Crohn’s disease is a chronic inflammatory intestinal disease that is frequently accompanied by aberrant healing and stricturing complications. Crosstalk between activated myeloid and stromal cells is critical in the pathogenicity of Crohn’s disease1,2, and increases in intravasating monocytes are correlated with a lack of response to anti-TNF treatment3. The risk alleles with the highest effect on Crohn’s disease are loss-of-function mutations in NOD24,5, which increase the risk of stricturing6. However, the mechanisms that underlie pathogenicity driven by NOD2 mutations and the pathways that might rescue a lack of response to anti-TNF treatment remain largely uncharacterized. Here we use direct ex vivo analyses of patients who carry risk alleles of NOD2 to show that loss of NOD2 leads to dysregulated homeostasis of activated fibroblasts and macrophages. CD14+ peripheral blood mononuclear cells from carriers of NOD2 risk alleles produce cells that express high levels of collagen, and elevation of conserved signatures is observed in nod2-deficient zebrafish models of intestinal injury. The enrichment of STAT3 regulation and gp130 ligands in activated fibroblasts and macrophages suggested that gp130 blockade might rescue the activated program in NOD2-deficient cells. We show that post-treatment induction of the STAT3 pathway is correlated with a lack of response to anti-TNF treatment in patients, and demonstrate in vivo in zebrafish the amelioration of the activated myeloid–stromal niche using the specific gp130 inhibitor bazedoxifene. Our results provide insights into NOD2-driven fibrosis in Crohn’s disease, and suggest that gp130 blockade may benefit some patients with Crohn’s disease—potentially as a complement to anti-TNF therapy.
Crosstalk between activated macrophages and fibroblasts drives complications of ileal Crohn’s disease3. Stable macrophage–fibroblast systems involve growth factors (for example, CSF1 and platelet-derived growth factors)7, and perturbations to stable cellular systems include genetic variation—prominently loss-of-function alleles of NOD2, which are associated with an increased risk of fibrostenotic complications6,8. However, the mechanisms through which NOD2 drives pathogenicity are not fully elucidated. NOD2 mutations have not been reported to correlate with responses to anti-TNF (which is the major agent used to treat moderate-to-severe Crohn’s disease)9,10; the timing of the start of anti-TNF treatment6,11 may alter responses. Previous reports of precise cell types at the single-cell level has elucidated clusters that define a composite gene-set module correlated with a lack of response to anti-TNF, with most transcripts expressed in activated macrophages and fibroblasts3.
CD14+PDGFRA+ cells in the inflamed ileum
Tissue-resident macrophages in the gut are replenished from blood monocytes12 (Fig. 1a). Activated fibroblasts in inflamed tissue demonstrate increased expression of PDPN, CHI3L1, MMP3, IL11, CXCL13 and WT1 (Fig. 1a; Extended Data Fig. 1a).
Fig. 1 |. Inflamed CD14+PDGFRA+ cells and scRNA-seq orthologies suggest key roles for myeloid–stromal clusters.
a, Expression (log2(cluster expression/cluster averages)) of genes (columns) across myeloid and stromal cell subsets (rows) from scRNA-seq data of PBMCs and inflamed ileum from individuals with Crohn’s disease ileum (n = 11). DC, dendritic cells (including plasmacytoid and classical dendritic cells). b, Gene expression in CD14+PDGFRA+ versus CD14− fraction from uninflamed versus inflamed tissue (n = 3 biological replicates). Data are mean ± s.e.m. *P < 0.05, **P < 0.1; two-tailed paired Student’s t-test. c, scRNA-seq of DSS-treated and untreated larval intestines (n = 5 replicates; 25 pooled intestines per replicate (80,098 cells in total)), and innate immune, epithelial and stromal cells from individuals with Crohn’s disease (24,364 cells in total). Uniform manifold approximation and projection (UMAP) shows joint clustering of 28 clusters between zebrafish (F) and human (H) cells. ILCs, innate lymphoid cells; ILC3s, type-3 ILCs; LREs, lysosome-rich enterocytes. d, Confusion matrices of a human random-forest model applied to actual human (top) and zebrafish (bottom) scRNA-seq datasets, showing the proportion of actual and model-predicted cells. M, macrophages; S1, activated fibroblasts; S2, residential fibroblasts; S3, smooth muscle cells; S4, endothelial cells; S5, mesothelial cells (zebrafish only). e, Top, haematoxylin and eosin staining of 1× DSS-treated nod2+/+ (nod2WT) (left) and nod2mss13/mss13 (nod2mss13) (right) larvae. Red arrows, leukocyte staining in anterior intestine. Scale bar, 100 μm (main panels), 50 μm (insets). Bottom, quantification of the number of leukocytes per 1.5 × 3.5-cm area in larvae treated with 1× (left) or 2× (right) DSS or untreated controls (ctrl). For 1× DSS, n = 3 (nod2+/+, treated), 5 (nod2+/+, control), 6 (nod2mss13, control) or 8 (nod2mss13 treated); for 2× DSS, n = 4 (nod2+/+, control), 5 (nod2mss13, control), 6 (nod2mss13, treated) or 10 (nod2+/+, treated). Data are mean ± s.e.m. *P < 0.05, **P < 0.01; one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons.
We found that most activated fibroblasts (97.6%) expressed at least one unique molecular identifier of COL1A1; 40% of activated fibroblasts expressed the stromal cell growth-factor receptor gene PDGFRA. Notably, 6.5% of activated fibroblasts expressed CD14, and 4.3% of activated fibroblasts expressed both CD14 and PDGFRA (Extended Data Fig. 1d). Distributions of unique molecular identifiers in CD14+PDGFRA+ cells were characteristic of single cells, which indicates that they are unlikely to be doublets (Supplementary Tables 1, 2). We compared the expression of CD14+PDGFRA+ cells from inflamed and uninflamed resected ileum (Extended Data Fig. 1e), and observed increased transcripts of activated fibroblasts in inflamed double-positive cells (Fig. 1b); the profibrotic-mediator genes WT1 and IL1113,14 (encoding IL-11, which acts downstream of WT115) were significantly enriched.
We observed expression of WT1, PDGFRA and CD14 in stromal and myeloid cells of the muscularis mucosae, lymphoid aggregates and around blood vessels in strictured tissue (Extended Data Fig. 1g). Intravasating leukocytes in the muscularis mucosae and the deposition of the extracellular matrix protein MFAP4 around blood vessels (Extended Data Fig. 1f) suggest points of entry in a pathogenic niche16.
We performed single-cell RNA sequencing (scRNA-seq) of intestines that were dissected zebrafish larvae treated with dextran sodium sulfate (DSS)17. Our joint clustering of 30,069 cells revealed 32 clusters that comprise a single-cell atlas of the intestine of the zebrafish larvae (Extended Data Fig. 2a, Supplementary Table 3). We then performed integrated clustering between zebrafish and human innate immune, stromal and epithelial cells (Fig. 1c, Extended Data Fig. 2b), which revealed conservation of myeloid, stromal and epithelial clusters. Using a trained random-forest model, we found that the proportion of actual and predicted cells was highly correlated between species (Fig. 1d). Conserved populations also shared distinctive transcriptional signatures (Extended Data Fig. 2c). Activated fibroblasts from both species express COL1A1 (col1a1a), PDGFRA (pdgfra), WT1 (wt1a) and IL11 (il11a) (zebrafish gene symbols in parentheses), highlighting the ability of the zebrafish model to represent key mediators of ileal Crohn’s disease in humans.
We quantified leukocyte infiltration in the anterior intestine of wild-type (nod2+/+) and CRISPR-knockout nod2mss133 (which contain a frameshift mutation in exon 1 of nod2; full details in ‘CRISPR–Cas9 generation of nod2-mutant zebrafish line’ in Methods) zebrafish larvae that were treated with DSS, and observed significant infiltration in the intestine of nod2mss13/mss13 zebrafish larvae treated with one dose (1×) of DSS, and in nod2mss13/mss13 larvae treated with two doses (2×) of DSS compared to 2×DSS-treated nod2+/+ larvae (Fig. 1e, Extended Data Fig. 1h).
NOD2 loss enhances pathogenic activation
NOD2 can sense bacterial components in fibroblasts during infection18 and NOD2, RIPK2 and XIAP are expressed in inflamed activated fibroblasts (Fig. 1a, Extended Data Fig. 1b). We hypothesized that loss-of-function alleles of NOD2 drive aberrant differentiation of newly recruited CD14+ peripheral blood mononuclear cells (PBMCs), which results in increased numbers of activated cells that express high levels of collagen as compared to classical macrophages19. We used the top 200 genes that are differentially expressed between the activated and nonactivated clusters in the scRNA-seq data (Supplementary Table 4) to develop ‘activated fibroblast’ and ‘inflammatory macrophage’ scores from bulk RNA sequencing ileal samples from patients with Crohn’s disease in the inception, pretreatment, paediatric cohort of the ‘Risk Stratification and Identification of Immunogenetic and Microbial Markers of Rapid Disease Progression in Children with Crohn’s Disease’ (RISK) study1 (Methods, Supplementary Table 5). The most significant principal components show changing gene signatures on the basis of the number of NOD2 risk alleles (Fig. 2a). Consistent with greater monocyte recruitment, we observed significant increases in CD14 expression with increasing numbers of NOD2 risk alleles. We observed similar trends for PDGFRA, activated fibroblast markers (PDPN and CHI3L1), matrix metalloproteinases (MMP3 and MMP9) and genes in the gp130 family (IL11, IL6 and OSM) (Fig. 2a).
Fig. 2 |. NOD2 deficiency establishes an activated fibroblast signature that implicates IL-11 and WT1.
a, Left, Principal component (PC) analysis of the top 200 differentially expressed genes in activated fibroblasts (top) and inflammatory macrophages (bottom) in the NOD2WT/WT versus NOD2WT/MT and NOD2MT/MT individuals from the RISK cohort. Right, expression of the indicated individual genes (DESeq2 normalized counts). n = 140, 61 and 17 for WT/WT (0), MT/WT (1) and MT/MT (2) individuals, respectively. In the box plots, lower and upper hinges correspond to the first (25th percentile) and third (75th percentile) quartiles. Upper and lower whiskers extend from the hinge to the largest value no further than 1.5× interquartile range from the hinge. *P < 0.05, **P < 0.01; glm function in R for linear regression. b, COL1A1 from unstimulated PBMCs (black) or PBMCs stimulated by 0.5 μg ml−1 lipid A (green), 2.5 μg ml−1 Pam3Cys (blue) or 1 μg ml−1 MDP (red), by Luminex (in pg ml−1). n = 2 NOD2WT/WT, n = 3 NOD2MT/MT. Data are mean for n = 2, mean ± s.e.m. for n = 3. c, Reverse-transcription PCR (RT–PCR) of expression of the indicated genes from PBMCs stimulated with 1 μg ml−1 MDP after 2 weeks of differentiation, from NOD2WT/WT, NOD2WT/MT and NOD2MT/MT individuals. n = 4 biological replicates per genotype. Data are mean relative expression (left y axis; shown as fold change relative to expression in wild type after MDP treatment) and individual 2−ΔCt expression values (right y-axis); error bars are individual ΔCt values ± s.e.m. *P < 0.05, **P < 0.01; two-tailed unpaired Student’s t-test. d, RT–PCR of gene expression 24 h and 48 h after removal of MDP from zebrafish larvae treated with 1 μg ml−1 MDP. n = 5 biological replicates for 1× MDP: n = 5 clutches, 10–15 larvae per genotype per clutch. n = 4 biological replicates for 2× MDP: n = 5 clutches, 10–15 larvae per genotype per clutch. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-way ANOVA.
We adapted in vitro methods20,21 to model CD14+ PBMC differentiation from healthy volunteers who carried no risk alleles (NOD2WT/WT), one risk allele (NOD2WT/MT) or two risk alleles (NOD2MT/MT) of NOD2. The NOD2-activating ligand muramyl dipeptide (MDP) is a component of bacterial cell walls22, and polarizes monocyte differentiation towards macrophages in the short term23,24. We tested long-term stimulation with MDP for two weeks, and observed greater numbers of spindle-like cells expressing high levels of collagen upon differentiation of CD14+ PBMCs in NOD2MT/MT individuals than in NOD2WT/WT individuals (Extended Data Fig. 3a, b). We observed profibrotic deposition of MFAP4 and COL5A1 along the edges of spindle cells, and global COL1A1 expression (Extended Data Fig. 3c). By contrast, the myeloid marker MPEG1 mostly localized to macrophage cytoplasm, with some co-expression with MFAP4 in spindle cells (Extended Data Fig. 3c). In the absence of NOD2, signalling is increased by intracellular pattern recognition receptors18,25: long-term stimulation with lipopolysaccharide (LPS) induces TLR4 signalling in spindle cells26, and MDP enhances LPS-induced TLR4 expression on stromal cells27. Long-term stimulation with both lipid A and MDP induced differentiation of spindle cells (Extended Data Fig. 3a, b). In NOD2MT/MT individuals, COL1A1 secretion is induced by lipid A, Pam3Cys and MDP (Fig. 2b), whereas only MDP significantly induced secretion of IL-6 and CXCL13 (Extended Data Fig. 3f). With increasing numbers of NOD2 risk alleles, the activated fibroblast gene signature (most significantly, IL11) is enriched upon stimulation with MDP (Fig. 2c).
To investigate in vivo effects of recurrent exposure to MDP, we used a previously generated nod2-mutant zebrafish line (nod2sa21011)28 (generated via ENU mutagenesis in exon 1) and our nod2mss13 line, and measured transcripts at 24 and 48 h after MDP removal (Extended Data Fig. 3g). We found that, during recovery from MDP injury, activated fibroblast transcripts are upregulated when one or more copies of nod2 are lost. Transcripts recovered to baseline levels in nod2+/+ larvae, but not in nod2+/sa21011 or nod2sa21011/sa21011 backgrounds (Fig. 2d). The profibrotic transcription factor wt1a remained elevated in nod2sa21011/sa21011 larvae with repetitive MDP injury, which indicates sustained upstream transcriptional activation.
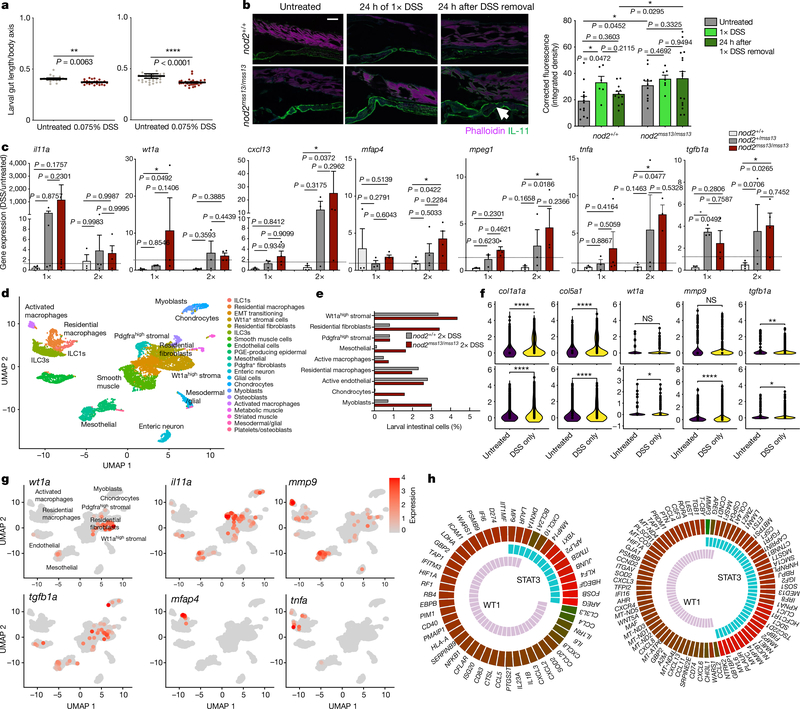
We next treated zebrafish larvae with 1× or 2× DSS; DSS significantly reduces intestinal length29 in both single and repetitive injuries (Fig. 3a). We observed marked differences in the kinetics of the profibrotic cytokine Il-11, which were stratified by nod2 genotype. In nod2+/+ larvae, we observed maximal expression immediately after treatment with DSS. By contrast, Il-11 was significantly increased in nod2mss13/mss13 larvae at 24 h after the removal of DSS, as compared to nod2+/+ larvae at this time (Fig. 3b). Activated fibroblast and macrophage transcripts were also increased with nod2 loss at 24 h after removal of DSS, after both single and repetitive injuries (Fig. 3c). These data suggest that the loss of Nod2 inhibits efficient amelioration of fibrotic and inflammatory activation at a molecular level.
Fig. 3 |. Single and recurrent DSS injury defines nod2-dependent gene activation, and myeloid and stromal pathway analyses implicate STAT3.
a, Zebrafish intestine length after a 24-h 1× (left) or 2× (right) DSS treatment. n = 3 biological replicates (3 clutches with 5–10 larvae per clutch). Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-sided Wilcoxon signed-rank test. b, Immunofluorescence of Il-11 (GFP) and phalloidin (for F-actin staining; far red) in middle intestines of DSS-treated or untreated nod2+/+ or nod2mss13/mss13 larvae. Right, quantification (corrected integrated density). n = 13 untreated nod2+/+ untreated; 6 1×DSS-treated nod2+/+; 12 nod2+/+ at 24 h after 1× DSS; 11 untreated nod2mss13; 7 1×DSS-treated nod2mss13; 16 nod2mss13 at 24 h after 1× DSS. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two way-ANOVA. Scale bar, 100 μm. c, Relative expression of the indicated genes at 24 h after removal of 1× or 2× DSS in nod2+/+, nod2+/mss13 and nod2mss13/mss13 larvae. n = 5 biological replicates for 1×DSS: n = 5 clutches, 10–15 larvae per genotype per clutch. n = 4 biological replicates for 2×DSS: n = 4 clutches, 10–15 larvae per genotype per clutch. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-way ANOVA. d, UMAP showing stromal and myeloid clusters from untreated and DSS-treated nod2+/+ and nod2mss13/mss13 larvae. EMT, epithelial–mesenchymal transition; ILC1s, type-1 ILCs; PGE, prostaglandin E. e, Percentage of stromal and myeloid populations out of total intestinal cells from nod2+/+ and nod2mss13/mss13 DSS-treated larvae. f, Violin plots for DSS-upregulated genes in nod2+/+ (top) and nod2mss13/mss13 (bottom) backgrounds. NS, not significant. g, Feature plots showing gene expression in nod2mss13/mss13 larvae in myeloid and stromal populations (log-normalized expression). h, Circos plots of WT1 and STAT3-regulated gene expression in activated fibroblasts (right) and inflammatory macrophages (left) from NOD2MT/MT and NOD2WT/MT individuals versus NOD2WT/WT individuals in scRNA-seq data from ileal samples (ingenuity pathway analysis).
Our joint clustering of scRNA-seq data from myeloid and stromal cells from nod2+/+ and nod2mss13/mss13 zebrafish larvae treated with DSS resolved 21 clusters (Fig. 3d, Extended Data Fig. 4b, e, Supplementary Table 7). The activated transcripts col1a1a, col5a1, wt1a, mmp9 and tgfb1a were significantly increased in DSS-treated compared to untreated larvae; of note, wt1a and mmp9 were significantly increased only in nod2mss13/mss13 larvae (Fig. 3f). These transcripts were uniquely expressed in Wt1ahigh stromal cells, Pdgfrahigh stromal cells, mesothelial cells and activated macrophages (Fig. 3g, Extended Data Fig. 4f), which expanded most in DSS-treated nod2mss13/mss13 larvae (Fig. 3e).
Upstream regulation of NOD2 signatures
We performed differential expression of activated fibroblasts and inflammatory macrophages from our ileal scRNA-seq data et comparing individuals with NOD2WT/WT, NOD2WT/MT and NOD2MT/MT, and observed increased expression of proinflammatory and profibrotic mediators in individuals who carry at least one risk allele of NOD2 (Extended Data Fig. 4c, Supplementary Table 8). Pathway analysis for upstream transcriptional regulators of differentially expressed genes revealed WT1, STAT3 (Fig. 3h), TWIST1 and CEBPB (Supplementary Table 9) as highly significant regulators of gene expression associated with NOD2 risk alleles; we observed more significant regulation by WT1 in activated fibroblasts than in inflammatory macrophages (Extended Data Fig. 4d). Among the top regulators that were shared between both cell populations, STAT3 demonstrated uniquely high enrichment (Extended Data Fig. 4d).
gp130 blockade alleviates pathogenicity
OSM signalling has a key role in the lack of response to anti-TNF therapy2, IL-11 is important in fibrosis13,14 and IL-6 is crucial in immune cell activation and proliferation30. Here we find a central role for STAT3 in regulating the expression of NOD2 risk alleles in the activated macrophage–fibroblast niche (Fig. 3h). We therefore reasoned that blockade of the common cytokine receptor subunit gp130 (encoded by IL6ST) (Fig. 4a) might precisely target aberrant or inappropriate fibrotic responses to injury. Bazedoxifene (BZA) is a small molecule gp130 inhibitor that has previously been used to treat colorectal and pancreatic cancers in preclinical models31–33.
Fig. 4 |. gp130 inhibition rescues activation of the myeloid–stromal niche.
a, gp130 signalling. IL6R, IL11R, OSMR and LIFR dimerize gp130. BZA competes with gp130 ligands to inhibit signalling. b, log2-transformed expression of the indicated genes from microarray data from a previous publication34 of patients before and after treatment with infliximab. Blue bars, patients who responded to infliximab (n = 18); red bars, patients who did not respond to infliximab (n = 16). Data are mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001; two-tailed paired Student’s t-test. c, Secretion of IL-6 (left), CXCL13 (middle) and MMP3 (right) from NOD2WT/WT versus NOD2MT/MT differentiated PBMCs by Luminex (in pg ml−1), treated with 0.5 μg ml−1 MDP (MDP) or 0.5 μg ml−1 MDP with 1 μM BZA (MDP + BZA), or left untreated (unt.). n = 2 replicates for NOD2WT/WT; n = 3 replicates for NOD2MT/MT (except MMP3, n = 2). *P < 0.05; one-tailed paired Student’s t-test. d, Violin plots for the upregulation or downregulation of the indicated genes by DSS or DSS + BZA, respectively (log-normalized expression). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; two-sided Wilcoxon signed-rank test. Red asterisks in the top right of the plots denote nod2mss13/mss13 differences. e, Top, haematoxylin and eosin staining of DSS + BZA-treated nod2+/+ (left) and nod2mss13/mss13 (right) larvae. Red arrows, leukocytes. Scale bar, 100 μm (main panels), 50 μm (insets). Bottom, quantification of leukocytes per 1.5 × 3.5 cm after 1× DSS + BZA (left) or 2× DSS + BZA (right) treatment. For 1× treatments, n = 5 untreated, 3 DSS and 14 DSS + BZA nod2+/+ larvae; and n = 6 untreated, 8 DSS and 10 DSS + BZA nod2mss13/mss13 larvae. For 2× treatments, n = 4 untreated, 10 DSS, 8 DSS + BZA nod2+/+ larvae; and n = 5 untreated, 6 DSS and 10 DSS + BZA nod2mss13/mss13 larvae. Data are mean ± s.e.m.. *P < 0.05, **P < 0.01, ***P < 0.001; one way-ANOVA test with Tukey’s multiple comparison. f, Intestine length 24 h after 1× (left) and 2× (right) DSS or BZA (10 μM) + DSS treatments. 1× DSS = 0.075%. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-sided Wilcoxon signed-rank test. n = 3 biological replicates; 3 clutches with 5–10 larvae per clutch. g, RT–PCR of expression of the indicated genes at 24 h after 1× DSS + BZA (left) and 2× DSS + BZA (right), relative to treatment with DSS only. n = 3 biological replicates for 1×DSS + BZA; n = 4 biological replicates for 2×DSS + BZA (3 and 4 clutches; 45 larvae per clutch for RNA). Data are mean ± s.e.m. *P < 0.05, **P < 0.01; one-tailed paired Student’s t-test.
We analysed data from a previous publication that compares bulk mucosal intestinal transcriptomes before and after anti-TNF treatment34. Comparing post-treatment gene expression along the gp130 pathway, we observed induction of IL11, OSM, IL6 and IL6R in patients who were refractory to anti-TNF treatment as compared to those who responded (Fig. 4b); we observed similar patterns with CHI3L1, CXCL13 (Extended Data Fig. 5a) and PDPN. NOD2 significantly decreased in patients who responded to anti-TNF as compared to those who did not, which reflects NOD2 induction in activated macrophages. Consistent with previous reports, pretreatment gene expression of OSM2, CHI3L1 and PDPN3 was higher in those who did not respond to anti-TNF, and remained higher even after the anti-TNF treatment course. TWIST1 demonstrated the same trends; TWIST1 is a transcriptional inducer of activated fibroblasts35, regulates genes that are differentially expressed by NOD2WT/MT and NOD2MT/MT individuals (Supplementary Table 9), and its expression decreases with BZA treatment in the livers of mice that are fed high-fat diets36.
We next differentiated NOD2WT/WT or NOD2MT/MT CD14+ PBMCs with MDP or with MDP in combination with BZA for two weeks, and observed a significant reduction in the secretion of CXCL13 and IL-6 (and a non-significant decrease in MMP3) in cotreated PBMCs, and a reversion from spindle to round cell morphology (Fig. 4c, Extended Data Fig. 5b). We confirmed the gp130 specificity of BZA inhibition by costimulating CD14+ PBMCs with MDP and a monoclonal gp130 antibody, and consistently observed the same reversion from spindle to round cells (Extended Data Fig. 5b), with reduced secretion of OSM, IL-6 and IL-11 (Extended Data Fig. 5c).
Finally, we observed gp130 (encoded by il6st) expression in all sub-types of stromal cell from zebrafish scRNA-seq data that were present transmurally (Extended Data Fig. 5d). Cotreatment with DSS and BZA predominantly contracted Wt1ahigh stromal cells, Pdgfrahigh stromal cells, mesothelial cells, activated macrophages, activated endothelial cells and myoblasts in nod2mss13/mss13 larvae (Extended Data Fig. 5e). Activated transcripts were significantly reduced in cotreated larvae compared to DSS-treated larvae (notably, cebpb, mmp9 and pdgfra in the nod2mss13/mss13 background) (Fig. 4d, Extended Data Fig. 5f, g). Histologically, we observed reduced infiltrating leukocytes and intestinal bulb hypertrophy in the anterior intestine with cotreatment with DSS and BZA; this was significantly decreased in cotreatment using 2× DSS and BZA of nod2mss13/mss13 larvae (Fig. 4e, Extended Data Fig. 5h). BZA also alleviated the shortening of intestinal lengths (Fig. 4f, Extended Data Fig. 5i) and significantly decreased activated transcripts (wt1a, il11a, tgfb1a, cxcl13 and tnfa); the reduction was more pronounced when 2× DSS (compared to 1× DSS) was used to cotreat zebrafish larvae (Fig. 4g). Although we observed some genotype-dependent trends of reduced gene expression with cotreatment (especially with 1× DSS) (Extended Data Fig. 5j), these were not significant. Instead, BZA demonstrated a generalized benefit that was most prominent with repeated injury.
Discussion
In this Article, we elucidate mechanisms of monocyte differentiation in which NOD2 deficiency drives a pathogenic macrophage–fibroblast program. WT1 regulates the transformation of myofibroblasts in fibrotic lung disease37, and mesothelial and fibroblastic stromal cells in homeostasis and disease38,39. WT1 is enriched in CD14+PDGFRA+ activated fibroblasts, was enhanced in our in vitro and in vivo systems in NOD2-deficient backgrounds, and is an upstream transcriptional regulator of genes that are increased in activated macrophages and fibroblasts from NOD2WT/MT and NOD2MT/MT individuals. STAT3 was highly significant among upstream transcriptional regulators of genes associated with NOD2 risk alleles in both cell types.
The early institution of anti-TNF treatment in Crohn’s disease maximizes its benefit40. A lack of response to this treatment may result from delayed institution, and from primary and secondary non-response (Extended Data Fig. 6). The specific gp130 inhibitor BZA is approved by the US Food and Drug Administration as a selective modulator of oestrogen receptor; it has previously been proposed as a candidate for repurposing for the treatment of colon and pancreatic cancers31,33 by dampening proliferative pathways. Therapeutic approaches that target IL-6 and IL-11 in Crohn’s disease have previously been reported, but these have not advanced towards approval for clinical use41,42. The potential advantage of targeting gp130 is through the simultaneous targeting of both the myeloid and stromal arms of the pathogenic module. Serial transcriptome analyses performed before and after anti-TNF treatment, combined with in vivo evidence for the amelioration of pathogenic myeloid–stromal activation, suggest that a gp130 signalling blockade may provide an important complementary pathway to anti-TNF therapy.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized. The researcher who analysed the histology of human ileal strictures and intestinal gut length was blinded to subject group. In addition, clinical coordinators were blinded to the genotype of the individuals who were recalled for PBMC isolation. For all experiments relating to zebrafish RNA and protein, the researcher was not blinded during data collection because dissected intestines needed to be tracked for downstream applications (RNA isolation, protein extraction and scRNA-seq). For the zebrafish-mutant analyses, genotypes were correlated after analysis.
RNA isolation and cDNA synthesis
We used two methods of RNA isolation. In one (which was used when there were >500,000 cells in a sample), cells or zebrafish larval intestines were homogenized in 1,000 μl Trizol and vigorously vortexed. RNA isolation was performed using chloroform isolation (Qiagen), and final pellet was resuspended in 20 μl RNase-free water. RNA was heated to 70 °C before cDNA synthesis to disrupt any secondary structures.
The other method of RNA isolation was used in cases in which there were <500,000 cells in a sample, the Invitrogen RNAqueous Micro Kit (AM1931) was used to isolate RNA. Cells after CD14+ differentiation were scraped and homogenized immediately in 200 μl of lysis buffer (provided in the kit). RNA isolation was performed via column methods as described in the protocol; in brief, this involved 2 wash buffer steps, and an elution phase. The final column wash was resuspended in 20 μl RNase free water. RNA was heated to 70 °C before cDNA synthesis to disrupt any secondary structures. cDNA was synthesized using Life Technologies SuperScript III First-Strand Synthesis SuperMix (18080400) for quantitative RT–PCR (RT–qPCR). Up to 1 μg of RNA was run per sample per reaction and final volume of cDNA generated was 20 μl.
In all cases in which DSS-treatment was used, an additional lithium chloride purification step43 was performed to inhibit interference with PCR amplification.
RT–qPCR
Applied Biosystems reagents and machinery were used in all RT–qPCR reactions. In brief, using the Power Sybr Green PCR Master Mix (Life Technologies, no. 4368577) 1 μl of cDNA concentrate was used in a 20-μl qPCR reaction. Five hundred nm of forward and reverse primers were used in each reaction. Plates were spun at 200g for 1 min before loading into the One-Step PCR thermocycler. The following conditions were used to run samples: 96 °C heat for 15 min, 96 °C, 72 °C, 54 °C (×40), 54 °C and 96 °C. Ct values from duplicated reactions were averaged and the 2Ct(target)/2Ct(reference) method was used to calculate expression, with RPL32 and rpl32 serving as reference genes for human cells and zebrafish larvae, respectively. In cases in which genotype and treatment both served as independent variables, values were first normalized to genotype and then to treatment, to calculate fold change.
Zebrafish primer sequences
The following primer sequences were used: mfap4 forward (_F), ATGGCAATCGTGCTGTTCTT; mfap4 reverse (_R), AACTTCTTGTGG CGTGTCA; mpeg1_F, TCACCTGCTGATGCTCTGCTG; mpeg1_R, TCTGTG GAATGACAAAGACCTC; col5a1_F, AAGATTTTAGATTTTCACAGTTTACCA GAC; col5a1_R, ATCGCTACACGGTGCCACTTTCC; tnfa_F, ACAACACTA TTTACCTCGGC; tnfa_R. ACCAAACACCCCAAAGAAGG; cxcl13_F, CCATCTCCACTTAGAGTAAATCAAT; cxcl13_R, TGCCAGCAGTAAAGAA GGTCG; il11a_F, CGTCCTGCTAATCTACCACAA; il11a_R, AGCGAATGTT CAAAGTCAGTCA; tgfb1a_F, GGTTGGTTTGCTTGGTGCTG; tgfb1a_R, ATCTTCTGTCCGTCGTCGTCC; wt1a_F, AGTTCCACCGCTGCCC; wt1a_R, CGTGCCGTAACCTTGATTCCT; rpl32_F, CCTCAGACCCCTAACCAAACCT AA; and rpl32_R, CGCACTCTGTTGTCAATACCT.
Human primer sequences
The following primer sequences were used: MFAP4_F, AGAGGGCACTAT GAAGGCACA; MFAP4_R, GCGGAAGAAACTTACTGAGC; MPEG1_F, ATCCTC TTCTGGGCAGCGGC; MPEG1_R, GTTTCTTACCTGAACTCGGG; COL5A1_F, CTGCTGCTGCTGCTGTGGG; COL5A1_R, GATGGAGAAGTCCTCGGG AAATG; TNF_F, CGCTCCCCAAGAAGACAG; TNF_R, AGACCCCTCCCAGA TAGATG; CXCL13_F, AAGGTGTTCTGGAGGTCTATTAC; CXCL13_R, GCATCAGGGAATCTTTCTCTT; IL11_F, GAACTGTGTTTGCCGCCTG; IL11_R, TGTCAGCACACCTGGGAGC; WT1_F, CTGTGCCCTGCCTGTGA; WT1_R, CGGCTGTGCCAGTGAAC; TGFB1_F, CTGCTGCCGCTGCTGCTA; TGFB1_R, AGATTTCGTTGTGGGTTTCC; RPL32_F, GACCCCTTGTGAAG CCCA; RPL32_R, ACGAACCCTGTTGTCAATGCC; PDPN_F, CCGCTCGGA AAGTTCTCAA; PDPN_R, ATCATCTTCGGCACCTGGC; CHI3L1_F, TGGGTGT GAAGGCGTCTC; CHI3L1_R, GTCAAGGGCATCTGGGAAG; CCL2_F, AGTCTCTGCCGCCCTTCTG; CCL2_R, CTTGCTGCTGGTGATTCTTCT AT; CD14_F, AATCCTTCCTGTTACGGTCCC; and CD14_R, ACACTTGTGAA CTCTTCGGCTGC.
Human specimens
Patients who were eligible for inclusion in the study were identified by screening surgical programmes at the Mount Sinai Hospital. Ileal resections from patients with Crohn’s disease, and venous blood was collected after surgical resection. Healthy volunteers with NOD2 mutations of interest were screened through the Charles Bronfman Institute for Personalized Medicine at Mount Sinai. Twenty-one ml of venous blood was collected from these patients for downstream PBMC isolation. Protocols were reviewed by the Institutional Review Board (IRB) at the Icahn School of Medicine at Mount Sinai (‘Mechanisms of intestinal inflammation following ileal resection for Crohn’s Disease’, HSM no. 13–00998; ‘Recall and Deep Phenotyping of BioMe Participants’, no. 17–02727)). All recruitment was performed after ethical approval and oversight from the IRB and informed consent was obtained from all participants before blood was drawn.
Single- cell isolation from terminal ileum plus CD14+PDGFRA+ sorting
Single-cell suspensions were prepared from 20–30 mucosal biopsies per resection, as previously described3. Cell suspensions from both tissues were subject to CD14+ magnetic selection, using the Miltenyi Biotec CD14 negative selection kit (no. 130–091-153). CD14+ cells (unstained at this point) were then washed and incubated with Miltenyi anti-biotin CD140a (PDGFRa) antibody (no. 130–115-335) for 10 min in the dark at 4 °C. After washing, cells were incubated with anti-biotin microbeads for 15 min at 4 °C and run through magnetic separation columns. Positive and negative fractions from each separation were pulled down, cells were counted and cells were dissociated in Trizol for RNA isolation.
CD14+ PBMC differentiation
Twenty-one ml of blood was isolated from healthy patients with 0× (WT/WT), 1× (WT/MT) or 2× (MT/MT) NOD2 genotype status. PBMCs were isolated using the BD Vacutainer CPT Mononuclear Cell Preparation Tube-Sodium Heparin (BD Biosciences) and centrifuged for 20 min at 1,800g at room temperature, with brakes off. PBMC were collected at the interphase and were subject to CD14+ separation (Miltenyi CD14+ negative selection kit). Final CD14+ cell suspensions were resuspended in DMEM + 20% HI-FBS. About 1 million cells were plated per well in a 6-well plate with or without stimuli (as described in ‘In vitro stimulants’). At 48 h after plating, unattached cells were aspirated and new medium was added. Cells were left to differentiate for two weeks following removal of nonadherent cells21.
In vitro stimulants
The following stimulants were used for in vitro experiments (concentration and vendor in parentheses): MDP (0.5 or 1 μg ml−1, Invivogen; 53678–77-6), lipid A (0.5 μg ml−1, innaxon IAX100–004-C250), Pam3Cys (2.5 μg ml−1, Sigma-Aldrich; 506350–2MG), BZA (1 or 2 μM, Selleck Chemicals; S2128); and human anti-gp130 monoclonal antibody (0.05 μg ml−1, R&D Systems; MAB628).
Quantification of CD14+ differentiated cells
Five images per well of cells were taken using the EVOS FL imaging system. Each cell of interest was selected by the freeform drawing selection tool of ImageJ. Measurements were set to ‘area, mean grey value, integrated density, and shape descriptors’. After all cells were outlined, ‘measure’ was applied on all selected cells to quantify the number of cells per morphological characterization on the basis of the roundness parameter: ‘spindle’ = 0–0.29, ‘intermediate’ = 0.3–0.64 and ‘round’ = 0.65–1.
Luminex assay of CD14+ PBMCs
Supernatants from NOD2WT/WT and NOD2MT/MT CD14+ differentiated PBMCs were collected two weeks after initial seeding. Custom Luminex plates were designed by R&D Systems to capture analytes of interest, and all analyses were performed at Mount Sinai’s Human Immune Monitoring Core, according to the manufacturer’s directions.
Immunofluorescence of CD14+ PBMCs
Cells were washed in 1× PBS for 1 min before fixing. Cells were fixed in paraformaldehyde (4% in PBS), and—when necessary—permeabilized using Triton-X (0.2% in PBS), and blocked in UltraCruz blocking reagent (Santa Cruz Biotechnology, sc51624) for 30 min at room temperature. Cells were incubated in 1:166 anti-MPEG1 antibody (LSBio, LS-C680878), 1:200 anti-MFAP4 antibody (SCBT, sc398438), or 1:100 anti-COL5A1–FITC antibody (SCBT, sc166155), diluted in UltraCruz blocking reagent overnight at 4 °C. Cells were then incubated in secondary antibodies (1:5,000, goat anti-rabbit or anti-mouse Alexafluor 568/388, Abcam) for 1 h at room temperature. Cells were washed and mounted with DAPI-mounting medium (EMS; no. 17984–24) and imaged either using EVOS upright microscope, or with the ZEISS LSM 800 Airyscan confocal microscope. All images were quantified using ImageJ via the corrected total cell fluorescence method.
Immunohistochemistry of stricture slides of the terminal ileum
Serial 4-μm-thick sections from uninflamed and inflamed regions of each block of stricture were taken to perform the following immunohistochemical analyses, as previously described44. For double stains, tissue specimens were fixed in 10% formalin and embedded in paraffin and 3-μm sections were used for immunohistochemistry.
Immunohistochemistry was performed using VENTANA DISCOVERY ULTRA from Roche. This system allows for automated baking, deparaffinization and cell conditioning. Semiautomatic dual staining was performed sequentially using WT1 at a 1:25 dilution (abcam ab89901) during 60 min. An automated one drop of a prediluted secondary antibody Discovery OMNIMap anti-rabbit-HRP from Roche (760–4310) was used and the signal was obtained using Discovery ChromoMap DAB RUO from Roche (760–2513) (brown signal). PDGFRA (Thermofisher TA804956) was used at a 1:50 dilution during 60 min and after this, secondary antibody (Discovery OMNIMap anti-mouse-NP from Roche (760–4816)) positive signal was obtained using Discovery Purple Kit (760–229)(purple signal). Tissues were counterstained with haematoxylin to visualize the nuclei.
Human antibodies
The antibodies against the following proteins were used (concentration and vendor in parentheses): MFAP4 (1:400, SCBT (no. sc-398438)), MPEG1 (1:500, LSbio (no. LS-C680878)), PDGFRA (1:50, Thermo Fisher (no. TA804956)), CD14 (1:25, Sigma (no. HPA001887–25UL)), WT1 (1:25, Abcam (no. ab89901)) and GP130 (1:100, Thermo Fisher (PA5–99526)).
Zebrafish maintenance
Adult zebrafish were maintained on a 14:10-h light:dark cycle at 28 °C. Wild-type (AB), nod2sa21011 (Sanger Mutation Database) and CRISPR-mediated nod2−/− knockout lines were used for all experiments. Fertilized eggs following natural spawning were cultured in 28-°C fish water (0.6g l−1 Crystal Sea Marinemix). In caes in which larvae were used for drug treatments, all larvae were maintained in 50-ml Petri dishes in a 28-°C incubator, and not in the fish facility. Larvae and adults were fed once a day with Hikari First Bites (Hikari) from 6 days post-fertilization (dpf) and Zeigler Zebrafish Diet with Hatching Brine Shrimp Eggs (Pentair Aquatic Eco-Systems, FL), respectively. The Mount Sinai School of Medicine Institutional Animal Care and Use Committee approved all protocols.
CRISPR–Cas9 generation of nod2-mutant zebrafish line
Fifty-eight pmol CRISPR RNA (crRNA) (sequences given in ‘Sequences of guide RNA and primers used in nod2-mutant line determination’), 58 pmol FP-labelled trans-crRNA (Sigma) and 6.75 pmol Cas9 protein were combined and allowed to complex for 30 min on ice before injection. Wild-type AB zebrafish embryos were injected with 4 nl of injection mix per embryo at the 1–4-cell stage. Embryos were screened for the presence of fluorescent guide RNA at 2–3 h after fertilization. At 5 dpf, larvae were collected for gDNA extraction. A region of genomic DNA containing the intended CRISPR target site was amplified by PCR. PCR products were purified using Qiaquick PCR Purification Kit (Qiagen). The EnGen Mutation Detection Kit (NEB) was used to identify mosaicism for nod2 mutations. Once T7 digestion indicated the presence of CRISPR–Cas9-induced mutations, injected fish were added to the system and raised to adulthood.
At 3 months of age, CRISPR F0 fish were outcrossed against wild-type AB zebrafish. gDNA was extracted from larvae at 5 dpf, and the EnGen Mutation Detection Kit was used to identify clutches with germline-transmitted mutations. Clutches positive for mutations in nod2 were added to the system and raised to adulthood. At 2–3 months of age, F1 fish were fin-clipped. High-resolution melting analysis was used to identify F1 fish that were heterozygous for a mutation in nod2 (sequences given in ‘Sequences of guide RNA and primers used in nod2-mutant line determination’). gDNA from mutant heterozygotes was amplified using PCR and sequenced by Sanger sequencing (Genewiz). CRISP-ID was used to predict the mutated sequence45. Fish that were predicted to carry the same mutation became founders for the mutant line and were in-crossed. The resultant embryos were sequenced with Sanger sequencing (Genewiz) to confirm the mutant sequence. The resulting mutation is a 4-bp deletion (CTTG) in exon 1, which causes a premature stop that results in a frameshift. The genomic location is chromosome 7: 37563204–37563207. These mutants were assigned the allele name ‘mss13’ by The Zebrafish Information Network (ZFIN).
Sequences of guide RNA and primers used in nod2-mutant line determination
The following sequences were used (given in order from the 5′ to 3′ end): nod2 crRNA, TTGGACCTGCTACTTGCTC; nod2 PCR forward, TTGTGACTGGGCTATGAGGC; nod2 PCR reverse, AGTTTAAGGTGGTA TTACCTGTTGT; nod2 high-resolution melting forward, GGAGAGT GTTTTGGACCTGC; and nod2 high-resolution melting reverse, CGTACCC TAAGGTAATCCTCCC.
Taqman genotyping of nod2sa21011 zebrafish larvae
After intestinal dissections of 6-dpf zebrafish larvae, carcasses were placed in 20 μl NaOH for gDNA isolation. Zebrafish carcasses in suspension were heated at 96 °C for 15 min, tubes were spun down and 2 μl of Tris-HCL was added to each reaction. Tubes were vortexed and gDNA was either immediately used or stored at −20 °C. All gDNA was nano-dropped for concentration before running Taqman assays.
The nod2sa21011 fish probe (TTTTGGCTGCGATTGAGCAAGAATTAGCT GAGGAG[C/T]AAAAGGCTGGACTATGTTTTGGTAATGGATGCGTT) was designed to perform Taqman genotyping. The Taqman genotyping protocol (96-well plate and 25-μl reaction) was followed, and plates were run on the Viia7 Applied Biosystems Taqman genotyping system.
DSS treatment of zebrafish larvae
Single and repetitive DSS treatments were conducted as previously described17, although in these experiments 0.075% DSS (MP Biomedicals #0216011010) was used in all cases and first-time treatments were conducted at 5 dpf. All incrossed larvae were subject to treatments; a single DSS treatment period was 24 h long. Larvae were collected for further experimental use (intestinal dissections, followed by genotyping and RNA extractions) at 24 h after removal of DSS, during the rescue phase. A second dose of DSS was administered 48 h after recovery from the first dose, at 8 dpf. DSS was administered for 24 h and the larvae recovered in egg water. Collection occurred 24 h after DSS removal.
MDP treatment of zebrafish larvae
A 10 mg ml−1 working stock of MDP (Invivogen no. 53678–77-6) was diluted to 1 μg ml−1 for all treatments. No more than 20 larvae at 5 dpf were placed in 1.5-ml Eppendorf tubes and incubated with 1 μg ml−1 MDP in 1 ml egg water with the cap open, in a 37-°C incubator for 1 h. Larvae were rinsed and moved to fresh egg water in a Petri dish for rescue. Larvae were collected for further experimental use (intestinal dissections, followed by genotyping and RNA extractions) at 1 h, 24 h and 48 h after removal of MDP, during the rescue phases. A second 1-h incubation of MDP was conducted at 8 dpf, and collection occurred at 24 h and 48 h after removal of the second MDP incubation.
BZA treatment of zebrafish larvae
BZA concentrations (Selleck Chemicals no. S2128) were titrated to obtain an optimal concentration of 10 μM (an adaptation from previous studies in mouse31; this showed a decrease in pSTAT3 upon treatment, and no toxicity to zebrafish larvae). BZA was either administered alone or with 0.075% DSS cotreatment to nod2+/+, nod2+/− or nod2−/− larvae at 5 dpf (1×) and 8 dpf (2×), by emulsion in 15 ml egg water. After 24 h of each administration, the zebrafish larvae were rescued to egg water without any additional chemicals. Larval intestines were collected for further experimental use at 24 h after removal of DSS and/or BZA.
STAT3-inhibitor treatment of zebrafish larvae
S3I-201 (Sigma-Aldrich SML0330–5MG) STAT3 inhibitor concentrations were titrated to obtain an optimal concentration of 20 μM. S3I-201was either administered alone, or with 10 μM BZA cotreatment to nod2+/+, nod2+/− or nod2−/− larvae at 5 dpf (1×) by emulsion in 15 ml egg water. After 24 h of each administration, the zebrafish larvae were rescued to egg water without any additional chemicals. Larval intestines were collected for further experimental use at 24 h after removal of DSS and/or BZA.
Western blotting
Protein lysates were prepared from 5-dpf zebrafish larvae. Twenty larvae were homogenized in lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 2 mM EDTA, 10% glycerol and protease inhibitors) by manual homogenization, followed by sonication. All lysates were centrifuged in a 1:5 volume of sample buffer (2% SDS, 5% 2-mercaptoethanol final concentration). Samples were boiled at 96 °C for 5 min before loading into Mini-Protean TGX 4–15% gels (Bio-Rad), and western-blotted. Membranes were blocked for 45 min in 5% BSA in TBST (0.1% Tween 20). Membranes were probed with a custom anti-zebrafish Nod2 antibody, raised in rabbit (1:500, Pocono Rabbit Farm) or anti-zebrafish phospho-Stat3 antibody (1:500, MBL no. D128–3), and anti-tubulin antibody (1:2,000, Cell Signaling no. 2148) in 5% BSA in TBST, at 4 °C overnight. Membranes were washed and incubated in HRP-conjugated secondary antibody (1:5,000 goat anti-rabbit, and 1:5,000 goat anti-mouse). Immunoblots were developed with SuperSignal West Pico (Life Technologies) and were visualized by chemiluminscence using standard film developing processes. Band intensities were quantified through ImageJ; fold change was normalized first to tubulin, and then to no-treatment controls.
Immunohistochemistry of zebrafish larval sections
The untreated or 1× or 2×DSS-treated and DSS+BZA cotreated zebrafish larvae were fixed in 4% PFA for 24 h at 4 °C. Larvae were then embedded at the Mount Sinai Biorepository and Pathology Core. Paraffin sections were cut at 4-μm using a Leica RM2125 RTS manual rotary microtome. The sections were placed on de-ionized water heated to a temperature of 40 °C using the Fisher Tissue Prep Flotation Bath, Model 134. Sections were picked up using StatLab Colorview Adhesion slides and placed in a 60 °C oven for 30 min. After removing from the oven, they were cooled and placed on the Leica Autostainer XL for haematoxylin and eosin staining.
All slides were analysed in collaboration with M. Ko; quantifications of leukocytes were performed in ImageJ by measuring number of leukocytes in a 1.5 × 3.5-cm square area of the anterior intestine.
Immunofluorescence of zebrafish larval sections
Zebrafish were fixed in 4% PFA in phosphate-buffered saline (PBS) overnight at 5 dpf and transferred to 30% sucrose in PBS overnight. Larvae were embedded in optimum cutting temperature compound (Tissue-Tek), and 10-μm serial sections were obtained using the Leica CM3050 S Research Cryostat. Sections were washed in PBS + 0.1% Tween-20 (PBST). Tissue sections were blocked with 5% FBS and 2% BSA in PBST for 1.5 h at room temperature. Sections were stained with Il-11 antibody (1:150, Thermo Fisher no. PA5–36544 or isotype control IgG rabbit antibody 1:1, Thermo Fisher no. 08–6199), overnight at 4 °C and then with 1:250 goat anti-rabbit Alexa Fluor 488 (Sigma Aldrich no. 16–237, for Il-11 non-fluorescently labelled) or Alex Fluor 647-conjugated phalloidin antibody (1:250, Thermo Fisher, no. A22287) for 1.5 h in the dark at room temperature. Sections were mounted with ProLong Gold Antifade Mountant with DAPI (Life Technologies) and imaged using a Leica SP5 DMI at 20× and 40×.
Intestinal dissections of larval zebrafish
Zebrafish larvae at 6 and 9 dpf were treated with DSS, DSS + BZA or MDP, or left untreated and subjected to intestinal dissections. Larvae were anaesthetized with Tricaine solution (1.25g per 500 ml water, pH 7.4) and an individual larva was placed in a depression slide with 3% methylcellulose in preparation, ensuring that the larva was laid anterior to posterior. One razor blade was used to keep the larva in place at the swim bladder, and the other razor blade was used to scoop the intestine free from the remaining carcass. Intestines were pooled and placed in Trizol for RNA extraction, or autoMACS Rinsing Solution (Miltenyi, no. 130–091-222) for single-cell suspension. In cases in which genotyping was conducted, individual carcasses were placed into NaOH in PCR strip tubes, and intestines were placed individually into tubes with Trizol.
Intestinal length measurements for larval zebrafish
Single course DSS treatments were administered to 5-dpf zebrafish larvae. DSS or DSS + BZA-treated zebrafish were washed and removed to clean egg water at 6 dpf. Larvae were individually mounted onto a depression slide with 3% methylcellulose and imaged under brightfield view, using a 4× objective on EVOS XL core microscope. Gut length measurements were characterized from the intestinal bulb until the completion of the intestine at the anal pore of the larva, and body length measurements were characterized from the tip of head to tail; measurements were quantified using ImageJ and the Wilcoxon signed-rank test was used to compare differences between control and treated zebrafish (not genotype-dependent) or two-way ANOVA (genotype variable included) to test for multiple comparisons.
scRNA-seq of zebrafish larvae
Single-cell suspension.
After intestinal dissections, 20–30 pooled larval intestines were mixed into an autoMACS blending tube (Miltenyi, no. 130–093-237) with 2 ml autoMACS Rinsing Solution (PBS + EDTA + 0.5% BSA). Tubes were rocked for 30 min at room temperature to exclude epithelial cell proportions. Tubes were spun at 4,200 rpm for 5 min. After removal of the supernatant, intestinal pellets were resuspended in 2 ml PBS–EDTA +Ca2+ + Mg2+ +1 mg DNaseI + 1 mg collagenase IV. Tubes were rocked for 35 min at 37 °C at 100 rpm. Mixtures were filtered with a 70-μm filter into a new tube and spun at 4,200 rpm for 5 min. Final suspensions were generated in 300 μl PBS + 0.5% BSA. Cells were counted with AO/PI dye using the Nexcelom Biosciences cell counter and viability was about 65%. Ten thousand cells were loaded onto the 10X Genomics Chromium Controller instrument within 30 min of completion of cell suspension using GemCode Gel Bead and Chip (10X Genomics). Cells were partitioned into gel beads in emulsion in the controller in which cell lysis and reverse transcription occurred. Libraries were prepared using 10X Genomics library kits and sequenced on Illumina NextSeq500, according to manufacturer’s recommendations.
Alignment, transcriptome assembly and quality control.
Raw BCL (raw base call) files generated from Illumina were demultiplexed using cellranger v.4.0.0, which uses the bcl2fastq pipeline, into single-cell FASTQ files. FASTQ files were then aligned to an Ensembl GRCz11 zebrafish genome and transcript count matrices were generated using default parameters in cellranger count. The raw (unfiltered) output matrices were then used for the clustering and downstream analysis in Seurat v.3.1. Density plots for unique molecular identifiers (UMIs), number of genes and mitochondrial percentages were analysed for each sample. Data were filtered to include cells with >200 UMIs and 250> genes per cell >4,000. Clustering was performed using the R package Seurat v.3.1. Samples were individually normalized, and the 200 most variable genes were identified for each sample. Integration of all zebrafish samples was performed using ‘FindIntegrationAnchors’, ensuring appropriate metadata annotation for each sample (genotype, treatment and stage). The data were scaled in Seurat, and dimensionality reduction was then performed; the first 18 principal components were used to generate clustering, with a resolution of 0.8. Characterization and marker designation of clusters is given in Supplementary Table 3. Cluster marker genes were identified using the ‘FindAllMarkers’ function.
Differential expression between clusters/samples.
In all cases in which differential expression testing was performed on zebrafish samples, the ‘FindMarkers’ function was used, with test.use = ‘negbinom’, to account for the one-tailed distribution of UMIs in each cluster.
Stromal and myeloid subclustering.
Cells annotated in ‘stromal’ or ‘myeloid’ families were divided into subsets; normalization and identification of variable features was then conducted. After dimensionality reduction, the first 15 principal components were used to generate clustering, with a resolution of 0.8. Characterization and marker designation of clusters is provided in Supplementary Table 7.
Human–zebrafish integrated clustering.
Human-to-fish orthologues list were generated using ZFIN zebrafish–human orthologues database (20 August 2020). The orthologues in the database were curated considering three factors: conserved genome location, amino acid sequence comparison and the phylogenetics tree. Raw (unfiltered) matrices for human single-cell data from 22 paired samples that were previously published3 were filtered to contain only cells with genes detected >500, number of UMI >300 and a minimum of at least 5 cells. The gene names from zebrafish data were changed to their corresponding orthologous pair in the human–zebrafish orthologue database. Joint clustering of human and zebrafish data was performed using Seurat v.3.1. The dataset was inspected for the number of significant principal components using the 200 most variable genes on an elbow plot. For the integrated clustering, the first 15 principal components were used to perform clustering with a resolution of 1.2. Characterization and marker designations of clusters are provided in Supplementary Table 3.
Random-forest models.
Cross-species random-forest models were trained using the ‘randomForest’ R package with normalized gene expression values from myeloid and stromal subsets from the zebrafish dataset and a dataset of ileal samples from individuals with Crohn’s disease. Gene names in the zebrafish dataset were renamed to their corresponding human orthologous gene names from the human–zebrafish orthologues dataset. The following parameters were used to run the training on the human dataset (a total of 9,157 cells): ntree = 1,000, mtry = square root of total gene (9,740). The zebrafish dataset (a total of 7,983 cells) was used as the test dataset. The ‘Predict’ function was used for the prediction in the test data using the random-forest model.
Principal component analyses of activated fibroblast and inflammatory macrophages from patients in the RISK study
RNA-seq data collected from terminal ileum biopsies by the RISK consortium1,46,47 were normalized (median-of-ratios) using DESeq248 on the basis of calculated size factors.
Differential expression was performed between the following cellular subsets in the scRNA-seq dataset for ileal samples from patients with Crohn’s disease (FindMarkers in Seurat v.3): (1) ident.1 = activated fibroblasts versus ident.2 = residential fibroblasts; and (2) ident.1 = inflammatory macrophages versus ident.2 = residential macrophages, from clusters that were previously reported3 from an ileal scRNA-seq cohort. The top 200 pattern genes for macrophages and fibroblasts were then used to calculate the first 10 principle components across each of the RISK samples, and each component was tested along with sex in a series of linear (for NOD2 minor allele count) or logistic (for stricturing status and anti-TNF response) regression models to identify relationships between cell-specific principal component scores and NOD2 minor allele count, stricture formation and response to anti-TNF treatment. Supplementary Table 5 provides the details of the results of the principal component analysis.
Statistical analysis
Statistical comparisons were performed as indicated in figure legends, and using GraphPad Prism v.8.3.1 or Microsoft Excel v.16.37. After determining normality, differences for means were tested for statistical significance with either Student’s t-test or Wilcoxon signed-rank test (paired analyses) in the event that multiple comparisons did not need to be accounted for. In cases in which variance analyses between genotypes and treatment conditions were to be tested, tests were performed using two-way ANOVA tests. P values < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All data and supporting findings of this study are available within the Article and its Supplementary Information. All scRNA-seq data for human ileal samples have been deposited in the Gene Expression Omnibus (GEO) repository with accession code GSE134809. All 10X Chromium zebrafish scRNA-seq data have been deposited in the GEO repository with accession code GSE150498. The previously published34 dataset can also be found in the GEO repository with accession code GSE16879. The ZFIN genomic databases can be found at https://zfin.atlassian.net/wiki/spaces/general/pages/1891412257/Genomic+Resources+for+Zebrafish. Source data are provided with this paper.
Code availability
Scripts to reproduce zebrafish scRNA-seq clustering and differential expression analyses, as well as any other scripts used to generate computational figures, are available on GitHub at https://github.com/Cho-lab-Sinai/Scripts_Nayar_et_al.
Extended Data
Extended Data Fig. 1 |. scRNA-seq exploration of ileum of patients with Crohn’s disease and validation by protein staining.
a, Genes (columns) across myeloid and stromal cells (rows) from scRNA-seq of uninflamed ileum of patients with Crohn’s disease (n = 11). Expression defined as log2(gene expression/average) across all shown clusters. b, Relative expression of NOD2, RIPK2 and XIAP (columns) across myeloid and stromal cells (rows) from scRNA-seq of PBMCs and inflamed ileum of patients with Crohn’s disease (n = 11). c, Gene ontology analysis from upregulated genes in activated fibroblasts from inflamed ileum. The number of genes per biological process is shown in parentheses. Processes ranked from top to bottom in decreasing order of −log(P value). d, Percentage of COL1A1+, PDGFRA+, CD14+ and CD14+PDGFRA+ double-positive cells of total activated fibroblasts (1,367 cells total) in inflamed ileum. e, Number of cells from direct ex vivo sorting of CD14+PDGFRA+ cells of uninflamed and inflamed ileum of patients with Crohn’s disease. n = 3 biological replicates. Data are mean ± s.e.m. f, Full-thickness sections of inflamed strictures stained with MPEG1 and MFAP4. n = 3 biological replicates per stain. g, Full-thickness sections of inflamed strictures showing expression of WT1, PDGFRA and CD14. n = 2 patients per stain (patient 7 and 8 denoted from a previous publication3) (left). In inflamed strictures from samples from patients with Crohn’s disease, WT1, PDGFRA and CD14 expression is seen in muscularis mucosae and lymphoid aggregates; and around blood vessels (right). n = 2 patients per stain; 5 images per patient. Scale bar, approximately 50 μm. h, RT–PCR of gene expression at 24 h after 1× DSS-treatment of zebrafish nod2+/+ and nod2sa21011/sa21011 larvae, comparing intestinal and carcass expression. n = 3 biological replicates; 3 clutches, 10–15 larvae per genotype per clutch. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-tailed paired Student’s t-test.
Extended Data Fig. 2 |. scRNA-seq data from intestines of DSS-treated zebrafish larvae.
a, scRNA-seq of intestines of untreated and DSS-treated zebrafish larvae from nod2+/+ and nod2mss13/mss13 backgrounds. UMAP shows joint clustering of 30,069 cells, revealing 32 unique clusters (left). Top 5 markers for each cluster (rows) (right). b, UMAP showing human–zebrafish clusters from species integration via scRNA-seq (top); top 3 markers for each cluster (rows) (bottom). Expression is UMI counts per gene per cell. c, Homologous expression between zebrafish and human clusters (expression is log-normalized).
Extended Data Fig. 3 |. NOD2 deficiency enhances differentiation of spindle cells from CD14+ monocytes.
a, Morphological quantification of PBMC differentiation from NOD2WT/WT or NOD2MT/MT cells, with 2.5 μg ml−1 Pam3Cys, 0.5 μg ml−1 lipid A and 1 μg ml−1 MDP. Classification of categories in Methods. n = 6 biological replicates for NOD2WT/WT, n = 3 biological replicates for NOD2MT/MT for each treatment condition. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-sided Wilcoxon signed-rank t-test. b, Representative morphological images of spindle, intermediate and round cells from NOD2WT/WT, NOD2WT/MT and NOD2MT/MT individuals, unstimulated or treated with 1 μg ml−1 MDP, 0.5 μg ml−1 lipid A or 2.5 μg ml−1 Pam3Cys. n = 5 images per well condition per patient. Scale bar, 100 μm. c, Representative images of myeloid (MPEG1) and stromal (MFAP4, COL5A1 and COL1A1) expression by immunofluorescence (left). Quantification of staining (right). Data are mean ± s.e.m. of corrected cellular fluorescence. ***P < 0.001; two-way ANOVA test with Sidak correction. n = 4 images per fluorescent marker; individual values, individual cells per image. d, Schematic of CD14+CD16− PBMCs isolated from healthy NOD2-mutant carriers and non-carriers, and differentiation assay (left). RT–PCR of gene expression from 1 μg ml−1 MDP-treated PBMCs after 2 weeks of differentiation from NOD2WT/WT or NOD2MT/MT individuals. n = 3 biological replicates (right). Data are mean MDP relative expression (left y axis) and individual 2−ΔCt expression values (right y axis) with error bars representing individual ΔCt values ± s.e.m.. e, RT–PCR of gene expression of unstimulated CD14+CD16+ PBMCs after two weeks of differentiation from NOD2WT/WT or NOD2MT/MT individuals. n = 3 biological replicates. Data are mean untreated relative expression (left y axis) and individual 2−ΔCt expression values (right y axis) with error bars representing individual ΔCt values ± s.e.m. f, Secreted protein (pg ml−1) by Luminex from unstimulated PBMCs or PBMCs stimulated with 0.5 μg ml−1 lipid A, 2.5 μg ml−1 Pam3Cys or 1 μg ml−1 MDP. n = 2 NOD2WT/WT, n = 3 NOD2MT/MT. Data are mean ± s.e.m. *P < 0.05; two-way ANOVA. g, Schematic of nod2sa21011 mutant from Zebrafish Sanger Mutation Project and nod2mss13/mss/13 CRISPR-knockout zebrafish line (left); timeline of 1× and 2× MDP stimulation of zebrafish larvae (right).
Extended Data Fig. 4 |. Staining of key fibrotic protein deposition in zebrafish larvae and human ileal stricture resections.
a, Nod2 expression as assessed by western blotting to show loss of protein levels in nod2sa21011 and nod2mss13/mss13 CRISPR-mutant zebrafish larvae. Larvae were untreated and protein was collected at 6 dpf. n = 1 biological replicate per nod2 mutation for protein-knockout validation (1 clutch per mutation line with 20 larvae for each genotype). For gel source data, see Supplementary Fig. 1. b, UMAP of myeloid and stromal clusters from joint clustering of scRNA-seq data for zebrafish larval cells, grouped by nod2+/+ and nod2mss13/mss13 genotypes. c, NOD2 mutation information of patients in the ileal Crohn’s disease scRNA-seq cohort, used for differential expression analysis between activated and nonactivated fibroblast and macrophage clusters. d, Transcription factors from ingenuity pathway analysis upstream of genes enriched in activated fibroblasts and inflammatory macrophages from individuals with NOD2 risk alleles versus individuals without these alleles. P values were determined by ingenuity pathway analysis for transcription-factor regulation of genes in differential expression analysis. e, Top 5 genes (rows) in each cluster (columns). Expression is in UMI counts. f, Feature plots to show localized expression of key transcripts in nod2WT/WT larvae from scRNA-seq myeloid and stromal populations (log-normalized expression).
Extended Data Fig. 5 |. gp130 blockade by BZA ameliorates fibrotic activation and may provide a supplementary approach to anti-TNF therapy.
a, log2-transformed microarray expression from a previous publication34, of gp130-associated genes in patients treated with infliximab. Blue bars, patients who responded to infliximab (n = 18); red bars, patients who did not respond to infliximab (n = 16). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; two-tailed paired Student’s t-test. b, Morphological quantification of NOD2WT/WT or NOD2MT/MT-differentiated PBMCs that were unstimulated or stimulated with 0.5 μg ml−1 MDP or 0.5 μg ml−1 MDP + 1 μM BZA. n = 6 biological replicates for NOD2WT/WT (n = 4 for MDP + BZA costimulation) and n = 3 biological replicates for NOD2MT/MT per each treatment condition (top); unstimulated, 0.5 μg ml−1 MDP, 0.5 μg ml−1 MDP + 1 μM BZA or MDP + 0.05 μg ml−1 monoclonal anti-gp130 antibody (bottom). n = 2 biological replicates for each genotype. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-sided Wilcoxon signed-rank test. c, Secreted protein (pg ml−1) by Luminex from unstimulated PBMCs or PBMCs stimulated with 0.5 μg ml−1 MDP, 0.5 μg ml−1 MDP + 1 μM BZA or 0.5 μg ml−1 MDP + 0.05 μg ml−1 anti-gp130. n = 2 individuals per genotype. data are mean values. d, BZA target-gene expression in stromal and myeloid cells of scRNA-seq data from DSS-treated larvae (log-normalized expression). e, Percentage of stromal and myeloid populations in total intestinal cells sequenced in DSS-treated versus DSS + BZA-treated larvae. Red boxes, nod2mss13/mss13 differences. f, g, Violin plots for upregulated genes upon DSS treatment, or for genes downregulated by DSS + BZA cotreatment in nod2+/+ (f) and nod2mss13/mss13 (g) larvae (log-normalized expression). h, Haematoxylin and eosin staining of DSS-treated and DSS + BZA-treated nod2+/+ and nod2mss13/mss13 zebrafish larvae. Intestinal bulb hypertrophy measured as indicated by scale bars, and quantified after 1× (left) and 2× (right) treatments. n = 5–10 biological replicates (individual larvae) per condition. Data are mean ± s.e.m. For nod2+/+, 5 untreated (1×), 3 1×DSS, 4 untreated (2×), 10 2×DSS; for nod2mss13, n = 6 untreated (1×, 8 1×DSS, 5 untreated (2×), 5 2×DSS. *P < 0.05, **P < 0.01; one way-ANOVA with Tukey’s multiple comparison. i, Intestine length of 2×DSS-treated or 2×DSS + BZA-cotreated nod2+/+, nod2+/− and nod2−/− larvae. n = 3 clutches; 10–15 larvae per group. Data are mean ± s.e.m. *P < 0.05, **P < 0.01; two-way ANOVA test with Tukey’s multiple comparison. j, RT–PCR of gene expression at 24 h after 1× or 2×DSS + BZA cotreatment of nod2+/+, nod2+/− or nod2−/− zebrafish larvae. n = 4 biological replicates for 1× treatment; 4 clutches, 10–15 larvae per genotype per clutch. n = 4 biological replicates for 2× treatment; 4 clutches, 10–15 larvae per genotype per clutch. Data are mean ± s.e.m.
Extended Data Fig. 6 |. Pathogenesis of Crohn’s disease and treatment pathways.
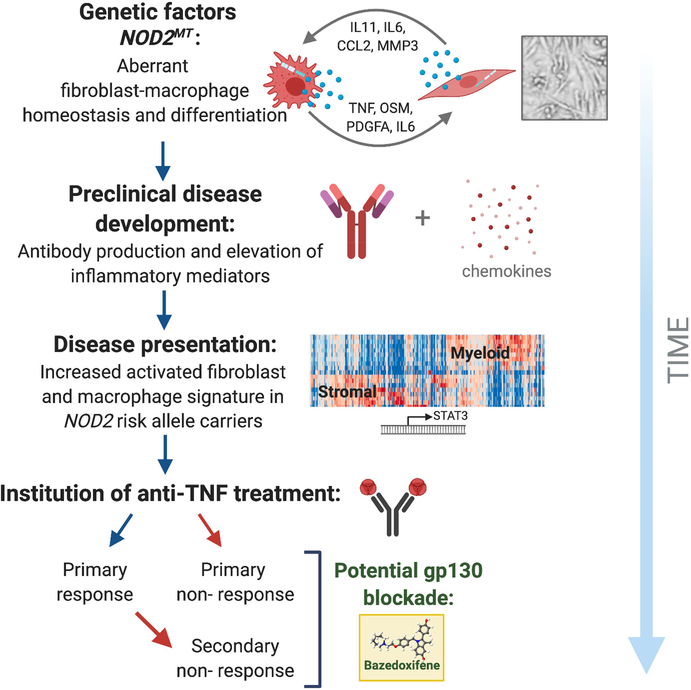
Model to show proposed mechanism of pathophysiology of Crohn’s disease under specific genetic and cellular constraints. Individuals who carry a NOD2 risk allele develop aberrant fibroblast–macrophage homeostasis and differentiation over time. At preclinical stages of disease development, patients with Crohn’s disease increase production of antibodies (ASCA and CBir) to contain infection caused by increased bacterial load and elevate inflammatory mediators (cytokines and chemokines such as IL11, CXCL13, IL6, CCL2 and OSM). Patients who carry NOD2 risk alleles will upregulate a pathogenic activated fibroblast and macrophage signature with disease development. This results in downstream activation of STAT3, among other chronic inflammatory and fibrotic consequences that we have shown in this Article. Some patients administered anti-TNF therapy will not respond, and often develop complications such as stricturing disease. Furthermore, over time, initial primary responders may become secondary nonresponders. Here we show that patients who are treatment-refractory have increased gp130 and activated fibroblast signatures; these would be patients for whom supplementing anti-TNF treatment with the gp130 inhibitor BZA could be beneficial. This model summarizes a two-step candidate selection approach: first by patients with Crohn’s disease who carrying NOD2 risk alleles with elevation of key expression signatures, and then by a lack of response to anti-TNF) to inform personalized therapeutic decision-making for Crohn’s disease.
Supplementary Material
Acknowledgements
We thank S. Grossman of the Dr Sanford J. Grossman Center for Integrative Studies in IBD, which is supported by the generosity of The Sanford J. Grossman Charitable Trust; the ISMMS Department of Pathology for providing human tissue samples; D. Derose, K. Svoboda, C. Chasteau and U. Korie for sample collection and coordination; A. Saha and H. Wen for recall of healthy volunteers with NOD2 mutations via the ISMMS BioMe Biobank; F. Avila and M. Garcia-Barros from the Biorepository and Pathology core for assistance with staining human ileal strictures; N. Hsu and N. Villaverde (members of the laboratory of J.H.C.) for discussions; and F. Marlow and B. Rosenberg for discussions relating to zebrafish and single-cell experiments. This work was also supported by National Institutes of Health (NIH) grants R01 DK106593, R01 DK123758-01 and U01 DK062422 to J.H.C.
Footnotes
Competing interests S.N. and J.H.C., through the Icahn School of Medicine at Mount Sinai, have filed a provisional US patent application (no. 63/130,035) on repurposing BZA for clinical use in a subset of patients with Crohn’s disease. All other authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-03484-5.
Peer review information Nature thanks Alison Simmons and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-03484-5.
References
- 1.Kugathasan S et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 389, 1710–1718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West NR et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med 23, 579–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin JC et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogura Y et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Hugot JP et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Lesage S et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet 70, 845–857 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X et al. Circuit design features of a stable two-cell system. Cell 172, 744–757.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verstockt B & Cleynen I Genetic influences on the development of fibrosis in Crohn’s disease. Front. Med 3, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermeire S et al. NOD2/CARD15 does not influence response to infliximab in Crohn’s disease. Gastroenterology 123, 106–111 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Mascheretti S et al. Pharmacogenetic investigation of the TNF/TNF-receptor system in patients with chronic active Crohn’s disease treated with infliximab. Pharmacogenomics J. 2, 127–136 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Brant SR et al. Defining complex contributions of NOD2/CARD15 gene mutations, age at onset, and tobacco use on Crohn’s disease phenotypes. Inflamm. Bowel Dis 9, 281–289 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Bain CC et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol 15, 929–937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schafer S et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim W-W et al. Transgenic interleukin 11 expression causes cross-tissue fibro-inflammation and an inflammatory bowel phenotype in mice. PLoS ONE 15, e0227505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H-S et al. Identification of novel Wilms’ tumor suppressor gene target genes implicated in kidney development. J. Biol. Chem 282, 16278–16287 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Tamplin OJ et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuang L-S et al. Zebrafish modeling of intestinal injury, bacterial exposures and medications defines epithelial in vivo responses relevant to human inflammatory bowel disease. Dis. Model. Mech 12, dmm037432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y-G et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity 34, 769–780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reilkoff RA, Bucala R & Herzog EL Fibrocytes: emerging effector cells in chronic inflammation. Nat. Rev. Immunol 11, 427–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H et al. Netrin-1 regulates fibrocyte accumulation in the decellularized fibrotic scleroderma lung microenvironment and in bleomycin induced pulmonary fibrosis. Arthritis Rheumatol. 68, 1251–1261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-de-Alba C et al. Expression of matrix metalloproteases by fibrocytes: possible role in migration and homing. Am. J. Respir. Crit. Care Med 182, 1144–1152 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Girardin SE et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem 278, 8869–8872 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Orekhov AN et al. Monocyte differentiation and macrophage polarization. Vessel Plus 3, 10 (2019). [Google Scholar]
- 24.Ma G et al. Nod2–Rip2 signaling contributes to intestinal injury induced by muramyl dipeptide via oligopeptide transporter in rats. Dig. Dis. Sci. 60, 3264–3270 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Strober W, Kitani A, Fuss I, Asano N & Watanabe T The molecular basis of NOD2 susceptibility mutations in Crohn’s disease. Mucosal Immunol. 1, S5–S9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y et al. Circulating fibrocytes are involved in inflammation and leukocyte trafficking in neonates with necrotizing enterocolitis. Medicine (Baltimore) 96, e7400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishida M et al. Muramyl dipeptide enhances lipopolysaccharide-induced osteoclast formation and bone resorption through increased RANKL expression in stromal cells. J. Immunol. Res 2015, 132765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kettleborough RNW et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496, 494–497 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chassaing B, Aitken JD, Malleshappa M & Vijay-Kumar M Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol 104, Unit 15.25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldner MJ & Neurath MF Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin. Immunol 26, 75–79 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Thilakasiri P et al. Repurposing the selective estrogen receptor modulator bazedoxifene to suppress gastrointestinal cancer growth. EMBO Mol. Med 11, e9539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei J et al. Bazedoxifene as a novel GP130 inhibitor for colon cancer therapy. J. Exp. Clin. Cancer Res. CR 38, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Cao Y, Xiao H, Li C & Lin J Bazedoxifene as a novel GP130 inhibitor for pancreatic cancer therapy. Mol. Cancer Ther 15, 2609–2619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arijs I et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE 4, e7984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo S-Y et al. A positive feedback loop bi-stably activates fibroblasts. Nat. Commun 9, 3016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen KLA, Zhao YC, Hieronymi K, Smith BP & Madak-Erdogan Z Bazedoxifene and conjugated estrogen combination maintains metabolic homeostasis and benefits liver health. PLoS ONE 12, e0189911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sontake V et al. Wilms’ tumor 1 drives fibroproliferation and myofibroblast transformation in severe fibrotic lung disease. JCI Insight 3, e121252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinchen J et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buechler MB et al. A stromal niche defined by expression of the transcription factor WT1 mediates programming and homeostasis of cavity-resident macrophages. Immunity 51, 119–130.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frei R et al. Early initiation of anti-TNF is associated with favourable long-term outcome in Crohn’s disease: 10-year-follow-up data from the Swiss IBD cohort study. J. Crohn’s Colitis 13, 1292–1301 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Danese S et al. Randomised trial and open-label extension study of an anti-interleukin-6 antibody in Crohn’s disease (ANDANTE I and II). Gut 68, 40–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sands BE et al. Randomized, controlled trial of recombinant human interleukin-11 in patients with active Crohn’s disease. Aliment. Pharmacol. Ther 16, 399–406 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Viennois E, Chen F, Laroui H, Baker MT & Merlin D Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res. Notes 6, 360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X et al. Luminally polarized mural and vascular remodeling in ileal strictures of Crohn’s disease. Hum. Pathol 79, 42–49 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Dehairs J, Talebi A, Cherifi Y & Swinnen JV CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep 6, 28973 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haberman Y et al. Pediatric Crohn’s disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest 124, 3617–3633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gettler L et al. Prioritizing Crohn’s disease genes by integrating association signals with gene expression implicates monocyte subsets. Genes Immun. 20, 577–588 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and supporting findings of this study are available within the Article and its Supplementary Information. All scRNA-seq data for human ileal samples have been deposited in the Gene Expression Omnibus (GEO) repository with accession code GSE134809. All 10X Chromium zebrafish scRNA-seq data have been deposited in the GEO repository with accession code GSE150498. The previously published34 dataset can also be found in the GEO repository with accession code GSE16879. The ZFIN genomic databases can be found at https://zfin.atlassian.net/wiki/spaces/general/pages/1891412257/Genomic+Resources+for+Zebrafish. Source data are provided with this paper.