Abstract
Within eukaryotic cells, biochemical reactions need to be organized on the surface of membrane compartments that use distinct lipid constituents to dynamically modulate the functions of integral proteins or influence the selective recruitment of peripheral membrane effectors. As a result of these complex interactions, a variety of human pathologies can be traced back to improper communication between proteins and membrane surfaces; either due to mutations that directly alter protein structure or as a result of changes in membrane lipid composition. Among the known structural lipids found in cellular membranes, phosphatidylinositol (PtdIns) is unique in that it also serves as the membrane-anchored precursor of low-abundance regulatory lipids, the polyphosphoinositides (PPIn), which have restricted distributions within specific subcellular compartments. The ability of PPIn lipids to function as signaling platforms relies on both non-specific electrostatic interactions and the selective stereospecific recognition of PPIn headgroups by specialized protein folds. In this chapter, we will attempt to summarize the structural diversity of modular PPIn-interacting domains that facilitate the reversible recruitment and conformational regulation of peripheral membrane proteins. Outside of protein folds capable of capturing PPIn headgroups at the membrane interface, recent studies detailing the selective binding and bilayer extraction of PPIn species by unique functional domains within specific families of lipid-transfer proteins will also be highlighted. Overall, this overview will help to outline the fundamental physiochemical mechanisms that facilitate localized interactions between PPIn lipids and the wide-variety of PPIn-binding proteins that are essential for the coordinate regulation of cellular metabolism and membrane dynamics.
Keywords: membrane biology, lipid-binding domains, signal transduction, cellular trafficking, phosphatidylinositol, phosphoinositides, pleckstrin homology, PTB, PDZ, GRAM, GLUE, FERM, phox homology, FYVE, C2, Tubby, PROPPINs, ENTH, ANTH, BAR, oxysterol-binding protein-related protein, phosphatidylinositol-transfer protein
1. Introduction
The structural integrity and dynamic remodeling of biological membranes relies on reciprocal interactions between membrane proteins and smaller amphipathic lipids. In general, membrane components exist as part of an asymmetric bilayer consisting of functionally distinct inner and outer leaflets; although, some subcellular compartments may function as lipid monolayers (Holthuis and Menon, 2010; Drin, 2014). Even small modifications to the relative abundance or identity of either the protein or lipid constituents present can significantly alter the intrinsic physiochemical properties of cellular membranes with important consequences for the activities of integral as well as peripheral membrane proteins (van Meer et al., 2008; Drin, 2014). Consequently, to perform specialized functions, eukaryotic cells have developed distinct membrane compartments with unique local properties that are characterized by specific protein and lipid compositions (Holthius and Menon, 2014). Despite the need for functional heterogeneity, throughout subcellular membranes, phospholipids are the most abundant structural components and are defined by the presence of a polar headgroup and two hydrophobic acyl tails, which can differ greatly in both their chain length as well as degree of hydrocarbon saturation across different membrane environments (Bigay and Antonny, 2012; Barelli and Antonny, 2016). Modifications to this general phospholipid structure, especially alterations to the surface-exposed headgroup, endow certain species with unique biophysical characteristics that have been shown to directly influence general membrane features such as fluidity, thickness, lipid-packing density, and surface charge (van Meer et al., 2008; Holthuis and Menon, 2010; Jackson et al., 2016). Overall, cellular membrane dynamics relies on the functional diversity of membrane lipids and the phospholipid composition, in particular, plays an important role in the coordinate regulation of signaling and trafficking functions throughout subcellular membrane compartments.
Of the known membrane lipid species, phosphatidylinositol (PtdIns) is unique in that is not only functions as an essential structural phospholipid, but it also serves as the precursor for important low-abundance regulatory lipids that are collectively referred to as polyphosphoinositides (PPIn; Balla, 2013). An essential mechanism involved in the regulation of diverse cellular functions depends on the reversible recruitment of peripheral cytosolic proteins or macromolecular complexes to the surface of specific subcellular membranes with high temporal resolution. Dynamic recruitment of peripheral protein effectors is orchestrated, in large part, by the local production of PPIn lipids through the addition of phosphate moieties to PtdIns at one or more of the hydroxyl-groups present at the 3-, 4-, or 5-position of the inositol ring. PtdIns-specific phosphorylation events are tightly controlled by highly conserved substrate-selective, as well as position-specific, lipid kinases and phosphatases that function to generate seven distinct membrane-embedded PPIn species; including mono-, bis-, or tris-phosphorylated derivatives (Balla, 2013). Although variability exists, unique PPIn lipids appear to localize to overlapping, as well as distinct, membrane surfaces and can recruit different intracellular effectors that not only contribute to the initiation of signaling responses, but can also function to define membrane identity or control local membrane dynamics (Hammond and Balla, 2015; Schink et al., 2016). In addition, many of the regulatory functions attributed to PPIn species can occur indirectly as a result of actions on cytoskeletal remodeling (Saarikangas et al., 2010; Bezanilla et al., 2015; Senju et al., 2017) or through the allosteric regulation of transmembrane-spanning receptors, ion channels, or transporters (Hilgemann et al., 2001; Gamper and Shapiro, 2007; Barrera et al., 2013; Hille et al., 2015; Hedger and Sansom, 2016).
Due to the expansive cellular roles performed by PPIn lipids, both as structural components and sites for protein-membrane interactions, it is not surprising that many human pathologies are the result of perturbations in PPIn production or clearance, including direct contributions of altered PPIn dynamics to: cancer, diabetes, degenerative myopathies and neuropathies (Pendaries et al., 2003; Wymann and Schneiter, 2008; McCrea and De Camilli, 2009; Bunney and Katan, 2010; Balla, 2013; Thapa et al., 2016), as well as being part of the invasion or evasion strategies employed by infectious agents (Altan-Bonnet and Balla, 2012; Payrastre et al., 2012; Pizzaro-Cerdá et al., 2015; Altan-Bonnet, 2017). To better understand how PPIn production regulates cellular functions, recent studies have tried to define the biosynthetic and inter-conversion pathways that modulate PPIn turnover as well as characterize the PPIn-binding domains responsible for recognizing distinct PPIn species within membrane compartments. Overall, this chapter will attempt to summarize the general mechanisms controlling PPIn recognition by peripheral membrane proteins and introduce the structural diversity of PPIn-interacting protein domains found in both prokaryotic and eukaryotic organisms. Collectively, by comparing the PPIn recognition systems used by peripheral proteins from bacterial and animal models, we hope to provide a foundation for understanding the general molecular processes that allow for inositol-containing lipids to function as membrane recognition sites that contribute to the dynamic regulation of diverse biological processes throughout evolution.
2. Synthesis and Subcellular Distribution of PPIn Lipids
The numerous PPIn kinases and phosphatases, as well as the reversible recruitment of numerous PPIn-binding effectors, all contribute to the steady-state availability of membrane PPIn species. The complex processes governing PPIn metabolism begins with the synthesis of PtdIns by a single enzyme, PtdIns synthase (PIS); which is present as an integral membrane protein within the endoplasmic reticulum (ER) and catalyzes the conjugation of the myo- stereoisomer of inositol to a cytidine diphosphate (CDP)-activated diacylglycerol (DAG) backbone (Agranoff et al., 1958; Agranoff et al., 1969; Agranoff, 2009). Despite localizing to membranes of the ER, work from our group has shown that catalytically-active PIS is concentrated within a mobile ER-derived sub-compartment that may function to actively distribute PtdIns to subcellular membranes (Kim et al., 2011). Within cellular membranes, PtdIns represents roughly 10–20 mol% of total phospholipids; whereas, despite their important cellular roles, PPIn species only represent an estimated 2–5% of the available PtdIns and therefore only contributes to 0.2–1 mol% of membrane phospholipids (Lemmon, 2008; Balla, 2013; Vance, 2015). However, it is important to mention that the relative amounts of PPIn lipids found within cells shows significant variations across species and even between cell types. Downstream of PtdIns production, PPIn lipids are continuously being turned over, but can also be concentrated within discrete subcellular compartments. Consequently, the rapid and localized production of PPIn lipids from the abundant membrane precursor PtdIns, can drastically increase the ratio of the target PPIn relative to the amount of a membrane-binding effector; making it possible to recruit a large amount of peripheral proteins without saturating the available PPIn headgroups. Sequential inter-conversion of PPIn species using substrate-selective enzymatic modifications may also confer a degree of biochemical processivity to the control of dynamic membrane signaling events (Cullen et al., 2001; Balla, 2005; Botelho, 2009). The coordinate production and targeted recognition of PPIn species is thought to be enhanced by recruiting macromolecular complexes containing PPIn kinases or phosphatases in close proximity to PPIn substrates or downstream effectors. An excellent example of this regulatory paradigm comes from a recent description of metabolic channeling of PPIn-mediated signaling by the multi-domain scaffold protein IQGAP1 (IQ motif-containing GTPase-activating protein 1; Choi et al., 2016). Specifically, IQGAP1 regulates the assembly and substrate presentation for three distinct PPIn kinases at the PM, which facilitates the sequential phosphorylation of PtdIns, to produce the important second messenger PtdIns 3,4,5-trisphosphate (PtdIns(3,4,5)P3), and controls the activation of additional PtdIns(3,4,5)P3-sensitive effectors that are also associated with IQGAP1 (Choi et al., 2016). Though of central importance for understanding PPIn biology, the regulation and cellular functions of the wide-variety of PPIn-modifying enzymes responsible for the production and inter-conversion of subcellular PPIn species will not be discussed at length in this chapter, but have been reviewed in depth elsewhere (Sasaki et al., 2009; Dyson et al., 2012; Balla, 2013; Hsu and Mao, 2015). However, it is clear that understanding the complexities associated with the control of PPIn metabolism will require additional investigations into the roles played by molecular scaffolds and regulatory protein-protein interactions on membrane surfaces.
Studies examining the subcellular localization of PPIn-modifying enzymes have provided some details on the potential landscape of PPIn species within subcellular compartments; however, to truly understand how dynamic changes in membrane PPIn composition occur, membrane-embedded PPIn species need to be visualized with high spatial and temporal resolution. In recent years, work from many laboratories, including our own, have contributed greatly to imaging breakthroughs that have been pivotal for the study of specific PPIn lipids in living cells. In particular, foundational studies using fluorescently-tagged constructs consisting of isolated PPIn-binding domains were able to selectively follow subcellular PtdIns 4,5-bisphosphate (PtdIns(4,5)P2; Varnai and Balla, 1998; Stauffer et al., 1998) or PtdIns(3,4,5)P3 (Kontos et al., 1998; Varnai et al., 1999; Watton and Downward, 1999; Servant et al., 2000) dynamics in real-time following receptor-dependent hydrolysis or class I PI3K activation, respectively. In addition to PtdIns(4,5)P2 and PtdIns(3,4,5)P3, selective lipid-binding probes have been established that can reliably visualize the PPIn species PtdIns 4-phosphate (PtdIns4P), PtdIns 3-phosphate (PtdIns3P), and PtdIns 3,4-bisphosphate (PtdIns(3,4)P2); as well as for other important structural or signaling lipids such as DAG, phosphatidic acid (PtdOH), and phosphatidylserine (PtdSer; Hammond and Balla, 2015; Varnai et al., 2017). Taken together, these studies not only revealed important details regarding cellular PPIn metabolism and turnover, but also provided proof of concept for the use of membrane-binding domains as specific biosensors to map subcellular phospholipid compartments. The utility of PPIn-binding domains as biosensors for visualizing and quantifying membrane PPIn lipids has been discussed at length by our group previously (Hammond and Balla, 2015; Varnai et al., 2017) and will not be the central focus of this chapter. Overall, work using selective lipid-binding probes, as well as more traditional biochemical approaches, reveal that PPIn lipids show a restricted subcellular distribution and that the enrichment of specific PPIn species occurs within distinct membrane compartments. Defining the localization of distinct PPIn species is of obvious importance for understanding the specialized functions of these lipids, and therefore we will briefly outline the PPIn-specific territories that have been mapped to discrete membrane compartments or larger organelles. Please be aware that although roles for nuclear PPIn lipids are emerging (Irvine, 2003; Barlow et al., 2010; Martelli et al., 2011; Shah et al., 2013; Crowder et al., 2017), and certainly represent an exciting new area of PPIn biology for investigation, we will restrict our discussion to the distribution of PPIn lipids in cytosolic membranes as these PPIn pools are more clearly defined and also appear to function independently from the unique system of PPIn metabolism that functions within the nucleus (Hammond and Balla, 2015).
Within mammalian cells, PtdIns4P and PtdIns(4,5)P2 are the most abundant PPIn species, constituting approximately 2–5% of the total cellular pool of PtdIns-containing lipids (Balla et al., 1988; Xu et al., 2003). The majority of PtdIns(4,5)P2 is found within the PM, although evidence for PtdIns(4,5)P2-mediated regulation of effectors at the Golgi (Watt et al., 2002; De Matteis et al., 2002) and within the endolysosomal system has also been presented (Choi et al., 2015; Tan et al., 2015). PtdIns4P pools appear to be generated in various membrane compartments using distinct PI4K enzymes (Balla and Balla, 2006; Boura and Nencka, 2015; Dornan et al., 2016). Specifically, the PM pool of PtdIns4P that serves as the precursor for PtdIns(4,5)P2 synthesis is generated primarily by the PI4KIIIα isoform (Balla et al., 2008; Nakatsu et al., 2012; Bojjireddy et al., 2014). Alternatively, Golgi pools of PtdIns4P are produced by PI4KIIIβ (Godi et al., 1999), with additional contributions from both type II enzymes, PI4KIIα and PI4KIIβ (Weixel et al., 2005; Wang et al., 2003), which are also responsible for producing PtdIns4P in the late endosomes (Hammond et al., 2014). Outside of PtdIns(4,5)P2 and PtdIns4P, the remaining PPIn species only contribute a small amount to the total cellular fraction of inositol-containing lipids. Minor amounts of PtdIns(3,4,5)P3 are found within the inner leaflet of the PM and only increases upon receptor-mediated activation of class I PI3Ks (Vanhaesebroeck et al., 2010; Burke and Williams, 2015); but, even at maximal levels, PtdIns(3,4,5)P3 only represent 2–5% of the PM PtdIns(4,5)P2 (Hawkins et al., 1992; Toker and Cantley, 1997). Dephosphorylation of PtdIns(3,4,5)P3 by 5-phosphatases is thought to generate PtdIns(3,4)P2 within the PM (Erneux et al., 2011; Ooms et al., 2015; Posor et al., 2013) and recent studies also suggest that PtdIns(3,4)P2 may persist within PM-derived vesicles internalized during endocytosis (Posor et al., 2013; Ketel et al., 2016; Marat et al., 2017; Malek et al., 2017). Interestingly, results in vitro as well as in vivo indicate that class II PI3Ks preferentially phosphorylate PtdIns and, to a lesser extent, PtdIns4P to generate PtdIns3P and PtdIns(3,4)P2, respectively (Arcaro, 1998; Misawa et al., 1998; Falasca et al., 2007; Franco et al., 2014; Braccini et al., 2015). Despite much debate about the relative importance of the kinase- and phosphatase-dependent metabolic pathways, cellular studies strongly suggest that local production of PtdIns(3,4)P2 in the endosomal system contributes to coordinate control of cellular signaling and membrane dynamics (Li and Marshall, 2015; Hawkins and Stephens, 2016; Marat and Haucke, 2016); including important roles during clathrin- and endophilin-mediated endocytosis (Posor et al., 2013; Boucrot et al., 2015; Renard et al., 2015). Mono-phosphorylated PtdIns3P represents 20–30% of the cellular PtdIns4P and is found primarily within the early endosomes (Gillooly et a., 2000) and in autophagosomes (Funderburk et al., 2010); with an additional report describing the presence of PtdIns3P in membranes of the Golgi and ER (Sarkes and Rameh, 2010). The more enigmatic monophosphorylated PPIn species PtdIns 5-phosphate (PtdIns5P) is estimated at only 1% of the PtdIns4P levels in mammalian cells and subcellular fractionation suggests that the highest amount is localized to the PM (Sarkes and Rameh, 2010). Additional enrichments of PtdIns5P may also be found in ER and Golgi membrane fractions (Sarkes and Rameh, 2010) as well as in the early endosomes (Ramel et al., 2011). Lastly, PtdIns 3,5-bisphosphate (PtdIns(3,5)P2) has the lowest abundance of the PPIn species found within mammalian cells, making up 1% of cellular PtdIns(4,5)P2 (Zolov et al., 2012; Sbrissa et al., 2012). Attempts to localize PtdIns(3,5)P2 has been hampered by a lack of specificity in the available biosensors (Hammond et al., 2015), but functional studies suggest that this minor PPIn species is important for the proper sorting of cargoes within the late endosome (Gary et al., 1998; Bonangelino et al., 2002). Taken together, the unique spatial distribution and selective enrichment of subcellular PPIn species highlights the utility of these regulatory lipids as sites for coordinating the dynamic recruitment of peripheral proteins to discrete membrane compartments. The rapid inter-conversion of PPIn species by PPIn-modifying enzymes may also enhance the spatiotemporal specificity of cellular responses that are controlled by localized PPIn production.
3. General Features of Membrane Binding by Peripheral Proteins
Biological membranes contain a variety of lipids, including the seven distinct PPIn species, which function to coordinate reciprocal interactions with diverse families of intracellular proteins and any associated small molecules or ions. The interfacial regions surrounding membrane bilayers consist of a complex mixture of water, lipid headgroups, backbone phosphates, and any polar portions of the fatty acyl chains (Lee, 2003; Cho and Stahelin, 2005; Pasenkiewicz-Gierula et al., 2016). The kinetics and energetics of membrane interactions are locally governed by the physiochemical properties of both the membrane and protein surfaces (Marsh, 2008). In general, initial membrane association of proteins are driven by diffusional as well as electrostatic forces to establish transient collisional intermediates that can be reinforced to form tightly-bound intermolecular complexes by additional hydrogen-bonding or electrostatic interactions (Cho and Stahelin, 2005; Whited and Johs, 2015). While non-specific interactions with membrane surfaces based on charge complementarity are unlikely to be sufficient to anchor proteins with high affinity, the initial membrane adsorption during these relatively weak associations facilitate specific membrane-binding events by orienting the geometry of peripheral proteins relative to the interface and reducing the dimensionality of the interaction space to the simple two-dimensional membrane surface; effectively increasing the local protein concentration (Cho and Stahelin, 2005; Mulgrew-Nesbitt et al., 2006; Fernandes et al., 2015). In some instances, initial membrane attachment can also facilitate interfacial penetration of hydrophobic or aromatic residues that surround the lipid-binding pocket into the hydrocarbon core of the bilayer (Yau et al., 1998; Killian and von Heijne, 2000; Lomize et al., 2007). Without the added affinity provided by interactions with lipid headgroups, peripheral protein domains are unable to penetrate the interfacial or hydrocarbon regions of membrane leaflets due to the high energetic penalty of desolvation (Pogozheva et al., 2013; Stahelin et al., 2014). Although a combination of these membrane-targeting mechanisms are required, specific lipid coordination and any associated electrostatic or hydrophobic interactions are essential components that drive the membrane recruitment and activation of peripheral membrane proteins; especially those that are coordinated by the anionic and structurally-distinct PPIn lipids. However, it should also be mentioned that, in addition to selective interactions with membrane-embedded lipids, bulk compositions or structural features, such as the charge or degree of membrane curvature, also contribute to the recognition of specific membrane surfaces by peripheral proteins (Lee, 2003; McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006; Marsh, 2008; Baumgart et al., 2011). As we will discuss below, PPIn lipids have emerged as an essential platform for the specific interaction between a wide array of lipid-binding protein domains in almost all membrane compartments.
4. Principles of PPIn-Protein Interactions
Given the variety of regulatory mechanisms that contribute to the local control of PPIn metabolism and turnover, it is not difficult to imagine that the PPIn-binding surfaces utilized by protein effectors for the reversible recruitment to specific PPIn isomers are similarly diverse. The unique structures that have been described for peripheral membrane protein domains have revealed many diverse modes for membrane binding that result in different PPIn specificities and membrane-binding orientations. Despite these complexities, it is possible to uncover specific themes that control the dynamic regulation of cytosolic effectors by membrane-embedded PPIn lipids. As the title of this chapter would suggest, the predominant structural features of PPIn-regulated proteins are specialized membrane-binding modules that allow for the selective recognition of individual PPIn species. However, before discussing the molecular diversity of PPIn-interacting protein domains in more detail, we will first highlight some of the general principles that guide protein interactions with membrane PPIn lipids. Fundamental features contributing to communication between PPIn lipids and peripheral proteins were recently detailed by our group (Hammond and Balla, 2015), as well as others (Kutateladze, 2010; Moravcevic et al., 2012; Stahelin et al., 2014; Choy et al., 2017), and will be summarized below.
To reliably regulate protein functions in time and space, the interactions between membrane PPIn species and proteins should be governed by high-affinity and stoichiometric PPIn-binding; most characteristically through a dedicated PPIn-recognition domain. However, this principle is not universal and not all PPIn-binding domains have the requisite affinity to sufficiently dictate protein localization in isolation. Consequently, although some PPIn interactions that have been identified are not solely responsible for dictating membrane localization, PPIn-binding may add the necessary avidity to a secondary or co-incident interaction(s) that can act together to facilitate peripheral membrane protein recruitment. The idea that PPIn- or other lipid-binding modules can function to complement other protein-protein or protein-lipid interactions at membrane surfaces has been characterized in a variety of signaling contexts (Balla, 2005; Carlton and Cullen, 2005; Moravcevic et al., 2012). In particular, PPInassisted membrane binding, which capitalizes on combinatorial interactions or scaffolding functions, is best exemplified by the regulation of the Arf (ADP-ribosylation factor) and Arl (Arf-like) superfamily of small guanine nucleotide-binding proteins (Godi et al., 2004; DiNitto et al., 2007; Liu et al., 2014; Jian et al., 2015). In diverse membrane compartments, the integration of coincident signals from PPIn- and protein-interactions can be used to effectively tune the regulatory functions of peripheral proteins or macromolecular complexes bound at the membrane interface. However, in addition to simple roles as membrane scaffolds, PPIn-coordination can also contribute to the complex control of protein conformational dynamics. Allosteric regulation of protein effectors by PPIn lipids has been characterized in detailed mechanistic studies of protein kinase B (Akt; Calleja et al., 2007; Calleja et al., 2009a, b; Calleja et al., 2012; Ebner et al., 2017), PTEN (phosphatase and tensin homolog deleted on chromosome ten; Campbell et al., 2003; Iijima et al., 2004; Walker et al., 2004; Redfern et al., 2008; Wei et al., 2015), BTK (Bruton’s tyrosine kinase; Joseph et al., 2017), Arf GTPase-activating proteins (GAPs; Kam et al., 2000; Campa et al., 2009), and Arf guanine nucleotide exchange factors (GEFs; Malaby et al., 2013); as well as several examples describing regulatory interactions between PPIn lipids and transmembrane-spanning ion channels or receptors (Hilgemann and Ball, 1996; Huang et al., 1998; Rohacs et al., 2003; Whorton and Mackinnon, 2011; Barrera et al., 2013; Laganowsky et al., 2014). Conformational gating by membrane PPIn species is an emerging field that might be most important for controlling the functions of lipid transfer proteins that use PPIn lipids for membrane recognition and as transport cargoes. However, the regulatory role for PPIn recognition in non-vesicular lipid transport is not yet fully understood, but the communication between membrane PPIn species and the binding domains found within the cellular lipid transfer machinery will be addressed further in Section 7 of this chapter.
Independent of stereospecific lipid coordination, anionic membrane lipids, including PPIn species and PtdSer, can contribute to the membrane targeting of proteins possessing functionalized regions enriched with basic amino acid residues through non-specific electrostatic interactions (Heo et al., 2006; Hammond et al., 2012). Classical examples of proteins that interact with PPIn lipids using unstructured polybasic stretches, which are not organized within a characteristic motif, include the MARCKS (myristoylated alanine-rich C-kinase substrate) proteins (Wang et al., 2002; Wang et al., 2004; Gambhir et al., 2004), c-Src (cellular-sarcoma non-receptor protein tyrosine kinase; Yeung et al., 2006), K-Ras (Kirsten-rat sarcoma; Heo et al., 2006; Gulyas et al., 2017), and GAP43 (growth-associated protein 43; McLaughlin and Murray, 2005). Interestingly, more recently, a unique structured membrane-binding module found at the C-terminus of the eukaryotic MARK (MAP/microtubule affinity-regulating kinases) family of kinases, called the KA1 (kinase-associated 1) domain, has also been shown to effectively sense membrane charge through cooperation between distinct basic regions on the membrane-binding surface (Moravcevic et al., 2010; Emptage et al., 2017a,b). Similar to unstructured polybasic segments, KA1 domains do not appear to distinguish between different anionic phospholipids in vitro or in vivo (Moravcevic et al., 2010; Hammond et al., 2012). Consequently, although important for controlling protein localization during diverse cellular processes, particularly within the unique electrostatic environment of the PM, these simple charge-based interactions capitalize on the general character of PPIn headgroups and will not be discussed at length in this chapter. Overall, using a combination of electrostatic interactions and PPIn-specific recognition, membrane-embedded PPIn species, though rare among phospholipids, function as central regulators of cellular physiology by orchestrating the dynamic recruitment and activation of diverse families of protein effectors at the membrane interface.
5. The Diversity of Eukaryotic PPIn-Binding Domains
There are a wide variety of well-folded modular domains that have evolved to selectively interact with PPIn-containing membranes through a combination of non-specific electrostatic interactions and the stereoselective coordination of PPIn headgroups. In this section, we will introduce the diversity of PPIn-recognizing protein folds, including descriptions of the PH (Pleckstrin Homology), PTB (phosphotyrosine-binding), PDZ (PSD-95, Discs Large, and ZO-1), GRAM (glucosyltransferases, Rablike GTPase activators, and myotubularins), GLUE (GRAM-like ubiquitin-binding in EAP45), FERM (4.1, ezrin, radixin, and moesin), PX (phox homology), FYVE (Fab1p, YOTB, Vac1p, and EEA1), C2 (protein kinase C (PKC) conserved 2), Tubby, PROPPINs (β–propellers that bind phosphoinositides), ENTH (Epsin N-terminal homology), ANTH (AP180 N-terminal homology), and BAR (Bin, Amphiphysin, and Rvs) domain families. Importantly, detailed discussions of the structural and biophysical characteristics of the PH, PX, ENTH, ANTH, and BAR domains will be provided within other chapters of this volume; therefore, our goal with this brief overview is to demonstrate the diversity of PPIn-interacting modules found in peripheral membrane proteins and highlight some of the molecular properties that contribute to the specificity exhibited by these domains during interactions with PPIn lipids.
5.1. PH Domains
PH domains typically consist of 100–120 amino acids and were the first protein fold shown to selectively recognize and coordinate membrane-embedded PPIn lipids (Harlan et al., 1994; Lemmon et al., 1995). Since their initial discovery based on sequence homology with two regions found within the major PKC substrate pleckstrin (Haslam et al., 1993; Mayer et al., 1993; Musacchio et al., 1993), PH domains have been identified in approximately 280 different human proteins; making them among the most commonly-occurring defined sequence motif within the eukaryotic proteome (Lemmon, 2008). Subsequent studies have shown PH domains to be versatile structures that are not only involved in PPIn recognition, but are also used for mediating protein-protein interactions (Maffucci and Falasca, 2001; Lemmon, 2004; DiNitto and Lambright, 2006; Lemmon, 2007). In fact, most PH domains weakly bind to PPIn lipids with limited specificity, and only a small fraction, estimated at between 10–15%, exhibit high affinity and selective binding to PPIn headgroups (Rameh et al., 1997; Takeuchi et al., 1997; Isakoff et al., 1998; Kavran et al., 1998; Yu et al., 2004). Of the seven PPIn species found within cells, to date, PH domains that specifically recognize PtdIns4P, PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 have been described; including detailed characterizations of the structural features that determine the PPIn-binding specificities (Cozier et al., 2004; Balla, 2005; DiNitto and Lambright, 2006; Kutateladze, 2010). In addition to the recognition of anionic PPIn species, many PH domains have been shown to cooperatively target membranes through additional interactions with other lipid species (Knight and Falke, 2009; Vonkova et al., 2015); as well as a growing number of examples defining independent binding sites for non-PPIn lipids, including PtdSer (Uchida et al., 2011; Jian et al., 2015).
In general, although PH domains show relatively low sequence homology (~30%; Lemmon et al., 2002), they adopt a characteristic fold consisting of two nearly orthogonal β-sheets formed by seven β-strands, splayed into a group of three (β5-β7) and four (β1-β4), that are capped by a C-terminal α-helix (Figure 1a; Ferguson et al., 1994; Ferguson et al., 1995). Within the PH domain fold, there are six loops connecting the β-strands and, overall, the β-sheets are tightly packed, especially at the closed corners of the β-sandwich. Three extended loops connecting the β1-β2, β3-β4, and β6-β7 strands project into the membrane-binding interface, at the open end of the β-sandwich, and show considerable sequence variation across PH domains; which likely contributes to the differences observed in PPIn-binding specificities (Lemmon and Ferguson, 2001; DiNitto and Lambright, 2006). The canonical binding pocket for the PPIn headgroup is formed by the β1-β2 and β3-β4 strands, as well as the variable loops that connect them. In particular, a basic sequence motif in the β1-β2 loop, defined as Kxn(K/R)xR, has been proposed to serve as a general interaction platform for PPIn headgroups by recognizing vicinal phosphate pairs present in stereospecific positions on the inositol ring (Lemmon, 2007; Moravcevic et al., 2012). PH domains that contain the Kxn(K/R)xR motif all bind to PPIn lipids and this motif is retained in more complex sequence features that have previously been shown to determine the selective coordination of bis- and tris-phosphorylated PPIn species; specifically those with paired phosphate groups at adjacent 4- and 5- or 3- and 4-positions (Lietzke et al., 2000; Yu et al., 2004; Park et al., 2008). Interestingly, non-canonical binding modes of PPIn lipids have been demonstrated for PH domains lacking the Kxn(K/R)xR motif, including those from β-spectrin (Figure 1b; Macias et al., 1994; Hyvonen et al., 1995), the p62 subunit of TFIIH (general transcription factor IIH; Di Lello et al., 2005), ArhGAP9 (Rho GTPase-activating protein 9; Ceccarelli et al., 2007), Tiam1 (T-lymphoma and metastasis 1; Ceccarelli et al., 2007), and the yeast protein Slm1 (synthetic lethal with MSS4 protein 1; Anand et al., 2012). Binding of PPIn lipids to each of these non-canonical PH domains occurs on the opposite face of the β1-”2 loop, with the bound headgroup positioned on the side of the core β-barrel and between the loops that connect the β1-β2 and β5-β6 strands (Balla, 2005; DiNitto and Lambright, 2006). Outside of variations to the location of the PPIn-binding site, unique sequence features have also been shown to influence the selectivity of certain PH domains for PPIn species. In particular, a subclass of PH domains recognize one or both of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 with a remarkable degree of specificity and affinity, including those found in BTK, PKB, and the cytohesin family Arf GEF Grp1 (general receptor for phosphoinositides 1; Lemmon, 2008). The structural features contributing to this binding selectivity primarily involve sequence-specific elaborations of the variable loops. Specifically, the solved structure of Grp1 reveals a long twenty-residue insertion within the β6-β7 loop, which adopts a twisted β-hairpin structure and essentially extends the β-barrel from 7 to 9 strands (Figure 1c; Lietzke et al., 2000; Ferguson et al., 2000). The headgroup of PtdIns(3,4,5)P3 is contacted by conserved residues within the canonical PH domain pocket that is formed at the top of the β1-β2 and β3-β4 strands and lined by the β1-β2 loop (Lietzke et al., 2000; Ferguson et al., 2000). However, residues from the β-hairpin insertion replace the β3-β4 loop to form the second wall of the PPIn-binding pocket and provide two additional hydrogen bonds with the 5-phosphate that account for the high specificity of the Grp1 PH domains towards PtdIns(3,4,5)P3 (Lietzke et al., 2000; Ferguson et al., 2000). A clear pocket for the 5-phosphate group is also seen in the PH domain of BTK, but, unlike the unique insertion found within the Grp1 PH domain, this pocket forms from an extension of the β1-β2 loop that is able to envelop the 5-phosphate (Baraldi et al., 1999; Ferguson et al., 2000). In addition to structural studies, recent efforts using molecular dynamics simulations of diverse PH domains have revealed important new insights into the binding orientation (Psachoulia and Sansom, 2008; Lumb and Sansom, 2012; Lenoir et al., 2015; Naughton et al., 2016; Yamamoto et al., 2016) and the dynamics associated with the diffusivity of the domain (Yamamoto et al., 2017) during PPIn-dependent membrane recognition events. Taken together, it is apparent that sequence differences in the PPIn-binding interface are responsible for the observed heterogeneity in the PPIn selectivity of distinct PH domains and may also influence membrane residency or conformational dynamics during PPIn interactions.
Figure 1.
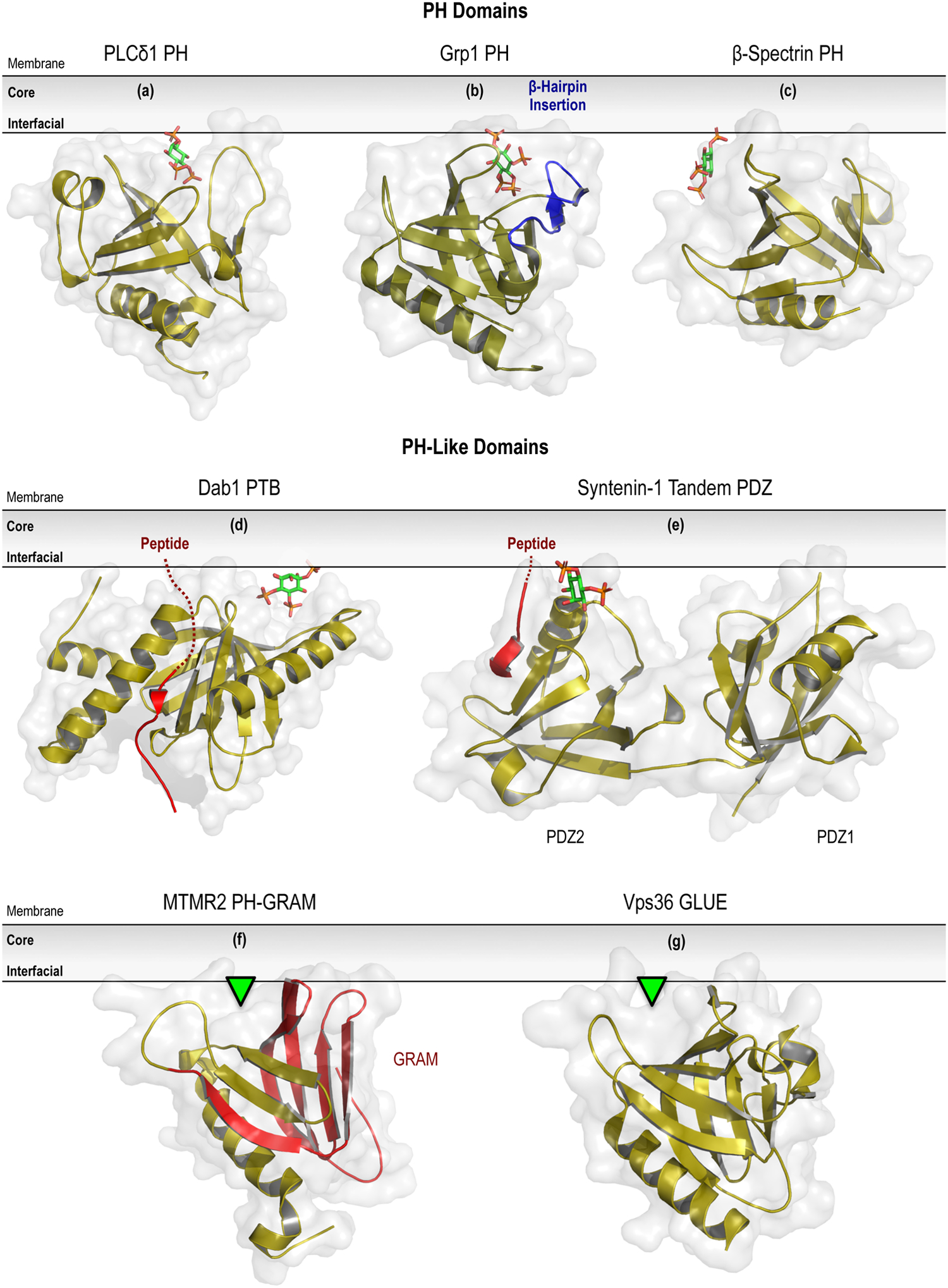
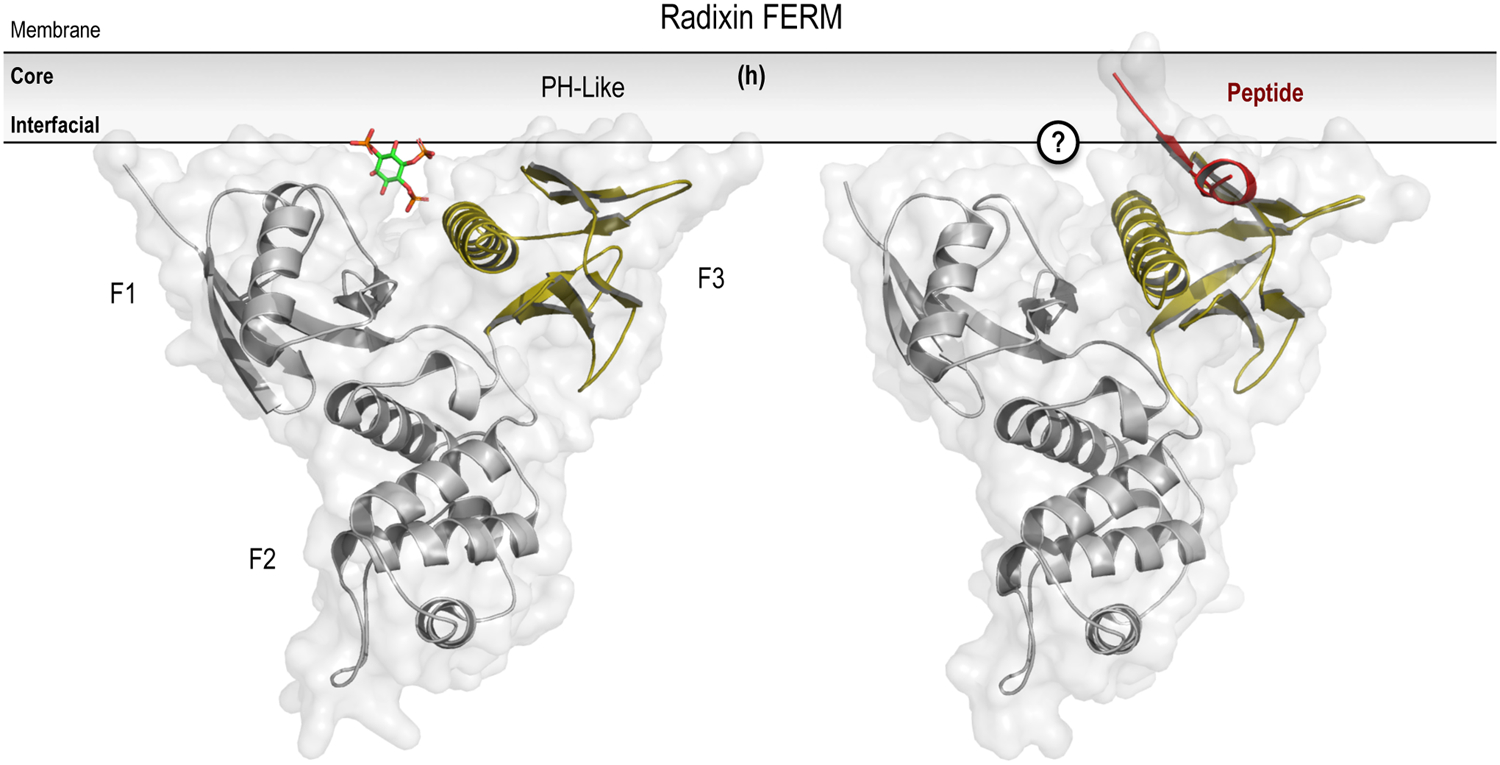
Structural variations on the PH superfold. Crystal structures of representative PH domains are shown, including examples of canonical (a; PLCδ1 PH domain in complex with Ins(1,4,5)P3; PDB entry 1MAI), elaborated (b; Grp1 PH domain in complex with Ins(1,3,4,5)P4; PDB entry 1FHX), and atypical (c; β-Spectrin PH domain in complex with Ins(1,4,5)P3; PDB entry 1BTN) PPIn-recognition modes. Coincident peptide (highlighted in red) and PPIn coordination is depicted for the PH-like PTB (d; Dab1 PTB domain ternary complex with ApoER2 peptide and PtdIns(4,5)P2; PDB entry 1NU2) and PDZ domains (e; Syntenin PDZ1 and PDZ2 tandem domains in a ternary complex with the Frizzled 7 C-terminal fragment and PtdIns(4,5)P2; PDB entry 4Z33). The structurally-related GRAM (f; isolated from within the structure of MTMR2; PDB entry 1LW3) and GLUE domains (g; Vps36 N-terminal domain; PDB entry 2CAY) are shown with their putative membrane-binding pose and PPIn-coordinating pockets highlighted by the green arrowhead. Notice that both may adopt an atypical PPIn-binding mode that is similar to that shown above in (c). Lastly, the unique inter-domain PPIn-binding surface of the Radixin FERM domain is depicted in association with either the PPIn lipid (left side; complex with Ins(1,4,5)P3; PDB entry 1GC6) or cognate peptide (shown in red) ligand (right side; complexed with ICAM-2 cytoplasmic peptide; PDB entry 1J19). For further details, please refer to Sections 5.1 (PH domains) and 5.2 (PH-like superfolds) of the text. Prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
Regardless of the sequence-specific variations that have been documented, overall, PH domains that exhibit PPIn-binding generally show pronounced electrostatic polarization; with strongly positive amino acid residues located at the membrane-binding surface (Macias et al., 1994; Blomberg and Nilges, 1997; Moravcevic et al., 2012). Following initial electrostatic interactions at the membrane interface, specific PPIn-binding is likely the primary tool for membrane-selective targeting and increased membrane residence. Interestingly, despite a general lack of hydrophobic or aromatic residues around the PPIn-coordinating pocket (Cho and Stahelin, 2005), evidence for penetration of surface-oriented hydrophobic residues into the interfacial region has been presented for the PLCδ1 PH domain using solid state NMR (Tuzi et al., 2003) and surface plasmon resonance (Flesch et al., 2005). Molecular dynamics simulations of other PH domains also suggest varying degrees of insertion of the PH domain fold into PPIn-containing monolayers (Manna et al., 2007; He et al., 2008; Lumb et al., 2011). However, overall, significant interfacial penetration does not seem to be a general feature of PH domains and is not a major driving force for membrane binding by PH domains; which are clearly more reliant on electrostatic attraction. Additionally, as mentioned above, despite examples of monovalent membrane recruitment, it is important to recognize that membrane binding of PH domains commonly require coincident binding to protein effectors. Coincidence detection by PH domains is not always a simple membrane localization signal, as growing evidence suggests that PH domains can also function at membrane interfaces as highly specific modules for communicating allosteric regulatory signals; including PPIn-mediated conformational switches (DiNitto and Lambright, 2006; Nawrotek et al., 2016; Roy et al., 2016). Both of these unique regulatory features exhibited by PH domains have been recently proposed to participate in the dynamic regulation of the Dbl superfamily Rho-GEF PREX1 (PtdIns(3,4,5)P3-dependent Rac exchanger 1). Briefly, allosteric regulation of PREX1-dependent GTP exchange activity by PtdIns(3,4,5)P3 binding is thought to occur in a stepwise fashion. Initial recruitment of PREX1 to the membrane involves transient electrostatic interactions with basic residues in the β3-β4 loop of the PH domain, which are stabilized by the coincident association of the fold to PM-anchored Gβγ heterodimers that are released from activated GPCRs (Cash et al., 2016). Gβγ-binding unmasks the PtdIns(3,4,5)P3-binding site within the PH domain fold, while subsequent conformational changes induced by PtdIns(3,4,5)P3 binding are transmitted through a hinge region at the junction between the PH and catalytic Dbl homology (DH) domain that results in the full activation of PREX1 (Cash et al., 2016). These mechanistic insights are consistent with previous data demonstrating synergistic regulation of PREX1 by PtdIns(3,4,5)P3- and Gβγ-mediated signals (Welch et al., 2002; Hill et al., 2005). Understanding how protein dynamics and inter-domain communication can be regulated at the membrane interface are extremely important areas for ongoing research. Descriptions of the molecular interactions between PPIn species and PH domains have provided some of the best demonstrations of stereoselective PPIn recognition as well as reveal how membrane-binding domains can function as integration centers that relay coincident protein- and lipid-derived signals.
5.2. PH Domain-Like Folds: PTB, PDZ, GRAM, GLUE, and FERM Domains
The slightly splayed β-sandwich superfold originally described for the PH domain has since been found in a series of structural homologs that are collectively referred to here as PH-like domains (Blomberg et al., 1999; Balla, 2005; Scheffzek and Welti, 2012). Despite limited sequence similarities, the conserved structural core of PH-like domains is commonly found in modular proteins implicated in the regulation of signal transduction and similarly possess binding sites for PPIn lipids, as well as surfaces that are involved in mediating protein-protein interactions (Lemmon, 2007; Scheffzek and Welti, 2012). A growing number of PH-like domains appear within proteins with activities in diverse subcellular compartments show variable lipid- as well as protein-binding partners. The identification of the structural conservation of the PH domain superfold included the early descriptions of the EVH1 (Enabled/VASP homology 1; Prehoda et al., 1999) and Ran-binding domain families (Vetter et al., 1999); however, neither of these folds are reported to exhibit PPIn-dependent regulation. Consequently, for the sake of this chapter, we will focus our discussion on representative PH-like domains with established PPIn-binding sites.
Originally characterized as protein modules that interact with tyrosine-phosphorylated peptides, specifically those containing the consensus sequence NPxY or other variants of this motif, a subset of PTB domains can also independently or simultaneously recognize PPIn headgroups with a broad range of affinities (DiNitto and Lambright, 2006; Kaneko et al., 2012). Compared to canonical PH domains, the PTB domain fold possesses a variable helical loop inserted between the β1 and β2 strands (Zhou et al., 1995; Zhou and Fesik, 1995). Solution structures of the Dab1 (disabled-1) PTB domain show that stereospecific PtdIns(4,5)P2 binding occurs at the outer surface of the helical insertion, distinct from the binding pockets described for both the canonical and atypical PH domains; whereas phosphorylated peptides associate within an elongated hydrophobic cleft contoured by the C-terminal α3 helix and the parallel β5 strand (Figure 1d; Yun et al., 2003; Stolt et al., 2003). Rationale mutagenesis and biophysical investigations of the Dab1 PTB domain demonstrate that PPIn-binding is requisite for the membrane localization and catalytic function of Dab1 in vitro as well as in vivo (Stolt et al., 2005; Huang et al., 2005; Xu et al., 2005). Furthermore, PPIn and peptide binding to the Dab1 PTB domain are energetically independent, and therefore do not exhibit any apparent cooperativity (Stolt et al., 2004). Outside of Dab1, the PTB domains of IRS-1 (insulin receptor substrate-1; Takeuchi et al., 1998; Dhe-Paganon et al., 1999) and Shc (Src homology 2 domain-containing transforming protein; Rameh et al., 1997; Ravichandran et al., 1997) have also been shown to selectively bind PPIn lipids, with some apparent selectivity for PtdIns(4,5)P2. In terms of the location for PPIn headgroup coordination, interactions with the Shc PTB domain are thought to involve a cluster of exposed basic residues that are located on the same side of the domain as the inserted helical loop; however, the PPIn-binding sites mapped in the Dab1 and Shc PTB domains do not appear to overlap and the residues implicated in PPIn recognition are not conserved across PTB domains (DiNitto and Lambright, 2006). Where investigated in depth, the ability of some PTB domains to coordinate PPIn lipids clearly contributes to the spatial organization and membrane adsorption of PTB domain-containing, particularly at the PM.
Similar to the PTB domain, the PDZ domain is another PH-like fold that typically functions during protein-protein interactions involving adaptor proteins and short peptide sequences, generally the last four to five residues, at the C-terminus of transmembrane proteins (Saras and Heldin, 1996; Nourry et al., 2003). However, more recent studies have shown that many PDZ domains can also bind internal peptide sequences as well as membrane phospholipids (Chang et al., 2011; Ivarsson, 2012; Mu et al., 2014); including an important role for PtdIns(4,5)P2 (Zimmermann, 2006; Wawrzyniak et al., 2013). Early studies of PPIn-PDZ interactions demonstrated that PtdIns(4,5)P2-binding to conserved tandem PDZ domains controlled the cellular localization of the molecular scaffolds syntenin-1 (Zimmermann et al., 2002) and syntenin-2 (Mortier et al., 2005). More recently, a series of large-scale screens of human PDZ domains, using a combination of in silico analyses and high-throughput binding assays, found that membrane association is a common property of PDZ domains, found in roughly 20–40%, and that PPIn lipids likely contribute to the cellular localization of a broad collection of PDZ domains; including roles for PPIn interactions with PDZ domain-containing effectors within the nucleus (Mortier et al., 2005; Wu et al., 2007; Chen et al., 2012; Ivarsson et al., 2013). Additionally, a small subset of PDZ domains were shown to bind PPIn lipids with relatively high-affinity; although in vitro binding studies, as well as prior characterizations of PDZ function, suggest that the stereospecificity for PPIn headgroups is limited (Zimmermann et al., 2002; Mortier et al., 2005; Ivarsson et al., 2011). Despite the generally low affinity interactions that have been described, in the few cases investigated in detail, PPIn-binding does appear to be important for controlling the cellular functions of PDZ domain-containing protein adaptors (Wawrzyniak et al., 2013). Additionally, where established, it is apparent that PDZ domains can interact with PPIn lipids through different and complex membrane-binding modes (Gallardo et al., 2010); including biophysical investigations identifying surface-exposed electrostatic or hydrophobic residues that facilitate competitive as well as cooperative binding of PDZ domains to PPIn and peptide ligands (Ivarsson, 2012; Wawrzyniak et al., 2013; Ernst et al., 2014). Although generally thought to lack a well-defined PPIn-binding pocket, a recent crystal structure of the tandem PDZ domains of syntenin in complex with both PtdIns(4,5)P2 and a cognate C-terminal peptide fragment, shows that the backbone of the bound peptide actually provides direct contacts that help to form the PtdIns(4,5)P2-binding interface and function to stabilize the inositol headgroup (Figure 1e; Egea-Jimenez et al., 2016). These new structural studies support evidence for synergistic binding of PPIn lipids and peptides to the tandem PDZ domain and suggest that peptide binding likely reinforces the interaction with membrane-embedded lipids (Egea-Jimenez et al., 2016). Additional structures of intact PDZ domain complexes will be extremely informative for understanding the extent to which the seemingly variable PPIn-binding sites can communicate with the well-mapped peptide-binding groove. Overall, as highlighted previously for the classical PH domains, coincident recognition of lipid headgroups and membrane-localized binding partners represents an important regulatory principle that is utilized by diverse families of adaptor proteins to mediate a wide range of cellular processes. The presence of unique variations on the PH superfold, and the PTB and PDZ scaffolds in particular, facilitate diverse PPIn- and peptide-binding activities during membrane-initiated signaling events.
In addition to membrane-binding domains that simply incorporate the PH superfold, other examples of PH-like domains include the reorganization or assembly of the canonical PH module from unique sequence-specific variats. For instance, the GRAM domain was originally identified as small motif predicted based on sequence homology to occur in approximately 180 eukaryotic proteins, including several important membrane-associated proteins such as the myotubularin (MTM) family of PPIn phosphatases (Doerks et al., 2000). The solved structure of MTMR2 (myotubularin-related protein 2) subsequently revealed that the GRAM domain motif consists of five β-strands, but is part of a larger protein fold that incorporates adjacent sequence features to form a fold with a topology that was extremely similar to the canonical PH domain from pleckstrin (Figure 1f; Begley et al., 2003). Sequence alignments of other representative members of the GRAM domain family showed high conservation of the residues involved in forming the hydrophobic core of the extended GRAM-PH motif, which is also referred to by some as the PH/G domain (Begley et al., 2003). Binding studies have shown that the PH/G domains of MTM1 (Tsujita et al., 2004) and MTMR3 (Lorenzo et al., 2005) preferentially bind to PtdIns(3,5)P2 and PtdIns5P, respectively; whereas the PH/G domain of MTMR2 interacts with both PtdIns(3,5)P2 and PtdIns5P (Berger et al., 2003). Crystallographic and deuterium exchange studies of MTMR2 show that the PH/G domain is strongly electropositive along the surface-exposed β5-β6 and β7-α1 loops (Begley et al., 2006). Although this electrostatically polarized surface represents the most likely interface for the recognition of PPIn-containing membranes, structural studies have yet to detect PPIn lipids associated with the PH/G domain. Alternatively, independent of PPIn-binding, the PH/G domain clearly plays important roles for mediating protein-protein interactions as well as during the allosteric control of MTM catalytic activity (Begley and Dixon, 2005; Clague and Lorenzo, 2005; Hnia et al., 2012). Interestingly, following the description of the PH/G domain, a novel GRAM-like motif dubbed the GLUE domain, was identified as a conserved sequence feature within the N-terminal region of the metazoan Vps36 (vacuolar protein-sorting-associated protein 36) family of ubiquitin-binding proteins (Slagsvold et al., 2005). Notably, Vps36 and its mammalian ortholog, Eap45 (ELL-associated protein of 45 kDa), are components of the ESCRT-II (endosomal sorting complex required for transport II) complex, which plays an essential role during diverse membrane trafficking events, including the biogenesis of multi-vesicular bodies (MVBs; Saksena et al., 2007; Williams and Urbe, 2007; Hurley, 2008). Functional studies revealed that the GLUE domain binds to both ubiquitin and various 3-phosphorylated PPIn species; suggesting a possible role for PPIn lipids during the coordination of membrane-targeting and cargo recognition within the endosomal system (Slagsvold et al., 2005). The solved structure of the GLUE domain of Vps36 shows that it has a split PH domain architecture, with a yeast-specific insertion of two NZF (Npl4-like zinc finger) domains that are oriented away from the membrane-binding surface (Teo et al., 2006). The walls of the PPIn-binding pocket, which shows high selectivity for mono-phosphorylated PtdIns3P, are built by the β1-β2, β5-β6, and β7-α1 loops and therefore forms outside of the canonical PH domain binding pocket (Figure 1g; Teo et al., 2006), on the opposite face of the β1-β2 loop, in a manner analogous to the atypical PPIn-binding site originally characterized for the β-spectrin PH domain (Macias et al., 1994; Hyvonen et al., 1995). The location of the PPIn-binding pocketed has since been confirmed by subsequent structures of human Eap45; however, the missing insertion of the NZF domains in the mammalian GLUE domain, reveals an alternative site for ubiquitin binding that lies along one edge of the core β-sandwich and distinct from the PPIn-binding pocket (Alam et al., 2006; Hirano et al., 2006). Overall, these collected structures of the GLUE domain help to demonstrate how recognition of ubiquitinated cargoes and endosomal membranes can be coupled during protein sorting in MVBs; once again pointing to the PH-like superfold as a common substrate for coincidence detection throughout biological systems. In fact, additional protein-protein contacts have been mapped between the GLUE domain and adjacent components of the ESCRT-II machinery, which highlight the critical role for multivalent membrane binding initiated by the PPIn-binding GLUE domain during ESCRT-II actions on protein and lipid sorting (Im and Hurley, 2008).
Lastly, unlike the other PH-like domains discussed, the FERM domain incorporates the PH superfold into a much larger multi-domain structure with clear deviations in site utilized for PPIn-binding. Briefly, FERM domains are present is a variety of mammalian proteins (Chishti et al., 1998) that function as important macromolecular scaffolds that link the PM with the cytoskeleton through complex binding interactions with both proteins and lipids (Frame et al., 2010; Moleirinho et al., 2013; Baines et al., 2014). FERM domains are organized by intimate inter-domain contacts and consist of three globular lobes (F1, F2, and F3), including a PH-like domain fold that forms the F3 subdomain and participates in the selective recognition of PtdIns(4,5)P2 (Hamada et al., 2000). Binding to PtdIns(4,5)P2 is thought to release an auto-inhibitory intermolecular interaction between the FERM domain and the C-terminal tail of ERM (ezrin, radixin, and moesin) proteins (Pearson et al., 2000; Edwards and Keep, 2001; Jayasundar et al., 2012). Interestingly, the PtdIns(4,5)P2 headgroup is coordinated within a shallow basic cleft located between the F1 and F3 subdomains, distinct from any of the binding surfaces mapped on other PH-like domains, by a relatively small number of hydrogen bonds that primarily target the 4-position phosphate group (Figure 1h; Hamada et al., 2000; Smith et al., 2003). The relative lack of stereospecificity observed within this binding cleft, coupled to reports of FERM domains with an altered F1-F3 cleft (Ceccarelli et al., 2006) or possessing multiple non-specific PtdIns(4,5)P2-binding motifs (Bompard et al., 2003; Zhao et al., 2010), raised the possibility that FERM domains may actually sense the density of PPIn lipids within membranes. Subsequent studies using fluorescence anisotropy measurements and molecular dynamics simulations suggest that the FERM domain from moesin can associate simultaneously with multiple PPIn headgroups and provide no evidence for the presence of discrete PPIn-binding pockets or for interactions of the FERM domain with the acyl chains of the lipid bilayer (Senju et al., 2017). These data support the idea that membrane binding of FERM domains is dependent on multivalent electrostatic interactions, potentiated by anionic PtdIns(4,5)P2, and is also in agreement with the general absence of hydrophobic or aromatic residues at the binding interface (Cho and Stahelin et al., 2005). Consequently, although interactions with PtdIns(4,5)P2 are requisite for PM anchoring, the membrane-targeting of the FERM domain module does not appear to require a defined stereospecific PPIn-binding pocket.
5.3. PX Domains
PX domains were originally identified in the p40phox and p47phox subunits of the phagocyte NADPH oxidase complex (Ponting, 1996; Sato et al., 2001; Wishart et a., 2001) and have subsequently been described in a variety of proteins involved in membrane trafficking and cellular signaling; including numerous sorting nexins (SNXs) as well as the class II PI3Ks (Seet and Hong, 2006). Functional studies quickly characterized PX domains as short membrane-binding modules, consisting of 100–140 residues, which show specificity for 3-phosphorylated PPIn lipids (Cheever et al., 2001; Ellson et al., 2001; Hiroaki et al., 2001; Kanai et al., 2001; Song et al., 2001; Yu and Lemmon, 2001) and a general preference for PtdIns3P (Seet and Hong, 2006). Explicit examples of PX domain recognition of substrates outside of PtdIns3P include: PtdIns(3,4)P2 binding to p47phox (Karathanassis et al., 2002), PtdIns(3,4,5)P3 selectivity for CISK (cytokine-independent survival kinase; Xu et al., 2001; Xing et al., 2004), as well as preferential recognition of PtdIns4P by the yeast protein Bem1 (bud emergence Protein 1; Ago et al., 2001; Stahelin et al., 2007) and PtdIns(4,5)P2-specific coordination by Class II PI3K-C2α (Song et al., 2001; Stahelin et al., 2006; Parkinson et al., 2008). Despite some heterogeneity in the PPIn species that are recognized, crystal structures of the p40phox (Bravo et al., 2001) and p47phox (Karathanassis et al., 2002) subunits both revealed a characteristic PX domain fold, which consists of an N-terminal three-stranded β-meander that is followed by a C-terminal α-helical subdomain consisting of three or four α helices; two of which are linked by an elongated poly-proline loop (Figure 2a). The PPIn isomer binds within a relatively narrow and positively charged groove that is formed by a β-bulge in the β1 strand that twists the β-sheet to form one wall of the binding pocket, as well as through specific contacts with the elongated loop joining the α1 and α2 helices (Cheever et al., 2006; Moravcevic et al., 2012). Specific recognition of the PPIn headgroup is facilitated by acidic membrane environments and is accompanied by the insertion of hydrophobic and aromatic residues within the flexible α1-α2 loop, also referred to as the membrane insertion loop (MIL), into the bilayer (Seet and Hong, 2006). Although the alignment of PX domains shows considerable variability in the sequence of the MIL, the presence of a clear hydrophobic motif and membrane penetration of this region appears to be highly conserved across PX domains (Seet and Hong, 2006; Kutateladze, 2010). Conserved basic residues surrounding the deep PPIn-binding groove and variable loop are involved in electrostatic interactions that facilitate substrate recognition and also enhance affinity of the PX domain for PPIn substrates by inducing insertion of the MIL (Stahelin et al., 2003a; Stahelin et al., 2004; Malkova et al., 2006; Stahelin et al., 2006). Three core motifs essential for PPIn binding, including RRYx2Fx2Lx3L of the β3-α1 loop, Px2PxK within the MIL, and RR/Kx2L of α2 are present within most PX domain sequences (Kutateladze, 2010). Interestingly, adjacent to the PPIn-binding pocket, an additional well-defined binding site for PtdSer or PtdOH has been described in the p47phox PX domain (Karathanassis et al., 2002; Stahelin et al., 2003a). Simultaneous occupation of both lipid-binding pockets is thought to modulate the local electrostatic potential and induce a conformational change within the MIL that promotes the insertion of hydrophobic residues into the membrane (Zhou et al., 2003; Cho and Stahelin, 2005). This cooperative binding mechanism appears to be an exaggeration of the common non-specific electrostatic interactions that normally initiates the adsorption of PPIn-binding domains onto anionic membrane surfaces. Outside of interactions with additional lipid substrates, PX domains are also involved in protein-protein interactions; including structural descriptions of the intramolecular binding between a C-terminal SH3 domain and the conserved Px2PxK motif within PX domain of p47phox that functions to prevent PtdIns(3,4)P2 binding (Hiroaki et al., 2001; Karathanassis et al., 2002). Similar intramolecular interactions have been demonstrated between the PX and SH3 domains of the fission yeast protein Scd2 (Endo et al., 2003), as well as intermolecular binding of p40phox or p47phox with the cytoskeletal scaffold moesin (Wientje et al., 2001). An unbiased genome-wide two-hybrid screen using isolated PX domains from yeast was also able to identified several putative PX domain-binding proteins that included known membrane-interacting effectors with roles in vesicular trafficking (Vollert and Uetz, 2004). However, overall, it remains unclear the extent to which protein-protein interactions influence or reinforce the PPIn-binding roles of PX domains.
Figure 2.
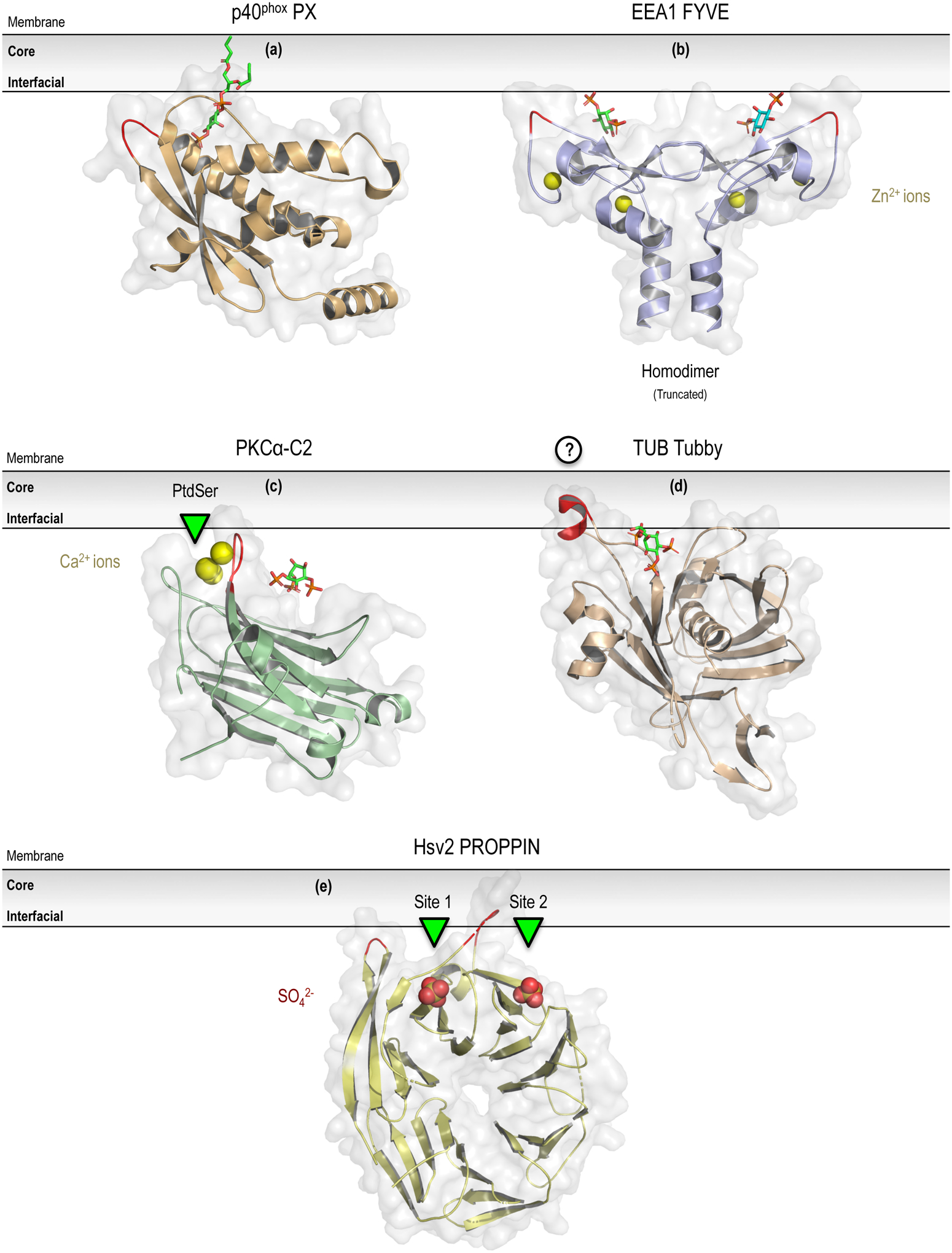
Diverse domains within peripheral membrane protein exhibiting stereospecific PPIn-binding. Selected examples of unique PPIn-binding domains from the PX (a; p40phox PX domain in complex with PtdIns3P; PDB entry 1H6H), FYVE (b; EEA1-FYVE domain homodimer bound to Ins(1,3)P2; PDB entry 1JOC), C2 (c; PKCα-C2 domain in complex with Ca2+ and PtdIns(4,5)P2; PDB entry 3GPE), Tubby (d; C-terminal domain of Tubby bound to PtdIns(4,5)P2; PDB entry 1I7E), and PROPPINs (e; yeast PROPPIN Hsv2; PDB entry 4EXV) domain families that exhibit stereospecific coordination of the target PPIn lipid. In each of these structures, sequences features identified as defined membrane insertion elements are highlighted in red. For further details about each of these domains, please refer to Sections 5.3 (PX), 5.4 (FYVE), 5.5 (C2), 5.6 (Tubby), and 5.7 (PROPPINs) of the text. Prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
5.4. FYVE Domains
FYVE domains are highly homologous cysteine-rich domains of 70–80 amino acids that are found in around 30 human proteins that have been shown to broadly participate in vacuolar sorting or endocytosis through direct binding to PtdIns3P (Stenmark et al., 1996; Gaullier et al., 1998; Simonsen et al., 1998; Burd and Emr, 1998); although the FYVE domain from protrudin has been proposed to associate with PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 both in vitro as well as in cells (Gil et al., 2012). The overall architecture of the FYVE domain is comprised of two anti-parallel β-hairpins and a small C-terminal α-helix, which is stabilized by two zinc-binding clusters containing four CxxC motifs in a cross-braced topology (Figure 2b; Kutateladze, 1999; Misra and Hurley, 1999). A conserved basic motif, defined as RR/KHHCR, in the first β-strand surrounding the third zinc-coordinating cysteine forms a shallow positively-charged PtdIns3P-binding pocket (Dumas et al., 2001; Kutateladze, 2006). Additional WxxD and RVC signature motifs not only help to distinguish FYVE domains within the larger family of zinc-coordinating RING fingers, but also, along with the RR/KHHCR motif, are centrally involved in the coordination of the PtdIns3P headgroup (Kutateladze, 2010). Due to the relatively shallow PtdIns3P-binding pocket and coordination of only a single phosphate, FYVE domains bind the monomeric PtdIns3P headgroup rather weakly and FYVE domain-containing effectors tend to require multivalent mechanisms for membrane localization (Dumas et al., 2001; Kutateladze, 2004). Importantly, a variable-length turret loop next to the PtdIns3P-binding pocket contains hydrophobic residues that insert into the lipid bilayer and stabilize membrane-bound complexes (Misra and Hurley, 1999; Kutateladze and Overduin, 2001; Stahelin et al., 2002; Diraviyam et al., 2003; Kutateladze et al., 2004; Brunecky et al., 2005). Basic and polar residues flank the turret loop and play an important role in non-specific electrostatic interactions with acidic lipids, including PtdSer and PtdOH, at the membrane interface and, similar to the PX domain, can be used to drive initial membrane docking (Kutateladze, 2010). Based on homology with the C1B domain of PKCδ, FYVE domains were originally proposed to bind with the long axis of the domain perpendicular to the membrane surface, which would facilitate the simultaneous recognition of PtdIns3P within the binding pocket and membrane insertion of the tip of the turret loop (Misra and Hurley, 1999). Biophysical and computational studies provide support for this mechanism and show that electrostatic association and PtdIns3P binding drive the membrane insertion of the hydrophobic and aromatic residues within the turret loop (Stahelin et al., 2002; Diraviyam et al., 2003). Interestingly, membrane association of the FYVE domain exhibits pH sensitivity, which is regulated by the adjacent histidine residues found within the core RR/KHHCR motif responsible for coordinating the 3-position phosphate group of PtdIns3P (Lee et al., 2005; He et al., 2009). Protonation of the motif occurs at acidic pH and reinforces the interactions between the PtdIns3P headgroup and the positively charged histidine pair, whereas deprotonation promotes the release of the PtdIns3P ligand and causes rapid membrane dissociation (Lee et al., 2005; He et al., 2009). Membrane avidity of FYVE domain interactions can also be enhanced by dimerization (Callaghan et al., 1999; Lawe et al., 2000; Dumas et al., 2001; Mao et al., 2000; Hayakawa et al., 2004); including a structural characterization of concurrent binding to two PtdIns3P headgroups through a parallel coiled-coil homodimer that juxtaposes the two FYVE domains of EEA1 (early endosomal antigen 1; Dumas et al., 2001). However, sequence analysis of FYVE domains show heterogeneity in their relative hydrophobicity at the putative dimer interface, as well as in residues within the membrane-penetrating turret loop, suggesting that individual FYVE domains are likely to show substantial variance with regards to their propensity for dimerization and orientation at the membrane interface (Stahelin et al., 2014). Regardless, the exquisite selectivity of the FYVE domain helps to demonstrate the importance of coordinate electrostatic and hydrophobic interactions with membrane surfaces during the specific recognition of PPIn lipids. More generally, coincident or multivalent membrane recognition modes are likely requisite for establishing high-affinity binding interactions with mono-phosphorylated PPIns.
5.5. C2 Domains
Originally identified as one of two regulatory domains in PKC (Ono et al., 1989; Osada et al., 1990), C2 domains have since been characterized as versatile membrane-interacting modules that are found in close to 150 different human proteins (Cho and Stahelin, 2006; Corbalan-Garcia and Gomez-Fernandez, 2014a,b). Canonical C2 domains show a common fold that consists of an eight-stranded antiparallel β-sandwich that is connected by variable surface loops (Shao et al., 1996). The majority of C2 domains show Ca2+-dependent binding to common anionic or zwitterionic membrane lipids, including PtdSer (Verdaguer et al., 1999; Stahelin et al., 2003c) or PtdCho (Perisic et al., 1998; Nalefski et al., 1998), through a lipid-binding site that is acidic in character rather than basic; as in PH, PX, and FYVE domains (Moravcevic et al., 2012). In general, Ca2+ ions influence C2 domain binding by enhancing the positive electrostatic potential around the Ca2+-binding loops to accelerate association with anionic membranes (Rizo and Sudhof, 1998; Murray and Honig, 2002) and Ca2+ can also induce local structural rearrangements that facilitate membrane binding (Sutton et al., 1995; Grobler et al., 1996; Davletov et al., 1998; Bittova et al., 1999; Kulkarni et al., 2002; Lai et al., 2010; Alwarawrah and Wereszczynski, 2017). Additionally, in some C2 domains, Ca2+ may slow membrane dissociation by directly coordinating lipid headgroups through Ca2+-mediated bridging (Verdaguer et al., 1999) or induce partial membrane penetration of aromatic residues surrounding the binding interface (Frazier et al., 2002; Kulkarni et al., 2002; Kohout et al., 2003; Stahelin et al., 2003c; Morales et al., 2016). Irrespective of the Ca2+ involvement, membrane recognition by C2 domains appears to occur without a great degree of specificity, which is not surprising since C2 domains lack a well-defined lipid-binding pocket (Stahelin et al., 2014; Corbalan-Garcia and Gomez-Fernandez, 2014a,b). However, outside of the canonical lipid- and Ca2+-coordinating surface, a small patch of positively-charged residues on the concave side of the β-sandwich, termed the polybasic cluster or cationic β-groove, has been shown to play an important role in the specific recognition of membrane lipids, including PPIn species (Cho and Stahelin, 2006; Guerrero-Valero et al., 2009; Corbalan-Garcia and Gomez-Fernandez, 2014a,b). The length and net electrostatics of the cationic β-groove, as well as the surface loops, are highly variable across C2 domains and the relative contribution of the two lipid-coordinating sites can be altered as a function of the intracellular Ca2+ concentration (Cho and Stahelin, 2006; Stahelin et al., 2014). The stereospecific recognition of PPIn species has been best characterized by studies of the PKCα-C2 domain, which binds predominantly to PtdIns(4,5)P2, but also other PPIn lipids, with nanomolar affinity (Figure 2c; Sanchez-Bautista et al., 2006; Manna et al., 2008). Binding of PtdIns(4,5)P2 to the PKCα-C2 occurs through interactions of the inositol headgroup with three lysine (K197, K209, and K211), one asparagine (N253), and two aromatic (Y197 and W245) residues within the cationic β-groove (Guerrero-Valero et al., 2009). Structure-based alignments of C2 domains suggests that these six residues likely form a consensus PtdIns(4,5)P2-interaction motif; although variability in the polybasic cluster exists, especially with regards to the conservation of K211 (Corbalan-Garcia and Gomez-Fernandez, 2014a,b). Interestingly, despite some controversies in the sequence of interactions, PPIn-association with the C2 domain appears to augment PtdSer binding by increasing the duration of membrane residency (Manna et al., 2008; Honigmann et al., 2013). Cooperative binding as a result of coincident Ca2+ and PtdIns(4,5)P2 signals has also been demonstrated for the C2B domain of synaptotagmin 1 (van den Bogaart et al., 2012; Guillen et al., 2013), C2C domain of ESyt1 (extended-synaptotagmin 1; Giordano et al., 2013), as well as for the C2A and C2B domains of rabphilin3A (Chung et al., 1998; Coudevylle et al., 2008; Montaville et al., 2008; Guillen et al., 2013). In addition, Ca2+-independent binding PtdIns(4,5)P2 has been described for the C2C domains of ESyt2 and Esyt3 (Giordano et al., 2013). Other examples of C2 domain recognition of PPIn species include promiscuous recognition of PPIn lipids by the Rasal C2B domain (Sot et al., 2013) and Ca2+-dependent interactions between mono-phosphorylated PPIn species and the KIBRA (KIdney/BRAin protein) C2 domain (Duning et al., 2013). Selective binding of PtdIns(3,4,5)P3 has been demonstrated for the DHR-1 (dock homology region-1) domain of the Dock family of atypical Rho-GEFs; which uses an elaborated C2 domain scaffold (Premkumar et al., 2010). Interestingly, structural and functional studies suggest that coordination of the PtdIns(3,4,5)P3 headgroup occurs within a basic pocket generated by extended surface loops, rather than through contacts with the cationic β-groove (Premkumar et al., 2010). Taken together, these studies clearly demonstrate the diversity of PPIn interactions with C2 domains and also highlight the how the complexity of multivalent binding events can be integrated at the membrane interface. Future studies should look to establish functional correlates between the diverse membrane-bound states of C2 domains, which appear to be highly sensitive to distinct combinations of anionic membrane lipids and Ca2+ ions, and their abilities to coordinate conformational dynamics within or between diverse macromolecular protein complexes.
5.6. Tubby Domains
The tubby domain consists of a roughly 260 amino acid module that is found within the C-terminus of members from the tubby-like protein (TULP) family of transcription factors (Kleyn et al., 1996; Noben-Trauth et al., 1996; Carroll et al., 2004). The isolated tubby domain displays PtdIns(4,5)P2-dependent membrane association in vitro as well as PtdIns(4,5)P2-mediated targeting to the PM within intact cells (Santagata et al., 2001; Szentpetery et al., 2009). Structural descriptions of the tubby C-terminal domain reveal a unique fold comprised of a closed β-barrel, consisting of 12 antiparallel β-strands that surround a central hydrophobic α-helix (Figure 2d; Boggon et al., 1999; Santagata et al., 2001). Coordination of the PtdIns(4,5)P2 headgroup occurs within a relatively shallow and positively charged cavity that results in a general lack of stereospecificity for PtdIns(3,5)P2 or monophosphorylated PPIn lipids relative to PtdIns(4,5)P2, PtdIns(3,4)P2, or PtdIns(3,4,5)P5 (Santagata et al., 2001). Recognition of the bound PtdIns(4,5)P2 requires specific interactions between the 4-position phosphate with conserved basic residues K330 and R332, as well as coordination of the inositol ring at the 3-position hydroxyl group by R363 (Santagata et al., 2001; Mukhopadhyay and Jackson, 2011). Of these PPIn-coordinating residues, K330 is positioned to interact with adjacent phosphate groups, which may help to explain the high selectivity of the tubby domain for bis- or tris-phosphorylated PPIn lipids with adjacent phosphate groups; including clear selectivity for PPIn species phosphorylated at both the 4- and 5-positions (Santagata et al., 2001). An adjacent loop that flanks the binding cavity, as well as polybasic patches on the tubby protein surface, may assist with high-affinity membrane interactions by associating with the interfacial region or through inserting into the membrane (Moravcevic et al., 2012). It is also important to realize that in addition to the selective recognition of PtdIns(4,5)P2, the tubby domain functions as an important transcriptional regulator by directly binding to double-stranded DNA; a process that once again capitalizes on the positively-charged binding surface described above (Boggon et al., 1999). Consequently, targeting of TULP proteins to PtdIns(4,5)P2 within the PM has been suggested to prevent nuclear localization and sequester TULP away from effectors within the nucleus (Santagata et al., 2001; Carroll et al., 2004). Although unlikely to be subject to coincident-binding within the PM, given the growing roles for nuclear PPIn lipids, understanding the relationship between selective PPIn coordination and the DNA-binding activity of TULPs within the nucleus could be an interesting area to investigate.
5.7. PROPPINs
The PROPPINs fold was originally described within a family of eukaryotic membrane-binding proteins that includes the important yeast macroautophagy effector Atg18 (autophagy-related protein 18; Michell et al., 2006). In general, PROPPINs consist of a seven-bladed β-propeller (Krick et al., 2012; Baskaran et al., 2012) and contain a conserved FRRG motif that is responsible for the specific recognition of PtdIns3P or PtdIns(3,5)P2 (Dove et al., 2004; Stromhaug et al., 2004; Krick et al., 2006; Obara et al., 2008). Recent solution structures show that PROPPINs contain two PPIn-binding sites, which are both localized at the rim of the β-propeller, and the side chains of each arginine within the conserved FRRG motif participate in the coordination of PPIn lipids within both binding pockets (Figure 2e; Krick et al., 2012; Baskaran et al., 2012). Interestingly, PROPPINs are thought to bind to membranes with an edge-on geometry that involves the insertion of aromatic residues into the membrane from within a flexible and exposed loop that protrudes from the β-propeller core and connects the two outer strands of blade six (6CD loop; Baskaran et al., 2012). Due to penetration into the membrane bilayer, PROPPINs such as Atg18 have been shown to bind more strongly to membrane-embedded PPIn lipids compared to short chain analogs or isolated headgroups (Lemmon, 2008). Membrane recognition is also thought to be curvature dependent and the initial targeting of the PROPPINs fold likely requires non-specific electrostatic interactions that are reinforced by the selective coordination of PPIn species and insertion of the flexible 6CD loop (Busse et al., 2015). Overall, the presence of two PPIn-binding sites, as well as a defined loop for membrane penetration, confer PROPPINs with the ability to interact with PtdIns3P- or PtdIns(3,5)P2-containing membranes with high avidity and affinity. Following membrane association, the exposure of the relatively large PROPPINs fold beyond the membrane interface could facilitate protein-protein interactions, including a recent report describing oligomerization of Atg18 upon binding to the membrane surface (Scacioc et al., 2017), which are likely to contribute to the membrane-targeting and function of PROPPINs in vivo (Michell et al., 2006; Busse et al., 2015).
5.8. ENTH, ANTH, and BAR Domains
In addition to selective interactions with discrete lipid headgroups, a subset of PPIn-binding domains are able to coordinately recognize or directly influence the local degree of membrane curvature; an important general feature of biological membranes (Antonny et al., 2011; Baumgart et al., 2011; Jarsch et al., 2016). Examples of these specialized curvature-sensitive binding modules include the ENTH, ANTH, and BAR domain families; which all play important roles during complex biological processes that involve substantial membrane deformation events (Itoh and De Camilli, 2006; Lemmon, 2008). The structurally-related ENTH, and later ANTH, domains were originally identified based on homology to an N-terminal region of epsin (Chen et al., 1998; Kay et al., 1999) and have since been identified within a small family of clathrin adaptor proteins that function as important regulators of membrane endocytosis as well as participate in additional aspects of vesicular trafficking (Itoh et al., 2001; De Camilli et al., 2002; Legendre-Guillemin et al., 2004). The ENTH and ANTH domains both consist of a superhelical solenoid of α-helices that are connected by loops of varying lengths; with the ANTH domain C-terminally extended by one or more α-helices compared with the ENTH domain (Itoh and De Camilli, 2006). Despite structural similarities and a shared preference for PtdIns(4,5)P2, the PPIn-binding modes observed for the ENTH and ANTH domains are quite distinct. Briefly, the structure of the epsin ENTH domain shows that binding of the PtdIns(4,5)P2 occurs within a well-defined pocket that makes extensive contacts with both the PPIn headgroup and glycerol backbone (Figure 3a,b; Ford et al., 2002); whereas the coordination of PtdIns(4,5)P2 by the ANTH domain relies on interactions between the PPIn phosphate groups and a surface-exposed patch of basic residues, formed by helices α1 and α2, that appears to be part of a consensus Kx9Kx(K/R)(H/Y) PPIn-binding motif (Ford et al., 2001; Mao et al., 2001; Itoh and De Camilli, 2006). An amphipathic α-helix located at the N-terminus, referred to as helix α0, was unseen in previous crystal structures of the ANTH domain (Ford et al., 2001; Miller et al., 2011) and therefore, was thought to be a specific structural feature of the ENTH domain (Ford et al., 2002). However, recent studies have identified helix α0, which turns out to be an extension of helix α1, within a variety of ANTH domains (Silkov et al., 2011); including detailed functional characterizations of helix α0 in the ANTH domain of the ubiquitous mammalian clathrin adaptor CALM (clathrin-assembly lymphoid myeloid leukaemia protein; Miller et al., 2015). Interestingly, helix α0 is unstructured in both the ENTH and ANTH domains, but becomes ordered upon binding to PtdIns(4,5)P2 (Ford et al., 2002; Miller et al., 2015). PtdIns(4,5)P2-induced structural rearrangements allow the hydrophobic face of helix α0 to penetrate deeply into targeted membranes to promote positive curvature through localized deformation of the membrane leaflet (Ford et al., 2002; Stahelin et al., 2003b; Kweon et al., 2006; Yoon et al., 2010; Lai et al., 2012; Miller et al., 2015). Unlike the ANTH domain, the ordered helix α0 of the ENTH domain contacts the PPIn headgroup within the binding pocket; seemingly to confer additional stereospecificity and possibly slow membrane dissociation kinetics (Hyman et al., 2000; Ford et al., 2002; Stahelin et al., 2003b; Lemmon, 2008). Consequently, relative to ENTH domains, the ANTH domain possesses relatively low affinity for PtdIns(4,5)P2 and is also quite promiscuous in terms of PPIn selectivity (Lemmon, 2003; Stahelin et al., 2003b). The apparent differences in the PPIn-binding modalities, as well as subtle discrepancies in the depth of the helix α0 insertion, support the idea that membrane-binding of proteins containing the ENTH and ANTH domains are likely to serve distinct functional roles during endocytosis. However, a recent cryo-electron microscopy study of clathrin adaptors from yeast suggests that ENTH and ANTH domains may co-assemble in a PtdIns(4,5)P2-dependent manner and form an organized oligomeric lattice that links polymerized clathrin to the membrane during remodeling events essential for endocytosis (Skruzny et al., 2015). The regular patterning of the ENTH and ANTH domains appears to require ENTH-mediated contact with the membrane leaflet through both its amphipathic α0 helix and PtdIns(4,5)P2-binding pocket, while the ANTH domain stabilizes the oligomer by contacting the ENTH domain but not the membrane (Skruzny et al., 2015). The extent to which this assembly occurs within other model systems has yet to be determined; although, in addition to a potential role in protein-protein interactions, membrane insertion of the ANTH domain from CALM appears to play an important role in promoting membrane curvature and defining the size of clathrin-coated vesicles in mammals (Miller et al., 2015). Overall, these studies identify an essential role for localized PPIn-binding by the ENTH and ANTH domains during the coordinate regulation of clathrin-mediated endocytosis. Future studies investigating the temporal relationship and interactions between ENTH and ANTH domain-containing effectors are still required to understand how the handling of PPIn lipids, and PtdIns(4,5)P2 in particular, is controlled during the formation of a clathrin-coated vesicle.
Figure 3.

PPIn-binding domains associated with the generation or recognition of membrane curvature. A comparison of the different PPIn-binding modes used by the membrane deforming ENTH (a; epsin ENTH bound to Ins(1,4,5)P3; PDB entry 1H0A) and ANTH (b; N-terminal domain of CALM bound to PtdIns(4,5)P2; PDB entry 1HFA) domains. The unstructured α0 helix that becomes structured upon interactions with targeted PPIn lipids, typically PtdIns(4,5)P2, is highlighted by blue. The canonical N-BAR domain of amphyphysin (c; PDB entry 1URU) is shown as the activate homodimer. For further details about each of these domains, please refer to Section 5.8 of the text. Prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
In addition to the ENTH and ANTH domains, proteins possessing domains from the BAR superfamily are also thought to promote as well as sense membrane curvature (Frost et al., 2009; Qualmann et al., 2011; Mim and Unger, 2012; Simunovic et al., 2015; Salzer et al., 2017). In general, several structurally-related groups exist within the BAR superfamily, with classifications based primarily on distinct elaborations of the classical BAR domain fold, and include the well-characterized N-BAR (N-terminal helix BAR), F-BAR (extended Fes/CIP4 homology BAR), and I-BAR (inverse-BAR) domain sub-types (Qualmann et al., 2011). Although these groupings share relatively little sequence similarities and lack signature motifs, all BAR superfamily domains possess a characteristic anti-parallel helical bundle of coiled-coils that interact to form a variety of curved dimeric modules (Figure 3c; Salzer, 2017). Depending on the oligomerization properties of the domain and the shape of the binding surface, BAR superfamily domains can generate positive (N-BAR, and F-BAR; Peter et al., 2004; Shimada et al., 2007) or negative (I-BAR; Millard et al., 2005) membrane curvature, as well as, in relatively few cases, function to stabilize planar membrane sheets (I-BAR; Pykalainen et al., 2011). The membrane-binding interface of each BAR domain contains a series of positively-charged patches, each representing a relatively weak membrane-binding site, which only cooperate with one another if the geometry of the membrane conformers to the degree of curvature defined by the assembled BAR module (Moravcevic et al., 2012). This method of binding relies on delocalized electrostatic attraction, rather than specific coordination of lipid headgroups, and most BAR domains show a general preference for anionic lipids; including targeting to membranes enriched with PtdIns(4,5)P2 and PtdSer (Moravcevic et al., 2012; Salzer, 2017). However, functionally-distinct binding modes and different lipid sensitivities are apparent across the BAR superfamily; including the description of a selective PPIn-binding site within the F-BAR domain from a conserved yeast RhoGAP (Moravcevic et al., 2015). Additionally, analogous to the ENTH and ANTH domains, the elaborated canonical BAR domain fold of N-BAR, as well as certain I-BAR domains (Saarikangas et al., 2009), possess amphipathic α-helices that can be inserted into membranes to potently induce curvature (Masuda et al., 2006; Gallop et al., 2006; Mim et al., 2012). Penetration of the amphipathic α-helix, and therefore the membrane deformation activity, is thought to be controlled by the local concentration of PtdIns(4,5)P2 (Mattila et al., 2007; Yoon et al., 2012). Interestingly, the ability of BAR domains to function as diffusion barriers that can restrict membrane PPIn dynamics has also been proposed to be a general feature of the BAR superfamily (Zhao et al., 2013) and may serve to coordinate the scaffolding of additional effectors at sites of membrane deformation. Independent of the recruitment of additional effectors, the presence of flanking PPIn-binding PX (PX-BAR domains; Pylypenko et al., 2007) or PH (BAR-PH domains; Li et al., 2007; Zhu et al., 2007) domains can also direct a subset of BAR domains towards membranes enriched with anionic PPIn lipids (Frost et al., 2009). In fact, recent evidence for coincidence detection and intermolecular communication between BAR domains and the classical PPIn-coordinating modules in PX-BAR (Pyplypenko et al., 2007; Daste et al., 2017; Lo et al., 2017; Schöneberg et al., 2017) and BAR-PH domains (Pang et al., 2014; Chan et al., 2017), suggests that PPIn lipids are likely to function as complex regulators of BAR domain activities. Consequently, despite the relatively non-specific interactions of BAR superfamily domains with PPIn lipids, the ability of BAR domains to couple curvature-sensing and scaffolding roles with the recruitment of additional PPIn-binding elements allows for these domains to shape the complex inter-relationship between membrane PPIn levels and the local architecture of the membrane. Importantly, in addition to this short overview, the structural and regulatory features of the ENTH, ANTH, and BAR domains, as well as the relationship between these domain families and the regulation and sensing of membrane curvature, will be explored in further detail within other chapters of this volume.
6. PPIn-Interacting Domains from Prokaryotic Effector Proteins
The reversible recruitment of peripheral proteins using membrane-embedded PPIn lipids is not a unique feature of eukaryotes and many pathogens target host cell membranes using PPIn-mediated interactions (Ham et al., 2011; Altan-Bonnet and Balla, 2012). The demonstrated ability of secreted bacterial effectors to target specific PPIn lipids within defined subcellular compartments highlights the need to better understand the structural features that control such selective and high affinity membrane interactions. Although new evidence for PPIn-dependent membrane targeting by prokaryotic peripheral proteins continues to emerge, the binding motifs identified in secreted bacterial proteins to date lack significant sequence or structural homology with the eukaryotic PPIn-binding domains described above. That said, it is interesting to note that a prokaryotic origin for the PH domain superfold has been suggested from sequence analysis and structural studies (Xu et al., 2010); however, the role of bacterial PH-like domains remains unclear and appears to primarily involve mediating protein-protein interactions rather than functioning as PPIn-targeting modules. Consequently, rather than exploring distant homology with known eukaryotic PPIn-coordinating domains, within this section we will describe the unique protein folds used by known PPIn-binding effectors from prokaryotes and focus specifically on those with descriptions of stereospecific PPIn coordination. In particular, virulence factors SidC and SidM of the intracellular parasite Legionella pneumophila have been shown to anchor to the replication-permissive Legionella-containing vacuole (LCV) through direct interactions with PtdIns4P (Ragaz et al., 2008; Brombacher et al., 2009; Schoebel et al., 2010). The exquisite specificity of the PtdIns4P-binding domains from SidC and SidM have both been exploited to generate unbiased probes that can be used to detect the major steady-state pools of PtdIns4P in living cells (Hammond et al., 2014; Luo et al., 2015). The recent descriptions of highly specific bacterial PPIn-binding domains, as well as their obvious therapeutic relevance, has reinforced the need to understand the unique features that define the selective recognition of host membranes by secreted bacterial effectors.
Structural analyses of SidC revealed a novel PtdIns4P-binding fold, called the P4C (PtdIns4P binding of SidC) domain, which was comprised of a four α-helical bundle with the PtdIns4-Pcoordinating pocket forming from a collection of cationic residues at one end of the bundled domain (Figure 4a; Luo et al., 2015). Two conserved arginine residues, R652 and R638, significantly contribute to the overall charge of the P4C pocket and likely coordinate the PtdIns4P headgroup directly (Luo et al., 2015). In addition to the electrostatic potential, two hydrophobic patches present on the L1 (W642, W643, and F644) and L2 (W704 and F705) loops that surround the PtdIns4P-binding pocket may also facilitate membrane insertion of the P4C (Luo et al., 2015). Importantly, mutation of the electrostatic or hydrophobic features of the P4C significantly reduced membrane binding in vitro and could abolish localization to the LCV within cells (Luo et al., 2015). PtdIns4P-binding by the P4C domain was not only requisite for the localization of SidC to the LCV, but, interactions between P4C and PtdIns4P also stimulated the E3 ligase activity of SidC; presumably through a conformational switch that functions to extend the P4C domain and uncover the ubiquitin ligase catalytic site of the SNL (SidC N-terminal E3 ligase) domain (Luo et al., 2015). As discussed previously, this type of allosteric regulation by interactions with membrane-embedded PPIn lipids is well characterized in eukaryotes; although, the need for secreted bacterial proteins to communicate directly with the host cell machinery appears to prioritize dynamic domain reorganizations and coincident-binding regulation during the membrane recruitment of many bacterial effectors.
Figure 4.
Prokaryotic PPIn-binding domains. The prokaryotic PPIn-binding P4C (a; unbound structure isolated from within the full-length SidC structure; PDB entry 4ZUZ) and P4M (b; in complex with di-butyl-PtdIns4P; PDB entry 4MXP) modules are shown with their predicted membrane-bound orientations. The PPIn-binding site (green arrowhead) of the P4C has been mapped by functional and mutagenesis studies, whereas the structure of the P4M module has been solved in complex with the PtdIns4P headgroup. An elaborated membrane insertion motif that significantly penetrates the membrane, as well as contributes to the coordination of the PPIn headgroup within the binding pocket, is shown in red. For further details, please refer to Section 6 of the text. Prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
Another secreted Legionella effector SidM, which functions as a GEF and adenylytransferase that is specific for the host Rab1 GTPase (Machner and Isberg, 2006; Ingmundson et al., 2007), also contains a novel PtdIns4P-binding module (P4M) that has high affinity and specificity for PtdIns4P (Schoebel et al., 2010; Zhu et al., 2010; Del Campo et al., 2014). Unlike the P4C domain, the structural basis for stereospecific recognition of PtdIns4P by the P4M has been determined explicitly (Figure 4b; Del Campo et al., 2014). The structure of the P4M fold consists of six α-helices (α10- α15) and an ordered loop (LC) that connects the lipid-binding module to the catalytic GEF domain (Del Campo et al., 2014). The base of the electropositive PtdIns4P-binding pocket is supported by three parallel helices (α11, α12, and α15), while, at the top of the domain, residues from helices α10, α13, α15, and the LC contact the DAG backbone to envelope the PtdIns4P headgroup (Del Campo et al., 2014). Coordination of the 4-position phosphate includes contributions from basic and polar residues that define a deep and narrow cavity that shows significant complementarity for the PtdIns4P headgroup while also excluding optimal binding modalities for other PPIn species (Del Campo et al., 2014). PH and PX domains have also been identified with deep PPIn-binding pockets, while the stereospecific coordination of the 1- and 4-position phosphate groups by the P4M certainly resembles the basic and polar networks characterized for many PH domains (Moravcevic et al., 2012; Del Campo et al., 2014). Alongside the constricted PtdIns4P-binding pocket, the α14 helix also extends well above the binding pocket and contains several leucine residues (L610, L614, L615, and L617) that appear to function as an elaborated version of the putative membrane insertion elements found in the P4C domain or those present in other eukaryotic PPIn-coordinating domains (Del Campo et al., 2014); including similarity to the examples discussed above for the PX and FYVE domain families. Importantly, penetration of SidM into PtdIns4P-containing monolayers was dependent on the density of PtdIns4P present within the membrane and not the general electrostatic character of the membrane interface (Del Campo et al., 2014). This suggests that stereospecific recognition of the PtdIns4P headgroup by P4M likely determines the extent of interfacial insertion and subsequent hydrophobic anchoring of SidM within cellular membranes (Del Campo et al., 2014). Overall, the unique structure and exquisite selectivity of the helical P4M fold suggests that the added depth of the binding pocket, resulting from an exaggerated membrane insertion, may help to convey enhanced specificity during PPIn-binding interactions. Understanding the complex inter-relationships between electrostatic and hydrophobic features that support selective PPIn-binding, especially across diverse model systems, represents an important step for defining conserved sequence features that contribute to PPIn-selective membrane interactions during the dynamic regulation of peripheral protein functions.
Outside of the detailed investigations into SidC and SidM interactions with PtdIns4P, relatively few studies have characterized stereospecific coordination of PPIn species by bacterial protein domains. Though lacking structural descriptions, other Legionella effectors have also been shown to require PPIn-binding, including PtdIns3P-specific binding during the recruitment of SetA (subversion of eukaryotic traffic A) to early endosomes (Jank et al., 2012) as well as for dynamic associations of LpnE (Legionella pneumophila entry) with the LCV (Weber et al., 2009). The cytoskeletal effector ActA (actin assembly-inducing protein) from Listeria monocytogenes, which binds to monomeric actin (Skoble et al., 2000; Zalevsky et al., 2001) and the Arp2/3 actin nucleation complex (Welch et al., 1998) to drive actin-dependent motility of the bacteria inside of host cells, is also able to interact with PtdIns(4,5)P2 or PtdIns(3,4,5)P3 using a small sub-region within the N-terminal domain (Cicchetti et al., 1999; Steffen et al., 2000; Sidhu et al., 2005). More generally, a broad series of bioinformatic and functional analyses identified a putative family of bacterial PPIn-binding domains (BPDs) within functionally diverse effectors from the type III secretion systems of both animal and plant pathogens (Salomon et al., 2013). Secondary structure predictions and NMR analysis identified a common BPD fold consisting of two β-strands followed by two α-helices; which is somewhat similar to the topology of the eukaryotic PX domain structure of three short β-strands followed by three or four α-helices (Salomon et al., 2013). Predicted BPD domains from phylogenetically-distinct effectors were shown to bind diverse PPIn species, although the most thorough evidence for BPD-mediated interactions with PPIn lipids emerge from studies showing the selective recognition of PtdIns(4,5)P2 by the Vibrio parahaemolyticus effectors VopR (Vibrio outer protein R) and VopS (Vibrio outer protein S). Interestingly, biophysical studies of VopR showed that the BPD domain is unfolded in solution and significantly increases in secondary structure in the presence of PtdIns(4,5)P2, but not other PPIn species; outlining a possible role for PPIn lipids during the refolding of bacterial effectors after entering into host cells (Salomon et al., 2013). Another conserved bacterial domain capable of associating with PtdIns(4,5)P2 was also described in the Pseudomonas cytotoxin ExoU (exoenzyme U; Tyson et al., 2015). Briefly, solution structures obtained in the absence of the PPIn headgroup show that the C-terminal membrane localization domain (MLD) of ExoU is organized as a four-helical bundle and possesses a conserved arginine residue (R616) protruding from the cap of the bundle that is required for PtdIns(4,5)P2 binding in vitro as well as for PM targeting of ExoU within cells (Tyson et al., 2015). Additional polar and charged residues that line surface-exposed pockets formed by the intervening loops of the helical bundle may also assist in PtdIns(4,5)P2 coordination; however, it is interesting to note that any conserved hydrophobic residues are buried within the MLD and are not likely to assist with membrane binding (Tyson et al., 2015).
Taken together, the novel bacterial domain structures that have been described help to reinforce the importance of electrostatic and polar residues for coordination of the anionic PPIn headgroup, as well as highlight the potential for hydrophobic contacts and membrane insertion for enhanced PPIn-binding specificity. Future structural and biophysical studies of the putative membrane-binding regions described in other prokaryotic effectors should help to identify a wider array of PPIn-interacting folds and may also lead to the identification of conserved prokaryotic PPIn-binding domains. These efforts will require high-throughput screening to more efficiently identify PPIn-interacting bacterial effectors. Along these lines, a recent study has carried out a systematic characterization of the membrane-binding properties and subcellular localization of close to 200 different type III and IV secreted effectors from six bacterial pathogens using yeast as a cellular model (Weigele et al., 2017). This screen identified 57 membrane-associating effectors, including 23 proteins that exhibited changes in localization following isogenic knockout or conditional repression of endogenous yeast PPIn kinases (Weigele et al., 2017). Additional in vitro binding studies identified 10 effectors with high affinity for PPIn lipids, but most effectors associated with multiple PPIn species; suggesting non-specific electrostatic interactions rather than PPIn-selective recognition (Weigele et al., 2017). Indeed, of the identified PPIn-bindings proteins, detailed studies of the Shigella flexneri factor IpgB1, a known membrane effector and functional mimetic of Rho-family GTPases (Alto et al., 2006; Handa et al., 2007), showed that PPIn recognition occurred through an N-terminal amphipathic helix enriched with basic residues (Weigele et al., 2017). Consequently, although this screen failed to identify effectors with clearly defined PPIn-coordinating pockets, these studies continue to highlight the breadth and diversity of the PPIn-binding strategies employed by prokaryotic effectors, including a conserved strategy that capitalizes on polybasic targeting of anionic PPIn headgroups. More expansive screens that incorporate PPIn-specific immobilization strategies and mass spectrometry, as described for the unbiased characterization of the mammalian PPIn-interacting proteome (Jungmichel et al., 2014), may help to reveal additional bacterial PPIn-binding effectors with more selective membrane recognition strategies. Nonetheless, these collected examples help to demonstrate the convergence of membrane-targeting strategies employed by eukaryotic and prokaryotic peripheral proteins, which also reflects the absolute conservation of PPIn lipids as integral regulatory components within biological membranes.
7. Non-vesicular Lipid Transport and PPIn-Coordinating Lipid-Transfer Domains
The appearance and redistribution of unique PPIn species within distinct subcellular membranes strongly suggests that local production or dynamic trafficking of inositol-containing lipids must occur to maintain PPIn availability. How newly synthesized lipids, and PtdIns in particular, are transported between membrane compartments has become a central question with significant implications for understanding fundamental questions throughout cell biology. Communication between the ER, the primary site of phospholipid biosynthesis, and many distant membrane compartments, including the distal PM, was long thought to be regulated by bulk membrane transfer through the vesicular trafficking system; but, it has recently been demonstrated that specialized lipid-transfer proteins (LTPs) can act to transport lipid monomers across aqueous spaces and between membranes independent of budding vesicles (Lev, 2010; Prinz, 2010; Drin, 2014). Non-vesicular lipid transport occurs primarily at specialized membrane interfaces, enriched with high-specificity molecular tethers, which are referred to as membrane contact sites (MCSs; Eisenberg-Bord et al., 2016; Jain and Holthuis, 2017; Muallem et al., 2017). The formation and dynamics of MCSs has become an increasingly important component of the regulatory mechanisms contributing to cellular membrane remodeling as they allow for the exchange of lipid isomers between organelles possessing different bulk properties. Directional movement of lipids at MCSs occurs by using intrinsic concentration gradients that are maintained through a combination of bulk vesicular trafficking and lipid transfer cycles, which ultimately function as part of the machinery used to define subcellular membrane identity. The lipid transport actions of LTPs are mediated by complex interactions with membrane surfaces and the associated transitions between distinct conformational states appear to confer the selectivity as well as directionality of the lipid transfer process (Chiapparino et al., 2016; Tong et al., 2016). In this section, we will describe the molecular features controlling the specific recognition of PPIn species by lipid transfer domains from eukaryotic families of LTPs. Despite the apparent structural diversity of LTPs, the functional lipid-binding cavities involved in lipid transfer exhibit similarities in their overall architecture and general mode of cargo recognition (Chiapparino et al., 2016). By examining the PPIn-binding modalities used across different families of LTPs, we will attempt to identify conserved strategies for PPIn recognition that emerge from comparing PPIn-selective lipid-transfer domains (LTDs) with the diversity of PPIn-binding folds already described.
7.1. PtdIns4P Transport by ORPs
PtdIns4P has emerged as an important cargo that can be used to drive the counter-exchange of other lipid species at MCSs, including foundational studies demonstrating lipid fluxes at ER-Golgi (Mesmin et al., 2013) and ER-PM contacts (Chung et al., 2015; Moser von Filseck et al., 2015a). Movement of PtdIns4P into the ER can be coupled with PtdIns recycling through the activity of the ER-resident PPIn phosphatase Sac1 (suppressor of actin mutations 1-like), which dephosphorylates PtdIns4P to produce PtdIns (Stefan et al., 2011). The extent to which Sac1 contributes to steady state PtdIns levels or metabolic-tunneling of PtdIns towards PPIn re-synthesis remains an active area of research. However, most importantly, the ability of LTPs to solubilize and transport PtdIns4P across membrane compartments that are defined by different PPIn-modifying enzymes provided a molecular mechanism for cells to generate PPIn lipid gradients (Kim et al., 2013b; Jackson et al., 2016; Wong et al., 2017). Throughout these studies, the molecular driving force responsible for the non-vesicular movement of PtdIns4P was shown to involve members of the oxysterol-binding protein (OSBP)-related protein (ORP) family of eukaryotic LTPs. As implied from their name, the ORP family of LTPs were originally thought to regulate the transport of sterols, but can also transfer other lipid cargoes; including a conserved role in the recognition and extraction of PtdIns4P (Kentala et al., 2016; Tong et al., 2016). Among the best-studies ORPs include the seven yeast ORP homologs (Osh1–Osh7), but more recent work has been focused on the repertoire of 12 ORP-encoding genes in humans; which give rise to 16 human ORP variants through alternative translation or splicing (Jaworski et al., 2001; Lehto et al., 2001). In general, ORPs are cytosolic proteins that possess a combination of sequence features that are collectively used to control protein-protein interactions or membrane targeting; including representative ORPs with N-terminal PH domains (Levine and Munro, 1998) or centralized FFAT (two phenylalanines (FF) in an acidic tract) motifs (Loewen and Levine, 2005), as well as some homologs possessing ankyrin repeats (Johansson et al., 2003; Tong et al., 2016). Not surprisingly, as discussed above in Section 5.1, the PH domains of ORPs are used for membrane targeting, including specific binding to PPIn species (Tong et al., 2016); whereas the FFAT motif present in some ORPs directly binds to ER-resident proteins called VAPs (vesicle associated membrane protein (VAMP)-associated proteins; Loewen and Levine, 2005; Weber-Boyvat et al., 2015; Murphy and Levine, 2016) that function to attach ORPs to the surface of ER membranes. Interestingly, two closely-related human ORPs, ORP5 and ORP8, are unique in that they contain a C-terminal transmembrane domain that anchors them within the ER (Yan and Olkkonen, 2008; Du et al., 2011); although the lipid-interacting PH and lipid-transfer domains are still capable of accessing lipids in adjacent membrane compartments when localized to MCSs (Chung et al., 2015).
Despite diversity in their domain organizations and recruitment to subcellular membranes, ORPs can all be identified at the sequence level by a C-terminal OSBP-related domain (ORD) containing a conserved N-terminal oriented signature motif, defined as EQVSHHPP, which is important for controlling cellular functions (Tong et al., 2016). The fold of the ORD from ORPs is unique among the known eukaryotic LTPs and although the structures determined to date are limited to Osh homologs from yeast, sequence conservation and functional studies of mammalian ORPs strongly support the ability of these proteins to facilitate lipid extraction and transport. The first solved structure reported from the yeast ORP homolog Osh4 shows the characteristic topology of the ORD, consisting of a hydrophobic tunnel that runs through a near complete central β-barrel that is built around an anti-parallel β-sheet of 15 β-strands (Figure 5a; Im et al., 2005). The extreme N-terminus contains a small amphipathic α-helix (α1) that is connected to a flexible loop, which attaches to an elongated antiparallel bundle consisting of a two stranded β-sheet (β1-β2) and three α-helices (α2-α4), and functions to close the incomplete β-barrel by forming one wall of the central lipid-binding tunnel (Im et al., 2005). An elongated C-terminal subdomain follows the base of the central β-barrel and projects to exterior surface where the α5, α6, and α7 helices line one-side of the tunnel opening (Im et al., 2005). Interestingly, the region comprising the N-terminal lid is thought to remain highly flexible in the unbound state, but is stabilized in a closed conformation through direct interactions with lipid cargoes (Im et al., 2005). Compared with the sterol-bound structure, the loss of interactions between the lipid cargo and residues lining the lid causes a conformational change that not only opens the tunnel but also reorganizes the α7 helix and β1 strand, which significantly shifts a conserved cluster of basic residues that is present at the tunnel entrance (Im et al., 2005). The conformational dynamics linking ligand-binding and the movements of the N-terminal lid help to define early models for membrane-binding cycles that would facilitate the acquisition and deposition of lipid cargoes by ORPs. However, these initial studies defining the ORD fold from Osh4 also showed that the sterol binding site was poorly conserved throughout the ORPs, but that the N-terminal lid and basic cluster were present in all ORP homologs and might function during the transport of non-sterol ligands (Im et al., 2005).
Figure 5.
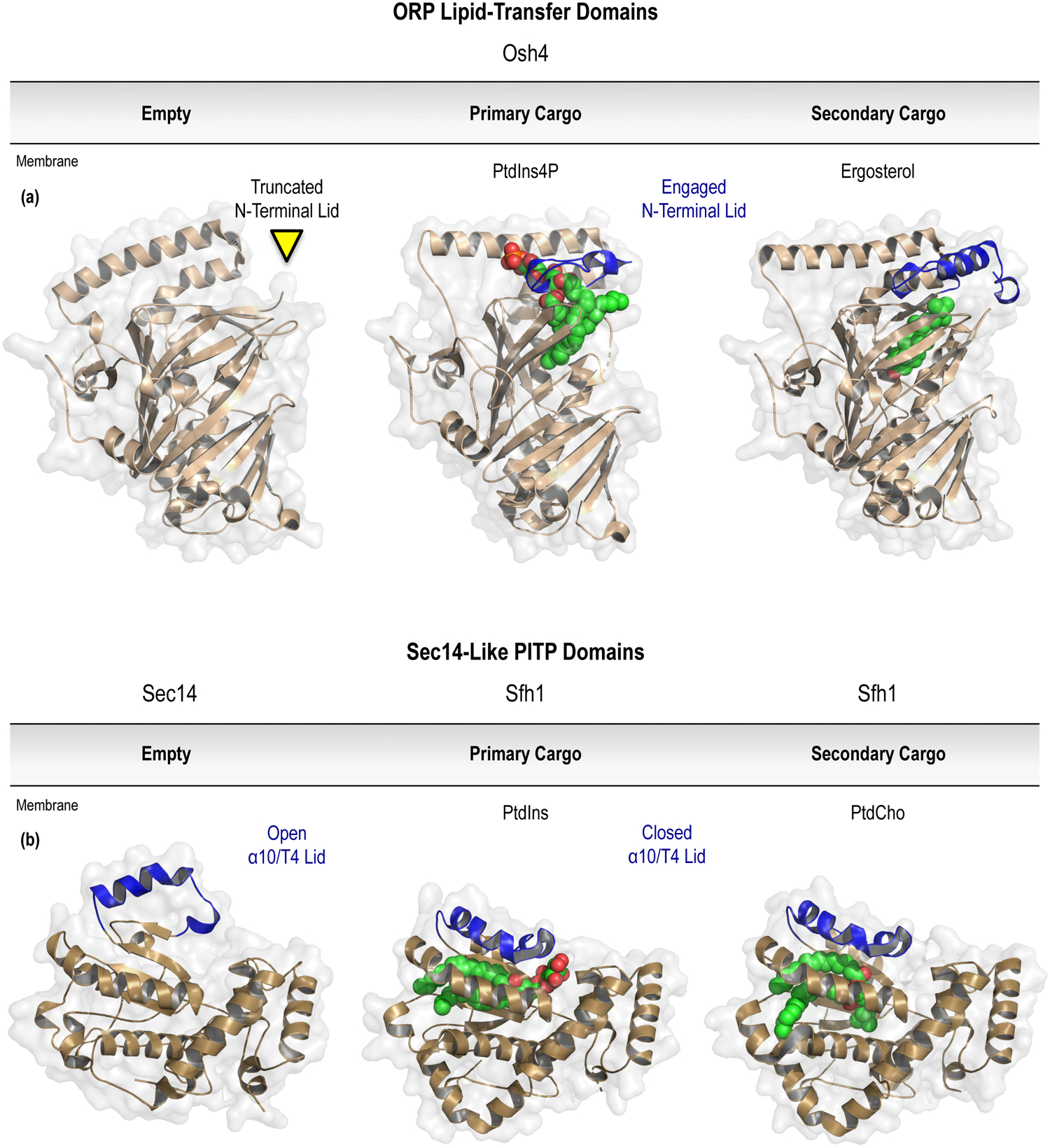
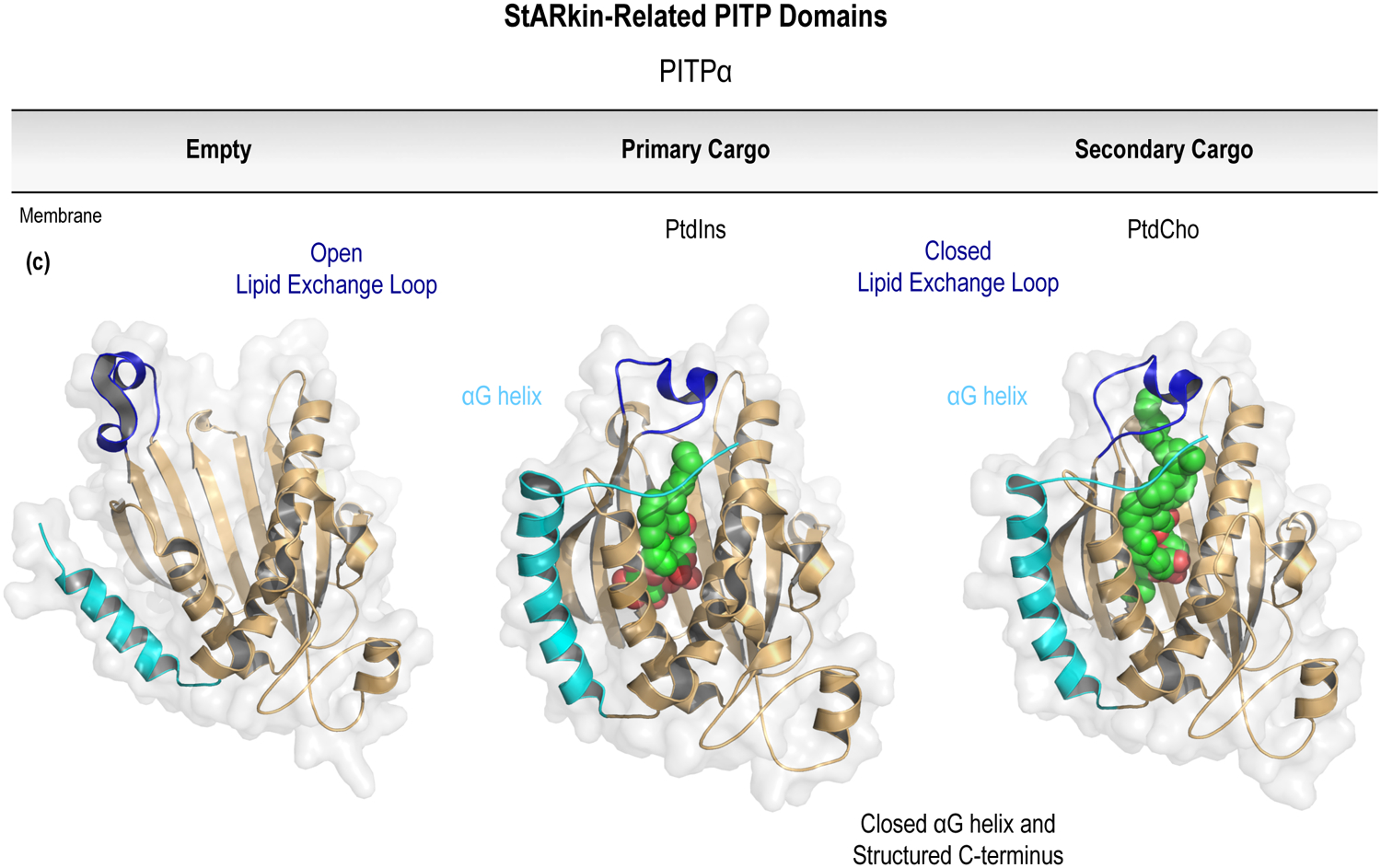
Lipid-transfer domains with selectivity for PPIn species. For each of the known families of PPIn-coordinating LTPs, the structure of the open LTD fold is shown in relationship to structures with either the primary or secondary lipid cargo bound. Of note, each of the LTPs are shown with the opening of the LTD pocket located at the top of the molecule. Unlike the ORP or Sec14-like LTDs, the PPIn headgroup is buried within the domain, with the fatty acyl chains projecting upwards to the top of the binding pocket. (a) The well-studied family of ORPs are important regulators of non-vesicular lipid transport across eukaryotes; including a conserved function for transporting PtdIns4P (PDB entry 3SPW) and a variety of secondary lipid cargoes, including sterols (PDB entry 1ZHZ). Please note that the open fold (PDB entry 1ZI7) could only be crystallized following truncation of the N-terminal lid (shown in blue) that gates the hydrophobic lipid-binding pocket. (b) The large family of Sec14-like PITPs might play more diverse roles outside of lipid transport to control intracellular signaling responses. The recognition of the conserved PtdIns (PDB entry 3B7N) and PtdCho (PDB entry 3B7Q) cargoes clearly involves unique binding surfaces within the PITP, as well as the dynamic reorganization of the α10/T4 lid (highlighted in blue) relative to the unbound structure (PDB entry 1AUA). (c) The prototypical StARkin-related PITP domain is also shown bound to PtdIns (PDB entry 1UW5) and PtdCho (PDB entry 1T27). Compared to the open fold (PDB entry 1KCM), closure of the lipid exchange loop (shown in blue) also stabilizes the elongated C-terminus to pin the αG helix (cyan) in the closed conformation. For further details, please refer to Sections 7.1 (ORPs) and 7.2 (7.2.1, Sec14-like; 7.2.2, StARkin-related) of the text. Prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
Subsequent experiments using in vitro liposome transfer assays demonstrated an interesting relationship between the rate of Osh4-mediated sterol transport and the presence of PPIn lipids (Raychaudhuri et al., 2006; Schulz et al., 2009). PtdIns4P-dependent inhibition of sterol transfer activity led to the eventual discovery of competitive binding between PtdIns4P and sterols for the Osh4 internal binding site (Saint-Jean et al., 2011). As predicted from the in vitro lipid transfer data, the solved structure of Osh4 bound to PtdIns4P showed that the PtdIns4P-coordinating pocket overlaps with the defined sterol-binding site (Figure 5a; Saint-Jean et al., 2011). PtdIns4P-recognition occurs with the backbone acyl chains occupying the central hydrophobic tunnel and the phosphorylated inositol headgroup being recognized by a shallow pocket at the tunnel entrance that is covered by the N-terminal lid, which also wraps the glycerol moiety (Saint-Jean et al., 2011). The 4-position phosphate group makes direct hydrogen bonds with the side chains from a conserved arginine (R344) in the α7 helix as well as with H143 and H144 within the β4-β5 sheets; which are now known to define the conserved OSBP-specific EQVSHHPP signature motif (Saint-Jean et al., 2011). Alternatively, the 1-position phosphate group that bridges the inositol ring and glycerol backbone forms hydrogen bonds to conserved lysine residues in the β4 sheet (K109) and α7 helix (K336; Saint-Jean et al., 2011). This general model for the specific recognition of PtdIns4P is also observed in the subsequent PtdIns4P-bound ORD structures obtained from Osh3 (Tong et al., 2013) and Osh6 (Moser von Filseck et al., 2015a). The strict sequence conservation of the residues that contact the PtdIns4P headgroup in the ORP homologs strongly suggests that PtdIns4P is a common ligand for the ORP family of LTPs (Saint-Jean et al., 2011; Tong et al., 2016). However, despite the demonstrated structural and functional conservation for PtdIns4P recognition, variations in the topology of the central hydrophobic binding pockets of Osh3, Osh4, and Osh6 together indicate that the identity of secondary ligands, such as sterols or other phospholipids, may be unique to individual ORPs (Tong et al., 2016). As outlined above, Osh4 shows clear structural features that facilitate the coordination of sterols along with PtdIns4P (Im et al., 2005). Alternatively, the Osh3 ORD shows a constricted pocket that excludes sterols (Tong et al., 2013), while Osh6 possesses a deeper hydrophobic tunnel that can accommodate the elongated acyl chains from an alternative secondary ligand, PtdSer (Moser von Filseck et al., 2015a). Regardless of the cargoes involved, the structural dynamics associated with the loading of lipids into the ORD remains unknown. Recent studies of Osh4 suggest that the directionality of lipid transfer is influenced by intermolecular interactions with the bound cargo, as well as the anionic character of the local membrane environment; which both regulate the movements of the N-terminal lid (Moser von Filseck et al., 2015b). Unfortunately, results from molecular dynamics simulations of Osh4 have been unable to investigate the role of dynamic gating of the ORD gating during membrane interactions, but these approaches do identify complex contacts with the membrane surface that involve multiple binding regions, including some that run along the length of the molecule and anti-parallel to the entrance of the ORD, which appear to result from a rotation of the fold once it is anchored at the membrane (Rogaski and Klauda, 2012; Monje-Galvan and Klauda et al., 2016). Clearly, additional biophysical descriptions of ORPs at the membrane interface will be required to elucidate the orientation of the binding pocket as well as for defining the role of the N-terminal lid during cargo loading. The relative lack of information regarding the initial events contributing cargo recognition, in particular, make it difficult to understand how PtdIns4P binding can occur with the anionic headgroup oriented at the top of the binding pocket and fatty acyl chains buried within the hydrophobic core of the ORD.
As outlined above, while the majority of the early studies characterizing ORPs have emerged from yeast, PtdIns4P-specific binding and transport activity has been established for many mammalian ORP homologs, including: OSBP (Mesmin et al., 2013), ORP4 (Charman et al., 2014), ORP5 (Chung et al., 2015), ORP8 (Chung et al., 2015), and ORP9 (Liu and Ridgway, 2014). However, the identity of the secondary ligands used by most ORPs remains unclear; especially considering the demonstrated roles for ligand-induced conformational dynamics in ORP functions as well as the importance of localized lipid gradients during the control of transfer activity. One of the best examples for the heterotypic exchange of lipid cargoes by mammalian ORPs comes from elegant studies demonstrating the bi-directional movement of PtdIns4P and PtdSer by ORP5 and ORP8 at MCSs between the ER and PM (Chung et al., 2015). This counter-transport mechanism was shown to be important for the homeostatic regulation of PM composition and revealed an intricate relationship between PtdIns4P gradients and PtdSer metabolism that was dependent on ORP5- and ORP8-mediated lipid exchange (Chung et al., 2015; Sohn et al., 2016). A more recent study of ORP5 and ORP8 functions has suggested that their ORD domains interact with multiple PPIn species and identify PtdIns(4,5)P2, in particular, as the primary lipid-transfer substrate for ORP8 (Ghai et al., 2017). At this time, it is unclear how the structural descriptions of the ORD fold, including those from the closest yeast homolog Osh6, can be reconciled with the proposed PtdIns(4,5)P2-transfer function of ORP8. It also remains unclear whether the transfer of PtdIns(4,5)P2 is as universal as the ability of ORPs to bind and transport PtdIns4P, as well as whether inter-domain communication within ORPs during the lipid-recognition process contributes to the regulation of the lipid exchange cycle.
7.2. PtdIns Interactions with LTPs
Studies examining PPIn metabolism and redistribution have been hampered by the fact that there is still a general lack of understanding with regards to how the synthesis as well as transport of PtdIns occurs. Interestingly, putative PtdIns-transfer proteins (PITPs) have been identified that are thought to rapidly exchange PtdIns, as well as other lipid cargoes, from donor to acceptor membranes and may also contribute to the allosteric control of other membrane-associated proteins or as modules for presenting bound cargo to PtdIns-modifying enzymes (Ile et al., 2006; Grabon et al., 2015; Wong et al., 2017). Outside of the molecular mechanisms involved, the proposed cellular roles for PITPs include the regulation of membrane trafficking and organelle biogenesis, as well as important roles within intracellular signal transduction networks (Kim et al., 2013b). In particular, two major families of eukaryotic PITPs have been identified based on either homology to the yeast protein Sec14 (Sec14-like; Grabon et al., 2015) or as part of a larger superfamily of related proteins that have a conserved StARkin (related (kin) to steroidogenic acute regulatory protein-related and Bet v 1) lipid-transfer domain (StARkin-related or Class I and II PITPs; Wong and Levine, 2016; Wong et al., 2017). Within this section, we will review the unique structural characteristics of each of these families of PITPs and attempt to highlight the features responsible for the selective recognition of lipid cargoes.
7.2.1. Sec14-Like PITP Domains
Studies examining the structure and functions of PITPs have been centered around the Sec14-like homologs (Grabon et al., 2015). The yeast Sec14 protein was originally described as a major regulator of phospholipid composition in membranes of the trans-Golgi network, making them permissive for vesicular transport (Bankaitis et al., 1989; Bankaitis et al., 1990; Salama et al., 1990; Ile et al., 2006). In addition to Sec14, five Sec14-like homologs are found in yeast (Sfh1-Sfh5) that all perform cellular functions related to lipid metabolism (Grabon et al., 2015). Many proteins containing Sec14-like domains are also present in higher eukaryotes and several phylogenetically-distinct proteins with sequence similarities to the conserved Sec14-like domain also show lipid transfer activities (Bankaitis et al., 2010; Kim et al., 2013b). Despite some controversy regarding their activities, the Sec14-like domain fold is thought to function as a dual lipid recognition module, similar in function to the ORPs, with selectivity and in vitro lipid transfer activity towards PtdIns and the amino-phospholipid phosphatidylcholine (PtdCho; Cleves et al., 1991; Bankaitis et al., 2010). Structural descriptions of the Sec14-like domain come predominantly from the yeast, including an open structure of the founding Sec14 protein, which lacks a lipid cargo, and lipid-bound structures from the Sec14 homolog Sfh1. The unbound Sec14 structure shows that the functional core of the fold is composed of two distinct subdomains consisting of a C-terminal hydrophobic pocket and helical N-terminal domain that are held together by hydrophobic stacking interactions (Figure 5b; Sha et al., 1998). The large hydrophobic pocket at the C-terminus is formed by six β-strands (β1-β6) that constitute the floor of the binding pocket with the sides of the binding cavity being lined by helices α8 and α9 on one side, as well as helices α10, 310 (T)4, and α11 on the other (Sha et al., 1998). The hydrophobic pocket is supported by an extended string motif that stretches around the floor of the central β-sheet (Sha et al., 1998). Within the N-terminal domain, a tripod-like motif comprised of helices α2, α3, and α4 is essential for membrane targeting, while the adjoining α5 helix may also help to support the hydrophobic core by surrounding central helices α8 and α9 (Sha et al., 1998). In this open conformation, core helices α9 and α10/T4 that line the lipid-binding pocket are bent away from one another to increase the volume of the cavity (Sha et al., 1998). Furthermore, a string of hydrophobic residues are found along the solvent-exposed surface of helices α10/T4, with their side chains oriented away towards the membrane interface (Sha et al., 1998). Movement of the α10/T4 helices away from the surface of Sec14 was proposed to facilitate insertion of the hydrophobic side chains into the inner leaflet of the membrane bilayer, which would open the hydrophobic pocket and allow for the deposition of phospholipid cargoes (Sha et al., 1998). Retraction of the α10/T4 helices from the membrane and toward the protein core would subsequently facilitate phospholipid extraction as a result of direct contacts between the inner face of the helices with the fatty acyl chains of the new lipid cargo (Sha et al., 1998).
Phospholipid-bound structures of the Sec14 homolog Sfh1 confirmed the dramatic repositioning of the α10/T4 helices (α9/T3 helices in Sfh1) to gate the hydrophobic cavity and explicitly define the closed conformation of the Sec14-like lipid-binding pocket (Figure 5b; Schaaf et al., 2008). However, it is important to note that despite sequence similarities, Sfh1 lacks true Sec14-like biological functions in yeast (Schaaf et al., 2011) and possesses reduced lipid-transfer activities in vitro (Li et al., 2000). That said, sequence analyses suggest that Sfh1 and Sec14 likely share the structural motifs required for phospholipid recognition (Schaaf et al., 2008; Schaaf et al., 2011). Interestingly, relatively few changes are observed in the core backbone of the Sec14-like PITP domain with different lipid occupancies (Schaaf et al., 2008). Binding of two structurally similar amino-phospholipids, PtdCho and phosphatidylethanolamine (PtdEtn), show a conserved orientation within the binding pocket; supported by extensive contacts between the lipid cargo and residues along the entire face of the β-sheet as well as in the helical pillars on either side of the cavity (Schaaf et al., 2008). However, as expected, incorporation of PtdIns within the Sfh1 lipid-binding pocket occurred with an orientation that was distinct from the amino-phospholipids (Schaaf et al., 2008). Although the distal regions of the fatty acyl chains are configured similarly, rather than the headgroup of PtdIns binding deep within the pocket, the inositol headgroup is positioned near the protein surface; but is still shielded from the solvent by the closed helical gate (Schaaf et al., 2008). Extensive hydrogen bonding is distributed across the PtdIns backbone and coordination of the headgroup phosphate occurs through an electrostatic interaction with K241 (K239 in Sec14; Schaaf et al., 2008). The hydroxyl groups on the inositol ring also forms direct hydrogen bonds or H2O-mediated contacts with side chains from the N-terminal tripod motif (R61 and K62, helix α2) and the helical walls of the cavity (D209, helix α8; D235, α9/T3 helical gate; Schaaf et al., 2008). Overall, the extensive hydrogen bonding network and Van der Waals contacts observed in the phospholipid-bound Sfh1 structures are in agreement with the experimental data demonstrating a much higher affinity of Sec14-like PITPs for PtdIns relative to PtdCho (Schaaf et al., 2008; Bankaitis et al., 2010).
Outside of the clear differences in the coordination of the lipid headgroups, a more general feature of the closed conformation of Sfh1, as well as the lipid-interacting surfaces mapped within Sec14 (Smirnova et al., 2006; Smirnova et al., 2007), is the apparent immobilization of the fatty acyl chains within the binding cavity. Interestingly, the intricate bonding network within hydrophobic pocket is much different than that observed for the ORD domain of ORPs, which show relatively loose nonspecific interactions between the acyl chains of lipid cargoes and the central hydrophobic tunnel (Tong et al., 2016). The top-down binding of the ORD, wherein the headgroup coordinating residues that line the top of the lipid-binding tunnel and lid confer selectivity towards PtdIns4P, but do not place as many restrictions on the identities of potential secondary ligands; including sequence-selective coordination of sterols and PtdSer that have already been described for different ORP homologs. Conversely, the much more restrictive hydrophobic pocket observed in Sec14 and Sfh1 clearly defines binding modalities for both the high-affinity primary ligand, PtdIns, and the secondary cargo; which is almost always PtdCho. These differences in binding are also reflected in the relatively small changes in backbone structure observed during the internal occupancy of different lipid cargoes in the Sec14-like PITP domains, compared to rather large changes in the conformation of the mobile lid observed in the PtdIns4P- and sterol-bound structures of the Osh4 ORD and other ORPs. That said, in terms of the functional communication between lipid cargoes, although the binding pocket of the Sec14-like PITP domain only accommodates a single bound phospholipid (Schaaf et al., 2008), studies using structure-guided design of headgroup-specific binding mutants showed that selective binding of PtdIns and PtdCho within the same molecule is required to maintain the cellular functions performed by intact Sec14 (Schaaf et al., 2008). These data suggest that the functions of Sec14 are defined by sequential lipid exchange cycles of PtdIns and PtdCho, rather than by the strict lipid-selectivity or conformational transitions associated with the lipid-transfer domain. However, the importance of conformational dynamics during Sec14-related lipid transfer activities have been highlighted by unique experiments that used rationale mutagenesis and directed evolution to alter the gating properties of the lipid-binding cavity to enhance phospholipid cycling and resurrect the Sec14-like biological functions of Sfh1; which, despite its utility for structural studies, is relatively inactive as a putative PITP (Schaaf et al., 2011). Molecular dynamics simulations have also been used to track conformational rearrangements during lipid binding (Ryan et al., 2007), but were unable to relate how the motions observed correlate with phospholipid exchange. Consequently, despite immense effort, further experiments are still required to uncover the relationship between the selective recognition of lipid cargoes and the explicit transport or presentation activities of Sec14-like PITPs bound to PtdIns.
7.2.2. StARkin-Related PITP Domains
Outside of the Sec14-like PITPs, the prototypical and first-described mammalian PITPs were the soluble Class I PITP isoforms PITPα and PITPβ from the StARkin-related superfamily (Cockcroft and Carvou, 2007; Kim et al., 2013b; Wong et al., 2017). Details regarding the structure and potential PtdIns-transfer characteristics of the Class I PITPs have been uncovered, however the cellular function of these proteins remains obscure. In general, Class I PITPs are relatively small proteins that have high sequence identity, but show different cellular localizations; PITPα is mostly present within the cytosol and nucleus, whereas PITPβ is found associated with membranes of the Golgi (de Vries et al., 1995; de Vries et al., 1996; Larijani et al., 2003). The StARkin-like PITP domain of both PITPα and PITPβ are capable of transferring PtdIns, and to a lesser extent PtdCho, between natural membranes and artificial liposomes (van Paridon et al., 1987; Wirtz, 1991; Wirtz, 1997; Segui et al., 2002); making them functionally similar to the Sec14-like PITP domains, despite almost no sequence homology (Hsuan and Cockcroft, 2001; Kim et al., 2013b). Also analogous to the Sec14-like PITPs, independent of any lipid-transfer functions, PITPα and PITPβ have been proposed to present PtdIns to modifying enzymes such as PPIn kinases (Cockcroft, 1999; Kular et al., 2002) and PPIn-specific phospholipases (Snoek et al., 1999). However, as their sequences would suggest, there are major differences in the overall structure of the Sec14-like and StARkin-related PITP domains; including clear deviations in the orientation of bound phospholipid cargoes.
Comprehensive descriptions of the StARkin-related PITP fold come from the solved structures of PITPα in the unbound conformation as well as in complex with PtdIns or PtdCho lipid cargoes; although, it should be mentioned, prior structures of other StAR-related lipid-transfer domains were also instructive (Tsujishita and Hurley, 2000; Roderick et al., 2002; Romanowski et al., 2002). The overall architecture of the open PITP fold shows a single phospholipid-binding pocket that is formed by the surface of a concave β-sheet made by eight β-strands (β1-β8) and two long α-helices (αA and αF) that are tethered by a regulatory loop and flank the core of the fold (Figure 5c; Schouten et al., 2002). Additional functional regions include a small lipid exchange loop, containing the short αB helix, that acts as a lid to gate access to the lipid-binding pocket as well as a C-terminal region containing the elongated αG helix (Schouten et al., 2002). The transport-competent conformations of PITPα, as well as PtdCho-bound PITPβ (Vordtriede et al., 2005), show the same overall orientation and accommodate a single phospholipid within the enclosed central cavity. Binding of PtdIns (Tilley et al., 2004) or PtdCho (Yoder et al., 2001; Vordtriede et al., 2005) to PITPα is characterized by similar structural rearrangements of the PITP fold; however, it is interesting to note that, opposite to the PPIn-binding LTDs of either the ORPs or Sec14-like PITPs, the phospholipid headgroups are buried within the hydrophobic core of the PITP fold, while the fatty acyl chains oriented outwards within two central channels (Figure 5c; Yoder et al., 2001; Tilley et al., 2004). The similarities between the lipid-bound structures of PITPα have made it difficult to define the dynamics and intermediate structural steps that contribute to the lipid exchange mechanism. In general, based on the open and closed structures, lipid-binding results in a downward movement of the αB helix that is associated with flattening of the peripheral regions within the β2-β4 strands to close off the hydrophobic cavity at the membrane interface (Yoder et al., 2001). The movement of the lipid exchange loop is also accompanied by an inward swing of the αG helix towards the core β-sheet and stabilization of the C-terminal tail (Yoder et al., 2001; Tilley et al., 2004). Interestingly, within the unbound state, the widening of the core hydrophobic pocket between the central β-sheet and helices αA and αF, as well as the formation of a smaller entrance between the N-terminus of helix αG and the β2-β3 strands, results in the opening of both ends of the lipid-binding cavity; effectively creating a central channel that is supported by the relatively immobile half of the core PITP β-sheet (Yoder et al., 2001). The presence of this hydrophobic tunnel could be relevant during the exchange of lipid cargoes at the membrane interface, or perhaps for the presentation of bound PtdIns to PPIn-modifying enzymes.
Despite the similarities of the transport-competent conformations of PITPs bound to PtdIns and PtdCho, there are clear differences in the coordination of the phospholipid headgroups that appear to explain the differences observed in the binding affinities and transfer rates described for the class I PITPs. Coordination of the inositol ring occurs through specific hydrogen bonds, including selective recognition of the PtdIns headgroup by K61, N90, and Q22 (Tilley et al., 2004). Selective binding to PtdCho and PtdIns involves recognition of the shared phosphate moiety in both lipids by K195, which binds to one phosphate oxygen, and residues T114 and T97 that interact with the other (Tilley et al., 2004). Residues T59 and E86 also contact both PtdCho and PtdIns, but do so in distinctive ways; making only van der Waals contacts with PtdCho, but facilitating hydrogen-bonding to the PtdIns headgroup (Tilley et al., 2004). Overall, studies using site-directed mutagenesis are in agreement with the structural data and demonstrate inhibition of PtdIns and PtdCho binding within cells as well as in lipid transfer assays performed in vitro. Selective reductions of PtdIns binding or transfer, without detrimental effects on PtdCho, could be achieved by mutations to the important inositol-coordinating residues T59, K61, E86, and N90 (Tilley et al., 2004). However, mutations significantly inhibiting PtdCho coordination were always followed by similar reductions in PtdIns-related binding (Tilley et al., 2004). These data suggest that the higher affinity of the PITPs towards PtdIns likely results from the additional hydrogen bonding network that forms between the polar inositol headgroup and the residues that line the internal binding cavity. The mechanisms contributing to the specificity of PITPs towards PtdCho need to be investigated in more detail and it is possible that additional secondary lipid cargoes may be relevant in vivo. Nevertheless, the residues that line the PITP lipid-binding cavity are highly conserved, including a number aromatic residues that have the potential to contribute membrane interactions during the lipid-exchange process. In fact, two conserved aromatic residues (W203 and W204) located at the end of the αF helix and adjacent to helix αG, have been shown to modulate the membrane association of Class I PITPs and are also thought to play a role during lipid exchange (Tilley et al., 2004; Phillips et al., 2006; Shadan et al., 2008; Yadav et al., 2015). Specifically, membrane insertion of this hydrophobic motif has also been postulated to initiate the opening of the binding cavity by disrupting the C-terminal tail and dislodging the αG helix; but no biophysical evidence for this mechanism has been provided (Shadan et al., 2008). A more recent study using molecular dynamics has attempted to further resolve features of the lipid exchange cycle utilized by the StARkin-related PITPs (Grabon et al., 2017). All-atom simulations using the open or membrane-docked conformation of PITPα identified overlapping as well as unique regions of the PITP domain interaction surface during binding to PtdCho or PtdIns (Grabon et al., 2017). Interestingly, after membrane association, bilayer insertion of the lipid exchange loop was shown to facilitate the partial loading of a single PtdCho into the hydrophobic pocket and coincident shielding of the fatty acyl chains from the bulk membrane (Grabon et al., 2017). These studies were unable to simulate the complete extraction of the phospholipid cargo into the PITP binding pocket, but do highlight the inherent complexities associated with the membrane recruitment and headgoup-specific coordination that are required for the selective transfer of lipids by PITPs.
Outside of the Class I PITPs, studies from Drosophila (Vihtelic et al., 1991; Vihtelic et al., 1993) and, more recently, mammals have characterized three homologs of class II PITPs: PITPNC1 (Class IIB), Nir2 (PITPNM1, Class IIA), and Nir3 (PITPNM2, Class IIA; Ocaka et al., 2005; Wyckoff et al., 2010). The only member of the Class IIB PITPs, PITPNC1, is a soluble protein that possesses a Class II-like PITP domain, but more closely resemble the Class I PITPs in overall architecture (Cockcroft, 2012; Kim et al., 2013b). Alternatively, the Class IIA PITPs are multi-domain proteins that possess an N-terminal PITP domain that is flanked by an acidic stretch containing an FFAT motif, which, similar to some ORPs, tethers the proteins to the ER membrane through interactions with ER-resident VAPs (Amarilio et al., 2005). The Class IIA PITPs also contain a DDHD domain that possesses some homology to sequence features found within a small group of intracellular phospholipase A1 (PLA1) proteins (PA-PLA1/DDHD1, p125/Sec23-Interacting Protein, and KIAA0725p/DDHD2; Inoue et al., 2012; Tani et al., 2012), as well as a C-terminal LNS2 (Lipin, Ned1, and Smp2) domain that was originally described in the well-characterized family of PtdOH phosphatases called lipins (Reue, 2009). Unfortunately, unlike the Class I PITPs, there are currently no structural descriptions of the PITP domains from the Class II PITPs; however extensive functional analyses have identified important roles for these proteins in the control of cellular signal transduction. In particular, the Class IIA PITPs, Nir2 and Nir3, have been shown to maintain PPIn signaling competence in response to PM PtdIns(4,5)P2 hydrolysis through the counter-exchange of PtdIns and PtdOH at ER-PM contact sites (Chang et al., 2013; Kim et al., 2013a; Chang and Liou, 2015; Kim et al., 2015; Yadav et al., 2015). The soluble Class IIB PITP, PITPNC1, has also been suggested to bind and transfer PtdOH in vitro (Garner et al., 2012); although our own studies using intact cells were unable to detect an enhanced clearance of PtdOH from the PM following over-expression of PITPNC1 (Kim et al., 2015; 2016). The identification of PtdOH transfer activity by the Class IIA PITPs, in particular, provides important evidence that phospholipids other than PtdIns or PtdCho can be used as cargoes by StARkin-related PITP domains. There are also reports of Class I PITPs binding to and transporting sphingomyelin in vitro (Li et al., 2002; Vordtriede et al., 2005), however, cellular studies do not support a role for PITPβ in the regulation of sphingomyelin biosynthesis or trafficking (Segui et al., 2002). Despite the possibility of binding to alternate cargoes, it is important to note that the overall topology of the distinct sequence motifs present within the StARkin-related PITP domains (Wyckoff et al., 2010), as well as the important residues directly involved in coordinating the inositol headgroup (T59, K61, E86, and N90; PITPα numbering), are conserved across the eukaryotic Class I and II PITPs (Tilley et al., 2004; Grabon et al., 2017). Taken together, these sequence features and the accompanying functional analyses suggest that the primary cargo of the Class I and II PITPs is likely to be PtdIns; whereas, similar to the ORPs, heterogeneity in the identity of the secondary cargo during the lipid exchange cycle could result from the limited specific contacts formed between phospholipids lacking the inositol headgroup. Consequently, although PtdIns transfer activity by Class II PITPs has been demonstrated in vitro and inferred in vivo, the complexities of the lipid exchange cycle, including the molecular determinants for cargo selectivity during lipid loading and unloading, still need to be determined.
8. Summary and Perspectives
PPIn lipids function as universal regulators of metabolism and membrane biology in part by orchestrating the spatial organization or activity of proteins within defined subcellular compartments. The goal of this chapter is to provide an overview of the structurally-diverse PPIn-binding protein folds and highlight the unique binding modalities that can be used for the specific recognition of PPIn lipids by peripheral binding proteins. Specific domain families exhibit clear preferences in their general methods for membrane association, although all of the examples presented here require that stereospecific headgroup recognition is coupled to a combination of electrostatic attraction and interfacial penetration of membrane-anchoring structural elements. Additionally, binding specificity is influenced not only by interactions with PPIn headgroups, but is also sensitive to the physical properties of the targeted membrane; including clear roles for membrane charge and curvature. It is still not clear to what extent heterogeneity in acyl chain composition contributes to the regulation of binding interactions between peripheral membrane proteins and PPIn species. The requirement for membrane insertion of specific residues or subdomains in many PPIn-binding folds suggests that features of the lipid backbone might be sampled during binding events. In particular, differences in acyl chain length could influence the presentation of the PPIn headgroup relative to other lipids within the bilayer (Choy et al., 2017), while the degree saturation within the local lipid environment may alter the relative ability of specific domain features to penetrate into the membrane leaflet. As outlined in the introduction, lipid saturation also has the potential to function as a general feature of organelle membrane identity (Bigay and Antonny, 2012; Barelli and Antonny, 2016) that likely functions in concert with PPIn-selective recognition modules for the selective targeting of proteins to specific subcellular compartments. Independent of general alterations to membrane composition, most of the PPIn-binding folds that have been identified show a high degree of conservation in their strategies for PPIn headgroup coordination. However, despite clear examples of high-affinity and univalent recognition of specific PPIn isomers, many of the domains discussed here interact with membrane-embedded PPIn species too weakly to drive membrane association of proteins in isolation. This reality highlights the need to understand how PPIn-binding domains communicate with other structural features to integrate multivalent interactions that not only increase membrane avidity of multi-domain proteins, but may also function to regulate the catalytic activity or coincident binding of additional protein effectors or lipids. New biophysical approaches, including advances in mass spectrometry (Konermann et al., 2014; Vadas and Burke, 2015) and spectroscopy (Chergui, 2016; Liang and Tamm, 2016), as well as the adoption of more sophisticated computational approaches (Lindahl and Sansom, 2008; Dror et al., 2012; Hospital et al., 2015; Hertig et al., 2016), will be required to map the conformational dynamics that relay PPIn-induced movements within individual proteins or membrane-binding macromolecular complexes. Investigations targeted at identifying molecular transitions will be vital for describing the structural intermediates associated with the coordinate regulation of membrane recognition and lipid exchange by the variety of PPIn-binding LTPs and should also provide novel functional insights into the metabolic coupling of PtdIns trafficking and PPIn production. Considering that many LTPs also possess additional membrane-targeting motifs or other structured folds, including some with PPIn-binding PH domains, LTPs offer the unique opportunity to study the potential role of PPIn-sensing during the conformation gating of the LTD for targeted lipid exchange. More generally, the presence of distinct PPIn-binding motifs within proteins with PPIn-modifying activity provides an interesting platform for investigating how patterns of PPIn recognition and metabolism function to regulate the spatiotemporal organization of PPIn species within subcellular compartments. Given the central importance of PPIn lipids for controlling membrane trafficking and signal transduction, and the clear links already identified between the dysregulation of PPIn metabolism and numerous human diseases, additional mechanistic insights into the interactions between PPIn species and peripheral protein effectors will be essential for defining the molecular pathways controlling cellular PtdIns metabolism. Just as importantly, these types of investigations, which uncover detailed molecular information regarding protein and membrane association, have the potential to inform novel therapeutic approaches that might be able to selectively target PPIn-dependent binding interactions or influence defined PPIn-mediated allosteric switches within specific membrane contexts.
Sources of Research Support:
TB is supported by the National Institutes of Health (NIH) Intramural Research Program (IRP). JGP is supported by an NICHD Visiting Fellowship and a Natural Sciences and Engineering Research Council of Canada (NSERC) Postdoctoral Fellowship.
Footnotes
Disclosures Statement:
The authors have nothing to disclose.
References
- Ago T, Takeya R, Hiroaki H, Kuribayashi F, Ito T, Kohda D, Sumimoto H (2001) The PX domain as a novel phosphoinositide-binding module. Biochem Biophys Res Commun 287 (3):733–738. doi: 10.1006/bbrc.2001.5629 [DOI] [PubMed] [Google Scholar]
- Agranoff BW (2009) Turtles All the Way: Reflections on myo-Inositol. J Biol Chem 284 (32):21121–21126. doi: 10.1074/jbc.X109.004747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranoff BW, Benjamin JA, Hajra AK (1969) Biosynthesis of phosphatidylinositol. Ann N Y Acad Sci 165 (2):755–760 [DOI] [PubMed] [Google Scholar]
- Agranoff BW, Bradley RM, Brady RO (1958) The enzymatic synthesis of inositol phosphatide. J Biol Chem 233 (5):1077–1083 [PubMed] [Google Scholar]
- Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI (2006) Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol 13 (11):1029–1030. doi: 10.1038/nsmb1160 [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet N (2017) Lipid Tales of Viral Replication and Transmission. Trends Cell Biol 27 (3):201–213. doi: 10.1016/j.tcb.2016.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet N, Balla T (2012) Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem Sci 37 (7):293–302. doi: 10.1016/j.tibs.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, Dixon JE (2006) Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124 (1):133–145. doi: 10.1016/j.cell.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Alwarawrah M, Wereszczynski J (2017) Investigation of the Effect of Bilayer Composition on PKCalpha-C2 Domain Docking Using Molecular Dynamics Simulations. J Phys Chem B 121 (1):78–88. doi: 10.1021/acs.jpcb.6b10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarilio R, Ramachandran S, Sabanay H, Lev S (2005) Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J Biol Chem 280 (7):5934–5944. doi: 10.1074/jbc.M409566200 [DOI] [PubMed] [Google Scholar]
- Anand K, Maeda K, Gavin AC (2012) Structural analyses of the Slm1-PH domain demonstrate ligand binding in the non-canonical site. PLoS One 7 (5):e36526. doi: 10.1371/journal.pone.0036526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B (2011) Mechanisms of membrane curvature sensing. Annu Rev Biochem 80:101–123. doi: 10.1146/annurev-biochem-052809-155121 [DOI] [PubMed] [Google Scholar]
- Arcaro A (1998) The small GTP-binding protein Rac promotes the dissociation of gelsolin from actin filaments in neutrophils. J Biol Chem 273 (2):805–813 [DOI] [PubMed] [Google Scholar]
- Baines AJ, Lu HC, Bennett PM (2014) The Protein 4.1 family: hub proteins in animals for organizing membrane proteins. Biochim Biophys Acta 1838 (2):605–619. doi: 10.1016/j.bbamem.2013.05.030 [DOI] [PubMed] [Google Scholar]
- Balla A, Balla T (2006) Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol 16 (7):351–361. doi: 10.1016/j.tcb.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Balla A, Kim YJ, Varnai P, Szentpetery Z, Knight Z, Shokat KM, Balla T (2008) Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIalpha. Mol Biol Cell 19 (2):711–721. doi: 10.1091/mbc.E07-07-0713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T (2005) Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J Cell Sci 118 (Pt 10):2093–2104. doi: 10.1242/jcs.02387 [DOI] [PubMed] [Google Scholar]
- Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93 (3):1019–1137. doi: 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Baukal AJ, Guillemette G, Catt KJ (1988) Multiple pathways of inositol polyphosphate metabolism in angiotensin-stimulated adrenal glomerulosa cells. J Biol Chem 263 (9):4083–4091 [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R (1989) The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol 108 (4):1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W (1990) An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347 (6293): 561–562. doi: 10.1083/347561a0 [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G (2010) The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci 35 (3):150–160. doi: 10.1016/j.tibs.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraldi E, Djinovic Carugo K, Hyvonen M, Surdo PL, Riley AM, Potter BV, O’Brien R, Ladbury JE, Saraste M (1999) Structure of the PH domain from Bruton’s tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure 7 (4):449–460 [DOI] [PubMed] [Google Scholar]
- Barelli H, Antonny B (2016) Lipid unsaturation and organelle dynamics. Curr Opin Cell Biol 41:25–32. doi: 10.1016/j.ceb.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, Anderson RA (2010) Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol 20 (1):25–35. doi: 10.1016/j.tcb.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera NP, Zhou M, Robinson CV (2013) The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends Cell Biol 23 (1):1–8. doi: 10.1016/j.tcb.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH (2012) Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell 47 (3):339–348. doi: 10.1016/j.molcel.2012.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T, Capraro BR, Zhu C, Das SL (2011) Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu Rev Phys Chem 62:483–506. doi: 10.1146/annurev.physchem.012809.103450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley MJ, Dixon JE (2005) The structure and regulation of myotubularin phosphatases. Curr Opin Struct Biol 15 (6):614–620. doi: 10.1016/j.sbi.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Begley MJ, Taylor GS, Brock MA, Ghosh P, Woods VL, Dixon JE (2006) Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc Natl Acad Sci U S A 103 (4):927–932. doi: 10.1073/pnas.0510006103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley MJ, Taylor GS, Kim SA, Veine DM, Dixon JE, Stuckey JA (2003) Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol Cell 12 (6):1391–1402 [DOI] [PubMed] [Google Scholar]
- Berger P, Schaffitzel C, Berger I, Ban N, Suter U (2003) Membrane association of myotubularin-related protein 2 is mediated by a pleckstrin homology-GRAM domain and a coiled-coil dimerization module. Proc Natl Acad Sci U S A 100 (21):12177–12182. doi: 10.1073/pnas.2132732100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Gladfelter AS, Kovar DR, Lee WL (2015) Cytoskeletal dynamics: a view from the membrane. J Cell Biol 209 (3):329–337. doi: 10.1083/jcb.201502062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Antonny B (2012) Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell 23 (5):886–895. doi: 10.1016/j.devcel.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Bittova L, Sumandea M, Cho W (1999) A structure-function study of the C2 domain of cytosolic phospholipase A2. Identification of essential calcium ligands and hydrophobic membrane binding residues. J Biol Chem 274 (14):9665–9672 [DOI] [PubMed] [Google Scholar]
- Blomberg N, Baraldi E, Nilges M, Saraste M (1999) The PH superfold: a structural scaffold for multiple functions. Trends Biochem Sci 24 (11):441–445 [DOI] [PubMed] [Google Scholar]
- Blomberg N, Nilges M (1997) Functional diversity of PH domains: an exhaustive modelling study. Fold Des 2 (6):343–355. doi: 10.1016/S1359-0278(97)00048-5 [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L (1999) Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286 (5447):2119–2125 [DOI] [PubMed] [Google Scholar]
- Bojjireddy N, Botyanszki J, Hammond G, Creech D, Peterson R, Kemp DC, Snead M, Brown R, Morrison A, Wilson S, Harrison S, Moore C, Balla T (2014) Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem 289 (9):6120–6132. doi: 10.1074/jbc.M113.531426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompard G, Martin M, Roy C, Vignon F, Freiss G (2003) Membrane targeting of protein tyrosine phosphatase PTPL1 through its FERM domain via binding to phosphatidylinositol 4,5-biphosphate. J Cell Sci 116 (Pt 12):2519–2530. doi: 10.1242/jcs.00448 [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS (2002) Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol 156 (6):1015–1028. doi: 10.1083/jcb.200201002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ (2009) Changing phosphoinositides “on the fly”: how trafficking vesicles avoid an identity crisis. Bioessays 31 (10):1127–1136. doi: 10.1002/bies.200900060 [DOI] [PubMed] [Google Scholar]
- Boucrot E, Ferreira AP, Almeida-Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, McMahon HT (2015) Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517 (7535):460–465. doi: 10.1038/nature14067 [DOI] [PubMed] [Google Scholar]
- Boura E, Nencka R (2015) Phosphatidylinositol 4-kinases: Function, structure, and inhibition. Exp Cell Res 337 (2):136–145. doi: 10.1016/j.yexcr.2015.03.028 [DOI] [PubMed] [Google Scholar]
- Braccini L, Ciraolo E, Campa CC, Perino A, Longo DL, Tibolla G, Pregnolato M, Cao Y, Tassone B, Damilano F, Laffargue M, Calautti E, Falasca M, Norata GD, Backer JM, Hirsch E (2015) PI3K-C2gamma is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat Commun 6:7400. doi: 10.1038/ncomms8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL (2001) The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell 8 (4):829–839 [DOI] [PubMed] [Google Scholar]
- Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H (2009) Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem 284 (8):4846–4856. doi: 10.1074/jbc.M807505200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunecky R, Lee S, Rzepecki PW, Overduin M, Prestwich GD, Kutateladze AG, Kutateladze TG (2005) Investigation of the binding geometry of a peripheral membrane protein. Biochemistry 44 (49):16064–16071. doi: 10.1021/bi051127+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney TD, Katan M (2010) Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer 10 (5):342–352. doi: 10.1038/nrc2842 [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD (1998) Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell 2 (1):157–162 [DOI] [PubMed] [Google Scholar]
- Burke JE, Williams RL (2015) Synergy in activating class I PI3Ks. Trends Biochem Sci 40 (2):88–100. doi: 10.1016/j.tibs.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Busse RA, Scacioc A, Krick R, Perez-Lara A, Thumm M, Kuhnel K (2015) Characterization of PROPPIN-Phosphoinositide Binding and Role of Loop 6CD in PROPPIN-Membrane Binding. Biophys J 108 (9):2223–2234. doi: 10.1016/j.bpj.2015.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan J, Simonsen A, Gaullier JM, Toh BH, Stenmark H (1999) The endosome fusion regulator early-endosomal autoantigen 1 (EEA1) is a dimer. Biochem J 338 (Pt 2):539–543 [PMC free article] [PubMed] [Google Scholar]
- Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B (2007) Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol 5 (4):e95. doi: 10.1371/journal.pbio.0050095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja V, Laguerre M, Larijani B (2009) 3-D structure and dynamics of protein kinase B-new mechanism for the allosteric regulation of an AGC kinase. J Chem Biol 2 (1):11–25. doi: 10.1007/s12154-009-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja V, Laguerre M, Larijani B (2012) Role of the C-terminal regulatory domain in the allosteric inhibition of PKB/Akt. Adv Biol Regul 52 (1):46–57. doi: 10.1016/j.advenzreg.2011.09.009 [DOI] [PubMed] [Google Scholar]
- Calleja V, Laguerre M, Parker PJ, Larijani B (2009) Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol 7 (1):e17. doi: 10.1371/journal.pbio.1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa F, Yoon HY, Ha VL, Szentpetery Z, Balla T, Randazzo PA (2009) A PH domain in the Arf GTPase-activating protein (GAP) ARAP1 binds phosphatidylinositol 3,4,5-trisphosphate and regulates Arf GAP activity independently of recruitment to the plasma membranes. J Biol Chem 284 (41):28069–28083. doi: 10.1074/jbc.M109.028266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RB, Liu F, Ross AH (2003) Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J Biol Chem 278 (36):33617–33620. doi: 10.1074/jbc.C300296200 [DOI] [PubMed] [Google Scholar]
- Carlton JG, Cullen PJ (2005) Coincidence detection in phosphoinositide signaling. Trends Cell Biol 15 (10):540–547. doi: 10.1016/j.tcb.2005.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K, Gomez C, Shapiro L (2004) Tubby proteins: the plot thickens. Nat Rev Mol Cell Biol 5 (1):55–63. doi: 10.1038/nrm1278 [DOI] [PubMed] [Google Scholar]
- Cash JN, Davis EM, Tesmer JJG (2016) Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3. Structure 24 (5):730–740. doi: 10.1016/j.str.2016.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli DF, Blasutig IM, Goudreault M, Li Z, Ruston J, Pawson T, Sicheri F (2007) Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J Biol Chem 282 (18):13864–13874. doi: 10.1074/jbc.M700505200 [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ (2006) Crystal structure of the FERM domain of focal adhesion kinase. J Biol Chem 281 (1):252–259. doi: 10.1074/jbc.M509188200 [DOI] [PubMed] [Google Scholar]
- Chan KC, Lu L, Sun F, Fan J (2017) Molecular Details of the PH Domain of ACAP1(BAR-PH) Protein Binding to PIP-Containing Membrane. J Phys Chem B 121 (15):3586–3596. doi: 10.1021/acs.jpcb.6b09563 [DOI] [PubMed] [Google Scholar]
- Chang BH, Gujral TS, Karp ES, BuKhalid R, Grantcharova VP, MacBeath G (2011) A systematic family-wide investigation reveals that ~30% of mammalian PDZ domains engage in PDZ-PDZ interactions. Chem Biol 18 (9):1143–1152. doi: 10.1016/j.chembiol.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC, Liou J (2013) Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep 5 (3):813–825. doi: 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- Chang CL, Liou J (2015) Phosphatidylinositol 4,5-Bisphosphate Homeostasis Regulated by Nir2 and Nir3 Proteins at Endoplasmic Reticulum-Plasma Membrane Junctions. J Biol Chem 290 (23):14289–14301. doi: 10.1074/jbc.M114.621375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charman M, Colbourne TR, Pietrangelo A, Kreplak L, Ridgway ND (2014) Oxysterol-binding protein (OSBP)-related protein 4 (ORP4) is essential for cell proliferation and survival. J Biol Chem 289 (22):15705–15717. doi: 10.1074/jbc.M114.571216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Kutateladze TG, Overduin M (2006) Increased mobility in the membrane targeting PX domain induced by phosphatidylinositol 3-phosphate. Protein Sci 15 (8):1873–1882. doi: 10.1110/ps.062194906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M (2001) Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol 3 (7):613–618. doi: 10.1038/35083000 [DOI] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P (1998) Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 394 (6695):793–797. doi: 10.1038/29555 [DOI] [PubMed] [Google Scholar]
- Chen Y, Sheng R, Kallberg M, Silkov A, Tun MP, Bhardwaj N, Kurilova S, Hall RA, Honig B, Lu H, Cho W (2012) Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol Cell 46 (2):226–237. doi: 10.1016/j.molcel.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chergui M (2016) Time-resolved X-ray spectroscopies of chemical systems: New perspectives. Struct Dyn 3 (3):031001. doi: 10.1063/1.4953104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiapparino A, Maeda K, Turei D, Saez-Rodriguez J, Gavin AC (2016) The orchestra of lipid-transfer proteins at the crossroads between metabolism and signaling. Prog Lipid Res 61:30–39. doi: 10.1016/j.plipres.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ Jr., Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Tsukita S, Hoover KB, et al. (1998) The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci 23 (8):281–282 [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV (2005) Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct 34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337 [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV (2006) Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta 1761 (8):838–849. doi: 10.1016/j.bbalip.2006.06.014 [DOI] [PubMed] [Google Scholar]
- Choi S, Hedman AC, Sayedyahossein S, Thapa N, Sacks DB, Anderson RA (2016) Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat Cell Biol 18 (12):1324–1335. doi: 10.1038/ncb3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Thapa N, Tan X, Hedman AC, Anderson RA (2015) PIP kinases define PI4,5P(2)signaling specificity by association with effectors. Biochim Biophys Acta 1851 (6):711–723. doi: 10.1016/j.bbalip.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy CH, Han BK, Botelho RJ (2017) Phosphoinositide Diversity, Distribution, and Effector Function: Stepping Out of the Box. Bioessays 39 (12). doi: 10.1002/bies.201700121 [DOI] [PubMed] [Google Scholar]
- Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P (2015) INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349 (6246):428–432. doi: 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Song WJ, Kim K, Bednarski JJ, Chen J, Prestwich GD, Holz RW (1998) The C2 domains of Rabphilin3A specifically bind phosphatidylinositol 4,5-bisphosphate containing vesicles in a Ca2+-dependent manner. In vitro characteristics and possible significance. J Biol Chem 273 (17):10240–10248 [DOI] [PubMed] [Google Scholar]
- Cicchetti G, Maurer P, Wagener P, Kocks C (1999) Actin and phosphoinositide binding by the ActA protein of the bacterial pathogen Listeria monocytogenes. J Biol Chem 274 (47):33616–33626 [DOI] [PubMed] [Google Scholar]
- Clague MJ, Lorenzo O (2005) The myotubularin family of lipid phosphatases. Traffic 6 (12):1063–1069. doi: 10.1111/j.1600-0854.2005.00338.x [DOI] [PubMed] [Google Scholar]
- Cleves A, McGee T, Bankaitis V (1991) Phospholipid transfer proteins: a biological debut. Trends Cell Biol 1 (1):30–34 [DOI] [PubMed] [Google Scholar]
- Cockcroft S (1999) Mammalian phosphatidylinositol transfer proteins: emerging roles in signal transduction and vesicular traffic. Chem Phys Lipids 98 (1–2):23–33 [DOI] [PubMed] [Google Scholar]
- Cockcroft S (2012) The diverse functions of phosphatidylinositol transfer proteins. Curr Top Microbiol Immunol 362:185–208. doi: 10.1007/978-94-007-5025-8_9 [DOI] [PubMed] [Google Scholar]
- Cockcroft S, Carvou N (2007) Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta 1771 (6):677–691. doi: 10.1016/j.bbalip.2007.03.009 [DOI] [PubMed] [Google Scholar]
- Corbalan-Garcia S, Gomez-Fernandez JC (2014a) Classical protein kinases C are regulated by concerted interaction with lipids: the importance of phosphatidylinositol-4,5-bisphosphate. Biophys Rev 6 (1):3–14. doi: 10.1007/s12551-013-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbalan-Garcia S, Gomez-Fernandez JC (2014b) Signaling through C2 domains: more than one lipid target. Biochim Biophys Acta 1838 (6):1536–1547. doi: 10.1016/j.bbamem.2014.01.008 [DOI] [PubMed] [Google Scholar]
- Coudevylle N, Montaville P, Leonov A, Zweckstetter M, Becker S (2008) Structural determinants for Ca2+ and phosphatidylinositol 4,5-bisphosphate binding by the C2A domain of rabphilin-3A. J Biol Chem 283 (51): 35918–35928. doi: 10.1074/jbc.M804094200 [DOI] [PubMed] [Google Scholar]
- Cozier GE, Carlton J, Bouyoucef D, Cullen PJ (2004) Membrane targeting by pleckstrin homology domains. Curr Top Microbiol Immunol 282:49–88 [DOI] [PubMed] [Google Scholar]
- Crowder MK, Seacrist CD, Blind RD (2017) Phospholipid regulation of the nuclear receptor superfamily. Adv Biol Regul 63:6–14. doi: 10.1016/j.jbior.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Cozier GE, Banting G, Mellor H (2001) Modular phosphoinositide-binding domains--their role in signalling and membrane trafficking. Curr Biol 11 (21):R882–893 [DOI] [PubMed] [Google Scholar]
- Daste F, Walrant A, Holst MR, Gadsby JR, Mason J, Lee JE, Brook D, Mettlen M, Larsson E, Lee SF, Lundmark R, Gallop JL (2017) Control of actin polymerization via the coincidence of phosphoinositides and high membrane curvature. J Cell Biol 216 (11):3745–3765. doi: 10.1083/jcb.201704061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletov B, Perisic O, Williams RL (1998) Calcium-dependent membrane penetration is a hallmark of the C2 domain of cytosolic phospholipase A2 whereas the C2A domain of synaptotagmin binds membranes electrostatically. J Biol Chem 273 (30): 19093–19096. doi: 10.1074/jbc.273.30.19093 [DOI] [PubMed] [Google Scholar]
- De Camilli P, Chen H, Hyman J, Panepucci E, Bateman A, Brunger AT (2002) The ENTH domain. FEBS Lett 513 (1):11–18 [DOI] [PubMed] [Google Scholar]
- De Matteis M, Godi A, Corda D (2002) Phosphoinositides and the golgi complex. Curr Opin Cell Biol 14 (4):434–447 [DOI] [PubMed] [Google Scholar]
- de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G (2011) Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol 195 (6):965–978. doi: 10.1083/jcb.201104062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries KJ, Heinrichs AA, Cunningham E, Brunink F, Westerman J, Somerharju PJ, Cockcroft S, Wirtz KW, Snoek GT (1995) An isoform of the phosphatidylinositol-transfer protein transfers sphingomyelin and is associated with the Golgi system. Biochem J 310 (Pt 2):643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries KJ, Westerman J, Bastiaens PI, Jovin TM, Wirtz KW, Snoek GT (1996) Fluorescently labeled phosphatidylinositol transfer protein isoforms (alpha and beta), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp Cell Res 227 (1):33–39. doi: 10.1006/excr.1996.0246 [DOI] [PubMed] [Google Scholar]
- Del Campo CM, Mishra AK, Wang YH, Roy CR, Janmey PA, Lambright DG (2014) Structural basis for PI(4)P-specific membrane recruitment of the Legionella pneumophila effector DrrA/SidM. Structure 22 (3):397–408. doi: 10.1016/j.str.2013.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Ottinger EA, Nolte RT, Eck MJ, Shoelson SE (1999) Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proc Natl Acad Sci U S A 96 (15):8378–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lello P, Nguyen BD, Jones TN, Potempa K, Kobor MS, Legault P, Omichinski JG (2005) NMR structure of the amino-terminal domain from the Tfb1 subunit of TFIIH and characterization of its phosphoinositide and VP16 binding sites. Biochemistry 44 (21):7678–7686. doi: 10.1021/bi050099s [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Delprato A, Gabe Lee MT, Cronin TC, Huang S, Guilherme A, Czech MP, Lambright DG (2007) Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol Cell 28 (4):569–583. doi: 10.1016/j.molcel.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNitto JP, Lambright DG (2006) Membrane and juxtamembrane targeting by PH and PTB domains. Biochim Biophys Acta 1761 (8):850–867. doi: 10.1016/j.bbalip.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Diraviyam K, Stahelin RV, Cho W, Murray D (2003) Computer modeling of the membrane interaction of FYVE domains. J Mol Biol 328 (3):721–736 [DOI] [PubMed] [Google Scholar]
- Doerks T, Strauss M, Brendel M, Bork P (2000) GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem Sci 25 (10):483–485 [DOI] [PubMed] [Google Scholar]
- Dornan GL, McPhail JA, Burke JE (2016) Type III phosphatidylinositol 4 kinases: structure, function, regulation, signalling and involvement in disease. Biochem Soc Trans 44 (1):260–266. doi: 10.1042/BST20150219 [DOI] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, Parker PJ, Lemmon MA (2004) Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J 23 (9):1922–1933. doi: 10.1038/sj.emboj.7600203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G (2014) Topological regulation of lipid balance in cells. Annu Rev Biochem 83:51–77. doi: 10.1146/annurev-biochem-060713-035307 [DOI] [PubMed] [Google Scholar]
- Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE (2012) Biomolecular simulation: a computational microscope for molecular biology. Annu Rev Biophys 41:429–452. doi: 10.1146/annurev-biophys-042910-155245 [DOI] [PubMed] [Google Scholar]
- Du X, Kumar J, Ferguson C, Schulz TA, Ong YS, Hong W, Prinz WA, Parton RG, Brown AJ, Yang H (2011) A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol 192 (1):121–135. doi: 10.1083/jcb.201004142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG (2001) Multivalent endosome targeting by homodimeric EEA1. Mol Cell 8 (5):947–958 [DOI] [PubMed] [Google Scholar]
- Duning K, Wennmann DO, Bokemeyer A, Reissner C, Wersching H, Thomas C, Buschert J, Guske K, Franzke V, Floel A, Lohmann H, Knecht S, Brand SM, Poter M, Rescher U, Missler M, Seelheim P, Propper C, Boeckers TM, Makuch L, Huganir R, Weide T, Brand E, Pavenstadt H, Kremerskothen J (2013) Common exonic missense variants in the C2 domain of the human KIBRA protein modify lipid binding and cognitive performance. Transl Psychiatry 3:e272. doi: 10.1038/tp.2013.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson JM, Fedele CG, Davies EM, Becanovic J, Mitchell CA (2012) Phosphoinositide phosphatases: just as important as the kinases. Subcell Biochem 58:215–279. doi: 10.1007/978-94-007-3012-0_7 [DOI] [PubMed] [Google Scholar]
- Ebner M, Lucic I, Leonard TA, Yudushkin I (2017) PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol Cell 65 (3):416–431 e416. doi: 10.1016/j.molcel.2016.12.028 [DOI] [PubMed] [Google Scholar]
- Edwards SD, Keep NH (2001) The 2.7 A crystal structure of the activated FERM domain of moesin: an analysis of structural changes on activation. Biochemistry 40 (24):7061–7068 [DOI] [PubMed] [Google Scholar]
- Egea-Jimenez AL, Gallardo R, Garcia-Pino A, Ivarsson Y, Wawrzyniak AM, Kashyap R, Loris R, Schymkowitz J, Rousseau F, Zimmermann P (2016) Frizzled 7 and PIP2 binding by syntenin PDZ2 domain supports Frizzled 7 trafficking and signalling. Nat Commun 7:12101. doi: 10.1038/ncomms12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Shai N, Schuldiner M, Bohnert M (2016) A Tether Is a Tether Is a Tether: Tethering at Membrane Contact Sites. Dev Cell 39 (4):395–409. doi: 10.1016/j.devcel.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PR, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR (2001) PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox). Nat Cell Biol 3 (7):679–682. doi: 10.1038/35083076 [DOI] [PubMed] [Google Scholar]
- Emptage RP, Lemmon MA, Ferguson KM (2017) Molecular determinants of KA1 domain-mediated autoinhibition and phospholipid activation of MARK1 kinase. Biochem J 474 (3):385–398. doi: 10.1042/BCJ20160792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emptage RP, Schoenberger MJ, Ferguson KM, Marmorstein R (2017) Intramolecular autoinhibition of Checkpoint Kinase 1 is mediated by conserved basic motifs of the C-terminal Kinase Associated-1 domain. J Biol Chem. doi: 10.1074/jbc.M117.811265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shirouzu M, Yokoyama S (2003) The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J Biol Chem 278 (2):843–852. doi: 10.1074/jbc.M209714200 [DOI] [PubMed] [Google Scholar]
- Erneux C, Edimo WE, Deneubourg L, Pirson I (2011) SHIP2 multiple functions: a balance between a negative control of PtdIns(3,4,5)P(3) level, a positive control of PtdIns(3,4)P(2) production, and intrinsic docking properties. J Cell Biochem 112 (9):2203–2209. doi: 10.1002/jcb.23146 [DOI] [PubMed] [Google Scholar]
- Ernst A, Appleton BA, Ivarsson Y, Zhang Y, Gfeller D, Wiesmann C, Sidhu SS (2014) A structural portrait of the PDZ domain family. J Mol Biol 426 (21):3509–3519. doi: 10.1016/j.jmb.2014.08.012 [DOI] [PubMed] [Google Scholar]
- Falasca M, Hughes WE, Dominguez V, Sala G, Fostira F, Fang MQ, Cazzolli R, Shepherd PR, James DE, Maffucci T (2007) The role of phosphoinositide 3-kinase C2alpha in insulin signaling. J Biol Chem 282 (38):28226–28236. doi: 10.1074/jbc.M704357200 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell 6 (2):373–384 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1994) Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell 79 (2):199–209 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83 (6):1037–1046 [DOI] [PubMed] [Google Scholar]
- Fernandes F, Coutinho A, Prieto M, Loura LM (2015) Electrostatically driven lipid-protein interaction: Answers from FRET. Biochim Biophys Acta 1848 (9):1837–1848. doi: 10.1016/j.bbamem.2015.02.023 [DOI] [PubMed] [Google Scholar]
- Flesch FM, Yu JW, Lemmon MA, Burger KN (2005) Membrane activity of the phospholipase C-delta1 pleckstrin homology (PH) domain. Biochem J 389 (Pt 2):435–441. doi: 10.1042/BJ20041721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419 (6905):361–366. doi: 10.1038/nature01020 [DOI] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT (2001) Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291 (5506):1051–1055. doi: 10.1126/science.291.5506.1051 [DOI] [PubMed] [Google Scholar]
- Frame MC, Patel H, Serrels B, Lietha D, Eck MJ (2010) The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol 11 (11):802–814. doi: 10.1038/nrm2996 [DOI] [PubMed] [Google Scholar]
- Franco I, Gulluni F, Campa CC, Costa C, Margaria JP, Ciraolo E, Martini M, Monteyne D, De Luca E, Germena G, Posor Y, Maffucci T, Marengo S, Haucke V, Falasca M, Perez-Morga D, Boletta A, Merlo GR, Hirsch E (2014) PI3K class II alpha controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev Cell 28 (6):647–658. doi: 10.1016/j.devcel.2014.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, Falke JJ, Cafiso DS (2002) Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry 41 (20):6282–6292 [DOI] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P (2009) The BAR domain superfamily: membrane-molding macromolecules. Cell 137 (2):191–196. doi: 10.1016/j.cell.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z (2010) The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol 20 (6):355–362. doi: 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo R, Ivarsson Y, Schymkowitz J, Rousseau F, Zimmermann P (2010) Structural diversity of PDZ-lipid interactions. Chembiochem 11 (4):456–467. doi: 10.1002/cbic.200900616 [DOI] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT (2006) Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J 25 (12):2898–2910. doi: 10.1038/sj.emboj.7601174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhir A, Hangyas-Mihalyne G, Zaitseva I, Cafiso DS, Wang J, Murray D, Pentyala SN, Smith SO, McLaughlin S (2004) Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys J 86 (4):2188–2207. doi: 10.1016/S0006-3495(04)74278-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper N, Shapiro MS (2007) Regulation of ion transport proteins by membrane phosphoinositides. Nat Rev Neurosci 8 (12):921–934. doi: 10.1038/nrn2257 [DOI] [PubMed] [Google Scholar]
- Garner K, Hunt AN, Koster G, Somerharju P, Groves E, Li M, Raghu P, Holic R, Cockcroft S (2012) Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J Biol Chem 287 (38):32263–32276. doi: 10.1074/jbc.M112.375840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD (1998) Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol 143 (1):65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R (1998) FYVE fingers bind PtdIns(3)P. Nature 394 (6692):432–433. doi: 10.1038/28767 [DOI] [PubMed] [Google Scholar]
- Ghai R, Du X, Wang H, Dong J, Ferguson C, Brown AJ, Parton RG, Wu JW, Yang H (2017) ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane. Nat Commun 8 (1):757. doi: 10.1038/s41467-017-00861-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil JE, Kim E, Kim IS, Ku B, Park WS, Oh BH, Ryu SH, Cho W, Heo WD (2012) Phosphoinositides differentially regulate protrudin localization through the FYVE domain. J Biol Chem 287 (49):41268–41276. doi: 10.1074/jbc.M112.419127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 19 (17):4577–4588. doi: 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P (2013) PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153 (7):1494–1509. doi: 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA (2004) FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6 (5):393–404. doi: 10.1038/ncb1119 [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA (1999) ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol 1 (5):280–287. doi: 10.1038/12993 [DOI] [PubMed] [Google Scholar]
- Grabon A, Khan D, Bankaitis VA (2015) Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim Biophys Acta 1851 (6):724–735. doi: 10.1016/j.bbalip.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabon A, Orlowski A, Tripathi A, Vuorio J, Javanainen M, Rog T, Lonnfors M, McDermott MI, Siebert G, Somerharju P, Vattulainen I, Bankaitis VA (2017) Dynamics and energetics of the mammalian phosphatidylinositol transfer protein phospholipid exchange cycle. J Biol Chem 292 (35):14438–14455. doi: 10.1074/jbc.M117.791467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler JA, Essen LO, Williams RL, Hurley JH (1996) C2 domain conformational changes in phospholipase C-delta 1. Nat Struct Biol 3 (9):788–795 [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S (2009) Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc Natl Acad Sci U S A 106 (16):6603–6607. doi: 10.1073/pnas.0813099106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen J, Ferrer-Orta C, Buxaderas M, Perez-Sanchez D, Guerrero-Valero M, Luengo-Gil G, Pous J, Guerra P, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S (2013) Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1. Proc Natl Acad Sci U S A 110 (51):20503–20508. doi: 10.1073/pnas.1316179110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyas G, Radvanszki G, Matuska R, Balla A, Hunyady L, Balla T, Varnai P (2017) Plasma membrane phosphatidylinositol 4-phosphate and 4,5-bisphosphate determine the distribution and function of K-Ras4B but not H-Ras proteins. J Biol Chem. doi: 10.1074/jbc.M117.806679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham H, Sreelatha A, Orth K (2011) Manipulation of host membranes by bacterial effectors. Nat Rev Microbiol 9 (9):635–646. doi: 10.1038/nrmicro2602 [DOI] [PubMed] [Google Scholar]
- Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T (2000) Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J 19 (17):4449–4462. doi: 10.1093/emboj/19.17.4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Balla T (2015) Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim Biophys Acta 1851 (6):746–758. doi: 10.1016/j.bbalip.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337 (6095):727–730. doi: 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, Balla T (2014) A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol 205 (1):113–126. doi: 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Takasuga S, Sasaki T, Balla T (2015) The ML1Nx2 Phosphatidylinositol 3,5-Bisphosphate Probe Shows Poor Selectivity in Cells. PLoS One 10 (10):e0139957. doi: 10.1371/journal.pone.0139957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, Fukui Y, Sasakawa C (2007) Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat Cell Biol 9 (1):121–128. doi: 10.1038/ncb1526 [DOI] [PubMed] [Google Scholar]
- Harlan JE, Hajduk PJ, Yoon HS, Fesik SW (1994) Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 371 (6493):168–170. doi: 10.1038/371168a0 [DOI] [PubMed] [Google Scholar]
- Haslam RJ, Koide HB, Hemmings BA (1993) Pleckstrin domain homology. Nature 363 (6427):309–310. doi: 10.1038/363309b0 [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Jackson TR, Stephens LR (1992) Platelet-derived growth factor stimulates synthesis of PtdIns(3,4,5)P3 by activating a PtdIns(4,5)P2 3-OH kinase. Nature 358 (6382):157–159. doi: 10.1038/358157a0 [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Stephens LR (2016) Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K-regulated pathways. Biochem Soc Trans 44 (1):307–314. doi: 10.1042/BST20150248 [DOI] [PubMed] [Google Scholar]
- Hayakawa A, Hayes SJ, Lawe DC, Sudharshan E, Tuft R, Fogarty K, Lambright D, Corvera S (2004) Structural basis for endosomal targeting by FYVE domains. J Biol Chem 279 (7):5958–5966. doi: 10.1074/jbc.M310503200 [DOI] [PubMed] [Google Scholar]
- He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG (2008) Molecular mechanism of membrane targeting by the GRP1 PH domain. J Lipid Res 49 (8):1807–1815. doi: 10.1194/jlr.M800150-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Vora M, Haney RM, Filonov GS, Musselman CA, Burd CG, Kutateladze AG, Verkhusha VV, Stahelin RV, Kutateladze TG (2009) Membrane insertion of the FYVE domain is modulated by pH. Proteins 76 (4):852–860. doi: 10.1002/prot.22392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedger G, Sansom MSP (2016) Lipid interaction sites on channels, transporters and receptors: Recent insights from molecular dynamics simulations. Biochim Biophys Acta 1858 (10):2390–2400. doi: 10.1016/j.bbamem.2016.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T (2006) PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314 (5804):1458–1461. doi: 10.1126/science.1134389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig S, Latorraca NR, Dror RO (2016) Revealing Atomic-Level Mechanisms of Protein Allostery with Molecular Dynamics Simulations. PLoS Comput Biol 12 (6):e1004746. doi: 10.1371/journal.pcbi.1004746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgemann DW, Ball R (1996) Regulation of cardiac Na+,Ca2+ exchange and KATP potassium channels by PIP2. Science 273 (5277):956–959 [DOI] [PubMed] [Google Scholar]
- Hilgemann DW, Feng S, Nasuhoglu C (2001) The complex and intriguing lives of PIP2 with ion channels and transporters. Sci STKE 2001 (111):re19. doi: 10.1126/stke.2001.111.re19 [DOI] [PubMed] [Google Scholar]
- Hill K, Krugmann S, Andrews SR, Coadwell WJ, Finan P, Welch HC, Hawkins PT, Stephens LR (2005) Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and Gbetagamma subunits. J Biol Chem 280 (6):4166–4173. doi: 10.1074/jbc.M411262200 [DOI] [PubMed] [Google Scholar]
- Hille B, Dickson EJ, Kruse M, Vivas O, Suh BC (2015) Phosphoinositides regulate ion channels. Biochim Biophys Acta 1851 (6):844–856. doi: 10.1016/j.bbalip.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, Kato R, Stenmark H, Wakatsuki S (2006) Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol 13 (11):1031–1032. doi: 10.1038/nsmb1163 [DOI] [PubMed] [Google Scholar]
- Hiroaki H, Ago T, Ito T, Sumimoto H, Kohda D (2001) Solution structure of the PX domain, a target of the SH3 domain. Nat Struct Biol 8 (6):526–530. doi: 10.1038/88591 [DOI] [PubMed] [Google Scholar]
- Hnia K, Vaccari I, Bolino A, Laporte J (2012) Myotubularin phosphoinositide phosphatases: cellular functions and disease pathophysiology. Trends Mol Med 18 (6):317–327. doi: 10.1016/j.molmed.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Menon AK (2014) Lipid landscapes and pipelines in membrane homeostasis. Nature 510 (7503):48–57. doi: 10.1038/nature13474 [DOI] [PubMed] [Google Scholar]
- Honigmann A, van den Bogaart G, Iraheta E, Risselada HJ, Milovanovic D, Mueller V, Mullar S, Diederichsen U, Fasshauer D, Grubmuller H, Hell SW, Eggeling C, Kuhnel K, Jahn R (2013) Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat Struct Mol Biol 20 (6):679–686. doi: 10.1038/nsmb.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital A, Goni JR, Orozco M, Gelpi JL (2015) Molecular dynamics simulations: advances and applications. Adv Appl Bioinform Chem 8:37–47. doi: 10.2147/AABC.S70333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Mao Y (2015) The structure of phosphoinositide phosphatases: Insights into substrate specificity and catalysis. Biochim Biophys Acta 1851 (6):698–710. doi: 10.1016/j.bbalip.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsuan J, Cockcroft S (2001) The PITP family of phosphatidylinositol transfer proteins. Genome Biol 2 (9):REVIEWS3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL, Feng S, Hilgemann DW (1998) Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gbetagamma. Nature 391 (6669):803–806. doi: 10.1038/35882 [DOI] [PubMed] [Google Scholar]
- Huang Y, Shah V, Liu T, Keshvara L (2005) Signaling through Disabled 1 requires phosphoinositide binding. Biochem Biophys Res Commun 331 (4):1460–1468. doi: 10.1016/j.bbrc.2005.04.064 [DOI] [PubMed] [Google Scholar]
- Hurley JH (2008) ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 20 (1):4–11. doi: 10.1016/j.ceb.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman J, Chen H, Di Fiore PP, De Camilli P, Brunger AT (2000) Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF). J Cell Biol 149 (3):537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvonen M, Macias MJ, Nilges M, Oschkinat H, Saraste M, Wilmanns M (1995) Structure of the binding site for inositol phosphates in a PH domain. EMBO J 14 (19):4676–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN (2004) Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem 279 (16):16606–16613. doi: 10.1074/jbc.M312098200 [DOI] [PubMed] [Google Scholar]
- Ile KE, Schaaf G, Bankaitis VA (2006) Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat Chem Biol 2 (11):576–583. doi: 10.1038/nchembio835 [DOI] [PubMed] [Google Scholar]
- Im YJ, Hurley JH (2008) Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell 14 (6):902–913. doi: 10.1016/j.devcel.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Raychaudhuri S, Prinz WA, Hurley JH (2005) Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437 (7055):154–158. doi: 10.1038/nature03923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR (2007) Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450 (7168):365–369. doi: 10.1038/nature06336 [DOI] [PubMed] [Google Scholar]
- Inoue H, Baba T, Sato S, Ohtsuki R, Takemori A, Watanabe T, Tagaya M, Tani K (2012) Roles of SAM and DDHD domains in mammalian intracellular phospholipase A1 KIAA0725p. Biochim Biophys Acta 1823 (4):930–939. doi: 10.1016/j.bbamcr.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Irvine RF (2003) Nuclear lipid signalling. Nat Rev Mol Cell Biol 4 (5):349–360. doi: 10.1038/nrm1100 [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY (1998) Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J 17 (18):5374–5387. doi: 10.1093/emboj/17.18.5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, De Camilli P (2006) BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta 1761 (8):897–912. doi: 10.1016/j.bbalip.2006.06.015 [DOI] [PubMed] [Google Scholar]
- Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T (2001) Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291 (5506):1047–1051. doi: 10.1126/science.291.5506.1047 [DOI] [PubMed] [Google Scholar]
- Ivarsson Y (2012) Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett 586 (17):2638–2647. doi: 10.1016/j.febslet.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarsson Y, Wawrzyniak AM, Kashyap R, Polanowska J, Betzi S, Lembo F, Vermeiren E, Chiheb D, Lenfant N, Morelli X, Borg JP, Reboul J, Zimmermann P (2013) Prevalence, specificity and determinants of lipid-interacting PDZ domains from an in-cell screen and in vitro binding experiments. PLoS One 8 (2):e54581. doi: 10.1371/journal.pone.0054581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarsson Y, Wawrzyniak AM, Wuytens G, Kosloff M, Vermeiren E, Raport M, Zimmermann P (2011) Cooperative phosphoinositide and peptide binding by PSD-95/discs large/ZO-1 (PDZ) domain of polychaetoid, Drosophila zonulin. J Biol Chem 286 (52):44669–44678. doi: 10.1074/jbc.M111.285734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Walch L, Verbavatz JM (2016) Lipids and Their Trafficking: An Integral Part of Cellular Organization. Dev Cell 39 (2):139–153. doi: 10.1016/j.devcel.2016.09.030 [DOI] [PubMed] [Google Scholar]
- Jain A, Holthuis JCM (2017) Membrane contact sites, ancient and central hubs of cellular lipid logistics. Biochim Biophys Acta 1864 (9):1450–1458. doi: 10.1016/j.bbamcr.2017.05.017 [DOI] [PubMed] [Google Scholar]
- Jank T, Bohmer KE, Tzivelekidis T, Schwan C, Belyi Y, Aktories K (2012) Domain organization of Legionella effector SetA. Cell Microbiol 14 (6):852–868. doi: 10.1111/j.1462-5822.2012.01761.x [DOI] [PubMed] [Google Scholar]
- Jarsch IK, Daste F, Gallop JL (2016) Membrane curvature in cell biology: An integration of molecular mechanisms. J Cell Biol 214 (4):375–387. doi: 10.1083/jcb.201604003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski CJ, Moreira E, Li A, Lee R, Rodriguez IR (2001) A family of 12 human genes containing oxysterol-binding domains. Genomics 78 (3):185–196. doi: 10.1006/geno.2001.6663 [DOI] [PubMed] [Google Scholar]
- Jayasundar JJ, Ju JH, He L, Liu D, Meilleur F, Zhao J, Callaway DJ, Bu Z (2012) Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J Biol Chem 287 (44):37119–37133. doi: 10.1074/jbc.M112.380972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Tang WK, Zhai P, Roy NS, Luo R, Gruschus JM, Yohe ME, Chen PW, Li Y, Byrd RA, Xia D, Randazzo PA (2015) Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1. Structure 23 (11):1977–1988. doi: 10.1016/j.str.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Bocher V, Lehto M, Chinetti G, Kuismanen E, Ehnholm C, Staels B, Olkkonen VM (2003) The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol Biol Cell 14 (3):903–915. doi: 10.1091/mbc.E02-08-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RE, Wales TE, Fulton DB, Engen JR, Andreotti AH (2017) Achieving a Graded Immune Response: BTK Adopts a Range of Active/Inactive Conformations Dictated by Multiple Interdomain Contacts. Structure 25 (10):1481–1494 e1484. doi: 10.1016/j.str.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Sylvestersen KB, Choudhary C, Nguyen S, Mann M, Nielsen ML (2014) Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry. Cell Rep 6 (3):578–591. doi: 10.1016/j.celrep.2013.12.038 [DOI] [PubMed] [Google Scholar]
- Kam JL, Miura K, Jackson TR, Gruschus J, Roller P, Stauffer S, Clark J, Aneja R, Randazzo PA (2000) Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1. Evidence for the pleckstrin homology domain functioning as an allosteric site. J Biol Chem 275 (13):9653–9663 [DOI] [PubMed] [Google Scholar]
- Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB (2001) The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol 3 (7):675–678. doi: 10.1038/35083070 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Joshi R, Feller SM, Li SS (2012) Phosphotyrosine recognition domains: the typical, the atypical and the versatile. Cell Commun Signal 10 (1):32. doi: 10.1186/1478-811X-10-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL (2002) Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J 21 (19):5057–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA (1998) Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem 273 (46):30497–30508 [DOI] [PubMed] [Google Scholar]
- Kay BK, Yamabhai M, Wendland B, Emr SD (1999) Identification of a novel domain shared by putative components of the endocytic and cytoskeletal machinery. Protein Sci 8 (2):435–438. doi: 10.1110/ps.8.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentala H, Weber-Boyvat M, Olkkonen VM (2016) OSBP-Related Protein Family: Mediators of Lipid Transport and Signaling at Membrane Contact Sites. Int Rev Cell Mol Biol 321:299–340. doi: 10.1016/bs.ircmb.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Ketel K, Krauss M, Nicot AS, Puchkov D, Wieffer M, Muller R, Subramanian D, Schultz C, Laporte J, Haucke V (2016) A phosphoinositide conversion mechanism for exit from endosomes. Nature 529 (7586):408–412. doi: 10.1038/nature16516 [DOI] [PubMed] [Google Scholar]
- Killian JA, von Heijne G (2000) How proteins adapt to a membrane-water interface. Trends Biochem Sci 25 (9):429–434 [DOI] [PubMed] [Google Scholar]
- Kim S, Kedan A, Marom M, Gavert N, Keinan O, Selitrennik M, Laufman O, Lev S (2013a) The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep 14 (10):891–899. doi: 10.1038/embor.2013.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Balla T (2011) A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev Cell 21 (5):813–824. doi: 10.1016/j.devcel.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T (2015) Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev Cell 33 (5):549–561. doi: 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Wisniewski E, Echeverria N, Balla T (2016) Phosphatidylinositol and phosphatidic acid transport between the ER and plasma membrane during PLC activation requires the Nir2 protein. Biochem Soc Trans 44 (1):197–201. doi: 10.1042/BST20150187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Hernandez ML, Balla T (2013b) Inositol lipid regulation of lipid transfer in specialized membrane domains. Trends Cell Biol 23 (6):270–278. doi: 10.1016/j.tcb.2013.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, Woolf EA, Tayber O, Brody T, Shu P, Hawkins F, Kennedy B, Baldini L, Ebeling C, Alperin GD, Deeds J, Lakey ND, Culpepper J, Chen H, Glucksmann-Kuis MA, Carlson GA, Duyk GM, Moore KJ (1996) Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell 85 (2):281–290 [DOI] [PubMed] [Google Scholar]
- Knight JD, Falke JJ (2009) Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys J 96 (2):566–582. doi: 10.1016/j.bpj.2008.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout SC, Corbalan-Garcia S, Gomez-Fernandez JC, Falke JJ (2003) C2 domain of protein kinase C alpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry 42 (5):1254–1265. doi: 10.1021/bi026596f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann L, Vahidi S, Sowole MA (2014) Mass spectrometry methods for studying structure and dynamics of biological macromolecules. Anal Chem 86 (1):213–232. doi: 10.1021/ac4039306 [DOI] [PubMed] [Google Scholar]
- Kontos CD, Stauffer TP, Yang WP, York JD, Huang L, Blanar MA, Meyer T, Peters KG (1998) Tyrosine 1101 of Tie2 is the major site of association of p85 and is required for activation of phosphatidylinositol 3-kinase and Akt. Mol Cell Biol 18 (7):4131–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kuhnel K (2012) Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc Natl Acad Sci U S A 109 (30):E2042–2049. doi: 10.1073/pnas.1205128109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Tolstrup J, Appelles A, Henke S, Thumm M (2006) The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett 580 (19):4632–4638. doi: 10.1016/j.febslet.2006.07.041 [DOI] [PubMed] [Google Scholar]
- Kular GS, Chaudhary A, Prestwich G, Swigart P, Wetzker R, Cockcroft S (2002) Co-operation of phosphatidylinositol transfer protein with phosphoinositide 3-kinase gamma in vitro. Adv Enzyme Regul 42:53–61 [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Das S, Funk CD, Murray D, Cho W (2002) Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J Biol Chem 277 (15):13167–13174. doi: 10.1074/jbc.M112393200 [DOI] [PubMed] [Google Scholar]
- Kutateladze T, Overduin M (2001) Structural mechanism of endosome docking by the FYVE domain. Science 291 (5509):1793–1796. doi: 10.1126/science.291.5509.1793 [DOI] [PubMed] [Google Scholar]
- Kutateladze TG (2006) Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim Biophys Acta 1761 (8):868–877. doi: 10.1016/j.bbalip.2006.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG (2010) Translation of the phosphoinositide code by PI effectors. Nat Chem Biol 6 (7):507–513. doi: 10.1038/nchembio.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG, Capelluto DG, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M (2004) Multivalent mechanism of membrane insertion by the FYVE domain. J Biol Chem 279 (4):3050–3057. doi: 10.1074/jbc.M309007200 [DOI] [PubMed] [Google Scholar]
- Kutateladze TG, Ogburn KD, Watson WT, de Beer T, Emr SD, Burd CG, Overduin M (1999) Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol Cell 3 (6):805–811 [DOI] [PubMed] [Google Scholar]
- Kweon DH, Shin YK, Shin JY, Lee JH, Lee JB, Seo JH, Kim YS (2006) Membrane topology of helix 0 of the Epsin N-terminal homology domain. Mol Cells 21 (3):428–435 [PubMed] [Google Scholar]
- Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV (2014) Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510 (7503):172–175. doi: 10.1038/nature13419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CL, Jao CC, Lyman E, Gallop JL, Peter BJ, McMahon HT, Langen R, Voth GA (2012) Membrane binding and self-association of the epsin N-terminal homology domain. J Mol Biol 423 (5):800–817. doi: 10.1016/j.jmb.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CL, Landgraf KE, Voth GA, Falke JJ (2010) Membrane docking geometry and target lipid stoichiometry of membrane-bound PKCalpha C2 domain: a combined molecular dynamics and experimental study. J Mol Biol 402 (2):301–310. doi: 10.1016/j.jmb.2010.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larijani B, Allen-Baume V, Morgan CP, Li M, Cockcroft S (2003) EGF regulation of PITP dynamics is blocked by inhibitors of phospholipase C and of the Ras-MAP kinase pathway. Curr Biol 13 (1):78–84 [DOI] [PubMed] [Google Scholar]
- Lawe DC, Patki V, Heller-Harrison R, Lambright D, Corvera S (2000) The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J Biol Chem 275 (5):3699–3705 [DOI] [PubMed] [Google Scholar]
- Lee AG (2003) Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta 1612 (1):1–40 [DOI] [PubMed] [Google Scholar]
- Lee SA, Eyeson R, Cheever ML, Geng J, Verkhusha VV, Burd C, Overduin M, Kutateladze TG (2005) Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc Natl Acad Sci U S A 102 (37):13052–13057. doi: 10.1073/pnas.0503900102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS (2004) ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci 117 (Pt 1):9–18. doi: 10.1242/jcs.00928 [DOI] [PubMed] [Google Scholar]
- Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, Ikonen E, Olkkonen VM (2001) The OSBP-related protein family in humans. J Lipid Res 42 (8):1203–1213 [PubMed] [Google Scholar]
- Lemmon MA (2003) Phosphoinositide recognition domains. Traffic 4 (4):201–213 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2004) Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans 32 (Pt 5):707–711. doi: 10.1042/BST0320707 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2007) Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp (74):81–93. doi: 10.1042/BSS0740081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA (2008) Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9 (2):99–111. doi: 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM (2001) Molecular determinants in pleckstrin homology domains that allow specific recognition of phosphoinositides. Biochem Soc Trans 29 (Pt 4):377–384 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS (2002) Pleckstrin homology domains and the cytoskeleton. FEBS Lett 513 (1):71–76 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, O’Brien R, Sigler PB, Schlessinger J (1995) Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A 92 (23):10472–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir M, Kufareva I, Abagyan R, Overduin M (2015) Membrane and Protein Interactions of the Pleckstrin Homology Domain Superfamily. Membranes (Basel) 5 (4):646–663. doi: 10.3390/membranes5040646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S (2010) Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol 11 (10):739–750. doi: 10.1038/nrm2971 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (1998) The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol 8 (13):729–739 [DOI] [PubMed] [Google Scholar]
- Li H, Marshall AJ (2015) Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling. Cell Signal 27 (9):1789–1798. doi: 10.1016/j.cellsig.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Li H, Tremblay JM, Yarbrough LR, Helmkamp GM Jr. (2002) Both isoforms of mammalian phosphatidylinositol transfer protein are capable of binding and transporting sphingomyelin. Biochim Biophys Acta 1580 (1):67–76 [DOI] [PubMed] [Google Scholar]
- Li J, Mao X, Dong LQ, Liu F, Tong L (2007) Crystal structures of the BAR-PH and PTB domains of human APPL1. Structure 15 (5):525–533. doi: 10.1016/j.str.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Li X, Routt SM, Xie Z, Cui X, Fang M, Kearns MA, Bard M, Kirsch DR, Bankaitis VA (2000) Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol Biol Cell 11 (6):1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Tamm LK (2016) NMR as a tool to investigate the structure, dynamics and function of membrane proteins. Nat Struct Mol Biol 23 (6):468–474. doi: 10.1038/nsmb.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG (2000) Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell 6 (2):385–394 [DOI] [PubMed] [Google Scholar]
- Lindahl E, Sansom MS (2008) Membrane proteins: molecular dynamics simulations. Curr Opin Struct Biol 18 (4):425–431. doi: 10.1016/j.sbi.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Liu X, Ridgway ND (2014) Characterization of the sterol and phosphatidylinositol 4-phosphate binding properties of Golgi-associated OSBP-related protein 9 (ORP9). PLoS One 9 (9):e108368. doi: 10.1371/journal.pone.0108368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kahn RA, Prestegard JH (2014) Interaction of Fapp1 with Arf1 and PI4P at a membrane surface: an example of coincidence detection. Structure 22 (3):421–430. doi: 10.1016/j.str.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WT, Vujicic Zagar A, Gerth F, Lehmann M, Puchkov D, Krylova O, Freund C, Scapozza L, Vadas O, Haucke V (2017) A Coincidence Detection Mechanism Controls PX-BAR Domain-Mediated Endocytic Membrane Remodeling via an Allosteric Structural Switch. Dev Cell 43 (4):522–529 e524. doi: 10.1016/j.devcel.2017.10.019 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Levine TP (2005) A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J Biol Chem 280 (14):14097–14104. doi: 10.1074/jbc.M500147200 [DOI] [PubMed] [Google Scholar]
- Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI (2007) The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct Biol 7:44. doi: 10.1186/1472-6807-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Urbe S, Clague MJ (2005) Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J Cell Sci 118 (Pt 9):2005–2012. doi: 10.1242/jcs.02325 [DOI] [PubMed] [Google Scholar]
- Lumb CN, He J, Xue Y, Stansfeld PJ, Stahelin RV, Kutateladze TG, Sansom MS (2011) Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain. Structure 19 (9):1338–1346. doi: 10.1016/j.str.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb CN, Sansom MS (2012) Finding a needle in a haystack: the role of electrostatics in target lipid recognition by PH domains. PLoS Comput Biol 8 (7):e1002617. doi: 10.1371/journal.pcbi.1002617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Wasilko DJ, Liu Y, Sun J, Wu X, Luo ZQ, Mao Y (2015) Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome. PLoS Pathog 11 (6):e1004965. doi: 10.1371/journal.ppat.1004965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR (2006) Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11 (1):47–56. doi: 10.1016/j.devcel.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Macias MJ, Musacchio A, Ponstingl H, Nilges M, Saraste M, Oschkinat H (1994) Structure of the pleckstrin homology domain from beta-spectrin. Nature 369 (6482):675–677. doi: 10.1038/369675a0 [DOI] [PubMed] [Google Scholar]
- Maffucci T, Falasca M (2001) Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett 506 (3):173–179 [DOI] [PubMed] [Google Scholar]
- Malaby AW, van den Berg B, Lambright DG (2013) Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors. Proc Natl Acad Sci U S A 110 (35):14213–14218. doi: 10.1073/pnas.1301883110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek M, Kielkowska A, Chessa T, Anderson KE, Barneda D, Pir P, Nakanishi H, Eguchi S, Koizumi A, Sasaki J, Juvin V, Kiselev VY, Niewczas I, Gray A, Valayer A, Spensberger D, Imbert M, Felisbino S, Habuchi T, Beinke S, Cosulich S, Le Novere N, Sasaki T, Clark J, Hawkins PT, Stephens LR (2017) PTEN Regulates PI(3,4)P2 Signaling Downstream of Class I PI3K. Mol Cell 68 (3):566–580 e510. doi: 10.1016/j.molcel.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova S, Stahelin RV, Pingali SV, Cho W, Schlossman ML (2006) Orientation and penetration depth of monolayer-bound p40phox-PX. Biochemistry 45 (45):13566–13575. doi: 10.1021/bi061133l [DOI] [PubMed] [Google Scholar]
- Manna D, Albanese A, Park WS, Cho W (2007) Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J Biol Chem 282 (44):32093–32105. doi: 10.1074/jbc.M703517200 [DOI] [PubMed] [Google Scholar]
- Manna D, Bhardwaj N, Vora MS, Stahelin RV, Lu H, Cho W (2008) Differential roles of phosphatidylserine, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in plasma membrane targeting of C2 domains. Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKCalpha C2 domain. J Biol Chem 283 (38):26047–26058. doi: 10.1074/jbc.M802617200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Chen J, Maynard JA, Zhang B, Quiocho FA (2001) A novel all helix fold of the AP180 amino-terminal domain for phosphoinositide binding and clathrin assembly in synaptic vesicle endocytosis. Cell 104 (3):433–440 [DOI] [PubMed] [Google Scholar]
- Mao Y, Nickitenko A, Duan X, Lloyd TE, Wu MN, Bellen H, Quiocho FA (2000) Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell 100 (4):447–456 [DOI] [PubMed] [Google Scholar]
- Marat AL, Haucke V (2016) Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic. EMBO J 35 (6):561–579. doi: 10.15252/embj.201593564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat AL, Wallroth A, Lo WT, Muller R, Norata GD, Falasca M, Schultz C, Haucke V (2017) mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 356 (6341):968–972. doi: 10.1126/science.aaf8310 [DOI] [PubMed] [Google Scholar]
- Marsh D (2008) Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta 1778 (7–8):1545–1575. doi: 10.1016/j.bbamem.2008.01.015 [DOI] [PubMed] [Google Scholar]
- Martelli AM, Ognibene A, Buontempo F, Fini M, Bressanin D, Goto K, McCubrey JA, Cocco L, Evangelisti C (2011) Nuclear phosphoinositides and their roles in cell biology and disease. Crit Rev Biochem Mol Biol 46 (5):436–457. doi: 10.3109/10409238.2011.609530 [DOI] [PubMed] [Google Scholar]
- Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N (2006) Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J 25 (12):2889–2897. doi: 10.1038/sj.emboj.7601176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Pykäläinen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P (2007) Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain–like mechanism. J Cell Biol 176 (7): 953–964. doi: 10.1083/jcb.200609176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ, Ren R, Clark KL, Baltimore D (1993) A putative modular domain present in diverse signaling proteins. Cell 73 (4):629–630 [DOI] [PubMed] [Google Scholar]
- McCrea HJ, De Camilli P (2009) Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 24:8–16. doi: 10.1152/physiol.00035.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Murray D (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438 (7068):605–611. doi: 10.1038/nature04398 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438 (7068):590–596. doi: 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B (2013) A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155 (4):830–843. doi: 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Michell RH, Heath VL, Lemmon MA, Dove SK (2006) Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci 31 (1):52–63. doi: 10.1016/j.tibs.2005.11.013 [DOI] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Futterer K (2005) Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J 24 (2):240–250. doi: 10.1038/sj.emboj.7600535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Mathiasen S, Bright NA, Pierre F, Kelly BT, Kladt N, Schauss A, Merrifield CJ, Stamou D, Honing S, Owen DJ (2015) CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev Cell 33 (2):163–175. doi: 10.1016/j.devcel.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Honing S, Robinson MS, Peden AA, Owen DJ (2011) The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell 147 (5):1118–1131. doi: 10.1016/j.cell.2011.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM (2012) Structural basis of membrane bending by the N-BAR protein endophilin. Cell 149 (1):137–145. doi: 10.1016/j.cell.2012.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Unger VM (2012) Membrane curvature and its generation by BAR proteins. Trends Biochem Sci 37 (12):526–533. doi: 10.1016/j.tibs.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa H, Ohtsubo M, Copeland NG, Gilbert DJ, Jenkins NA, Yoshimura A (1998) Cloning and characterization of a novel class II phosphoinositide 3-kinase containing C2 domain. Biochem Biophys Res Commun 244 (2):531–539. doi: 10.1006/bbrc.1998.8294 [DOI] [PubMed] [Google Scholar]
- Misra S, Hurley JH (1999) Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell 97 (5):657–666 [DOI] [PubMed] [Google Scholar]
- Moleirinho S, Tilston-Lunel A, Angus L, Gunn-Moore F, Reynolds PA (2013) The expanding family of FERM proteins. Biochem J 452 (2):183–193. doi: 10.1042/BJ20121642 [DOI] [PubMed] [Google Scholar]
- Monje-Galvan V, Klauda JB (2016) Peripheral membrane proteins: Tying the knot between experiment and computation. Biochim Biophys Acta 1858 (7 Pt B):1584–1593. doi: 10.1016/j.bbamem.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Montaville P, Coudevylle N, Radhakrishnan A, Leonov A, Zweckstetter M, Becker S (2008) The PIP2 binding mode of the C2 domains of rabphilin-3A. Protein Sci 17 (6):1025–1034. doi: 10.1110/ps.073326608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales KA, Yang Y, Cole TR, Igumenova TI (2016) Dynamic Response of the C2 Domain of Protein Kinase Calpha to Ca(2+) Binding. Biophys J 111 (8):1655–1667. doi: 10.1016/j.bpj.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Alvarado D, Schmitz KR, Kenniston JA, Mendrola JM, Ferguson KM, Lemmon MA (2015) Comparison of Saccharomyces cerevisiae F-BAR domain structures reveals a conserved inositol phosphate binding site. Structure 23 (2):352–363. doi: 10.1016/j.str.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA (2010) Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell 143 (6):966–977. doi: 10.1016/j.cell.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA (2012) Conditional peripheral membrane proteins: facing up to limited specificity. Structure 20 (1):15–27. doi: 10.1016/j.str.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier E, Wuytens G, Leenaerts I, Hannes F, Heung MY, Degeest G, David G, Zimmermann P (2005) Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J 24 (14):2556–2565. doi: 10.1038/sj.emboj.7600722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser von Filseck J, Copic A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G (2015a) INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science 349 (6246):432–436. doi: 10.1126/science.aab1346 [DOI] [PubMed] [Google Scholar]
- Moser von Filseck J, Vanni S, Mesmin B, Antonny B, Drin G (2015b) A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun 6:6671. doi: 10.1038/ncomms7671 [DOI] [PubMed] [Google Scholar]
- Mu Y, Cai P, Hu S, Ma S, Gao Y (2014) Characterization of diverse internal binding specificities of PDZ domains by yeast two-hybrid screening of a special peptide library. PLoS One 9 (2):e88286. doi: 10.1371/journal.pone.0088286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muallem S, Chung WY, Jha A, Ahuja M (2017) Lipids at membrane contact sites: cell signaling and ion transport. EMBO Rep 18 (11):1893–1904. doi: 10.15252/embr.201744331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Jackson PK (2011) The tubby family proteins. Genome Biol 12 (6):225. doi: 10.1186/gb-2011-12-6-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulgrew-Nesbitt A, Diraviyam K, Wang J, Singh S, Murray P, Li Z, Rogers L, Mirkovic N, Murray D (2006) The role of electrostatics in protein-membrane interactions. Biochim Biophys Acta 1761 (8):812–826. doi: 10.1016/j.bbalip.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Murphy SE, Levine TP (2016) VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta 1861 (8 Pt B):952–961. doi: 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Murray D, Honig B (2002) Electrostatic control of the membrane targeting of C2 domains. Mol Cell 9 (1):145–154 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Gibson T, Rice P, Thompson J, Saraste M (1993) The PH domain: a common piece in the structural patchwork of signalling proteins. Trends Biochem Sci 18 (9):343–348 [DOI] [PubMed] [Google Scholar]
- Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR, De Camilli P (2012) PtdIns4P synthesis by PI4KIIIalpha at the plasma membrane and its impact on plasma membrane identity. J Cell Biol 199 (6):1003–1016. doi: 10.1083/jcb.201206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, McDonagh T, Somers W, Seehra J, Falke JJ, Clark JD (1998) Independent folding and ligand specificity of the C2 calcium-dependent lipid binding domain of cytosolic phospholipase A2. J Biol Chem 273 (3):1365–1372 [DOI] [PubMed] [Google Scholar]
- Naughton FB, Kalli AC, Sansom MS (2016) Association of Peripheral Membrane Proteins with Membranes: Free Energy of Binding of GRP1 PH Domain with Phosphatidylinositol Phosphate-Containing Model Bilayers. J Phys Chem Lett 7 (7):1219–1224. doi: 10.1021/acs.jpclett.6b00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrotek A, Zeghouf M, Cherfils J (2016) Allosteric regulation of Arf GTPases and their GEFs at the membrane interface. Small GTPases 7 (4):283–296. doi: 10.1080/21541248.2016.1215778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Naggert JK, North MA, Nishina PM (1996) A candidate gene for the mouse mutation tubby. Nature 380 (6574):534–538. doi: 10.1038/380534a0 [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP (2003) PDZ domain proteins: plug and playα Sci STKE 2003 (179):RE7. doi: 10.1126/stke.2003.179.re7 [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y (2008) The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem 283 (35):23972–23980. doi: 10.1074/jbc.M803180200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocaka L, Spalluto C, Wilson DI, Hunt DM, Halford S (2005) Chromosomal localization, genomic organization and evolution of the genes encoding human phosphatidylinositol transfer protein membrane-associated (PITPNM) 1, 2 and 3. Cytogenet Genome Res 108 (4):293–302. doi: 10.1159/000081519 [DOI] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y (1989) Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci U S A 86 (13):4868–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms LM, Binge LC, Davies EM, Rahman P, Conway JR, Gurung R, Ferguson DT, Papa A, Fedele CG, Vieusseux JL, Chai RC, Koentgen F, Price JT, Tiganis T, Timpson P, McLean CA, Mitchell CA (2015) The Inositol Polyphosphate 5-Phosphatase PIPP Regulates AKT1-Dependent Breast Cancer Growth and Metastasis. Cancer Cell 28 (2):155–169. doi: 10.1016/j.ccell.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Osada S, Mizuno K, Saido TC, Akita Y, Suzuki K, Kuroki T, Ohno S (1990) A phorbol ester receptor/protein kinase, nPKC eta, a new member of the protein kinase C family predominantly expressed in lung and skin. J Biol Chem 265 (36):22434–22440 [PubMed] [Google Scholar]
- Pang X, Fan J, Zhang Y, Zhang K, Gao B, Ma J, Li J, Deng Y, Zhou Q, Egelman EH, Hsu VW, Sun F (2014) A PH domain in ACAP1 possesses key features of the BAR domain in promoting membrane curvature. Dev Cell 31 (1):73–86. doi: 10.1016/j.devcel.2014.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WS, Heo WD, Whalen JH, O’Rourke NA, Bryan HM, Meyer T, Teruel MN (2008) Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell 30 (3):381–392. doi: 10.1016/j.molcel.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson GN, Vines D, Driscoll PC, Djordjevic S (2008) Crystal structures of PI3K-C2alpha PX domain indicate conformational change associated with ligand binding. BMC Struct Biol 8:13. doi: 10.1186/1472-6807-8-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasenkiewicz-Gierula M, Baczynski K, Markiewicz M, Murzyn K (2016) Computer modelling studies of the bilayer/water interface. Biochim Biophys Acta 1858 (10):2305–2321. doi: 10.1016/j.bbamem.2016.01.024 [DOI] [PubMed] [Google Scholar]
- Payrastre B, Gaits-Iacovoni F, Sansonetti P, Tronchere H (2012) Phosphoinositides and cellular pathogens. Subcell Biochem 59:363–388. doi: 10.1007/978-94-007-3015-1_12 [DOI] [PubMed] [Google Scholar]
- Pearson MA, Reczek D, Bretscher A, Karplus PA (2000) Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 101 (3):259–270 [DOI] [PubMed] [Google Scholar]
- Pendaries C, Tronchere H, Plantavid M, Payrastre B (2003) Phosphoinositide signaling disorders in human diseases. FEBS Lett 546 (1):25–31 [DOI] [PubMed] [Google Scholar]
- Perisic O, Fong S, Lynch DE, Bycroft M, Williams RL (1998) Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A2. J Biol Chem 273 (3):1596–1604 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303 (5657):495–499. doi: 10.1126/science.1092586 [DOI] [PubMed] [Google Scholar]
- Phillips SE, Ile KE, Boukhelifa M, Huijbregts RP, Bankaitis VA (2006) Specific and nonspecific membrane-binding determinants cooperate in targeting phosphatidylinositol transfer protein beta-isoform to the mammalian trans-Golgi network. Mol Biol Cell 17 (6):2498–2512. doi: 10.1091/mbc.E06-01-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Kuhbacher A, Cossart P (2015) Phosphoinositides and host-pathogen interactions. Biochim Biophys Acta 1851 (6):911–918. doi: 10.1016/j.bbalip.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Pogozheva ID, Tristram-Nagle S, Mosberg HI, Lomize AL (2013) Structural adaptations of proteins to different biological membranes. Biochim Biophys Acta 1828 (11):2592–2608. doi: 10.1016/j.bbamem.2013.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP (1996) Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci 5 (11):2353–2357. doi: 10.1002/pro.5560051122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y, Eichhorn-Gruenig M, Puchkov D, Schoneberg J, Ullrich A, Lampe A, Muller R, Zarbakhsh S, Gulluni F, Hirsch E, Krauss M, Schultz C, Schmoranzer J, Noe F, Haucke V (2013) Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 499 (7457):233–237. doi: 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Lee DJ, Lim WA (1999) Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell 97 (4):471–480 [DOI] [PubMed] [Google Scholar]
- Premkumar L, Bobkov AA, Patel M, Jaroszewski L, Bankston LA, Stec B, Vuori K, Cote JF, Liddington RC (2010) Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs). J Biol Chem 285 (17):13211–13222. doi: 10.1074/jbc.M110.102517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA (2010) Lipid trafficking sans vesicles: where, why, how? Cell 143 (6):870–874. doi: 10.1016/j.cell.2010.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psachoulia E, Sansom MS (2008) Interactions of the pleckstrin homology domain with phosphatidylinositol phosphate and membranes: characterization via molecular dynamics simulations. Biochemistry 47 (14):4211–4220. doi: 10.1021/bi702319k [DOI] [PubMed] [Google Scholar]
- Pykalainen A, Boczkowska M, Zhao H, Saarikangas J, Rebowski G, Jansen M, Hakanen J, Koskela EV, Peranen J, Vihinen H, Jokitalo E, Salminen M, Ikonen E, Dominguez R, Lappalainen P (2011) Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol 18 (8):902–907. doi: 10.1038/nsmb.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A (2007) The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J 26 (22):4788–4800. doi: 10.1038/sj.emboj.7601889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM (2011) Let’s go bananas: revisiting the endocytic BAR code. EMBO J 30 (17):3501–3515. doi: 10.1038/emboj.2011.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H (2008) The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol 10 (12):2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x [DOI] [PubMed] [Google Scholar]
- Rameh LE, Arvidsson A, Carraway KL 3rd, Couvillon AD, Rathbun G, Crompton A, VanRenterghem B, Czech MP, Ravichandran KS, Burakoff SJ, Wang DS, Chen CS, Cantley LC (1997) A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem 272 (35):22059–22066 [DOI] [PubMed] [Google Scholar]
- Ramel D, Lagarrigue F, Pons V, Mounier J, Dupuis-Coronas S, Chicanne G, Sansonetti PJ, Gaits-Iacovoni F, Tronchere H, Payrastre B (2011) Shigella flexneri infection generates the lipid PI5P to alter endocytosis and prevent termination of EGFR signaling. Sci Signal 4 (191):ra61. doi: 10.1126/scisignal.2001619 [DOI] [PubMed] [Google Scholar]
- Ravichandran KS, Zhou MM, Pratt JC, Harlan JE, Walk SF, Fesik SW, Burakoff SJ (1997) Evidence for a requirement for both phospholipid and phosphotyrosine binding via the Shc phosphotyrosine-binding domain in vivo. Mol Cell Biol 17 (9):5540–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Im YJ, Hurley JH, Prinz WA (2006) Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol 173 (1):107–119. doi: 10.1083/jcb.200510084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern RE, Redfern D, Furgason ML, Munson M, Ross AH, Gericke A (2008) PTEN phosphatase selectively binds phosphoinositides and undergoes structural changes. Biochemistry 47 (7):2162–2171. doi: 10.1021/bi702114w [DOI] [PubMed] [Google Scholar]
- Renard HF, Simunovic M, Lemiere J, Boucrot E, Garcia-Castillo MD, Arumugam S, Chambon V, Lamaze C, Wunder C, Kenworthy AK, Schmidt AA, McMahon HT, Sykes C, Bassereau P, Johannes L (2015) Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 517 (7535):493–496. doi: 10.1038/nature14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue K (2009) The lipin family: mutations and metabolism. Curr Opin Lipidol 20 (3):165–170. doi: 10.1097/MOL.0b013e32832adee5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC (1998) C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem 273 (26):15879–15882 [DOI] [PubMed] [Google Scholar]
- Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE (2002) Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol 9 (7):507–511. doi: 10.1038/nsb812 [DOI] [PubMed] [Google Scholar]
- Rogaski B, Klauda JB (2012) Membrane-binding mechanism of a peripheral membrane protein through microsecond molecular dynamics simulations. J Mol Biol 423 (5):847–861. doi: 10.1016/j.jmb.2012.08.015 [DOI] [PubMed] [Google Scholar]
- Rohacs T, Lopes CM, Jin T, Ramdya PP, Molnar Z, Logothetis DE (2003) Specificity of activation by phosphoinositides determines lipid regulation of Kir channels. Proc Natl Acad Sci U S A 100 (2):745–750. doi: 10.1073/pnas.0236364100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski MJ, Soccio RE, Breslow JL, Burley SK (2002) Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc Natl Acad Sci U S A 99 (10):6949–6954. doi: 10.1073/pnas.052140699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy NS, Yohe ME, Randazzo PA, Gruschus JM (2016) Allosteric properties of PH domains in Arf regulatory proteins. Cell Logist 6 (2):e1181700. doi: 10.1080/21592799.2016.1181700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MM, Temple BR, Phillips SE, Bankaitis VA (2007) Conformational dynamics of the major yeast phosphatidylinositol transfer protein sec14p: insight into the mechanisms of phospholipid exchange and diseases of sec14p-like protein deficiencies. Mol Biol Cell 18 (5):1928–1942. doi: 10.1091/mbc.E06-11-1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Lappalainen P (2010) Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev 90 (1):259–289. doi: 10.1152/physrev.00036.2009 [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P (2009) Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol 19 (2):95–107. doi: 10.1016/j.cub.2008.12.029 [DOI] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD (2007) ESCRTing proteins in the endocytic pathway. Trends Biochem Sci 32 (12):561–573. doi: 10.1016/j.tibs.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Salama SR, Cleves AE, Malehorn DE, Whitters EA, Bankaitis VA (1990) Cloning and characterization of Kluyveromyces lactis SEC14, a gene whose product stimulates Golgi secretory function in Saccharomyces cerevisiae. J Bacteriol 172 (8):4510–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D, Guo Y, Kinch LN, Grishin NV, Gardner KH, Orth K (2013) Effectors of animal and plant pathogens use a common domain to bind host phosphoinositides. Nat Commun 4:2973. doi: 10.1038/ncomms3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer U, Kostan J, Djinovic-Carugo K (2017) Deciphering the BAR code of membrane modulators. Cell Mol Life Sci 74 (13):2413–2438. doi: 10.1007/s00018-017-2478-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Bautista S, Marin-Vicente C, Gomez-Fernandez JC, Corbalan-Garcia S (2006) The C2 domain of PKCalpha is a Ca2+ -dependent PtdIns(4,5)P2 sensing domain: a new insight into an old pathway. J Mol Biol 362 (5):901–914. doi: 10.1016/j.jmb.2006.07.093 [DOI] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L (2001) G-protein signaling through tubby proteins. Science 292 (5524):2041–2050. doi: 10.1126/science.1061233 [DOI] [PubMed] [Google Scholar]
- Saras J, Heldin CH (1996) PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci 21 (12):455–458 [DOI] [PubMed] [Google Scholar]
- Sarkes D, Rameh LE (2010) A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J 428 (3):375–384. doi: 10.1042/BJ20100129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Takasuga S, Sasaki J, Kofuji S, Eguchi S, Yamazaki M, Suzuki A (2009) Mammalian phosphoinositide kinases and phosphatases. Prog Lipid Res 48 (6):307–343. doi: 10.1016/j.plipres.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Sato TK, Overduin M, Emr SD (2001) Location, location, location: membrane targeting directed by PX domains. Science 294 (5548):1881–1885. doi: 10.1126/science.1065763 [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Filios C, Delvecchio K, Shisheva A (2012) Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am J Physiol Cell Physiol 303 (4):C436–446. doi: 10.1152/ajpcell.00105.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacioc A, Schmidt C, Hofmann T, Urlaub H, Kuhnel K, Perez-Lara A (2017) Structure based biophysical characterization of the PROPPIN Atg18 shows Atg18 oligomerization upon membrane binding. Sci Rep 7 (1):14008. doi: 10.1038/s41598-017-14337-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Dynowski M, Mousley CJ, Shah SD, Yuan P, Winklbauer EM, de Campos MK, Trettin K, Quinones MC, Smirnova TI, Yanagisawa LL, Ortlund EA, Bankaitis VA (2011) Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol Biol Cell 22 (6):892–905. doi: 10.1091/mbc.E10-11-0903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA (2008) Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell 29 (2):191–206. doi: 10.1016/j.molcel.2007.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Welti S (2012) Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett 586 (17):2662–2673. doi: 10.1016/j.febslet.2012.06.006 [DOI] [PubMed] [Google Scholar]
- Schink KO, Tan KW, Stenmark H (2016) Phosphoinositides in Control of Membrane Dynamics. Annu Rev Cell Dev Biol 32:143–171. doi: 10.1146/annurev-cellbio-111315-125349 [DOI] [PubMed] [Google Scholar]
- Schoebel S, Blankenfeldt W, Goody RS, Itzen A (2010) High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA. EMBO Rep 11 (8):598–604. doi: 10.1038/embor.2010.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoneberg J, Lehmann M, Ullrich A, Posor Y, Lo WT, Lichtner G, Schmoranzer J, Haucke V, Noe F (2017) Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat Commun 8:15873. doi: 10.1038/ncomms15873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten A, Agianian B, Westerman J, Kroon J, Wirtz KW, Gros P (2002) Structure of apo-phosphatidylinositol transfer protein alpha provides insight into membrane association. EMBO J 21 (9):2117–2121. doi: 10.1093/emboj/21.9.2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA (2009) Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol 187 (6):889–903. doi: 10.1083/jcb.200905007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet LF, Hong W (2006) The Phox (PX) domain proteins and membrane traffic. Biochim Biophys Acta 1761 (8):878–896. doi: 10.1016/j.bbalip.2006.04.011 [DOI] [PubMed] [Google Scholar]
- Segui B, Allen-Baume V, Cockcroft S (2002) Phosphatidylinositol transfer protein beta displays minimal sphingomyelin transfer activity and is not required for biosynthesis and trafficking of sphingomyelin. Biochem J 366 (Pt 1):23–34. doi: 10.1042/BJ20020317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senju Y, Kalimeri M, Koskela EV, Somerharju P, Zhao H, Vattulainen I, Lappalainen P (2017) Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc Natl Acad Sci U S A 114 (43):E8977–E8986. doi: 10.1073/pnas.1705032114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR (2000) Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287 (5455):1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B, Phillips SE, Bankaitis VA, Luo M (1998) Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391 (6666):506–510. doi: 10.1038/35179 [DOI] [PubMed] [Google Scholar]
- Shadan S, Holic R, Carvou N, Ee P, Li M, Murray-Rust J, Cockcroft S (2008) Dynamics of lipid transfer by phosphatidylinositol transfer proteins in cells. Traffic 9 (10):1743–1756. doi: 10.1111/j.1600-0854.2008.00794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah ZH, Jones DR, Sommer L, Foulger R, Bultsma Y, D’Santos C, Divecha N (2013) Nuclear phosphoinositides and their impact on nuclear functions. FEBS J 280 (24):6295–6310. doi: 10.1111/febs.12543 [DOI] [PubMed] [Google Scholar]
- Shao X, Davletov BA, Sutton RB, Sudhof TC, Rizo J (1996) Bipartite Ca2+-binding motif in C2 domains of synaptotagmin and protein kinase C. Science 273 (5272):248–251 [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T, Yokoyama S (2007) Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129 (4):761–772. doi: 10.1016/j.cell.2007.03.040 [DOI] [PubMed] [Google Scholar]
- Sidhu G, Li W, Laryngakis N, Bishai E, Balla T, Southwick F (2005) Phosphoinositide 3-kinase is required for intracellular Listeria monocytogenes actin-based motility and filopod formation. J Biol Chem 280 (12):11379–11386. doi: 10.1074/jbc.M414533200 [DOI] [PubMed] [Google Scholar]
- Silkov A, Yoon Y, Lee H, Gokhale N, Adu-Gyamfi E, Stahelin RV, Cho W, Murray D (2011) Genome-wide structural analysis reveals novel membrane binding properties of AP180 N-terminal homology (ANTH) domains. J Biol Chem 286 (39):34155–34163. doi: 10.1074/jbc.M111.265611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394 (6692):494–498. doi: 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- Simunovic M, Voth GA, Callan-Jones A, Bassereau P (2015) When Physics Takes Over: BAR Proteins and Membrane Curvature. Trends Cell Biol 25 (12):780–792. doi: 10.1016/j.tcb.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD (2000) Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol 150 (3):527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJ, Schulz J, Meijers R, Rybin V, Briggs JA, Sachse C, Kaksonen M (2015) An organized co-assembly of clathrin adaptors is essential for endocytosis. Dev Cell 33 (2):150–162. doi: 10.1016/j.devcel.2015.02.023 [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiolo D, Wakatsuki S, Stenmark H (2005) Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem 280 (20):19600–19606. doi: 10.1074/jbc.M501510200 [DOI] [PubMed] [Google Scholar]
- Smirnova TI, Chadwick TG, MacArthur R, Poluektov O, Song L, Ryan MM, Schaaf G, Bankaitis VA (2006) The chemistry of phospholipid binding by the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14p as determined by EPR spectroscopy. J Biol Chem 281 (46):34897–34908. doi: 10.1074/jbc.M603054200 [DOI] [PubMed] [Google Scholar]
- Smirnova TI, Chadwick TG, Voinov MA, Poluektov O, van Tol J, Ozarowski A, Schaaf G, Ryan MM, Bankaitis VA (2007) Local polarity and hydrogen bonding inside the Sec14p phospholipid-binding cavity: high-field multi-frequency electron paramagnetic resonance studies. Biophys J 92 (10):3686–3695. doi: 10.1529/biophysj.106.097899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WJ, Nassar N, Bretscher A, Cerione RA, Karplus PA (2003) Structure of the active N-terminal domain of Ezrin. Conformational and mobility changes identify keystone interactions. J Biol Chem 278 (7):4949–4956. doi: 10.1074/jbc.M210601200 [DOI] [PubMed] [Google Scholar]
- Snoek GT, Berrie CP, Geijtenbeek TB, van der Helm HA, Cadee JA, Iurisci C, Corda D, Wirtz KW (1999) Overexpression of phosphatidylinositol transfer protein alpha in NIH3T3 cells activates a phospholipase A. J Biol Chem 274 (50):35393–35399 [DOI] [PubMed] [Google Scholar]
- Sohn M, Ivanova P, Brown HA, Toth DJ, Varnai P, Kim YJ, Balla T (2016) Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER-PM and ER-Golgi junctions. Proc Natl Acad Sci U S A 113 (16):4314–4319. doi: 10.1073/pnas.1525719113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xu W, Zhang A, Huang G, Liang X, Virbasius JV, Czech MP, Zhou GW (2001) Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry 40 (30):8940–8944 [DOI] [PubMed] [Google Scholar]
- Sot B, Behrmann E, Raunser S, Wittinghofer A (2013) Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc Natl Acad Sci U S A 110 (1):111–116. doi: 10.1073/pnas.1201658110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Ananthanarayanan B, Blatner NR, Singh S, Bruzik KS, Murray D, Cho W (2004) Mechanism of membrane binding of the phospholipase D1 PX domain. J Biol Chem 279 (52):54918–54926. doi: 10.1074/jbc.M407798200 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W (2003a) Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J Biol Chem 278 (16):14469–14479. doi: 10.1074/jbc.M212579200 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Bruzik KS, Waterfield MD, Bravo J, Williams RL, Cho W (2006) Structural and membrane binding analysis of the Phox homology domain of phosphoinositide 3-kinase-C2alpha. J Biol Chem 281 (51):39396–39406. doi: 10.1074/jbc.M607079200 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Murray D, Williams RL, Cho W (2007) Structural and membrane binding analysis of the Phox homology domain of Bem1p: basis of phosphatidylinositol 4-phosphate specificity. J Biol Chem 282 (35):25737–25747. doi: 10.1074/jbc.M702861200 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W (2002) Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem 277 (29):26379–26388. doi: 10.1074/jbc.M201106200 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W (2003b) Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J Biol Chem 278 (31):28993–28999. doi: 10.1074/jbc.M302865200 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Rafter JD, Das S, Cho W (2003c) The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J Biol Chem 278 (14):12452–12460. doi: 10.1074/jbc.M212864200 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Scott JL, Frick CT (2014) Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chem Phys Lipids 182:3–18. doi: 10.1016/j.chemphyslip.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T (1998) Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol 8 (6):343–346 [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144 (3):389–401. doi: 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Steffen P, Schafer DA, David V, Gouin E, Cooper JA, Cossart P (2000) Listeria monocytogenes ActA protein interacts with phosphatidylinositol 4,5-bisphosphate in vitro. Cell Motil Cytoskeleton 45 (1):58–66. doi: [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh BH, D’Arrigo A (1996) Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem 271 (39):24048–24054 [DOI] [PubMed] [Google Scholar]
- Stolt PC, Chen Y, Liu P, Bock HH, Blacklow SC, Herz J (2005) Phosphoinositide binding by the disabled-1 PTB domain is necessary for membrane localization and Reelin signal transduction. J Biol Chem 280 (10):9671–9677. doi: 10.1074/jbc.M413356200 [DOI] [PubMed] [Google Scholar]
- Stolt PC, Jeon H, Song HK, Herz J, Eck MJ, Blacklow SC (2003) Origins of peptide selectivity and phosphoinositide binding revealed by structures of disabled-1 PTB domain complexes. Structure 11 (5):569–579 [DOI] [PubMed] [Google Scholar]
- Stolt PC, Vardar D, Blacklow SC (2004) The dual-function disabled-1 PTB domain exhibits site independence in binding phosphoinositide and peptide ligands. Biochemistry 43 (34):10979–10987. doi: 10.1021/bi049092l [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ (2004) Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell 15 (8):3553–3566. doi: 10.1091/mbc.E04-02-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR (1995) Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell 80 (6):929–938 [DOI] [PubMed] [Google Scholar]
- Szentpetery Z, Balla A, Kim YJ, Lemmon MA, Balla T (2009) Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol 10:67. doi: 10.1186/1471-2121-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Kanematsu T, Misumi Y, Sakane F, Konishi H, Kikkawa U, Watanabe Y, Katan M, Hirata M (1997) Distinct specificity in the binding of inositol phosphates by pleckstrin homology domains of pleckstrin, RAC-protein kinase, diacylglycerol kinase and a new 130 kDa protein. Biochim Biophys Acta 1359 (3):275–285 [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Matsuda M, Yamamoto T, Kanematsu T, Kikkawa U, Yagisawa H, Watanabe Y, Hirata M (1998) PTB domain of insulin receptor substrate-1 binds inositol compounds. Biochem J 334 (Pt 1):211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Thapa N, Choi S, Anderson RA (2015) Emerging roles of PtdIns(4,5)P2--beyond the plasma membrane. J Cell Sci 128 (22):4047–4056. doi: 10.1242/jcs.175208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani K, Kogure T, Inoue H (2012) The intracellular phospholipase A1 protein family. Biomol Concepts 3 (5):471–478. doi: 10.1515/bmc-2012-0014 [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL (2006) ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125 (1):99–111. doi: 10.1016/j.cell.2006.01.047 [DOI] [PubMed] [Google Scholar]
- Thapa N, Tan X, Choi S, Lambert PF, Rapraeger AC, Anderson RA (2016) The Hidden Conundrum of Phosphoinositide Signaling in Cancer. Trends Cancer 2 (7):378–390. doi: 10.1016/j.trecan.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley SJ, Skippen A, Murray-Rust J, Swigart PM, Stewart A, Morgan CP, Cockcroft S, McDonald NQ (2004) Structure-function analysis of human [corrected] phosphatidylinositol transfer protein alpha bound to phosphatidylinositol. Structure 12 (2):317–326. doi: 10.1016/j.str.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Toker A, Cantley LC (1997) Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 387 (6634):673–676. doi: 10.1038/42648 [DOI] [PubMed] [Google Scholar]
- Tong J, Manik MK, Yang H, Im YJ (2016) Structural insights into nonvesicular lipid transport by the oxysterol binding protein homologue family. Biochim Biophys Acta 1861 (8 Pt B):928–939. doi: 10.1016/j.bbalip.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Tong J, Yang H, Yang H, Eom SH, Im YJ (2013) Structure of Osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure 21 (7):1203–1213. doi: 10.1016/j.str.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Tsujishita Y, Hurley JH (2000) Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol 7 (5):408–414. doi: 10.1038/75192 [DOI] [PubMed] [Google Scholar]
- Tsujita K, Itoh T, Ijuin T, Yamamoto A, Shisheva A, Laporte J, Takenawa T (2004) Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J Biol Chem 279 (14):13817–13824. doi: 10.1074/jbc.M312294200 [DOI] [PubMed] [Google Scholar]
- Tuzi S, Uekama N, Okada M, Yamaguchi S, Saito H, Yagisawa H (2003) Structure and dynamics of the phospholipase C-delta1 pleckstrin homology domain located at the lipid bilayer surface. J Biol Chem 278 (30):28019–28025. doi: 10.1074/jbc.M300101200 [DOI] [PubMed] [Google Scholar]
- Tyson GH, Halavaty AS, Kim H, Geissler B, Agard M, Satchell KJ, Cho W, Anderson WF, Hauser AR (2015) A novel phosphatidylinositol 4,5-bisphosphate binding domain mediates plasma membrane localization of ExoU and other patatin-like phospholipases. J Biol Chem 290 (5):2919–2937. doi: 10.1074/jbc.M114.611251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, Wakatsuki S, Misaki R, Koike M, Uchiyama Y, Iemura S, Natsume T, Kuwahara R, Nakagawa T, Nishikawa K, Mukai K, Miyoshi E, Taniguchi N, Sheff D, Lencer WI, Taguchi T, Arai H (2011) Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc Natl Acad Sci U S A 108 (38):15846–15851. doi: 10.1073/pnas.1109101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadas O, Burke JE (2015) Probing the dynamic regulation of peripheral membrane proteins using hydrogen deuterium exchange-MS (HDX-MS). Biochem Soc Trans 43 (5):773–786. doi: 10.1042/BST20150065 [DOI] [PubMed] [Google Scholar]
- van den Bogaart G, Meyenberg K, Diederichsen U, Jahn R (2012) Phosphatidylinositol 4,5-bisphosphate increases Ca2+ affinity of synaptotagmin-1 by 40-fold. J Biol Chem 287 (20):16447–16453. doi: 10.1074/jbc.M112.343418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9 (2):112–124. doi: 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Paridon PA, Somerharju P, Wirtz KW (1987) Phosphatidylinositol-transfer protein and cellular phosphatidylinositol metabolism. Biochem Soc Trans 15 (3):321–323 [DOI] [PubMed] [Google Scholar]
- Vance JE (2015) Phospholipid synthesis and transport in mammalian cells. Traffic 16 (1):1–18. doi: 10.1111/tra.12230 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 11 (5):329–341. doi: 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 143 (2):501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Gulyas G, Toth DJ, Sohn M, Sengupta N, Balla T (2017) Quantifying lipid changes in various membrane compartments using lipid binding protein domains. Cell Calcium 64:72–82. doi: 10.1016/j.ceca.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Rother KI, Balla T (1999) Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton’s tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem 274 (16):10983–10989 [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC (1999) Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J 18 (22):6329–6338. doi: 10.1093/emboj/18.22.6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A (1999) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398 (6722):39–46. doi: 10.1038/17969 [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Goebl M, Milligan S, O’Tousa JE, Hyde DR (1993) Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J Cell Biol 122 (5):1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR, O’Tousa JE (1991) Isolation and characterization of the Drosophila retinal degeneration B (rdgB) gene. Genetics 127 (4):761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollert CS, Uetz P (2004) The phox homology (PX) domain protein interaction network in yeast. Mol Cell Proteomics 3 (11):1053–1064. doi: 10.1074/mcp.M400081-MCP200 [DOI] [PubMed] [Google Scholar]
- Vonkova I, Saliba AE, Deghou S, Anand K, Ceschia S, Doerks T, Galih A, Kugler KG, Maeda K, Rybin V, van Noort V, Ellenberg J, Bork P, Gavin AC (2015) Lipid Cooperativity as a General Membrane-Recruitment Principle for PH Domains. Cell Rep 12 (9):1519–1530. doi: 10.1016/j.celrep.2015.07.054 [DOI] [PubMed] [Google Scholar]
- Vordtriede PB, Doan CN, Tremblay JM, Helmkamp GM Jr., Yoder MD (2005) Structure of PITPbeta in complex with phosphatidylcholine: comparison of structure and lipid transfer to other PITP isoforms. Biochemistry 44 (45):14760–14771. doi: 10.1021/bi051191r [DOI] [PubMed] [Google Scholar]
- Walker SM, Leslie NR, Perera NM, Batty IH, Downes CP (2004) The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem J 379 (Pt 2):301–307. doi: 10.1042/BJ20031839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gambhir A, Hangyas-Mihalyne G, Murray D, Golebiewska U, McLaughlin S (2002) Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J Biol Chem 277 (37):34401–34412. doi: 10.1074/jbc.M203954200 [DOI] [PubMed] [Google Scholar]
- Wang J, Gambhir A, McLaughlin S, Murray D (2004) A computational model for the electrostatic sequestration of PI(4,5)P2 by membrane-adsorbed basic peptides. Biophys J 86 (4):1969–1986. doi: 10.1016/S0006-3495(04)74260-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL (2003) Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114 (3):299–310 [DOI] [PubMed] [Google Scholar]
- Watt SA, Kular G, Fleming IN, Downes CP, Lucocq JM (2002) Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem J 363 (Pt 3):657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watton SJ, Downward J (1999) Akt/PKB localisation and 3’ phosphoinositide generation at sites of epithelial cell-matrix and cell-cell interaction. Curr Biol 9 (8):433–436 [DOI] [PubMed] [Google Scholar]
- Wawrzyniak AM, Kashyap R, Zimmermann P (2013) Phosphoinositides and PDZ domain scaffolds. Adv Exp Med Biol 991:41–57. doi: 10.1007/978-94-007-6331-9_4 [DOI] [PubMed] [Google Scholar]
- Weber SS, Ragaz C, Hilbi H (2009) The inositol polyphosphate 5-phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole and binds to the bacterial effector LpnE. Cell Microbiol 11 (3):442–460. doi: 10.1111/j.1462-5822.2008.01266.x [DOI] [PubMed] [Google Scholar]
- Weber-Boyvat M, Kentala H, Peranen J, Olkkonen VM (2015) Ligand-dependent localization and function of ORP-VAP complexes at membrane contact sites. Cell Mol Life Sci 72 (10):1967–1987. doi: 10.1007/s00018-014-1786-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Stec B, Redfield AG, Weerapana E, Roberts MF (2015) Phospholipid-binding sites of phosphatase and tensin homolog (PTEN): exploring the mechanism of phosphatidylinositol 4,5-bisphosphate activation. J Biol Chem 290 (3):1592–1606. doi: 10.1074/jbc.M114.588590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele BA, Orchard RC, Jimenez A, Cox GW, Alto NM (2017) A systematic exploration of the interactions between bacterial effector proteins and host cell membranes. Nat Commun 8 (1):532. doi: 10.1038/s41467-017-00700-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA (2005) Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem 280 (11):10501–10508. doi: 10.1074/jbc.M414304200 [DOI] [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR (2002) P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell 108 (6):809–821 [DOI] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ (1998) Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science 281 (5373):105–108 [DOI] [PubMed] [Google Scholar]
- Whited AM, Johs A (2015) The interactions of peripheral membrane proteins with biological membranes. Chem Phys Lipids 192:51–59. doi: 10.1016/j.chemphyslip.2015.07.015 [DOI] [PubMed] [Google Scholar]
- Whorton MR, MacKinnon R (2011) Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147 (1):199–208. doi: 10.1016/j.cell.2011.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes FB, Reeves EP, Soskic V, Furthmayr H, Segal AW (2001) The NADPH oxidase components p47(phox) and p40(phox) bind to moesin through their PX domain. Biochem Biophys Res Commun 289 (2):382–388. doi: 10.1006/bbrc.2001.5982 [DOI] [PubMed] [Google Scholar]
- Williams RL, Urbe S (2007) The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol 8 (5):355–368. doi: 10.1038/nrm2162 [DOI] [PubMed] [Google Scholar]
- Wirtz KW (1991) Phospholipid transfer proteins. Annu Rev Biochem 60:73–99. doi: 10.1146/annurev.bi.60.070191.000445 [DOI] [PubMed] [Google Scholar]
- Wirtz KW (1997) Phospholipid transfer proteins revisited. Biochem J 324 (Pt 2):353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart MJ, Taylor GS, Dixon JE (2001) Phoxy lipids: revealing PX domains as phosphoinositide binding modules. Cell 105 (7):817–820 [DOI] [PubMed] [Google Scholar]
- Wong LH, Copic A, Levine TP (2017) Advances on the Transfer of Lipids by Lipid Transfer Proteins. Trends Biochem Sci 42 (7):516–530. doi: 10.1016/j.tibs.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Levine TP (2016) Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing? Biochem Soc Trans 44 (2):517–527. doi: 10.1042/BST20150275 [DOI] [PubMed] [Google Scholar]
- Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M (2007) PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell 28 (5):886–898. doi: 10.1016/j.molcel.2007.10.028 [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Solidar A, Yoden MD (2010) Phosphatidylinositol transfer proteins: sequence motifs in structural and evolutionary analyses. J Biomed Sci Eng 3 (1):65–77. doi: 10.4236/jbise.2010.31010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymann MP, Schneiter R (2008) Lipid signalling in disease. Nat Rev Mol Cell Biol 9 (2):162–176. doi: 10.1038/nrm2335 [DOI] [PubMed] [Google Scholar]
- Xing Y, Liu D, Zhang R, Joachimiak A, Songyang Z, Xu W (2004) Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX). J Biol Chem 279 (29):30662–30669. doi: 10.1074/jbc.M404107200 [DOI] [PubMed] [Google Scholar]
- Xu C, Watras J, Loew LM (2003) Kinetic analysis of receptor-activated phosphoinositide turnover. J Cell Biol 161 (4):779–791. doi: 10.1083/jcb.200301070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu D, Gill G, Songyang Z (2001) Regulation of cytokine-independent survival kinase (CISK) by the Phox homology domain and phosphoinositides. J Cell Biol 154 (4):699–705. doi: 10.1083/jcb.200105089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Arnaud L, Cooper JA (2005) Both the phosphoinositide and receptor binding activities of Dab1 are required for Reelin-stimulated Dab1 tyrosine phosphorylation. Brain Res Mol Brain Res 139 (2):300–305. doi: 10.1016/j.molbrainres.2005.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Bateman A, Finn RD, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Carlton D, Chen C, Chiu HJ, Chiu M, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Ernst D, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Marciano D, McMullan D, Miller MD, Morse AT, Nigoghossian E, Nopakun A, Okach L, Puckett C, Reyes R, Rife CL, Sefcovic N, Tien HJ, Trame CB, van den Bedem H, Weekes D, Wooten T, Hodgson KO, Wooley J, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wilson IA (2010) Bacterial pleckstrin homology domains: a prokaryotic origin for the PH domain. J Mol Biol 396 (1):31–46. doi: 10.1016/j.jmb.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Garner K, Georgiev P, Li M, Gomez-Espinosa E, Panda A, Mathre S, Okkenhaug H, Cockcroft S, Raghu P (2015) RDGBalpha, a PtdIns-PtdOH transfer protein, regulates G-protein-coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. J Cell Sci 128 (17):3330–3344. doi: 10.1242/jcs.173476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Akimoto T, Kalli AC, Yasuoka K, Sansom MS (2017) Dynamic interactions between a membrane binding protein and lipids induce fluctuating diffusivity. Sci Adv 3 (1):e1601871. doi: 10.1126/sciadv.1601871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Kalli AC, Yasuoka K, Sansom MSP (2016) Interactions of Pleckstrin Homology Domains with Membranes: Adding Back the Bilayer via High-Throughput Molecular Dynamics. Structure 24 (8):1421–1431. doi: 10.1016/j.str.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Olkkonen VM (2008) Characteristics of oxysterol binding proteins. Int Rev Cytol 265:253–285. doi: 10.1016/S0074-7696(07)65007-4 [DOI] [PubMed] [Google Scholar]
- Yau WM, Wimley WC, Gawrisch K, White SH (1998) The preference of tryptophan for membrane interfaces. Biochemistry 37 (42):14713–14718. doi: 10.1021/bi980809c [DOI] [PubMed] [Google Scholar]
- Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S (2006) Receptor activation alters inner surface potential during phagocytosis. Science 313 (5785):347–351. doi: 10.1126/science.1129551 [DOI] [PubMed] [Google Scholar]
- Yoder MD, Thomas LM, Tremblay JM, Oliver RL, Yarbrough LR, Helmkamp GM Jr. (2001) Structure of a multifunctional protein. Mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J Biol Chem 276 (12):9246–9252. doi: 10.1074/jbc.M010131200 [DOI] [PubMed] [Google Scholar]
- Yoon Y, Tong J, Lee PJ, Albanese A, Bhardwaj N, Kallberg M, Digman MA, Lu H, Gratton E, Shin YK, Cho W (2010) Molecular basis of the potent membrane-remodeling activity of the epsin 1 N-terminal homology domain. J Biol Chem 285 (1):531–540. doi: 10.1074/jbc.M109.068015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Zhang X, Cho W (2012) Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) specifically induces membrane penetration and deformation by Bin/amphiphysin/Rvs (BAR) domains. J Biol Chem 287 (41):34078–34090. doi: 10.1074/jbc.M112.372789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Lemmon MA (2001) All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem 276 (47):44179–44184. doi: 10.1074/jbc.M108811200 [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13 (5):677–688 [DOI] [PubMed] [Google Scholar]
- Yun M, Keshvara L, Park CG, Zhang YM, Dickerson JB, Zheng J, Rock CO, Curran T, Park HW (2003) Crystal structures of the Dab homology domains of mouse disabled 1 and 2. J Biol Chem 278 (38):36572–36581. doi: 10.1074/jbc.M304384200 [DOI] [PubMed] [Google Scholar]
- Zalevsky J, Grigorova I, Mullins RD (2001) Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem 276 (5):3468–3475. doi: 10.1074/jbc.M006407200 [DOI] [PubMed] [Google Scholar]
- Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P (2013) Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep 4 (6):1213–1223. doi: 10.1016/j.celrep.2013.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ma Y, Seemann J, Huang LJ (2010) A regulating role of the JAK2 FERM domain in hyperactivation of JAK2(V617F). Biochem J 426 (1):91–98. doi: 10.1042/BJ20090615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CZ, Li de La Sierra-Gallay I, Quevillon-Cheruel S, Collinet B, Minard P, Blondeau K, Henckes G, Aufrere R, Leulliot N, Graille M, Sorel I, Savarin P, de la Torre F, Poupon A, Janin J, van Tilbeurgh H (2003) Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J Biol Chem 278 (50):50371–50376. doi: 10.1074/jbc.M304392200 [DOI] [PubMed] [Google Scholar]
- Zhou MM, Fesik SW (1995) Structure and function of the phosphotyrosine binding (PTB) domain. Prog Biophys Mol Biol 64 (2–3):221–235 [DOI] [PubMed] [Google Scholar]
- Zhou MM, Ravichandran KS, Olejniczak EF, Petros AM, Meadows RP, Sattler M, Harlan JE, Wade WS, Burakoff SJ, Fesik SW (1995) Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature 378 (6557):584–592. doi: 10.1038/378584a0 [DOI] [PubMed] [Google Scholar]
- Zhu G, Chen J, Liu J, Brunzelle JS, Huang B, Wakeham N, Terzyan S, Li X, Rao Z, Li G, Zhang XC (2007) Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J 26 (14):3484–3493. doi: 10.1038/sj.emboj.7601771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hu L, Zhou Y, Yao Q, Liu L, Shao F (2010) Structural mechanism of host Rab1 activation by the bifunctional Legionella type IV effector SidM/DrrA. Proc Natl Acad Sci U S A 107 (10):4699–4704. doi: 10.1073/pnas.0914231107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM (2006) How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7 (1):9–19. doi: 10.1038/nrm1784 [DOI] [PubMed] [Google Scholar]
- Zimmermann P (2006) The prevalence and significance of PDZ domain-phosphoinositide interactions. Biochim Biophys Acta 1761 (8):947–956. doi: 10.1016/j.bbalip.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J (2002) PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell 9 (6):1215–1225 [DOI] [PubMed] [Google Scholar]
- Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, Lenk GM, Converso-Baran K, Weide T, Albin RL, Saltiel AR, Meisler MH, Russell MW, Weisman LS (2012) In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci U S A 109 (43):17472–17477. doi: 10.1073/pnas.1203106109 [DOI] [PMC free article] [PubMed] [Google Scholar]


