Abstract
Klebsiella aerogenes is a multidrug-resistant Gram-negative bacterium that causes nosocomial infections. The organism showed resistance to most of the conventional antibiotics available. Because of the high resistance of the species, the treatment of K. aerogenes is difficult. These species are resistant to third-generation cephalosporins due to the production of chromosomal beta-lactams with cephalosporin activity. The lack of better treatment and the development of therapeutic resistance in hospitals hinders better/new broad-spectrum-based treatment against this pathogen. This study identifies potential drug targets/vaccine candidates through a computational subtractive proteome-driven approach. This method is used to predict proteins that are not homologous to humans and human symbiotic intestinal flora. The resultant proteome of K. aerogenes was further searched for proteins, which are essential, virulent, and determinants of antibiotic/drug resistance. Subsequently, their druggability properties were also studied. The data set was reduced based on its presence in the pathogen-specific metabolic pathways. The subtractive proteome analysis predicted 13 proteins as potential drug targets for K. aerogenes. Furthermore, these target proteins were annotated based on their spectrum of activity, cellular localization, and antigenicity properties, which ensured that they are potent candidates for broad-spectrum antibiotic and vaccine design. The results open up new opportunities for designing and manufacturing powerful antigenic vaccines against K. aerogenes and the detection and release of new and active drugs against K. aerogenes without altering the gut microbiome.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42485-021-00068-9.
Keywords: Enterobacter/Klebsiella infections, Subtractive proteome analysis, Drug targets, Vaccine candidates
Introduction
Klebsiella aerogenes from the Enterobacteriaceae family is a multi-resistant, Gram-negative bacterial pathogen that is commonly found and is part of the normal human intestinal tract flora. It is the most common cause of hospital ward infection and has also been identified as a significant and versatile opportunistic pathogen for humans (Fok et al. 1998; Davin-Regli 2015). In the recent past, it has been reported that this species is the primary cause of high mortality and morbidity (Jacoby 2009). It mainly originates in intensive care patients, especially those with mechanical ventilation. It is known for its role in hospital infections, such as bacteremia, pneumonia, surgical infections, meningitis, skin and soft tissue infections, infections of the lower respiratory tract, endocarditis, intra-abdominal infections, urinary tract infections, infections of the central nervous system, infections of the eye, osteomyelitis, and septic arthritis. Enterobacter infection management options are relatively limited (Mezzatesta et al.2012; Langley et al. 2001; Sanders and Sanders 1997).
The overuse of antibiotics has increased K. aerogenes infections in hospital wards (Arpin et al.1996; Anastay et al. 2013). It is mainly known for its resistance to many drugs against a wide variety of antimicrobials. K. aerogenes has progressively been reported, leading to MDR isolates (Chang et al. 1990). According to previous studies, K. aerogenes is resistant to beta-lactam antibiotics and extended-spectrum antibiotics (Arpin et al. 2003; Cantón et al. 2002; Charrel et al. 1996). Membrane permeability, efflux mechanisms, and enzyme degradation are the primary reasons for increased resistance to antibiotics (Nikaido 2003; Pagès et al. 2008). With the emergence of multidrug resistance and the lack of optimal treatment, there is a constant need to discover new drugs to prevent these infections. Identifying new drug targets is one of the key steps in the drug discovery process. The current study identifies promising drug/vaccine targets using a systematic "in silico" approach that can be extended to other life-threatening pathogens. Recently, species-specific targets of drugs and vaccines for many pathogenic bacteria have been identified using subtractive proteomics approaches (Uddin and Jamil 2018; Sarangi et al. 2009; Mondal et al. 2015).
Traditional drug detection methods are time-consuming and yield just a few drug targets. In recent times, computational methods and omics data have been an exciting way to find new drug targets and reduce drug failure rates in the last stages of clinical trials (Reker et al. 2018). Access to the entire genome sequence of specific pathogens and humans dramatically simplifies the search for new target candidates. Subtractive and comparative proteomics has become the method of choice for identifying potential drug targets against various life-threatening pathogenic bacteria (Bottacini et al. 2014; Cava et al. 2017; Perumal et al. 2007). These goal-setting methods consider essential characteristics as the major criteria for selecting medical candidates. To prevent disruptive interactions, passive therapeutic targets are unique to the bacterial pathogen. Targeting pathogen-specific proteins will aid in the prevention of bacterial infection.
This manuscript presents a new "in silico" approach that incorporates various computer technologies to identify and characterize therapeutic candidates for K. aerogenes. The first list of proteins for K. aerogenes was determined by scanning their entire proteome against the human gut microbiome. This was then filtered to find a potential drug target. Various "in silico" tools have been used to identify proteins important for pathogen survival. Non homologous and metabolic pathway analysis was performed to prevent both pathogenic proteins and pathogens from being involved in the host’s metabolism. The characteristics of the candidate targets predict their location within the bacterial cell, their ability to function as a broad-spectrum target, their cellular function, and their antigenic properties. Current approaches allow the identification of vaccine and drug candidates based on essential and favorable properties.
Materials and methods
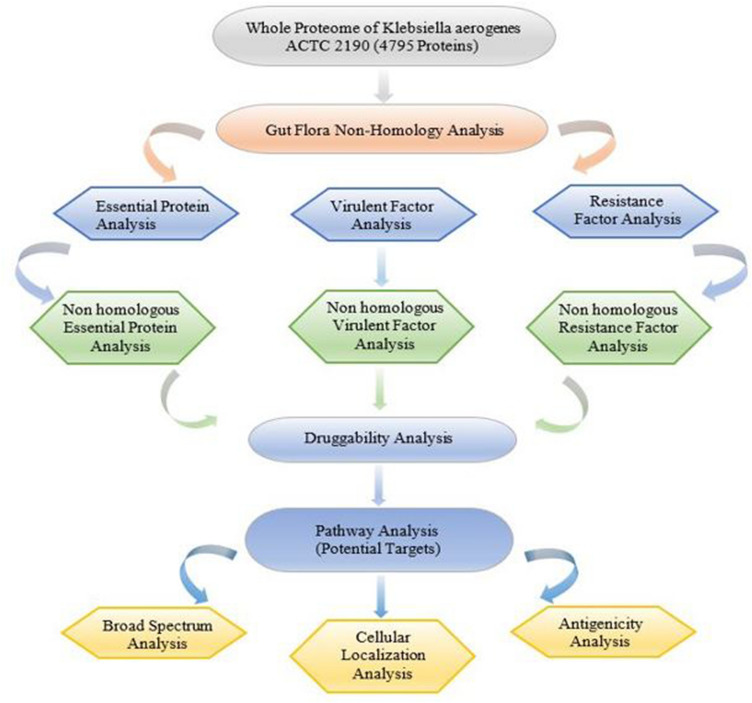
The entire K. aerogenes KCTC 2190 proteome was investigated using subtractive proteomics techniques to understand novel drug targets and vaccine applicants. The overall workflow is outlined in Fig. 1.
Fig. 1.
Schematic representation of the workflow. The flowchart summarizes the various steps that were used to identify and characterize potential drug targets in the study
Data collection of proteome
The complete protein sequence or proteome of K. aerogenes strain KCTC 2190, containing 4795 protein sequences, was downloaded from the database of the National Center for Biotechnology Information (NCBI) (Pruitt et al. 2007).
Non-homology sequence analysis against gut microflora
The proteome of K. aerogenes was subjected to BLASTP (Altschul et al. 1990) against the proteome of 83 gut microflora reported in the literature (Shanmugham and Pan 2013). K. aerogenes proteins with a similarity of less than 50% and a cutoff E value of 0.001 were regarded as non-homologous with the proteome of the intestinal microflora (Shende et al. 2017) and were subjected to further analysis to find essential, virulent, and resistant proteins.
Essentiality analysis
The Essential Genes Database (Luo et al. 2014) (DEG) (http://www.essentialgene.org/) contained more than 22,343 experimentally identified essential/necessary genes and was used to extract proteins of K. aerogenes. The default BLASTP against DEG was done by placing non-homologous proteins obtained in the previous step using an E value cutoff value of 0.0001 (Sharma and Pan 2012), based on the fact that similar proteins in different organisms are probably equally required.
Virulence factor analysis
The Virulence Factor Database (Liu et al. 2019) (http://www.mgc.ac.cn/VFs/search_VFs.htm) comprises four classes of Virulence Factors (VFs), specifically offensive, defensive, non-specific, and pathogenic. Bacterial virulence-related genes were used to retrieve the virulence factors of K. aerogenes. For screening virulent factors, the non-homologous gut microflora protein subset was subjected to a predetermined BLASTP against virulence factors in VFDB with an E value cutoff of 0.0001.
Resistance factor analysis
The BacMet2.0 database (Pal et al. 2014) (http://bacmet.biomedicine.gu.se/) is a manually organized bacterial gene/protein database, and contains experimentally confirmed resistance genes/proteins and was used to extract resistant proteins of K. aerogenes. Non-homologous gut microflora proteins were subjected to default BLASTP screening against antibiotic-resistant sequences in the BacMet database with an E value cutoff of 0.0001 to classify resistant proteins.
Non-homology analysis against the human host
To prevent drug binding to human hosts and highly undesirable cross-reactivity, the generated output contains non-redundant proteins subjected to a sequence similarity search against human proteins using BLASTP. BLAST screening is set by default to an E value range of 0.001 (Anishetty et al. 2005; Sarkar et al. 2012).
Druggability analysis
DrugBank 5.1.6 (Wishart et al. 2018) (https://www.drugbank.ca/releases/5-1-6) includes 13,577 drugs, including 2634 approved low molecular weight drugs, 131 nutraceuticals, 1377 and about 6375 investigational drugs (discovery phase). The druggability of the resulting data set of K. aerogenes was analyzed using BLASTP search against the DrugBank 5.1.6, with a cutoff E value of 0.0001.
Analyzing the metabolic pathway of K. aerogenes
KEGG (Kanehisa et al. 2017) (https://www.genome.jp/kegg/) is a pathway database that contains curated metabolic pathways of proteins of all organisms. KEGG was used to retrieve all specific pathways of K. aerogenes and H. sapiens with their corresponding organism codes; ‘eae’ and ‘hsa’ for K. aerogenes and H. sapiens. Then, by manually comparing pathogen and host metabolic pathways, unique and common pathways were listed separately. The identified K. aerogenes drug targets were then submitted to BLASTP via the KAAS server to obtain information on the various biological processes and metabolic pathways involving putative drug targets (Moriya et al. 2007). Proteins present only in unique metabolic pathways of pathogens were detected, and proteins involved in both or only the host's pathways were not included.
Broad-spectrum analysis
A list of 240 disease-causing microorganisms from distinct classes that are detailed in the literature (Raman et al. 2008) was utilized for the broad-spectrum analysis. A subset of proteins acquired from the pathway analysis was analyzed using BLASTP search against these pathogenic micro-organisms with an E value of 0.0001, which can be used in broad-spectrum drug/vaccine design.
Proteins subcellular localization prediction
Since K. aerogenes is a Gram-negative bacterium, the protein of this bacterium can be distinguished in five cell locations: extracellular space, cytoplasmic, periplasmic, outer membrane, and inner membrane. To predict the intracellular localization of pre-selected proteins, PSORTbv3.0.2 (Yu et al. 2010) and CELLOv.2.5 (Yu et al. 2006), servers were used. The highest score generated by these two servers was used to recognize the subcellular localization of the proteins.
Antigenicity prediction
To predict possible antigens and subunit vaccines, the identified target proteins were analyzed by the VaxiJenv2.0 server. We chose VaxiJen as it is a reliable and consistent tool for the prediction of protective antigens with a reported accuracy of 82% (Doytchinova and Flower 2007). This server was developed to classify antigens based on physicochemical properties. Protein sequences with antigenicity greater than 0.5 were considered antigens.
Results and discussion
In this study, we adopted subtractive proteome-based analysis to categorize novel drug targets as well as vaccine candidates against K. aerogenes KCTC 2190 using various databases and computational tools. The phases of the sequential analysis are listed below (Table 1).
Table 1.
Subtractive proteome analysis for finding drug targets
| S. no | Steps | K. aerogenes ACTC 2190 |
|---|---|---|
| 1 | Total number of proteins | 4795 |
| 2 | Number of non-homologous proteins to gut microflora using BLASTP | 1142 |
| 3 | Essential proteins | 478 |
| 4 | Virulent proteins | 332 |
| 5 | Resistance causing proteins | 131 |
| 6 | Non-homologous gut flora essential proteins | 347 |
| 7 | Non-homologous gut flora virulent proteins | 256 |
| 8 | Non-homologous gut flora resistance proteins | 97 |
| 9 | A total non-redundant set of proteins non-homologous to H. sapiens | 445 |
| 10 | Druggable proteins | 144 |
| 11 | Proteins involved in pathogen metabolic pathways (KAAS) | 13 |
| 12 | Broad-spectrum analysis | 13 |
| 13 | Localization using PSORTb, CELLO | 13 |
| 14 | Proteins show antigenicity using VaxiJen v2.0/vaccine candidates | 5 |
Non-homology analysis against gut microflora
The NCBI was used to download the full proteome of K. aerogenes, which included 4795 protein sequences, and compare it to the proteome of the human gut microbiota. Gut microflora plays an essential part in metabolism by fermenting particles of food and protecting the intestine from colonization by pathogenic bacteria (Rabizadeh and Sears 2008). Intestinal obstruction of the gut microflora proteins has an unfavorable impact on the host. To maintain a strategic distance from such conditions, the proteome was subjected to a homology-based search against 83 intestine gut-flora proteomes utilizing BLASTP with an E value edge of 0.0001. Within the data set of 4795 proteins, we found 1142 non-homologous proteins for the gut microflora proteome, based on an E value estimation of 0.0001 and an identity of < 50 percent. The proteins following the above models were selected for further examination and the remaining were excluded from the investigation.
Essentiality analysis
Antibacterial drugs are typically intended to dock with essential bacterial proteins, which have proven to be the most effective therapeutic targets. 1142 non-homologous proteins were screened with an E value cutoff of 0.0001 using a DEG database for BLASTP searches. Out of 1142 proteins, only 478 were found to be essential for K. aerogenes. The rest of the proteins without any hits against the DEG database were then considered non-essential and were excluded from further analysis.
Virulence factor analysis
Virulence factors produced by pathogenic microorganisms are essential proteins that cause undesired harm to the host. This protein serves as a "guard unit" for assaulting the immune system of the host (Segovia et al. 2018). Hence, these proteins should be prioritized as they serve as possible drug and immunogenic-based vaccine candidates (Mora et al. 2006). Here, we tried predicting VFs by searching the database for virulence factors. 332 proteins were found to be virulence factors with a bit score of 100 out of 1142 non-homologous proteins and were considered for further analysis.
Resistance factor analysis
Pathogenic bacteria are developing resistance to antibiotics due to the indiscriminate use of traditional antibiotics. Proteins that enhance resistance can serve as good therapeutic drug targets (Padiadpu et al. 2010). Here, we got 131 antibiotics-resistant proteins in K. aerogenes with a cutoff value of 0.0001.
Non-homology analysis against humans
For human hosts to avoid side effects of the drug complex, it is critical to elucidate remedial targets specific to the pathogen. For the identification of such proteins, listed data sets of essential, virulent, and antimicrobial-resistant proteins were exposed to homology-based search over the entire protein set of H. sapiens utilizing BLASTP with an E value cutoff of 0.001. Therefore, proteins that tend to be identical to host human proteins were omitted from the list. There were 347 non-homologous proteins out of 478 essential proteins, 256 non-homologous proteins out of 332 virulence factors, and 97 non-homologous antibiotic-resistant proteins out of 131 antibiotic-resistant proteins. A total of 445 non-redundant non-homologous proteins were found.
Druggability analysis
The current study was further expanded by listing the potential efficacy of non-homologous proteins on the candidate list. 445 non-redundant proteins from the shortlisted ones were screened against the DrugBank database to identify druggable proteins. Only those proteins that tend to bind to a small molecule that ultimately inhibits or modulates its function are considered druggable proteins (Uddin and Saeed 2014). In total, 144 proteins were similar to the drug targets already available in the database. All these 144 proteins were considered potential drug targets, because they all share the same homolog with at least 30% sequence similarity at Drugbank. The remaining proteins that were not found in the database were excluded from further analysis.
Pathway analysis
There are 117 metabolic pathways for K. aerogenes and 337 pathways in humans on the KEGG server. Pathways that are displayed in K. aerogenes but are missing in humans are identified as unique pathways, while those displayed in both the pathogen and the host are common pathways. Among the 117 pathways, 39 were found to be unique K. aerogenes pathways and 78 pathways were common to both humans and K. aerogenes. These 39 unique pathogen-specific pathways are listed in (Supplementary Data). In addition, 144 druggable proteins were subjected to BLASTP on the KAAS server in KEGG. KAAS reported the pathway annotation for 86 proteins, out of 144. The remaining 58 proteins had no information available on their signaling pathway. To avoid unwanted cross-reaction with the human pathway, out of the 86 proteins, only 13 were selected and proposed as new targets that were associated with pathogen-specific or unique pathways (Table 2). The distribution of these 13 proteins in the unique metabolic pathway of K. aerogenes is illustrated in (Fig. 2).
Table 2.
Drug targets predicted from the unique pathway of K. aerogenes
| Protein accession ID | Pathway Name | Protein Name | KO assignment |
|---|---|---|---|
| WP_015706103.1 | Benzoate degradation, microbial metabolism in diverse environments | 3-Hydroxybenzoate 6-monooxygenase | K22270 |
| WP_015705999.1 | Biosynthesis of secondary metabolites, biosynthesis of siderophore group nonribosomal peptides | 4′-Phosphopantetheinyl transferase superfamily protein | K02362 |
| WP_015705506.1 | Quorum sensing | TIGR04028 family ABC transporter substrate-binding protein | K02035 |
| WP_015705462.1 | Bacterial secretion system | DotU family type VI secretion system protein | K11892 |
| WP_015705202.1 | Quorum sensing | ABC transporter substrate-binding protein | K02035 |
| WP_015705001.1 | Beta-lactam resistance, two-component system | Porin | K09476 |
| WP_015704800.1 | Cationic antimicrobial peptide (CAMP) resistance, two-component system | Two-component system sensor histidine kinase PmrB | K07643 |
| WP_015704359.1 | Two-component system | Sensor histidine kinase | K07700 |
| WP_015704248.1 | Two-component system | Cu (+)/Ag (+) sensor histidine kinase | K07644 |
| WP_015704019.1 | Two-component system | Two-component system sensor histidine kinase ZraS | K07709 |
| WP_015703856.1 | Beta-Lactam resistance, quorum sensing | Multidrug efflux RND transporter outer membrane subunit EefC | K18139 |
| WP_015370099.1 | Quorum sensing | ABC transporter substrate-binding protein | K02035 |
| WP_015368314.1 | Two-component system | Response regulator | K07702 |
Fig. 2.
Pie-chart depicts the distribution of the 13 drug targets in unique metabolic pathways of K. aerogenes
Analysis of broad-spectrum targets
Comparison of test target sequences with therapeutically necessary pathogenic bacteria facilitates the assessment of proteins that serve as perfect broad-spectrum targets. BLASTP related homology searches against 240 pathogenic bacteria identified broad-spectrum targets. The homology search found that all predicted targets have close homologs to more than 40 pathogens and are shown in Table 3. The analysis also showed that the list of proteins included clinically critical bacterial pathogens, such as Klebsiella pneumoniae, Shigella dysenteriae serotype 1, Salmonella typhi, Shigella flexneri, Yersinia enterocolitica, Helicobacter pylori, and Bacillus cereus. The drug particles designed to prevent such broad-spectrum targets are easy to use to eradicate pathogens.
Table 3.
Broad-spectrum analysis results
| Protein ID | Protein name | Broad-spectrum targets | No of homologs |
|---|---|---|---|
| WP_015706103.1 | 3-Hydroxybenzoate 6-monooxygenase | Yes | 178 |
| WP_015705999.1 | 4′-Phosphopantetheinyl transferase superfamily protein | Yes | 41 |
| WP_015705506.1 | TIGR04028 family ABC transporter substrate-binding protein | Yes | 487 |
| WP_015705462.1 | DotU family type VI secretion system protein | Yes | 380 |
| WP_015705202.1 | ABC transporter substrate-binding protein | Yes | 757 |
| WP_015705001.1 | Porin | Yes | 103 |
| WP_015704800.1 | Two-component system sensor histidine kinase PmrB | Yes | 1000 |
| WP_015704359.1 | Sensor histidine kinase | Yes | 476 |
| WP_015704248.1 | Cu (+)/Ag (+) sensor histidine kinase | Yes | 1005 |
| WP_015704019.1 | Two-component system sensor histidine kinase ZraS | Yes | 1014 |
| WP_015703856.1 | Multidrug efflux RND transporter outer membrane subunit EefC | Yes | 493 |
| WP_015370099.1 | ABC transporter substrate-binding protein | Yes | 752 |
| WP_015368314.1 | Response regulator | Yes | 664 |
Cellular location prediction
Protein sub-cellular analysis helps to understand the details of the location as well as to determine its function. The location of the proteins within the cell is a significant factor in the characterization of suitable and effective drug/vaccine targets. Based on localization scores, 13 targets were divided into two cytoplasmic proteins, 7 membrane proteins, and 4 periplasmic proteins (Table 4). Intracellular localization of recognized targets was predicted using PSORTb and double-checked using CELLO.
Table 4.
Subcellular localization of shortlisted proteins
| Protein accession ID | Protein name | Subcellular localization |
|---|---|---|
| WP_015706103.1 | 3-Hydroxybenzoate 6-monooxygenase | Cytoplasmic |
| WP_015705999.1 | 4′-Phosphopantetheinyl transferase superfamily protein | Periplasmic |
| WP_015705506.1 | TIGR04028 family ABC transporter substrate-binding protein | Periplasmic |
| WP_015705462.1 | DotU family type VI secretion system protein | Cytoplasmic membrane |
| WP_015705202.1 | ABC transporter substrate-binding protein | Periplasmic |
| WP_015705001.1 | Porin | Outer membrane |
| WP_015704800.1 | Two-component system sensor histidine kinase PmrB | Cytoplasmic membrane |
| WP_015704359.1 | Sensor histidine kinase | Cytoplasmic membrane |
| WP_015704248.1 | Cu (+)/Ag (+) sensor histidine kinase | Cytoplasmic membrane |
| WP_015704019.1 | Two-component system sensor histidine kinase ZraS | Cytoplasmic membrane |
| WP_015703856.1 | Multidrug efflux RND transporter outer membrane subunit EefC | Outer membrane |
| WP_015370099.1 | ABC transporter substrate-binding protein | Periplasmic |
| WP_015368314.1 | Response regulator | Cytoplasmic |
Antigenicity prediction
Although still in the development stage, safe recombinant vaccine-based antigenic protein formulations are proving to be the most attractive and effective way to fight infectious diseases. The development of vaccines in contradiction to membrane proteins will help to control and eradicate infectious diseases in humans. The VaxiJen server was used to predict antigenicity with a cutoff value of 0.5 (Solanki and Tiwari 2018; Satyam et al. 2020; Vakili et al. 2018). Of the seven membrane proteins, five (WP_015705001.1, WP_015704800.1, WP_015704248.1, WP_015704019.1 and WP_015703856.1) have a vaccine score greater than 0.5 and can be considered as vaccine candidates. The resulting antigen proteins are shown in Table 5.
Table 5.
Antigenicity of the predicted membrane proteins
| Protein accession ID | Protein name | Antigenicity property |
|---|---|---|
| WP_015705462.1 | DotU family type VI secretion system protein | 0.4598 |
| WP_015705001.1 | Porin | 0.6709 |
| WP_015704800.1 | Two-component system sensor histidine kinase PmrB | 0.5062 |
| WP_015704359.1 | Sensor histidine kinase | 0.4131 |
| WP_015704248.1 | Cu (+)/Ag (+) sensor histidine kinase | 0.5321 |
| WP_015704019.1 | Two-component system sensor histidine kinase ZraS | 0.5989 |
| WP_015703856.1 | Multidrug efflux RND transporter outer membrane subunit EefC | 0.5809 |
Discussion
The increasing frequency of adverse reactions accompanying different antibiotic therapy measures makes it necessary to develop significant drug targets and candidate vaccines for many pathogens. Recently, "in silico" subtractive and comparative proteomics has been used extensively to predict and identify potential novel therapeutic targets and antigenic vaccine candidates in several infective bacteria (Amineni et al. 2010; Chong et al. 2006; Doyle et al. 2010). In this study, the computer analyses, combined with the accessibility of total-genome sequence, suggest the possibility of performing the first "in silico" comparative and subtractive proteome analysis to identify putative therapeutic targets and vaccine candidates for K. aerogenes. Investigation and identification of therapeutic targets are conducted using several target priority parameters.
The first challenge in drug development against pathogenic related bacteria is that humans have a wide diversity of bacteria in their gut or intestinal microbiome, including commensal or symbiotic bacteria. The suppression of the intestinal flora can adversely affect the host by promoting pathogenic colonization of the intestine (Levy 2000). Ideally, these beneficial flora proteins should not be targeted. To evade this problem, BLASTP was performed on these proteins to detect those proteins that were non-homologous to the intestinal flora. We identified 1142 non-homologous proteins out of 4795 for the gut microflora proteome. Since essential proteins are involved in the basic cellular processes of pathogenic bacteria, they are suitable antibiotic targets when developing new antibacterial drugs. Among these, 478 proteins were identified as essential proteins. Furthermore, 131 resistant and 332 virulent proteins were considered parallel in terms of potential drug and antigenic vaccine targets. To avoid severe cross-reactions and adverse effects on humans, the identification of proteins non-homologous to humans is another critical step that is being considered in this study. Therefore, proteins that tend to be identical to host human proteins have been omitted from the list. There were 347 non-homologous proteins out of 478 essential proteins, 256 non-homologous proteins out of 332 virulence factors, and 97 non-homologous antibiotic-resistant proteins out of 131 antibiotic-resistant proteins. There are a total of 445 proteins that are non-redundant and non-homologous to human proteins.
Furthermore, the druggability analysis by the DrugBank database allows the detection of target proteins corresponding to the active compounds of the drug. 144 druggable target proteins were discovered among the 445 non-redundant non-homologous proteins. Pathway annotation of 144 proteins was carried out using the KAAS server. To avoid unwanted cross-reaction with the human pathway, only 13 proteins associated with pathogen-specific or unique pathways were selected and proposed as new targets. These 13 targets are associated with 11 unique pathways, namely, a two-component system, cationic antimicrobial peptide (CAMP) resistance, beta-lactam resistance, bacterial secretion system, biosynthesis of siderophore group non-ribosomal peptides, quorum sensing, biosynthesis of secondary metabolites, biosynthesis of benzoate degradation and microbial metabolism in diverse environments (Supplementary Data: S1, S2, S3, S4, S5, S6).
By analyzing the results of metabolic pathway analysis of K. aerogenes, scientists get a glimpse of a list of unique proteins in different pathways. Proteins shared in several pathways are thought to be highly effective targetable candidates, as blocking the action of particular targets can have the toxic effect of inhibiting the role of various pathogens. Of the 13 listed candidates, 5 candidates, namely, 3-hydroxybenzoate 6-monooxygenase, 4′-phosphopantetheinyl transferase superfamily protein, porin, two-component system sensor histidine kinase PmrB and multidrug efflux RND transporter outer membrane subunit EefC were involved in multiple pathways. The present drug identification method focuses on the lipopolysaccharide and peptidoglycan biosynthetic pathways (Zoeiby et al. 2003). This limited approach to targeting specific pathways is a necessary factor in the advancement of multidrug resistance among pathogenic bacteria (Stephenson and Hoch 2002). The current process takes all the various metabolic pathways, including a two-component system, quorum sensing, benzoate degradation, cationic antimicrobial peptide (CAMP) resistance, beta-lactam resistance, biosynthesis of siderophore group non-ribosomal peptides, microbial metabolism of diverse environments, and biosynthesis of secondary metabolites. New pathways can facilitate the discovery of antimicrobial agents (Brown and Wright 2005).
13 proteins were chosen and proposed as new targets based on the subtractive analysis described above, which included non-homology analysis against gut microflora, essentiality analysis, virulent factor analysis, resistance factor analysis, non-homology analysis against humans, druggability analysis, and pathway analysis. Then the short-listed 13 new target proteins were characterised using broad-spectrum analysis, cellular localization analysis, and antigenicity prediction. Broad-spectrum drug target analysis revealed many proteins whose domains are found in at least ten species, including K. pneumoniae, S. dysenteriae serotype 1, S. typhi, S. flexneri, Y. enterocolitica, H. pylori, B. cereus, Streptococcus pyogenes STAB902, etc. Thus, 13 predicted drug targets were identified as broad-spectrum drug targets. A wide variety of pathogens can be destroyed by drug particles assumed to obstruct these broad-spectrum targets. Due to the growing occurrence of coinfection, the use of drugs effective against a wide variety of pathogens has been strongly suggested.
Further localization was analyzed to identify proteins that are either the membrane, cytoplasm, or periplasm. As a result, the seven membrane proteins found can be exploited as vaccine targets. Effective vaccine candidates have high antigenicity, which is critical for inducing an immune response in the host and protecting against infections (Monterrubio-Lopez et al. 2015). Therefore, determining the highest antigenicity value for selecting the suitable vaccine candidates is a significant phase used for vaccine design. Using the VaxiJen server, we selected five antigenic proteins with an antigenicity level (0.5) above the threshold. Thus, these five proteins, namely, porin, sensor histidine kinase PmrB, two-component system, Cu (+)/Ag (+) sensor histidine kinase, sensor histidine kinase ZraS, and multidrug efflux RND transporter outer membrane subunit EefC, were considered as vaccine candidates. To the best of our knowledge, this work is the first computational investigation of the K. aerogenes proteome to identify new drug and vaccine candidates.
Conclusion
The comparative and subtractive proteomic analysis of K. aerogenes was performed, and several proteins of the K. aerogenes proteome were prioritized as potential drug/vaccine candidates. The identified therapeutic candidates were discovered to be involved in important metabolic processes that control nutrient absorption and bacterial pathogenicity. Further characterization of the final list of proteins will lead to the identification of drug and vaccine candidates. The identified candidates are widely represented in various clinically significant bacterial pathogens and are potential candidates for broad-spectrum drug development. Therefore, our current study will help develop an appropriate treatment. Future advancements in vaccine development against K. aerogenes will necessitate fewer trials and fewer errors, saving time and money in "in vitro" research. The current study opens up new opportunities in the design and manufacture of potential drugs/powerful antigenic vaccines that target only the pathogenic system without affecting the physiology or biology of the hosts.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
There is no separate funding available for this research.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amineni U, Pradhan D, Marisetty H. In silico identification of common putative drug targets in Leptospira interrogans. J Chem Biol. 2010;3:165–173. doi: 10.1007/s12154-010-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastay M, Lagier E, Blanc V, Chardon H. Épidémiologie des bêtalactamases à spectre étendu (BLSE) chez les entérobactéries dans un hôpital du sud de la France, 1999–2007. Pathol Biol (paris) 2013;61:38–43. doi: 10.1016/j.patbio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Anishetty S, Pulimi M, Pennathur G. Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Comput Biol Chem. 2005;29:368–378. doi: 10.1016/j.compbiolchem.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Arpin C, Coze C, Rogues AM, Gachie JP, Bebear C, Quentin C. Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit. J Clin Microbiol. 1996;34:2163–2169. doi: 10.1128/jcm.34.9.2163-2169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpin C, Dubois V, Coulange L, André C, Fischer I, Noury P, Larribet G. Extended-spectrum β-lactamase-producing Enterobacteriaceae in community and private health care centers. Antimicrob Agents Chemother. 2003;47:3506–3514. doi: 10.1128/AAC.47.11.3506-3514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottacini F, Motherway MOC, Kuczynski J, O’Connell KJ, Serafini F, Duranti S, Zhurina D. Comparative genomics of the Bifidobacterium brevetaxon. BMC Genom. 2014;15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ED, Wright GD. New targets and screening approaches in antimicrobial drug discovery. Chem Rev. 2005;105:759–774. doi: 10.1021/cr030116o. [DOI] [PubMed] [Google Scholar]
- Cantón R, Oliver A, Coque TM, del Carmen VM, Pérez-Díaz JC, Baquero F. Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J Clin Microbiol. 2002;40:1237–1243. doi: 10.1128/JCM.40.4.1237-1243.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava C, Colaprico A, Bertoli G, Graudenzi A, Silva TC, Olsen C, Castiglioni I. SpidermiR: an R/bioconductor package for integrative analysis with miRNA data. Int J Mol Sci. 2017;18:274. doi: 10.3390/ijms18020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Chen YC, Hsu LY. Epidemiologic study of pathogens causing nosocomial infections. J Formos Med Assoc. 1990;89:1023–1030. [PubMed] [Google Scholar]
- Charrel RN, Pagès JM, De Micco PHILIPPE, Mallea M. Prevalence of outer membrane porin alteration in beta-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother. 1996;40:2854–2858. doi: 10.1128/AAC.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong CE, Lim BS, Nathan S, Mohamed R. “In silico” analysis of Burkholderia pseudomallei genome sequence for potential drug targets. In Silico Biol. 2006;6:341–346. [PubMed] [Google Scholar]
- Davin-Regli A. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol. 2015;6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MA, Gasser RB, Woodcroft BJ, Hall RS, Ralph SA. Drug target prediction and prioritization: using orthology to predict essentiality in parasite genomes. BMC Genom. 2010;11:1–14. doi: 10.1186/1471-2164-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova IA, Flower DR. Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine. 2007;25:856–866. doi: 10.1016/j.vaccine.2006.09.032. [DOI] [PubMed] [Google Scholar]
- El Zoeiby A, Sanschagrin F, Levesque RC. Structure and function of the Mur enzymes: development of novel inhibitors. Mol Microbiol. 2003;47:1–12. doi: 10.1046/j.1365-2958.2003.03289.x. [DOI] [PubMed] [Google Scholar]
- Fok TF, Lee CH, Wong EM, Lyon DJ, Wong W, Ng PC, Cheng AF. Risk factors for Enterobacter septicemia in a neonatal unit: case-control study. Clin Infect Dis. 1998;27:1204–1209. doi: 10.1086/514974. [DOI] [PubMed] [Google Scholar]
- Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev. 2009;22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley JM, Hanakowski M, LeBlanc JC. Unique epidemiology of nosocomial urinary tract infection in children. Am J Infect Control. 2001;29:94–98. doi: 10.1067/mic.2001.111537. [DOI] [PubMed] [Google Scholar]
- Levy J. The effects of antibiotic use on gastrointestinal function. Am J Gastroenterol. 2000;95:S8–S10. doi: 10.1016/S0002-9270(99)00808-4. [DOI] [PubMed] [Google Scholar]
- Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42:D574–D580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- Mondal SI, Ferdous S, Jewel NA, Akter A, Mahmud Z, Islam MM, Karim N. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: an in-silico approach. Adv Appl Bioinform Chem. 2015;8:49. doi: 10.2147/AABC.S88522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterrubio-Lopez GP, Gloria P, Ribas-Aparicio RM (2015) Identification of novel potential vaccine candidates against tuberculosis based on reverse vaccinology. BioMed Res Int 2015 [DOI] [PMC free article] [PubMed]
- Mora M, Donati C, Medini D, Covacci A, Rappuoli R. Microbial genomes and vaccine design: refinements to the classical reverse vaccinology approach. Curr Opin Microbiol. 2006;9:532–536. doi: 10.1016/j.mib.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padiadpu J, Vashisht R, Chandra N. Protein–protein interaction networks suggest different targets have different propensities for triggering drug resistance. Syst Synth Biol. 2010;4:311–322. doi: 10.1007/s11693-011-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès JM, James CE, Winter halter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DJ. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014;42:D737–D743. doi: 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal D, Lim CS, Sakharkar KR, Sakharkar MK. Differential genome analyses of metabolic enzymes in Pseudomonas aeruginosa for drug target identification. In Silico Biol. 2007;7:453–465. [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabizadeh S, Sears C. New horizons for the infectious diseases specialist: how gut microflora promote health and disease. Curr Infect Dis Rep. 2008;10:92–98. doi: 10.1007/s11908-008-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K, Yeturu K, Chandra N. targetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Syst Biol. 2008;2:109. doi: 10.1186/1752-0509-2-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reker D, Rodrigues T, Schneider P, Schneider G. Identifying the macromolecular targets of de novo-designed chemical entities through self-organizing map consensus. Proc Natl Acad Sci. 2014;111:4067–4072. doi: 10.1073/pnas.1320001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders WE, Sanders CC. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev. 1997;10:220–241. doi: 10.1128/CMR.10.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi AN, Aggarwal R, Rahman Q, Trivedi N. Subtractive genomics approach for “in silico” identification and characterization of novel drug targets in Neisseria Meningitides Serogroup B. J Comput Sci Syst Biol. 2009;2:255–258. [Google Scholar]
- Sarkar M, Maganti L, Ghoshal N, Dutta C. “In silico” quest for putative drug targets in Helicobacter pylori HPAG1: molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J Mol Model. 2012;18:1855–1866. doi: 10.1007/s00894-011-1204-3. [DOI] [PubMed] [Google Scholar]
- Satyam R, Bhardwaj T, Jha NK, Jha SK, Nand P. Toward a chimeric vaccine against multiple isolates of Mycobacteroides—an integrative approach. Life Sci. 2020;150:117541. doi: 10.1016/j.lfs.2020.117541. [DOI] [PubMed] [Google Scholar]
- Segovia JA, Chang TH, Winter VT, Coalson JJ, Cagle MP, Pandranki L, Kannan TR. NLRP3 is a critical regulator of inflammation and innate immune cell response during Mycoplasma pneumoniae infection. Infect Immun. 2018;86:e00548-17. doi: 10.1128/IAI.00548-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugham B, Pan A. Identification and characterization of potential therapeutic candidates in emerging human pathogen Mycobacterium abscessus: a novel hierarchical “in silico” approach. PLoS ONE. 2013;8:e59126. doi: 10.1371/journal.pone.0059126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Pan A. Identification of potential drug targets in Yersinia pestis using metabolic pathway analysis: MurE ligase as a case study. Eur J Med Chem. 2012;57:185–195. doi: 10.1016/j.ejmech.2012.09.018. [DOI] [PubMed] [Google Scholar]
- Shende G, Haldankar H, Barai RS, Bharmal MH, Shetty V, Idicula-Thomas S. PBIT: pipeline builder for identification of drug targets for infectious diseases. Bioinformatics. 2017;33:929–931. doi: 10.1093/bioinformatics/btw760. [DOI] [PubMed] [Google Scholar]
- Solanki V, Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep. 2018;8:1–19. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson K, Hoch JA. Two-component and phosphorelay signal-transduction systems as therapeutic targets. Curr Opin Pharmacol. 2002;2:507–512. doi: 10.1016/S1471-4892(02)00194-7. [DOI] [PubMed] [Google Scholar]
- Uddin R, Saeed K. Identification and characterization of potential drug targets by subtractive genome analyses of methicillin resistant Staphylococcus aureus. Comput Biol Chem. 2014;48:55–63. doi: 10.1016/j.compbiolchem.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Uddin R, Jamil F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and protein–protein interaction network. Comput Biol Chem. 2018;74:115–122. doi: 10.1016/j.compbiolchem.2018.02.017. [DOI] [PubMed] [Google Scholar]
- Vakili B, Nezafat N, Hatam GR, Zare B, Erfani N, Ghasemi Y. Proteome-scale identification of Leishmania infantum for novel vaccine candidates: a hierarchical subtractive approach. Comput Biol Chem. 2018;72:16–25. doi: 10.1016/j.compbiolchem.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Assempour N. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins Struct Funct Bioinform. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Brinkman FS. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.