17:e88–e100. 10.1016/S1473-3099(16)30473-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinney RM, Butrapet S, Chang GJ, Tsuchiya KR, Roehrig JT, Bhamarapravati N, Gubler DJ. 1997. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230:300–308. 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- 18.Keestra-Gounder AM, Tsolis RM. 2017. NOD1 and NOD2: beyond peptidoglycan sensing. Trends Immunol 38:758–767. 10.1016/j.it.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratajczak MZ, Bujko K, Ciechanowicz A, Sielatycka K, Cymer M, Marlicz W, Kucia M. 2021. SARS-CoV-2 entry receptor ACE2 is expressed on very small CD45− precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 inflammasome. Stem Cell Rev Rep 17:266–277. 10.1007/s12015-020-10010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoel H, Heggelund L, Reikvam DH, Stiksrud B, Ueland T, Michelsen AE, Otterdal K, Muller KE, Lind A, Muller F, Dudman S, Aukrust P, Dyrhol-Riise AM, Holter JC, Trøseid M. 2021. Elevated markers of gut leakage and inflammasome activation in COVID-19 patients with cardiac involvement. J Intern Med 289:523–531. 10.1111/joim.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toldo S, Bussani R, Nuzzi V, Bonaventura A, Mauro AG, Cannatà A, Pillappa R, Sinagra G, Nana-Sinkam P, Sime P, Abbate A. 2021. Inflammasome formation in the lungs of patients with fatal COVID-19. Inflamm Res 70:7–10. 10.1007/s00011-020-01413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphries F, Yang S, Wang B, Moynagh PN. 2015. RIP kinases: key decision makers in cell death and innate immunity. Cell Death Differ 22:225–236. 10.1038/cdd.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haile PA, Votta BJ, Marquis RW, Bury MJ, Mehlmann JF, Singhaus R, Charnley AK, Lakdawala AS, Convery MA, Lipshutz DB, Desai BM, Swift B, Capriotti CA, Berger SB, Mahajan MK, Reilly MA, Rivera EJ, Sun HH, Nagilla R, Beal AM, Finger JN, Cook MN, King BW, Ouellette MT, Totoritis RD, Pierdomenico M, Negroni A, Stronati L, Cucchiara S, Ziółkowski B, Vossenkämper A, MacDonald TT, Gough PJ, Bertin J, Casillas LN. 2016. The identification and pharmacological characterization of 6-(tert-butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a highly potent and selective inhibitor of RIP2 kinase. J Med Chem 59:4867–4880. 10.1021/acs.jmedchem.6b00211. [DOI] [PubMed] [Google Scholar]
- 24.Acosta-Ampudia Y, Monsalve DM, Rodríguez Y, Pacheco Y, Anaya J-M, Ramírez-Santana C. 2018. Mayaro: an emerging viral threat? Emerg Microbes Infect 7:163. 10.1038/s41426-018-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierson TC, Diamond MS. 2018. The emergence of Zika virus and its new clinical syndromes. Nature 560:573–581. 10.1038/s41586-018-0446-y. [DOI] [PubMed] [Google Scholar]
- 26.Pons-Salort M, Parker EP, Grassly NC. 2015. The epidemiology of non-polio enteroviruses: recent advances and outstanding questions. Curr Opin Infect Dis 28:479–487. 10.1097/QCO.0000000000000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baggen J, Thibaut HJ, Strating JRPM, van Kuppeveld FJM. 2018. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol 16:368–381. 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 28.Rosas IO, Bräu N, Waters M, Go RC, Hunter BD, Bhagani S, Skiest D, Aziz MS, Cooper N, Douglas IS, Savic S, Youngstein T, Del Sorbo L, Cubillo Gracian A, De La Zerda DJ, Ustianowski A, Bao M, Dimonaco S, Graham E, Matharu B, Spotswood H, Tsai L, Malhotra A. 2021. Tocilizumab in hospitalized patients with severe Covid-19 pneumonia. N Engl J Med 384:1503–1516. 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glasgow A, Glasgow J, Limonta D, Solomon P, Lui I, Zhang Y, Nix MA, Rettko NJ, Zha S, Yamin R, Kao K, Rosenberg OS, Ravetch JV, Wiita AP, Leung KK, Lim SA, Zhou XX, Hobman TC, Kortemme T, Wells JA. 2020. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc Natl Acad Sci U S A 117:28046–28055. 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh M-D, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC, ACTT-1 Study Group Members. 2020. Remdesivir for the treatment of Covid-19—final report. N Engl J Med 383:1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.WHO Solidarity Trial Consortium, Pan H, Peto R, Henao-Restrepo A-M, Preziosi M-P, Sathiyamoorthy V, Abdool Karim Q, Alejandria MM, Hernández García C, Kieny M-P, Malekzadeh R, Murthy S, Reddy KS, Roses Periago M, Abi Hanna P, Ader F, Al-Bader AM, Alhasawi A, Allum E, Alotaibi A, Alvarez-Moreno CA, Appadoo S, Asiri A, Aukrust P, Barratt-Due A, Bellani S, Branca M, Cappel-Porter HBC, Cerrato N, Chow TS, Como N, Eustace J, García PJ, Godbole S, Gotuzzo E, Griskevicius L, Hamra R, Hassan M, Hassany M, Hutton D, Irmansyah I, Jancoriene L, Kirwan J, Kumar S, Lennon P, Lopardo G, Lydon P, Magrini N, Maguire T, Manevska S, Manuel O, et al. 2021. Repurposed antiviral drugs for Covid-19—interim WHO Solidarity Trial results. N Engl J Med 384:497–511. 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial Investigators. 2021. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med 384:238–251. 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, Shawa I, Adams AC, Van Naarden J, Custer KL, Shen L, Durante M, Oakley G, Schade AE, Sabo J, Patel DR, Klekotka P, Skovronsky DM, BLAZE-1 Investigators. 2021. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med 384:229–237. 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO. 2021. Draft landscape of COVID-19 candidate vaccines. WHO, Geneva, Switzerland. [Google Scholar]
- 35.WHO Ad Hoc Expert Group on the Next Steps for Covid-19 Vaccine Evaluation, Krause PR, Fleming TR, Longini IM, Peto R, Beral V, Bhargava B, Cravioto A, Cramer JP, Ellenberg SS, Figueroa JP, Halloran E, Henao-Restrepo AM, Ryan MJ, Levine MM, Nason M, Nohynek HM, Plotkin S, Rees H, Singh JA, Swaminathan S. 2021. Placebo-controlled trials of Covid-19 vaccines—why we still need them. N Engl J Med 384:e2. 10.1056/NEJMp2033538. [DOI] [PubMed] [Google Scholar]
- 36.Hodgson SH, Mansatta K, Mallett G, Harris V, Emary KRW, Pollard AJ. 2021. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect Dis 21:e26–e35. 10.1016/S1473-3099(20)30773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tigno-Aranjuez JT, Asara JM, Abbott DW. 2010. Inhibition of RIP2’s tyrosine kinase activity limits NOD2-driven cytokine responses. Genes Dev 24:2666–2677. 10.1101/gad.1964410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duran A, Valero N, Mosquera J, Fuenmayor E, Alvarez-Mon M. 2017. Gefitinib and pyrrolidine dithiocarbamate decrease viral replication and cytokine production in dengue virus infected human monocyte cultures. Life Sci 191:180–185. 10.1016/j.lfs.2017.10.027. [DOI] [PubMed] [Google Scholar]
- 39.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. 2020. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA 324:782–793. 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 40.Gutjahr A, Papagno L, Vernejoul F, Lioux T, Jospin F, Chanut B, Perouzel E, Rochereau N, Appay V, Verrier B, Paul S. 2020. New chimeric TLR7/NOD2 agonist is a potent adjuvant to induce mucosal immune responses. EBioMedicine 58:102922. 10.1016/j.ebiom.2020.102922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shafique M, Wilschut J, de Haan A. 2012. Induction of mucosal and systemic immunity against respiratory syncytial virus by inactivated virus supplemented with TLR9 and NOD2 ligands. Vaccine 30:597–606. 10.1016/j.vaccine.2011.11.054. [DOI] [PubMed] [Google Scholar]
- 42.Limonta D, Branton W, Wong CP, Saito L, Power C, Hobman TC. 2020. Use of primary human fetal astrocytes and tissue explants as ex vivo models to study Zika virus infection of the developing brain. Methods Mol Biol 2142:251–259. 10.1007/978-1-0716-0581-3_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC00491-21_Supp_S1_seq5.pdf, PDF file, 0.9 MB (910.9KB, pdf)