ABSTRACT
Movement of patients in a health care network poses challenges for the control of carbapenemase-producing Enterobacteriaceae (CPE). We aimed to identify intra- and interfacility transmission events and facility type-specific risk factors of CPE in an acute-care hospital (ACH) and its intermediate-term and long-term-care facilities (ILTCFs). Serial cross-sectional studies were conducted in June and July of 2014 to 2016 to screen for CPE. Whole-genome sequencing was done to identify strain relatedness and CPE genes (blaIMI, blaIMP-1, blaKPC-2, blaNDM-1, and blaOXA-48). Multivariable logistic regression models, stratified by facility type, were used to determine independent risk factors. Of 5,357 patients, half (55%) were from the ACH. CPE prevalence was 1.3% in the ACH and 0.7% in ILTCFs (P = 0.029). After adjusting for sociodemographics, screening year, and facility type, the odds of CPE colonization increased significantly with a hospital stay of ≥3 weeks (adjusted odds ratio [aOR], 2.67; 95% confidence interval [CI], 1.17 to 6.05), penicillin use (aOR, 3.00; 95% CI, 1.05 to 8.56), proton pump inhibitor use (aOR, 3.20; 95% CI, 1.05 to 9.80), dementia (aOR, 3.42; 95% CI, 1.38 to 8.49), connective tissue disease (aOR, 5.10; 95% CI, 1.19 to 21.81), and prior carbapenem-resistant Enterobacteriaceae (CRE) carriage (aOR, 109.02; 95% CI, 28.47 to 417.44) in the ACH. For ILTCFs, presence of wounds (aOR, 5.30; 95% CI, 1.01 to 27.72), respiratory procedures (aOR, 4.97; 95% CI, 1.09 to 22.71), vancomycin-resistant enterococcus carriage (aOR, 16.42; 95% CI, 1.52 to 177.48), and CRE carriage (aOR, 758.30; 95% CI, 33.86 to 16,982.52) showed significant association. Genomic analysis revealed only possible intra-ACH transmission and no evidence for ACH-to-ILTCF transmission. Although CPE colonization was predominantly in the ACH, risk factors varied between facilities. Targeted screening and precautionary measures are warranted.
KEYWORDS: blaIMI, blaIMP-1, blaKPC-2, blaNDM-1, blaOXA-48, beta-lactam resistant, bla, carbapenem-resistant Enterobacteriaceae, carbapenemase-producing Enterobacteriaceae, epidemiology, health care facilities, molecular epidemiology, risk factors, transmission
INTRODUCTION
The carbapenem-resistant Enterobacteriaceae (CRE) are a group of Gram-negative bacteria in the family Enterobacteriaceae that are phenotypically resistant to the carbapenem class of antibiotics. They are resistant to a wide range of antibiotics, mainly as a result of the production of carbapenemases encoded by carbapenemase genes. In recent years, carbapenemase-producing Enterobacteriaceae (CPE) have become notable causes of nosocomial infections and outbreaks in acute-care hospitals (ACHs), resulting in high morbidity and mortality (1–4). However, knowledge about CPE colonization in intermediate-care facilities (ITCFs) and long-term-care facilities (LTCFs) remains limited (5–7). Because of frequent bidirectional movement of patients between ACHs and affiliated ITCFs and LTCFs, interfacility transmission of health care-associated infections is possible (6, 7). Knowledge of the epidemiology of CPE colonization and risk profiling of patients can provide valuable guidance for targeted screening and proactive measures to prevent nosocomial transmission and outbreaks (8). The high resolution afforded by modern molecular techniques potentially allows understanding of the epidemiology and mechanisms of transmission of CPE in an interconnected health care network, enabling development of control strategies for intra- and interfacility transmission (9).
In this study, we compared the epidemiology of CPE colonization in an ACH and its affiliated intermediate- and long-term-care facilities (ILTCFs) in a single health care network to identify intra- and interfacility transmission events and facility type-specific risk factors to tailor infection prevention and control strategies and guide their implementation.
RESULTS
Epidemiology.
A total of 5,357 patients were screened for CRE, with about half (2,956 [55.2%]) being from the ACH (Table 1). The median length of hospital stay prior to screening was 10 days (interquartile range [IQR], 6 to 18) in the ACH, 22 days (IQR, 11 to 37) in the ITCFs, and 503 days (IQR, 222 to 1,665) in the LTCFs. The median age was 73 years (IQR, 62 to 81), with a slight preponderance of males (2,876 [53.7%]). Patients in the ACH had more comorbidities, with 59.7% having a Charlson comorbidity index (CCI) score of >3, compared with 50.6% of patients in ILTCFs (P < 0.001). Patients in the ACH also had more exposures to antibiotics and medical procedures.
TABLE 1.
Comparison and univariate analysis of epidemiological and clinical factors associated with CPE colonization in the ACH and ILTCFsa
| Overall |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n = 5,357) |
CPE not detected (n = 5,301) |
CPE detected (n = 56) |
ACH (n = 2,956) |
ILTCFs (n = 2,401) |
||||||||||||
| Factor | No. | Range or % | No. | Range or % | No. | Range or % | OR | 95% CI | No. | Range or % | OR | 95% CI | No. | Range or % | OR | 95% CI |
| Atge (yr)b | 73 | 62–81 | 73 | 62–82 | 71 | 62.5–80 | 0.99 | 0.97–1.01 | 74 | 62–82 | 0.99 | 0.97–1.01 | 72 | 62–82 | 0.99 | 0.95–1.02 |
| Male gender | 2,876 | 53.7 | 2,846 | 53.7 | 30 | 53.6 | 1 | 0.59–1.69 | 1,626 | 55 | 1.18 | 0.62–2.24 | 1,250 | 52.1 | 0.64 | 0.24–1.69 |
| Length of hospital stay (days)b | 17 | 8–64.5 | 17 | 8–66 | 16.5 | 7.5–33.5 | 0.99 | 1.00–1.00 | 10 | 6–18 | 1 | 0.99–1.01 | 67 | 22–482 | 1 | 1.00–1.00 |
| Hospital stay ≥3 wks | 2,489 | 46.5 | 2,463 | 46.5 | 26 | 46.4 | 0.91 | 0.54–1.55 | 662 | 22.4 | 1.58 | 0.84–2.97 | 1,827 | 76.1 | 0.57 | 0.19–1.77 |
| ≤4 beds in the same cubicle | 838 | 15.6 | 819 | 15.4 | 19 | 33.9 | 2.81 | 1.61–4.91 | 705 | 23.8 | 2.02 | 1.05–3.87 | 133 | 5.5 | 5.38 | 1.73–16.73 |
| Current admission in ACH (vs ILTCFs) | 2,956 | 55.2 | 2,917 | 55 | 39 | 69.6 | 1.87 | 1.06–3.32 | ||||||||
| Current location | ||||||||||||||||
| LTCFs | 1,157 | 21.6 | 1,153 | 21.8 | 4 | 7.1 | Ref | |||||||||
| ITCFs | 1,244 | 23.2 | 1,231 | 23.2 | 13 | 23.2 | 3.04 | 0.99–9.36 | ||||||||
| ACH | 2,956 | 55.2 | 2,917 | 55 | 39 | 69.6 | 3.85 | 1.37–10.81 | ||||||||
| Yr of sample collection | ||||||||||||||||
| 2014 | 1,673 | 31.2 | 1,662 | 31.4 | 11 | 19.6 | Ref | 976 | 33 | Ref | 697 | 29 | Ref | |||
| 2015 | 1,794 | 33.5 | 1,773 | 33.4 | 21 | 37.5 | 1.79 | 0.86–3.72 | 969 | 32.8 | 1.58 | 0.68–3.66 | 825 | 34.4 | 2.97 | 0.62–14.36 |
| 2016 | 1,890 | 35.3 | 1,866 | 35.2 | 24 | 42.9 | 1.94 | 0.95–3.98 | 1,011 | 34.2 | 1.73 | 0.76–3.93 | 879 | 36.6 | 3.19 | 0.68–15.08 |
| Prior admission to any facilities | 3,027 | 56.5 | 2,994 | 56.5 | 33 | 58.9 | 1.11 | 0.65–1.89 | 1,359 | 46 | 1.01 | 0.53–1.90 | 1,668 | 69.5 | 3.32 | 0.76–14.54 |
| Admission to ACH | 3,012 | 56.2 | 2,979 | 56.2 | 33 | 58.9 | 1.12 | 0.65–1.91 | 1,359 | 46 | 1.01 | 0.53–1.90 | 1,653 | 68.8 | 3.42 | 0.78–14.97 |
| Admission to ILTCFs | 186 | 3.5 | 183 | 3.5 | 3 | 5.4 | 1.58 | 0.49–5.11 | 0 | NA | 186 | 7.7 | 2.58 | 0.73–9.05 | ||
| Prior ICU admission | 133 | 2.5 | 129 | 2.4 | 4 | 7.1 | 3.08 | 1.10–8.65 | 133 | 4.5 | 2.47 | 0.86–7.05 | 0 | NA | ||
| Prior wound | 2,284 | 42.6 | 2,255 | 42.5 | 29 | 51.8 | 1.45 | 0.86–2.46 | 1,162 | 39.3 | 0.96 | 0.50–1.85 | 1,122 | 46.7 | 5.37 | 1.54–18.75 |
| Prior surgical operations | 2,398 | 44.8 | 2,367 | 44.7 | 31 | 55.4 | 1.54 | 0.91–2.61 | 1,514 | 51.2 | 1.11 | 0.59–2.10 | 884 | 36.8 | 2.47 | 0.94–6.51 |
| Prior vascular access procedures | 4,291 | 80.1 | 4,239 | 80 | 52 | 92.9 | 3.26 | 1.18–9.02 | 2,771 | 93.7 | 2.56 | 0.35–18.74 | 1,520 | 63.3 | 2.72 | 0.78–9.49 |
| Arterial line | 755 | 14.1 | 743 | 14 | 12 | 21.4 | 1.67 | 0.88–3.18 | 601 | 20.3 | 1.76 | 0.88–3.49 | 154 | 6.4 | NA | |
| CVP line | 408 | 7.6 | 400 | 7.5 | 8 | 14.3 | 2.04 | 0.96–4.35 | 320 | 10.8 | 1.21 | 0.47–3.13 | 88 | 3.7 | 5.8 | 1.63–20.55 |
| Hemodialysis line | 303 | 5.7 | 295 | 5.6 | 8 | 14.3 | 2.83 | 1.33–6.03 | 216 | 7.3 | 2.34 | 0.97–5.66 | 87 | 3.6 | 3.61 | 0.81–16.02 |
| Peripheral line | 4,279 | 79.9 | 4,227 | 79.7 | 52 | 92.9 | 3.3 | 1.19–9.15 | 2,766 | 93.6 | 2.63 | 0.36–19.28 | 1,513 | 63 | 2.76 | 0.79–9.61 |
| PICC line | 236 | 4.4 | 231 | 4.4 | 5 | 8.9 | 2.15 | 0.85–5.44 | 141 | 4.8 | 2.32 | 0.81–6.62 | 95 | 4 | 1.52 | 0.20–11.60 |
| Prior respiratory procedures | 858 | 16 | 842 | 15.9 | 16 | 28.6 | 2.12 | 1.18–3.80 | 539 | 18.2 | 1.16 | 0.53–2.54 | 319 | 13.3 | 5.92 | 2.27–15.47 |
| Endotracheal tube | 631 | 11.8 | 619 | 11.7 | 12 | 21.4 | 2.06 | 1.08–3.93 | 441 | 1.8 | 1.25 | 0.55–2.85 | 190 | 7.9 | 4.95 | 1.73–14.21 |
| Chest tube | 87 | 1.6 | 86 | 1.6 | 1 | 1.8 | 1.1 | 0.15–8.06 | 71 | 0.1 | 1.07 | 0.14–7.91 | 16 | 0.7 | NA | |
| Tracheostomy | 309 | 5.8 | 302 | 5.7 | 7 | 12.5 | 2.36 | 1.06–5.26 | 152 | 5.1 | 1.55 | 0.47–5.09 | 157 | 6.5 | 4.49 | 1.45–13.92 |
| Prior gastrointestinal procedures | 1,690 | 31.5 | 1,666 | 31.4 | 24 | 42.9 | 1.64 | 0.96–2.79 | 932 | 31.5 | 1.22 | 0.63–2.36 | 758 | 31.6 | 3.12 | 1.18–8.24 |
| Nasogastric tube | 1,620 | 30.2 | 1,597 | 30.1 | 23 | 41.1 | 1.62 | 0.95–2.76 | 903 | 30.5 | 1.28 | 0.66–2.47 | 717 | 29.9 | 2.66 | 1.02–6.93 |
| PEG tube | 513 | 9.6 | 506 | 9.5 | 7 | 12.5 | 1.35 | 0.61–3.00 | 357 | 12.1 | 1.07 | 0.42–2.76 | 156 | 6.5 | 1.93 | 0.44–8.52 |
| Colostomy | 67 | 1.3 | 67 | 1.3 | 0 | 44 | 1.5 | 23 | 1 | |||||||
| Prior urinary procedures | 1,657 | 30.9 | 1,635 | 30.8 | 22 | 39.3 | 1.45 | 0.85–2.49 | 1,012 | 34.2 | 1.34 | 0.71–2.55 | 645 | 26.9 | 1.49 | 0.55–4.04 |
| Suprapubic catheter | 38 | 0.7 | 38 | 0.7 | 0 | 16 | 0.5 | 22 | 0.9 | |||||||
| Urethral catheter | 1,638 | 30.6 | 1,616 | 30.5 | 22 | 39.3 | 1.48 | 0.86–2.53 | 1,008 | 34.1 | 1.35 | 0.71–2.57 | 630 | 26.2 | 1.54 | 0.57–4.18 |
| Prior use of: | ||||||||||||||||
| Any antibiotics | 4,123 | 77 | 4,072 | 76.8 | 51 | 91.1 | 3.08 | 1.23–7.73 | 2,477 | 83.8 | 1.7 | 0.60–4.81 | 1,646 | 68.6 | 7.4 | 0.98–55.91 |
| Aminoglycosides | 1,253 | 23.4 | 1,235 | 23.3 | 18 | 32.1 | 1.56 | 0.89–2.74 | 971 | 32.8 | 1.28 | 0.67–2.46 | 282 | 11.7 | 1.62 | 0.46–5.66 |
| Carbapenems | 670 | 12.5 | 656 | 12.4 | 14 | 25 | 2.36 | 1.28–4.35 | 485 | 16.4 | 1.32 | 0.60–2.89 | 185 | 7.7 | 6.72 | 2.46–18.38 |
| Cephalosporins | 1,467 | 27.4 | 1,444 | 27.2 | 23 | 41.1 | 1.86 | 1.09–3.18 | 937 | 31.7 | 1.68 | 0.89–3.17 | 530 | 22.1 | 1.94 | 0.71–5.26 |
| Fluoroquinolones | 1,173 | 21.9 | 1,157 | 21.8 | 16 | 28.6 | 1.43 | 0.80–2.57 | 610 | 20.6 | 1.16 | 0.55–2.45 | 563 | 23.4 | 2.3 | 0.87–6.07 |
| Penicillins | 3,429 | 64 | 3,383 | 63.8 | 46 | 82.1 | 2.61 | 1.31–5.18 | 2,105 | 71.2 | 2.78 | 1.08–7.13 | 1,324 | 55.1 | 1.96 | 0.69–5.58 |
| Vancomycins | 1,350 | 25.2 | 1,330 | 25.1 | 20 | 35.7 | 1.66 | 0.96–2.88 | 1,038 | 35.1 | 0.82 | 0.41–1.62 | 312 | 13 | 6.08 | 2.33–15.88 |
| Any others antibiotics | 904 | 16.9 | 888 | 16.8 | 16 | 28.6 | 1.99 | 1.11–3.57 | 526 | 17.8 | 1.39 | 0.66–2.95 | 378 | 15.7 | 3.8 | 1.44–10.04 |
| Corticosteroids | 857 | 16 | 845 | 15.9 | 12 | 21.4 | 1.44 | 0.76–2.73 | 563 | 19 | 1.47 | 0.71–3.04 | 294 | 12.2 | 0.96 | 0.22–4.20 |
| Antacids | 154 | 2.9 | 153 | 2.9 | 1 | 1.8 | 0.61 | 0.08–4.45 | 78 | 2.6 | NA | 76 | 3.2 | 1.92 | 0.25–14.70 | |
| PPI | 3,461 | 64.6 | 3,414 | 64.4 | 47 | 83.9 | 2.89 | 1.41–5.90 | 2,166 | 73.3 | 3.23 | 1.14–9.11 | 1,295 | 53.9 | 2.06 | 0.72–5.86 |
| H2 receptor blockers | 791 | 14.8 | 782 | 14.8 | 9 | 16.1 | 1.11 | 0.54–2.27 | 428 | 14.5 | 1.3 | 0.57–2.96 | 363 | 15.1 | 0.75 | 0.17–3.28 |
| Comorbidities | ||||||||||||||||
| HIV | 39 | 0.7 | 38 | 0.7 | 1 | 1.8 | 2.52 | 0.34–18.67 | 32 | 1.1 | 2.45 | 0.33–18.41 | 7 | 0.3 | NA | |
| Cerebrovascular disease | 2,026 | 37.8 | 2,003 | 37.8 | 23 | 41.1 | 1.15 | 0.67–1.96 | 831 | 28.1 | 1.28 | 0.66–2.51 | 1,195 | 49.8 | 1.45 | 0.55–3.81 |
| Myocardial infarction | 991 | 18.5 | 978 | 18.4 | 13 | 23.2 | 1.34 | 0.72–2.49 | 509 | 17.2 | 1.45 | 0.68–3.07 | 482 | 20.1 | 1.23 | 0.40–3.78 |
| Peripheral vascular disease | 610 | 11.4 | 603 | 11.4 | 7 | 12.5 | 1.11 | 0.50–2.47 | 320 | 10.8 | 1.21 | 0.47–3.13 | 290 | 12.1 | 0.97 | 0.22–4.27 |
| Hemiplegia or paraplegia | 722 | 13.5 | 713 | 13.5 | 9 | 16.1 | 1.23 | 0.60–2.53 | 293 | 9.9 | 1.34 | 0.52–3.46 | 429 | 17.9 | 1.42 | 0.46–4.37 |
| Malignant lymphoma | 47 | 0.9 | 46 | 0.9 | 1 | 1.8 | 2.08 | 0.28–15.33 | 30 | 1 | 2.62 | 0.35–19.74 | 17 | 0.7 | NA | |
| Leukemia | 27 | 0.5 | 27 | 0.5 | 0 | 22 | 0.7 | 5 | 0.2 | |||||||
| Congestive heart failure | 664 | 12.4 | 657 | 12.4 | 7 | 12.5 | 1.01 | 0.46–2.24 | 484 | 16.4 | 1.12 | 0.49–2.55 | 180 | 7.5 | NA | |
| Malignancy | 703 | 13.1 | 697 | 13.1 | 6 | 10.7 | 0.79 | 0.34–1.86 | 493 | 16.7 | 0.73 | 0.28–1.88 | 210 | 8.7 | 0.65 | 0.09–4.93 |
| Diabetes | 2,257 | 42.1 | 2,235 | 42.2 | 22 | 39.3 | 0.89 | 0.52–1.52 | 1,310 | 44.3 | 1.08 | 0.57–2.03 | 947 | 39.4 | 0.47 | 0.15–1.45 |
| Dementia | 949 | 17.7 | 937 | 17.7 | 12 | 21.4 | 1.27 | 0.67–2.41 | 521 | 17.6 | 1.62 | 0.79–3.35 | 428 | 17.8 | 0.61 | 0.14–2.69 |
| Peptic ulcer disease | 341 | 6.4 | 336 | 6.3 | 5 | 8.9 | 1.45 | 0.57–3.65 | 219 | 7.4 | 1.44 | 0.51–4.08 | 122 | 5.1 | 1.17 | 0.15–8.89 |
| Connective tissue disease | 75 | 1.4 | 72 | 1.4 | 3 | 5.4 | 4.11 | 1.26–13.46 | 57 | 1.9 | 4.42 | 1.32–14.79 | 18 | 0.7 | NA | |
| Chronic pulmonary disease | 562 | 10.5 | 559 | 10.5 | 3 | 5.4 | 0.48 | 0.15–1.54 | 373 | 12.6 | 0.37 | 0.09–1.55 | 189 | 7.9 | 0.73 | 0.10–5.54 |
| Renal disease | 1,338 | 25 | 1,319 | 24.9 | 19 | 33.9 | 1.55 | 0.89–2.71 | 906 | 30.6 | 1.27 | 0.66–2.46 | 432 | 18 | 1.91 | 0.67–5.45 |
| Liver disease | 356 | 6.6 | 351 | 6.6 | 5 | 8.9 | 1.38 | 0.55–3.49 | 256 | 8.7 | 1.21 | 0.43–3.43 | 100 | 4.2 | 1.44 | 0.19–10.99 |
| Immunocompromised statusc | 786 | 14.7 | 779 | 14.7 | 7 | 12.5 | 0.83 | 0.37–1.84 | 552 | 18.7 | 0.79 | 0.33–1.89 | 234 | 9.7 | 0.58 | 0.08–4.37 |
| Charlson comorbidity indexb | 3 | 1–5 | 3 | 1–5 | 3 | 1–5 | 1.04 | 0.95–1.15 | 3 | 1–6 | 1.04 | 0.94–1.16 | 3 | 1–4 | 0.97 | 0.77–1.21 |
| Charlson comorbidity index >3 | 2,979 | 55.6 | 2,945 | 55.6 | 34 | 60.7 | 1.24 | 0.71–2.17 | 1,765 | 59.7 | 1.1 | 0.58–2.12 | 1,214 | 50.6 | 1.17 | 0.38–3.61 |
| Prior carriage of: | ||||||||||||||||
| MRSA | 985 | 18.4 | 976 | 18.4 | 9 | 16.1 | 0.85 | 0.41–1.74 | 630 | 21.3 | 0.81 | 0.35–1.83 | 355 | 14.8 | 0.77 | 0.17–3.37 |
| VRE | 85 | 1.6 | 83 | 1.6 | 2 | 3.6 | 2.33 | 0.56–9.71 | 62 | 3.2 | NA | 23 | 1 | 15 | 3.23–69.75 | |
| CRE | 35 | 0.7 | 23 | 0.4 | 12 | 21.4 | 62.58 | 29.32–133.61 | 27 | 49 | 39.36 | 16.02–96.69 | 8 | 0.3 | 183.08 | 41.29–811.71 |
| MDRO | 1,985 | 37.1 | 1,955 | 36.9 | 30 | 53.6 | 1.97 | 1.16–3.35 | 1,449 | 1.2 | 1.22 | 0.65–2.29 | 536 | 22.3 | 3.96 | 1.52–10.33 |
Ref, reference category; NA, not applicable; MDRO, multidrug-resistant organisms.
Values are medians and IQR.
Includes patients with any malignancy, including lymphoma or leukemia, or AIDS.
From 5,357 patients screened, a total of 237 Enterobacteriaceae isolates (from 206 patients) were retrieved from ChromID Carba selective chromogenic agar. The identities of all Enterobacteriaceae species isolates were verified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) (see Table S1 in the supplemental material). Among them, 99 (41.8%) isolates from 82 patients were found to be phenotypically resistant to both meropenem and ertapenem according to a Vitek 2 sensitivity test. After sequencing all 237 Enterobacteriaceae isolates, we identified 68 isolates (from 56 patients) carrying at least one CPE gene of interest, and 64/68 of them were carbapenem resistant (Fig. 1). Instances of other important carbapenemase genes, such as blaVIM and blaSME, as well as less common ones, such as blaGES and blaFRI, were not identified among the isolates. A previous study of carbapenemase-producing Enterobacteriaceae in Singapore noted the common presence of four plasmids carrying the KPC-1 or various NDM alleles. Only two of these plasmids, pNDM-ECS01 (carrying NDM-1) and pHS102707 (carrying KPC-1), were found in our data (see Table S2).
FIG 1.
Flow chart of participants showing prevalence of meropenem susceptibility and carbapenemase genes among Enterobacteriaceae isolates obtained from patients screened in the acute care hospital and intermediate- and long-term-care facilities. For ertapenem, resistant is defined as a MIC of ≥2 mg/liter, intermediate as 1 mg/liter, and susceptible as ≤0.5 mg/liter; for meropenem, resistant is defined as a MIC of ≥4 mg/liter, intermediate as 2 mg/liter, and susceptible as ≤1 mg/liter. Intermediate isolates were considered susceptible to carbapenems. ACH, acute-care hospital; ILTCFs, intermediate- and long-term-care facilities.
The overall prevalence of CPE colonization was low in the ACH (1.32%) and even lower in ILTCFs (0.71%). An increasing trend in prevalence was observed in the ACH, from 0.92% in 2014 to 1.44% in 2015 and 1.58% in 2016 (Ptrend = 0.201). A similar nonsignificant increasing trend was noticed in ILTCFs (from 0.29% in 2014 to 0.85% in 2015 and 0.91% in 2016; Ptrend = 0.162) (see Fig. S1 in the supplemental material).
Genomic analysis.
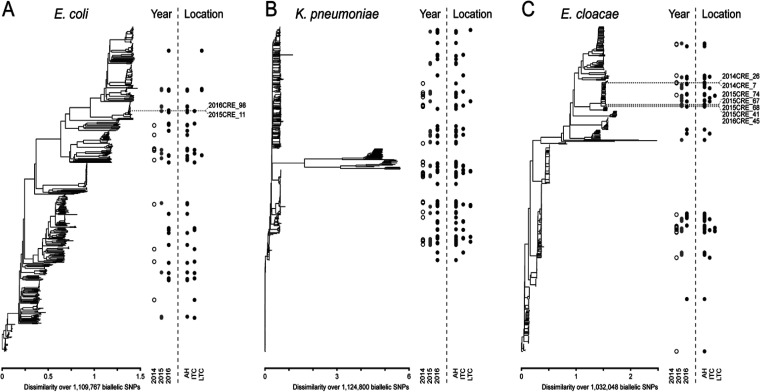
By multilocus sequence typing (MLST) and whole-genome single nucleotide polymorphism (SNP) trees, diverse representatives of Klebsiella pneumoniae (102 [43.0%]), Enterobacter cloacae (66 [27.9%]), and Escherichia coli (44 [18.6%]) were isolated, with no strong overall pattern of clustering by year or location. However, a subset of strains for E. coli and E. cloacae, which were potentially informative about intra- or interfacility spread, were clustered together (Fig. 2).
FIG 2.
Phylogenetic tree diagram of E. coli, K. pneumoniae, and E. cloacae showing the locations of CPE genes on each strain identified. Whole-genome phylogenetic trees for E. coli (A), K. pneumoniae (B), and E. cloacae (C) strains isolated in this study. For each species, 500 randomly sequenced complete genomes were selected from the GenBank RefSeq database to provide context for the overall species diversity. Neighbor-joining phylogenetic trees, based on SNPs called against a common reference sequence (see Materials and Methods), are shown in the left portion of each panel. The x axis shows the scale in terms of dissimilarity, as calculated by the SNPRelate package in R. The total number of biallelic SNPs used in the alignment is indicated below the x axis. The year and location for each isolate sequenced in this study are indicated by the circles in the right portion of each panel, which are placed at the vertical location where the strain is found in the phylogenetic tree. Dots indicate different years or locations, indicated by the labels at the bottom. Dotted lines and text labels in panels A and C indicate sets of strains that are further discussed in the main text.
On further analysis of these clusters, we found two E. cloacae isolates, both carrying blaNDM-1 and separated by 16 SNPs (Table 2). They were from different wards from the ACH in 2014, suggesting a possible (though limited) intra-ACH transmission. Another cluster of five blaIMI-carrying E. cloacae isolates (four in 2015 and one in 2016) from the ACH revealed pairwise SNP distances from 0 to 5 among 2015 strains and 19 to 23 between the 2015 and 2016 strains, suggesting a potentially longer-term circulation of this strain from 2015 to 2016 (10). Finally, there was one example of two closely related (22 SNPs) E. coli isolates from an ITCF in 2015 and the ACH in 2016. Based on the isolation years, this was clearly not a transmission from the ACH; however, further examination of the patient records showed that both patients had had recent prior admissions to the same ACH. Unfortunately, additional E. coli samples from the hospital during these times were not available for analysis in this study.
TABLE 2.
Details of individual CPE clusters in the ACH and ILTCFs screened in June and July of 2014 to 2016
| Cluster and strain | SNP distance from strain | CPE gene | Current location | Yr of identification | Prior admission within 1 yr | Current ward | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 (E. cloacae) | 2014CRE_26 | 2014CRE_7 | ||||||||
| 2014CRE_26 | 16 | blaNDM-1 | ACH | 2014 | No | Different ward | ||||
| 2014CRE_7 | 16 | blaNDM-1 | ACH | 2014 | No | Different ward | ||||
| Cluster 2 (E. cloacae) | 2015CRE_41 | 2015CRE_67 | 2015CRE_68 | 2015CRE_74 | 2016CRE_45 | |||||
| 2015CRE_41 | 4 | 5 | 5 | 19 | blaIMI | ACH | 2015 | No | Different ward | |
| 2015CRE_67 | 4 | 1 | 1 | 22 | blaIMI | ACH | 2015 | No | Same ward | |
| 2015CRE_68 | 5 | 1 | 0 | 23 | blaIMI | ACH | 2015 | No | Same ward | |
| 2015CRE_74 | 5 | 1 | 0 | 21 | blaIMI | ACH | 2015 | No | Different ward | |
| 2016CRE_45 | 19 | 22 | 23 | 21 | blaIMI | ACH | 2016 | No | Different ward | |
| Cluster 3 (E. coli) | 2015CRE_11 | 2016CRE_98 | ||||||||
| 2015CRE_11 | 22 | blaNDM-1 | ITCF | 2015 | ACH | Different ward | ||||
| 2016CRE_98 | 22 | blaNDM-1 | ACH | 2016 | ACH | Different ward | ||||
Univariate analysis.
The type of health care facility was significantly associated with CPE colonization, with patients in the ACH being 4 times as likely as those from the LTCFs to be CPE colonized (odds ratio [OR], 3.85; 95% confidence interval [CI], 1.37 to 10.81) (Table 1). In the ACH, a history of connective tissue disease increased the odds of CPE colonization by 4.4 times (OR, 4.42; 95% CI, 1.32 to 14.79), and exposures to the penicillin group of antibiotics and proton pump inhibitors (PPI) approximately tripled the odds of colonization (OR, 2.78 [95% CI, 1.08 to 7.13], and OR, 3.23 [95% CI, 1.14 to 9.11], respectively). In ILTCFs, exposures to carbapenems (OR, 6.72; 95% CI, 2.46 to 18.38) and vancomycins (OR, 6.08; 95% CI, 2.33 to 15.88), presence of wounds (OR, 5.37; 95% CI, 1.54 to 18.75), respiratory procedures (OR, 5.92; 95% CI, 2.27 to 15.47), and gastrointestinal procedures (OR, 3.12; 95% CI, 1.18 to 8.24) were significantly associated with CPE colonization. In both the ACH and ILTCFs, prior CRE carriage was significantly associated with CPE colonization (OR, 39.36 [95% CI, 16.02 to 96.69] for ACH; OR, 183.08 [95% CI, 41.29 to 811.71] for ILTCFs).
Multivariate analysis.
After adjusting for sociodemographics, year of screening, and the type of health care facility, prior CRE carriage (adjusted OR [aOR], 95.86; 95% CI, 31.99 to 287.21) was the strongest predictor of CPE colonization (Table 3). The modifying effect by the type of health care facility was further assessed by stratification (6, 11). As very few patients in LTCFs were CPE colonized (4 [0.35%]), a combined analysis of ILTCFs was performed.
TABLE 3.
Multivariable logistic regression analysis of epidemiological and clinical factors associated with CPE colonization in the ACH and ILTCFsa
| Factorb | Overall (n = 5,357) |
ACH (n = 2,956) |
ILTCFs (n = 2,401) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| aOR | 95% CI | P | aOR | 95% CI | P | aOR | 95% CI | P | |
| Age (yr) | 0.99 | 0.97–1.01 | 0.313 | 0.99 | 0.96–1.01 | 0.347 | 0.99 | 0.94–1.04 | 0.609 |
| Male gender | 1.04 | 0.58–1.88 | 0.898 | 1.60 | 0.76–3.36 | 0.217 | 0.27 | 0.07–1.12 | 0.072 |
| ≤4 beds in the same cubicle | 0.95 | 0.42–2.17 | 0.907 | 1.03 | 0.41–2.56 | 0.949 | 0.94 | 0.12–7.08 | 0.951 |
| Hospital stay ≥3 wks | 1.37 | 0.69–2.74 | 0.372 | 2.67 | 1.17–6.05 | 0.019 | 0.65 | 0.16–2.73 | 0.558 |
| Current location | |||||||||
| LTCFs | Ref | Ref | |||||||
| ITCFs | 4.13 | 1.08–15.76 | 0.038 | 4.32 | 0.60–31.08 | 0.146 | |||
| ACH | 4.57 | 1.14–18.44 | 0.033 | ||||||
| Yr of sample collection | |||||||||
| 2014 | Ref | Ref | Ref | ||||||
| 2015 | 2.61 | 1.13–6.04 | 0.025 | 2.52 | 0.95–6.64 | 0.062 | 2.12 | 0.34–13.20 | 0.422 |
| 2016 | 2.98 | 1.30–6.80 | 0.010 | 2.99 | 1.15–7.75 | 0.024 | 2.13 | 0.32–14.12 | 0.434 |
| Prior admission to any facilities | 0.78 | 0.38–1.61 | 0.504 | 0.70 | 0.31–1.61 | 0.405 | 1.42 | 0.14–14.20 | 0.767 |
| Prior ICU admission | 1.36 | 0.33–5.68 | 0.672 | 2.84 | 0.59–13.58 | 0.192 | NA | ||
| Prior surgical operations | 0.94 | 0.48–1.86 | 0.860 | 0.95 | 0.42–2.11 | 0.893 | 1.14 | 0.22–5.86 | 0.873 |
| Prior wound | 0.96 | 0.52–1.79 | 0.909 | 0.57 | 0.26–1.25 | 0.158 | 5.30 | 1.01–27.72 | 0.048 |
| Prior vascular access procedures | 1.29 | 0.39–4.25 | 0.681 | 2.11 | 0.26–16.95 | 0.483 | 0.39 | 0.06–2.77 | 0.350 |
| Prior respiratory procedures | 1.73 | 0.75–3.97 | 0.198 | 0.86 | 0.27–2.76 | 0.795 | 4.97 | 1.09–22.71 | 0.038 |
| Prior gastrointestinal procedures | 0.94 | 0.45–1.95 | 0.864 | 0.64 | 0.25–1.64 | 0.355 | 2.59 | 0.51–13.05 | 0.249 |
| Prior urinary procedures | 0.88 | 0.46–1.69 | 0.700 | 1.29 | 0.60–2.77 | 0.511 | 0.33 | 0.07–1.54 | 0.157 |
| Aminoglycosides | 0.72 | 0.35–1.47 | 0.362 | 0.84 | 0.38–1.84 | 0.656 | 0.15 | 0.01–1.84 | 0.139 |
| Carbapenems | 1.09 | 0.46–2.57 | 0.851 | 0.74 | 0.24–2.27 | 0.600 | 3.49 | 0.66–18.57 | 0.143 |
| Cephalosporins | 1.08 | 0.56–2.09 | 0.817 | 1.46 | 0.67–3.21 | 0.341 | 0.63 | 0.14–2.72 | 0.533 |
| Fluoroquinolones | 1.55 | 0.77–3.13 | 0.221 | 1.68 | 0.67–4.19 | 0.267 | 1.58 | 0.37–6.74 | 0.536 |
| Penicillins | 1.96 | 0.90–4.26 | 0.088 | 3.00 | 1.05–8.56 | 0.040 | 0.69 | 0.17–2.90 | 0.614 |
| Vancomycins | 0.71 | 0.33–1.51 | 0.372 | 0.47 | 0.19–1.18 | 0.110 | 2.41 | 0.49–11.99 | 0.281 |
| Any others antibiotics | 1.36 | 0.65–2.84 | 0.413 | 1.01 | 0.37–2.72 | 0.992 | 2.34 | 0.55–9.85 | 0.248 |
| Corticosteroids | 1.49 | 0.70–3.19 | 0.300 | 1.51 | 0.62–3.69 | 0.370 | 0.48 | 0.05–4.17 | 0.503 |
| Antacids | 0.37 | 0.05–2.99 | 0.353 | NA | 0.46 | 0.03–7.55 | 0.583 | ||
| PPI | 2.06 | 0.93–4.57 | 0.074 | 3.20 | 1.05–9.80 | 0.041 | 1.15 | 0.27–4.83 | 0.853 |
| H2 receptor blockers | 1.26 | 0.57–2.80 | 0.568 | 1.55 | 0.61–3.95 | 0.360 | 1.15 | 0.20–6.81 | 0.874 |
| Diabetes | 0.71 | 0.38–1.31 | 0.274 | 0.99 | 0.48–2.05 | 0.983 | 0.21 | 0.04–1.09 | 0.063 |
| Dementia | 2.52 | 1.16–5.50 | 0.020 | 3.42 | 1.38–8.49 | 0.008 | 1.52 | 0.18–12.47 | 0.699 |
| Peptic ulcer disease | 1.40 | 0.52–3.81 | 0.508 | 1.26 | 0.38–4.14 | 0.701 | 1.76 | 0.16–18.89 | 0.642 |
| Connective tissue disease | 2.87 | 0.71–11.64 | 0.140 | 5.10 | 1.19–21.81 | 0.028 | NA | ||
| Chronic pulmonary disease | 0.29 | 0.07–1.11 | 0.070 | 0.24 | 0.05–1.18 | 0.079 | 0.33 | 0.01–7.14 | 0.477 |
| Renal disease | 1.23 | 0.63–2.42 | 0.544 | 1.10 | 0.49–2.44 | 0.821 | 1.62 | 0.31–8.34 | 0.567 |
| Liver disease | 1.17 | 0.53–2.59 | 0.705 | 1.02 | 0.41–2.52 | 0.971 | 2.76 | 0.22–34.72 | 0.433 |
| Immunocompromised status | 0.56 | 0.23–1.37 | 0.200 | 0.62 | 0.22–1.71 | 0.353 | 0.10 | 0.00–2.92 | 0.180 |
| Prior carriage of: | |||||||||
| MRSA | 0.46 | 0.19–1.07 | 0.072 | 0.6 | 0.22–1.62 | 0.310 | 0.25 | 0.03–2.14 | 0.204 |
| VRE | 1.78 | 0.36–8.78 | 0.479 | NA | 16.42 | 1.52–177.48 | 0.021 | ||
| CRE | 95.86 | 31.99–287.21 | <0.001 | 109.02 | 28.47–417.44 | <0.001 | 758.30 | 33.86–16,982.52 | <0.001 |
| MDRO | 0.74 | 0.37–1.47 | 0.387 | 0.56 | 0.24–1.29 | 0.174 | 1.25 | 0.31–5.13 | 0.753 |
Ref, reference; NA, not applicable. Significant P values of <0.05 are in bold.
For clinical procedures and explanation of immunocompromised status, see Table 1.
In the ACH, the odds of CPE colonization tripled in patients with prior exposures to penicillins (aOR, 3.00; 95% CI, 1.05 to 8.56) and PPI (aOR, 3.20; 95% CI, 1.05 to 9.80). A hospital stay of at least 3 weeks (aOR, 2.69; 95% CI, 1.17 to 6.05), dementia (aOR, 3.42; 95% CI, 1.38 to 8.49), connective tissue disease (aOR, 5.10; 95% CI, 1.19 to 21.81), and prior carriage of CRE (aOR, 109.02; 95% CI, 28.47 to 417.44) were independently associated with CPE colonization in the ACH. For patients from ILTCFs, prior histories of wounds (aOR, 5.30; 95% CI, 1.01 to 27.72), respiratory procedures (aOR, 4.97; 95% CI, 1.09 to 22.71), vancomycin-resistant enterococci (VRE) carriage (aOR, 16.42; 95% CI, 1.52 to 177.48), and CRE carriage (aOR, 758.30; 95% CI, 33.86 to 16,982.52) were significantly associated with CPE colonization. Prior hospital admission was not associated with CPE colonization in either the ACH or ILTCFs.
DISCUSSION
Prevalence and transmission of CPE in the health care network.
In Singapore, different types of carbapenemase genes have been identified (12–16); this is suspected to be a consequence of its being a highly connected international travel hub (1, 17). In our study, the proportion of CPE among meropenem-resistant Enterobacteriaceae in the ACH was 47/78 (60.3%) which was similar to the CPE prevalence of 64.7% (2010 to 2015) in six public hospitals reported by the Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group (12). Our study further observed that three of the ertapenem-susceptible isolates harbored CPE genes (two blaIMP-1 and one blaKPC-2). blaOXA-48 was also identified in one meropenem-susceptible isolate. Additionally, we identified the presence of plasmid pNDM-ECS01 in two E. cloacae strains, which has not been previously reported. It is therefore prudent to include the use of molecular and genomic methods, in addition to conventional cultures, for the active surveillance of CPE in all facilities.
The ACH was the main reservoir of CPE in the health care network, with patients in the ACH being 1.3 and 4 times as likely to be CPE colonized as patients in ITCFs and LTCFs, respectively. ACH patients with ≥3 weeks of hospital stay were 2.7 times as likely to be CPE colonized as those with shorter stays (aOR, 2.67; 95% CI, 1.17 to 6.05). In contrast, ILTCFs patients with a <3-week stay were 53% more likely to be CPE colonized (aOR, 1.53; 95% CI, 0.37 to 6.41), suggesting possible colonization due to recent admissions to ACHs. The effect of duration of stay on CPE colonization differed by the type of health care facility (18). Our findings support CPE screening when long stayers are transferred from the ACH to ILTCFs to prevent interfacility transmission of CPE (19). Moreover, the gradual increase in CPE prevalence in both the ACH and ILTCFs over the 3 years highlights the need for enhancing infection prevention and control strategies in all health care facilities.
Interfacility transmission of CPE would result in hospital outbreaks (20, 21). The estimated cost for a single CPE outbreak (assuming it affects 40 patients) within a network of health care facilities is 1 to 1.6 million U.S. dollars (22). Of 983 patients who overlapped based on year of admission in the same facility, we found only one potential example of a phenotypically identical and genotypically nearly identical strain isolated from two patients. These patients were on different wards, which could suggest a widespread distribution of this strain; alternatively, as we found no additional closely related CPE strains from that facility that year, this could indicate that any intrafacility spread, if present, was limited and sporadic. There was one additional cluster of five closely related E. cloacae strains, all isolated from the ACH. The SNP distances for the strains isolated in 2015 were suggestive of direct transmission within the ACH, with potential persistence of that strain at least until 2016.
Acquisition of CPE could happen within a facility as well as between the different types of facilities during step-up or step-down treatments (23). We found only one potential example of two E. coli strains (with the same carbapenem resistance phenotype) that were genomically separated by 22 SNPs. These were isolated in different years, and the isolate from 2015 was from an LTCF, while the isolate from 2016 was from the ACH. Therefore, we had no evidence of direct interfacility transmission and concluded that such transmission events must be too rare for us to have captured them in our data set. Patients transferred from ACHs to long-term acute-care hospitals (LTACHs) in the United States tended to be more severely ill and possibly more prone to CPE colonization (19). Although only half of the LTACH residents had prior ACH admissions, carbapenem resistance in LTACHs was 9 times higher than that in ACHs, suggesting that ACH-to-LTACH transmission was uncommon (11). As LTACHs provide just as intense clinical care as ACHs and for a much longer duration (average length of stay in LTACHs is ≥25 days [24]), it is no surprise that they are major reservoirs of carbapenem and other antibiotic resistance. Unlike LTACHs in the United States, which provide rigorous clinical care and observation, including the prolonged use of ventilators, the ILTCFs in Singapore provide more rehabilitative and much less intense clinical care, as well as long-term residential nursing care for their patients and residents. As such, ILTCFs are likely to play a much smaller role in the CPE epidemic. Moreover, the prevalence of CPE in ILTCFs was very low, supporting our suggestion that ACH-to-ILTCF transmission is rare. Prior hospital admission was also not found to be associated with CPE colonization in ILTCFs.
Risk factors for CPE colonization.
Several risk factors for colonization of carbapenem-resistant Enterobacteriaceae have been previously identified (6, 25, 26). As a well-known risk factor for antibiotic resistance (27, 28), history of use of any antibiotics increased the prevalence of CPE colonization in all health care facilities overall. In multivariate analysis, we observed that exposure to penicillins tripled the odds of CPE colonization in ACH patients (29, 30). Hence, antibiotic stewardship is crucial in the control of CPE in ACHs. We observed that prior PPI exposure tripled the odds of CPE colonization in ACH patients; interestingly, this association was not observed among ILTCF patients. This could possibly be due to the combined effect of PPI and antibiotics on the gut bioflora reducing the population of commensal bacteria and increasing the risk of colonization with pathogenic ones (31). We found only a marginal interaction between penicillins and PPI (data not shown).
We also observed that a history of wounds increased the odds of CPE colonization in ILTCFs by 5 times. Presence of wound in patients indirectly reflects functional status, underlying medical conditions and nursing care required by a patient. Prevention of wounds and proper wound care in ILTCFs cannot be overemphasized (32). The finding of an association between dementia and higher CPE colonization in the ACH might be due to dementia patients requiring intensive nursing care, which could increase their risk for acquisition of CPE in ACHs. We further identified an association of connective tissue diseases with CPE colonization in the ACH, possibly due to the immunosuppressive effects of their medications.
We found that prior exposures to respiratory procedures were independent risk factors for CPE colonization in ILTCFs. Although respiratory procedures were likely to have been performed in ACHs, care for these devices continues even after patients are transferred to ILTCFs. Hence, the proper handling of medical devices is more important than the type of medical devices used, irrespective of the health care facility (27). Training of health care staff in the proper handling and cleaning of devices, good hand hygiene after handling of devices, and contact precautions of patients after identification of patients with CPE are recommended for all types of health care facility (11).
In our study population, as with those in other studies (33), prior CRE carriage was the factor most strongly associated with current CPE colonization, regardless of facility type. The time from prior CRE colonization to current CPE colonization ranged from 4 to 493 days (median, 24; IQR, 10 to 66). We further observed that prior VRE carriage was an independent risk factor for CPE colonization in ILTCFs. This could be due to the similar mode of transmission by VRE and CRE. These findings support our current hospital policy of isolation and contact precautions for prior VRE and CRE carriers from the point of admission (34).
Strengths and limitations.
A major strength of our study was the inclusion of a large sample of patients hospitalized at various health care facility types, representing a participation rate of 87%. Hence, any selection bias was likely to be minimal. Moreover, the comprehensive, systematic, and standardized manner in which the rectal swabs/stool samples and clinical data were collected reduced any potential measurement error. Furthermore, any potential confounding was adjusted for in the multivariable regression models. Nonetheless, the study had some limitations. Antibiotic exposures outside the respective health care facilities, if not documented in the medical records, would have been missed. However, any information bias was likely to be nondifferential, thereby attenuating observed effects; hence, the associations observed in our study are likely to be conservative estimates. Another limitation is that the health care system-specific risk factors in the Singapore population, particularly under the provision of subsidized care, might not be generalizable to other health care systems. Regardless, the advanced medical care provided to patients receiving subsidized care makes the study findings applicable to other developed health care systems. Unfortunately, the epidemiology and risk factors for specific carbapenemase genes could not be examined in our study population due to the small sample size.
Conclusion.
In conclusion, CPE colonization was low in the ACH and very low in ILTCFs. Indications of CPE transmission within the ACH were seen, but no examples of transfer between ACH and ILTCFs were observed. As CPE prevalence increases with time, CPE screening of long stayers being transferred from ACHs to ILTCFs can prevent interfacility transmission. Furthermore, preemptive isolation and contact precautions for prior CRE carriers should be undertaken on admission to any health care facility.
MATERIALS AND METHODS
Study design and setting.
We conducted serial cross-sectional studies in an ACH and three each of its closely affiliated ITCFs and LTCFs over a 6-week period during June and July in 2014 to 2016. Over each 6-week period annually, the study was conducted serially in the ACH and ILTCFs. The ACH was a 1,700-bed tertiary-care hospital which provided emergency, inpatient, and intensive care services for adults in general medicine, infectious diseases, cancer chemo- and radiotherapies, and general surgery, as well as trauma, neurosurgical, and spinal cord injury care. The ITCFs involved were a 100-bed rehabilitation center, a 116-bed community hospital, and a 360-bed community hospital. These facilities provide rehabilitative and subacute care for a period of 1 to 2 months for patients who require such care after admission to acute-care hospitals. In comparison, LTCFs are residential facilities that provide long-term nursing care for individuals who are unable to be cared for in their own homes. The LTCFs included in the study comprised a 164-bed chronic sick unit and two nursing homes with 234 and 236 beds, respectively. About 50 to 80% of patients from the ACH are transferred to the respective ITCFs and LTCFs, and vice versa.
Ethical approval was received from the Domain Specific Research Board, National Healthcare Group, Singapore (DSRB-2013/00965 and 2014/01139).
Study participants.
All inpatients and residents of the ITCFs and LTCFs were included in the study, and 3,357 in-patients with a stay in the ACH of >48 h were randomly selected to participate in the study. Stratified sampling of inpatients in the ACH wards proportional to the ward’s bed census was performed, with all wards in the ACH systematically covered over 5 days and each ward sampled three times over 15 days each year.
Microbiological methods.
Rectal swabs or stool samples were collected and inoculated onto ChromID Carba-selective chromogenic agar. The identities of all isolates as Enterobacteriaceae species were confirmed by MALDI-TOF. Using Vitek 2, organisms were classified as resistant, intermediate, and susceptible to meropenem and ertapenem according to Clinical and Laboratory Standards Institute (CLSI) M100 breakpoints (for ertapenem, resistant is defined as a MIC of ≥2 mg/liter, intermediate as 1 mg/liter, and susceptible as ≤0.5 mg/liter; for meropenem, resistant is defined as a MIC of ≥4 mg/liter, intermediate as 2 mg/liter, and susceptible as ≤1 mg/liter). Intermediate isolates were considered susceptible to carbapenems.
Genome sequencing and analysis.
A single colony of each strain from all Enterobacteriaceae isolates was inoculated into Luria-Bertani broth (Gibco) and cultured. Genomic DNA was isolated using the QIAamp DNA minikit (Qiagen) and quantified using a QUBIT 2.0 fluorometer (Invitrogen). Sequencing libraries were prepared with the Nextera XT library prep kit (Illumina). The adapters were indexed using either the Nextera XT Index kit or the Nextera XT Index kit v2 (Illumina). Finally, all sample DNA sequencing libraries (10 nM each) were pooled and sequenced on a HiSeq 4000 (Illumina) with a 2 × 151-bp run. Resistance genes and multilocus sequence types (MLST) were called using the SRST2 program (v0.2.0) using the ARG-ANNOT database as provided in the SRST2 distribution (35, 36) and MLST alleles and profiles from https://pubmlst.org. In this study, we focused on β-lactamase genes, including blaIMI, blaIMP-1, blaKPC-2, blaNDM-1, and blaOXA-48. Single-nucleotide polymorphisms (SNPs) were initially called using a reference-based analysis. Briefly, FASTQ files were mapped using BWA (Burrows-Wheeler Aligner, version 0.7.10) (37). Reference sequences were ATCC 13047 for E. cloacae (GCF_000025565.1), EC958 for E. coli (GCF_000285655.3), and HS11286 for K. pneumoniae (GCF_00024185.1). Indel realignment and SNP calling were done with LoFreq version 2.1.2 with default parameters (38). De novo assemblies were performed with velvet (version 1.2.10) (39). Initial screening was done on SNP differences as called by LoFreq; manual inspection of reference-based alignments and the corresponding sequences within de novo assemblies was then performed to obtain final SNP counts for the clusters described in the text. Assemblies were tested for the presence of plasmids previously identified to be common among carbapenemase-producing strains in Singapore (12) pHS102707 (NC_023907.1; carrying KPC-1), pNDM-ECS01 (NC_024954.1; carrying NDM-1), pNDM_MGR194 (NC_022740.1; potentially carrying different NDM alleles), and pSg1-NDM (CP011839.1; mostly carrying pNDM-1). Plasmids were predicted to be present according to the criteria used in reference 12. In brief, assembled contigs were aligned to the plasmid sequences using BLASTn with default parameters; the plasmid was predicted to be present when overall coverage of the plasmid reference sequence was >90% using a cutoff of >80% nucleotide identity. None of the sequenced isolates in this study were predicted to carry pNDM_MGR194 or pSg1-NDM.
Epidemiological and clinical data.
Sociodemographic data such as age, gender, race, class of admission, and duration of stay in health care facilities were obtained from administrative databases. Clinical data extracted from medical records in the prior 12 months included colonization/infection with multidrug-resistant organisms (including methicillin-resistant Staphylococcus aureus [MRSA], carbapenem-resistant Enterobacteriaceae [CRE], and vancomycin-resistant enterococci [VRE]); exposure to antibiotics, steroids, antacids, H2 receptor blockers, and proton pump inhibitors (PPI); and hospital admissions, presence of wounds, and surgical operations. Procedures were later grouped into vascular access procedures, including insertion of an arterial line, central venous pressure (CVP) line, hemodialysis line, peripheral line, or peripherally inserted central catheter (PICC); respiratory procedures, including chest tube insertion, endotracheal tube insertion, and tracheostomy; gastrointestinal procedures, including colostomy, nasogastric tube insertion, and insertion of a percutaneous endoscopic gastrostomy (PEG) tube; and urinary procedures, including insertion of suprapubic or urethral catheters. The Charlson comorbidity index (CCI) was computed from the 16 categories of comorbidities identified from medical records (40).
Data analysis.
Student's t test or Wilcoxon rank-sum test was used to compare differences in means or medians, and chi-square or Fisher’s exact test was used to compare differences in proportions. Simple logistic regression models were constructed to test for associations between individual factors and CPE colonization. Factors with a P value of <0.05 and those based on literature review were entered into multivariable logistic regression models as probable predictor variables. Finally, stratified analyses were performed to assess for facility type-specific risk factors. Odds ratios (OR) with 95% confidence intervals (CIs) are presented. A two-tailed P value of <0.05 was considered statistically significant. All statistical analyses were performed with STATA/SE-13.0 (StataCorp LP, USA).
Data availability.
Raw sequencing reads for whole-genome sequencing (WGS) have been deposited in the GenBank Short Read Archive under BioProject no. PRJNA674942.
ACKNOWLEDGMENTS
We thank all patients who participated in the study and the staff who assisted in the study conduct and data collection. We thank Balamurugan Periaswamy for assistance with DNA extraction and sequencing library construction.
This study was funded by a Singapore Ministry of Health’s Communicable Diseases Public Health Research Grant (CDPHRG/0008/2014) and an Agency for Science, Technology, and Research (A*STAR) grant (IAF311018). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no conflicts of interest to report.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, and, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q, Zhang Y, Yao X, Xian H, Liu Y, Li H, Chen H, Wang X, Wang R, Zhao C, Cao B, Wang H. 2016. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis 35:1679–1689. doi: 10.1007/s10096-016-2710-0. [DOI] [PubMed] [Google Scholar]
- 3.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cienfuegos-Gallet AV, Ocampo de Los Rios AM, Sierra Viana P, Ramirez Brinez F, Restrepo Castro C, Roncancio Villamil G, Del Corral Londono H, Jimenez JN. 2019. Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: a case-control and cohort study. BMC Infect Dis 19:830. doi: 10.1186/s12879-019-4461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. 2014. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis 20:1170–1175. doi: 10.3201/eid2007.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhargava A, Hayakawa K, Silverman E, Haider S, Alluri KC, Datla S, Diviti S, Kuchipudi V, Muppavarapu KS, Lephart PR, Marchaim D, Kaye KS. 2014. Risk factors for colonization due to carbapenem-resistant Enterobacteriaceae among patients exposed to long-term acute care and acute care facilities. Infect Control Hosp Epidemiol 35:398–405. doi: 10.1086/675614. [DOI] [PubMed] [Google Scholar]
- 7.Giannella M, Trecarichi EM, De Rosa FG, Del Bono V, Bassetti M, Lewis RE, Losito AR, Corcione S, Saffioti C, Bartoletti M, Maiuro G, Cardellino CS, Tedeschi S, Cauda R, Viscoli C, Viale P, Tumbarello M. 2014. Risk factors for carbapenem-resistant Klebsiella pneumoniae bloodstream infection among rectal carriers: a prospective observational multicentre study. Clin Microbiol Infect 20:1357–1362. doi: 10.1111/1469-0691.12747. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MK, Lin MY, Lolans K, Weiner S, Blom D, Moore NM, Fogg L, Henry D, Lyles R, Thurlow C, Sikka M, Hines D, Weinstein RA, for the Centers for Disease Control and Prevention Epicenters Program. 2015. Prevention of colonization and infection by Klebsiella pneumoniae carbapenemase-producing enterobacteriaceae in long-term acute-care hospitals. Clin Infect Dis 60:1153–1161. doi: 10.1093/cid/ciu1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trepanier P, Mallard K, Meunier D, Pike R, Brown D, Ashby JP, Donaldson H, Awad-El-Kariem FM, Balakrishnan I, Cubbon M, Chadwick PR, Doughton M, Doughton R, Hardiman F, Harvey G, Horner C, Lee J, Lewis J, Loughrey A, Manuel R, Parsons H, Perry JD, Vanstone G, White G, Shetty N, Coia J, Wiuff C, Hopkins KL, Woodford N. 2017. Carbapenemase-producing Enterobacteriaceae in the UK: a national study (EuSCAPE-UK) on prevalence, incidence, laboratory detection methods and infection control measures. J Antimicrob Chemother 72:596–603. doi: 10.1093/jac/dkw414. [DOI] [PubMed] [Google Scholar]
- 10.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H, ESGEM Study Group. 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han JH, Goldstein EJC, Wise J, Bilker WB, Tolomeo P, Lautenbach E. 2017. Epidemiology of carbapenem-resistant Klebsiella pneumoniae in a network of long-term acute care hospitals. Clin Infect Dis 64:839–844. doi: 10.1093/cid/ciw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marimuthu K, Venkatachalam I, Khong WX, Koh TH, Cherng BPZ, Van La M, De PP, Krishnan PU, Tan TY, Choon RFK, Pada SK, Lam CW, Ooi ST, Deepak RN, Smitasin N, Tan EL, Lee JJ, Kurup A, Young B, Sim NTW, Thoon KC, Fisher D, Ling ML, Peng BAS, Teo YY, Hsu LY, Lin RTP, Ong RT, Teo J, Ng OT, Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group. 2017. Clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae among adult inpatients in Singapore. Clin Infect Dis 64:S68–s75. doi: 10.1093/cid/cix113. [DOI] [PubMed] [Google Scholar]
- 13.Molton JS, Tambyah PA, Ang BS, Ling ML, Fisher DA. 2013. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin Infect Dis 56:1310–1318. doi: 10.1093/cid/cit020. [DOI] [PubMed] [Google Scholar]
- 14.Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. 2012. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac Surveill Response J 3:19–24. doi: 10.5365/wpsar.2011.2.4.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatachalam I, Teo J, Balm MN, Fisher DA, Jureen R, Lin RT. 2012. Klebsiella pneumoniae carbapenemase-producing enterobacteria in hospital, Singapore. Emerg Infect Dis 18:1381–1383. doi: 10.3201/eid1808.110893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balm MN, Ngan G, Jureen R, Lin RT, Teo JW. 2013. OXA-181-producing Klebsiella pneumoniae establishing in Singapore. BMC Infect Dis 13:58. doi: 10.1186/1471-2334-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher JC, Kuriakose S, Haynes K, Axelrod P. 2014. Case-case-control study of patients with carbapenem-resistant and third-generation-cephalosporin-resistant Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother 58:5732–5735. doi: 10.1128/AAC.03564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toth DJA, Khader K, Slayton RB, Kallen AJ, Gundlapalli AV, O’Hagan JJ, Fiore AE, Rubin MA, Jernigan JA, Samore MH. 2017. The potential for interventions in a long-term acute care hospital to reduce transmission of carbapenem-resistant Enterobacteriaceae in affiliated healthcare facilities. Clin Infect Dis 65:581–587. doi: 10.1093/cid/cix370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho HJ, Toh CY, Ang B, Krishnan P, Lin RT, La MV, Chow A. 2016. Outbreak of New Delhi metallo-beta-lactamase-1-producing Enterobacter cloacae in an acute care hospital general ward in Singapore. Am J Infect Control 44:177–182. doi: 10.1016/j.ajic.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 21.Khong WX, Marimuthu K, Teo J, Ding Y, Xia E, Lee JJ, Ong RT, Venkatachalam I, Cherng B, Pada SK, Choong WL, Smitasin N, Ooi ST, Deepak RN, Kurup A, Fong R, Van La M, Tan TY, Koh TH, Lin RT, Tan EL, Krishnan PU, Singh S, Pitout JD, Teo YY, Yang L, Ng OT, Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group. 2016. Tracking inter-institutional spread of NDM and identification of a novel NDM-positive plasmid, pSg1-NDM, using next-generation sequencing approaches. J Antimicrob Chemother 71:3081–3089. doi: 10.1093/jac/dkw277. [DOI] [PubMed] [Google Scholar]
- 22.Otter JA, Burgess P, Davies F, Mookerjee S, Singleton J, Gilchrist M, Parsons D, Brannigan ET, Robotham J, Holmes AH. 2017. Counting the cost of an outbreak of carbapenemase-producing Enterobacteriaceae: an economic evaluation from a hospital perspective. Clin Microbiol Infect 23:188–196. doi: 10.1016/j.cmi.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Han JH, Lapp Z, Bushman F, Lautenbach E, Goldstein EJC, Mattei L, Hofstaedter CE, Kim D, Nachamkin I, Garrigan C, Jain T, Bilker W, Wolford HM, Slayton RB, Wise J, Tolomeo P, Snitkin ES. 2019. Whole-genome sequencing to identify drivers of carbapenem-resistant Klebsiella pneumoniae transmission within and between regional long-term acute-care hospitals. Antimicrob Agents Chemother 63:e01622-19. doi: 10.1128/AAC.01622-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahn JM, Barnato AE, Lave JR, Pike F, Weissfeld LA, Le TQ, Angus DC. 2015. A comparison of free-standing versus co-located long-term acute care hospitals. PLoS One 10:e0139742. doi: 10.1371/journal.pone.0139742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills JP, Talati NJ, Alby K, Han JH. 2016. The epidemiology of carbapenem-resistant Klebsiella pneumoniae colonization and infection among long-term acute care hospital residents. Infect Control Hosp Epidemiol 37:55–60. doi: 10.1017/ice.2015.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Duin D, Doi Y. 2017. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Loon K, Voor In ‘T Holt AF, Vos MC. 2017. A systematic review and meta-analyses of the clinical epidemiology of carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 62:e01730-17. doi: 10.1128/AAC.01730-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz-Neiderman A, Braun T, Fallach N, Schwartz D, Carmeli Y, Schechner V. 2016. Risk factors for carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CP-CRE) acquisition among contacts of newly diagnosed CP-CRE patients. Infect Control Hosp Epidemiol 37:1219–1225. doi: 10.1017/ice.2016.153. [DOI] [PubMed] [Google Scholar]
- 29.Ling ML, Tee YM, Tan SG, Amin IM, How KB, Tan KY, Lee LC. 2015. Risk factors for acquisition of carbapenem resistant Enterobacteriaceae in an acute tertiary care hospital in Singapore. Antimicrob Resist Infect Control 4:26. doi: 10.1186/s13756-015-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariappan S, Sekar U, Kamalanathan A. 2017. Carbapenemase-producing Enterobacteriaceae: risk factors for infection and impact of resistance on outcomes. Int J Appl Basic Med Res 7:32–39. doi: 10.4103/2229-516X.198520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corleto VD, Festa S, Giulio ED, Annibale B. 2014. Proton pump inhibitor therapy and potential long-term harm. Curr Opin Endocrinol Diabetes Obes 21:3–8. doi: 10.1097/MED.0000000000000031. [DOI] [PubMed] [Google Scholar]
- 32.McKinnell JA, Miller LG, Singh R, Kleinman K, Peterson EM, Evans KD, Dutciuc TD, Heim L, Gombosev A, Estevez M, Launer B, Tjoa T, Tam S, Bolaris MA, Huang SS. 2016. Prevalence of and factors associated with multidrug resistant organism (MDRO) colonization in 3 nursing homes. Infect Control Hosp Epidemiol 37:1485–1488. doi: 10.1017/ice.2016.215. [DOI] [PubMed] [Google Scholar]
- 33.Giacobbe DR, Del Bono V, Bruzzi P, Corcione S, Giannella M, Marchese A, Magnasco L, Maraolo AE, Pagani N, Saffioti C, Ambretti S, Cardellino CS, Coppo E, De Rosa FG, Viale P, Viscoli C, on behalf of ISGRI-SITA (Italian Study Group on Resistant Infections of the Società Italiana Terapia Antinfettiva). 2017. Previous bloodstream infections due to other pathogens as predictors of carbapenem-resistant Klebsiella pneumoniae bacteraemia in colonized patients: results from a retrospective multicentre study. Eur J Clin Microbiol Infect Dis 36:663–669. doi: 10.1007/s10096-016-2843-1. [DOI] [PubMed] [Google Scholar]
- 34.Otter JA, Dyakova E, Bisnauthsing KN, Querol-Rubiera A, Patel A, Ahanonu C, Tosas Auguet O, Edgeworth JD, Goldenberg SD. 2016. Universal hospital admission screening for carbapenemase-producing organisms in a low-prevalence setting. J Antimicrob Chemother 71:3556–3561. doi: 10.1093/jac/dkw309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain J-M. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1 and Tables S1 and S2. Download AAC02584-20_Supp_1_seq6.pdf, PDF file, 1.2 MB (1,005.9KB, pdf)
Data Availability Statement
Raw sequencing reads for whole-genome sequencing (WGS) have been deposited in the GenBank Short Read Archive under BioProject no. PRJNA674942.