Abstract
Extended-spectrum β-lactamase (ESBL)-producing Gram-negative pathogens are a major cause of resistance to expanded-spectrum β-lactam antibiotics. Since their discovery in the early 1980s, they have spread worldwide and an are now endemic in Enterobacterales isolated from both hospital-associated and community-acquired infections. As a result, they are a global public health concern. In the past, TEM- and SHV-type ESBLs were the predominant families of ESBLs. Today CTX-M-type enzymes are the most commonly found ESBL type with the CTX-M-15 variant dominating worldwide, followed in prevalence by CTX-M-14, and CTX-M-27 is emerging in certain parts of the world. The genes encoding ESBLs are often found on plasmids and harboured within transposons or insertion sequences, which has enabled their spread. In addition, the population of ESBL-producing Escherichia coli is dominated globally by a highly virulent and successful clone belonging to ST131. Today, there are many diagnostic tools available to the clinical microbiology laboratory and include both phenotypic and genotypic tests to detect β-lactamases. Unfortunately, when ESBLs are not identified in a timely manner, appropriate antimicrobial therapy is frequently delayed, resulting in poor clinical outcomes. Several analyses of clinical trials have shown mixed results with regards to whether a carbapenem must be used to treat serious infections caused by ESBLs or whether some of the older β-lactam-β-lactamase combinations such as piperacillin/tazobactam are appropriate. Some of the newer combinations such as ceftazidime/avibactam have demonstrated efficacy in patients. ESBL-producing Gram-negative pathogens will continue to be major contributor to antimicrobial resistance worldwide. It is essential that we remain vigilant about identifying them both in patient isolates and through surveillance studies.
1. Introduction
Although naturally occurring in some species of bacteria, β-lactamases have become mobilized on plasmids and have become widespread in response to the use and overuse of β-lactam antibiotics. In Gram-negative bacteria, broad-spectrum enzymes such as TEM-1 and SHV-1 arose following the introduction of first- and second-generation cephalosporins.1 Subsequently, expanded-spectrum β-lactam antibiotics were introduced that were refractory to hydrolysis by these enzymes. In particular, the oxyimino-cephalosporins such as ceftazidime and cefotaxime became widely used. This led to evolution of new β-lactamases that hydrolysed these new drugs.2 The most epidemiologically important group of such enzymes is the extended-spectrum β-lactamases, which have become endemic worldwide. ESBLs are serine β-lactamases, belonging to Ambler molecular and structural classification as class A. They are biochemically characterized by their ability to hydrolyse expanded spectrum β-lactam antibiotics, and inhibition by β-lactamase inhibitors, specifically clavulanate.3 ESBLs have been found in many genera of Enterobacterales as well as in Pseudomonas aeruginosa. They confer resistance to most β-lactam antibiotics, including expanded-spectrum cephalosporins and monobactams, but not to carbapenems and cephamycins.
The original ESBL enzymes were variants of TEM and SHV variants that had amino acid substitutions leading to a change in their substrate profile to include the expanded-spectrum cephalosporins. With the widespread usage of gene sequencing to identify β-lactamase genes in clinical isolates, multiple variants of the common TEM and SHV enzymes have been identified. As of this writing, 243 variants of TEM and 228 variants of SHV have been identified, although not all of these possess the ESBL phenotype (https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/TEM; https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/SHV).
ESBL-producing Gram-negative pathogens are now commonplace in both the hospital and community settings.4 The impact of ESBL-positive Enterobacterales on the choice of empirical and definitive antimicrobial therapy has been substantial, resulting in the increased use of carbapenems in many institutions, which led to the increase of carbapenem resistance in these organisms.5,6 This review will focus on the phenotypic and genetic characterization of ESBLs, their epidemiology, the state of the art of detection of these enzymes, and therapeutic options.
2. ESBL placement in β-lactamase classification scheme
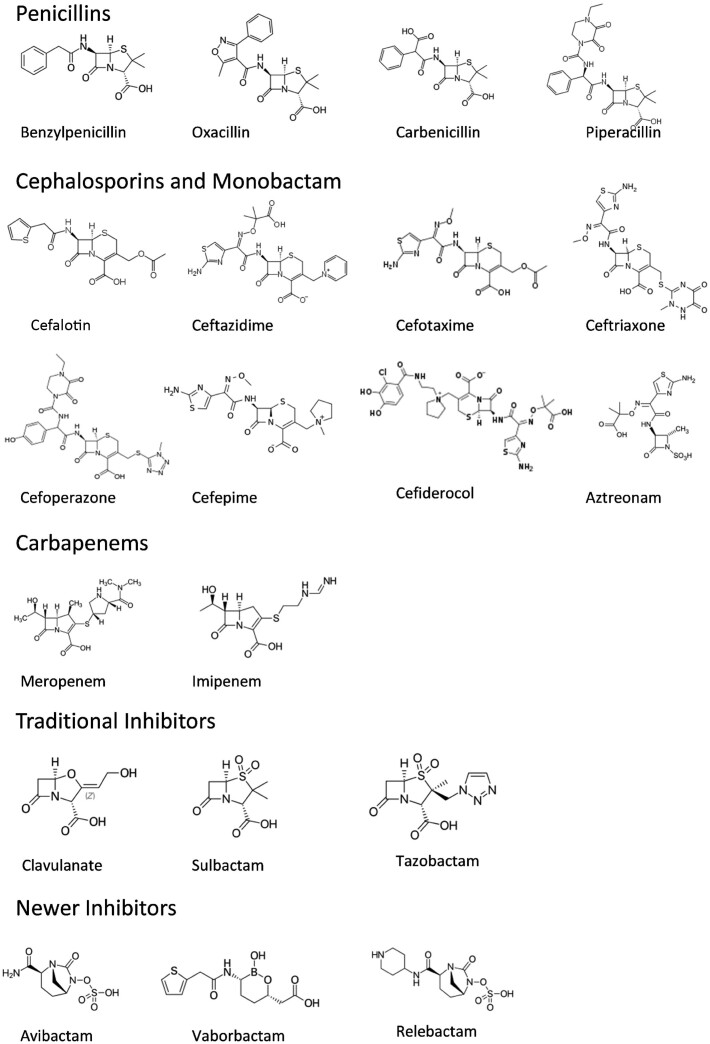
The first classification scheme for β-lactamases that recognized ESBLs was established in 1989 by Karen Bush,7 in which group 2b' was defined as β-lactamase enzymes that can hydrolyse oxyimino-β-lactams such as cefotaxime, ceftazidime and aztreonam at rates at least 10% that of benzylpenicillin and that are strongly inhibited by clavulanate (Figure 1). Subsequently, these enzymes were designated as group 2be in the functional classification scheme of developed by Bush, Jacoby and Mederios.8 In this scheme, ESBLs retained the strict definition of class A β-lactamases that could hydrolyse these expanded-spectrum β-lactam antibiotics and are also susceptible to inhibition by the original β-lactamase inhibitors clavulanate, sulbactam and tazobactam. The original classification scheme also included only plasmid-mediated enzymes, however the updated scheme now recognizes the fluidity of genes expressing ESBLs between plasmids and chromosomes.3 Traditional ESBLs are inhibited by all of the β-lactamase inhibitors including the older inhibitors clavulanate, sulbactam and tazobactam as well as by the newer inhibitors such as avibactam, relebactam and vaborbactam. Although the notion of including any enzyme that can hydrolyse the oxyimino-β-lactams in the classification as an ESBL has been proposed, the strict definition of an ESBL remains that the inhibition by clavulanate is a requirement for designation in this group.3,9–11
Figure 1.
Structures of β-lactam antibiotics and β-lactamase inhibitors.
3. ESBL families
Although ESBLs have common biochemical properties with regards to the hydrolysis of expanded-spectrum β-lactam antibiotics and inhibition by clavulanate, the genes encoding these enzymes are diverse in nature and can be grouped into several families (Table 1).1 Some of these families such as the TEM- and SHV-type ESBLs are highly related, with variants differing by only a few amino acid substitutions. Other families such as the CTX-M-type ESBLs are much more genetically diverse. Each of the ESBL families have some unique characteristics.
Table 1.
ESBL families
| Family | Nomenclature | Characteristics |
|---|---|---|
| TEM | Temoneira, the patient infected with the first isolate expressing TEM-1 | Point mutation variants of TEM-1 or TEM-2 |
| SHV | Sulfhydryl reagent variable | Point mutation variants of SHV-1 |
| IRT | Inhibitor-resistant TEM | TEM variants that are resistant to inhibition by clavulanate and sulbactam, but do not have ESBL phenotype |
| CMT | Complex mutant derived from TEM-1 | TEM variants that are resistant to inhibition by clavulanate and sulbactam and also have ESBL phenotype |
| CTX-M | Cefotaxime-hydrolysing β-lactamase isolated in Munich | Derived from the chromosomal β-lactamase from Kluyvera spp. |
| Preferentially hydrolyses cefotaxime | ||
| GES | Guiana-extended spectrum | More prevalent in P. aeruginosa than Enterobacterales |
| Some variants also hydrolyse carbapenems | ||
| PER | Pseudomonas extended resistant | More prevalent in P. aeruginosa and A. baumannii than Enterobacterales |
| Inhibition by newer β-lactamase inhibitors is variable | ||
| VEB | Vietnam extended-spectrum β-lactamase | Preferentially hydrolyses ceftazidime and aztreonam compared with cefotaxime |
| Inhibition by newer β-lactamase inhibitors is variable | ||
| BEL | Belgium extended β-lactamase | Preferentially hydrolyses ceftazidime and aztreonam compared with cefotaxime |
| TLA | Named after the Tlahuica Indians (Mexico), from whom the first isolate was obtained | Preferentially hydrolyses ceftazidime and aztreonam compared with cefotaxime |
| SFO | From Serratia fonticola | Inducible |
| OXY | From Klebsiella oxytoca | Chromosomally encoded |
Adapted from Jacoby.89
3.1 TEM
TEM-type ESBLS are variants of the original plasmid mediated β-lactamase, TEM-1, which was described in the early 1960s.12 This enzyme was so named because it was originally found in an isolate of Escherichia coli isolate that came from a blood culture from a Greek patient named Temoneira.13 The first derivative of TEM, TEM-2, has a single amino acid substitution of Gln39Lys from the original TEM-1 β-lactamase.14 This change did not alter the substrate profile from TEM-1, however TEM-2 served as the progenitor for many of the TEM-type ESBLs.1 The first TEM-type variant that showed the ESBL phenotype was TEM-3, which was reported in 1989.15 As of this writing, 243 different TEM variants have been described, although not all are ESBLs (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#TEM).
The amino acid substitutions that occur within the TEM enzyme occur at a limited number of positions1 The amino acid residues (Ambler numbering) are most frequently involved in conferring the ESBL phenotype to TEM-type enzymes are Gly238 and Glu240 located on the b3 β-pleated sheet; Arg164 located on the neck of the Ω loop; and Glu104 located directly across from Gly238 Glu240 at the opening of the active-site cavity (Figure 2).16,17 Of these, the substitutions Gly238Ser and Glu240Lys appear to be have the most impact on producing the ESBL phenotype.1 Some of the newer TEM variants have subtle changes in the substrate profile. For example, TEM-184 (amino acid substitutions at Q6K, E104K, I127V, R164S and M182T) hydrolysed aztreonam more efficiently than ceftazidime or cefotaxime.18 Although so many new variants are being discovered by WGS, few of these are being phenotypically characterized to determine if they have properties of an ESBL. However, computer modelling and network analysis has enabled the prediction of whether a particular sequence is likely to belong to functional groups 2 b (original broad spectrum), 2be (ESBL) or 2br (inhibitor resistant).19
Figure 2.
Amino acid alignments of TEM-1, SHV-1 and CTX-M-1. The amino acid sequences WP_000027057.1 (TEM-1), WP_001620095.1 (SHV-1) and WP_013188473.1 (CTX-M-1) were obtained from NCBI and aligned using Clustal Omega.224–226 Numbering according to Ambler.227 Asterisk (*) indicates positions that have a single, fully conserved residue. Colon (:) indicates conservation between groups of strongly similar properties. Period (.) indicates conservation between groups of weakly similar properties. The yellow highlights show the active site Ser70-X-X-Lys active site common to all serine β-lactamases. Red amino acids denote residues where substitutions provide ESBL phenotype (TEM and SHV). Blue amino acids denote where substitutions provide inhibitor resistance phenotype. Green indicates position 240 in CTX-M-1, which has been identified as being associated with increased hydrolysis of cefotaxime.
At the height of prominence for TEM-type ESBLs, the prevalence of some of the variants were regional in nature. For example, TEM-3 was very common in France, but rarely seen in the USA.20 In contrast, TEM-10 was the most prevalent TEM-type ESBL in the USA.21 Interestingly, TEM-26 was detected in isolates from across the globe.20,22–24 As the CTX-M-type β-lactamases became the most prevalent ESBL worldwide, TEM-type enzymes became more infrequent. In a recent survey of European isolates, TEM-type ESBLs were detected in less than 1% of ESBL-producing E. coli and Klebsiella pneumoniae.25
3.2 SHV
The SHV-type β-lactamases (so named for sulfhydryl reagent variable) originated as chromosomally encoded enzymes in K. pneumoniae.26 The first ESBL described in 1985 was SHV-2 and was found in a single strain of Klebsiella ozaenae isolated in Germany that differed from SHV-1 by a single amino acid substitution of Gly to Ser at position 238.27 Similar to what is seen in TEM-type ESBLs, the majority of SHV-type ESBLs also have mutations at Ambler positions 238 (Gly to Ser) and 240 (Lys to Glu) (Figure 2).1 The substitution of serine at position 238 appears to be critical for the efficient hydrolysis of ceftazidime, whereas the substitution of Lys at residue 240 is critical for the efficient hydrolysis of cefotaxime.27 The relevance of the various amino acid substitutions with regards to phenotypic changes in substrate profile has recently been investigated using a mathematical model.28 To date, 228 sequence variants of SHV have been detected, although not all have been functionally characterized to determine if they possess the ESBL phenotype (https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/SHV). Worldwide, SHV-5 and SHV-12 have been the most common ESBL variants found in Enterobacterales.29,30 SHV-type ESBLs are most often found in clinical isolates of K. pneumoniae, however, these enzymes have also been found in other genera of Enterobacterales and P. aeruginosa as well.29,31 In recent European surveillance, SHV-type ESBLs were found in 3.1%–17.0% of clinical isolates of K. pneumoniae, depending on region.25 However, in a clinical trial that targeted ceftazidime-resistant pathogens from complicated intra-abdominal infections (cIAI) and complicated urinary tract infections (cUTI), SHV-type ESBLs were rarely encountered and were only found in strains that also produced a plasmid-mediated AmpC or carbapenemase.32 Although TEM- and SHV-type ESBLs are still encountered, it appears that the impact of their presence among clinical isolates is minimal.
3.3 Inhibitor-resistant β-lactamases
Inhibitor-resistant β-lactamases are derivatives of TEM and SHV enzymes that have amino acid substitutions that confer resistance to inhibition by the β-lactamase inhibitors clavulanate and sulbactam. In the functional classification scheme, they belong to functional group 2br.3,8 Most of these remain susceptible to inhibition by tazobactam and avibactam.33,34 The majority of inhibitor-resistant β-lactamases are derivatives of TEM-1 and were formerly called IRT (for inhibitor resistant TEM), but are now given sequential TEM numbering.35 Common substitutions in the TEM variants have been characterized at amino acid positions Met69, Ser130, Arg244, Arg275 and Asn276 (Figure 2).36 It appears that the cost of the mutations resulting in resistance to clavulanate and sulbactam is a reduction in the efficiency of hydrolysing some penicillins and cephalosporins such as cefalotin.37 Although these mutants are rarely detected, a strain of K. pneumoniae expressing the inhibitor-resistant TEM-30 was identified in several KPC-producing isolates from an outbreak of carbapenem-resistant Enterobacterales (CRE) in New York City.33 Several SHV-type β-lactamases have been characterized as inhibitor resistant, including SHV-49, -56 and -107, which were identified in K. pneumoniae clinical isolates from patients in Europe.38–40
A few complex TEM mutant (CMT) β-lactamases have been described that are mutants of TEM β-lactamases that have both the ESBL phenotype and inhibitor resistance.36 These CMT variants will not be detected with any of the screening methods used to detect ESBLs because those tests rely on inhibition with clavulanate. One such complex mutant, TEM-152, was found in an isolate of E. coli in a patient hospitalized in France.41 This mutant harboured amino acid substitutions Arg164His and Glu240Lys, previously observed in ESBLs, plus Met69Val and Asn276Asp, previously observed in the inhibitor-resistant enzyme TEM-36, which resulted in efficient hydrolysis of ceftazidime and a 50% reduction in inhibition by clavulanate. Because these complex mutants are not resistant to avibactam, ceftazidime/avibactam or one of the other new β-lactamase inhibitor combinations may be a therapeutic option to treat infections caused by organisms expressing one of these enzymes.34 It is likely that the prevalence of TEM- or SHV-type inhibitor-resistant β-lactamases is underestimated because there is not a phenotypic test that laboratories can routinely use to identify these strains.42
3.4 CTX-M
CTX-M-type β-lactamase enzymes were initially reported in the late 1980s, emerging concomitantly in several locations. The nomenclature CTX-M (cefotaximase from Munich) was initially used in a report from Germany.43 However, CTX-M-type enzymes identified in other regions received different names, including FEC-1 (Japan), Toho-1 (Japan) and MEN-1 (France in an Italian patient).44 These initial reports were followed by outbreaks in several countries. The worldwide expansion of isolates carrying these ESBLs later would be referred as the ‘CTX-M pandemic’. Since the early 2000s, CTX-M-type enzymes have been recognized as the most common ESBL group, replacing TEM and SHV as the dominant ESBL type. CTX-M variants have been reported among several members of the order Enterobacterales and in P. aeruginosa and Acinetobacter spp.45–47 Isolates carrying CTX-M-encoding genes have been detected in nosocomial and community settings as well as in companion animals, the environment, food products and livestock.48
Most CTX-M enzymes can be clustered into five groups based on sequence homologies: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9 and CTX-M-25. By far the most common CTX-M-1 group is CTX-M-15, followed by CTX-M-3 and CTX-M-1.44 In the CTX-M-9 group, CTX-M-9 and CTX-M-14 were the most common enzymes, but more recently CTX-M-27 has often been reported.49–53 CTX-M-2, CTX-M-8, and CTX-M-25 are the most frequent variants within their own groups. The analysis of the upstream sequences flanking the genes encoding CTX-M-2 groups belonged to Kluyvera spp. Further analysis demonstrated that this group derived from KLUA-1, an enzyme from Kluyvera ascorbate.54,55 Similarly, CTX-M-134 (CTX-M-1 group) is derived from KLUC-1 from Kluyvera cryocrescens and the CTX-M-9 group have similarity with KLUG-1 from Kluyvera georgiana.56,57 Notably, CTX-M enzymes also exhibit structural similarity and hydrolytic profiles with other class A β-lactamases from environmental organisms, such as Erwinia persicina and Rahnella aquatilis.58,59
The early CTX-M variants efficiently hydrolysed cefotaxime and ceftriaxone, hence the name cefotaximase.44 Contrary to the TEM- and SHV-type ESBLs reported to that point, the early CTX-M enzymes had limited activity against ceftazidime, however CTX-M variants with enhanced ceftazidime hydrolytic activity were later described. Important examples of CTX-M enzymes displaying ceftazidime hydrolysis are CTX-M-15 and CTX-M-27, which are from the CTX-M-1 and CTX-M-9 groups, respectively.
CTX-M-15 is derived from CTX-M-3 and displays a single amino acid change in position 240 (Asp to Gly) when compared with its ancestor (Figure 2).60 The Asp240 residue is located in the terminal part of the B3 β-strand and is responsible for the flexibility of this structure and the accommodation of ceftazidime, which is a bulkier molecule than cefotaxime.61 Despite the modest increase in hydrolytic activity observed against ceftazidime, this change significantly increased ceftazidime MIC values of constructs carrying CTX-M-15.60,62 CTX-M-27 has the same residue in position 240 that is present in CTX-M-15. This residue confers elevated ceftazidime MIC values, despite the overall poor activity of CTX-M-27 against other substrates compared with its ancestor CTX-M-14.63 Data from clinical isolates collected from the SENTRY Antimicrobial Surveillance Program showed that the ceftazidime MIC for CTX-M-producing isolates varies, with MIC values ranging from 0.25 mg/L to >32 mg/L (M. Castanheira, JMI Laboratories, unpublished data).
In 2019, Poirel et al.64 described a new CTX-M variant, CTX-M-33, that had an alteration in position 109 (Asp to Ser) compared with CTX-M-15. This enzyme displayed decreased ceftazidime hydrolysis, but significant meropenem hydrolysis, although this translated into only a modest increase in meropenem MIC in an isogenic pair. However, the clinical isolate of K. pneumoniae isolate carrying this new variant also had impaired permeability resulting in a meropenem MIC of 8 mg/L. CTX-M-type β-lactamases are widespread enzymes. Although there are still treatment options for isolates carrying these enzymes alone, the combination of these enzymes in isolates with other resistance mechanisms and the expansion of hydrolytic profiles with single amino acid mutations could limit the activity of meropenem and newer agents.
3.5 ESBL phenotype OXA-type β-lactamases
The OXA-type β-lactamases hydrolyse oxacillin and are grouped as Ambler class D and Bush-Jacoby-Medeiros functional group 2d enzymes.8 In general, OXA-type enzymes are a broad group that displays variability in substrate profiles and amino acid sequences. However, several OXA-type variants have been noted to hydrolyse cephalosporins, cephems, and/or monobactams. These OXA enzymes with an ESBL phenotype are categorized in Bush functional subgroup 2de.3 Whether or not these oxacillinases with activity against expanded-spectrum cephalosporins are defined as ESBLs is debatable.9 Many researchers do not apply the ESBL terminology to oxacillinases because these enzymes are not classified in the 2be group and are refractory to inhibition by clavulanate or other inhibitors in the same manner as the true ESBLs.
According to a recent review, there are 27 oxacillinase enzymes described as extended spectrum. These enzymes’ substrates include third- and/or fourth-generation cephalosporins in addition to penicillins and early cephalosporins.65 Most extended-spectrum oxacillinases derive from OXA-10 (also named PSE-2) and OXA-2. The OXA-10 derivatives include OXA-11, OXA-13, OXA-14, OXA-16, OXA-17, OXA-19 and OXA-28.66 In addition, OXA-16 has only a partial sequence submitted as its first description (GenBank #AF043100). Among the OXA-2 derivatives, OXA-15, OXA-32, OXA-34, OXA-36 (partial sequence), OXA-53, OXA-141, OXA-161, OXA-210 and OXA-226 have been described.65 Many OXA-2 and OXA-10 derivatives are detected in isolates of P. aeruginosa.
Despite not being considered as extended-spectrum oxacillinases, OXA-1 and OXA-30 have been named for their ability to hydrolyse cefepime.67–69 OXA-1 and OXA-30 were initially reported to differ by one amino acid; however, it was corrected later that these enzymes were identical.70 OXA-1 combined with loss of porins has been implicated in false-ESBL phenotypes among E. coli isolates and resistance to β-lactamase inhibitor combinations.71 Contrary to most oxacillinases, which have a dimeric described structure, OXA-1 was reported to be a monomer.72 An OXA-31 that was detected in an isolate of P. aeruginosa had three amino acid substitutions compared with OXA-1, including the amino acid differences from OXA-4 and also displayed activity against cefepime.73 OXA-48 derivatives, namely OXA-163 and OXA-405, have been described to display activity against extended-spectrum β-lactams with or without many of the OXA-48-like enzyme’s characteristic carbapenemase activity.74,75
3.6 Other ESBL families
The GES (Guiana extended-spectrum β-lactamase) family is the most prevalent group of the less common ESBLs. The gene encoding GES-1 is not closely related to any other plasmid-mediated β-lactamase but does show 36% homology to a carbenicillin-hydrolysing enzyme from Proteus mirabilis.1 Despite initially being reported among species of Enterobacterales, GES enzymes are more common among isolates of P. aeruginosa and A. baumannii isolates.76–80 GES enzymes are notable for their ability to acquire single or double amino acid substitutions and expand their spectrum to carbapenems.
The ESBL GES-1 was first described in 1998 in a K. pneumoniae isolate collected in France from a patient who had recently been hospitalized in French Guiana.81 At the same time, another group described a similar enzyme, named IBC, from an E. cloacae isolate from Greece.82 Subsequent enzymes GES-2 and IBC-2 were both found in isolates of P. aeruginosa isolates.83,84 IBC-1 was later renamed GES-7 and IBC-2, GES-8.
Interestingly, GES-2 had a single amino acid substitution (Gly170Asp) compared with GES-1 and displayed some hydrolytic activity against carbapenems.83 Later described GES β-lactamases fell into two categories: enzymes that were ESBLs and those that showed some modest carbapenemase activity. The original GES enzymes were ESBLs that hydrolyse penicillins and cephalosporins well, but not aztreonam.81 These enzymes are inhibited by clavulanate, tazobactam, and the newer β-lactamase inhibitors such as avibactam, relebactam and vaborbactam.85,86 This means that isolates expressing GES enzymes are often susceptible to ceftazidime/avibactam, but not ceftolozane/tazobactam.85 GES-1 hydrolyses ceftazidime better than cefotaxime. Amino acid substitutions of Glu104Lys or Gly243Ala/Ser that were detected in GES variants described later have been shown to confer greater resistance to cephalosporins and aztreonam.87
The PER-1 β-lactamase (Pseudomonasextended resistant) was initially described from an isolate of P. aeruginosa displaying resistance to cephalosporins and inhibition to clavulanate.88,89 This enzyme hydrolysed most penicillins well and cephalosporins including cefalotin, cefoperazone, cefuroxime, ceftriaxone and ceftazidime. PER-1 did not hydrolyse oxacillin, cephamycins or imipenem. Only a few years later, PER-2 was described in a P. aeruginosa isolate from Argentina, which was 86.4% homologous with PER-1.90 PER enzymes have since been described from A. baumannii and Aeromonas spp., and in various species of Enterobacterales.
PER-1 and PER-2 are the most common members of the PER family. These enzymes have been reported to be inhibited by avibactam to a lesser extent than other class A β-lactamases, with significant differences in MIC values for avibactam and relebactam when tested in combination with other β-lactams.85,91 More detailed studies are warranted due to the difference in activity of these two inhibitors of the same class. Recent analysis demonstrated that A. baumannii isolates harbouring PER enzymes can display elevated MIC values against cefiderocol, a siderophore cephalosporin.92 PER enzymes are most commonly found in isolates from Turkey and Mediterranean countries.93,94
VEB-1 (Vietnamese extended-spectrum β-lactamase) was first detected from an E. coli isolate recovered from a Vietnamese infant.95 VEB-1 conferred high MIC values for ceftazidime and aztreonam, but only modest elevation of MIC values for cefotaxime when expressed in an E. coli background. A 4-fold increase in cefepime MIC values and no activity against imipenem was observed. This enzyme was well inhibited by clavulanate, but avibactam was initially reported to not reduce the ceftazidime MIC values for P. aeruginosa isolates harbouring these enzymes.96 Further studies demonstrated that when various VEB enzymes were expressed in an E. coli isogenic background, the ceftazidime/avibactam MIC values were reduced in a concentration-dependent manner, lowering the ceftazidime MIC values >8-fold when 4 mg/L of inhibitor was used.97 VEB-1 and other VEB variants have been described among various Gram-negative pathogens of including multiple species of Enterobacterales, Vibrio spp., Achromobacter xylosoxidans and more clinically relevant species such as P. aeruginosa and A. baumannii.98–100
Less common ESBLs have been described, but their occurrence is limited. These less common ESBLs include SFO-1 from Serratia fonticola, TLA-1 from the Mexican indigenous people group Tlahuicas, TLA-2 from Germany that displays only 51% homology to TLA-1, BES-1 from Brazil, and BEL-1 from Belgium.98 In a review that addresses these rare ESBLs, Naas et al.98 summarized the MIC values for isolates carrying these enzymes against various β-lactams. The cefotaxime MIC value for a blaSFO-1 transconjugant was 8 mg/L and the result for ceftazidime was 4 mg/L. For a blaBEL-1-harbouring recombinant strain, the cefotaxime MIC results was 1 mg/L, but ceftazidime was 4 mg/L. Higher ceftazidime MIC values were noted for clinical isolates carrying blaBES-1 (16 mg/L) or blaTLA-1 (>256 mg/L).
Several other ESBLs have been detected in the chromosome of Enterobacterales and non-fermentative species. Among those, the OXY β-lactamases in Klebsiella oxytoca are probably the most common cause of resistance in clinical isolates.101,102
4. Molecular characterization of ESBL-producing isolates
4.1 Genetic environment of ESBL genes
Mobile genetic elements (MGEs) such as plasmids, transposons, insertion sequences, integrons and bacteriophages contribute to the dissemination of various ESBL-encoding genes. MGEs can move themselves and/or genes from one location to another within the cell or be transferred from cell to cell horizontally by conjugation, transformation or, in the case of bacteriophages, by transduction.103 More often than not, MGEs carry multiple resistance genes that confer an MDR phenotype to their hosts.104 Some of the main elements of MGEs that carry different ESBL types are highlighted in the section below.
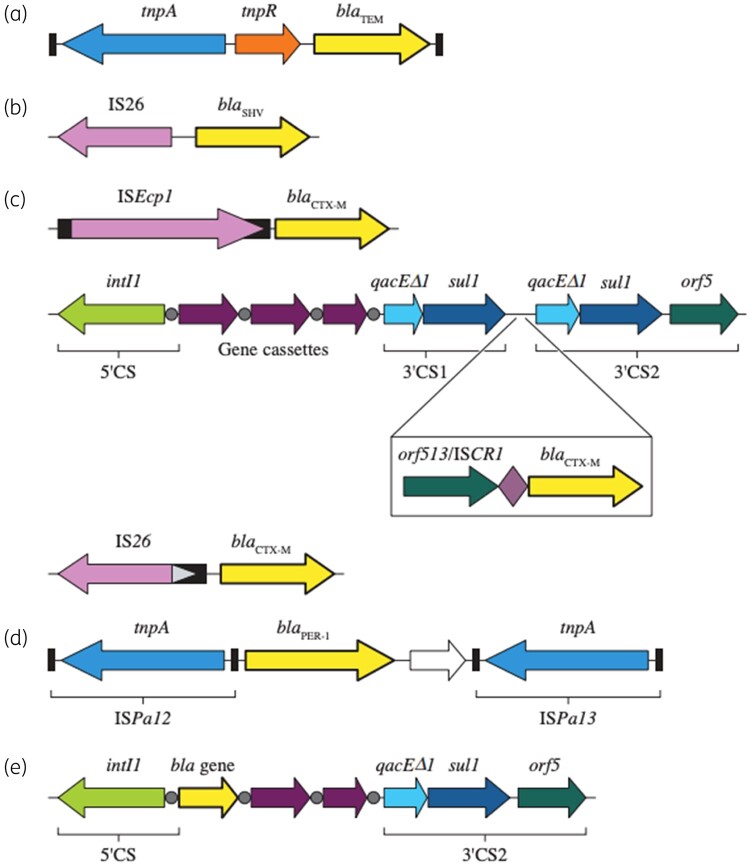
Genes encoding TEM-1, TEM-2 and their ESBL derivatives are usually carried by Tn1-, Tn2-, or Tn3-like transposons (Figure 3).105 These structures were initially named TnA and display 99% nucleotide homology, with most nucleotide differences identified close to their resolvase (res) site.105 A limited number of studies specifically report on the MGE-carrying, blaTEM-encoding ESBL enzymes. In an early study, blaTEM-12 was reported to be part of Tn841, which exhibits homology to Tn3.106 The gene encoding TEM-3 was located on an interrupted copy on Tn1. Tn2 was reported to carry blaTEM-10 whereas blaTEM-24 was associated with Tn1.107–109 In all cases, these structures were embedded in plasmids.110 A study by Marcadé et al.111 that evaluated replicon types of conjugative plasmids carrying ESBL genes revealed that 67% of the plasmids harbouring TEM-type ESBL genes belonged to the IncA/C type. Most of these plasmids carried blaTEM-24, but the plasmids also carried blaTEM-3, blaTEM-10 and blaTEM-21. Others confirmed the occurrence of blaTEM-encoding ESBLs in IncA/C plasmids.108,112,113 Notably, the blaTEM-52 reported by Marcadé et al.111 was embedded in IncI1 plasmids.
Figure 3.
Genetic structures harbouring genes encoding ESBLs. Genetic structures most commonly reported to harbour (a) blaTEM, (b) blaSHV, (c) blaCTX-M, (d) blaPER or (e) class 1 integrons that can carry uncommon ESBL genes. Schematic representations were adapted from Rossolini et al.,60,62 Poirel et al.110 and Diestra et al.62,110,127
The presence of IS26 flanking blaSHV was initially described in the early 1990s. In the first report of blaSHV, IS26 was identified as the mobilizing element for multiple resistance genes and provided a promoter for the expression of blaSHV (Figure 3).114 Intact copies of IS26 have been reported in the plasmids or the chromosome of various bacterial species flanking blaSHV, portions of its 5′ proximal termini or defective IS26 elements.115–118 Genes encoding SHV-type ESBLs can be found either in plasmids or the chromosome. Seven plasmid replicon types that predominantly carry blaSHV-encoding ESBL enzymes—IncA/C, IncF, IncHI2, IncI1, IncL/M, IncN and IncX3—have been identified.115,119 Different blaSHV variants have been detected in each of these plasmid types, with the exception of IncX3, which has only been detected carrying blaSHV-12.115 Additionally, Billard-Pomares et al.120 reported a blaSHV-2-carrying E. coli where this gene was embedded in a P1 bacteriophage structure.
ISEcp1 has been identified upstream of several blaCTX-M types (Figure 3).121 Lartigue et al.122 observed an ISEcp1 upstream of the genes belonging to the CTX-M groups 1, 2, and 9. In another study, Eckert et al.123 analysed the genetic environment of 28 isolates carrying 7 unique blaCTX-M types and observed ISEcp1 in 23 of them. Analysis of the sequences surrounding ISEcp1 and blaCTX-M types revealed signature sequences indicating that transposition events were responsible for the mobilization of blaCTX-M.121 Beyond promoting the dissemination of these genes, ISEcp1 provided a strong promoter for the expression of blaCTX-M.121 ISEcp1 also has been detected flanking other β-lactamase genes, including KLU enzymes in the Kluyvera spp.124
In addition to ISEcp1, blaCTX-M have been detected in the 3′ end of complex class 1 integrons between two qacED1/sul1 elements.123 The ESBL gene was not part of a gene cassette like the genes upstream of qacED1/sul1, but rather in all cases the ESBL gene was flanked upstream by orf513. This structure has been named ISCR1 and was postulated to mobilize genes by rolling circle. Notably, orf513 might function as a transposase that displays similarities to IS91-like transposases.125 Structures harbouring blaCTX-M, including combinations involving IS26, can be observed in several combinations, most likely due to the development of multiple recombination exchanges over time.126,127
Elements harbouring blaCTX-M are usually carried by conjugative plasmids. In a study evaluating CTX-M-15-producing isolates from seven countries located on four continents, Coque et al.128 observed that blaCTX-M-15 was embedded in the narrow host range plasmid IncF with replicon types FII alone or in association with FIA or FIB. Subsequent analysis demonstrated that this was true for various other isolates that harboured blaCTX-M-15. The dissemination of blaCTX-M group 9 genes seems to be associated with an IncHI2-type plasmid, but there have also been reports of IncFII-types.115 Other blaCTX-M carried various incompatibility-type plasmids, including narrow and broad range conjugative plasmids that may also carry additional resistance genes.
4.2 Common strain types for ESBL-producing isolates
MLST has been used extensively to track and monitor the spread of resistance determinants in bacterial pathogens. Several widely disseminated sequence types have been found in epidemics and outbreaks due to resistant clones that are highly associated with specific resistance mechanisms. Until the mid-2000s, it appeared that CTX-M enzymes spread in an seemingly random pattern, with no major clones responsible for their dissemination.129 However, in the last two decades, the dissemination of CTX-M-producing enzymes has been mainly associated with spread of E. coli belonging to a new clonal group ST131.130
E. coli ST131 derives from the phylogenetic group B2 and serotype O25b: H4 and exhibits multiple virulence factors such as adhesins, siderophores, toxins and a group 2 capsule. ST131 isolates differ from most other MDR E. coli by being quite pathogenic.131E. coli belonging to ST131 causes a wide variety of infections, but is most commonly found in urinary tract infections including cystitis, pyelonephritis and urosepsis.132
E. coli ST131 isolates have been reported to carry a variety of β-lactamases and several CTX-M types, most commonly CTX-M-15.133 Five groups (A through E) have been described according to the virulence factors identified among E. coli ST131 isolates. These clones vary according to geographic region. Interestingly, virotypes A, B and over half of virotype C carry blaCTX-M-15, whereas isolates from virotype D carry other β-lactamase genes, including blaCTX-M group 9 genes and blaSHV-12.131,134 In addition to blaCTX-M-15, other characteristics of virotypes A, B and C include resistance to fluoroquinolones and the Type 1 fimbria gene fimH30.130 This group also carries a ISL3-like transposase within its fimH gene. Typing fimH highlighted that the subgroups H30, H30-R, and H30-Rx are associated with MDR clones of E. coli ST131.130 These groups seem to have evolved in a stepwise manner, first by acquiring fluoroquinolone resistance for H30-R and then incorporating blaCTX-M-15 for the H30-Rx group.130
The occurrence of ST131 E. coli isolates carrying blaCTX-M-15 have been well documented globally. In an early survey, Coque et al.128 reported that ST131 E. coli isolates producing CTX-M-15 and belonging to ST131 were detected in all seven countries for which isolates were analysed. Among E. coli clinical isolates collected as part of the SENTRY and MYSTIC programmes in 2007, it was found that 54/127 (47.1%) isolates belonged to ST131.135 These isolates were estimated to correspond to 17% of the overall isolates. Almost 70% of the ST131 isolates were resistant to fluoroquinolones or broad-spectrum cephalosporins that was mediated mainly by CTX-M-15.135 Peirano et al.133 reported that 46% of the ESBL-producing E. coli isolates collected in 11 Canadian hospitals belonged to ST131. Most of these isolates harboured blaCTX-M-15, but other blaCTX-M types were also observed. More recently, Mendes et al.136 reported that 53.6% of the bloodstream and 58.2% of the urinary tract infection isolates collected in US hospitals as part of the SENTRY programme belonged to ST131 or to clonal complex (CC) 131. These isolates were collected during 2016 in 36 US states and were screened using WGS after displaying elevated MIC values of ceftazidime, ceftriaxone, aztreonam or the carbapenems. A recent study from Colombia showed that E. coli isolates from patients with urinary tract infections that expressed CTX-M-15 all belonged to ST131 and the epidemic subclone O25b: H4-B2-H30-Rx.137 The emergence of other E. coli and K. pneumoniae STs disseminating blaCTX-M genes has been recently documented, with blaCTX-M-15 being the most prevalent.138,139 Among these, ST1193 E. coli appears to have rapidly emerged worldwide.140–143
Among the less common ESBLs, GES- and VEB-encoding genes are usually gene cassettes within class I integron structures.79,81,95 These structures can be mobilized as single genes and often are carried alongside other resistance genes that confer resistance to aminoglycosides, quinolones and/or trimethoprim/sulfamethoxazole. The genes encoding PER are a part of a composite transposons such as Tn1213, Tn4176 and ISCR. TLA-1 and PME-1 are carried by ISCR structures.104,115,144,145 Lastly, IS26 has been detected flanking both ends of blaBES-1 and blaSFO-1.115
5. Epidemiology
In the early 2000s, reports suggested that CTX-M-producing isolates were becoming widespread in Europe, Latin America and the Asia-Pacific region.129,146–148 Previously, TEM- and SHV-type enzymes had been the most predominant ESBLs worldwide.149 Later, this shift in the ESBL population toward higher numbers of CTX-M-producing isolates was observed in the USA with two studies: first CTX-M-producing isolates were found in a single hospital and then these isolates were found in 80% of the hospitals participating in the MYSTIC surveillance programme, with CTX-M-15 and CTX-M-14 being the most prevalent types identified.150,151 Since the year 2000, the incidence of ESBL infections has risen in the USA with an increase of 53% between 2012 and 2017, largely due to an increase in community-onset cases.152
Evaluating the epidemiology of ESBLs from a literature review is challenging. As studies use varying isolate selection criteria and a range of methodologies to detect genes, remarkable disparities in outcomes are generated. Unpublished data (M. Castanheira, JMI Laboratories) from the SENTRY Antimicrobial Surveillance Program demonstrated that among 22 548 non-carbapenem-resistant E. coli and K. pneumoniae clinical isolates consecutively collected in US hospitals, 3363 isolates exhibited an MIC value ≥2 mg/L for two of the following agents: ceftazidime, ceftriaxone or aztreonam. These isolates were screened for β-lactamases using previously described methods.102,136 An ESBL gene was detected in 2059 (13.3%) E. coli and 836 (11.8%) K. pneumoniae. Of these, 92.5% carried CTX-M-encoding genes belonging to the CTX-M group 1 (70.0%) or CTX group 9 (22.8%). SHV genes encoding ESBL enzymes were noted among 8.6% of sequenced isolates, mostly in K. pneumoniae (6.5%). TEM ESBLs were only detected among 20 isolates, including 16 E. coli isolates. The prevalence of ESBLs can vary with geographical location, even within one country. A recent study of Gram-negative blood culture isolates taken across the USA showed an overall prevalence of 11% for blaCTX-M, however the percentages ranged from 5% (Michigan) to 26% (Washington, DC).153
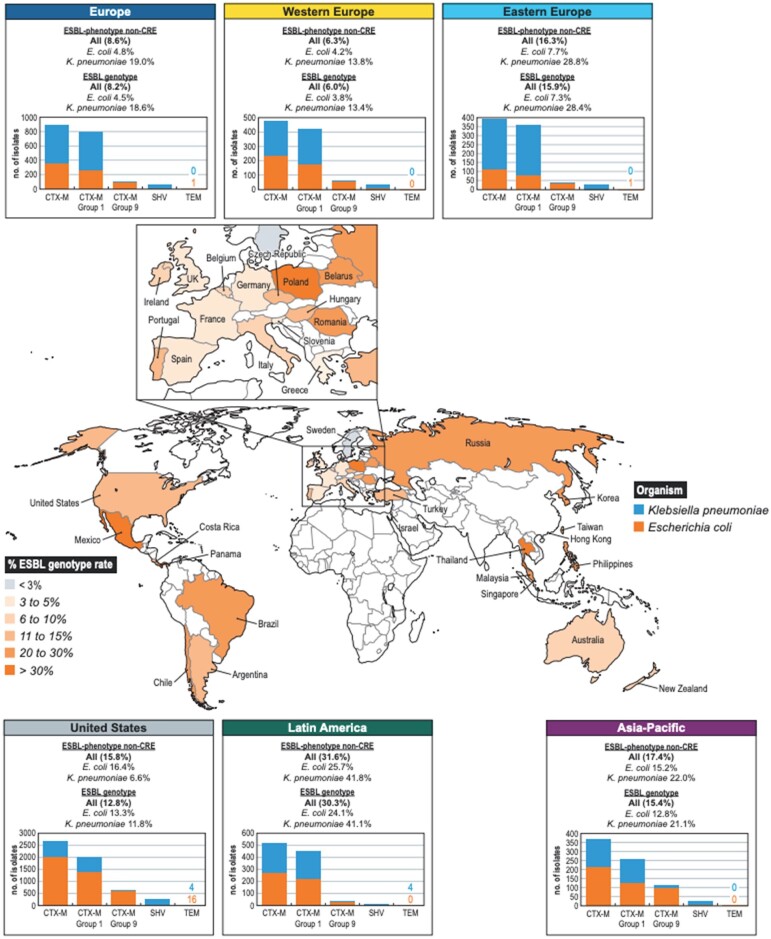
Among 15 449 non-CRE E. coli and K. pneumoniae clinical isolates collected as a part of the SENTRY programme in Europe, Asia-Pacific and Latin America, an ESBL gene was detected among 8.2%, 15.4% and 30.3% of the isolates, respectively (M. Castanheira, JMI Laboratories, unpublished data). These rates varied among individual countries (Figure 4). Similar to the scenario in the USA, most isolates carrying an ESBL gene from the Europe, the Asia-Pacific region and Latin America harboured a CTX-M gene (95.1%, 85.2%, and 98.1%, respectively). Genes belonging to CTX-M group 1 and CTX-M group 9 were the most common.
Figure 4.
Distribution of CTX-M-, TEM- and SHV-producing isolates in the USA, Asia-Pacific, Europe and Latin America.
Canton and Coque129 reported endemicity of CTX-M-producing isolates in various geographic areas. The authors highlighted a drastic increase among E. coli isolates producing CTX-M in the early 2000s. In their analysis, CTX-M-3 and CTX-M-15 were the most common genes detected among the CTX-M group 1 and CTX-M-9. Additionally, CTX-M-14 was most frequent gene observed among group 9. Livermore et al.154 reported similar observations when evaluating several European countries. A literature search by Bevan et al.149 revealed a significant increase in ESBLs in all of the WHO regions analysed. This increase in the prevalence of ESBLs was mainly caused by the dissemination of CTX-M genes. blaCTX-M-15 was the most common gene in all regions except Latin America, where blaCTX-M-2 was the most common gene observed. Among other groups, blaCTX-M-9,blaCTX-M-14 and blaCTX-M-27 have also spread globally.
A study evaluating the epidemiology of ESBL-producing E. coli in Spain demonstrated a decrease of TEM-producing isolates from >19% in 2000 to 1.2% in 2006.155 These investigators also highlighted the dominance of CTX-M-producing E. coli, and noted that CTX-M-14 was the most common type of ESBL. Subsequent studies by the same group highlighted an increase of CTX-M-15-producing isolates that appeared to replace the CTX-M-14-producing population.156,157 Rodriguez-Villalobos et al.158 highlighted an increase in ESBL production and differences in ESBL types when comparing clinical isolates from 2008 to 2006. These authors evaluated Enterobacterales isolates collected in 118 Belgium clinical laboratories and noted that ESBL rates and CTX-M production increased in E. coli, K. pneumoniae and E. cloacae isolates. Similar trends were not observed among K. aerogenes. Similarly, Peirano et al.159 reported CTX-M enzymes replacing SHV-type ESBLs among K. pneumoniae when surveying Canadian isolates, which has also been observed in other regions.159,160
ESBLs are less common in P. aeruginosa than in isolates of Enterobacterales. Croughs et al.161 evaluated 1528 P. aeruginosa isolates from referral hospitals in the Netherlands: 113 isolates displaying ceftazidime MIC values >8 mg/L were screened for ESBL genes and only 6 isolates (0.4% overall; 5.3% among ceftazidime-resistant isolates) possessed ESBLs. These Dutch ESBL-carrying P. aeruginosa isolates harboured blaTEM-12 (2 isolates), blaVEB-2 (2), blaBEL-1 (1) and oxacillinase genes (2). Laudy et al.162 reported that among 900 P. aeruginosa isolates recovered during 2010–14 from four hospitals in Poland, 99 carried (11.0%) ESBL genes. Among the ESBLs, 69 isolates had VEB-9 and 14 isolates had GES (6 with GES-1, 1 with GES-5, 5 with GES-13 and 2 with GES-15). In Brazil, CTX-M-2 was detected among 19.6% of carbapenem-resistant P. aeruginosa screened for ESBLs.163 In this study, 2/56 isolates carried GES-encoding genes. In Greece, PER-1-producing P. aeruginosa isolates belonging to the international high-risk clonal complex 11 were identified among the isolates that displayed a ceftazidime MIC >8 mg/L and a positive ESBL phenotypic test.94 Additionally, outbreaks of blaSHV-2a-producing P. aeruginosa were described in France and Tunisia.164,165 Data regarding P. aeruginosa producing of ESBLs in US isolates is scarce. The analysis of 155 P. aeruginosa isolates reported as part of a previous study evaluating resistance mechanisms against various antipseudomonal β-lactams revealed that only 3 (1.9%) isolates harboured ESBLs.166
ESBL-producing A. baumannii isolates have been described in specific locations and/or as part of outbreaks. ESBL genes that have been reported in A. baumannii include GES, VEB, PER, TEM and CTX-M-15, among others.167–170 A study by Endimiani et al.168 screened 407 A. baumannii isolates collected in an Italian hospital during a 7 year period for resistance to ceftazidime. Of the 119 that had MIC values >8 mg/L, 31 isolates were found to possess blaTEM-92. In a study from Celenza et al.,46 150 A. baumannii isolates from a Bolivian hospital were screened for ESBLs and found that 106 carried blaCTX-M-2, 32 carried blaCTX-43 and 12 carried blaPER-2. Many other studies highlight single occurrences or groups of isolates harbouring PER, VEB and GES in A. baumannii isolates suggesting these are the most common ESBLs in this species.170–172
The dissemination of ESBL-producing bacterial pathogens is likely due to many factors such as geographical location, population density, hygiene and usage of antibiotics. For example, the prevalence of ESBLs in E. coli is low in Europe but is very high in Southeast Asia, Africa, and Central America.173 There is even country to country variation with regions. For example, the prevalence of Enterobacterales expressing ESBLs is higher in Mediterranean countries but is very low in the Netherlands and Scandinavia.174 Today, our global society is quite mobile, whether it be as vacationers, medical tourists or refugees. All of these factors have contributed to outbreaks and the overall global dissemination of ESBL-mediated resistance.
The epidemiology of ESBL-mediated resistance primarily follows the type of infections where the pathogen encountered often require heavy usage of expanded-spectrum β-lactam antibiotics. For ESBL-producing Enterobacterales, the main source of these pathogens is the genitourinary tract of patients, with infections most often caused by strains with which the patient is already colonized.175 The transmission of ESBL-producing Enterobacterales can occur between patients with or without the involvement of a healthcare worker as an intermediate vector. The rate of transmission is also likely to vary based on differences in various species due to differences in virulence factors. The transmission rate from patients colonized with ESBL-producing K. pneumoniae was shown to be 2-fold higher than from those colonized with ESBL-producing E. coli.176
Initially, ESBL-producing clinical isolates were found only in the hospital setting, however, they quickly spread into nursing homes and then into the community.4,21 Although most outbreaks of ESBL-producing Enterobacterales occur in the ICU or in immunocompromised patients, other patient populations can also be affected. In Japan, there was an outbreak of ESBL-producing E. coli in neonates that was traced to shared breast milk from donor mothers.177 A 2017 survey of US hospital infections caused by ESBL-producing E. coli and K. pneumoniae showed that the rates of these infections were increasing.178
Both TEM- and SHV-type ESBLs were detected throughout the USA and Europe in the late 1980s and 1990s with specific variants noted to have variations in regional prevalence.154,179,180 For example, TEM-10 was identified in several unrelated outbreaks of ESBL-producing Enterobacterales in the USA, but was rarely seen in Europe.21 The prevalence of both TEM- and SHV-type ESBLs has now diminished at the same time as the worldwide dominance of isolates producing CTX-M-type β-lactamases has occurred.181 ESBLs have also been reported from many environmental, food and veterinary samples.182
6. Detection of ESBL-producing Gram-negative organisms in clinical microbiology laboratories
Detection of ESBL-producing Enterobacterales has traditionally relied on phenotypic methods for detection in clinical microbiology laboratories. These methods exploit the fact that ESBLs are inhibited by traditional β-lactamase inhibitors such as clavulanate. Both CLSI and EUCAST have endorsed screening and confirmatory tests for detection of ESBL producers and guidance on use of these tests varies based on the cephalosporin breakpoints applied by the laboratory.183,184 Although the methods described by these standards setting organizations are similar, differences exist in the recommended organisms to test, screening and confirmatory test methods and interpretations (Table 2). These ESBL methods require overnight incubation and have known limitations that affect both sensitivity (e.g. false negatives due to the co-production of an AmpC β-lactamase) and specificity (e.g. false positivity due to hyperproduction of narrower-spectrum β-lactamases combined with altered permeability). Commercially available automated antimicrobial susceptibility testing systems have adopted comparable built-in ESBL screening and confirmation tests on their panels. However, these systems are known to report false positive ESBL results and some lack US FDA clearance for P. mirabilis due to poor performance.185–187
Table 2.
ESBL screen and confirmatory tests as recommended by CLSI and EUCAST
| Criteria | CLSI | EUCAST |
|---|---|---|
| Organisms | E. coli, K. oxytoca, K. pneumoniae and P. mirabilis | Group 1: E. coli, Klebsiella spp. [not including Klebsiella (formerly Enterobacter) aerogenes], P. mirabilis, Raoultella spp., Salmonella spp. and Shigella spp. |
| Group 2 (Enterobacterales with inducible chromosomal AmpC): Enterobacter spp., Citrobacter freundii, Morganella morganii, Providencia stuartii, Serratia spp., Hafnia alvei | ||
| Screening test methods | Disc diffusion and BMD methods | Broth dilution, agar dilution or disc diffusion |
| Screening agents and cutoffs | Aztreonam, cefotaxime, ceftazidime and ceftriaxone MIC of ≥2 mg/L | Cefpodoxime, cefotaxime, ceftazidime and ceftriaxone MIC of ≥2 mg/L |
| Cefpodoxime MIC of ≥2 mg/L for P. mirabilis or MIC ≥8 mg/L for E. coli, K. pneumoniae and K. oxytoca | ||
| Positive screening results | Either (i) cefpodoxime alone | Either (i) cefpodoxime alone |
| Or (ii) aztreonam (excluding P. mirabilis), cefotaxime, ceftazidime or ceftriaxone screen positive | Or (ii) cefotaxime or ceftriaxone AND ceftazidime screen positive | |
| Confirmatory test methods | Disc diffusion and BMD methods | CDT, DDST, ESBL gradient test and BMD test |
| Test | Ceftazidime and cefotaxime ± clavulanate | Group 1: Ceftazidime and cefotaxime ± clavulanate; |
| add cefepime ± clavulanate if cefoxitin has been tested and has an MIC of ≥16 mg/L | ||
| Group 2: Cefepime ± clavulanate | ||
| Positive interpretation | Disc diffusion: ≥5 mm increase in zone diameter for either agent tested in combination with clavulanate versus the zone diameter of the agent tested alone | CDT: Same interpretation as the CLSI disc diffusion test |
| DDST: Zones of inhibition around cephalosporin discs are augmented or there is a keyhole in the direction of the disc containing clavulanate | ||
| BMD: ≥3 2-fold concentration decreases in an MIC for either agent tested in combination with clavulanate versus the MIC of the agent tested alone | BMD: ≥8-fold reduction is observed in the MIC of the cephalosporin combined with clavulanate compared with the MIC of the cephalosporin alone | |
| Gradient diffusion: The same as above for BMD or if a phantom zone or deformed ellipse is present | ||
| Reporting cephalosporin results for ESBL-producing isolates | ||
| use of obsolete cephalosporin breakpoints | Report all penicillins, cephalosporins and aztreonam as resistant | |
| use of current cephalosporin breakpoints | Report the MICs and interpretations as tested | |
Over a decade ago, both CLSI and EUCAST lowered the cephalosporin breakpoints to increase the sensitivity of identifying ESBL-producing organisms, to decrease the burden confirmatory testing placed on microbiology laboratories and because of updated pharmacokinetics (PK)/pharmacodynamics (PD) data, MIC distributions and limited clinical outcome data suggesting improved patient outcomes with lower breakpoints.183,188 With the lowering of the breakpoints, they revised the recommendations to perform ESBL confirmatory tests for epidemiological or infection control purposes only.183,189 Based on this guidance, many laboratories updated their cephalosporin breakpoints and stopped performing routine ESBL confirmatory testing. As such, the MICs and interpretations are reported as tested for the penicillins, cephalosporins and aztreonam without identifying the mechanism leading to third-generation cephalosporin resistance.
The MERINO trial was the first randomized clinical trial comparing the outcomes of patients receiving piperacillin/tazobactam and meropenem for the treatment of presumed ESBL-producing bloodstream infections.190 The original analyses found inferior outcomes for patients treated with piperacillin/tazobactam (although they were later modified due to inaccurate susceptibility testing). This has led to a renewed interest in understanding the role of ESBL tests in clinical practice. In the absence of ESBL confirmation testing, some clinicians and more recently the IDSA antimicrobial resistance treatment guidance recommended using a ceftriaxone MIC ≥2 mg/L (not susceptible) as a proxy for predicting ESBL production to guide treatment-based decisions.191,192 In one study, the use of a ceftriaxone MIC ≥2 mg/L to predict ESBL production resulted in overestimation of ESBL production due to a less than ideal specificity which led to increased prescribing of carbapenems.193 Thus, further guidance may be on the horizon for use of ESBL tests to not only guide infection control practices but to identify ESBL producers to help guide therapeutic decision-making. It remains controversial whether ESBL testing should occur or not, although international thought leaders agree that if an accurate, timely and comprehensive ESBL test was available it could be helpful in clinical decision-making.194–196
In contrast to traditional phenotypic ESBL tests, rapid phenotypic methods have more recently been developed with same day results for the detection of ESBL producers including colorimetric and immunological lateral flow assays. The rapid colorimetric methods provide results within 15 min to 2 h and include methods that specifically detect ESBL producers [Rapid ESBL NDP or the Rosco Diagnostica Rapid ESBL Screen (Taastrup, Denmark)] or more broadly detect ESBL, AmpC and carbapenemases without distinction due to cleavage of an expanded-spectrum chromogenic cephalosporin (β Lacta Test; Bio-Rad, Marnes-La-Coquette, France).197 These tests have been evaluated from cultured isolates and directly from various specimen types (e.g. blood, urine, respiratory specimens) with good sensitivity (>90%) and variable specificity depending on the test.197–201 The rapid calorimetric tests detect a phenotype broadly associated with ESBL production without discriminating between the various enzymes. Recently, a lateral flow immunoassay (NG-Test CTX-M MULTI assay, NG Biotech, Guipry, France) was developed to detect and differentiate the five groups of CTX-M enzymes (i.e. groups 1, 2, 8, 9, 25) from colonies and from positive blood cultures within 15 min with excellent sensitivity and specificity (≥98%).202,203
In addition to phenotypic methods, molecular methods that target specific ESBL genes have been developed and implemented in clinical microbiology laboratories. The most widely adopted include the commercially available syndromic sepsis panels performed from positive blood culture broths that include CTX-M as the sole ESBL target associated with the detection of the Enterobacterales, P. aeruginosa and/or A. baumannii. The detection of the globally dominant ESBL gene helps with more rapid selection of appropriate therapy. These rapid panels (1 to 4 h) have good sensitivity and specificity for detection of blaCTX-M and include the GenMark Dx ePlex® Blood Culture Identification Gram-Negative (BCID-GN) Panel (Carlsbad, CA, USA), BioFire BCID2 Panel (Salt Lake City, UT, USA) and the Verigene Gram-Negative Blood Culture Nucleic Acid Test BC-GN panel (Austin, TX, USA).204,205 More recently, multiplex pneumonia panels that include blaCTX-M as a marker have been introduced, including the Unyvero LRT panel (Curetis, Holzgerlingen, Germany) and BioFire FilmArray pneumonia panel (Salt Lake City, UT, USA), for detection directly from respiratory specimens. The Unyvero LRT demonstrated 95.7% sensitivity for the detection of blaCTX-M from bronchoalveolar lavage (BAL) specimens, whereas evaluation of the BioFire pneumonia panel showed 85.7% (6/7 specimens) and 80% (8/10 specimens) sensitivity from BAL and sputum specimens, respectively.206,207 The lack of inclusion of TEM- and SHV-type ESBL variants on these panels likely reflect the challenge that only a few single nucleotide polymorphisms differentiate narrower-spectrum variants from ESBL variants. However, future molecular diagnostic panels would benefit from the inclusion of SHV- and/or TEM-type ESBL targets as geographic and species-specific differences occur in the distribution of ESBL genes as highlighted in the epidemiology section of this review.208,209 Last, several research use only molecular assays such as DNA microarray assays, PCR and/or sequencing and WGS have been described for the characterization of ESBL genes and/or variants.210 However, they are not broadly implemented in clinical laboratories due to the complexity of the methods. The limitations of molecular methods include the expense, requirement of instrumentation and/or requirement of highly trained staff with molecular and/or bioinformatics expertise.
7. Current therapies
The presence of ESBL-producing Enterobacterales in serious infections has had a significant impact on the choice of empirical antimicrobial therapy and is associated with a delay in the initiation of appropriate therapy.211 The failure to initiate appropriate antibiotic therapy from the start is associated with prolonged hospital stays, increasing hospital costs and higher patient mortality. This has led to the increased use of carbapenems in many institutions, which has subsequently resulted in increased resistance to carbapenems.5,6 There are few randomized controlled clinical trials that study the treatment of infections due to ESBL-producing bacteria. However, there have been a number of observational studies such as retrospective cohorts, case series and anecdotal reports that examine different treatment regimens for infections caused by ESBL-producing organisms. Several of these studies have been focused on the β-lactam/β-lactamase inhibitor (BL/BLI) combinations amoxicillin/clavulanate and piperacillin/tazobactam.212–214 Each of these retrospective examinations of clinical data concluded that retrospective data that included patients with urosepsis and other bloodstream infections (BSI), BL/BLI combinations were non-inferior to the carbapenems and could be used as carbapenem-sparing therapy. In another study, the INCREMENT project developed a scoring tool to predict whether or not a patient was at high or low risk for mortality following a BSI caused by an ESBL-producing organism.215 This study collected data on over 1000 patients from 7 tertiary hospitals in 11 different countries. With regards to therapy, they found that when the isolates demonstrated in vitro susceptibility to BL/BLI combinations (mainly piperacillin/tazobactam), aminoglycosides (mainly amikacin) or fluoroquinolones (mainly ciprofloxacin), these agents appeared to be as effective as the carbapenems for both the empirical and targeted therapy.
In 2018 the results of the MERINO trial—an international, non-inferiority, open-label randomized controlled study that compared piperacillin/tazobactam (4.5 g every 6 h) with meropenem (1 g every 8 h) for the treatment of BSIs due to cephalosporin-resistant Enterobacterales—were published.190 The trial included 379 patients in 26 hospitals in 9 countries. The results showed that BSI patients infected with ceftriaxone-resistant E. coli or K. pneumoniae, treatment with piperacillin/tazobactam was not shown to be non-inferior when compared with meropenem with the endpoint of 30 day mortality. The authors concluded that piperacillin/tazobactam should not be used in this patient population.190 This study received a great deal of attention with many interpreting the results to mean that no infections caused by any ESBL-positive pathogen, regardless of body site, should be treated with piperacillin/tazobactam. As a consequence, there has been a large increase in the usage of carbapenems to treat ESBLs. Unfortunately, there were many limitations of the study design and subsequent analysis. For example, empirical treatments were allowed prior to randomization, acceptable therapies for both empirical and step-down were not pre-specified, crossover of patients from one group to the other was allowed, and there was an unusually low mortality of patients in the meropenem arm of the study.211 In addition, it was an open-labelled trial; piperacillin/tazobactam was administered with a 30 min infusion, even though the current recommendations are for extended infusion for serious infections, the trial was prematurely stopped, and the imbalance of groups for some variables might not have been corrected in the analysis, and finally, the endpoint of all-cause mortality was skewed as most of them were unrelated to the infection but occurred mostly in patients with advanced cancer.216
From a microbiology standpoint, the trial included piperacillin/tazobactam resistant organisms, but none for meropenem and although the authors claim that there was no correlation of MIC with mortality, no MIC by outcome data were provided. Furthermore, susceptibility tests were conducted using the Vitek automated susceptibility testing system, disc diffusion or gradient strip devices at the site lab, then confirmed by in a central lab using Etest strips. Unfortunately, none of these is the standard reference method. This is especially important to note, as Etest strips had previously been noted to be unreliable for testing piperacillin/tazobactam.217
To address some of the concerns with regards to the microbiological data from the MERINO trial, the authors then performed a post hoc analysis of MIC values and resistance genes detected comparative to the 30 day mortality of patients treated with both piperacillin/tazobactam and meropenem.218 MICs of both test drugs for all organisms isolated from BSI in the MERINO trial were retested using the reference method of broth microdilution and all of the β-lactamase genes present were determined. In retesting, they found that a significant number of isolates that had previously been reported as susceptible to piperacillin/tazobactam were non-susceptible (i.e. intermediate or resistant). This, in turn, had a significant impact on the 30 day all-cause mortality endpoint for the trial, as when patients infected with these non-susceptible isolates were removed from the analysis, the absolute difference from meropenem was reduced. The authors found that a piperacillin/tazobactam MIC of >16 mg/L was the strongest predictor of mortality. Many of the piperacillin/tazobactam-resistant isolates expressed AmpC or OXA-1 in addition to the ESBL, which are not expected to be susceptible to piperacillin/tazobactam. Therefore, the previous conclusion that all ceftriaxone-resistant organisms should be treated with carbapenems is not valid. The authors revised their conclusion to the recommendation of allowing susceptibility testing (reference method) to guide therapy with piperacillin/tazobactam for ESBL-producing strains.218 Despite these findings and the revised mortality assessment, the authors of the MERINO trial continue to be proponents of the notion that all infections caused by ceftriaxone-resistant organisms (and by association ESBL-positive) should be treated with a carbapenem.219 Other experts have concluded that there has been an overinterpretation of the MERINO trial results, which has caused an overuse of carbapenems and potentially contributed to the dramatic increase of carbapenem-resistant organisms.216 They agree that patients with severe or difficult-to-treat infections caused by ceftriaxone-resistant Enterobacterales should be treated with carbapenems. However, they believe that many infections can be safely treated with other options if therapy is guided by strong microbiological data. Clinicians should carefully weigh both arguments as they navigate the management of these increasingly common infections.220
Some of the new BL/BLI combinations may be suitable therapies for ESBL-producing organisms. In randomized clinical trials, ceftolozane/tazobactam showed comparable efficacy to levofloxacin (cUTI) or meropenem (cIAI) against ESBL-producing Enterobacterales.221 A recent meta-analysis of five randomized controlled trials with ESBL- and AmpC-specific outcome data showed that of the 246 patients infected with an ESBL-producing pathogen in the ceftazidime/avibactam treatment arm 91% had a favourable clinical response at test of cure compared with 89% in the carbapenem arm.222 The authors warned that this dataset largely consisted of patients with cUTI or cIAI, therefore caution should be taken before extrapolating to more serious infections. Although ESBL-specific clinical experience in the literature is scarce, the carbapenem-based BL/BLI combinations of meropenem/vaborbactam and imipenem/relebactam show in vitro activity against ESBL-producing strains.223
With regards to choosing therapy for less severe infections such as community-onset urinary tract infections due to ESBL-producing Enterobacterales, agents such as ciprofloxacin, amoxicillin/clavulanate, nitrofurantoin and fosfomycin show good in vitro activity against ESBL-producing bacteria and may be good options.211 However, resistance is also increasing among these isolates, therefore susceptibility testing is essential for guiding therapy.
8. Conclusions
Antimicrobial resistance is a significant problem worldwide that has been an unwelcome result of modern medical care. ESBL-producing Enterobacterales remain the most commonly encountered mechanism providing resistance to expanded-spectrum cephalosporins in these pathogens, both in healthcare and community settings. ESBLs have spread in virulent clones such as E. coli ST131. With today’s modern technologies, the detection and molecular characterization of ESBLs has become commonplace. However, with so many ESBL variants that are often produced in combination with other β-lactamases, decoding this information is not always easy. Unfortunately, appropriate therapy is frequently delayed in these patients, who then may suffer clinical outcomes. Although there have been recent debates about whether a carbapenem must be used to treat serious infections caused by ESBLs or whether some of the BL/BLI combinations are appropriate, the fact remains that we have at our disposal a good armamentarium to treat these infections. As ESBLs are now endemic in clinical isolates of Enterobacterales, it will continue to be essential that we remain vigilant about identifying them both in patient isolates and in surveillance.
Transparency declarations
M.C. is an employee of JMI Laboratories, which was contracted to perform services during 2020–21 for Affinity Biosensors, Allergan, Amicrobe Inc., Amplyx Pharma, Artugen Therapeutics USA Inc., Astellas, Basilea, Beth Israel Deaconess Medical Center, BIDMC, bioMérieux Inc., BioVersys Ag, Bugworks, Cidara, Cipla, Contrafect, Cormedix, Crestone Inc., Curza, CXC7, Entasis, Fedora Pharmaceutical, Fimbrion Therapeutics, Fox Chase, GlaxoSmithKline, Guardian Therapeutics, Hardy Diagnostics, IHMA, Janssen Research & Development, Johnson & Johnson, Kaleido Biosciences, KBP Biosciences, Luminex, Matrivax, Mayo Clinic, Medpace, Meiji Seika Pharma Co. Ltd, Melinta, Menarini, Merck, Meridian Bioscience Inc., Micromyx, MicuRx, N8 Medical, Nabriva, NIH, National University of Singapore, North Bristol NHS Trust, Novome Biotechnologies, Paratek, Pfizer, Prokaryotics Inc., QPEX Biopharma, Rhode Island Hospital, RIHML, Roche, Roivant, Salvat, Scynexis, SeLux Diagnostics, Shionogi, Specific Diagnostics, Spero, SuperTrans Medical LT, T2 Biosystems, The University of Queensland, Thermo Fisher Scientific, Tufts Medical Center, Universite de Sherbrooke, University of Iowa, University of Iowa Hospitals and Clinics, University of Wisconsin, UNT System College of Pharmacy, URMC, UT Southwestern, VenatoRx, Viosera Therapeutics, and Wayne State University. There are no speakers’ bureaus or stock options to declare.
P.J.S. reports grants and personal fees from Accelerate Diagnostics, OpGen Inc and BD Diagnostics, grants from bioMérieux Inc., Affinity Biosensors and Hardy Diagnostics; and personal fees from Roche Diagnostics, Shionogi Inc. and GeneCapture, outside the submitted work.
P.A.B. has received consulting fees during 2020–21 from Boston Pharmaceuticals, ContraFect, Emergent Biosolutions, Entasis, Genentech, ICPD, Prokaryotics, Recreo Pharmaceuticals, Sihuan Pharmaceuticals, Sinovent, SuperTrans Medical, X-Biotix and Zai Labs and owns stock in Pfizer.
References
- 1. Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 2001; 14: 933–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bush K, Fisher JF.. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu Rev Microbiol 2011; 65: 455–78. [DOI] [PubMed] [Google Scholar]
- 3. Bush K, Jacoby GA.. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 2010; 54: 969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pitout JD, Nordmann P, Laupland KB. et al. Emergence of Enterobacteriaceae producing extended-spectrum b-lactamases (ESBLs) in the community. J Antimicrob Chemother 2005; 56: 52–9. [DOI] [PubMed] [Google Scholar]
- 5. Perez F, Endimiani A, Hujer KM. et al. The continuing challenge of ESBLs. Curr Opin Pharmacol 2007; 7: 459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paterson DL, Bonomo RA.. Extended-spectrum b-lactamases: a clinical update. Clin Microbiol Rev 2005; 18: 657–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bush K. Classification of β-lactamases: Groups 1, 2a, 2b, and 2b'. Antimicrob Agents Chemother 1989; 33: 264–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bush K, Jacoby GA, Medeiros AAA.. functional classification scheme for b-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 1995; 39: 1211–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Livermore DM. Defining an extended-spectrum b-lactamase. Clin Microbiol Infect 2008; 14 Suppl 1: 3–10. [DOI] [PubMed] [Google Scholar]
- 10. Giske CG, Sundsfjord AS, Kahlmeter G. et al. Redefining extended-spectrum β-lactamases: balancing science and clinical need. J Antimicrob Chemother 2009; 63: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bush K, Jacoby GA, Amicosante G. et al. Comment on: Redefining extended-spectrum β-lactamases: balancing science and clinical need. J Antimicrob Chemother 2009; 64: 212–3. [DOI] [PubMed] [Google Scholar]
- 12. Datta N, Kontomichalou P.. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 1965; 208: 239–41. [DOI] [PubMed] [Google Scholar]
- 13. Medeiros AA. βlactamases. Br Med Bull 1984; 40: 18–27. [DOI] [PubMed] [Google Scholar]
- 14. Barthélémy M, Peduzzi J, Labia R.. [Distinction between the primary structures of TEM-1 and TEM-2 β-lactamases]. Ann Inst Pasteur Microbiol (1985) 1985; 136a: 311–21. [DOI] [PubMed] [Google Scholar]
- 15. Sougakoff W, Goussard S, Gerbaud G. et al. Plasmid-mediated resistance to third-generation cephalosporins caused by point mutations in TEM-type penicillinase genes. Rev Infect Dis 1988; 10: 879–84. [DOI] [PubMed] [Google Scholar]
- 16. Knox JR. Extended-spectrum and inhibitor-resistant TEM-type b-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother 1995; 39: 2593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raquet X, Lamotte-Brasseur J, Fonze E. et al. TEM β-lactamase mutants hydrolysing third-generation cephalosporins. A kinetic and molecular modelling analysis. J Mol Biol 1994; 244: 625–39. [DOI] [PubMed] [Google Scholar]
- 18. Piccirilli A, Perilli M, Amicosante G. et al. TEM-184, a novel TEM-derived extended-spectrum β-lactamase with enhanced activity against aztreonam. Antimicrob Agents Chemother 2018; 62: e00688-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeil C, Widmann M, Fademrecht S. et al. Network analysis of sequence-function relationships and exploration of sequence space of TEM β-lactamases. Antimicrob Agents Chemother 2016; 60: 2709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soilleux MJ, Morand AM, Arlet GJ. et al. Survey of Klebsiella pneumoniae producing extended-spectrum β-lactamases: prevalence of TEM-3 and first identification of TEM-26 in France. Antimicrob Agents Chemother 1996; 40: 1027–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiener J, Quinn JP, Bradford PA. et al. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA 1999; 281: 517–23. [DOI] [PubMed] [Google Scholar]
- 22. Urban C, Mariano N, Rahman N. et al. Detection of multiresistant ceftazidime-susceptible Klebsiella pneumoniae isolates lacking TEM-26 after class restriction of cephalosporins. Microb Drug Resist 2000; 6: 297–303. [DOI] [PubMed] [Google Scholar]
- 23. Shannon K, Stapleton P, Xiang X. et al. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains causing nosocomial outbreaks of infection in the United Kingdom. J Clin Microbiol 1998; 36: 3105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pitout JD, Thomson KS, Hanson ND. et al. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob Agents Chemother 1998; 42: 1350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kazmierczak KM, de Jonge BLM, Stone GG. et al. Longitudinal analysis of ESBL and carbapenemase carriage among Enterobacterales and Pseudomonas aeruginosa isolates collected in Europe as part of the International Network for Optimal Resistance Monitoring (INFORM) global surveillance programme, 2013. J Antimicrob Chemother 2020; 75: 1165–73. [DOI] [PubMed] [Google Scholar]
- 26. Livermore DM. b-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev 1995; 8: 557–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huletsky A, Knox JR, Levesque RC.. Role of Ser-238 and Lys-240 in the hydrolysis of third-generation cephalosporins by SHV-type β-lactamases probed by site-directed mutagenesis and three-dimensional modeling. J Biol Chem 1993; 268: 3690–7. [PubMed] [Google Scholar]
- 28. Neubauer S, Madzgalla S, Marquet M. et al. A genotype-phenotype correlation study of SHV β-lactamases offers new insight into SHV resistance profiles. Antimicrob Agents Chemother 2020; 64: e02293–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perilli M, Dell'Amico E, Segatore B. et al. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J Clin Microbiol 2002; 40: 611–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yan J-J, Wu S-M, Tsai S-H. et al. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae Producing extended-spectrum β-lactamases and identification of a novel AmpC Enzyme (CMY-8) in Southern Taiwan. Antimicrob Agents Chemother 2000; 44: 1438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coque TM, Baquero F, Cantón R.. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Europe Euro Surveill 2008; 13: pii=19044. [PubMed] [Google Scholar]
- 32. Mendes RE, Castanheira M, Woosley LN. et al. Characterization of β-lactamase content of ceftazidime-resistant pathogens recovered during the pathogen-directed phase 3 REPRISE trial for ceftazidime-avibactam: correlation of efficacy against β-lactamase producers. Antimicrob Agents Chemother 2019; 63: e02655-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bradford PA, Bratu S, Urban C. et al. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 b-lactamases in New York City. Clin Infect Dis 2004; 39: 55–60. [DOI] [PubMed] [Google Scholar]
- 34. Lahiri SD, Bradford PA, Nichols WW. et al. Structural and sequence analysis of class A b-lactamases with respect to avibactam inhibition: impact of Omega-loop variations. J Antimicrob Chemother 2016; 71: 2848–55. [DOI] [PubMed] [Google Scholar]
- 35. Bush K, Jacoby G.. Nomenclature of TEM β-lactamases. J Antimicrob Chemother 1997; 39: 1–3. [DOI] [PubMed] [Google Scholar]
- 36. Canton R, Morosini MI, de la Maza OM. et al. IRT and CMT b-lactamases and inhibitor resistance. Clin Microbiol Infect 2008; 14 Suppl 1: 53–62. [DOI] [PubMed] [Google Scholar]
- 37. Bret L, Chaibi EB, Chanal-Claris C. et al. Inhibitor-resistant TEM (IRT) β-lactamases with different substitutions at position 244. Antimicrob Agents Chemother 1997; 41: 2547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dubois V, Poirel L, Arpin C. et al. SHV-49, a novel inhibitor-resistant β-lactamase in a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother 2004; 48: 4466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dubois V, Poirel L, Demarthe F. et al. Molecular and biochemical characterization of SHV-56, a novel inhibitor-resistant b-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 2008; 52: 3792–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mendonca N, Ferreira E, Louro D. et al. Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int J Antimicrob Agents 2009; 34: 29–37. [DOI] [PubMed] [Google Scholar]
- 41. Robin F, Delmas J, Schweitzer C. et al. Evolution of TEM-Type enzymes: biochemical and genetic characterization of two new complex mutant TEM enzymes, TEM-151 and TEM-152, from a single patient. Antimicrob Agents Chemother 2007; 51: 1304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaye KS, Gold HS, Schwaber MJ. et al. Variety of β-lactamases produced by amoxicillin-clavulanate-resistant Escherichia coli isolated in the Northeastern United States. Antimicrob Agents Chemother 2004; 48: 1520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bauernfeind A, Grimm H, Schweighart S.. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 1990; 18: 294–8. [DOI] [PubMed] [Google Scholar]
- 44. Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 2004; 48: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Picão RC, Poirel L, Gales AC. et al. Further identification of CTX-M-2 extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53: 2225–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Celenza G, Pellegrini C, Caccamo M. et al. Spread of bla(CTX-M-type) and bla(PER-2) β-lactamase genes in clinical isolates from Bolivian hospitals. J Antimicrob Chemother 2006; 57: 975–8. [DOI] [PubMed] [Google Scholar]
- 47. Walther-Rasmussen J, Høiby N.. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum β-lactamases. Can J Microbiol 2004; 50: 137–65. [DOI] [PubMed] [Google Scholar]
- 48. Liu CM, Stegger M, Aziz M. et al. Escherichia coli ST131-H22 as a Foodborne Uropathogen. mBio 2018; 9: e00470-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peirano G, Lynch T, Matsumara Y. et al. Trends in population dynamics of Escherichia coli sequence type 131, Calgary, Alberta, Canada, 2006-20161. Emerg Infect Dis 2020; 26: 2907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colmenarejo C, Hernández-García M, Muñoz-Rodríguez JR. et al. Prevalence and risks factors associated with ESBL-producing faecal carriage in a single long-term-care facility in Spain: emergence of CTX-M-24- and CTX-M-27-producing Escherichia coli ST131-H30R. J Antimicrob Chemother 2020; 75: 2480–4. [DOI] [PubMed] [Google Scholar]
- 51. Ghosh H, Doijad S, Falgenhauer L. et al. bla(CTX-M-27)-encoding Escherichia coli sequence type 131 lineage C1-M27 clone in clinical isolates, Germany. Emerg Infect Dis 2017; 23: 1754–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matsumura Y, Pitout JD, Gomi R. et al. Global Escherichia coli sequence type 131 clade with bla(CTX-M-27) gene. Emerg Infect Dis 2016; 22: 1900–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Flament-Simon SC, García V, Duprilot M. et al. High prevalence of ST131 subclades C2-H30Rx and C1-M27 among extended-spectrum β-lactamase-producing Escherichia coli causing human extraintestinal infections in patients from two hospitals of Spain and France during 2015. Front Cell Infect Microbiol 2020; 10: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oliver A, Perez-Diaz JC, Coque TM. et al. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing b-lactamase (CTX-M-10) isolated in Spain. Antimicrob Agents Chemother 2001; 45: 616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Humeniuk C, Arlet G, Gautier V. et al. Β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob Agents Chemother 2002; 46: 3045–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Decousser JW, Poirel L, Nordmann P.. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob Agents Chemother 2001; 45: 3595–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Poirel L, Kämpfer P, Nordmann P.. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob Agents Chemother 2002; 46: 4038–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vimont S, Poirel L, Naas T. et al. Identification of a chromosome-borne expanded-spectrum class a β-lactamase from Erwinia persicina. Antimicrob Agents Chemother 2002; 46: 3401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bellais S, Poirel L, Fortineau N. et al. Biochemical-genetic characterization of the chromosomally encoded extended-spectrum class A β-lactamase from Rahnella aquatilis. Antimicrob Agents Chemother 2001; 45: 2965–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Poirel L, Gniadkowski M, Nordmann P.. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J Antimicrob Chemother 2002; 50: 1031–4. [DOI] [PubMed] [Google Scholar]
- 61. Chen Y, Delmas J, Sirot J. et al. Atomic resolution structures of CTX-M β-lactamases: extended spectrum activities from increased mobility and decreased stability. J Mol Biol 2005; 348: 349–62. [DOI] [PubMed] [Google Scholar]
- 62. Rossolini GM, D'Andrea MM, Mugnaioli C.. The spread of CTX-M-type extended-spectrum β-lactamases. Clin Microbiol Infect 2008; 14: 33–41. [DOI] [PubMed] [Google Scholar]
- 63. Bonnet R, Recule C, Baraduc R. et al. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother 2003; 52: 29–35. [DOI] [PubMed] [Google Scholar]
- 64. Poirel L, de la Rosa J-MO, Richard A. et al. CTX-M-33 is a CTX-M-15 derivative conferring reduced susceptibility to carbapenems. Antimicrob Agents Chemother 2019; 63: e01515-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yoon EJ, Jeong SH.. Class D β-lactamases. J Antimicrob Chemother 2020; 76: 836–64. [DOI] [PubMed] [Google Scholar]
- 66. Evans BA, Amyes SGB.. OXA β-lactamases. Clin Microbiol Rev 2014; 27: 241–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Beceiro A, Maharjan S, Gaulton T. et al. False extended-spectrum b-lactamase phenotype in clinical isolates of Escherichia coli associated with increased expression of OXA-1 or TEM-1 penicillinases and loss of porins. J Antimicrob Chemother 2011; 66: 2006–10. [DOI] [PubMed] [Google Scholar]
- 68. Siu LK, Lo JY, Yuen KY. et al. β-Lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like β-lactamase, OXA-30. Antimicrob Agents Chemother 2000; 44: 2034–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dubois V, Arpin C, Quentin C. et al. Decreased susceptibility to cefepime in a clinical strain of Escherichia coli related to plasmid- and integron-encoded OXA-30 β-lactamase. Antimicrob Agents Chemother 2003; 47: 2380–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boyd DA, Mulvey MR.. OXA-1 is OXA-30 is OXA-1. J Antimicrob Chemother 2006; 58: 224–5. [DOI] [PubMed] [Google Scholar]
- 71. Livermore DM, Day M, Cleary P. et al. OXA-1 β-lactamase and non-susceptibility to penicillin/β-lactamase inhibitor combinations among ESBL-producing Escherichia coli. J Antimicrob Chemother 2019; 74: 326–33. [DOI] [PubMed] [Google Scholar]
- 72. Sun T, Nukaga M, Mayama K. et al. Comparison of β-lactamases of classes A and D: 1.5-A crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci 2003; 12: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aubert D, Poirel L, Chevalier J. et al. Oxacillinase-Mediated Resistance to Cefepime and Susceptibility to Ceftazidime in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2001; 45: 1615–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Poirel L, Castanheira M, Carrër A. et al. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother 2011; 55: 2546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dortet L, Oueslati S, Jeannot K. et al. Genetic and biochemical characterization of OXA-405, an OXA-48-type extended-spectrum β-lactamase without significant carbapenemase activity. Antimicrob Agents Chemother 2015; 59: 3823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zeka AN, Poirel L, Sipahi OR. et al. GES-type and OXA-23 carbapenemase-producing Acinetobacter baumannii in Turkey. J Antimicrob Chemother 2014; 69: 1145–6. [DOI] [PubMed] [Google Scholar]
- 77. Bonnin RA, Nordmann P, Potron A. et al. Carbapenem-hydrolyzing GES-type extended-spectrum β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother 2011; 55: 349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Picão RC, Poirel L, Gales AC. et al. Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob Agents Chemother 2009; 53: 3908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Castanheira M, Mendes RE, Walsh TR. et al. Emergence of the extended-spectrum β-lactamase GES-1 in a Pseudomonas aeruginosa strain from Brazil: report from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother 2004; 48: 2344–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Castanheira M, Costello SE, Woosley LN. et al. Evaluation of clonality and carbapenem resistance mechanisms among Acinetobacter baumannii-Acinetobacter calcoaceticus complex and Enterobacteriaceae isolates collected in European and Mediterranean countries and detection of two novel β-lactamases, GES-22 and VIM-35. Antimicrob Agents Chemother 2014; 58: 7358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Poirel L, Le Thomas I, Naas T. et al. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother 2000; 44: 622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Giakkoupi P, Tzouvelekis LS, Tsakris A. et al. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob Agents Chemother 2000; 44: 2247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Poirel L, Weldhagen GF, Naas T. et al. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother 2001; 45: 2598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mavroidi A, Tzelepi E, Tsakris A. et al. An integron-associated β-lactamase (IBC-2) from Pseudomonas aeruginosa is a variant of the extended-spectrum β-lactamase IBC-1. J Antimicrob Chemother 2001; 48: 627–30. [DOI] [PubMed] [Google Scholar]
- 85. Ortiz de la Rosa JM, Nordmann P, Poirel L.. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother 2019; 74: 1934–9. [DOI] [PubMed] [Google Scholar]
- 86. Sader HS, Rhomberg PR, Flamm RK. et al. WCK 5222 (cefepime/zidebactam) antimicrobial activity tested against Gram-negative organisms producing clinically relevant β-lactamases. J Antimicrob Chemother 2017; 72: 1696–703. [DOI] [PubMed] [Google Scholar]
- 87. Bontron S, Poirel L, Nordmann P.. In vitro prediction of the evolution of GES-1 β-lactamase hydrolytic activity. Antimicrob Agents Chemother 2015; 59: 1664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nordmann P, Ronco E, Naas T. et al. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 1993; 37: 962–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jacoby GA. Β-Lactamase nomenclature. Antimicrob Agents Chemother 2006; 50: 1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bauernfeind A, Stemplinger I, Jungwirth R. et al. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob Agents Chemother 1996; 40: 616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ruggiero M, Papp-Wallace KM, Brunetti F. et al. Structural insights into the inhibition of the extended-spectrum β-lactamase PER-2 by avibactam. Antimicrob Agents Chemother 2019; 63: e00487-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kohira N, Hackel MA, Ishioka Y. et al. Reduced susceptibility mechanism to cefiderocol, a siderophore cephalosporin, among clinical isolates from a global surveillance programme (SIDERO-WT-2014). J Glob Antimicrob Resistance 2020; 22: 738–41. [DOI] [PubMed] [Google Scholar]
- 93. Kolayli F, Gacar G, Karadenizli A. et al. PER-1 is still widespread in Turkish hospitals among Pseudomonas aeruginosa and Acinetobacter spp. FEMS Microbiol Lett 2005; 249: 241–5. [DOI] [PubMed] [Google Scholar]
- 94. Ranellou K, Kadlec K, Poulou A. et al. Detection of Pseudomonas aeruginosa isolates of the international clonal complex 11 carrying the blaPER-1 extended-spectrum β-lactamase gene in Greece. J Antimicrob Chemother 2012; 67: 357–61. [DOI] [PubMed] [Google Scholar]
- 95. Poirel L, Naas T, Guibert M. et al. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother 1999; 43: 573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mushtaq S, Warner M, Livermore DM.. In vitro activity of ceftazidime+NXL104 against Pseudomonas aeruginosa and other non-fermenters. J Antimicrob Chemother 2010; 65: 2376–81. [DOI] [PubMed] [Google Scholar]
- 97. Lahiri SD, Alm RA.. Identification of novel VEB β-lactamase enzymes and their impact on avibactam inhibition. Antimicrob Agents Chemother 2016; 60: 3183–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Naas T, Poirel L, Nordmann P.. Minor extended-spectrum b-lactamases. Clin Microbiol Infect 2008; 14 Suppl 1: 42–52. [DOI] [PubMed] [Google Scholar]
- 99. Li R, Ye L, Zheng Z. et al. Genetic characterization of a blaVEB-2-carrying plasmid in Vibrio parahaemolyticus. Antimicrob Agents Chemother 2016; 60: 6965–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jain S, Gaind R, Kothari C. et al. VEB-1 extended-spectrum β-lactamase-producing multidrug-resistant Proteus mirabilis sepsis outbreak in a neonatal intensive care unit in India: clinical and diagnostic implications. JMM Case Rep 2016; 3: e005056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fevre C, Jbel M, Passet V. et al. Six groups of the OXY β-Lactamase evolved over millions of years in Klebsiella oxytoca. Antimicrob Agents Chemother 2005; 49: 3453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Castanheira M, Farrell SE, Deshpande LM. et al. Prevalence of β-lactamase-encoding genes among Enterobacteriaceae bacteremia isolates collected in 26 U.S. Hospitals: report from the SENTRY antimicrobial surveillance program (2010). Antimicrob Agents Chemother 2013; 57: 3012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Partridge SR, Kwong SM, Firth N. et al. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 2018; 31: e00088–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rodríguez-Baño J, Pascual A.. Clinical significance of extended-spectrum β-lactamases. Expert Rev Anti Infect Ther 2008; 6: 671–83. [DOI] [PubMed] [Google Scholar]
- 105. Partridge SR, Hall RM.. Evolution of transposons containing blaTEM genes. Antimicrob Agents Chemother 2005; 49: 1267–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Heritage J, Hawkey PM, Todd N. et al. Transposition of the gene encoding a TEM-12 extended-spectrum b-lactamase. Antimicrob Agents Chemother 1992; 36: 1981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rasmussen BA, Bradford PA, Quinn JP. et al. Genetically diverse ceftazidime-resistant isolates from a single center: biochemical and genetic characterization of TEM-10 b-lactamases encoded by different nucleotide sequences. Antimicrob Agents Chemother 1993; 37: 1989–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Novais Â, Baquero F, Machado E. et al. International spread and persistence of TEM-24 is caused by the confluence of highly penetrating Enterobacteriaceae clones and an IncA/C2 plasmid containing Tn1696::Tn1 and IS5075-Tn21. Antimicrob Agents Chemother 2010; 54: 825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mabilat C, Lourençao-Vital J, Goussard S. et al. A new example of physical linkage between Tn1 and Tn21: the antibiotic multiple-resistance region of plasmid pCFF04 encoding extended-spectrum β-lactamase TEM-3. Mol Gen Genet 1992; 235: 113–21. [DOI] [PubMed] [Google Scholar]
- 110. Poirel L, Bonnin RA, Nordmann P.. Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods. Infect Genet Evol 2012; 12: 883–93. [DOI] [PubMed] [Google Scholar]
- 111. Marcadé G, Deschamps C, Boyd A. et al. Replicon typing of plasmids in Escherichia coli producing extended-spectrum β-lactamases. J Antimicrob Chemother 2008; 63: 67–71. [DOI] [PubMed] [Google Scholar]
- 112. Chouchani C, El Salabi A, Marrakchi R. et al. Characterization of IncA/C conjugative plasmid harboring bla TEM-52 and bla CTX-M-15 extended-spectrum β-lactamases in clinical isolates of Escherichia coli in Tunisia. Eur J Clin Microbiol Infect Dis 2012; 31: 1081–7. [DOI] [PubMed] [Google Scholar]
- 113. Bielak E, Bergenholtz RD, Jørgensen MS. et al. Investigation of diversity of plasmids carrying the blaTEM-52 gene. J Antimicrob Chemother 2011; 66: 2465–74. [DOI] [PubMed] [Google Scholar]
- 114. Lee KY, Hopkins JD, Syvanen M.. Direct involvement of IS26 in an antibiotic resistance operon. J Bacteriol 1990; 172: 3229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Poirel L, Naas T, Nordmann P.. Genetic support of extended-spectrum b-lactamases. Clin Microbiol Infect 2008; 14 Suppl 1: 75–81. [DOI] [PubMed] [Google Scholar]
- 116. Ford PJ, Avison MB.. Evolutionary mapping of the SHV b-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J Antimicrob Chemother 2004; 54: 69–75. [DOI] [PubMed] [Google Scholar]
- 117. Preston KE, Venezia RA, Stellrecht KA.. The SHV-5 extended-spectrum β-lactamase gene of pACM1 is located on the remnant of a compound transposon. Plasmid 2004; 51: 48–53. [DOI] [PubMed] [Google Scholar]
- 118. Garza-Ramos U, Davila G, Gonzalez V. et al. The blaSHV-5 gene is encoded in a compound transposon duplicated in tandem in Enterobacter cloacae. Clin Microbiol Infect 2009; 15: 878–80. [DOI] [PubMed] [Google Scholar]
- 119. Liakopoulos A, Mevius D, Ceccarelli D.. A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front Microbiol 2016; 7: 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Billard-Pomares T, Fouteau S, Jacquet ME. et al. Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob Agents Chemother 2014; 58: 6550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Poirel L, Decousser J-W, Nordmann P.. Insertion sequence IS Ecp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother 2003; 47: 2938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lartigue M-F, Poirel L, Aubert D. et al. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob Agents Chemother 2006; 50: 1282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Eckert C, Gautier V, Arlet G.. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother 2006; 57: 14–23. [DOI] [PubMed] [Google Scholar]
- 124. Rodriguez MM, Power P, Radice M. et al. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob Agents Chemother 2004; 48: 4895–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Toleman MA, Bennett PM, Walsh TR.. ISCR elements: novel gene-capturing systems of the 21st Century? Microbiol Mol Biol Rev 2006; 70: 296–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Partridge SR, Zong Z, Iredell JR.. Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli. Antimicrob Agents Chemother 2011; 55: 4971–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Diestra K, Juan C, Curiao T. et al. Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother 2009; 63: 60–6. [DOI] [PubMed] [Google Scholar]
- 128. Coque TM, Novais A, Carattoli A. et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis 2008; 14: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cantón R, Coque TM.. The CTX-M β-lactamase pandemic. Curr Opin Microbiol 2006; 9: 466–75. [DOI] [PubMed] [Google Scholar]
- 130. Banerjee R, Johnson JR.. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 2014; 58: 4997–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Nicolas-Chanoine M-H, Bertrand X, Madec J-Y.. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 2014; 27: 543–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Courpon-Claudinon A, Lefort A, Panhard X. et al. Bacteraemia caused by third-generation cephalosporin-resistant Escherichia coli in France: prevalence, molecular epidemiology and clinical features. Clin Microbiol Infect 2011; 17: 557–65. [DOI] [PubMed] [Google Scholar]
- 133. Peirano G, Richardson D, Nigrin J. et al. High prevalence of st131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-β-lactamase-producing Escherichia coli isolates from Canada. Antimicrob Agents Chemother 2010; 54: 1327–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pitout JDD, Laupland KB, Church DL. et al. Virulence factors of Escherichia coli isolates that produce CTX-M-type extended-spectrum β-lactamases. Antimicrob Agents Chemother 2005; 49: 4667–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Johnson JR, Johnston B, Clabots C. et al. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 2010; 51: 286–94. [DOI] [PubMed] [Google Scholar]
- 136. Mendes RE, Jones RN, Woosley LN. et al. Application of next-generation sequencing for characterization of surveillance and clinical trial isolates: analysis of the distribution of β-lactamase resistance genes and lineage background in the United States. Open Forum Infect Dis 2019; 6: S69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. De La Cadena E, Mojica MF, Castillo N. et al. Genomic analysis of CTX-M-group-1-producing extraintestinal pathogenic E. coli (ExPEc) from patients with urinary tract infections (UTI) from Colombia. Antibiotics (Basel) 2020; 9: 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Johnson TJ, Elnekave E, Miller EA. et al. Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. Antimicrob Agents Chemother 2019; 63: e01913–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Peirano G, Chen L, Kreiswirth BN. et al. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob Agents Chemother 2020; 64: e01148–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tchesnokova VL, Rechkina E, Larson L. et al. Rapid and extensive expansion in the United States of a new multidrug-resistant Escherichia coli clonal group, sequence type 1193. Clin Infect Dis 2019; 68: 334–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Xia L, Liu Y, Xia S. et al. Prevalence of ST1193 clone and IncI1/ST16 plasmid in E. coli isolates carrying bla(CTX-M-55) gene from urinary tract infections patients in China. Sci Rep 2017; 7: 44866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kim Y, Oh T, Nam YS. et al. Prevalence of ST131 and ST1193 among bloodstream isolates of Escherichia coli not susceptible to ciprofloxacin in a tertiary care university hospital in Korea, 2013 - 2014. Clin Lab 2017; 63: 1541–3. [DOI] [PubMed] [Google Scholar]
- 143. Valenza G, Werner M, Eisenberger D. et al. First report of the new emerging global clone ST1193 among clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Germany. J Glob Antimicrob Resist 2019; 17: 305–8. [DOI] [PubMed] [Google Scholar]
- 144. Mantengoli E, Rossolini GM.. Tn5393d, a complex Tn5393 derivative carrying the PER-1 extended-spectrum β-lactamase gene and other resistance determinants. Antimicrob Agents Chemother 2005; 49: 3289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bonnin RA, Potron A, Poirel L. et al. PER-7, an extended-spectrum β-lactamase with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob Agents Chemother 2011; 55: 2424–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Villegas MV, Correa A, Perez F. et al. Prevalence and characterization of extended-spectrum b-lactamases in Klebsiella pneumoniae and Escherichia coli isolates from Colombian hospitals. Diagn Microbiol Infect Dis 2004; 49: 217–22. [DOI] [PubMed] [Google Scholar]
- 147. Hawkey PM. Prevalence and clonality of extended-spectrum b-lactamases in Asia. Clin Microbiol Infect 2008; 14 Suppl 1: 159–65. [DOI] [PubMed] [Google Scholar]
- 148. Rojas LJ, Weinstock GM, De La Cadena E. et al. An analysis of the epidemic of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: convergence of two evolutionary mechanisms creates the "Perfect Storm". J Infect Dis 2017; 217: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bevan ER, Jones AM, Hawkey PM.. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 2017; 72: 2145–55. [DOI] [PubMed] [Google Scholar]
- 150. Lewis JS, Herrera M, Wickes B. et al. First report of the emergence of CTX-M-type Extended-Spectrum β-Lactamases (ESBLs) as the predominant ESBL isolated in a U.S. Health care system. Antimicrob Agents Chemother 2007; 51: 4015–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Castanheira M, Mendes RE, Rhomberg PR. et al. Rapid emergence of blaCTX-M among Enterobacteriaceae in U.S. Medical Centers: molecular evaluation from the MYSTIC program (2007). Microb Drug Resist 2008; 14: 211–6. [DOI] [PubMed] [Google Scholar]
- 152. Jernigan JA, Hatfield KM, Wolford H. et al. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012-2017. N Engl J Med 2020; 382: 1309–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Tamma PD, Smith TT, Adebayo A. et al. Prevalence of blaCTX-M genes in Gram-negative bloodstream isolates across 66 hospitals in the United States. J Clin Microbiol 2021; 59: e00127–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Livermore DM, Canton R, Gniadkowski M. et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother 2007; 59: 165–74. [DOI] [PubMed] [Google Scholar]
- 155. Díaz MA, Hernández-Bello JR, Rodríguez-Baño J. et al. Diversity of Escherichia coli strains producing extended-spectrum β-lactamases in Spain: second nationwide study. J Clin Microbiol 2010; 48: 2840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Blanco J, Mora A, Mamani R. et al. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother 2011; 66: 2011–21. [DOI] [PubMed] [Google Scholar]
- 157. Blanco J, Mora A, Mamani R. et al. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 2013; 51: 3358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Rodriguez-Villalobos H, Bogaerts P, Berhin C. et al. Trends in production of extended-spectrum b-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J Antimicrob Chemother 2011; 66: 37–47. [DOI] [PubMed] [Google Scholar]
- 159. Peirano G, Sang JH, Pitondo-Silva A. et al. Molecular epidemiology of extended-spectrum-β-lactamase-producing Klebsiella pneumoniae over a 10 year period in Calgary, Canada. J Antimicrob Chemother 2012; 67: 1114–20. [DOI] [PubMed] [Google Scholar]
- 160. Castanheira M, Mendes RE, Jones RN. et al. Changes in the frequencies of β-lactamase genes among Enterobacteriaceae isolates in U.S. Hospitals, 2012 to 2014: activity of ceftazidime-avibactam tested against β-lactamase-producing isolates. Antimicrob Agents Chemother 2016; 60: 4770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Croughs PD, Klaassen CHW, van Rosmalen J. et al. Unexpected mechanisms of resistance in Dutch Pseudomonas aeruginosa isolates collected during 14 years of surveillance. Int J Antimicrob Agents 2018; 52: 407–10. [DOI] [PubMed] [Google Scholar]
- 162. Laudy AE, Róg P, Smolińska-Król K. et al. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS One 2017; 12: e0180121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Polotto M, Casella T, de Lucca Oliveira MG. et al. Detection of P. aeruginosa harboring bla CTX-M-2, bla GES-1 and bla GES-5, bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis 2012; 12: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Royer G, Fourreau F, Boulanger B. et al. Local outbreak of extended-spectrum β-lactamase SHV2a-producing Pseudomonas aeruginosa reveals the emergence of a new specific sub-lineage of the international ST235 high-risk clone. J Hosp Infect 2020; 104: 33–9. [DOI] [PubMed] [Google Scholar]
- 165. Mansour W, Dahmen S, Poirel L. et al. Emergence of SHV-2a extended-spectrum b-lactamases in clinical isolates of Pseudomonas aeruginosa in a university hospital in Tunisia. Microb Drug Resist 2009; 15: 295–301. [DOI] [PubMed] [Google Scholar]
- 166. Castanheira M, Doyle TB, Smith CJ. et al. Combination of MexAB-OprM overexpression and mutations in efflux regulators, PBPs and chaperone proteins is responsible for ceftazidime/avibactam resistance in Pseudomonas aeruginosa clinical isolates from US hospitals. J Antimicrob Chemother 2019; 74: 2588–95. [DOI] [PubMed] [Google Scholar]
- 167. Vahaboglu H, Coskunkan F, Tansel O. et al. Clinical importance of extended-spectrum b-lactamase (PER-1-type)-producing Acinetobacter spp. and Pseudomonas aeruginosa strains. J Med Microbiol 2001; 50: 642–5. [DOI] [PubMed] [Google Scholar]
- 168. Endimiani A, Luzzaro F, Migliavacca R. et al. Spread in an Italian hospital of a clonal Acinetobacter baumannii strain producing the TEM-92 extended-spectrum β-lactamase. Antimicrob Agents Chemother 2007; 51: 2211–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Potron A, Munoz-Price LS, Nordmann P. et al. Genetic features of CTX-M-15-producing Acinetobacter baumannii from Haiti. Antimicrob Agents Chemother 2011; 55: 5946–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Poirel L, Menuteau O, Agoli N. et al. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J Clin Microbiol 2003; 41: 3542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Feng Y, Yang P, Wang X. et al. Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J Antimicrob Chemother 2016; 71: 71–5. [DOI] [PubMed] [Google Scholar]
- 172. Naas T, Bogaerts P, Bauraing C. et al. Emergence of PER and VEB extended-spectrum b-lactamases in Acinetobacter baumannii in Belgium. J Antimicrob Chemother 2006; 58: 178–82. [DOI] [PubMed] [Google Scholar]
- 173. Woerther PL, Burdet C, Chachaty E. et al. Trends in human fecal carriage of extended-spectrum b-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 2013; 26: 744–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Reuland EA, Al Naiemi N, Kaiser AM. et al. Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in Amsterdam. J Antimicrob Chemother 2016; 71: 1076–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Tschudin-Sutter S, Lucet J-C, Mutters NT. et al. Contact precautions for preventing nosocomial transmission of extended-spectrum β lactamase–producing Escherichia coli: a point/counterpoint review. Clin Infect Dis 2017; 65: 342–7. [DOI] [PubMed] [Google Scholar]
- 176. Hilty M, Betsch BY, Bogli-Stuber K. et al. Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis 2012; 55: 967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Nakamura K, Kaneko M, Abe Y. et al. Outbreak of extended-spectrum b-lactamase-producing Escherichia coli transmitted through breast milk sharing in a neonatal intensive care unit. J Hosp Infect 2016; 92: 42–6. [DOI] [PubMed] [Google Scholar]
- 178. McDanel J, Schweizer M, Crabb V. et al. Incidence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella infections in the United States: a systematic literature review. Infect Control Hosp Epidemiol 2017; 38: 1209–15. [DOI] [PubMed] [Google Scholar]
- 179. Sader HS, Pfaller MA, Jones RN.. Prevalence of important pathogens and the antimicrobial activity of parenteral drugs at numerous medical centers in the United States. II. Study of the intra- and interlaboratory dissemination of extended-spectrum β-lactamase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 1994; 20: 203–8. [DOI] [PubMed] [Google Scholar]
- 180. De Champs C, Sirot D, Chanal C. et al. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. The French Study Group. Antimicrob Agents Chemother 2000; 44: 3177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Doi Y, Iovleva A, Bonomo RA.. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J Travel Med 2017; 24: S44–S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Bush K, Bradford PA.. Epidemiology of β-lactamase-producing pathogens. Clin Microbiol Rev 2020; 33: e00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Thirtieth Edition: M100. 2020.
- 184. EUCAST. Guidelines for Detection of Resistance Mechanisms and Specific Resistance of Clinical and/or Epidemiological Importance, Version 2.0, 2017. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf.
- 185. El-Jade MR, Parcina M, Schmithausen RM. et al. ESBL detection: comparison of a commercially available chromogenic test for third generation cephalosporine resistance and automated susceptibility testing in Enterobactericeae. PLoS One 2016; 11: e0160203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Spanu T, Sanguinetti M, Tumbarello M. et al. Evaluation of the new VITEK 2 Extended-Spectrum Beta-Lactamase (ESBL) test for rapid detection of ESBL production in Enterobacteriaceae isolates. J Clin Microbiol 2006; 44: 3257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Thomson KS, Cornish NE, Hong SG. et al. Comparison of phoenix and VITEK 2 Extended-Spectrum-β-Lactamase Detection Tests for Analysis of Escherichia coli and Klebsiella isolates with well-characterized β-lactamases. J Clin Microbiol 2007; 45: 2380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Dudley MN, Ambrose PG, Bhavnani SM. et al. Background and rationale for revised clinical and laboratory standards institute interpretive criteria (Breakpoints) for Enterobacteriaceae and Pseudomonas aeruginosa: I. Cephalosporins and Aztreonam. Clin Infect Dis 2013; 56: 1301–9. [DOI] [PubMed] [Google Scholar]
- 189. EUCAST. Clinical breakpoints. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf.
- 190. Harris PA, Tambyah PA, Lye DC. et al. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E. coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial. JAMA 2018; 320: 984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Huang Y, Carroll KC, Cosgrove SE. et al. Determining the optimal ceftriaxone MIC for triggering extended-spectrum β-lactamase confirmatory testing. J Clin Microbiol 2014; 52: 2228–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Tamma PD, Aitken SL, Bonomo RA. et al. Infectious diseases society of America guidance on the treatment of extended-spectrum β-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin Infect Dis 2021; 72: e169–83. [DOI] [PubMed] [Google Scholar]
- 193. Tamma PD, Cosgrove SE, Avdic E. et al. Reporting extended-spectrum β-lactamase positivity may reduce carbapenem overuse. Open Forum Infect Dis 2019; 6: ofz064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Villegas MV, Esparza G, Reyes J.. Should ceftriaxone-resistant Enterobacterales be tested for ESBLs? A PRO/CON debate. JAC-Antimicrob Resist 2021; 3: dlab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Tamma PD, Humphries RM.. PRO: Testing for ESBL production is necessary for ceftriaxone-non-susceptible Enterobacterales: perfect should not be the enemy of progress. JAC-Antimicrob Resist 2021; 3: dlab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Mathers AJ, Lewis JS II. CON: Testing for ESBL production is unnecessary for ceftriaxone-resistant Enterobacterales. JAC-Antimicrob Resist 2021; 3: dlab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Nordmann P, Dortet L, Poirel L.. Rapid detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 2012; 50: 3016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Gallah S, Benzerara Y, Tankovic J. et al. β LACTA test performance for detection of extended-spectrum β-lactamase-producing Gram-negative bacilli directly on bronchial aspirates samples: a validation study. Clin Microbiol Infect 2018; 24: 402–8. [DOI] [PubMed] [Google Scholar]
- 199. Morosini MI, García-Castillo M, Tato M. et al. Rapid detection of β-lactamase-hydrolyzing extended-spectrum cephalosporins in Enterobacteriaceae by use of the new chromogenic βLacta test. J Clini Microbiol 2014; 52: 1741–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Poirel L, Fernández J, Nordmann P.. Comparison of three biochemical tests for rapid detection of extended-spectrum-β-Lactamase-producing Enterobacteriaceae. J Clin Microbiol 2016; 54: 423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Renvoisé A, Decré D, Amarsy-Guerle R. et al. Evaluation of the βLacta Test, a rapid test detecting resistance to third-generation cephalosporins in clinical strains of Enterobacteriaceae. J Clin Microbiol 2013; 51: 4012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Bernabeu S, Ratnam KC, Boutal H. et al. A lateral flow immunoassay for the rapid identification of CTX-M-producing Enterobacterales from culture plates and positive blood cultures. Diagnostics (Basel) 2020; 10: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Bianco G, Boattini M, Iannaccone M. et al. Evaluation of the NG-Test CTX-M MULTI immunochromatographic assay for the rapid detection of CTX-M extended-spectrum-β-lactamase producers from positive blood cultures. J Hosp Infect 2020; 105: 341–3. [DOI] [PubMed] [Google Scholar]
- 204. Cortazzo V, D’Inzeo T, Giordano L. et al. Comparing BioFire FilmArray BCID2 and BCID panels for direct detection of bacterial pathogens and antimicrobial resistance genes from positive blood cultures. J Clinl Microbiol 2021; 59: e03163–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Rödel J, Karrasch M, Edel B. et al. Antibiotic treatment algorithm development based on a microarray nucleic acid assay for rapid bacterial identification and resistance determination from positive blood cultures. Diagn Microbiol Infect Dis 2016; 84: 252–7. [DOI] [PubMed] [Google Scholar]
- 206. Klein M, Bacher J, Barth S. et al. Multicenter evaluation of the Unyvero platform for testing bronchoalveolar lavage fluid. J Clin Microbiol 2021; 59: e02497–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Murphy CN, Fowler R, Balada-Llasat JM. et al. Multicenter evaluation of the BioFire FilmArray pneumonia/pneumonia plus panel for detection and quantification of agents of lower respiratory tract infection. J Clin Microbiol 2020; 58: e00128–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Sanguinetti M, Posteraro B, Spanu T. et al. Characterization of clinical isolates of Enterobacteriaceae from Italy by the BD phoenix extended-spectrum β-lactamase detection method. J Clin Microbiol 2003; 41: 1463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Tamma PD, Sharara SL, Pana ZD. et al. Molecular epidemiology of ceftriaxone non-susceptible Enterobacterales isolates in an academic medical center in the United States. Open Forum Infect Dis 2019; 6: ofz353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Rood IGH, Li Q.. Review: Molecular detection of extended spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae in a clinical setting. Diagn Microbiol Infect Dis 2017; 89: 245–50. [DOI] [PubMed] [Google Scholar]
- 211. Peirano G, Pitout JDD.. Extended-spectrum b-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 2019; 79: 1529–41. [DOI] [PubMed] [Google Scholar]
- 212. Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I. et al. Treatment of infections caused by extended-spectrum-β-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev 2018; 31: e00079–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Harris PNA, Tambyah PA, Paterson DL.. b-lactam and b-lactamase inhibitor combinations in the treatment of extended-spectrum b-lactamase producing Enterobacteriaceae: time for a reappraisal in the era of few antibiotic options? Lancet Infect Dis 2015; 15: 475–85 [DOI] [PubMed] [Google Scholar]
- 214. Muhammed M, Flokas ME, Detsis M. et al. Comparison between carbapenems and β-lactam/β-lactamase inhibitors in the treatment for bloodstream infections caused by extended-spectrum β-lactamase-producing Enterobacteriaceae: a systematic review and meta-analysis. Open Forum Infect Dis 2017; 4: ofx099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Palacios-Baena ZR, Gutiérrez-Gutiérrez B, De Cueto M. et al. Development and validation of the INCREMENT-ESBL predictive score for mortality in patients with bloodstream infections due to extended-spectrum-β-lactamase-producing Enterobacteriaceae. J Antimicrob Chemother 2016; 72: 906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Pascual A.. CON: carbapenems are NOT necessary for all infections caused by ceftriaxone-resistant Enterobacterales. JAC-Antimicrob Resist 2021; 3: dlaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Henderson A, Humphries R.. Building a better test for piperacillin-tazobactam susceptibility testing: would that it were so simple (it's complicated). J Dlin Microbiol 2020; 58: e01649–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Henderson A, Paterson DL, Chatfield MD. et al. Association between minimum inhibitory concentration, β-lactamase genes and mortality for patients treated with piperacillin/tazobactam or meropenem from the MERINO study. Clin Infect Dis 2020; ciaa1479. [DOI] [PubMed] [Google Scholar]
- 219. Paterson DL, Isler B, Harris PNA.. PRO: Carbapenems should be used for ALL infections caused by ceftriaxone-resistant Enterobacterales. JAC-Antimicrob Res 2021; 3: dlab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Tamma PD, Mathers AJ.. Navigating treatment approaches for presumed ESBL-producing infections. JAC-Antimicrob Res 2021; 3: dlaa111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Popejoy MW, Paterson DL, Cloutier D. et al. Efficacy of ceftolozane/tazobactam against urinary tract and intra-abdominal infections caused by ESBL-producing Escherichia coli and Klebsiella pneumoniae: a pooled analysis of Phase 3 clinical trials. J Antimicrob Chemother 2017; 72: 268–72. [DOI] [PubMed] [Google Scholar]
- 222. Isler B, Ezure Y, Romero J-F. et al. Is ceftazidime/avibactam an option for serious infections due to extended-spectrum-β-Lactamase- and AmpC-producing Enterobacterales: a systematic review and meta-analysis. Antimicrob Agents Chemother 2020; 65: e01052–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Zhanel GG, Lawrence CK, Adam H. et al. Imipenem-relebactam and meropenem-vaborbactam: two novel carbapenem-β-lactamase inhibitor combinations. Drugs 2018; 78: 65–98. [DOI] [PubMed] [Google Scholar]
- 224. Madeira F, Park Y, Lee J. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 2019; 47: W636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Sievers F, Wilm A, Dineen D. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Goujon M, McWilliam H, Li W. et al. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res 2010; 38: W695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Ambler RP. The Structure of b-Lactamases. Philos Trans R Soc Lond B, Biol Sci 1980; 289: 321–31. [DOI] [PubMed] [Google Scholar]