Abstract
Objective:
To explore whether rs4784227 polymorphism of CASC16 is correlated with risk of breast cancer.
Methods:
Relevant studies up to December 24, 2020 were searched in PubMed, Embase, Web of Science, CNKI, VIP, and WANFANG databases. Data were analyzed by using Stata 12.0. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated, and country-based subgroup analyses were conducted. Sensitivity analysis was conducted to assess the stability of the results. Publication bias was assessed by using the Egger regression asymmetry test and visualization of funnel plots.
Results:
Seven case-control studies enrolling 4055 breast cancer cases and 4229 controls were included. rs4784227 was found significantly associated with increased risk of breast cancer in a dominant (OR = 1.301, 95% CI = 1.190–1.423, P < .001), a recessive (OR = 1.431, 95% CI = 1.216–1.685, P < .001), and an allele model (OR = 1.257, 95% CI = 1.172–1.348, P < .001), while an over-dominant model showed that rs4784227 was correlated with decreased breast cancer risk (OR = 0.852, 95% CI = 0.778–0.933, P = .001).
Conclusion:
The rs4784227 polymorphism of CASC16 gene is correlated with breast cancer susceptibility.
Keywords: breast cancer, CASC16/ TOX high-mobility box protein group family member 3, meta-analysis, rs4784227, SNP
1. Introduction
Breast cancer is a malignant tumor occurring in the breast epithelium or ductal epithelium. It is one of the most common malignant tumors in women, accounting for 30% of female cancers, and the main cause of death in women aged 20 to 59 years.[1] In China, with the development of social economy and the change of people's lifestyle, the incidence and death rates of breast cancer are increasing year by year, and more than 300,000 women are diagnosed with breast cancer every year.[2] The etiology is not completely clear yet, and it may be ascribed to genetic factors, breast cancer related gene mutations, reproductive factors, sex hormones, nutrition, and dietary and environmental factors.
In 2007, the Breast Cancer Association Collabation reported 5 susceptibility sites associated with the occurrence of breast cancer for the first time through a three-stage GWAS study.[3] Thereafter, accumulating GWAS studies were carried out and risk single nucleotide polymorphic sites have been identified. The role of rs4784227 of CASC16, also known as TOX3 (TOX high-mobility box protein group family member 3), in the regulation of tumor biological function has attracted much attention.[4,5] The rs4784227 is located in the intron region of LOC643714, which is upstream of CASC16.[6]TOX3 encodes the TOX high-mobility box protein group family member 3, a protein involved in changes in chromatin structure, with 3 nucleotide repeat motifs and a putative gene LOC643714.[7] The clinical significance of CASC16 and its role in the development and invasion of breast cancer have been demonstrated in several studies.[8,9]
Studies have shown that rs4784227 T allele can reduce luciferase activity and change DNA-protein binding, regulate chromatin affinity for FOXA1 (forkhead box O1), and lead to allele-specific gene expression.[10–12] Similarly in breast cancer cell lines, different genotypes at rs4784227 can affect the affinity of CASC16 promoter for FOXA1 transcription factor and regulate the expression of CASC16 thus affecting the occurrence and development of breast cancer.[3,13–15] Zheng et al first identified rs4784227 as a genetic susceptibility locus for breast cancer in European and Asian populations,[10,16] However, the studies of Cai Q and Antoniou AC did not find an association between rs4784227 and breast cancer.[17,18] Therefore, here we conduct a meta-analysis of the published relevant articles to comprehensively analyze the correlation between CASC16 rs4784227 polymorphism and breast cancer risk.
2. Materials and methods
2.1. Literature retrieval strategy
We systematically searched the electronic databases (PubMed, Embase, Web of Science, CNKI, VIP, and WANFANG DATA) to identify related studies published up to December 24, 2020. The terms used for search were (“breast cancer ∗” OR “breast neoplasms ∗” OR “mammary cancer ∗”) AND (“rs4784227∗” OR “TOX3 ∗” OR “CASC16∗”) AND (“gene∗” OR “polymorphism∗” OR “mutation∗” OR “single nucleotide polymorphism∗” OR “SNP∗” OR “variant∗”). The reference lists of relevant articles were checked to identify additional eligible studies not included.
2.2. Inclusion and exclusion criteria
Eligible articles evaluating the association between CASC16 gene rs4784227 polymorphism and breast cancer risk should meet the following inclusion criteria:
-
1.
Patients in the case group should be diagnosed with breast cancer;
-
2.
Case-control studies analyzing CASC16 gene rs4784227 polymorphism and breast cancer susceptibility;
-
3.
Studies providing the frequency distribution or enough data to calculate the frequency distribution of rs4784227 genotype.
Exclusion criteria are as follows: not enough data are included. In the case of conference abstract, if the data of animal model, cell experiments and case reports overlap, studies with more subjects will be included.
2.3. Data extraction and quality assessment
Two investigators (Liang Xiongshun and Hu Liming) extracted data from the included studies independently, and when there were inconsistences in the data, the differences were resolved through discussion with a third investigator (Mo Junluan). Data extracted from eligible studies included: first author, year of publication, journal, country, sample size, mean age, number or frequency of genotypes and alleles, and Hardy-Weinberg equilibrium (HWE) status. Newcastle-Ottawa Scale (NOS) was used to evaluate the quality of the studies included in the Meta-analysis. Studies with NOS score ≥6 were considered of high quality.
2.4. Statistical analysis
The OR value and 95% CI of different gene models were calculated by STATA/SE 12.0 to evaluate the association between rs4784227 polymorphism of CASC16 and breast cancer susceptibility. The significance of OR was determined according to Z test. The combined results of the dominant, recessive, over-dominant and allele models were evaluated respectively. I2 statistics and corresponding P values were used to evaluate the heterogeneity between studies. When there was heterogeneity, a random effect model (P < .1 and/or I2 > 50%) was used to combine the results, otherwise a fixed effect model (P ≥ .1 and/or I2 ≤ 50%) was used.[19,20] The genotype Hardy-Weinberg equilibrium in the control group was calculated by the Chi-Squared test, and P > .05 was considered to meet the Hardy-Weinberg equilibrium. Funnel plots and Egger regression were used to assess publication bias, and sensitivity analysis was used to evaluate the stability of the results.
2.5. Ethics and dissemination
Since this is a protocol for meta-analysis, all data in this study come from published studies and do not involve patients, so ethical approval is not required. Findings of this research will be disseminated in a peer-reviewed journal or conference presentations.
3. Result
3.1. Basic data of the included literature
According to the above inclusion criteria, 7 case-control studies were finally included in this meta-analysis[21–27] (Fig. 1). All 7 studies were conducted in Asian populations, including 5 in China and 2 in Iran. A total of 4055 cases and 4229 controls were included for analysis. The basic data of the included studies are shown in Table 1.
Figure 1.
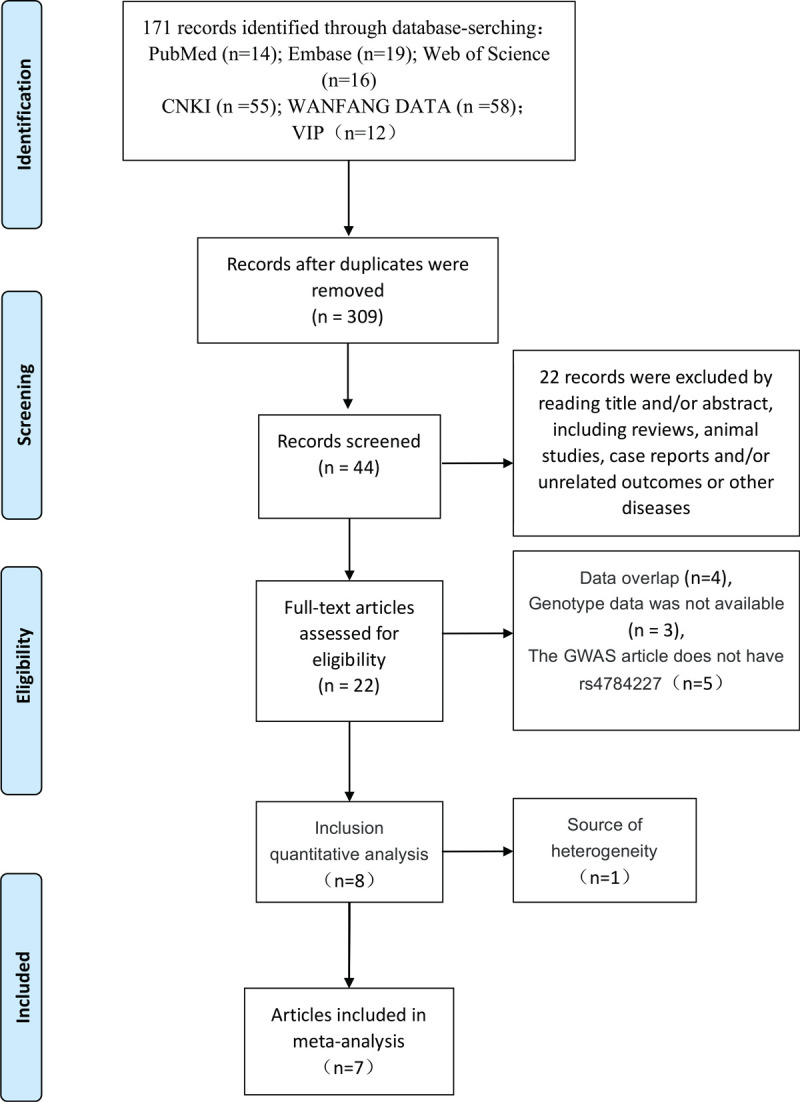
Flowchart of literature selection.
Table 1.
Characteristics of studies included in the meta-analysis.
| Sample size | Mean age (years) | Genotype | Allele (M/m)∗ | OR (95% CI) | ||||||||||||
| SNPs | Author and reference | Year | Country | Ethnicity | Case | Control | Case | Control | Case | Control | Case | Control | Dominant model | Allelic model | NOS | HWE |
| rs4784227 | Yao Sun | 2020 | China | Asian | 503 | 503 | 52.04 ± 9.52 | 51.90 ± 9.55 | 266/199/38 | 292/180/31 | 275/731 | 242/764 | 1.23 (0.96–1.58) | 1.19 (0.97–1.45) | 6 | 0.644135 |
| (C > T) | Nehle Jamali | 2020 | Iran | Asian | 107 | 91 | 53.51 ± 5.16 | 52.51 ± 5.97 | 31/46/30 | 36/49/6 | 106/108 | 61/121 | 1.61 (0.89–2.90) | 1.95 (1.30–2.93) | 7 | 0.046983 |
| Xiaoxiao Zuo | 2020 | China | Asian | 681 | 680 | 50.58 ± 9.84 | 50.63 ± 9.71 | 353/270/52 | 394/240/41 | 374/976 | 322/1028 | 1.28 (1.03–1.59) | 1.22 (1.03–1.45) | 6 | 0.581859 | |
| Amir Tajbakhsh | 2019 | Iran | Asian | 505 | 567 | 50.52 ± 12.29 | 43.45 ± 12.21 | 209/218/78 | 285/222/60 | 374/636 | 342/792 | 0.52 (0.42–0.66) | 1.49 (1.26–1.77) | 6 | 0.092861 | |
| Zhiping Deng | 2016 | China | Asian | 551 | 577 | 49.09 ± 11.022 | 48.790 ± 8.294 | 123/102/13 | 340/205/36 | 128/348 | 277/885 | 1.38 (1.02–1.86) | 1.19 (0.98–1.44) | 6 | 0.495311 | |
| Yuxiang Lin | 2014 | China | Asian | 702 | 794 | 47.8 ± 10.9 | 48.7 ± 9.7 | 331/302/68 | 424/313/57 | 438/964 | 427/1161 | 1.27 (1.03–1.56) | 1.24 (1.06–1.45) | 7 | 0.941233 | |
| Ali Hajizadeh | 2017 | Iranian | Asian | 126 | 160 | – | – | 75/35/16 | 54/96/10 | 67/185 | 116/204 | 0.22 (0.095–0.549) | – | 6 | 0.000162 | |
| Furu Wang | 2020 | China | Asian | 1006 | 1017 | 51.59 ± 11.14 | 51.54 ± 11.18 | 500/415/83 | 569/385/61 | 581/1415 | 507/1523 | 1.27 (1.06–1.52) | – | 7 | 0.69847 | |
M/m: major/minor allele
Genotype presented as wild type/heterozygous/homozygous; −, not available; SNP = single nucleotide polymorphisms, OR = odds ratio, 95% CI = 95% confidence interval, NOS = Newcastle–Ottawa quality assessment scale, HWE = Hardy–Weinberg equilibrium.
3.2. Heterogeneity assessment of included literature
As can be seen from the comparison of results between Figure 2 and Figure 3, the study by Ali Hajizadeh et al[28] was the source of heterogeneity. Before the study was excluded, the subgroup in which the study was located P = .274, I2 = 92.9%. After the exclusion of this study, P < .001, I2 = 0.
Figure 2.
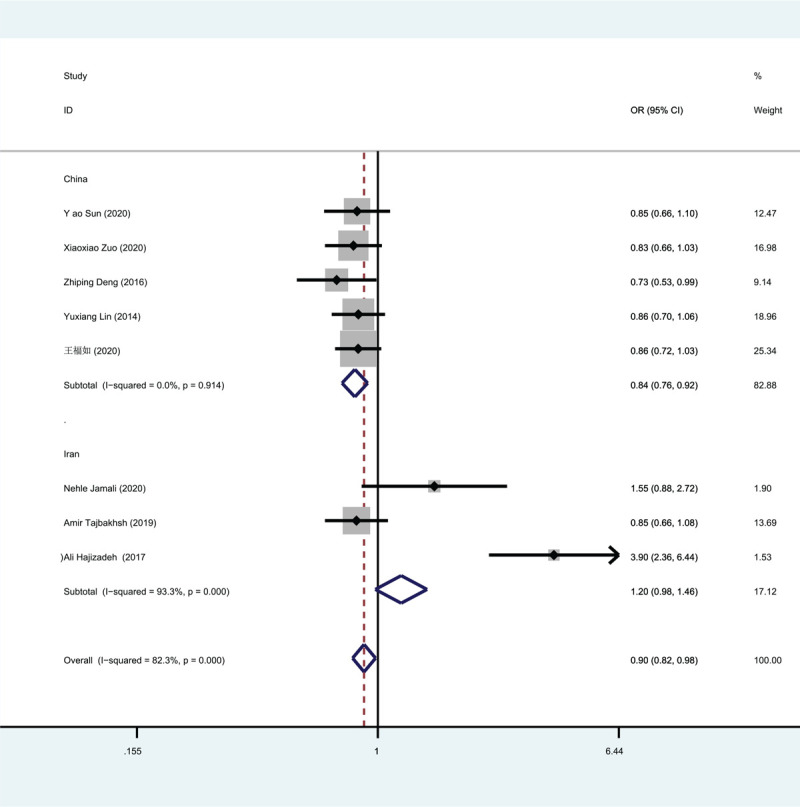
Meta-analysis forest map.
Figure 3.
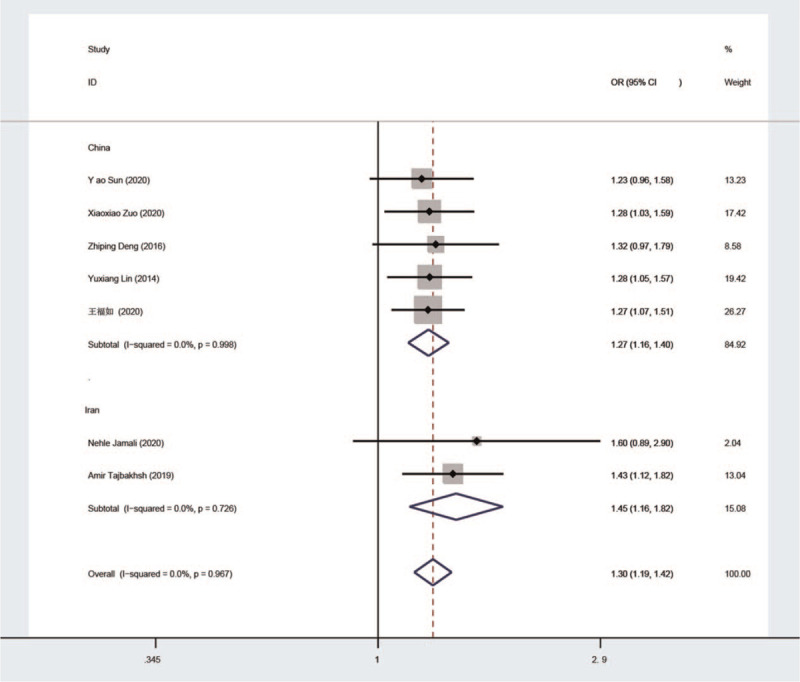
Eliminate a paper with heterogeneity.
3.3. Meta-analysis of association between rs4784227 polymorphism of CASC16 gene and breast cancer risk
Rs4784227 was significantly associated with breast cancer in a dominant model (OR = 1.301, 95% CI = 1.190–1.423, P < .001), recessive model (OR = 1.431, 95% CI = 1.216–1.685, P < .001), allele model (R = 1.257, 95% CI = 1.172–1.348, P < .001) and over-dominant model (OR = 0.852, 95% CI = 0.778–0.933, P = .001). The results of the meta-analysis are shown in Table 2. From the analysis of the total study population, the 4 models were statistically significant. The results showed that the rs4784227 T allele of CASC16 gene significantly increased the risk of breast cancer compared with the C allele.
Table 2.
Association of rs4784227 polymorphism in CASC16 gene with breast cancer risk.
| Sample size | Dominant model | Recessive model | Allelic model | Over-dominant model | |||||||||||||||
| SNPs | Number of study | case | control | OR (95% CI) | I2 (%) | PH | PZ | OR (95% CI) | I2 (%) | PH | PZ | OR (95% CI) | I2 (%) | PH | PZ | OR (95% CI) | I2 (%) | PH | PZ |
| rs4784227 | |||||||||||||||||||
| Total | 7 | 4055 | 4229 | 1.301 (1.190–1.423) | 0.0 | .998 | <.001 | 1.431 (1.216–1.685) | 45.60% | 0.088 | <0.001 | 1.257 (1.172–1.348) | 0.0 | .424 | <.001 | 0.852 (0.778–0.933) | 0.0 | .491 | .001 |
| China | 5 | 3443 | 3571 | 1.274 (1.155–1.404) | 0.0 | .726 | <.001 | 1.303 ( 1.079–1.572) | 0.00% | 0.769 | 0.006 | 1.219 (1.128–1.317) | 0.0 | .995 | <.001 | 0.837 (0.758–0.924) | 0.0 | .914 | <.001 |
| Iran | 2 | 612 | 658 | 1.455 (1.163–1.820 ) | 0.0 | .726 | .001 | 1.897 ( 1.365–2.636) | 84.20% | 0.012 | <0.001 | 1.444 (1.225–1.702) | 59.5 | .116 | <.001 | 0.933 (0.746–1.166) | 73.0 | .054 | .54 |
M/m∗: major/minor allele
Genotype presented as wild type/heterozygous/homozygous; −, not available; 95% CI = 95% confidence interval, HWE = Hardy–Weinberg equilibrium, NOS = Newcastle–Ottawa quality assessment scale, OR = odds ratio, SNPs = single nucleotide polymorphisms.
3.4. Subgroup analysis
Subgroup analysis by country was performed (Table 2). In the Chinese population, 5 studies were included with a total of 3443 cases and 3571 controls. Rs4784227 was significantly associated with breast cancer in dominant model (OR = 1.274, 95% CI = 1.155–1.404, P < .001), recessive model (OR = 1.303, 95% CI = 1.079–1.572, P = .006), allele (OR = 1.219, 95% CI = 1.128–1.317, P < .001) and over-dominant model (OR = 0.837, 95% CI = 0.758–0.924, P < .001). These results suggest that the polymorphism of rs4784227 locus of CASC16 gene may be a risk factor for breast cancer in Chinese women. In the Iranian population, 2 studies were included with a total of 612 cases and 658 controls. According to results from the dominant model (OR = 1.455, 95% CI = 1.163–1.820, P = .001), the recessive model (OR = 1.897, 95% CI = 1.365–2.636, P < .001), the allele (OR = 1.444, 95%CI = 1.225–1.702, P < .001), and the hyperdominant model (OR = 0.933, 95%CI = 0.746–1.166, P < .54), rs4784227 was significantly associated with breast cancer susceptibility, suggesting that the polymorphism of rs4784227 locus of CASC16 gene may be a risk factor for breast cancer.
3.5. Sensitivity analysis
Sensitivity analysis was used to assess the impact of individual studies on the pooled results. Removing any of the studies had no significant effect on the results, indicating that the results were statistically stable and reliable.
3.6. Publication bias
Begg funnel plot and Egger regression asymmetry test were used to evaluate publication bias. No publication bias was found in the dominant model (P > .05), as shown in Figure 4.
Figure 4.
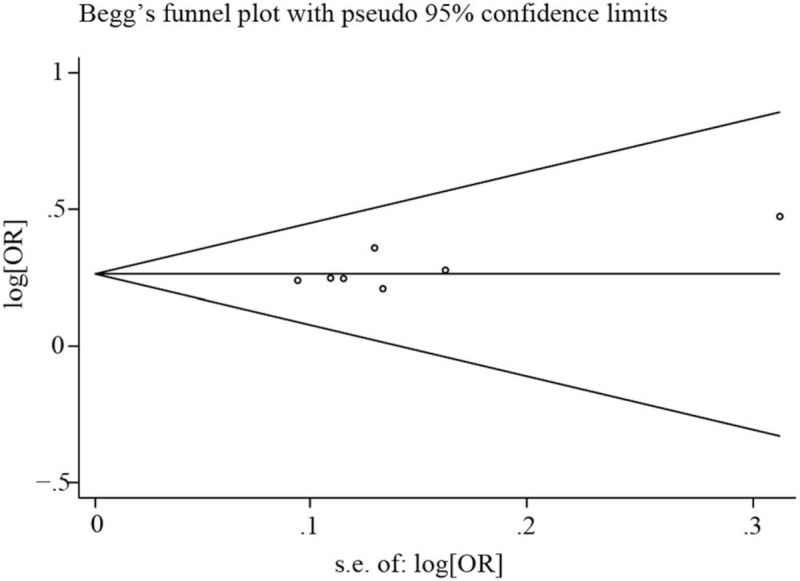
Publication bias analysis of included literatures.
4. Discussion
Breast cancer has replaced lung cancer as the world's leading cancer, according to the latest data on the global cancer burden for 2020 released by International Agency for Research on Cancer. Behind the increasing incidence rate of breast cancer, Single nucleotide polymorphisms are a major factor for the occurrence of cancer, accounting for 27% of the total risk of breast cancer.[29]
CASC16 is mainly expressed in the brain, but studies have shown high CASC16 expression in tumor tissues. It induces transcription of the estrogen response (ER) and Bcl-2.[7,8] To date, studies have shown contradictory results in terms of the association of CASC16 rs4784227 with breast cancer susceptibility.[22] In this meta-analysis, a total of 7 studies were included, including 4055 cases and 4229 controls. The results of combination and subgroup analysis from dominant model, recessive model, allele model and over-dominant model were statistically significant. These results suggest that gene polymorphism at rs4784227 of CASC16 gene may increase the risk of breast cancer, which is consistent with the research results of Zheng et al.[10,16] The study by Ali et al is the source of heterogeneity. This heterogeneity may be due to the relatively small sample size of the study, the genetic differences among different regions and nationalities, and the other possibility is the difference of linkage imbalance, so this study was not included in the final meta-analysis.
Single nucleotide polymorphisms in related risk genes are one of the important causes of disease susceptibility. In recent years, the association between CASC16 rs4784227 C > T gene polymorphism and the risk of breast cancer has been increasingly studied.[3] For example, Udler et al found that rs4784227 polymorphism was associated with the expression of Retinoblastoma-like protein 2 protein, increasing the risk of breast cancer.[30] Meyer et al found that rs4782447 could lead to changes in FOXA1-binding sequence, which may increase its affinity and thus enhance the binding of FOXA1 to CASC16 gene promoter. In various in vitro experiments, this SNP has been identified as a functional genetic risk mutation site for breast cancer.[31,32] Zou et al found that rs4784227 was associated with an increased risk of lymph node metastasis in individuals with breast cancer.[26] RS4784227 is enriched in FOXA1 and regulates the affinity between chromatin and FOXA1 on the distal regulatory elements, leading to changes in the ability of FOXA1 to bind DNA. The binding site of FOXA1 can form chromatin rings with the promoter of CASC16 gene, and alterations in FOXA1-binding DNA sequence will directly affect the expression of CASC16 gene.[22,32]CASC16 is a nuclear protein chromatin structure that can be modified,[33] and its high expression can affect the methylation of breast cancer 1 promoter, resulting in the reduction of breast cancer 1expression, thus improving proliferation, invasion, and survival ability of breast cancer cells in vivo.[34] Therefore, we predict that rs4784227 C > T gene polymorphism changes the affinity between FOXA1 and CASC16 promoter, thereby promoting the high expression of CASC16 and accelerating lymph node metastasis of breast cancer.
In general, genetic polymorphisms at rs4784227 of CASC16 gene are associated with risk of breast cancer. However, the biological mechanism by which rs4784227 is involved in the occurrence of breast cancer is still unclear. More functional studies may help to increase our understanding of the biological characteristics of breast cancer. This study has certain limitations, the sample size is not large enough and only studies from China and Iran were included, thus our study fails to answer whether there is a similar association of rs4784227 with breast cancer susceptibility in patients with other ethnicities. In addition, the impact of lifestyle, environmental exposures, and other diseases on the results could not be assessed.
Author contributions
Conceptualization: Xiong-Shun Liang, Jun-Luan Mo, Wen-Xu Hong.
Data curation: Li-Ming Hu, Ji-Ye Yin.
Formal analysis: Xiong-Shun Liang, Chun-Mei Gong.
Funding acquisition: Jun-Luan Mo.
Methodology: Xiong-Shun Liang, Jun-Luan Mo, Wen-Xu Hong.
Project administration: Xiong-Shun Liang, Jun-Luan Mo, Wen-Xu Hong.
Software: Xiong-Shun Liang, Li-Ming Hu, Ji-Ye Yin.
Supervision: Tao Liu, Zhao-Qian Liu, Hong-Hao Zhou.
Validation: Jun-Luan Mo, Li-Ming Hu.
Visualization: Tao Liu, Zhao-Qian Liu.
Writing – original draft: Xiong-Shun Liang.
Writing – review & editing: Xiong-Shun Liang, Wen-Xu Hong.
Footnotes
Abbreviations: FOXA1 = forkhead box O1, TOX3 = TOX high-mobility box protein group family member 3.
How to cite this article: Liang XS, Mo JL, Hu LM, Gong CM, Liu T, Hong WX, Yin JY, Liu ZQ, Zhou HH. Association between CASC16 rs4784227 polymorphism and breast cancer susceptibility: a meta-analysis. Medicine. 2021;100:28(e26215).
XSL and JLM contributed equally to this work.
Supported by the Sanming Project of Medicine in Shenzhen (No. SZSM201811057). Guangdong Medical Science and Technology Research Fund (No. B2021240).
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].MPH RLS, MPH KDM, DVM AJ, PhD. Cancer statistics, 2020%J CA: A Cancer Journal for Clinicians. 2020; 70(1). [DOI] [PubMed] [Google Scholar]
- [2].Hong-da. CW-qLNCM-mRJ-sSJ-fC, jie LJLC-qYX-yCHDMH Preliminary Analysis of Cancer Screening Program in Urban China from 2013 to 2017 2020;29(1):01–6. [Google Scholar]
- [3].Wen YC, Ong WS, Tan PH, et al. Validation of the Gail model for predicting individual breast cancer risk in a prospective nationwide study of 28,104 Singapore women. Breast Cancer Res 2012;14(1):R19–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].A MK, A GP, C B. Agreement between breast cancer risk estimation methods. J National Cancer Institute 1996;88(18): [DOI] [PubMed] [Google Scholar]
- [5].Fischer C, Kuchenbacker K, Engel C, et al. Evaluating the performance of the breast cancer genetic risk models BOADICEA, IBIS, BRCAPRO and Claus for predicting BRCA1/2 mutation carrier probabilities: a study based on 7352 families from the German Hereditary Breast and Ovarian Cancer Consortium. J Med Genet 2013;50(6):360–7. [DOI] [PubMed] [Google Scholar]
- [6].Han YJ, Zhang J, Zheng Y, Huo D, Olopade OI. Genetic and epigenetic regulation of TOX3 expression in breast cancer. PLoS One 2016;11(11):e0165559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tajbakhsh A, Javan FA, Rivandi M, et al. Significant association of TOX3/LOC643714 locus-rs3803662 and breast cancer risk in a cohort of Iranian population. Mol Biol Rep 2018. [DOI] [PubMed] [Google Scholar]
- [8].Chalabi N, Bernardgallon DJ, Bignon YJ, et al. Comparative clinical and transcriptomal profiles of breast cancer between french and south mediterranean patients show minor but significative biological differences. Cancer Genomics Proteomics 2008;5(5):253. [PubMed] [Google Scholar]
- [9].Mahfoudh W, Bouaouina N, Ahmed SB, et al. Hereditary breast cancer in Middle Eastern and North African (MENA) populations: identification of novel, recurrent and founder BRCA1 mutations in the Tunisian population. Mol Biol Rep 2012;39(2):1037–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet 2009;41(3):324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lin Y. Associations between SNPs on 6q25. 1 and in TOX3 with Risk of Breast Cancer in a Fujian Population. Interaction: Fujian Medical University; 2015. [Google Scholar]
- [12].Cowper-Sal·lari R, Zhang X, Wright JB, et al. Breast cancer risk–associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 2012;44(11): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet 2008;40(6):703. [DOI] [PubMed] [Google Scholar]
- [14].Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].J L, Q C, H S, et al. Genome-wide association study in East Asians identifies novel susceptibility loci for breast cancer. PLoS Genet 2012;8(2):e1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].J L, Q C, XO S, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet 2010;6(6):e1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].AC A, C K, OM S, et al. Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Human Mol Genet 2011;20(16):3304–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Qiuyin C, Wanqing W, Shimian Q, et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res 2011;71(4): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nathan M, William H. Statistical aspects of the analysis of data from retrospective studies of disease. Narnia 1959;22(4): [PubMed] [Google Scholar]
- [20].Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327(7414): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nanjing Medical University, Wang F. Association study of some regions of chromosomes in Jiangsu population with the susceptibility of breast cancer. 2010. [Google Scholar]
- [22].Lin Y, Fu F, Chen M, Meng H, Wang C, Coleman WB. Associations of two common genetic variants with breast cancer risk in a Chinese population: a stratified interaction analysis. Plos One 2014;9(12):e115707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhiping D, Hua Y, Qiufang L, et al. Identification of novel susceptibility markers for the risk of overall breast cancer as well as subtypes defined by hormone receptor status in the Chinese population. Journal of Human Genetics 2016;61(12): [DOI] [PubMed] [Google Scholar]
- [24].Jamali N, Nasiri M, Yavarian M. Association of the functional genetic variants of TOX3 gene with breast cancer in Iran: a case-control study. Gene Rep 2019;18:100511. [Google Scholar]
- [25].Tajbakhsh A, Farjami Z, Darroudi S, et al. Association of rs4784227-CASC16 (LOC643714 locus) and rs4782447-ACSF3 polymorphisms and their association with breast cancer risk among Iranian population. Excli J 2019. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sun Y, Chen P, Wu J, et al. Association of polymorphisms in LOC105377871 and CASC16 with breast cancer in the northwest Chinese Han population. J Gene Med 2020;22(3): [DOI] [PubMed] [Google Scholar]
- [27].Zuo X, Wang H, Mi Y, et al. The association of CASC16 variants with breast Cancer risk in a northwest Chinese female population. Mol Med 2020. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hajizadeh A, Houshmand M, Hosseini M. TNP1 non-gene region and influences tumor characteristics by low-risk alleles in breast cancer. Quid Investigación Ciencia Y Tecnología 2017. 669–73. [Google Scholar]
- [29].Alborzi A, Houshmand M, Hosseini M. ER and PR positive, or Her2 negative tumor of rs2363956 and rs3803662 GWAS in breast cancer. Gene Cell Tissue 2017;In Press (In Press). [Google Scholar]
- [30].S UM, Shahana A, S HC, et al. Fine scale mapping of the breast cancer 16q12 locus. Human Molecular Genet 2010;19(12): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cowper-Sal·Lari R, Zhang X, Wright JB, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 2012;44(11):1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Meyer KB, Carroll JS. FOXA1 and breast cancer risk. Nat Genet 2012;44(11):1176–7. [DOI] [PubMed] [Google Scholar]
- [33].Han Y-J, Zhang J, Zheng Y, Huo D, Olopade OI. Genetic and epigenetic regulation of TOX3 expression in breast cancer. PLoS ONE 2017;11(11): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jingxuan S, P DS, Sasha B, et al. TNRC9 downregulates BRCA1 expression and promotes breast cancer aggressiveness. Cancer Res 2013;73(9): [DOI] [PubMed] [Google Scholar]


