ABSTRACT
Antibiotic collateral sensitivity, in which acquired resistance to one drug leads to decreased resistance to a different drug, occurs in Burkholderia multivorans. Here, we observed that treatment of extensively drug-resistant variants evolved from a cystic fibrosis (CF) sputum sample isolate with either meropenem or sulfamethoxazole-trimethoprim, depending on past resistance phenotypes, resulted in increased sensitivity to five different classes of antibiotics. We further identified mutations, including putative resistance-nodulation-division efflux pump regulators and uncharacterized pumps, that may be involved in this phenotype in B. multivorans.
KEYWORDS: Burkholderia, antibiotic resistance, collateral sensitivity, combination antibiotic, sequential therapy, XDR
INTRODUCTION
Members of the Burkholderia cepacia complex (BCC), primarily Burkholderia multivorans and Burkholderia cenocepacia, are associated with increased morbidity due to unremitting airway infections exacerbated by failed antibiotic treatments and increased mortality by impeding life-extending therapies such as lung transplants in cystic fibrosis (CF) patients (1–5). The recommended chronic suppressive antimicrobial therapies for CF patients require sustained exposure to antibiotics, potentiating the development of multidrug resistance (MDR) or extensive drug resistance (XDR). One possible therapeutic strategy to combat MDR and XDR, antibiotic cycling, takes advantage of a phenomenon called collateral sensitivity (CS) (6). CS is observed when bacteria are exposed to and acquire resistance to one antibiotic (the “treatment drug” [TD]) and subsequently demonstrate increased sensitivities to other antibiotics (“nontreatment drugs” [NTDs]).
Our lab has previously reported that drug cycling in a B. multivorans cystic fibrosis isolate, AS149 (7), using six clinically relevant antibiotics with various mechanisms of action (meropenem [MEM], 10-μg disk; ceftazidime [CAZ], 30-μg disk; minocycline [MIN], 30-μg disk; levofloxacin [LVX], 5-μg disk; chloramphenicol [CHL], 30-μg disk; and sulfamethoxazole-trimethoprim [SXT], two 23.75/1.25-μg disks), can induce CS to at least one other antibiotic in a majority of the variants tested (8, 9). However, during the lab evolution experiment, we observed six variants that independently lost all previous CS phenotypes and acquired XDR (Table 1) (9). To determine if the XDR phenotype could be reverted back to susceptible, we performed an additional lab evolution experiment beginning with the six XDR variants using continued antibiotic cycling.
TABLE 1.
B. multivorans variants in this studya
| B. multivorans variant | Treatment drug | Drug resistance | Collateral sensitivity | Reference or source |
|---|---|---|---|---|
| AS149 | None | None | None | 7 |
| AS553 | CAZ | MEM, LVX, CAZ, MIN, CHL | None | 9 |
| AS625 | SXT | SXT | MEM, LVX, CHL, MIN, CAZ | This study |
| AS524 | CAZ | MEM, LVX, CAZ, MIN, CHL | None | 9 |
| AS647 | SXT | SXT | MEM, LVX, CHL, MIN, CAZ | This study |
| AS525 | CAZ | LVX, CAZ, MIN, CHL | None | 9 |
| AS626 | SXT | SXT | MEM, LVX, CHL, MIN, CAZ | This study |
| AS542 | MEM | MEM, LVX, MIN, CHL | None | 9 |
| AS581 | SXT | SXT | MEM, LVX, CHL, MIN, CAZ | This study |
| AS591 | CAZ | MEM, LVX, CAZ, MIN, CHL | None | 9 |
| AS697 | SXT | SXT | MEM, LVX, CHL, MIN, CAZ | This study |
| AS610 | MIN | LVX, CAZ, SXT, MIN, CHL | None | 9 |
| AS717 | MEM | MEM | LVX, SXT, CHL, MIN, CAZ | This study |
Extensively drug-resistant variants were derived in a previous lab evolution experiment (9) using resistance selection to a treatment drug (TD) in the ancestral cystic fibrosis (CF) B. multivorans isolate AS149 (7). To determine if extensive drug resistance (XDR) could be reversed by exposure to a drug to which the variants were still susceptible, a second in vitro lab evolution experiment was performed. Acquired resistance to the TD in XDR variants resulted in increased sensitivity to all five nontreatment drugs (NTD)—collateral sensitivity (CS). The XDR variant and resultant progeny pan-CS variant (bold) pair antibiograms are described in the table.
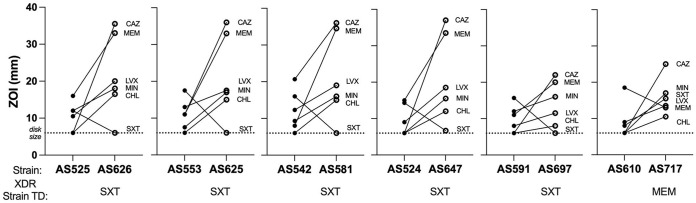
Each of the six XDR variants (AS553, AS524, AS525, AS542, AS591, and AS610) was cultured under the selective pressure of the antibiotic to which the bacteria remained susceptible (TD) for 20 to 25 exposures until resistance was reached. Once resistance was acquired to the TD, the antibiograms of the resultant progeny variants were determined as previously described (8, 9). Acquisition of resistance to SXT resulted in five variants (AS625, AS647, AS626, AS581, and AS697) showing increased sensitivity to all five NTD antibiotics (MEM, LVX, CHL, MIN, and CAZ), further referred to as pancollateral sensitivity (pan-CS). The sixth variant (AS717) also showed pan-CS to all five NTD antibiotics (LVX, SXT, CHL, MIN, and CAZ) after developing resistance to MEM (Table 1). We observed the intensity of CS in the pan-CS variants was unfixed and was independent of TD or parent XDR variant, with four pan-CS variants (AS626, AS625, AS581, and AS647) showing similar sizeable increases in sensitivity to NTDs (Fig. 1; see also Table S1 at https://pages.uncc.edu/todd-steck/aac00611-21-2/).
FIG 1.
Changes in antibiogram in XDR parent variant to the pan-CS progeny variant. XDR (left) and pan-CS (right) pairs are displayed together. The drug to which the XDR variant was exposed to during cyclic therapy, the “treatment drug” (TD), is given under each pan-CS variant. The antibiotic disk size, 6 mm (dotted line), represents confluent bacterial growth up to the antibiotic disk. Zone of inhibition (ZOI) values (in mm) for each variant in shown for XDR (•) and pan-CS (○). Lines represent the change in sensitivity to each antibiotic (meropenem [MEM], ceftazidime [CAZ], levofloxacin [LVX], minocycline [MIN], chloramphenicol [CHL], and sulfamethoxazole-trimethoprim [SXT]), labeled at the pan-CS value. Lines with positive trends represent an increase in sensitivity, while lines with negative trends represent an increase in resistance. The steeper the slope of the connecting line, the greater the change in antibiotic sensitivity. Raw values can be found in Table S1 at https://pages.uncc.edu/todd-steck/aac00611-21-2/.
To determine which mutational events contributed to the increase in sensitivity, the genomes of six XDR variants and respective six pan-CS variants were sequenced using the Illumina HiSeq 2500 platform with 151-bp paired-end reads (BioProject accession number PRJNA691610). Single-nucleotide polymorphisms (SNPs) and insertions/deletions (indels) were called against the ancestral B. multivorans CF isolate, AS149, as previously described (8, 9). Structural variations (>10 bp) were identified using the software packages Pindel (10), BreakDancer (11), GRIDSS (12), Manta (13), LUMPY (14), and DELLY (15). Sample calls from each program were merged at a threshold of 75% and kept if at least three callers identified a variant in that region.
RND-type efflux pumps in B. multivorans.
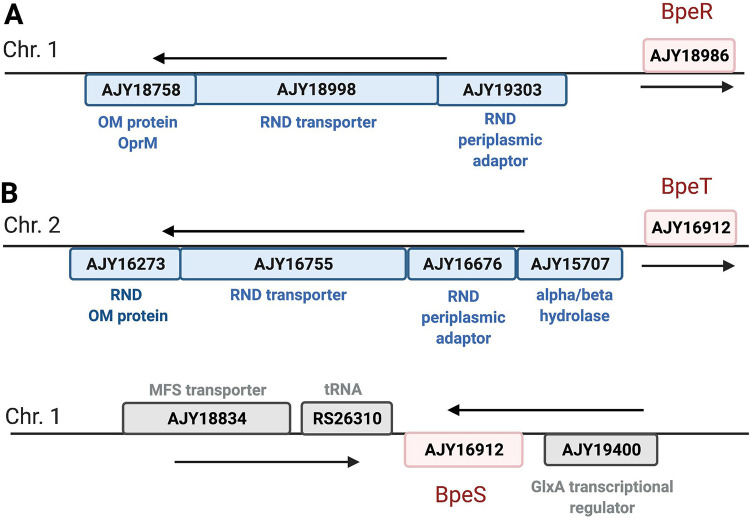
During the whole-genome sequence (WGS) analysis, we identified three putative B. multivorans efflux pump regulators either upstream of or related to hypothesized resistance-nodulation-division (RND) efflux pumps (Fig. 2). To characterize the putative regulator and its respective RND pump, we used EMBOSS Needle global alignments to determine similarities to known RND efflux pumps in B. cenocepacia J2315. We identified a TetR-type regulator protein, AJY18986, which showed 95.3% similarity to the BpeR repressor (NCBI accession number CAR53123) of the RND-4 RND efflux pump in B. cenocepacia J2315 (Fig. 2A) (1). We propose that the AJY19303-AJY18998-AJY18758 operon in B. multivorans encodes the orthologous RND efflux pump to the RND-4 efflux pump and is regulated by upstream AJY18986.
FIG 2.
Putative RND efflux gene operons in Burkholderia multivorans. Two schematic representations of the putative operons encoding RND efflux regulators and efflux pumps in Burkholderia multivorans ATCC BAA-247 using NCBI accession numbers. The putative regulator and respective operons are shown for (A) BpeR and AJY19303-AJY18998-AJY18758 and (B) BpeT, AJY15707-AJY16676-AJY16755-AJY16273, and the LTTR BpeS, found on chromosome 1. Regulators are designated in red, RND efflux pump operons in blue, and unrelated genes in gray. Genes shown above the line are located on the positive strand, and those shown underneath the line are on the negative strand. Figure created using BioRender.
Additionally, we found one putative LysR-type transcriptional regulator (LTTR), AJY16912, that is 98.2% similar to the CeoR regulator (NCBI accession number CAR56419) of the CeoAB-OpcM RND efflux pump in B. cenocepacia (Fig. 2B) and is an ortholog to BpeT in Burkholderia pseudomallei. In B. pseudomallei, the BpeT regulator is upstream of the known RND efflux pump, BpeEF-OprC, and this pump can also be controlled by an additional LTTR, BpeS (16–18). In our B. multivorans variants, we were able to identify a BpeS (NCBI accession number AFI67377) ortholog, AJY19458, on chromosome 1 with 90.1% similarity to BpeS in B. pseudomallei (Fig. 2B). We propose that the AJY15707-AJY16676-AJY16755-AJY16273 operon in B. multivorans is an ortholog to the llpE-bpeE-bpeF-oprC operon in B. pseudomallei, which encodes the BpeEF-OprC RND-type efflux pump (17), and that it is regulated by upstream LTTR AJY16912 and distant LTTR AJY19458.
XDR mutations.
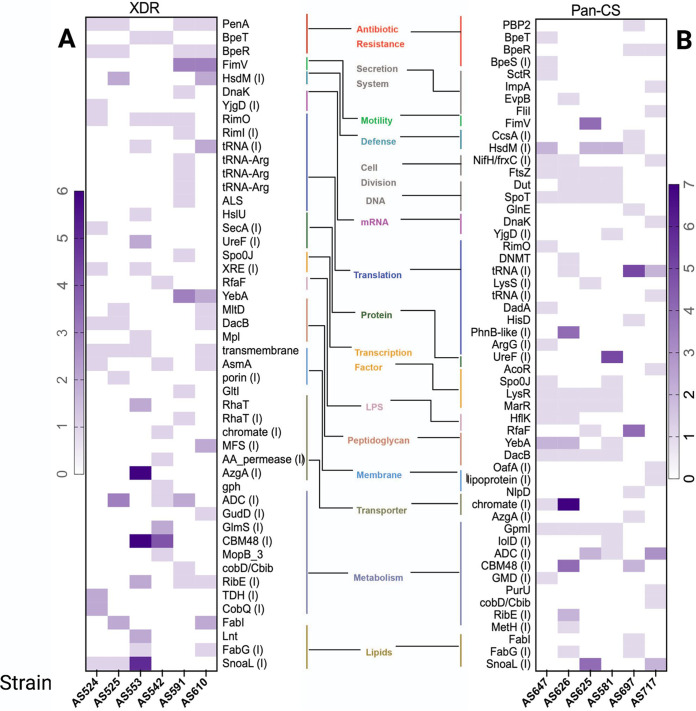
WGS analysis revealed that mutations called across all six XDR variants were commonly involved in antibiotic resistance, membrane proteins, and lipid synthesis (Fig. 3A). All variants acquired mutations within either putative RND efflux pump regulators bpeR or bpeT genes (Fig. 3A; see also Table S2 at https://pages.uncc.edu/todd-steck/aac00611-21-2/). BpeR mutants observed in this study either gained an early stop codon, resulting in aberrated protein, or a large disruptive in-frame deletion (Table S2 at https://pages.uncc.edu/todd-steck/aac00611-21-2/). Disruption of BpeR activity would result in complete or partial derepression of the respective RND efflux target, leading to hyperexpression and increased antibiotic resistance (19). It is noteworthy that the BpeEF-OprC efflux pump is only expressed in BpeT regulatory mutants and is induced by substrates (20). XDR variants with mutations in BpeT (AS553 and AS542), were evolved from the same ancestor variant, which was resistant to CHL, a substrate for this pump. The BpeT mutation acquired during this exposure to CHL was maintained in the subsequent evolved XDR variants.
FIG 3.
Mutation analysis of the XDR and pan-CS variants. Heat map of unique, nonsynonymous mutations (single-nucleotide polymorphisms, insertions, and deletions) in (A) the six XDR variants and (B) the six pan-CS variants. Genes are organized by functional category. Mutations in intergenic regions (I) were only considered if the mutation fell within 100 bp upstream of the downstream gene’s start site. Intensity of color increases with mutation number.
Ortholog pumps in B. pseudomallei (e.g., BpeEF-OprC and BpeAB-OprD) and B. cenocepacia (e.g., RND-4 and CeoAB-OpcM) have similar and distinct substrates, suggesting there is a slight species divergence in substrate specificity. Due to the proposed efflux pumps in this study being uncharacterized, it cannot be determined which are the preferred substrates for each pump. However, overlapping substrates, such as chloramphenicol and fluroquinolones, suggest that these may be substrates in the putative RND efflux pumps (19–21).
Of interest, we identified mutations in the class A β-lactamase gene penA (22) in four of the six XDR variants (Fig. 3A and Table S2 at https://pages.uncc.edu/todd-steck/aac00611-21-2/). Two variants, AS525 and AS610, acquired PenAD192G mutations, a mutation predicted to result in MEM sensitivity (9).
Pan-CS mutations.
WGS analysis showed there was no significant difference in the total number of mutations accumulated between the XDR parent variants and pan-CS progeny (see Table S3 at https://pages.uncc.edu/todd-steck/aac00611-21-2/). However, we observed complete reversion of all penA mutations and efflux regulator mutations in four variants (Fig. 3B and Table S2 at https://pages.uncc.edu/todd-steck/aac00611-21-2/). The two pan-CS variants that did not revert mutations in the repressor BpeR, AS697 and AS717, showed low to moderate increase in drug sensitivity (Fig. 1). However, in these variants the bpeR mutation did not result in aberrated protein and therefore, it is possible that BpeR maintains some regulatory function. Furthermore, variant AS647 acquired additional mutations in the bpeT gene and in the upstream region of bpeS, potentially in response to SXT exposure (16).
Most mutations identified in the pan-CS variants were in different functional categories compared to the parent XDR variants with those in secretion system, cell division, and DNA repair genes unique to the pan-CS progeny variants (Fig. 3B). All four pan-CS variants evolved under the TD SXT that exhibited higher sensitivity (AS625, AS647, AS626, and AS581) (Fig. 1) acquired the exact same mutations in LysRT245P, SpoTT41A, DacBL235P, MarRW8S, and FtsZA105stop, which have been associated with cell division, increased permeability, and stress response (23–26) (Fig. 3B and Table S2 at https://pages.uncc.edu/todd-steck/aac00611-21-2/). Together, these data suggest that the stress endured from the exposure to antibiotics activates alternative transcription strategies and DNA damage responses.
Concluding remarks.
Here, we have shown that when XDR has developed in B. multivorans, sequential drug therapy has the potential to resensitize bacteria and induce multiple CS pairs, a phenomenon recently observed in Pseudomonas aeruginosa (27). In five of six variants, the combination drug SXT caused XDR reversal; in the sixth variant, MEM led to XDR reversal in a variant previously resistant to SXT. Because the antibiotic disks used in the in vitro evolution experiment are at the concentrations necessary to determine clinical breakpoints, we hypothesize that drug cycling is an efficient strategy to enhance CS, even after XDR has occurred. However, the heterogeneity of the bacterial population in vivo and the effects of CS cycling on B. multivorans infections requires further investigation. Interestingly, the maintenance of BpeT regulator mutations after the substrate CHL of BpeEF-OprC is removed highlights the importance of considering past antibiotic exposures when deciding on drug cycles for chronic suppressive therapies, like those in CF. Furthermore, we have identified the presence of three putative regulators, BpeT, BpeS, and BpeR, that are related to operons containing predicted RND periplasmic adaptor, transporter, and outer membrane proteins that are involved in XDR in B. multivorans.
Data availability.
The whole-genome sequencing data used in this study are available at the NCBI Sequence Read Archive under BioProject accession number PRJNA691610.
ACKNOWLEDGMENTS
This work was supported by funds provided by the University of North Carolina at Charlotte and by National Institutes of Health grant 1R15HL126122-01 to T.R.S.
REFERENCES
- 1.Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sfeir MM. 2018. Burkholderia cepacia complex infections: more complex than the bacterium name suggest. J Infect 77:166–170. doi: 10.1016/j.jinf.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Lord R, Jones AM, Horsley A. 2020. Antibiotic treatment for Burkholderia cepacia complex in people with cystic fibrosis experiencing a pulmonary exacerbation. Cochrane Database Syst Rev doi: 10.1002/14651858.CD009529.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephenson AL, Sykes J, Stanojevic S, Quon BS, Marshall BC, Petren K, Ostrenga J, Fink AK, Elbert A, Goss CH. 2017. Survival comparison of patients with cystic fibrosis in Canada and the United States: a population-based cohort study. Ann Intern Med 166:537–546. doi: 10.7326/M16-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupont L. 2017. Lung transplantation in cystic fibrosis patients with difficult to treat lung infections. Curr Opin Pulm Med 23:574–579. doi: 10.1097/MCP.0000000000000431. [DOI] [PubMed] [Google Scholar]
- 6.Roemhild R, Andersson DI. 2021. Mechanisms and therapeutic potential of collateral sensitivity to antibiotics. PLoS Pathog 17:e1009172. doi: 10.1371/journal.ppat.1009172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stokell JR, Gharaibeh RZ, Steck TR. 2013. Rapid emergence of a ceftazidime-resistant Burkholderia multivorans strain in a cystic fibrosis patient. J Cyst Fibros 12:812–816. doi: 10.1016/j.jcf.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Flanagan JN, Kavanaugh L, Steck TR. 2020. Burkholderia multivorans exhibits antibiotic collateral sensitivity. Microb Drug Resist 26:1–8. doi: 10.1089/mdr.2019.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kavanaugh LG, Flanagan JN, Steck TR. 2020. Reciprocal antibiotic collateral sensitivity in Burkholderia multivorans. Int J Antimicrob Agents 56:105994. doi: 10.1016/j.ijantimicag.2020.105994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan X, Abbott TE, Larson D, Chen K. 2014. BreakDancer: identification of genomic structural variation from paired-end read mapping. Curr Protoc Bioinformatics 45:15.6.1–1. doi: 10.1002/0471250953.bi1506s45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron DL, Schröder J, Penington JS, Do H, Molania R, Dobrovic A, Speed TP, Papenfuss AT. 2017. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res 27:2050–2060. doi: 10.1101/gr.222109.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 14.Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Podnecky NL, Rhodes KA, Mima T, Drew HR, Chirakul S, Wuthiekanun V, Schupp JM, Sarovich DS, Currie BJ, Keim P, Schweizer HP. 2017. Mechanisms of resistance to folate pathway inhibitors in Burkholderia pseudomallei: deviation from the norm. mBio 8:e01357-17. doi: 10.1128/mBio.01357-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes KA, Somprasong N, Podnecky NL, Mima T, Chirakul S, Schweizer HP. 2018. Molecular determinants of Burkholderia pseudomallei BpeEF-OprC efflux pump expression. Microbiology (Reading) 164:1156–1167. doi: 10.1099/mic.0.000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair BM, Cheung KJ, Jr, Griffith A, Burns JL. 2004. Salicylate induces an antibiotic efflux pump in Burkholderia cepacia complex genomovar III (B. cenocepacia). J Clin Invest 113:464–473. doi: 10.1172/JCI19710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan YY, Tan TM, Ong YM, Chua KL. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother 48:1128–1135. doi: 10.1128/AAC.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schweizer HP. 2012. Mechanisms of Burkholderia pseudomallei antimicrobial resistance, p. 229–238. In Ketheesan N (ed), Melioidosis—a century of observation and research. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 21.Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. 2011. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS One 6:e18902. doi: 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papp-Wallace KM, Taracila MA, Gatta JA, Ohuchi N, Bonomo RA, Nukaga M. 2013. Insights into β-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants. J Biol Chem 288:19090–19102. doi: 10.1074/jbc.M113.458315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roghanian M, Semsey S, Løbner-Olesen A, Jalalvand F. 2019. (p)ppGpp-mediated stress response induced by defects in outer membrane biogenesis and ATP production promotes survival in Escherichia coli. Sci Rep 9:2934. doi: 10.1038/s41598-019-39371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Gorostiaga A, Palacios P, Martínez-Arteaga R, Sánchez M, Casanova M, Vicente M. 2016. Life without division: physiology of Escherichia coli FtsZ-deprived filaments. mBio 7:e01620-16. doi: 10.1128/mBio.01620-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Lu X, Hu J, Chen Y, Shen W, Liu L. 2018. Boosting secretion of extracellular protein by Escherichia coli via cell wall perturbation. Appl Environ Microbiol 84:e01382-18. doi: 10.1128/AEM.01382-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grove A. 2013. MarR family transcription factors. Curr Biol 23:R142–R143. doi: 10.1016/j.cub.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Imamovic L, Ellabaan MMH, Dantas Machado AM, Citterio L, Wulff T, Molin S, Krogh Johansen H, Sommer MOA. 2018. Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell 172:121–134.e14. doi: 10.1016/j.cell.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome sequencing data used in this study are available at the NCBI Sequence Read Archive under BioProject accession number PRJNA691610.